Abstract
Background
Identification of blood biomarkers that prospectively predict progression of Mycobacterium tuberculosis infection to tuberculosis disease may lead to interventions that impact the epidemic.
Methods
Healthy, M. tuberculosis infected South African adolescents were followed for 2 years; blood was collected every 6 months. A prospective signature of risk was derived from whole blood RNA-Sequencing data by comparing participants who ultimately developed active tuberculosis disease (progressors) with those who remained healthy (matched controls). After adaptation to multiplex qRT-PCR, the signature was used to predict tuberculosis disease in untouched adolescent samples and in samples from independent cohorts of South African and Gambian adult progressors and controls. The latter participants were household contacts of adults with active pulmonary tuberculosis disease.
Findings
Of 6,363 adolescents screened, 46 progressors and 107 matched controls were identified. A 16 gene signature of risk was identified. The signature predicted tuberculosis progression with a sensitivity of 66·1% (95% confidence interval, 63·2–68·9) and a specificity of 80·6% (79·2–82·0) in the 12 months preceding tuberculosis diagnosis. The risk signature was validated in an untouched group of adolescents (p=0·018 for RNA-Seq and p=0·0095 for qRT-PCR) and in the independent South African and Gambian cohorts (p values <0·0001 by qRT-PCR) with a sensitivity of 53·7% (42·6–64·3) and a specificity of 82·8% (76·7–86) in 12 months preceding tuberculosis.
Interpretation
The whole blood tuberculosis risk signature prospectively identified persons at risk of developing active tuberculosis, opening the possibility for targeted intervention to prevent the disease.
Funding
Bill and Melinda Gates Foundation, the National Institutes of Health, Aeras, the European Union and the South African Medical Research Council (detail at end of text).
Introduction
One-third of the global population is infected with Mycobacterium tuberculosis,1 but <10% will progress to active tuberculosis disease during their life time; the majority will remain healthy.2–6 Risk of progression is associated with young or old age, co-morbidities such as human immunodeficiency virus (HIV) infection and diabetes mellitus, socio-economic and nutritional compromise, and therapy with immune modulatory agents such as tumor necrosis factor inhibitors, among others.7,8 It is not possible to predict which M. tuberculosis infected individuals will develop active tuberculosis, given current tools. Our aim was to identify peripheral blood biomarkers of this disease risk. Knowledge gained could lead to targeted antimicrobial therapy to prevent tuberculosis disease, as treating all latently infected persons in endemic countries for 6–9 months is not feasible. Other potential applications of biomarkers of risk of tuberculosis disease include assessment of response to drug therapy and targeted enrollment into efficacy trials of new tuberculosis vaccines and drugs.
Our study evaluated whether global gene expression measured in whole blood of healthy persons allows identification of prospective signatures of risk of active tuberculosis disease. Previous systems biology approaches have identified diagnostic signatures that discriminate tuberculosis disease from latent M. tuberculosis infection and from other disease states.9–20 For example, Berry, et al., identified and validated a 393 gene signature that allowed differentiation of persons with active tuberculosis disease and latent infection.11 Anderson, et al., identified and validated a 53 gene signature that distinguished active tuberculosis from other diseases in African children with or without HIV infection.13 In contrast to the published diagnostic studies, our focus was on prospective signatures of risk that could be identified in healthy individuals, up to 2 years before clinical tuberculosis disease manifests. Given that one third of the world’s population is latently infected with M. tuberculosis, our approach constitutes an opportunity to impact the burden of disease.
Methods
Cohorts and blood collection
Participants from multiple cohorts were included in analysis. First, participants from the South African adolescent cohort study (ACS) were evaluated to identify and validate a tuberculosis risk signature (Figure 1A). Briefly, 6,363 healthy adolescents, aged 12–18 years, were enrolled between July 2005 and April 2007; follow-up was completed by February 2009 (exclusion criteria and more detail in the Supplementary Appendices). Approximately half the participants were evaluated at enrollment and every 6 months during 2 years follow-up; the other half was evaluated at baseline and at 2 years. At enrollment and at each visit, clinical data were collected and 2.5mL blood was collected directly into PAXgene blood RNA tubes (PreAnalytiX), which were stored at −20°C.
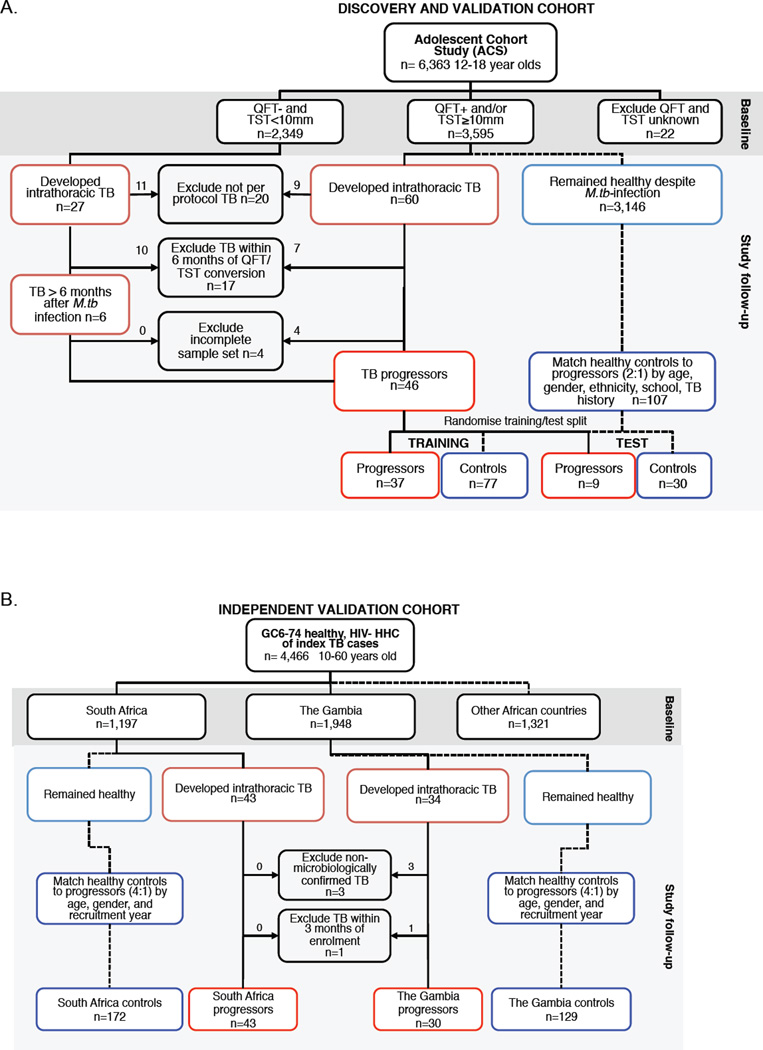
Figure 1. The Adolescent Cohort Study (ACS) and the Grand Challenges 6-74 Study (GC6-74) cohorts for the discovery and validation of the tuberculosis risk signature.
