Abstract
This study evaluates μNMR technology for molecular profiling of tumor fine needle aspirates and peripheral blood of melanoma patients. In vitro assessment of melanocyte (MART-1, HMB45) and MAP kinase signaling (pERK, pS6K) molecule expression was performed in human cell lines, while clinical validation was performed in an IRB-approved study of melanoma patients undergoing biopsy and blood sampling. Tumor FNA and blood specimens were compared with BRAF genetic analysis and cross-sectional imaging. μNMR in vitro analysis showed increased expression of melanocyte markers in melanoma cells as well as increased expression of phosphorylated MAP kinase targets in BRAF-mutant melanoma cells. Melanoma patient FNA samples showed increased pERK and pS6K levels in BRAF mutant compared with BRAF WT melanomas, with μNMR blood circulating tumor cell level increased with higher metastatic burden visible on imaging. These results indicate μNMR technology provides minimally invasive point-of-care evaluation of tumor signaling and metastatic burden in melanoma patients.
Keywords: melanoma, circulating tumor cells, treatment response, biopsy
Graphical abstract
BACKGROUND
Metastatic melanoma is an aggressive disease that traditionally demonstrated poor responsiveness to systemic therapies such as chemotherapy, with median survival times ranging from 6 to 10 months(1-3). Recently, a number of novel molecule-targeted and immune-targeted agents have shown remarkable clinical activity and improved survival compared with conventional cytotoxic agents (reviewed in(4-6)). The BRAF serine/threonine kinase is mutated in up to 50% of melanomas(7, 8) leading to MAP kinase pathway activation and increased tumor cell proliferation. BRAF inhibitor therapy is associated with improved clinical response and progression-free survival compared with conventional therapies in BRAF-mutant melanoma patients(9). However, tumors ultimately develop resistance to BRAF inhibition by acquiring mutations that circumvent BRAF(10-12), and recent data shows a survival benefit when BRAF-mutant melanoma patients are treated with inhibitors of both BRAF and MEK (the downstream target of BRAF in the MAP kinase pathway)(13).
As more molecule-targeted cancer therapies enter the clinic, it is increasingly important to develop methods to monitor the activity of cancer signaling pathways, as well as overall metastatic burden, in patients over time. Such methods would provide early evidence of both therapeutic efficacy and acquired resistance, before macroscopic changes would be detectable by conventional anatomic imaging with CT or MRI. Also, assessment of cancer signaling at the molecular level will be extremely important for treatment decisions regarding next-line therapy in refractory patients, particularly in the context of clinical trials that are increasingly adaptive in nature. A point of care technique, which is minimally invasive and could be performed during routine clinic visits when other lab tests are being performed, would be ideal for serial assessment.
In the current study, we apply micro-nuclear magnetic resonance (μNMR) technology(14, 15) to perform molecular profiling of tumor cells isolated from fine needle aspirates and peripheral blood of melanoma patients. μNMR is a novel point-of-care diagnostic method, which can provide rapid and accurate molecular analysis of intact cells from in vivo clinical specimens. Similar to clinical MRI, the technology uses magnetic resonance principles to quantify biomarker expression levels on cells in small sample volumes, with molecular specificity achieved through targeted magnetic nanoparticles(16, 17). The low background signal level from nontarget cells allows in vivo specimens to be analyzed with little preparation, maximizing sensitivity and enabling μNMR to provide rapid quantitation of multiple molecular targets. Previous studies have shown μNMR to be a robust method for diagnosing metastatic disease and profiling tumor expression from fine needle aspirates and ascites(18, 19), and for detecting circulating tumor cells in peripheral blood(20). However, μNMR analysis has not been applied to the detection or profiling of melanomas. The availability of superficial cutaneous lesions for serial fine needle aspirate sampling, as well as the availability of multiple molecule-targeted therapies in the clinical setting(21), are features of melanoma that make it well-suited for validation of the μNMR cell approach for serial tumor molecular assessment.
In this study, we apply μNMR technology to the evaluation of patients with melanoma. The first aim is to evaluate the ability of μNMR to discriminate tumor cell MAP kinase signaling activity in BRAF wild-type versus BRAF mutant melanomas from tumor cells isolated from fine needle aspirates. The second aim was to correlate levels of circulating tumor cells identified by μNMR with overall metastatic burden assessed by imaging.
