Abstract
Neuromyelitis optica (NMO) is a primary astrocyte disease associated with CNS inflammation, demyelination, and tissue injury. Brain lesions are frequently observed in regions enriched in expression of the aquaporin-4 (AQP4) water channel, an antigenic target of the NMO IgG serologic marker. Based on observations of disease reversibility and careful characterization of NMO lesion development, we propose that the NMO IgG may induce a dynamic immunological response in astrocytes. Using primary rat astrocyte-enriched cultures and treatment with NMO patient-derived serum or purified IgG, we observed a robust pattern of gene expression changes consistent with the induction of a reactive and inflammatory phenotype in astrocytes. The reactive astrocyte factor lipocalin-2 and a broad spectrum of chemokines, cytokines, and stress response factors were induced by either NMO patient serum or purified IgG. Treatment with IgG from healthy controls had no effect. The effect is disease-specific, as serum from patients with relapsing-remitting multiple sclerosis, Sjögren’s, or systemic lupus erythematosus did not induce a response in the cultures. We hypothesize that binding of the NMO IgG to AQP4 induces a cellular response that results in transcriptional and translational events within the astrocyte that are consistent with a reactive and inflammatory phenotype. Strategies aimed at reducing the inflammatory response of astrocytes may short circuit an amplification loop associated with NMO lesion development.
Keywords: Lipocalin-2, CCL2, CXCL1, CCL5, innate immunity, aquaporin-4, granulocyte, NFκB
Introduction
Neuromyelitis optica (NMO) is a primary astrocytopathy associated with central nervous system (CNS) inflammation, secondary demyelination, and variable tissue necrosis and cavitation (Popescu and Lucchinetti, 2012). The disease is characterized by optic neuritis, often severe, and transverse, longitudinally extensive myelitis, and is most frequently associated with a relapsing phenotype. Brain lesions, including diffuse cerebral white matter lesions that resemble acute disseminated encephalomyelitis and cerebral lesions associated with posterior reversible encephalopathy syndrome, are also a feature of NMO (Wingerchuk et al., 2007). Distinguishing brain lesions in NMO are typically observed in periventricular regions and include such symptomatic manifestions as intractable hiccups and nausea associated with ventral medullary periaqueductal gray lesions (Popescu et al., 2011), nacrolepsy-like hypersomnia associated with hypothalamic lesions (Baba et al., 2009), and nystagmus and diplopia associated with brainstem lesions (Kim et al., 2011). Of note, these regions are enriched in expression of the aquaporin-4 (AQP4) water channel (Pittock et al., 2006), an antigenic target of the NMO-IgG serologic marker (Lennon et al., 2004).
AQP4 is a bidirectional water channel that is predominantly expressed on the foot processes of astrocytes within the CNS (Rash et al., 1998). This channel plays a critical role in CNS water transport, particularly within the context of dyshomeostatic pathophysiological conditions such as trauma and ischemia (Papadopoulos and Verkman, 2013). Binding of the NMO-IgG to three-dimensional conformational epitopes in the extracellular loops of AQP4 on the surface of astrocytes may trigger the pathophysiology underlying NMO (Iorio et al., 2013; Melamud et al., 2012). Indeed, the high serum levels of NMO-IgG in many NMO patients coupled with the observations of functional recovery following plasma exchange (Magana et al., 2011) and anti-B cell therapy with rituximab (Cree et al., 2005) support a pathogenic role of IgG binding to the water channel (Hinson et al., 2007; Hinson et al., 2012; Melamud et al., 2012).
Despite the known interaction of NMO-IgG with AQP4, the mechanism(s) responsible for NMO lesion development and loss of function in patients remains unknown and contentious. Much of the field has emphasized astrocyte destruction mediated by complement fixation at the site of NMO-IgG interactions with AQP4 (Saadoun et al., 2010). While destructive lesions and complement deposition are certainly an important aspect of NMO pathophysiology (Pittock et al., 2013), current evidence gleaned from human tissue suggests that many NMO lesions are non-destructive and are biased toward an inflammatory phenotype (Popescu et al., 2011; Popescu and Lucchinetti, 2012). Indeed, from a therapeutic standpoint, resolution of both neurologic function and magnetic resonance imaging hallmarks in NMO patients is not consistent with widespread, overt lytic destruction (Magana et al., 2011; Magana et al., 2009). Therefore, a mechanism that links NMO-IgG binding to astrocytic AQP4 with the initiation of a reversible, inflammatory, accumulative cascade rather than an explosively destructive irreversible event is needed to understand NMO pathogenesis.
Based on our previous observation of granulocytic infiltrate present in early active NMO lesions (Almekhlafi et al., 2011; Lucchinetti et al., 2002) and the recognition that some lesions show loss of APQ4 coupled to preservation of GFAP (Popescu et al., 2011; Popescu and Lucchinetti, 2012; Roemer et al., 2007), we hypothesized that binding of the NMO-IgG to AQP4 triggers an immunologic response in astrocytes that results in the recruitment of innate immune cells into the CNS and the concomitant amplification and exacerbation of tissue dyshomeostasis and injury. To test this hypothesis we queried the genetic and proteomic responses of primary rodent astrocyte cultures to incubation with the NMO-IgG. We found that NMO-IgG induces rapid production of chemokines and cytokines by primary rat astrocyte-enriched cultures that are consistent with the creation of a pro-granulocytic recruitment milieu. We also found that NMO-IgG induces stereotypical stress pathways in primary astroglia characterized by NFκB activation. Finally, the initiation of such responses was unique to NMO patient-derived IgG, suggesting that an NMO-IgG-induced inflammatory response in astroglia is the underlying pathogenic event in this disease.
Materials and Methods
Patient-derived serum collection and processing
Blood was drawn from patients or healthy volunteers into SST tubes and held at 4°C until use. Serum was prepared by centrifugation for 10 min at 3000 rpm in a clinical centrifuge. Purified serum was heat-inactivated for 30 min at 56°C and clarified by centrifugation at 4500 rpm for 10 min in a clinical centrifuge. Samples were sterile filtered at 0.22 μm and working aliquots were stored at −80°C. Except for experiments involving analysis of individual serum responses, all treatments used sera pooled from more than 10 donors.
IgG purification
Human IgG was isolated from sterile-filtered, heat-inactivated serum samples using Ab SpinTrap G affinity columns (GE Healthcare). Serum samples were mixed 1:1 with sterile PBS at pH 7 and loaded onto equilibrated columns. IgG was allowed to bind for 4 min at 22°C, then columns were washed and antibody was eluted following manufacturer's directions. Eluate was dialyzed against PBS at pH 7.4 overnight at 4°C, then concentrated on Amicon Ultra-4 centrifugation units (Millipore) with 10,000 MW cut-offs. The concentrated IgG was sterile filtered at 0.22 μm, concentration was determined by absorbance at 280 nm, and working aliquots were stored at −20°C. All experiments used IgG purified from sera pooled from more than 10 donors.
Rat primary astrocyte cultures
Mixed astrocyte-enriched cultures were prepared from cerebral cortices of E18-21 Lewis rat pups (Harlan Labs, Madison, WI, USA), following established methods (McCarthy and de Vellis, 1980). Cells were cultivated for 25 days in vitro (DIV) prior to replating at 2x105 cells per mL per well in 12-well dishes coated with 25 μg/mL poly-D-lysine. Cells were used for experiments at 28 DIV. For immunofluorescence experiments cells were plated on poly-L-lysine-coated glass coverslips.
Immunostaining
Cultures were exposed to 2.5% human serum diluted in culture medium for 1 hr on ice to bind surface antigens on living cells. After washing, cells were fixed in 4% paraformaldehyde, washed, permeabilized in 0.5% TX-100, and blocked in 10% normal donkey serum and 2% bovine serum albumin. Cells were incubated overnight at 4°C with MAB360 mouse anti-GFAP (Millipore) at 1:200, washed, incubated for 1 hr at RT with FITC-conjugated donkey anti-human (Jackson IR, 709-096-149) and Alexa-594-conjugated donkey anti-mouse (Jackson IR, 715-515-150) secondaries at 1:200. Coverslips were washed in PBS and mounted under Vectashield containing DAPI.
RNA isolation and RTPCR
Total cellular RNA was isolated using Qiagen RNeasy kits, per manufacturer's directions. RNA purity and concentration were determined spectrophotometrically using a NanoDrop-2000. At least 100 ng RNA was subjected to cDNA synthesis with the Roche Transcriptor First Strand cDNA Synthesis kits using anchored oligo(dT) and random hexamer primers. Quantitative PCR was performed on a Roche LightCycler 480 using the LightCycler 480 SYBR Green 1 master mix. After activating the enzyme for 5 min at 95°C, 45 cycles of PCR were performed as follows: 10 sec denaturation at 94°C, 10 sec annealing at 60°C, 10 sec amplification at 72°C. Primers for CCL5 were: 5'-CAC TCC CTG CTG CTT TGC-3', 5'-CAC TTG GCG GTT CCT TCG-3'. Primers for Cfb were: 5'-AAC ACT CCA TCA AGG TCA AC-3', 5'-GGC AAC ATC ATA GTC ATA GAA C-3'. Standard curves were generated using serial dilutions of oligonulceotides synthesized over the entire length of the amplicon. Crossing points on the amplification curves for experimental samples were converted to log copies by comparison to the standard curve.