(A) Inclusion and exclusion of participants from the ACS and assignment of eligible progressors and controls to the training and test sets. QFT: Quantiferon Gold In-Tube. TST: tuberculin skin test. (B) Inclusion and exclusion of adult household contacts of patients with lung tuberculosis from the GC6-74 cohorts, and assignment of eligible progressors and controls. HHC: household contact.
Only adolescents with latent M. tuberculosis infection at enrollment were included in analysis aimed at identification of a tuberculosis risk signature. Latent M. tuberculosis infection was diagnosed by a positive QuantiFERON TB GOLD In-Tube Assay (QFT, Cellestis; >0·35 IU/mL) and/or a positive tuberculin skin test (TST, 0·1mL dose of Purified Protein Derivative RT-23, 2-TU, Staten Serum Institute; >10mm). According to South African policy, QFT and/or TST positive adolescents were not given therapy to prevent tuberculosis disease.21 Adolescents who developed active tuberculosis disease during follow-up were included as “progressors” (cases). Tuberculosis was defined as intrathoracic disease, with either two sputum smears positive for acid-fast bacilli or one positive sputum culture confirmed as M. tuberculosis complex (mycobacterial growth indicator tube, BD BioSciences). For each progressor, two matched controls that remained healthy during follow-up were selected and matched by age at enrolment, gender, ethnicity, school of attendance, and presence or absence of prior episodes of tuberculosis disease. Participants were excluded if they developed tuberculosis disease within 6 months of enrollment to exclude early asymptomatic disease that could have been present at the time of evaluation, or if they were HIV infected. Prior to analysis, the ACS progressors (cases) and controls were randomly divided into training and test sets, at a ratio of 3:1 using the randomization function in Microsoft Excel.
The other cohorts consisted of South African and Gambian participants from the Grand Challenges 6-74 Study (GC6-74; http://www.case.edu/affil/tbru/collaborations_gates.html), who were enrolled between February 2005 and December 2010 to independently validate the tuberculosis risk signature (Figure 1B). Briefly, from a parent GC6-74 cohort 4,466 healthy, HIV-negative persons aged 10–60 years who had household exposure to an adult with sputum smear positive tuberculosis disease, 1,197 and 1,948 were enrolled in South Africa and The Gambia, respectively (exclusion criteria and more detail in the Supplementary Appendices). At baseline, at 6 months (The Gambia only) and at 18 months (both sites), participants were evaluated clinically and blood was collected and stored in PAXgene tubes as above. Follow-up continued for 2 years, and concluded in November 2012. Among GC6-74 participants, progressors (cases) had intrathoracic tuberculosis, defined on the basis of sputum culture, smear microscopy and clinical signs. For each progressor, four controls were matched according to recruitment region, age category (≤18, 19–25, 26–35, ≥ 36 years), gender and year of enrolment.
The study protocols were approved by relevant human research ethics committees (Supplementary Appendix 3). Written informed consent was obtained from participants. For adolescents, consent was obtained from parents or legal guardians of adolescents, and written informed assent from each adolescent. In both studies, participants with diagnosed or suspected tuberculosis disease were referred to a study-independent public health physician for treatment according to national tuberculosis control programs of the country involved.
Overview of strategy for identifying and validating the tuberculosis risk signature
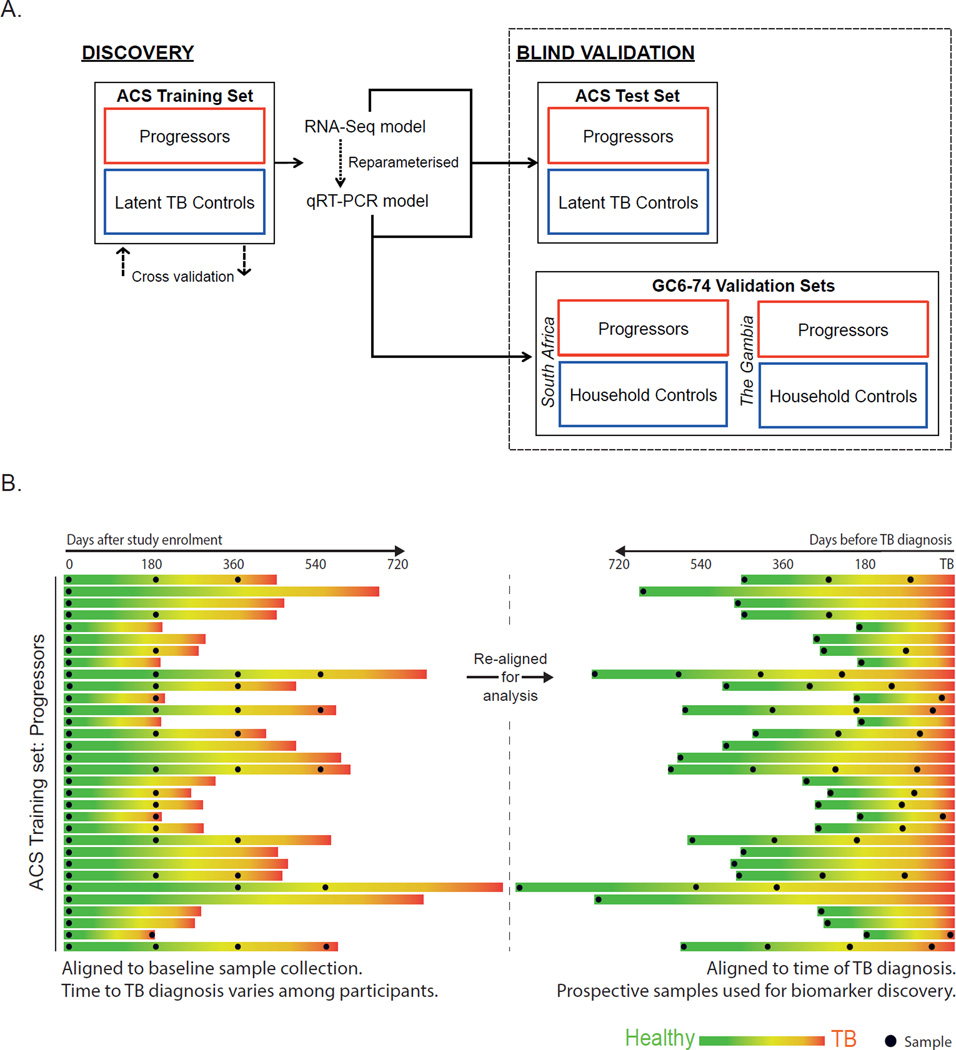
Figure 2A shows the analytical approach (detail in sections below). The tuberculosis risk signature was derived from mining RNA sequencing (RNA-Seq) data generated from the ACS training set. The RNA-Seq-based tuberculosis risk signature was then adapted to the quantitative real-time PCR (qRT-PCR) platform. The RNA-Seq- and qRT-PCR-based signature of risk was validated by blind prediction on untouched samples from the ACS test set. The qRT-PCR-based TB risk signature was also validated by blind prediction on independent GC6-74 cohort samples from South Africa and The Gambia.
Figure 2. Strategy for discovery and validation of the tuberculosis risk signature.