METHODS
Antibodies and cell lines
For assessment of MAP kinase signaling activity, antibodies directed against total S6K (rabbit anti-human total S6K, Cell Signaling), pS6K (rabbit anti-human pS6K, Cell Signaling), total ERK (mouse anti-human total ERK, R&D), and pERK (mouse anti-human pERK, R&D) were utilized. For detection of melanoma circulating tumor cells, a previously published four biomarker approach to detect MelanA/MART-1 (mouse anti-human MART-1, Thermo Fisher), gp100/HMB45 (rabbit anti-human gp100, Thermo Fisher), EpCAM (mouse anti-human IgG, R&D), and EGFR (humanized mouse anti-human EGFR, Imclone) was utilized, which previously was shown to be more accurate for CTC quantitation than single EpCAM marker-based methods (22). Secondary anti-mouse or anti-rabbit IgG antibodies (R&D) were used for nanoparticle labeling. Melanoma cell lines used for in vitro validation studies include the BRAF wild type cell lines MeWo, JuSo, and CHL and the BRAF V600E mutant lines A375, SKMel28, and UACC. HeLa cells were used as a non-melanoma control cell line. All cell lines were obtained from American Type Culture Collection.
Nanoparticle conjugation of antibodies
Secondary IgG antibodies were conjugated with (E)-cyclooct-4-enyl 2,5-dioxopyrrolidin-1-yl carbonate (TCO-NHS), as previously reported(22, 23). TCO conjugation was performed with 0.5 mg of anti-mouse or anti-rabbit IgG antibody in the presence of 1000 molar equivalents of TCO-NHS in phosphate-buffered saline (PBS) and 10% dimethylformamide at room temperature for 3 hours. Unreacted TCO-NHS was subsequently washed using 2 ml of Zeba desalting columns (Thermo Fisher, Rockford, IL), and antibody concentrations were determined by absorbance measurement. Cross-linked iron oxide (CLIO) nanoparticles were prepared as previously described(23). Tetrazine (Tz)-CLIO conjugation was performed in PBS containing 5% dimethylformamide for 3 hours at room temperature, as previously described. Excess Tz-NHS was removed by gel filtration using Sephadex G-50 (GE Healthcare, Pittsburgh, PA).
Cell labeling
Cell specimens (in vitro cultured cells, tumor fine needle aspirates, or peripheral blood) were centrifuged at 3500 rpm. Peripheral blood samples (7 ml) were lysed and cell pellet resuspended in buffer (100 μl of 1× PBS/1% FBS). Cells were fixed by incubation of cell pellets in 4% formaldehyde in PBS for 15 minutes at room temperature. For intracellular biomarker assays, cells were additionally permeabilized in saponin (Permeabilization/Wash buffer; Becton Dickinson Biosciences, San Jose, CA) for 15 minutes at room temperature. The resulting cell pellets were incubated with the above antibodies for 20 minutes in a total volume of 100 μL of PBS at room temperature. TCO-modified secondary IgG antibodies (10 μg/ml) were added to the pellet and incubated for an additional 20 minutes. Cell pellets were then washed twice with 1 X PBS and incubated with magnetic nanoparticles (100 nM Tz-CLIO) for 10 minutes. Excess Tz-CLIO was subsequently removed by washing the pellet twice with PBS, before being resuspended in 20 μl of PBS for μNMR measurements.
μNMR biomarker quantitation
μNMR measurements of cellular biomarker expression were performed as previously described(22) using a miniaturized μNMR device to assess transverse relaxation rate (R2). Briefly, T2 values were obtained in triplicate using the μNMR system, and R2 values were calculated as the inverse of the mean T2 values. To account the effect of media and background signal, the μNMR readout was calculated by subtracting the R2 value of the media from either the values obtained from the test (i.e., samples receiving antibody and CLIO for labeling) or from the control (i.e., samples receiving only CLIO for background signal determination). The final μNMR readout was then calculated by dividing ΔR2sample by ΔR2control. A Carr-Purcell-Meiboom-Gill pulse sequence was used with the following parameters: Repetition time 6000 msecs, echo time, 4 msecs; number of 180° pulses per scan, 500; number of scans, 8. R2 values were translated into normalized protein expression levels relative to control samples lacking primary antibody. Each sample consisted of 100 cells resuspended in a final volume of 1 μL for analysis, with all samples analyzed in triplicate.