Microarray
RNA samples were assessed by Agilent for integrity, purity, and concentration. Samples passing quality control were analyzed on Illumina RatRef-12 BeadChips in the Mayo Clinic Medical Genome Facility Gene Expression Core. Expression data were analyzed using the Stanford Statistical Analysis of Microarrays package (Tusher et al., 2001) as an Excel plugin.
ELISA
Following stimulation of cells, supernatants were collected, clarified by centrifugation at 14,000 g, and stored as aliquots at −80°C until analysis. Rat CCL5, CCL2, and CXCL1 were detected in the supernatants using ELISA construction kits from Antigenix America Inc.
Statistics
α=0.05 and β=0.2 were established a priori. Post hoc power analysis was performed for all experiments and significance was only considered when power≥0.8. Statistical analysis was performed using SigmaStat (Systat Software, Inc; San Jose, CA). Normality was determined by the Shapiro-Wilk test and normally distributed data were checked for equal variance. Parametric tests were only applied to data that were both normally distributed and of equal variance. The Student-Newman-Keuls pairwise comparison test was used for all post-hoc sequential comparisons. Curran-Everett guidelines were used for reporting statistical values (Curran-Everett and Benos, 2007). Statistical significance of the microarray data was determined using Storey’s positive false discovery rate (FDR) for multiple hypothesis testing and the q-value, as decribed (Storey, 2002).
Results
Serum antibodies from NMO patients bind the surface of live rat astrocytes
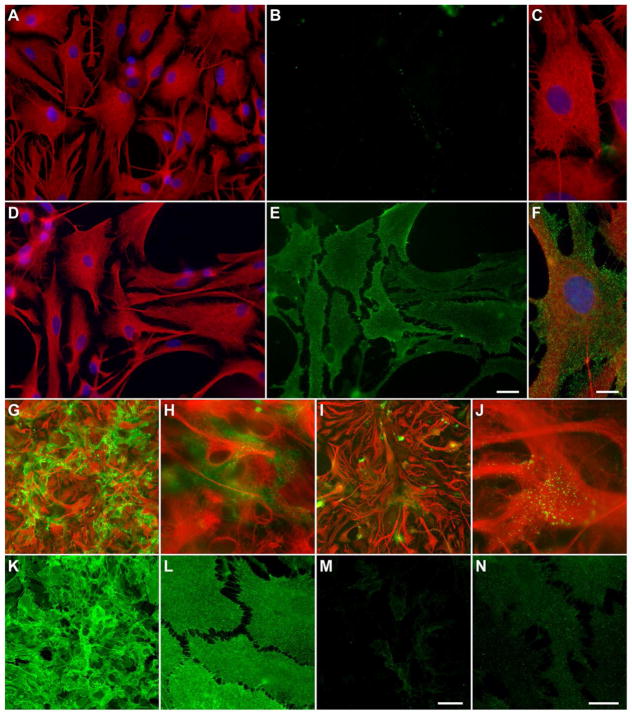
Primary rat astrocyte cultures were incubated with 2.5% normal human (CON) or NMO patient serum for 1 hr at 4°C to permit binding of immunoglobulins to the cell surface without internalization. Cells were then washed, fixed, and permeabilized, and subsequently stained overnight (O/N) with anti-GFAP, then incubated with FITC-conjugated donkey anti-human IgG to label surface-bound human serum antibodies and Alexa594-conjugated donkey anti-mouse to label GFAP. GFAP staining revealed dense fields of astrocytes with large, flat membranes (Figure 1A–F). Serum IgG from NMO patients (Figure 1E, 1F) but not from controls (Figure 1B, 1C) robustly bound the surface of astrocytes under these conditions. In order to characterize the impact of persistent exposure to NMO immunoglobulins under treatment conditions, the IgG fraction was column-purified from pooled NMO patient sera or pooled CON sera. This pooled, purifed IgG (100 μg/mL) was incubated on astrocytes for 1 hr at 37°C (Figure 1G, 1H) or 24 hr at 37°C (Figure 1I, 1J). Cells were washed, fixed, permeabilized, and stained as above to reveal GFAP and both internalized and surface human IgG (Figure 1G–1J). While robust labeling was still present after 1 hr incubation with NMO IgG (Figure 1G, 1H), by 24 hr the overall amount of signal had decreased and was mostly present in bright punctate structures (Figure 1I, 1J) that differed from the more even distribution observed after 1 hr. No labeling was observed at either timepoint in cultures incubated with CON IgG (data not shown). These observations suggested that the total level of IgG-bound AQP4 was decreased after 24 hr incubation at 37°C. To verify that expression of the antigenic target was, in fact, decreased under these conditions, cells were treated with purified NMO IgG for 24 hr, then washed, fixed, and labeled with rabbit anti-AQP4 (Sigma, #A5971, 1:500). While incubation with CON IgG had no effect on expression of AQP4 (data not shown), incubation with NMO IgG for 24 hr at 37°C induced a marked reduction in AQP4 (Figure 1M, 1N) as compared to untreated cells (Figure 1K, 1L). We conclude that serum-derived IgGs from NMO patients bind the surface of live astrocytes, triggering internalization and degradation of the antibody:antigen complexes.
Figure 1. Serum IgG from NMO patients but not from healthy controls binds the surface of live rat astrocytes.
Primary cultures of rat astroglia were incubated on ice for 1 hr with 2.5% serum collected and pooled from NMO patients (D–F) or from controls (CON) (A–C). Anti-GFAP, shown in red, revealed astrocytes (A, C, D, F). Anti-human IgG, shown in green, revealed surface binding on astrocytes in samples incubated with NMO serum (E, F) but not on astrocytes incubated with CON serum (B, C). Incubation with IgG (100 μg/mL) purified from pooled NMO patient sera showed robust labeling of astrocytes after 1 hr (G, H) at 37°C (red = GFAP; green = anti-human IgG). By 24 hr (I, J), the overall labeling intensity was greatly reduced, suggesting internalization and degradation of antigen-antibody complexes. Labeling of total AQP4 after 24 hr incubation with NMO IgG confirmed the marked loss of antigen (M, N) as compared to untreated cells (K, L). Scale bar in E is 25 μm and refers to A, B, D, E. Scale bar in F is 10 μm and refers to C. Scale bar in M is 100 μm and refers to G, I, K. Scale bar in N is 20 μm and refers to H, J, L.
Serum from NMO patients stimulates immune gene expression in rat astrocyte cultures
Primary rat astrocyte cultures were incubated with 10% CON or 10% NMO serum for 24 hr at 37°C. Triplicate experimental samples of RNA were isolated and gene expression was analyzed using Illumina RatRef-12 BeadChips. Genes failing to show significant expression in one or more of either the CON-treated or NMO-treated samples (as defined by “present” call at p>0.05) were excluded from further analysis. The dataset was then processed through the Stanford Significance Analysis of Microarrays (SAM) plugin for Excel 2010 (Tusher et al., 2001) and a curve was constructed (Supplemental Figure 1). With the false discovery rate (FDR) set to 0.2 (Storey, 2002) the SAM algorithm identified 270 genes significantly altered by treatment with NMO serum as compared to CON serum. The SAM score, calculated fold-change, and computed FDR are provided for the top 50 upregulated genes organized by fold-change (Table 1). In addition, all 270 significantly altered genes are listed in Supplemental Tables 1 and 2. Note that of the 28 genes upregulated at FDR=0, 15 are immune genes (Table 1 and Supplemental Table 1), including 49.8-fold upregulation of lipocalin 2 (LCN2), 39.5-fold upregulation of CXCL1, 16.1-fold upregulation of CCL2, and 6.6-fold upregulation of CCL5. In addition, Ingenuity Pathway Analysis (IPA) identified numerous immune-associated pathways as significantly regulated by NMO serum (Supplemental Figure 2), including the acute phase response, interferon, chemokine, cytokine, and complement signaling pathways, and NFκB signal transduction.
Table 1.