(A) Flow diagram for the discovery and validation of the tuberculosis risk signature. The tuberculosis risk signature was obtained by data mining of a whole blood RNA-Seq dataset generated from the ACS training set. The predictive potential of the risk signature was evaluated by rigorous cross-validation. The tuberculosis risk signature was adapted to qRT-PCR, and then the RNA-Seq and qRT-PCR versions of the signature were employed to predict tuberculosis progression using untouched blinded samples from the ACS test set. The qRT-PCR-based tuberculosis risk signature was then employed to predict tuberculosis progression using untouched blinded samples from the South African and Gambian cohorts of GC6-74. (B) Synchronization of the ACS training set in terms of the clinical outcome. To ensure optimal extraction of a tuberculosis risk signature from the ACS training set, the time scale of the RNA-Seq dataset was re-aligned according to tuberculosis diagnosis instead of study enrolment, allowing gene expression differences to be measured before disease diagnosis. Each progressor within the ACS training set is represented by a horizontal bar. The length of the bar represents the number of days between study enrolment and diagnosis with active tuberculosis. During follow-up, each progressor transitioned from an asymptomatic healthy state (green) to pulmonary disease (red). Left side: alignment of PAXgene sample collection (black points) with respect to study enrolment. Right side: alignment of PAXgene sample collection with respect to diagnosis with active tuberculosis, for use in analysis.
RNA-Seq analysis of the ACS training and test sets
RNA was extracted from PAXgene tubes of the ACS training set. Globin transcript depletion (GlobinClear, Life Technologies) was followed by cDNA library preparation using Illumina mRNA-Seq Sample Prep Kit. RNA-Seq was performed by Expression Analysis Inc., at 30 million 50bp paired-end reads, on Illumina HiSeq-2000 sequencers. Read pairs were aligned to the hg19 human genome using gsnap22, which generated a table of gene expression abundances for each sample. This gene expression abundance was measured at the level of splice junction counts, which quantifies the frequency of specific mRNA splicing events in expressed genes; this approach would facilitate translation to qRT-PCR. For simplicity, splice junction expression levels are referred to as “gene expression levels” throughout.
Generation of the tuberculosis risk signature
The computational strategy employed was an extension of the k-top-scoring pairs (k-TSP) methodology, which has been used successfully for identifying cancer biomarkers23,24. The k-TSP approach identifies pairs of genes that discriminate better than either gene would individually24. We replaced the k-TSP rank-based gene pair models with so-called support vector machine (SVM)–based gene pair models for greater flexibility in predictions. This modification is similar to the k-TSP modification proposed by Shi et al.25, but holds the advantage of retaining the fault-tolerance and parsimony of k-TSP.
For analysis, prospective RNA-Seq data of progressors was realigned to the time point at which active tuberculosis was diagnosed (Figure 2B), thereby synchronizing the cohort with respect to outcome.
The genes that comprise the final tuberculosis risk signature were selected in two stages, using data from the ACS training set. First, a large set of genes was identified by comparing gene expression in progressors at the most proximal time point to diagnosis with that in matched controls. SVM models were trained on these data points for all possible pairwise combinations of risk-associated genes. Second, the models were filtered for predictive accuracy using the remaining prospective progressor and control samples. Surviving SVM models comprised the tuberculosis risk signature, which computes a “tuberculosis risk score” based on blood gene expression levels measured at a single time point. The algorithm is fully described in Supplementary Appendix 4.
Adaptation of the tuberculosis risk signature to qRT-PCR
The tuberculosis risk signature was adapted from the original RNA-Seq-based platform to qRT-PCR by directly matching splice junctions in the signature to commercial TaqMan primer sets (Thermo Fisher Scientific, Supplementary Appendix 4). A complete set of qRT-PCR data for selected primers was generated for ACS training set samples, using the BioMark HD multiplex microfluidic instrument (Fluidigm). Parameters in the qRT-PCR-based version of the tuberculosis signature were then assigned by fitting the model to the dataset.