Western blotting
Western blot was performed according to a previously published protocol(24). Whole cell lysates were prepared in lysis buffer supplemented with protease and phosphatase inhibitors (Roche). Equal amounts of protein were resolved by electrophoresis on gradient gels (Bio-Rad). Primary antibodies specific for phospho-ERK1/2 and ERK1/2 were used along with secondary antibodies conjugated to horseradish peroxidase (Cell Signaling Technology) for chemiluminescent detection. Western blot band intensity was quantified using ImageJ software(25).
Clinical evaluation of μNMR in melanoma patients
In a single institution, institutional review board approved, HIPAA-compliant pilot study, 10 patients with known melanoma at a single institution (Massachusetts General Hospital/Harvard Cancer Center) scheduled for a clinical biopsy of a suspected metastatic lesion were underwent μNMR specimen collection and molecular analysis. All patients were at least 18 years of age and provided informed consent. At the time of biopsy, 7mL of peripheral blood and 25 gauge fine needle aspirates of the biopsied lesion were collected from each patient and analyzed by μNMR for MAP kinase signaling and circulating tumor cells.
Imaging and BRAF genetic reference
Electronic medical records and imaging studies obtained near the time of biopsy from the patients enrolled in the clinical study were reviewed by a board certified radiologist (M.G.) who was blinded to the μNMR results. BRAF status (wild type vs mutant) was recorded based on melanoma genetic testing performed as part of clinical workup and documented in the electronic medical record. Metastatic burden for each patient was calculated based on Response Evaluation Criteria in Solid Tumors (RECIST 1.1) analysis(26) of the most recent staging imaging studies performed before biopsy, which included brain MRI as well as CT of the chest, abdomen, and pelvis. The score for metastatic burden was the sum of the long axis diameters (SLD, measured in mm) of the RECIST target lesions seen on imaging. The number of target lesions for each patient on imaging was also recorded.
Statistical analysis
Student’s t test was performed to evaluate for statistical significance of μNMR values between different groups.
RESULTS
μNMR analysis of cell lines in vitro
To determine whether melanoma cells could be selectively analyzed, μNMR was first tested in vitro in human cell lines using antibodies specific for the melanocyte markers MART-1 and HMB45. Two melanoma lines were tested, BRAF wild-type MeWo and BRAF-mutant SKMel28 cells, as well as non-melanoma HeLa cells as a control. Expression of MART-1 (20.2 ± 1.8 for BRAF WT melanoma, 17.0 ± 1.5 for BRAF mutant melanoma) and HMB45 (10.0 ± 0.90 for BRAF WT, 15.9 ± 1.4 for BRAF mutant) were higher in both melanoma cell lines (Figure 1) compared with non-melanoma cells (2.4 ± 0.2 for MART1, 0.5 ± 0.04 HMB45), to a degree of statistical significance (P < 0.0001 for both MART-1 and HMB45, Student’s t test).
Figure 1.
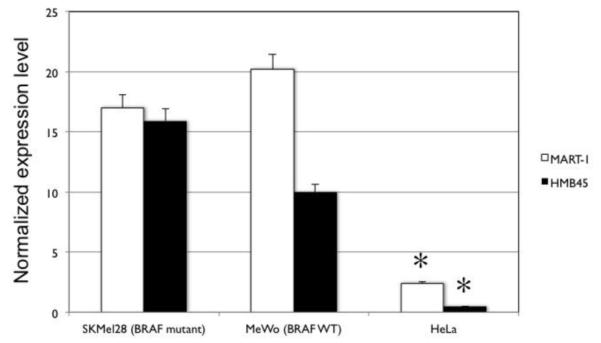
μNMR detection of melanoma markers in vitro. Two human melanoma cell lines (SKMel28 and MeWo) and one non-melanoma (HeLa) line were assayed for expression of melanocyte-specific cell markers MART-1 and HMB45 using μNMR technique. Expression of both markers was significantly higher in both melanoma lines compared with HeLa (asterisks indicate statistically significant difference by Student’s t test; P <0.0001).