NMO Serum vs Control Serum Top 50 Upregulated Genes
| Gene Symbol | Accession | Gene | SAM score1 | Fold Change2 | cutoff3 |
|---|---|---|---|---|---|
| CCL20 | NM_019233 | chemokine (C-C) ligand 20 | 3.79 | 52.1 | 0 |
| LCN2 | NM_130741 | lipocalin 2 | 4.03 | 49.8 | 0 |
| CXCL1 | NM_030845 | chemokine (C-X-C) ligand 1 | 8.82 | 39.5 | 0 |
| C3 | NM_016994 | complement component 3 | 4.14 | 33.7 | 0 |
| CCL2 | NM_031530 | chemokine (C-C) ligand 2 | 4.19 | 16.1 | 0 |
| CCL7 | NM_001007612 | chemokine (C-C) ligand 7 | 3.43 | 13.0 | 0 |
| CFB | NM_212466 | complement factor B | 2.69 | 6.9 | 9 |
| IL1RN | NM_022194 | interleukin 1 receptor antagonist | 4.77 | 6.8 | 0 |
| CCL5 | NM_031116 | chemokine (C-C) ligand 5 | 6.46 | 6.6 | 0 |
| CP | NM_012532 | ceruloplasmin | 2.44 | 5.8 | 13 |
| MT1A | NM_138826 | metallothionein 1a | 3.14 | 5.5 | 4 |
| C1S | NM_138900 | complement component 1s | 2.36 | 5.2 | 14 |
| ABCB1B | NM_012623 | ATP-binding cassette, subfamily B (MDR/TAP) member 1B | 3.04 | 4.9 | 4 |
| TF | NM_001013110 | transferrin | 3.25 | 4.8 | 2 |
| GDA | NM_031776 | guanine deaminase | 4.19 | 4.7 | 0 |
| SOD2 | NM_017051 | superoxide dismutase 2, mitochondrial | 2.48 | 4.6 | 12 |
| SULT1A1 | NM_031834 | sulfotransferase family, cytosolic, 1A | 2.09 | 4.4 | 22 |
| TLR2 | NM_198769 | toll-like receptor 2 | 3.15 | 4.3 | 4 |
| SLC11A2 | NM_013173 | solute carrier family 11, member 2 | 3.78 | 4.2 | 0 |
| HOPX | NM_133621 | HOP homeobox | 3.22 | 4.0 | 2 |
| RNF125 | NM_001108424 | ring finger protein 125 | 2.18 | 3.9 | 16 |
| NFKBIA | NM_001105720 | nuclear factor of kappa enhancer in B-cells inhibitor, alpha | 6.93 | 3.6 | 0 |
| PLAC8 | NM_001108353 | placenta-specific 8 | 2.21 | 3.4 | 16 |
| C1R | NM_001134555 | complement component 1r | 2.64 | 3.1 | 9 |
| CEBPB | NM_024125 | CCAAT/enhancer binding protein (C/EBP), beta | 2.53 | 3.0 | 11 |
| ADORA2B | NM_017161 | adenosine A2B receptor | 2.28 | 3.0 | 15 |
| RT1-A2 | NM_001008829 | MHC class I RT1-A2 | 2.91 | 2.9 | 6 |
| VCAM1 | NM_012889 | vascular cell adhesion molecule 1 | 2.17 | 2.9 | 18 |
| PLET1 | NM_001014209 | placenta-expressed transcript 1 | 3.78 | 2.8 | 0 |
| RAMP2 | NM_031646 | receptor (G protein-coupled) activity modifying protein 2 | 2.08 | 2.8 | 22 |
| ASS1 | NM_013157 | argininosuccinate synthetase 1 | 3.23 | 2.8 | 2 |
| ANGPTL4 | NM_199115 | angiopoietin-like 4 | 6.51 | 2.8 | 0 |
| AGTR1A | NM_030985 | angiotensin II receptor, type 1a | 2.24 | 2.8 | 16 |
| IFI204 | NM_001012029 | interferon activated gene 204 | 3.02 | 2.7 | 4 |
| CXCL11 | NM_182952 | chemokine (C-X-C) ligand 11 | 3.31 | 2.7 | 2 |
| RT1-A3 | NM_001008830 | MHC class I RT1-A3 | 2.52 | 2.6 | 11 |
| RT1-A1 | NM_001008827 | MHC class I RT1-A1 | 2.73 | 2.6 | 9 |
| ADAMTS2 | NM_001137622 | ADAM metallopeptidase with thrombospondin 1 motif, 2 | 2.86 | 2.4 | 6 |
| FKBP5 | NM_001012174 | FK506 binding protein 5 | 3.79 | 2.4 | 0 |
| RT1-M10-1 | NM_001008851 | MHC class I RT1-M10 | 2.87 | 2.4 | 6 |
| FIGF | NM_031761 | vascular endothelial growth factor D | 2.37 | 2.3 | 14 |
| TNIP1 | NM_001108826 | TNFAIP3 interacting protein 1 | 2.27 | 2.3 | 16 |
| RT1-149 | NM_001008826 | MHC class I RT1-T24 | 3.55 | 2.3 | 0 |
| ACSBG1 | NM_134389 | acyl-CoA synthetase bubblegum family member 1 | 2.69 | 2.3 | 9 |
| AMPD3 | NM_031544 | adenosine monophosphate deaminase 3 | 3.27 | 2.2 | 2 |
| RSPO2 | NM_001130575 | R-spondin 2 homolog | 2.42 | 2.2 | 13 |
| TESC | XM_213790 | tescalcin | 6.22 | 2.2 | 0 |
| PLA1A | NM_138882 | phospholipase A1 member A | 6.61 | 2.2 | 0 |
| NDRG2 | NM_133583 | N-myc downstream regulated gene 2 | 3.00 | 2.1 | 6 |
| ADM | NM_012715 | adrenomedullin | 2.61 | 2.1 | 9 |
SAM (significance analysis of microarray) score = d(i); similar to t-test t-score
Fold change NMO serum Tx vs CON serum Tx; generated by SAM
SAM false discovery rate cut-off value = q
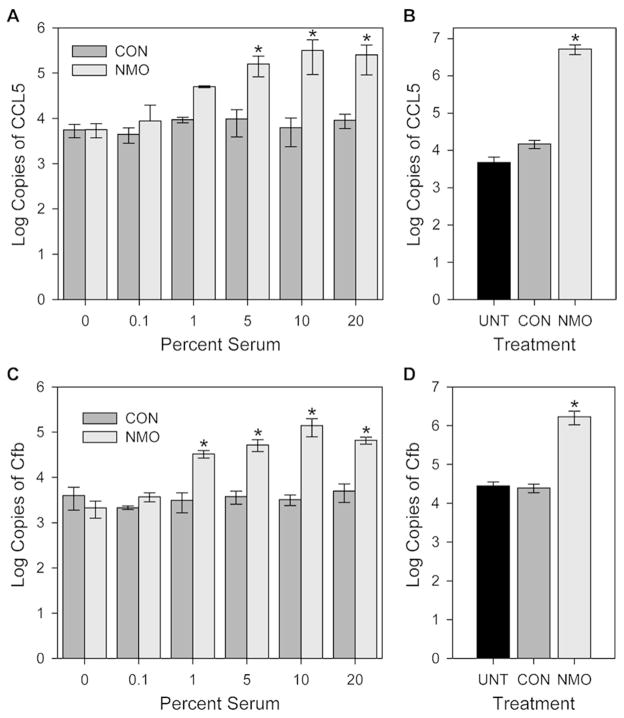
Based on the microarray analysis we selected two genes of interest, the granulocytic chemokine CCL5 and complement factor B (Cfb), and performed a dose response comparison for CON and NMO serum. Rat astrocyte cultures at 28 DIV were incubated for 6 hr at 37°C with different concentrations of serum and RNA was analyzed by RTPCR for expression of CCL5 and Cfb. Plasmid gene standards were used to provide real copy numbers for the two genes. As shown in Figure 2A and 2C, both genes were robustly upregulated only in the presence of NMO serum. For CCL5 induction, two-way ANOVA identified 5% NMO serum as the lowest concentration significantly different from CON serum (P=0.020). For Cfb, 1% NMO serum significantly upregulated expression (P=0.032). ANOVA revealed no effect of CON serum at any concentration for either gene product. The upregulation of CCL5 and Cfb following 24 hr treatment with 10% NMO serum was confirmed in separate experiments (Figure 2B and 2D). One-way ANOVA revealed that the roughly 400-fold increase in CCL5 expression in response to NMO serum was highly significant (P<0.001), while there was no difference between untreated and CON serum treated samples (P=0.992). The 100-fold induction in Cfb was also highly significant (P<0.001).
Figure 2. Serum from NMO patients but not controls induces expression of CCL5 and Cfb.
Rat astroglial cultures were treated for 6 hr (A, C) or 24 hr (B, D) at 37°C with various concentrations of pooled NMO or CON serum. Expression levels of CCL5 (A, B) and Cfb (C, D) were assessed by quantitative RT-PCR using standard curves to calculate gene copy numbers. (A) Treatment with 5% NMO serum for 6 hr induced a significant increase in CCL5 gene expression. Two-way ANOVA, F(11,24)=26.006, P<0.001 between treatments; SNK pairwise: 5% NMO vs 5% CON, q(24,2)=3.540, P=0.020. (B) Treatment for 24 hr with 10% NMO serum induced a 3-log increase in CCL5 gene expression compared to CON treated or untreated (UNT). One-way ANOVA, F(2,33)=45.318, P<0.001; SNK pairwise: NMO vs UNT, q(33,2)=11.668, P<0.001; NMO vs CON, q(33,2)=11.652, P<0.001; CON vs UNT, q(33,2)=0.0151, P=0.992. (C) Treatment with 1% NMO serum for 6 hr induced a significant increase in Cfb gene expression. Two-way ANOVA, F(11,2)=72.676, P<0.001 between treatments; SNK pairwise: 1% NMO vs 1% CON, q(24,2)=3.230, P=0.032. (D) Treatment for 24 hr with 10% NMO serum induced a 2-log increase in Cfb gene expression compared to CON or UNT. One-way ANOVA, F(2,81)=93.592, P<0.001; SNK pairwise: NMO vs UNT, q(81,2)=17.116, P<0.001; NMO vs CON, q(81,2)=17.002, P<0.001; CON vs UNT, q(81,2)=0.416, P=0.769. * = P<0.05 vs CON.
