Abstract
Glucocorticoids are primary stress hormones that regulate many physiological processes, and synthetic derivatives of these molecules are widely used in the clinic. The molecular factors that govern tissue specificity of glucocorticoids, however, are poorly understood. The actions of glucocorticoids are mediated by the glucocorticoid receptor (GR). To discover new proteins that interact with GR and modulate its function, we performed a yeast two-hybrid assay. The MyoD family inhibitor domain-containing protein (MDFIC) was identified as a binding partner for GR. MDFIC associated with GR in the cytoplasm of cells, and treatment with glucocorticoids resulted in the dissociation of the GR-MDFIC complex. To investigate the function of the GR-MDFIC interaction, we performed a genome-wide microarray in intact and MDFIC-deficient A549 cells that were treated with glucocorticoids. A large cohort of genes was differentially regulated by GR depending on the presence or absence of MDFIC. These gene changes were strongly associated with inflammation, and glucocorticoid regulation of the inflammatory response was altered in MDFIC-deficient cells. At a molecular level, the interaction of MDFIC with GR altered the phosphorylation status of the receptor. We demonstrate in COS-1 cells that changes in receptor phosphorylation underlie the ability of MDFIC to regulate the transcriptional activity of GR. Finally, we show that GR directly represses the MDFIC gene, revealing a negative feedback loop by which glucocorticoids limit MDFIC activity. These findings identify a new binding partner for cytoplasmic GR that modulates the receptor transcriptome and contributes to the tissue-specific actions of glucocorticoids.
Keywords: gene expression, gene regulation, glucocorticoid, glucocorticoid receptor, phosphorylation, MyoD family inhibitor domain-containing protein
Introduction
Glucocorticoids are released by the hypothalamic-pituitary-adrenal axis in a circadian manner and in response to stress (1). They act on nearly every tissue and organ of the body and function to maintain homeostasis. Biological processes regulated by these hormones include intermediary metabolism, cellular proliferation and differentiation, apoptosis, skeletal growth, cognition, cardiac function, development, reproduction, and immune function (2). The ability of glucocorticoids to inhibit inflammation and suppress the immune system has made them one of the most prescribed drugs in the world today. Synthetic glucocorticoids are used to treat inflammatory and autoimmune diseases, prevent organ transplant rejection, and combat cancers of the lymphoid system. In addition, glucocorticoids are routinely given to preterm babies to improve survival for their effects on lung maturation (3, 4). The therapeutic benefit of glucocorticoids, however, is limited by severe side effects that develop in patients chronically treated with these steroids. Adverse responses include hypertension, osteoporosis, glaucoma, abdominal obesity, diabetes, growth retardation in children, and depression. Many of these symptoms are also observed in patients with excessive glucocorticoid production due to chronic stress or Cushing's disease.
The physiological and pharmacological actions of glucocorticoids are mediated by the glucocorticoid receptor (GR;2 NR3C1), a member of the nuclear receptor superfamily of ligand-dependent transcription factors (5). GR is a modular protein composed of an amino-terminal transactivation domain (NTD), a central DNA binding domain (DBD), and a carboxyl-terminal ligand binding domain (LBD). Separating the DBD and LBD is a flexible linker called the hinge region. In the absence of glucocorticoids, GR is found predominantly in the cytoplasm of cells in a complex with various chaperone proteins that maintain the receptor in a conformation that binds glucocorticoids with high affinity (6, 7). Upon binding glucocorticoids, GR undergoes a conformational change that results in the dissociation of the chaperone proteins, the exposure of nuclear localization sequences, and receptor translocation into the nucleus (8). GR in the nucleus interacts with an assortment of co-activators and co-repressors to induce or repress the expression of thousands of genes. GR alters gene transcription by directly binding to specific sequences of DNA termed glucocorticoid-responsive elements (GREs) and/or by physically interacting with other DNA-bound transcription factors.
The cellular response to glucocorticoids is remarkably diverse across tissues and cell types (9–12). Glucocorticoids induce apoptosis in thymocytes and osteoblasts but promote the survival of hepatocytes and cardiomyocytes (13–15). Sensitivity to glucocorticoids varies among individuals, among tissues from the same individual, and even within the same cell depending on the phase of the cell cycle. The development of tissue-specific glucocorticoid resistance is a major limitation to effective long term glucocorticoid therapy (16, 17). Various factors have been shown to modulate the type and/or magnitude of the glucocorticoid response including ligand availability, the cellular composition of GR isoforms, post-translational modifications of GR, the availability of specific co-activators and co-repressors, epigenetic regulators, and the chromatin landscape (1, 18, 19). Understanding the molecular mechanisms that contribute to the heterogeneity and tissue specificity of glucocorticoid signaling will facilitate the development of novel glucocorticoids and treatment strategies with improved benefit/risk ratios.