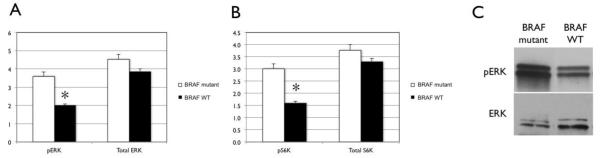
μNMR was also tested for its ability to distinguish BRAF signaling activity between BRAF WT and BRAF mutant human melanoma cells in vitro, using antibodies specific for the BRAF downstream signaling molecules ERK (Figure 2A) and S6K (Figure 2B). For each molecule, paired antibodies were used specific for total and phosphorylated (active) protein, respectively. Expression of total ERK (3.9 ± 0.23 for BRAF WT and 4.5 ± 0.41 for BRAF mutant) and total S6K (3.3 ± 0.20 for BRAF WT, 3.8 ± 0.34 for BRAF mutant) were similar between BRAF WT and BRAF mutant cells. However, phosphorylated ERK (pERK) and phosphorylated S6K (pS6K) were both elevated in BRAF mutant cells (3.6 ± 0.32 for pERK, 3.0 ± 0.32 for pS6K) compared with BRAF WT cells (2.0 ± 0.12 for pERK, 1.6 ± 0.23 for pS6K), differences that were statistically significant (P = 0.001 for pERK and 0.007 for pS6K). Western blot analysis of cell lysates was also performed to independently assess ERK phosphorylation (Figure 2C). The ERK phosphorylation percentages for BRAF mutant and BRAF WT melanoma cells based on quantitative Western blot analysis (91.7% and 47.6%, respectively) were concordant with the proportions observed by μNMR (mean pERK percentages 80.0% and 51.2%, respectively).
Figure 2.
μNMR discrimination of MAP kinase pathway signaling between BRAF wild-type and BRAF mutant human melanoma cells in vitro. μNMR analysis of relative expression of MAP kinase pathway molecules ERK (A) and S6K (B) show significant increases in phorphorylated (active) forms of both molecules in BRAF mutant melanoma cells compared with BRAF wild-type cells. Error bars indicate standard error. Asterisks indicate statistical significance (P < 0.0001, Student’s t test). (C) Western blot analysis also demonstrates an increase in phosphorylated ERK in BRAF-mutant melanoma cells compared with BRAF wild-type cells.
μNMR analysis of BRAF tumor signaling in melanoma patients
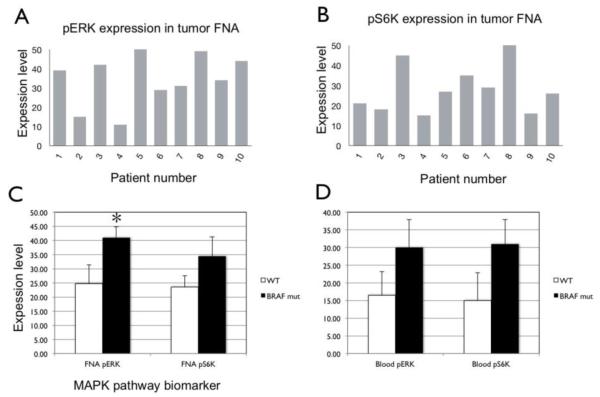
We next performed μNMR molecular analysis of melanomas in patients in a single institution, IRB-approved clinical study (Figure 3). 10 total patients with known melanoma who were scheduled to undergo biopsy of a suspected metastatic lesion were enrolled in the study. The age, sex, biopsy locations for all of the enrolled patients are listed in Table 1. The mean patient age was 57.1 ± 13.4 years. Genetic analysis of the 10 patients in the clinical study demonstrated 4 patients with BRAF wild-type and 5 with BRAF V600E mutant melanomas, with the BRAF status of each individual patient listed in Table 1. For each patient, fine needle aspiration was performed on the biopsied lesion and analyzed by μNMR for BRAF signaling activity based on expression of pERK and pS6K (Figure 4A-B). The BRAF status of each patient’s melanoma was determined based on tumor genetic analysis (listed in Table 1). μNMR analysis of tumor FNA samples demonstrated expression levels of pERK (41.0 ± 8.6) and pS6K (34.4 ± 15.5) in BRAF mutant melanomas that were both increased compared with those observed in BRAF wild type melanomas (24.8 ± 15.0 and 23.5 ± 9.0, respectively). The increased expression of pERK in BRAF mutant melanomas was statistically significant (P= 0.009, Student’s t test). A μNMR tumor FNA pERK threshold score of 30 for defining BRAF mutant signaling yielded 100% sensitivity and 75% specificity for detecting BRAF mutant signaling compared with genetic reference.
Figure 3.