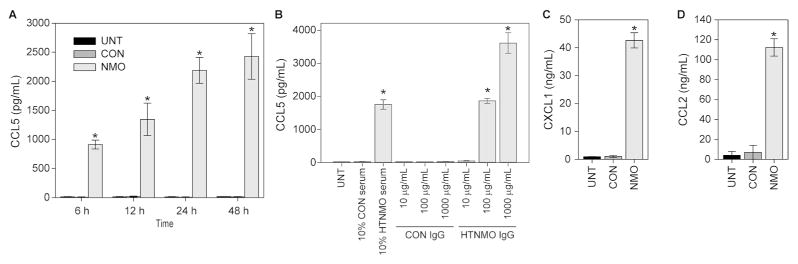
The large increase in CCL5 RNA resulted in production and release of CCL5 into the astrocyte culture supernatant following incubation with 10% NMO serum for 24 hr (Figure 3). While CCL5 was found at less than 20 pg/mL in untreated cultures and in supernatants from CON serum treated cultures, stimulation with NMO serum for 24 hr induced the release of more than 2000 pg/mL CCL5 (P<0.001) (Figure 3A). Moreover, we detected nearly 1000 pg/mL CCL5 by 6 hr in response to 10% NMO serum and this amount continued to rise through 48 hr (P<0.001) (Figure 3A).
Figure 3. Serum and purified IgG from NMO patients but not controls induces production and release of chemokines.
Rat astroglial cultures were treated with pooled serum or IgG purified from pooled serum for various times and chemokine protein production was measured in supernatants by ELISA. (A) Treatment with 10% serum for 24 hr induced the release of 2000 pg/mL CCL5, with significant production as early as 6 hr; treatment with CON serum had no effect. Two-way ANOVA, F(11,24)=623.478, P<0.001 between treatments; SNK pairwise at 6 hr (all timepoints P<0.001 for NMO vs CON or NMO vs UNT): NMO vs UNT, q(24,2)=11.385, P<0.001; NMO vs CON, q(24, 6)=11.462, P<0.001; CON vs UNT, q(24,6)=0.0765, P=0.957. (B) IgG purified from pooled NMO serum triggered CCL5 protein production at 100 μg/mL but not at 10 μg/mL. Even 1 mg/mL CON IgG did not induce CCL5 release. One-way ANOVA, F(8,18)=226.062, P<0.001 between treatments; SNK pairwise: 100 ug/mL NMO vs 100 ug/mL CON, q(18,2)=21.570, P<0.001. (C) Purified IgG (100 μg/mL) from NMO serum but not CON serum stimulated the release of more than 40 ng/mL CXCL1 over 24 hr. One-way ANOVA, F(2,7)=4270.225, P<0.001 between treatments; SNK pairwise: NMO vs UNT, q(7,2)=113.329, P<0.001; NMO vs CON, q(7,2)=113.039, P<0.001; CON vs UNT, q(7,2)=0.290, P=0.844. (D) Purified IgG (100 μg/mL) from NMO serum but not CON serum stimulated the release of more than 100 ng/mL CCL2 over 24 hr. One-way ANOVA, F(2,7)=1499.140, P<0.001 between treatments; SNK pairwise: NMO vs UNT, q(7,2)=68.021, P<0.001; NMO vs CON, q(7,2)=66.062, P<0.001; CON vs UNT, q(7,2)=1.959, P=0.216. * = P<0.001 vs CON.
Purified IgG from NMO patient serum stimulates immune gene expression in rat astrocyte cultures
To determine whether the NMO serum-induced effect was IgG dependent we incubated astrocyte cultures with 750 μg/mL IgG column-purified from pooled NMO or CON serum for 24 hr at 37°C, collected RNA, and analyzed gene expression by Illumina BeadChip. As above, we processed the data using SAM and found 3066 genes significantly regulated by NMO IgG at a FDR=0.2 (Supplemental Figure 3). While the overall magnitude of the responses was smaller than in the serum-treated experiments, the findings were highly significant, with 305 upregulated and 81 downregulated genes exhibiting a zero FDR. As with NMO serum treatment, many of the upregulated genes were immune-associated, as revealed by IPA (Supplemental Figure 4). The SAM score, calculated fold-change, and computed FDR for immune genes responding to NMO IgG are shown in Table 2. A striking pattern of CCL and CXCL chemokines was induced, as were numerous cytokines and immune response genes. For example, the neutrophilic chemokines CXCL1 and CXCL2 were both induced around 2-fold with a FDR=0 (indicating very high signficance), the monocytic chemokines CCL2 and CCL7 were also both induced around 2-fold at FDR=0, and the eosinophilic chemokine CCL5 was upregulated nearly 5-fold, again with FDR=0 (Table 2). The pro-inflammatory interleukins IL-1α and IL-1β were upregulated more than 3-fold at FDR=0 and the adhesion factors ICAM1 and VCAM were upregulated at more than 1.5-fold and FDR=0. Overall, stimulation of primary astrocytes with NMO IgG induced a transcriptional program marked by the upregulation of numerous leukocyte recruitment chemokines and cellular stress and immune activation markers.
Table 2.
Immune Genes Regulated by NMO IgG
| Gene Symbol | Accession | Gene | SAM score1 | Fold Change2 | FDR- cutoff3 |
|---|---|---|---|---|---|
| A2M | NM_012488 | alpha-2-macroglobulin | 2.1 | 1.4 | 0 |
| ADORA2A | NM_053294 | adenosine A2a receptor | 3.0 | 1.8 | 0 |
| AGT | NM_134432 | angiotensinogen | 1.3 | 1.3 | 2 |
| AGTR1A | NM_030985 | angiotensin II receptor 1a | 2.0 | 1.5 | 0 |
|
| |||||
| AP3D1 | XM_234908 | adaptor-related protein complex 3d1 | −1.1 | −1.2 | 5 |
| APOE | NM_138828 | apolipoprotein E | −1.4 | −1.4 | 2 |
| ARHGAP1 | XM_230284 | Rho GTPase activating protein 1 | −1.5 | −1.2 | 2 |
|
| |||||
| C1QB | NM_019262 | complement component 1qB | 1.0 | 1.2 | 6 |
| C1QTNF1 | NM_001007675 | C1q and tumor necrosis factor related protein 1 | 0.8 | 1.2 | 16 |
| C1R | NM_001134555 | complement component 1r | 2.6 | 1.5 | 0 |
| C1S | NM_138900 | complement component 1s | 1.3 | 1.3 | 2 |
| C2 | NM_172222 | complement component 2 | 3.5 | 2.0 | 0 |
| C3 | NM_016994 | complement component 3 | 2.6 | 2.1 | 0 |
| C4 | NM_001002805 | complement component 4 | 2.2 | 1.6 | 0 |
| C6 | NM_176074 | complement component 6 | 0.9 | 1.2 | 13 |
| CASP1 | NM_012762 | caspase 1 | 2.7 | 1.7 | 0 |
|
| |||||
| CBLB | NM_133601 | cas-br-m ectopic retroviral transforming b | −0.8 | −1.2 | 16 |
|
| |||||
| CCL2 | NM_031530 | chemokine (C-C) ligand 2 | 4.6 | 2.3 | 0 |
| CCL3 | NM_013025 | chemokine (C-C) ligand 3 | 6.2 | 3.5 | 0 |
| CCL4 | NM_053858 | chemokine (C-C) ligand 4 | 7.9 | 4.3 | 0 |
| CCL5 | NM_031116 | chemokine (C-C) ligand 5 | 6.7 | 4.7 | 0 |
| CCL6 | NM_001004202 | chemokine (C-C) ligand 6 | 2.4 | 1.6 | 0 |
| CCL7 | NM_001007612 | chemokine (C-C) ligand 7 | 5.0 | 2.3 | 0 |
| CCL12 | NM_001105822 | chemokine (C-C) ligand 12 | 6.1 | 5.3 | 0 |
| CCL19 | NM_001108661 | chemokine (C-C) ligand 19 | 3.5 | 4.2 | 0 |
| CCL20 | NM_019233 | chemokine (C-C) ligand 20 | 2.6 | 1.8 | 0 |
| CCND1 | NM_171992 | cyclin D1 | 1.2 | 1.2 | 3 |
| CCR5 | NM_053960 | chemokine (C-C) receptor 5 | 1.5 | 1.3 | 1 |
| CEBPB | NM_024125 | CCAAT/enhancer binding protein beta | 2.5 | 3.0 | 11 |
|
| |||||
| CD44 | NM_012924 | CD44 | −1.2 | −1.2 | 4 |
|
| |||||
| CD74 | NM_013069 | CD74 | 2.0 | 1.4 | 0 |
|
| |||||
| CDKN1A | NM_080782 | cyclin-dependent kinase inhibitor 1A | −1.1 | −1.2 | 5 |
|
| |||||
| CFB | NM_212466 | complement factor B | 8.7 | 5.0 | 0 |
| CFH | NM_130409 | complement factor H | 0.8 | 1.2 | 16 |
| CFLAR | NM_057138 | CASP8 and FADD-like apoptosis regulator | 1.0 | 1.2 | 8 |
|
| |||||