Blind prediction of tuberculosis risk in the ACS test set cohort
RNA-Seq and qRT-PCR analysis of samples from the ACS test set were performed as described above. Prior to analysis, all test set samples were assigned random numerical codes that masked study time points and progressor and control status. Prediction of tuberculosis risk on the masked ACS test set samples was then performed in a fully blinded manner, in parallel, using RNA-Seq and qRT-PCR-based versions of the signature.
Blind prediction on the independent GC6-74 validation cohorts using the qRT-PCR-based signature of risk
qRT-PCR analysis of samples from the South African and Gambian cohorts of GC6-74 was performed as described above, >1 year after ACS validation analysis. Prior to analysis, all samples were assigned random numerical codes. Fully blinded predictions were then made using the qRT-PCR-based signature of risk.
Applying the tuberculosis risk signature to published tuberculosis diagnosis datasets
To allow evaluation of the risk signature for diagnosis of active disease, results from published microarray-based studies of active tuberculosis vs. latent disease or other disease was used9–13. The signature was adapted from RNA-Seq to the Illumina platform and parameterized using tuberculosis cases and latently M. tuberculosis infected controls from the UK training cohort of Berry, et al11 (detail in Supplementary Appendix 4). The locked-down Illumina microarray based risk signature was used to make predictions on the independent test and validation cohorts from the Berry, et al. study,11 and from the subsequent studies.9,10,12,13
The funders played no role in data collection, analysis, interpretation or writing of the manuscript. DEZ, APN, TJS, ET, LMA, AA and WAH had access to all of the data. All authors read and approved submission of the manuscript.
Results
Participants
Forty-six ACS participants with microbiologically confirmed tuberculosis were identified as progressors (Figure 1A, Supplementary Table 1). For progressors, the time between sample collection and diagnosis with active tuberculosis (“time to diagnosis”) ranged from 1 to 894 days (Figure 2B, Supplementary Table 2). One hundred and seven controls who were infected with M. tuberculosis at enrollment but who remained healthy during 2 years of follow-up were matched to progressors. Prior to analysis, progressors and controls were randomly divided into a training set of 37 progressors and 77 controls, and a test set of 9 progressors and 30 controls (Figure 1A and 2A).
GC6-74 participants were household contacts of newly diagnosed index cases with pulmonary tuberculosis disease (Figure 1B). Two GC6-74 sites, South Africa and The Gambia, had sufficient numbers of progressors and controls to allow analysis, and were therefore included in this study (Figure 1B and 2A). A total of 43 progressors and 172 controls were identified at the South African site, while 30 progressors and 129 controls were identified at the Gambian site (Figure 1B, Supplementary Tables 3 and 4).
Construction of the tuberculosis risk signature from the ACS training set
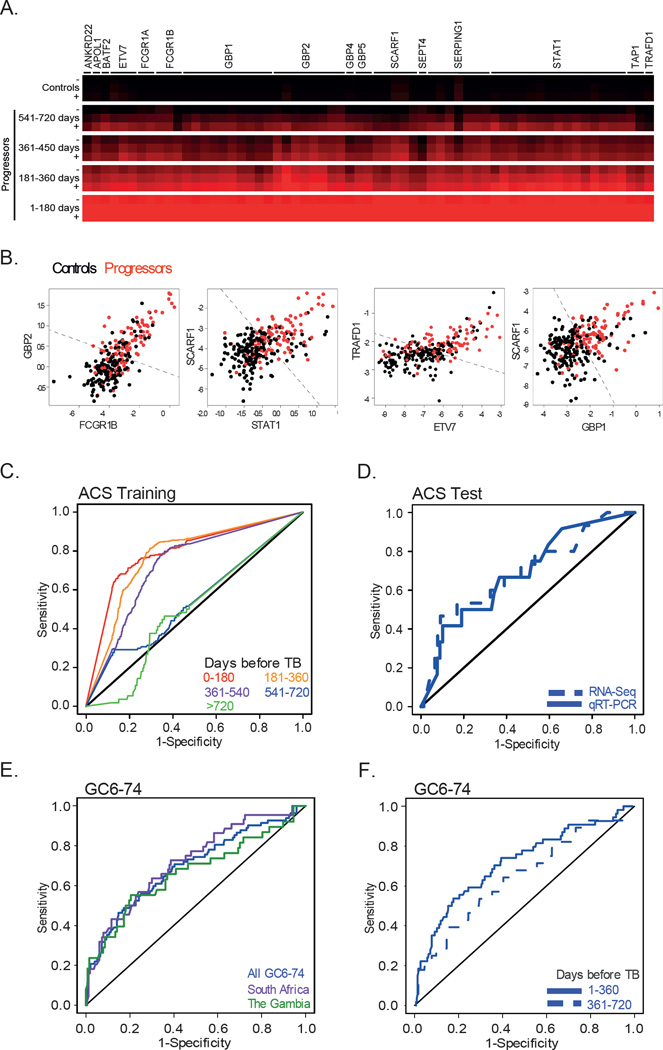
RNA was isolated from all progressor and matched control samples of the ACS training set and analyzed by RNA-Seq (Figure 2A and Supplementary Tables 5 and 6). Data mining of the RNA-Seq data derived a candidate signature of risk for tuberculosis disease progression. The signature comprised paired splice junction data from 16 genes (Supplementary Figure 1A and Supplementary Tables 7–9 and 11). Expression of signature genes in samples from progressors increased as tuberculosis diagnosis approached (Figure 3A). Robust discrimination between progressors and controls based on the expression of the gene pairs in the signature was readily apparent (Figure 3B).
Figure 3. The tuberculosis risk signature and validation by prediction of tuberculosis disease progression in the untouched ACS test set and the independent GC6-74 cohorts.