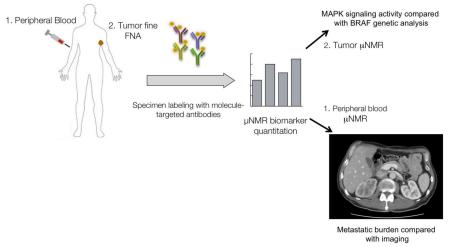
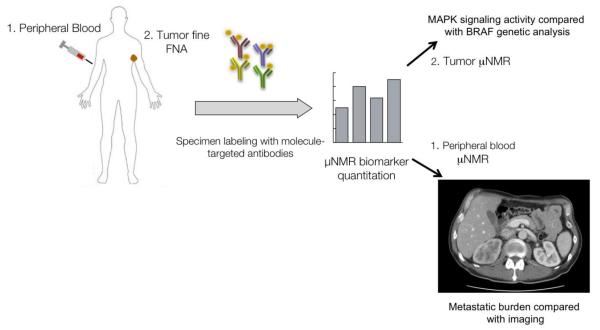
Overview of μNMR melanoma clinical study design. Peripheral blood and tumor fine needle aspiration (FNA) specimens are collected from melanoma patients. Specimens are labeled with molecule-targeted antibodies conjugated to magnetic nanoparticles and then analyzed on a miniaturized μNMR device at the point of care. Tumor cells from FNA are analyzed for MAP kinase signaling activity, while peripheral blood circulating tumor cells are identified and correlated with metastatic burden assessed on cross-sectional imaging.
Table 1.
Demographic information on patients enrolled in μNMR melanoma clinical trial. Sex, age, lesion location, and tumor BRAF status are listed.
| Patient | Sex | Age | Lesion location | BRAF status |
|---|---|---|---|---|
| 1 | M | 52 | Inguinal (R) | Unknown |
| 2 | M | 71 | Axillary (R) | Wild type |
| 3 | F | 56 | Liver | V600E mutant |
| 4 | F | 69 | Chest wall | Wild type |
| 5 | F | 53 | Arm (R) | V600E mutant |
| 6 | M | 64 | Liver | Wild type |
| 7 | M | 71 | Neck | V600E mutant |
| 8 | F | 32 | Retroperitoneum | V600E mutant |
| 9 | M | 64 | Pelvis | V600E mutant |
| 10 | F | 39 | Back | Wild type |
Figure 4.
μNMR assessment of MAPK activity from tumor FNA samples of melanoma pts. Tumor cell expression levels of pERK (A) and pS6K (B) are shown for each patient in the clinical study based on tumor FNA samples. (C) Mean expression levels of pERK and pS6K in BRAF wild-type (white bars) vs BRAF mutant (black bars) patients. Error bars indicate standard error. Asterisk indicates statistical significance (P = 0.0009, Student’s t test).
Comparison of μNMR circulating tumor cell quantification with metastatic burden on imaging
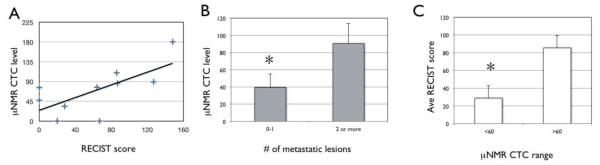
A specimen of peripheral blood was also taken from each melanoma patient undergoing biopsy, in order to detect circulating tumor cells (CTC) using μNMR based on a previously published four biomarker method including the melanoma markers MART-1 and HMB45, as well as EpCAM and EGFR(20). CTC levels from 7mL of peripheral blood were compared with metastatic burden assessed on clinical imaging (Figure 5) performed near the time of biopsy (mean time between biopsy and imaging 15.8 ± 11 days). Overall, there was a general trend of increasing μNMR CTC level with increased metastatic burden measured by RECIST SLD score for individual patients (Figure 6A). Overall, patients with a CTC level > 60 were associated with a significantly higher RECIST SLD score on imaging (85.3 ± 34.2; Figure 6B) compared with patients with CTC levels ≤ 60 (28.8 ± 28.1; P = 0.021, Student’s t test). In addition, CTC levels in patients showed significant differences based on number metastatic lesions with patients with multiple metastases on imaging exhibiting higher (90.3 ± 57.9) compared with those with either 0-1 lesions (39.3 ± 31.5; P = 0.045).
Figure 5.