| CKLF | NM_139111 | chemokine-like factor | −1.1 | −1.3 | 5 |
|
| |||||
| CORO1A | NM_130411 | coronin 1A | 0.9 | 1.2 | 11 |
| CSF1 | NM_023981 | colony stimulating factor 1 | 1.3 | 1.3 | 2 |
| CSF2 | XM_340799 | colony stimulating factor 2 | 1.3 | 1.2 | 2 |
| CSF3 | NM_017104 | colony stimulating factor 3 | 2.3 | 1.5 | 0 |
| CX3CL1 | NM_134455 | chemokine (C-X3-C) ligand 1 | 2.6 | 1.6 | 0 |
| CXCL1 | NM_030845 | chemokine (C-X-C) ligand 1 | 4.0 | 2.1 | 0 |
| CXCL2 | NM_053647 | chemokine (C-X-C) ligand 2 | 3.8 | 2.0 | 0 |
| CXCL6 | NM_022214 | chemokine (C-X-C) ligand 6 | 7.2 | 4.4 | 0 |
| CXCL9 | NM_145672 | chemokine (C-X-C) ligand 9 | 3.9 | 3.1 | 0 |
| CXCL11 | NM_182952 | chemokine (C-X-C) ligand 11 | 5.7 | 3.8 | 0 |
| CXCL13 | NM_001017496 | chemokine (C-X-C) ligand 13 | 3.1 | 2.0 | 0 |
|
| |||||
| DLG1 | NM_012788 | discs large homolog 1 | −1.0 | −1.2 | 6 |
|
| |||||
| FAS | NM_139194 | Fas | 2.4 | 1.5 | 0 |
| FGR | NM_024145 | fgr tyrosine kinase | 2.6 | 1.5 | 0 |
|
| |||||
| HMOX1 | NM_012580 | heme oxygenase 1 | −2.6 | −2.4 | 0 |
|
| |||||
| HP | NM_012582 | haptoglobin | 2.7 | 1.7 | 0 |
| HPX | NM_053318 | hemopexin | 6.9 | 4.0 | 0 |
| ICAM1 | NM_012967 | intercellular adhesion molecule a | 2.5 | 1.6 | 0 |
| IER3 | NM_212505 | immediate early response 3 | 1.0 | 1.3 | 6 |
| IFNGR1 | NM_053783 | interferon gamma receptor 1 | 1.0 | 1.2 | 6 |
| IL1A | NM_017019 | interleukin 1 alpha | 6.0 | 3.3 | 0 |
| IL1B | NM_031512 | interleukin 1 beta | 5.5 | 3.2 | 0 |
| IL1F8 | XM_342377 | interleukin 1 family member 8 | 1.4 | 1.2 | 2 |
| IL1RN | NM_022194 | interleukin 1 receptor antagonist | 1.4 | 1.4 | 2 |
| IL2RA | NM_013163 | interleukin 2 receptor alpha | 2.8 | 1.5 | 0 |
| IL6 | NM_012589 | interleukin 6 | 2.0 | 1.5 | 0 |
| IL10 | NM_012854 | interleukin 10 | 2.7 | 1.6 | 0 |
| IL13RA2 | NM_133538 | interleukin 13 receptor alpha 2 | 2.9 | 1.6 | 0 |
|
| |||||
| IL18 | NM_019165 | interleukin 18 | −0.8 | −1.2 | 13 |
|
| |||||
| IL18BP | NM_053374 | interleukin 18 binding protein | 2.1 | 1.5 | 0 |
| IL33 | NM_001014166 | interleukin 33 | 2.1 | 1.3 | 0 |
| IRAK3 | XM_235183 | interleukin 1 receptor-associated kinase 3 | 0.8 | 1.2 | 16 |
| IRF1 | NM_012591 | interferon regulatory factor 1 | 2.0 | 1.4 | 0 |
| IRF7 | NM_001033691 | interferon regulatory factor 7 | 2.7 | 2.1 | 0 |
| JAK2 | NM_031514 | janus kinase 2 | 2.6 | 1.7 | 0 |
| LBP | NM_017208 | lipopolysaccharide binding protein | 2.6 | 1.5 | 0 |
| LCN2 | NM_130741 | lipocalin 2 | 4.1 | 1.8 | 0 |
|
| |||||
| MDK | NM_030859 | midkine | −0.8 | −1.1 | 16 |
| MLL5 | XM_231287 | mixed-lineage leukemia 5 | −0.9 | −1.2 | 11 |
| MMP3 | NM_133523 | matrix metallopeptidase 3 | −1.1 | −1.2 | 6 |
|
| |||||
| MMP9 | NM_031055 | matrix metallopeptidase 9 | 2.9 | 1.5 | 0 |
| MMP12 | NM_053963 | matrix metallopeptidase 12 | 1.6 | 1.3 | 0 |
| MX1 | NM_173096 | myxovirus resistance 1 | 3.6 | 3.7 | 0 |
| MX2 | NM_134350 | myxovirus resistance 2 | 1.8 | 2.2 | 0 |
| NEDD9 | NM_001011922 | neural precursor cell expressed 9 | 2.0 | 1.4 | 0 |
| NFKB2 | NM_001008349 | nuclear factor of kappa B2, p49/p100 | 1.1 | 1.3 | 5 |
| NFKBIA | NM_001105720 | nuclear factor of kappa B inhibitor, A | 3.0 | 1.7 | 0 |
| NFKBIE | NM_199111 | nuclear factor of kappa B inhibitor, E | 2.3 | 1.5 | 0 |
|
| |||||
| NGF | XM_227525 | nerve growth factor | −0.9 | −1.2 | 11 |
| NOD1 | XM_575485 | nucleotide-binding oligomerization domain 1 | −0.9 | −1.2 | 11 |
|
| |||||
| NPY | NM_012614 | neuropeptide Y | 2.0 | 1.4 | 0 |
|
| |||||
| NQO1 | NM_017000 | NAD(P)H dehydrogenase quinone 1 | −1.5 | −1.4 | 1 |
|
| |||||
| NUP35 | NM_001004229 | nucleoporin 35 | 2.1 | 1.4 | 0 |
| NUP85 | NM_001025401 | nucleoporin 85 | 1.2 | 1.2 | 3 |
| NUP107 | NM_053830 | nucleoporin 107 | 2.1 | 1.4 | 0 |
| NUP205 | NM_001108620 | nucleoporin 205 | 1.6 | 1.3 | 0 |
| ORM1 | NM_053288 | orosomucoid 1 | 1.8 | 1.3 | 0 |
| PARP1 | NM_013063 | poly (ADP-ribose) polymerase 1 | 1.1 | 1.2 | 4 |
|
| |||||
| PDE4D | NM_017032 | phosphodiesterase 4D | −0.9 | −1.2 | 11 |
| PGF | NM_053595 | placental growth factor | −1.3 | −1.2 | 3 |
|
| |||||
| PLEC | NM_022401 | plectin | 1.3 | 1.4 | 2 |
| PLSCR1 | NM_057194 | phospholipid scramblase 1 | 2.3 | 1.8 | 0 |
| PRKCQ | XM_341553 | protein kinase C theta | 2.0 | 1.5 | 0 |
| PSMB8 | NM_080767 | proteasome subunit B8 | 1.4 | 1.3 | 1 |
| PSMB9 | NM_012708 | proteasome subunit B9 | 2.0 | 1.7 | 0 |
| PSMB10 | NM_001025637 | proteasome subunit B10 | 2.7 | 1.7 | 0 |
| PSMB10 | NM_001025637 | proteasome subunit B10 | 1.5 | 1.4 | 1 |
| PSMC3 | NM_031595 | proteasome 26S subunit ATPase 3 | 1.4 | 1.3 | 2 |
| PSME1 | NM_017264 | proteasome 28 subunit alpha | 1.7 | 1.3 | 0 |
| PSME2 | NM_017257 | proteasome activator subunit 2 | 1.6 | 1.4 | 1 |
|
| |||||
| PTEN | NM_031606 | phosphatase and tensin homolog | −1.0 | −1.2 | 8 |
| PYCARD | NM_172322 | PYD and CARD domain containing | −1.0 | −1.2 | 8 |
|
| |||||
| RAC2 | NM_001008384 | ras-related C3 botulinum toxin substrate 2 | 1.4 | 1.3 | 1 |
| RARRES2 | NM_001013427 | retinoic acid receptor responder 2 | 1.0 | 1.2 | 11 |
|
| |||||
| RASA1 | NM_013135 | RAS p21 protein activator 1 | −1.7 | −1.4 | 1 |
|
| |||||
| REG3A | NM_172077 | regenerating islet-derived 3 alpha | 1.8 | 1.4 | 0 |
| REG3B | NM_053289 | regenerating islet-derived 3 beta | 1.3 | 1.4 | 2 |
| REG3G | NM_173097 | regenerating islet-derived 3 gamma | 2.4 | 1.7 | 0 |
| RT1-A1 | NM_001008827 | MHC class I RT1-A1 | 1.9 | 1.5 | 0 |
| RT1-A2 | NM_001008829 | MHC class I RT1-A2 | 2.3 | 1.7 | 0 |
| RT1-A3 | NM_001008830 | MHC class I RT1-A3 | 2.4 | 2.3 | 0 |
| RT1-CE15 | NM_001008838 | MHC class I RT1-CE15 | 1.9 | 1.5 | 0 |
| RT1-DB1 | NM_001008884 | MHC class II RT1-Db1 | 2.4 | 1.5 | 0 |
| RT1-M10-1 | NM_001008851 | MHC class I RT1-M10.1 | 1.7 | 1.5 | 0 |
| RT1-M6-2 | NM_001008853 | MHC class I RT1-M6 | 2.7 | 1.7 | 0 |
| RT1-S3 | NM_001008886 | MHC class Ib locus S3 | 3.9 | 2.1 | 0 |
| RT1-T24 | NM_001008826 | MHC class I RT1-T24.4 | 2.1 | 1.7 | 0 |
| RT1-T24-1 | NM_001008858 | MHC class I RT1-T24.1 | 2.4 | 1.7 | 0 |
| RUNX3 | NM_130425 | runt-related transcription factor 3 | 0.9 | 1.2 | 13 |
| S100A1 | NM_011309 | S100 calcium binding protein A1 | 1.9 | 1.4 | 0 |
| S100A13 | NM_001191607 | S100 calcium binding protein A13 | 2.5 | 2.0 | 0 |
| S100A16 | NM_001108557 | S100 calcium binding protein A16 | 2.2 | 1.5 | 0 |
| S100A3 | NM_053681 | S100 calcium binding protein A3 | 2.9 | 2.1 | 0 |
| S100A8 | NM_053822 | S100 calcium binding protein A8 | 3.8 | 1.7 | 0 |
| SELP | NM_013114 | selectin P | 2.5 | 1.7 | 0 |