(A) Heatmap depicting relative expression level of genes comprising the tuberculosis risk signature in progressors, compared with controls. Higher expression in progressors relative to controls is indicated by intensity of red colour; the average and standard devations (+ and −) are shown. Individual heatmap rows represent distinct splice junctions of individual genes that comprise the signature. Relative expression in each of four 180-day time windows prior to tuberculosis diagnosis is shown. (B) The tuberculosis risk signature was generated by assessing multiple gene-pair interactions; two representative gene-pair signatures are shown. In each scatterplot, the normalized expression of one gene within the pair is plotted against that of the other gene, for all ACS training set data points. The black dots represent control samples, whereas the red dots represent progressor samples. The dotted black line indicates the optimal linear decision boundary for discriminating progressors from controls. (C) Receiver operating characteristic curves (ROCs) depicting the predictive potential of the tuberculosis risk signature for discriminating progressors from controls. Each ROC curve corresponds to a 180-day interval prior to tuberculosis diagnosis. Prediction performance was assessed by 100 four-to-one training-to-test splits of the ACS training set. (D) ROC curves for blind prediction of tuberculosis disease progression on untouched ACS test set samples using the RNA-Seq-based (dotted line) or qRT-PCR-based (solid line) signature. (E) Blind prediction on the combined GC6-74 cohort (blue), South African cohort (purple) or Gambian cohort (green); (F) Stratification of prediction on the overall GC6-74 cohort by time before tuberculosis diagnosis.
The predictive potential of the tuberculosis risk signature was demonstrated within the ACS training set by cross-validation (Figure 2A); the risk signature achieved 71·2% sensitivity in the 6 month period immediately prior to diagnosis, and 62·9% sensitivity 6–12 months before diagnosis, at a specificity of 80·6% (Figure 3C and Table 1). During the 12–18 month period prior to diagnosis, the signature achieved 47·7% sensitivity.
Table 1.
Cross-validation performance of the TB risk signature on the ACS training set.
| Days before TB | ROC AUC (95% CI) | Accuracy | Threshold |
|---|---|---|---|
| 1–180 | 0·791 (0·763, 0·820) | 71·2% (66·6, 75·2) | 61% |
| 181–360 | 0·771 (0·749, 0·794) | 62·9% (59·0, 66·4) | 61% |
| 361–540 | 0·726 (0·698, 0·755) | 47·7% (42·9, 52·5) | 61% |
| 541–720 | 0·540 (0·490, 0·591) | 29·1% (23·1, 35·9) | 61% |
| > 720 | 0·496 (0·433, 0·559) | 5·4% (2·4, 13·0) | 61% |
| 1–360 | 0·779 (0·761, 0·798) | 66·1% (63·2, 68·9) | 61% |
| 360–720 | 0·647 (0·621, 0·673) | 37·5% (33·9, 41·2) | 61% |
| Full ACS | 0·743 (0·729, 0·758) | 58·4% (56·1, 60·7) | 61% |
| Specificity | -- | 80·0% (78·6, 81·4) | 61% |
Validation of the signature of risk on the ACS test set
To facilitate broad application, the tuberculosis risk signature was adapted to a practical platform, qRT-PCR (Figure 2A and Supplementary Tables 13 and 14). The RNA-Seq and qRT-PCR versions of the tuberculosis risk signature were used to predict tuberculosis risk in the heretofore untouched ACS test set samples. This was done in a fully blinded manner, with all sample meta-data masked prior to making predictions. The ability of both versions of the signature to predict tuberculosis progression in healthy subjects was validated (Figure 3D and Table 2).
Table 2.
Blind prediction performance of the TB risk signature on the ACS test set by RNA-Seq and qRT-PCR, and on the GC6-74 cohorts from South Africa and The Gambia by qRT-PCR.
| Prediction accuracy (95% CI) | |||||||
|---|---|---|---|---|---|---|---|
| Cohort | Platform | Days before TB |
ROC AUC (95% CI) |
ROC p- value |
Sensitivity | Specificity | Threshold |
| ACS Test | |||||||
| All ACS Test |
RNA-Seq | 0·686 (0·523, 0·849) |
0·018 | 41·7% (22·3%, 64·5%) |
89·9% (82·6%, 94·0%) |
82% | |
| All ACS Test |
qRT-PCR | 0·693 (0·536, 0·850) |
0·0095 | 46·7% (27·8%, 66·6%) |
90·9% (83·8%, 94·7%) |
76% | |
| GC6-74 | |||||||
| All GC6- 74 |
qRT-PCR | 1–720 | 0·694 (0·625, 0·763) |
<0·0001 | 48·8% (39·9%, 57·7%) |
82·8% (78·7%, 86%) |
76% |
| South Africa |
qRT-PCR | 1–720 | 0·720 (0·633, 0·806) |
<0·0001 | 43·2% (31·7%, 55·5%) |
87·7% (82·7%, 91·2%) |
79% |
| The Gambia |
qRT-PCR | 1–720 | 0·665 (0·555, 0·775) |
0·001 | 50% (37·1%, 62·8%) |
81·9% (75·5%, 86·7%) |
78% |
| All GC6- 74 |
qRT-PCR | 1–360 | 0·718 (0·637, 0·800) |
<0·0001 | 53·7% (42·6%, 64·3%) |
82·8% (78·7%, 86%) |
76% |
| All GC6- 74 |
qRT-PCR | 361–720 | 0·648 (0·532, 0·764) |
0·0048 | 39·3% (25·8%, 54·8%) |
85·5% (81·7%, 88·5%) |
79% |
To determine whether inclusion of a larger number of genes would have increased accuracy of predictions, the performance of a random forest-based classifier comprised of 631 genes was assessed; the outcome was equivalent to when the tuberculosis risk signature was used for classification (Supplementary Figure 2 and Supplementary Table 15)
Validation of the tuberculosis risk signature on the independent GC6-74 cohorts
For independent validation, the qRT-PCR-based signature was used to predict tuberculosis progression using samples collected from healthy participants in the GC6-74 adult household contact cohorts from South Africa and The Gambia (Figure 2A). Predictions were made in a blinded manner. The ability of the signature to predict tuberculosis progression in healthy subjects did validate in these independent cohorts, regardless of whether these were analyzed individually or collectively (p<0·0001; Figure 3E and Table 2). As for the ACS, the signature had greater sensitivity for predicting tuberculosis in samples collected closer to the time of diagnosis (Figure 3F, Table 2).