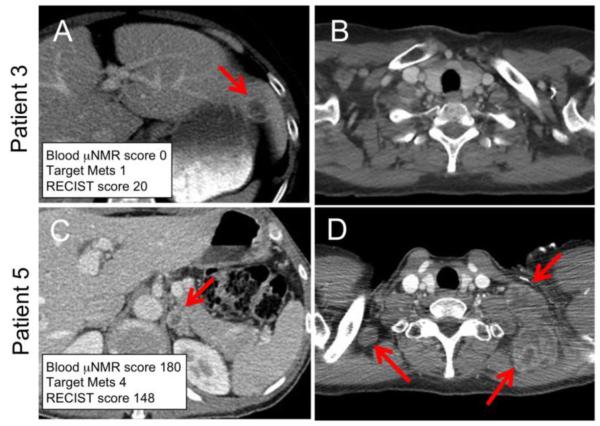
μNMR assessment of melanoma circulating tumor cells in patients. Representative axial CT images of the abdomen (A, C) and chest (B, D) from two melanoma patients obtained at the time of μNMR peripheral blood CTC analysis. Patient 3 (top row) had a μNMR score of zero which correlates with a single target lesion in the left hepatic lobe (A; arrow), no thoracic lesions, and a RECIST SLD score of 20. Patient (bottom row) had a μNMR score of 180 and was found to have four target lesions, including a left adrenal gland lesion (C; arrow) and multiple enlarged left cervical lymph nodes (D; arrows), associated with a RECIST SLD score of 148.
Figure 6.
μNMR assessment of melanoma circulating tumor cells in peripheral blood correlates with metastatic burden on imaging. (A) μNMR CTC level plotted as a function of RECIST metastatic score based on imaging for individual patients. (B) Mean CTC level based on number of metastatic lesions visualized on imaging. (C) Average RECIST metastatic score based on high (>60) or low (< 60) CTC level. Error bars indicate statistical significance. Asterisks indicate statistical significance (P <0.05, Student’s t test).
DISCUSSION
Since the initial identification of activating BRAF mutations in a high percentage of human melanomas(27), a number of molecule-targeted inhibitors of the MAP kinase signaling molecules BRAF and MEK have been demonstrated to improve survival of patients with metastatic melanoma compared standard chemotherapy(2, 28). However, while initial response rates of BRAF-mutant melanoma patients treated with BRAF and/or MEK inhibitors is high, acquired resistance is a frequent problem with patients developing resistance within the first year of treatment(2, 5). Mechanisms of acquired resistance include acquisition of additional mutations that restore MAP kinase signaling as well as activation of other signaling pathways such as PI3K/mTOR(5). With the continuing development of second line treatments including inhibitors of PI3K/mTOR and ERK(29) as well as immunotherapies(30), it is important to develop early detection methods of resistance to BRAF/MEK inhibitors to allow timely initiation of alternate therapies in order to maximize the chance of success.
In the current study, we apply μNMR technology(14, 15) to perform point of care molecular profiling of tumor cells isolated from melanoma patients. Similar to clinical MRI, μNMR quantifies biomarker expression levels based on changes in T2 relaxivity induced by binding of molecule-targeted magnetic nanoparticles(16). Previous studies have shown μNMR to be a robust method for diagnosing scarce cell populations such as malignant cells in ascites(19), fine needle aspirates(18), or peripheral blood(20). μNMR is capable of providing rapid molecular analysis of intact cells from in vivo clinical specimens from a hand-held device with minimal specimen processing, and is ideally suited for evaluation of melanoma patients in an outpatient clinic setting, where fine needle aspiration and peripheral blood sampling can be easily performed.
Our results indicate that μNMR is able to discriminate MAP kinase signaling activity, measured by levels of phosphorylated S6K and ERK, between BRAF wild type and BRAF mutant melanomas both in vitro and in vivo. The difference in S6K and ERK phosphorylation observed between BRAF wild-type and mutant melanomas (Figure 4) in patients was consistent with in vitro results observed with BRAF wild type and mutant human melanoma cell lines assessed by μNMR and Western blot (Figure 2). The in vivo analysis was based on tumor fine needle aspirates obtained with a 22 gauge needle, which can easily be obtained from dermal lesions in melanoma patients in the clinic setting with no or only local anesthetic. This technique offers great potential for serial evaluation of response to BRAF or MEK inhibitors in a minimally invasive fashion. The ability to monitor activity of different MAP kinase signaling molecules can help identify mechanisms of acquired resistance in individual patients and track genetic alterations impacting on personalized therapeutic decision-making(7).