| SERPING2 | NM_199093 | serine peptidase inhibitor G1 | 1.6 | 1.4 | 0 |
| SLPI | NM_053372 | secretory leukocyte peptidase inhibitor | 6.2 | 3.2 | 0 |
| SMPD1 | NM_001006997 | sphongomyelin phosphodiesterase 1 | 1.0 | 1.2 | 8 |
|
| |||||
| SPP1 | NM_012881 | secreted phosphoprotein 1 | −2.7 | −2.7 | 0 |
|
| |||||
| STAT1 | NM_032612 | signal transducer and activator of transcription 1 | 1.0 | 1.2 | 6 |
| THBD | NM_031771 | thrombomodulin | −0.8 | −1.2 | 16 |
| TIAM1 | NM_001100558 | T-cell lymphoma invasion and metastasis | −2.5 | −1.5 | 0 |
|
| |||||
| TF | NM_001013110 | transferrin | 3.0 | 1.7 | 0 |
| TGFA | NM_012671 | transforming growth factor alpha | 3.4 | 2.1 | 0 |
|
| |||||
| TGFB1 | NM_021578 | transforming growth factor beta 1 | −2.6 | −1.5 | 0 |
| TIMP2 | NM_021989 | TIMP metallopeptidase inhibitor 2 | −2.4 | −1.5 | 0 |
|
| |||||
| TLR2 | NM_198769 | toll-like receptor 2 | 3.6 | 1.7 | 0 |
| TLR6 | NM_207604 | toll-like receptor 6 | 1.5 | 1.3 | 1 |
| TNIP1 | NM_001108826 | TNFAIP3 interacting protein 1 | 2.4 | 1.6 | 0 |
|
| |||||
| TNFRSF1A | NM_013091 | tumor necrosis factor receptor superfamily 1A | −1.1 | −1.2 | 6 |
|
| |||||
| TNFRSF21 | NM_001108207 | tumor necrosis factor receptor superfamily 21 | 0.9 | 1.2 | 13 |
| TXNIP | NM_001008767 | thioredoxin-interacting protein | 2.7 | 2.0 | 0 |
| VCAM1 | NM_012889 | vascular cell adhesion molecule 1 | 2.2 | 1.7 | 0 |
|
| |||||
| VEGFA | NM_031836 | vascular endothelial growth factor A | −1.3 | −1.2 | 3 |
SAM (significance analysis of microarray) score = d(i); similar to t-test t-score
Fold change NMO serum Tx vs CON serum Tx; generated by SAM; color-coded up vs down
SAM false discovery rate cut-off value = q
The effect of NMO IgG on CCL5 production and release from astrocyte cultures was tested by ELISA (Figure 3B). While 100 μg/mL NMO IgG was sufficient to induce release of CCL5 equivalent to 10% NMO serum treatment (P<0.001), even 1000 μg/mL CON IgG did not result in CCL5 production (P=0.981). Likewise, stimulation with 100 μg/mL NMO IgG for 24 hr induced the release of more than 40 ng/mL CXCL1 (P<0.001) (Figure 3C) and more than 100 ng/mL CCL2 (P<0.001) (Figure 3D); CON IgG had no effect on the production of either chemokine (P=0.844, P=0.216, respectively). We conclude that the IgG fraction of NMO serum induces a specific immune gene program in rat astrocyte cultures and triggers the production and release of the neutrophilic chemokine CXCL1, the monocytic chemokine CCL2, and the eosinophilic chemokine CCL5.
Chemokine production in response to NMO serum is disease specific
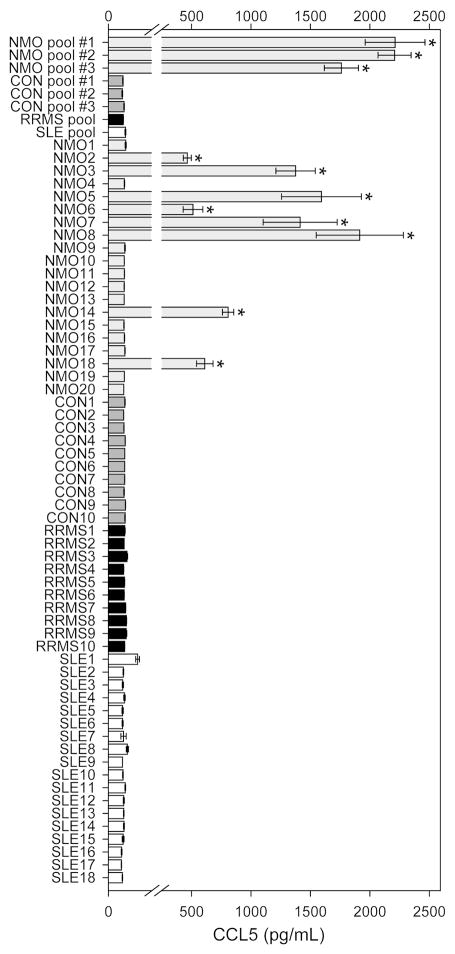
Finally, we tested the disease specificity of CCL5 induction by NMO serum. Astrocyte cultures were incubated for 24 hr at 37°C with 10% serum from 20 individual NMO patients, 10 individual control serum samples (CON), 10 serum samples collected from individual relapsing-remitting multiple sclerosis patients (RRMS), and 18 serum samples collected from individual Sjögren or systemic lupus erythematosus patients (SLE). Patient age and sex data are provided in Supplemental Table 3. Groups showed equivalent age and sex distributions. In addition, three NMO pools, three CON pools, one RRMS pool, and one SLE pool were prepared from separate patient samples and tested for induction of CCL5 release. As shown in Figure 4, we found that only samples or pools derived from NMO patients induced CCL5 release by astrocyte cultures. Surprisingly, while all three NMO pools induced a robust CCL5 response, only a subset of individual NMO serum samples stimulated the response (Figure 4). Indeed, 12 of 20 NMO samples had no effect on CCL5 release, equivalent to CON-treated cultures. Full statistical analyses are provided in Supplemental Table 4. In summary, there was no signficant difference between CON pools, RRMS pool, SLE pool, all CON samples, all RRMS samples, and all SLE samples and all of these were significant at P<0.001 from the three NMO pools. The 8 individual NMO samples that induced CCL5 release were significant at P<0.001 from every other treatment except the three NMO pools. We conclude that CCL5 induction in primary astrocyte cultures is unique to serum samples derived from NMO patients and is not merely a reflection of ongoing autoimmune disease. Furthermore, we conclude that CCL5 induction may stratify NMO patients into inducers and non-inducers and may serve as a potential biomarker for disease activity, pathogenesis, or stage.
Figure 4. The NMO serum-induced CCL5 response is disease specific.
Rat astroglial cultures were treated with pooled or individual patient serum at 10% concentration for 24 hr. CCL5 release was assessed by ELISA. No individual or pooled sample derived from controls (CON), relapsing-remitting multiple sclerosis (RRMS), or Sjögren's/Systemic Lupus Erythematosus (SLE) patients induced CCL5 release. All NMO pools induced CCL5 release, but only a subset of individual patient samples triggered a CCL5 response. One-way ANOVA, F(65,133)=164.376, P<0.001 between all treatments. * = P<0.001 vs CON; no other responses were significant. See Supplemental Table 4 for complete statistical analysis.
Discussion
The underlying pathogenic mechanisms responsible for CNS injury in patients with NMO is currently unknown. The most common model posits entry of the NMO IgG into the CNS at either naturally or pathologically open blood-brain barrier sites, binding of the IgG to AQP4 on the surface of astrocytes, recruitment of the lytic complement cascade, and irreversible destruction of astrocytes and neighboring cells. While complement deposition is a pathological feature of some NMO lesions (Lucchinetti et al., 2002) and while current evidence indicates therapeutic efficacy of complement inhibition in some NMO patients (Pittock et al., 2013), the apparent reversibility of functional and imaging parameters in patients coupled to the overwhelming evidence from human pathology specimens that NMO lesions have a far more complex spectrum of characteristics than previously appreciated suggests that complement-mediated destruction is only one pathogenic mechanism in NMO. Moreover, from a therapeutic perspective, complement-mediated destruction is a terminal step that must be prevented rather than interrupted in process. In contrast, the unique characteristics of NMO lesions suggests that a process of injury evolution occurs in situ that may present opportunities for therapeutic interruption and lesion resolution far upstream from terminal lytic events.