Performance of risk signatures for diagnosis of tuberculosis and for response to therapy
Since the sensitivity of the tuberculosis risk signature increased as the time of diagnosis approached, we evaluated performance of the risk signature for diagnosis of active tuberculosis disease. We performed these analyses after adapting the signature to Illumina microarrays, using data from the UK training cohort of Berry, et al.11 (Supplementary Figure 3A and Supplementary Table 17), which enabled use of published datasets9–13. The signature readily differentiated active tuberculosis from latent infection in adult cohorts from the UK, South Africa, and Malawi, including populations that were co-infected with HIV (Supplementary Figure 3B–C and Supplementary Table 17). The signature also discriminated active tuberculosis from other pulmonary diseases (Supplementary Figure 3D–E and Supplementary Table 17). Despite being derived from adolescents, the signature discriminated active, culture confirmed, tuberculosis from latent M. tuberculosis infection and from other diseases in childhood (Supplementary Figure 3F–G and Supplementary Table 17). Finally, applying the signature to data from a treatment study9 showed that the active tuberculosis signature gradually disappears during 6 months of therapy (Supplementary Figure 3H and Supplementary Table 17).
Discussion
Approximately one third of the world’s population may harbour latent M. tuberculosis infection and is at risk of active disease. We have identified a gene expression signature for predicting risk of tuberculosis disease progression. This signature was discovered in a longitudinal analysis of South African adolescents with latent M. tuberculosis infection who either developed tuberculosis disease or remained healthy. The signature was then validated on blinded samples from untouched adolescents of the same parent cohort. The signatures were again validated, in independent cohorts of longitudinally followed household contacts of tuberculosis disease patients from South Africa and The Gambia, who either developed tuberculosis disease or remained healthy. These results demonstrate that it is possible to predict progression from latent to active disease, using whole blood gene expression measurements at any single time point up to 18 months before tuberculosis disease manifests.
The tuberculosis risk signature was discovered using RNA-Seq, a transcriptome analysis technology that is quantitative, sensitive, and unbiased.26 The signature was formulated using a framework termed SVM, an extension of the k-TSP approach23 which robustly generates a tuberculosis risk score from gene expression data, using simple arithmetics (Supplementary Table 11). The signature was adapted from RNA-Seq to qRT-PCR, a more targeted and affordable technology. The power of the approach was demonstrated by blinded validation of the qRT-PCR-based signature in the independent cohorts.
The tuberculosis risk signature predicted tuberculosis disease progression despite marked diversity between the ACS and GC6-74 cohorts. This result is encouraging given the different age ranges (adolescents versus adults), different infection or exposure status, distinct ethnicity and genetic backgrounds,27,28 differing local epidemiology,1 and differing circulating strains of M. tuberculosis29 between South Africa and The Gambia. It is conceivable that distinct mechanisms of progression will be revealed when specific sub-groups of progressors are analysed (e.g., early vs. late progressors in GC6-74). Targeted analyses to identify distinct mechanisms of progression are underway.
To explore potential application of the signature for targeted preventive therapy, we estimated the relative risk for tuberculosis disease between signature positive and negative persons from a representative adult population from South Africa, where tuberculosis is endemic. The relative risk of tuberculosis disease is approximately 2 when IGRA or TST is used30, whereas the relative risk using our risk signature was between 6 and 14. Moreover, this risk signature would aid in detection of asymptomatic and or undiagnosed tuberculosis disease. For example, when applied to combined data from 4 studies of HIV uninfected South African adults9–12, involving 130 prevalent tuberculosis cases and 230 controls, the signature discriminated between active tuberculosis patients and uninfected or tuberculosis infected healthy controls with 87% sensitivity and 97% specificity.
Although our focus was on prospective prediction of tuberculosis disease, we also showed that the risk signature was excellent for differentiating tuberculosis disease from latent infection and from other disease states. This ability to diagnose tuberculosis disease was not markedly impacted by HIV status. The risk signature could also diagnose culture positive childhood tuberculosis, but not culture negative childhood disease.31 These results suggest that the risk signature might reflect bacterial load in the lung, as culture positive childhood tuberculosis is likely associated with higher bacterial loads, compared with culture negative disease. An association between the risk signature and bacterial load was further supported by meta-analysis of a published treatment study9, in which the signature relaxed during 6 months of antimicrobial therapy. It is presently not known whether the risk signature will be useful for predicting treatment failure or recurrence.
While enrichment analysis of published blood signatures for active tuberculosis implicates a variety of biological processes in the disease, the gene module “interferon response” was the sole module that was over-represented in the risk signature (Supplementary Tables 19–21). Overlap between 15 of the 16 genes in our prospective TB risk signature and the 393 gene signature of active tuberculosis disease from Berry, et al.11 suggests that chronic peripheral activation of the interferon response precedes the onset of active disease and the inflammatory manifestations of tuberculosis disease revealed by previously published gene expression studies9–13. Although additional research is required to understand the functional role of interferon responses during tuberculosis progression, pathogen induction of type I interferons and their detrimental effects on immunity to tuberculosis have been shown in several in vivo studies in mice32–34 and in in vitro experiments of human cells.35 Nevertheless, not all interferon response genes may be associated with a poor outcome, as we showed that genes such as GBP1, STAT1, and TAP1 may play a protective role during tuberculosis infection (Supplementary Table 18).