We also measured circulating tumor cells in peripheral blood using the μNMR technique and correlated with metastatic burden assessed by RECIST 1.1 criteria on cross-sectional imaging. Significant differences in μNMR CTC level were observed based on overall RECIST 1.1. metastatic burden as well as number of target lesions (Figure 6). These results build on prior studies demonstrating an association between CTC levels and survival in breast and lung cancer patients(31-33), as well as preclinical studies in mice suggesting an important role for circulating tumor cells in mediating melanoma invasiveness(34). An important result of our study is the establishment of a relationship between peripheral CTC levels and imaging assessment of metastatic burden. Because imaging plays such an important role in current clinical assessment of disease extent and treatment response in oncology patients, it is important for new emerging biomarkers of treatment response such as CTC levels to be validated against imaging assessment of disease(35). It is likely that cell-based biomarkers such as CTC level will serve a complementary role to cross-sectional imaging in the future. Imaging is noninvasive but less practical to perform at the frequent short intervals that can be achieved by FNA or peripheral blood sampling based methods. In addition, imaging is likely to be less sensitive to early acquisition of therapy resistance. At the same time, CTC quantitation may provide a more sensitive assessment of changes in metastatic burden but lacks the ability of cross-sectional imaging to identify new sites of metastasis and metastasis-associated complications. It is likely that both imaging and CTC quantitation will play important roles in oncologic surveillance in the future.
Limitations of this study include the small number of patients enrolled, which limits our ability to perform statistical inference. These results should be considered a promising pilot study requiring larger studies with more patients for validation. In addition, our study was limited by μNMR assessment at a single timepoint, as the research protocol utilized for this study required eligible patients to be undergoing a lesion biopsy as part of standard medical care. These biopsies were predominantly requested for suspected disease progression, and future studies will be needed to evaluate the μNMR technique in a surveillance setting with serial assessments performed at multiple timepoints during therapy.
In summary, our results indicate that the μNMR technique is a promising method for point of care assessment of tumor MAP kinase signaling in patients with melanoma. Our results also establish a link between μNMR peripheral blood circulating tumor cell levels and metastatic burden visualized on cross-sectional imaging, and suggest potentially complementary roles for the two tests. It is likely that point of care techniques such as μNMR will see an increasing role in oncologic surveillance as novel molecule-targeted primary and next-line therapies continue to undergo clinical development.
ACKNOWLEDGEMENTS
This work was supported in part by a Research Scholar Grant from the Radiological Society of North America (to M.S.G.) and NIH grant 2R01EB004626 (to R.W.).
Abbreviations
- NMR
nuclear magnetic resonance
- CTC
circulating tumor cells
- CT
computed tomography
- MRI
magnetic resonance imaging
- RECIST
Response Evaluation Criteria in Solid Tumors
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: One study author (R.W.) has a financial interest in T2 Biosystems, a biotechnology company that utilizes μNMR technology for medical applications. However, T2 Biosystems provided no support for this research and has no rights over any data resulting from this work. None of the other study authors has any competing conflicts of interest.
REFERENCES
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Carlino MS, Long GV, Kefford RF, Rizos H. Targeting oncogenic BRAF and aberrant MAPK activation in the treatment of cutaneous melanoma. Crit Rev Oncol Hematol. 2015 doi: 10.1016/j.critrevonc.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Korn EL, Liu PY, Lee SJ, Chapman JA, Niedzwiecki D, Suman VJ, et al. Meta-analysis of phase II cooperative group trials in metastatic stage IV melanoma to determine progression-free and overall survival benchmarks for future phase II trials. J Clin Oncol. 2008;26(4):527–34. doi: 10.1200/JCO.2007.12.7837. [DOI] [PubMed] [Google Scholar]
- 4.Puzanov I, Flaherty KT. Targeted molecular therapy in melanoma. Semin Cutan Med Surg. 2010;29(3):196–201. doi: 10.1016/j.sder.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan RJ, Flaherty KT. Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer. 2013;49(6):1297–304. doi: 10.1016/j.ejca.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan RJ, Flaherty KT. New strategies in melanoma: entering the era of combinatorial therapy. Clin Cancer Res. 2015;21(11):2424–35. doi: 10.1158/1078-0432.CCR-14-1650. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas N Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62(23):6997–7000. [PubMed] [Google Scholar]