We hypothesized that binding of the NMO IgG to AQP4 on the surface of astrocytes induces a cellular response that results in transcriptional and translational events within the astrocyte and leads to the creation of an environment around the astrocyte that recruits peripheral immune cells to amplify the pathogenic process. Not only does such a model create multiple intervention points, it also explains how a small amount of NMO IgG access to the CNS at naturally permeable sites such as the area postrema (Popescu et al., 2011) could lead to the formation of large and functionally relevant lesions. Amplification is a fundamental biological principle, whether amplification at the level of signaling through cascading kinases or amplification via recruitment of effector cells with expanded physical or functional repertoires. For example, the type I interferon response involves rapid activation of interferon receptor signaling that results in further production of type I interferons and feed-forward amplification of an antiviral response (Hall and Rosen, 2010). Likewise, allergen exposure in the lungs initiates a cascade of cellular responses in a small number of T cells and lung-resident cells that results in the recruitment of a large number of eosinophils, consequent release of leukotrienes, neuropeptides, metalloproteinases, and cytotoxic granules that drive bronchoconstriction, and the explosive induction of asthma (Possa et al., 2013). Amplification is therefore a potent pathogenic mechanism – it is also highly amenable to therapeutic strategies that short-circuit the amplification pathways.
Our current findings indicate that interaction of the NMO IgG with AQP4 initiates a cellular response that results in the acquisition of an inflammatory phenotype by astroglia. The 50-fold upregulation of Lcn2 is consistent with a reactive astrocyte phenotype – indeed, the pattern of gene induction in astrocyte cultures in response to either NMO serum or purified IgG is highly similar to that induced by LPS (Zamanian et al., 2012). In addition to upregulation of adhesion factors such as ICAM-1 and VCAM-1, the initiation of oxidative stress response pathways involving SOD2, ceruloplasmin, and NFκB, and the upregulation of inflammasome components such as caspase-1 and various proteasome subunits, the NMO IgG induced a massive cytokine and chemokine response (Table 1, Table 2). For example, IL-1α and IL-1β were both upregulated more than 5-fold and IL-6, IL-10, and IL-33 were upregulated more than 2-fold by NMO IgG (Table 2). Most notably, NMO IgG induced 9 CCL and 6 CXCL chemokines at levels over 2-fold and all with false discovery rates of zero (Table 2). The pattern of chemokine induction is heavily skewed toward monocytic (CCL2/3/4/5/7/12 on CCR2), neutrophilic (CXCL1/2/6 on CXCR1/2), and eosinophilic (CCL5/7 on CCR3) recruitment. This finding is particularly striking given the presence of eosinophils and neutrophils in NMO lesions (Lucchinetti et al., 2002) and the recent correlation between CSF levels of the human CXCR2 ligand CXCL8 (functionally homologous to CXCL1/2 in rodents) with disability score in NMO patients (Matsushita et al., 2013).
The mechanism(s) by which the NMO IgG induces a reactive and inflammatory response in primary astrocytes is currently unknown. The literature is divided with regard to the stimulation of an active perturbation in AQP4 function by the NMO IgG. While little doubt exists that binding of the IgG to the surface of astrocytes, within the appropriate molecular context, has the capacity to passively drive events such as complement deposition, some investigators have observed AQP4 internalization (Melamud et al., 2012) and acute blockade of AQP4 permeability (Hinson et al., 2012) in response to the NMO IgG, while others have failed to find such responses (Nicchia et al., 2009; Rossi et al., 2012). These findings must be interpreted catiously, as a number of uncontrolled variables exist that may preclude direct comparisons between studies. For example, some groups culture astrocytes in the presence of dibutyryl cAMP for several weeks (Solenov et al., 2004) while others culture only in the presence of serum and for shorter times in vitro (Hinson et al., 2012); some experiments were performed in cultures prepared from CD1 mice (Ratelade et al., 2011) while others used astrocytes derived from C57BL/6 mice (Sabater et al., 2009) or astrocyte cultures prepared from unidentified mouse or rat strains; experiments have used transfected human embryonic kidney cells (Hinson et al., 2008), glioblastoma-astrocytoma cells (Rossi et al., 2012), or Chinese hamster ovary cells (Phuan et al., 2012), each with different levels of AQP4 expression and different sub- and supra-molecular contexts. Ultimately, however, it is currently unclear how binding of the NMO IgG to surface AQP4 on astrocytes induces acute cellular signaling. Our findings indicate that the NMO IgG induces a specific transcriptional program in primary astrocytes that leads to the acquistion of a reactive and pro-inflammatory phenotype. Additional work is required to identify the mechanism by which such a program is induced.
Why did only a subset of NMO patient serum samples induce a CCL5 response in primary astrocyte cultures (Figure 4)? Given that no such response was induced by any RRMS, SLE, or control serum sample tested in our experiments, it is likely that the chemokine induction response is specific to NMO. But the clear segregation of individual NMO patient samples into CCL5 inducers and non-inducers suggests that this response may serve as a biomarker for some aspect of the disease associated with progression, relapse, or disability burden. At the present time, other than fulfilling the diagnostic criteria for NMO (Wingerchuk et al., 2006) and exhibiting an ELISA titer greater than 1 in 160 (Lennon et al., 2004), we lack patient-specific data. A prospective study is underway to assess any correlation between patient characteristics and chemokine induction in astrocytes.
The existence of CCL5 inducers and non-inducers despite the uniformity of high NMO IgG titer is consistent with the polyclonality of the NMO IgG (Iorio et al., 2013). However, preliminary observations indicate that CCL5 induction does not correlate with intensity of surface binding to astrocytes, ratio of M1 to M23 AQP4 isoform expression, or capacity to downregulate surface AQP4 (data not shown). While such assays are useful tools for screening NMO patient samples, they may lack the sensitivity necessary to detect salient biophysical events occurring within microdomains on the surface of astrocytes. Likewise, the total macroscopic cellular ratio of M1:23 AQP4 may not adequately reflect microscopic organization of these isoforms at the plasma membrane. For example, some NMO-IgG clones may recognize AQP4 or AQP4 isoforms only within the context of a specific lipid environment or may have a predilection for recruiting specific isoforms to lipid-enriched signaling domains (Tong et al., 2012). Alternatively, some NMO IgG clones may differentially alter the function of astrocytes in a manner that triggers more intracellular signaling (Hinson et al., 2012; Melamud et al., 2012), leading to the induction of reactive and stress response pathways in the target astrocyte. Finally, despite the clear presence of AQP4-specific IgG in NMO serum and the diagnostic utility of this interaction, other antigenic targets may exist that distinguish NMO patient subsets. Indeed, it is a logical fallacy to conclude that because most or all NMO patients have serum IgG that recognizes AQP4, such IgG is the only or even the primary pathogenic mediator. The presence of anti-AQP4 specificity does not rule out the possibility of additional pathogenically relevant targets. Because current animal models only prove that co-injection of NMO IgG with human complement into a context in which AQP4 is expressed triggers complement-dependent lysis and damage – a tautological outcome – the role of macro- and microdomain specificity and the contribution of other antigenic targets remain open questions. Nonetheless, our findings implicate NMO IgG-induced astrocytic reactivity and the concomitant induction of an inflammatory phenotype heavily skewed toward the recruitment of monocytes and granulocytes as a possible pathogenic mechanism in patients with NMO. A critical aspect of this model is the opportunity to employ novel therapeutic strategies to halt disease progression.
Supplementary Material
Acknowledgments
Dr. Vanda Lennon and Dr. Sean Pittock provided invaluable reagents and guidance for this project. Jim Fryer, Tom Kryzer, and John Schmeling provided technical assistance with collection and preparation of patient samples. Chris Kolbert and the Mayo Medical Genome Facility Gene Expression Core assisted with the microarray analyses. This work was funded by the Guthy-Jackson Charitable Foundation. The authors have no conflict of interest to report.