Our predictive signature was obtained from transcriptomic analysis of peripheral whole blood. This compartment, although conveniently sampled, may not accurately reflect pathogenic events in the lung, primarily affected by tuberculosis disease. Regardless, circulating white blood cells can serve as sentinels of lung pathophysiology, as transcriptional changes occur when the cells migrate through this organ. To explore a possible cell-type specific origin of the risk signature, we used data from published global gene expression in whole blood and sorted PBMC, monocytes, neutrophils, and T cells from healthy controls and tuberculosis patients10. Differential expression of the risk signature genes between healthy controls and tuberculosis patients was comparable in whole blood and PBMC (Supplementary Table 22), suggesting that contribution of granulocytes to the risk signature is redundant. Consistent with this result, comparable differential expression of risk signature genes was observed in monocytes and neutrophils. When compared to the diagnostic signature of Berry, et al.11, reported to be derived from neutrophils, these results suggest that progression to active tuberculosis involves more diverse cell types.
To date, Sloot, et al.36 published the only report of prospective associations between blood gene expression and tuberculosis disease risk. Using a pre-defined 141 gene panel, PBMC RNA expression in 15 HIV infected drug users who ultimately developed active tuberculosis disease was compared with 16 who did not develop tuberculosis. Four genes assayed exhibited nominal expression differences (unadjusted p<0·05) and, when combined, two genes, IL-13 and AIRE, fit the data (ROC AUC fit = 0·8). The association between these genes and tuberculosis progression was not validated in a test set or independent cohort; none of the four genes exhibited differences between progressors and controls in our whole blood RNA-Seq datasets.
Our results, demonstrating that blood-based signatures in healthy individuals can predict progression to active tuberculosis disease, pave the way for the establishment of diagnostic tools that are scalable and inexpensive. An important first step would be to test whether the signature can predict tuberculosis disease in the general population, rather than the select populations included in this project; for example, the risk of TB disease in our populations was much higher than the lifetime risk of 10% encountered in the general population. The newly described signature holds potential for highly targeted preventive therapy, and therefore for interrupting the global epidemic.
Supplementary Material
Acknowledgments
Funding, and role of the funding source
This work was supported by grants from the Bill and Melinda Gates Foundation (BMGF) Global Health grants OPP1021972 and OPP1023483 and Grand Challenges in Global Health (GC6-74 grant 37772), the National Institutes of Health (RO1-AI087915), the European Union FP7 (ADITEC, 280873), and the Strategic Health Innovation Partnerships (SHIP) Unit of the South African Medical Research Council with funds received from the South African Department of Science and Technology. The ACS study was also supported by Aeras and BMGF GC 6-74 (grant 37772) and BMGF GC 12 (grant 37885) for QuantiFERON testing. We acknowledge European Commission (EC) FP7 NEWTBVAC contract no. HEALTH.F3.2009 241745 and EC HOR2020 project TBVAC2020.
AP-N and SS were supported by Postdoctoral Research Awards from The Carnegie Corporation of New York. AP-N was also supported by The Claude Leon Foundation and the Columbia University-Southern African Fogarty AIDS International Training and Research Program (AITRP) through the Fogarty International Center, National Institutes of Health (grant # 5 D43 TW000231). SHEK also acknowledges support from European Union’s Seventh Framework Programme (EU FP7) project “ADITEC” (HEALTH-F4-2011-280873; the Innovative Medicines Initiative (IMI) Joint Undertaking (JU) “Biomarkers for Enhanced Vaccine Safety” project BioVacSafe (IMI JU Grant No. 115308); and the European Union’s Horizon 2020 project “TBVAC2020” (grant no. 643381).
The funding sources had no role in the writing or decision to submit this publication. No payment was received for writing this manuscript. The corresponding author has had full access to all the data of the study, and has had the final responsibility for submission for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ contributions
HM, GDH, TH, SV, EJH, MO, RH, HMD, WHB, BT, HM-K, MH, SKP, SHEK, GW and WAH contributed to the original design of the studies. AP-N, TJS, SS, HM, ME, WW, DA, FK, TH, EJH, MO, RH, HMD, WHB, BT, HM-K, AC, KD, MH, SKP, SHEK, GW and WAH contributed to data collection. DEZ, AP-N, TJS, ET, SS, LMA, SHEK, AA and WAH contributed to data analysis and interpretation of the results. The manuscript was written by DEZ, AP-N, TJS, AA and WAH. DEZ, APN, TJS, ET, LMA, AA and WAH had access to all of the data. All authors read and approved submission of the manuscript.
Declaration of interests
The authors have no potential conflicts to declare.
References
- 1.WHO. WHO. Global Tuberculosis Report. 2014 2014. http://www.who.int/tb/publications/global_report/en/ (accessed.