- 9.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18(6):683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468(7326):973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perna D, Karreth FA, Rust AG, Perez-Mancera PA, Rashid M, Iorio F, et al. BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. Proc Natl Acad Sci U S A. 2015;112(6):E536–45. doi: 10.1073/pnas.1418163112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371(20):1867–76. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 14.Lee H, Sun E, Ham D, Weissleder R. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008;14(8):869–74. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Issadore D, Chung J, Shao H, Liong M, Ghazani AA, Castro CM, et al. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-hall detector. Sci Transl Med. 2012;4(141):141ra92. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Issadore D, Park YI, Shao H, Min C, Lee K, Liong M, et al. Magnetic sensing technology for molecular analyses. Lab Chip. 2014;14(14):2385–97. doi: 10.1039/c4lc00314d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H, Shin TH, Cheon J, Weissleder R. Recent Developments in Magnetic Diagnostic Systems. Chem Rev. 2015 doi: 10.1021/cr500698d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haun JB, Castro CM, Wang R, Peterson VM, Marinelli BS, Lee H, et al. Micro-NMR for rapid molecular analysis of human tumor samples. Sci Transl Med. 2011;3(71):71ra16. doi: 10.1126/scitranslmed.3002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson VM, Castro CM, Chung J, Miller NC, Ullal AV, Castano MD, et al. Ascites analysis by a microfluidic chip allows tumor-cell profiling. Proc Natl Acad Sci U S A. 2013;110(51):E4978–86. doi: 10.1073/pnas.1315370110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghazani AA, McDermott S, Pectasides M, Sebas M, Mino-Kenudson M, Lee H, et al. Comparison of select cancer biomarkers in human circulating and bulk tumor cells using magnetic nanoparticles and a miniaturized micro-NMR system. Nanomedicine. 2013;9(7):1009–17. doi: 10.1016/j.nano.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov. 2014;13(12):928–42. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 22.Ghazani AA, Castro CM, Gorbatov R, Lee H, Weissleder R. Sensitive and direct detection of circulating tumor cells by multimarker micro-nuclear magnetic resonance. Neoplasia. 2012;14(5):388–95. doi: 10.1596/neo.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haun JB, Yoon TJ, Lee H, Weissleder R. Molecular detection of biomarkers and cells using magnetic nanoparticles and diagnostic magnetic resonance. Methods Mol Biol. 2011;726:33–49. doi: 10.1007/978-1-61779-052-2_3. [DOI] [PubMed] [Google Scholar]
- 24.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23(3):302–15. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schingelin J, Rueden CT, Hiner MC, Elceiri KW. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev. 2015;82(7-8):518–29. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 27.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 28.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367(18):1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatzivassiliou G, Liu B, O'Brien C, Spoerke JM, Hoeflich KP, Haverty PM, et al. ERK inhibition overcomes acquired resistance to MEK inhibitors. Mol Cancer Ther. 2012;11(5):1143–54. doi: 10.1158/1535-7163.MCT-11-1010. [DOI] [PubMed] [Google Scholar]
- 30.Improta G, Leone I, Donia M, Gieri S, Pelosi G, Fraggetta F. New developments in the management of advanced melanoma - role of pembrolizumab. Onco Targets Ther. 2015;8:2535–43. doi: 10.2147/OTT.S72823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucci A, Hall CS, Lodhi AK, Bhattacharyya A, Anderson AE, Xiao L, et al. Circulating tumour cells in non-metastatic breast cancer: a prospective study. Lancet Oncol. 2012;13(7):688–95. doi: 10.1016/S1470-2045(12)70209-7. [DOI] [PubMed] [Google Scholar]
- 32.Rack B, Schindlbeck C, Juckstock J, Andergassen U, Hepp P, Zwingers T, et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J Natl Cancer Inst. 2014;106(5) doi: 10.1093/jnci/dju066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol. 2011;29(12):1556–63. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 34.Luo X, Mitra D, Sullivan RJ, Wittner BS, Kimura AM, Pan S, et al. Isolation and molecular characterization of circulating melanoma cells. Cell Rep. 2014;7(3):645–53. doi: 10.1016/j.celrep.2014.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neal JW, Gainor JF, Shaw AT. Developing biomarker-specific end points in lung cancer clinical trials. Nat Rev Clin Oncol. 2015;12(3):135–46. doi: 10.1038/nrclinonc.2014.222. [DOI] [PubMed] [Google Scholar]