References
- Almekhlafi MA, Clark AW, Lucchinetti CF, Zhang Y, Power C, Bell RB. Neuromyelitis optica with extensive active brain involvement: an autopsy study. Arch Neurol. 2011;68:508–12. doi: 10.1001/archneurol.2011.62. [DOI] [PubMed] [Google Scholar]
- Baba T, Nakashima I, Kanbayashi T, Konno M, Takahashi T, Fujihara K, Misu T, Takeda A, Shiga Y, Ogawa H, et al. Narcolepsy as an initial manifestation of neuromyelitis optica with anti-aquaporin-4 antibody. J Neurol. 2009;256:287–8. doi: 10.1007/s00415-009-0139-4. [DOI] [PubMed] [Google Scholar]
- Cree BA, Lamb S, Morgan K, Chen A, Waubant E, Genain C. An open label study of the effects of rituximab in neuromyelitis optica. Neurology. 2005;64:1270–2. doi: 10.1212/01.WNL.0000159399.81861.D5. [DOI] [PubMed] [Google Scholar]
- Curran-Everett D, Benos DJ. Guidelines for reporting statistics in journals published by the American Physiological Society: the sequel. Adv Physiol Educ. 2007;31:295–8. doi: 10.1152/advan.00022.2007. [DOI] [PubMed] [Google Scholar]
- Hall JC, Rosen A. Type I interferons: crucial participants in disease amplification in autoimmunity. Nat Rev Rheumatol. 2010;6:40–9. doi: 10.1038/nrrheum.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson SR, Pittock SJ, Lucchinetti CF, Roemer SF, Fryer JP, Kryzer TJ, Lennon VA. Pathogenic potential of IgG binding to water channel extracellular domain in neuromyelitis optica. Neurology. 2007;69:2221–31. doi: 10.1212/01.WNL.0000289761.64862.ce. [DOI] [PubMed] [Google Scholar]
- Hinson SR, Roemer SF, Lucchinetti CF, Fryer JP, Kryzer TJ, Chamberlain JL, Howe CL, Pittock SJ, Lennon VA. Aquaporin-4-binding autoantibodies in patients with neuromyelitis optica impair glutamate transport by down-regulating EAAT2. J Exp Med. 2008;205:2473–81. doi: 10.1084/jem.20081241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson SR, Romero MF, Popescu BF, Lucchinetti CF, Fryer JP, Wolburg H, Fallier-Becker P, Noell S, Lennon VA. Molecular outcomes of neuromyelitis optica (NMO)-IgG binding to aquaporin-4 in astrocytes. Proc Natl Acad Sci U S A. 2012;109:1245–50. doi: 10.1073/pnas.1109980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio R, Fryer JP, Hinson SR, Fallier-Becker P, Wolburg H, Pittock SJ, Lennon VA. Astrocytic autoantibody of neuromyelitis optica (NMO-IgG) binds to aquaporin-4 extracellular loops, monomers, tetramers and high order arrays. J Autoimmun. 2013;40:21–7. doi: 10.1016/j.jaut.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Kim SH, Lee SH, Li XF, Kim HJ. Brain abnormalities as an initial manifestation of neuromyelitis optica spectrum disorder. Mult Scler. 2011;17:1107–12. doi: 10.1177/1352458511404917. [DOI] [PubMed] [Google Scholar]
- Lennon VA, Wingerchuk DM, Kryzer TJ, Pittock SJ, Lucchinetti CF, Fujihara K, Nakashima I, Weinshenker BG. A serum autoantibody marker of neuromyelitis optica: distinction from multiple sclerosis. Lancet. 2004;364:2106–12. doi: 10.1016/S0140-6736(04)17551-X. [DOI] [PubMed] [Google Scholar]
- Lucchinetti CF, Mandler RN, McGavern D, Bruck W, Gleich G, Ransohoff RM, Trebst C, Weinshenker B, Wingerchuk D, Parisi JE, et al. A role for humoral mechanisms in the pathogenesis of Devic's neuromyelitis optica. Brain. 2002;125:1450–61. doi: 10.1093/brain/awf151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magana SM, Keegan BM, Weinshenker BG, Erickson BJ, Pittock SJ, Lennon VA, Rodriguez M, Thomsen K, Weigand S, Mandrekar J, et al. Beneficial plasma exchange response in central nervous system inflammatory demyelination. Arch Neurol. 2011;68:870–8. doi: 10.1001/archneurol.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magana SM, Matiello M, Pittock SJ, McKeon A, Lennon VA, Rabinstein AA, Shuster E, Kantarci OH, Lucchinetti CF, Weinshenker BG. Posterior reversible encephalopathy syndrome in neuromyelitis optica spectrum disorders. Neurology. 2009;72:712–7. doi: 10.1212/01.wnl.0000343001.36493.ae. [DOI] [PubMed] [Google Scholar]
- Matsushita T, Tateishi T, Isobe N, Yonekawa T, Yamasaki R, Matsuse D, Murai H, Kira J. Characteristic cerebrospinal fluid cytokine/chemokine profiles in neuromyelitis optica, relapsing remitting or primary progressive multiple sclerosis. PLoS ONE. 2013;8:e61835. doi: 10.1371/journal.pone.0061835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamud L, Fernandez JM, Rivarola V, Di Giusto G, Ford P, Villa A, Capurro C. Neuromyelitis Optica Immunoglobulin G present in sera from neuromyelitis optica patients affects aquaporin-4 expression and water permeability of the astrocyte plasma membrane. J Neurosci Res. 2012;90:1240–8. doi: 10.1002/jnr.22822. [DOI] [PubMed] [Google Scholar]
- Nicchia GP, Mastrototaro M, Rossi A, Pisani F, Tortorella C, Ruggieri M, Lia A, Trojano M, Frigeri A, Svelto M. Aquaporin-4 orthogonal arrays of particles are the target for neuromyelitis optica autoantibodies. Glia. 2009;57:1363–73. doi: 10.1002/glia.20855. [DOI] [PubMed] [Google Scholar]
- Papadopoulos MC, Verkman AS. Aquaporin water channels in the nervous system. Nat Rev Neurosci. 2013;14:265–77. doi: 10.1038/nrn3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuan PW, Ratelade J, Rossi A, Tradtrantip L, Verkman AS. Complement-dependent cytotoxicity in neuromyelitis optica requires aquaporin-4 protein assembly in orthogonal arrays. J Biol Chem. 2012;287:13829–39. doi: 10.1074/jbc.M112.344325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittock SJ, Lennon VA, McKeon A, Mandrekar J, Weinshenker BG, Lucchinetti CF, O'Toole O, Wingerchuk DM. Eculizumab in AQP4-IgG-positive relapsing neuromyelitis optica spectrum disorders: an open-label pilot study. Lancet Neurol. 2013;12:554–62. doi: 10.1016/S1474-4422(13)70076-0. [DOI] [PubMed] [Google Scholar]
- Pittock SJ, Weinshenker BG, Lucchinetti CF, Wingerchuk DM, Corboy JR, Lennon VA. Neuromyelitis optica brain lesions localized at sites of high aquaporin 4 expression. Arch Neurol. 2006;63:964–8. doi: 10.1001/archneur.63.7.964. [DOI] [PubMed] [Google Scholar]
- Popescu BF, Lennon VA, Parisi JE, Howe CL, Weigand SD, Cabrera-Gomez JA, Newell K, Mandler RN, Pittock SJ, Weinshenker BG, et al. Neuromyelitis optica unique area postrema lesions: nausea, vomiting, and pathogenic implications. Neurology. 2011;76:1229–37. doi: 10.1212/WNL.0b013e318214332c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu BF, Lucchinetti CF. Pathology of demyelinating diseases. Annu Rev Pathol. 2012;7:185–217. doi: 10.1146/annurev-pathol-011811-132443. [DOI] [PubMed] [Google Scholar]
- Possa SS, Leick EA, Prado CM, Martins MA, Tiberio IF. Eosinophilic inflammation in allergic asthma. Front Pharmacol. 2013;4:46. doi: 10.3389/fphar.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash JE, Yasumura T, Hudson CS, Agre P, Nielsen S. Direct immunogold labeling of aquaporin-4 in square arrays of astrocyte and ependymocyte plasma membranes in rat brain and spinal cord. Proc Natl Acad Sci U S A. 1998;95:11981–6. doi: 10.1073/pnas.95.20.11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratelade J, Bennett JL, Verkman AS. Evidence against cellular internalization in vivo of NMO-IgG, aquaporin-4, and excitatory amino acid transporter 2 in neuromyelitis optica. J Biol Chem. 2011;286:45156–64. doi: 10.1074/jbc.M111.297275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer SF, Parisi JE, Lennon VA, Benarroch EE, Lassmann H, Bruck W, Mandler RN, Weinshenker BG, Pittock SJ, Wingerchuk DM, et al. Pattern-specific loss of aquaporin-4 immunoreactivity distinguishes neuromyelitis optica from multiple sclerosis. Brain. 2007;130:1194–205. doi: 10.1093/brain/awl371. [DOI] [PubMed] [Google Scholar]
- Rossi A, Ratelade J, Papadopoulos MC, Bennett JL, Verkman AS. Neuromyelitis optica IgG does not alter aquaporin-4 water permeability, plasma membrane M1/M23 isoform content, or supramolecular assembly. Glia. 2012;60:2027–39. doi: 10.1002/glia.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadoun S, Waters P, Anthony Bell B, Vincent A, Verkman AS, Papadopoulos MC. Intra-cerebral injection of neuromyelitis optica immunoglobin G and human complement produces neuromyelitis optica lesions in mice. Brain. 2010;133:349–361. doi: 10.1093/brain/awp309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabater L, Giralt A, Boronat A, Hankiewicz K, Blanco Y, Llufriu S, Alberch J, Graus F, Saiz A. Cytotoxic effect of neuromyelitis optica antibody (NMO-IgG) to astrocytes: an in vitro study. J Neuroimmunol. 2009;215:31–5. doi: 10.1016/j.jneuroim.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Solenov E, Watanabe H, Manley GT, Verkman AS. Sevenfold-reduced osmotic water permeability in primary astrocyte cultures from AQP-4-deficient mice, measured by a fluorescence quenching method. Am J Physiol Cell Physiol. 2004;286:C426–32. doi: 10.1152/ajpcell.00298.2003. [DOI] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J Roy Stat Soc Ser B. 2002;64:479–498. [Google Scholar]
- Tong J, Briggs MM, McIntosh TJ. Water permeability of aquaporin-4 channel depends on bilayer composition, thickness, and elasticity. Biophys J. 2012;103:1899–908. doi: 10.1016/j.bpj.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingerchuk DM, Lennon VA, Lucchinetti CF, Pittock SJ, Weinshenker BG. The spectrum of neuromyelitis optica. Lancet Neurol. 2007;6:805–15. doi: 10.1016/S1474-4422(07)70216-8. [DOI] [PubMed] [Google Scholar]
- Wingerchuk DM, Lennon VA, Pittock SJ, Lucchinetti CF, Weinshenker BG. Revised diagnostic criteria for neuromyelitis optica. Neurology. 2006;66:1485–9. doi: 10.1212/01.wnl.0000216139.44259.74. [DOI] [PubMed] [Google Scholar]
- Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, Barres BA. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32:6391–410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.