- 2.Comstock GW, Livesay VT, Woolpert SF. The prognosis of a positive tuberculin reaction in childhood and adolescence. American journal of epidemiology. 1974;99(2):131–138. doi: 10.1093/oxfordjournals.aje.a121593. [DOI] [PubMed] [Google Scholar]
- 3.Vynnycky E, Fine PE. Lifetime risks, incubation period, and serial interval of tuberculosis. American journal of epidemiology. 2000;152(3):247–263. doi: 10.1093/aje/152.3.247. [DOI] [PubMed] [Google Scholar]
- 4.Shea KM, Kammerer JS, Winston CA, Navin TR, Horsburgh CR., Jr Estimated rate of reactivation of latent tuberculosis infection in the United States, overall and by population subgroup. American journal of epidemiology. 2014;179(2):216–225. doi: 10.1093/aje/kwt246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horsburgh CR, Jr, O'Donnell M, Chamblee S, et al. Revisiting rates of reactivation tuberculosis: a population-based approach. American journal of respiratory and critical care medicine. 2010;182(3):420–425. doi: 10.1164/rccm.200909-1355OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horsburgh CR., Jr Priorities for the treatment of latent tuberculosis infection in the United States. The New England journal of medicine. 2004;350(20):2060–2067. doi: 10.1056/NEJMsa031667. [DOI] [PubMed] [Google Scholar]
- 7.Barry CE, 3rd, Boshoff HI, Dartois V, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nature reviews Microbiology. 2009;7(12):845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walzl G, Ronacher K, Hanekom W, Scriba TJ, Zumla A. Immunological biomarkers of tuberculosis. Nat Rev Immunol. 2011;11(5):343–354. doi: 10.1038/nri2960. [DOI] [PubMed] [Google Scholar]
- 9.Bloom CI, Graham CM, Berry MP, et al. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PloS one. 2012;7(10):e46191. doi: 10.1371/journal.pone.0046191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bloom CI, Graham CM, Berry MP, et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PloS one. 2013;8(8):e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry MP, Graham CM, McNab FW, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaforou M, Wright VJ, Oni T, et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS medicine. 2013;10(10):e1001538. doi: 10.1371/journal.pmed.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson ST, Kaforou M, Brent AJ, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. The New England journal of medicine. 2014;370(18):1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutherland JS, Loxton AG, Haks MC, et al. Differential gene expression of activating Fcgamma receptor classifies active tuberculosis regardless of human immunodeficiency virus status or ethnicity. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2014;20(4):O230–O238. doi: 10.1111/1469-0691.12383. [DOI] [PubMed] [Google Scholar]
- 15.Ottenhoff TH, Dass RH, Yang N, et al. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PloS one. 2012;7(9):e45839. doi: 10.1371/journal.pone.0045839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maertzdorf J, Weiner J, 3rd, Mollenkopf HJ, et al. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(20):7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maertzdorf J, Repsilber D, Parida SK, et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 2011;12(1):15–22. doi: 10.1038/gene.2010.51. [DOI] [PubMed] [Google Scholar]
- 18.Maertzdorf J, Ota M, Repsilber D, et al. Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PloS one. 2011;6(10):e26938. doi: 10.1371/journal.pone.0026938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joosten SA, Fletcher HA, Ottenhoff TH. A helicopter perspective on TB biomarkers: pathway and process based analysis of gene expression data provides new insight into TB pathogenesis. PloS one. 2013;8(9):e73230. doi: 10.1371/journal.pone.0073230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cliff JM, Lee JS, Constantinou N, et al. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. The Journal of infectious diseases. 2013;207(1):18–29. doi: 10.1093/infdis/jis499. [DOI] [PubMed] [Google Scholar]
- 21.SADOH. SADoH. National tuberculosis management guidelines. 2014 2014. http://www.tbonline.info/media/uploads/documents/ntcp_adult_tb-guidelines-27.5.2014.pdf (accessed. [Google Scholar]
- 22.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics (Oxford, England) 2010;26(7):873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan AC, Naiman DQ, Xu L, Winslow RL, Geman D. Simple decision rules for classifying human cancers from gene expression profiles. Bioinformatics. 2005;21(20):3896–3904. doi: 10.1093/bioinformatics/bti631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bo T, Jonassen I. New feature subset selection procedures for classification of expression profiles. Genome biology. 2002;3(4):RESEARCH0017. doi: 10.1186/gb-2002-3-4-research0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi P, Ray S, Zhu Q, Kon MA. Top scoring pairs for feature selection in machine learning and applications to cancer outcome prediction. BMC bioinformatics. 2011;12:375. doi: 10.1186/1471-2105-12-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tishkoff SA, Reed FA, Friedlaender FR, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324(5930):1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Black GF, Thiel BA, Ota MO, et al. Immunogenicity of novel DosR regulon-encoded candidate antigens of Mycobacterium tuberculosis in three high-burden populations in Africa. Clinical and vaccine immunology : CVI. 2009;16(8):1203–1212. doi: 10.1128/CVI.00111-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comas I, Coscolla M, Luo T, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nature genetics. 2013;45(10):1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahomed H, Ehrlich R, Hawkridge T, et al. TB incidence in an adolescent cohort in South Africa. PLoS One. 2013;8(3):e59652. doi: 10.1371/journal.pone.0059652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray JW. Childhood tuberculosis and its early diagnosis. Clinical biochemistry. 2004;37(6):450–455. doi: 10.1016/j.clinbiochem.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Manca C, Tsenova L, Freeman S, et al. Hypervirulent M. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak-Stat pathway. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2005;25(11):694–701. doi: 10.1089/jir.2005.25.694. [DOI] [PubMed] [Google Scholar]
- 33.Mayer-Barber KD, Andrade BB, Oland SD, et al. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature. 2014;511(7507):99–103. doi: 10.1038/nature13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorhoi A, Yeremeev V, Nouailles G, et al. Type I IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. European journal of immunology. 2014;44(8):2380–2393. doi: 10.1002/eji.201344219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teles RM, Graeber TG, Krutzik SR, et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science. 2013;339(6126):1448–1453. doi: 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sloot R, Schim van der Loeff MF, van Zwet EW, et al. Biomarkers Can Identify Pulmonary Tuberculosis in HIV-infected Drug Users Months Prior to Clinical Diagnosis. EBioMedicine. 2015;2:172–179. doi: 10.1016/j.ebiom.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahomed H, Hawkridge T, Verver S, et al. The tuberculin skin test versus QuantiFERON TB Gold(R) in predicting tuberculosis disease in an adolescent cohort study in South Africa. PloS one. 2011;6(3):e17984. doi: 10.1371/journal.pone.0017984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Machingaidze S, Wiysonge CS, Gonzalez-Angulo Y, et al. The utility of an interferon gamma release assay for diagnosis of latent tuberculosis infection and disease in children: a systematic review and meta-analysis. The Pediatric infectious disease journal. 2011;30(8):694–700. doi: 10.1097/INF.0b013e318214b915. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Ishwaran H. Random forests for genomic data analysis. Genomics. 2012;99(6):323–329. doi: 10.1016/j.ygeno.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owzar K, Barry WT, Jung SH. Statistical considerations for analysis of microarray experiments. Clinical and translational science. 2011;4(6):466–477. doi: 10.1111/j.1752-8062.2011.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Platt JC. Sequential Minimal Optimization: A Fast Algorithm for Training Support Vector Machines. Microsoft Research Technical Report. 1998 MSR-TR-98-14. [Google Scholar]
- 42.Liaw A, Wiener M. Classification and regression by randomForest. R news. 2002;2(3):18–22. [Google Scholar]
- 43.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic acids research. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obermoser G, Presnell S, Domico K, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38(4):831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.