In the following study we performed a yeast two-hybrid assay to discover new proteins that interact with GR and modulate its function. We show that the MyoD family inhibitor domain-containing protein (MDFIC) interacts with the hinge region of unliganded GR in the cytoplasm of cells. Binding of glucocorticoids promotes the dissociation of the GR-MDFIC complex. The interaction of MDFIC with the receptor alters the GR transcriptome and leads to unique cellular responses after glucocorticoid treatment. At a molecular level the presence of MDFIC modulates both the basal and glucocorticoid-induced phosphorylation status of GR. We also demonstrate that glucocorticoids directly repress MDFIC gene expression revealing a negative feedback loop by which GR can curb the modulatory actions of MDFIC. These findings suggest that alterations in the expression level of MDFIC provide a novel mechanism for generating tissue- and cell type-specific responses to glucocorticoids.
Results
MDFIC associates with GR in the cytoplasm of cells
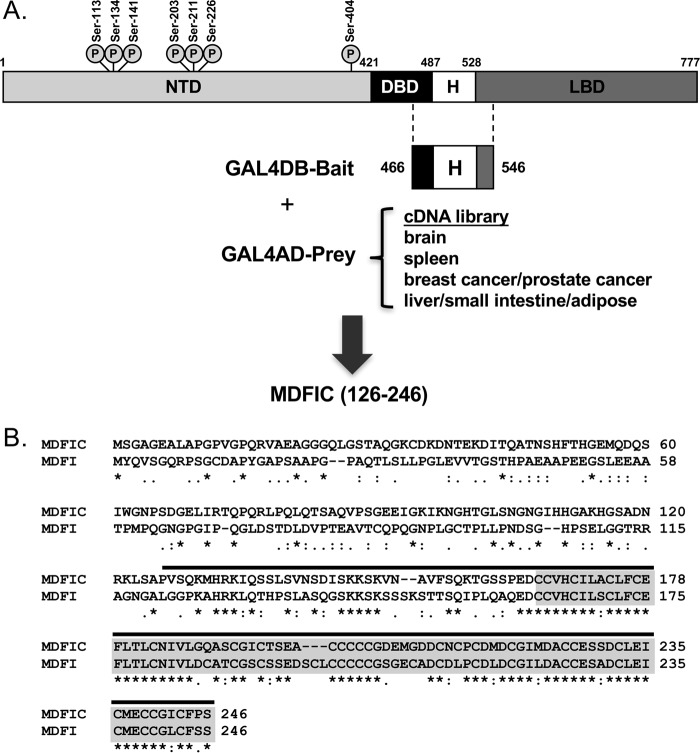
To discover new proteins that interact with GR, we performed a yeast two-hybrid screen using as bait a region of human GR spanning the hinge domain (Fig. 1A). The hinge domain was chosen because relatively little is known about how this region modulates GR activity compared with the well studied NTD, DBD, and LBD. cDNA libraries derived from human brain, spleen, breast cancer/prostate cancer, and liver/small intestine/adipose tissues were used as prey. One clone obtained from screening the breast cancer/prostate cancer cDNA library contained an open reading frame with 100% identity to the carboxyl-terminal half (amino acids 126–246) of MDFIC (Fig. 1B). MDFIC belongs to a small family of gene expression regulators characterized by having a unique cysteine-rich carboxyl-terminal domain (20, 21). The other member of this family is MyoD family inhibitor isoform 1 (MDFI) (Fig. 1B). The cysteine-rich carboxyl-terminal domain of MDFIC is composed of 81 amino acids (166–246) and shares 77% identity with the corresponding region of MDFI. A total of 24 and 26 cysteine residues are located within the cysteine-rich carboxyl-terminal domain for MDFIC and MDFI, respectively.
Figure 1.
Yeast two-hybrid screen identifies MDFIC as a GR-binding protein. A, schematic of human GR depicting domain structure and sites of phosphorylation. Amino acids 466–546 span the hinge (H) region. This sequence was fused to the GAL4 DNA binding domain (GAL4DB-bait) and used as bait in a yeast two-hybrid assay. The bait was screened against brain, spleen, pooled breast cancer/prostate cancer, and pooled liver/small intestine/adipose cDNA libraries fused to the GAL4 activation domain (GALAD-prey). An open reading frame encoding amino acids 126–246 of MDFIC was retrieved from the breast cancer/prostate cancer cDNA library. B, amino acid alignment of MDFIC and its related family member MDFI. The cysteine-rich carboxyl-terminal domain (highlighted in gray) is highly conserved between the two proteins (conserved residues are indicated with an asterisk). The MDFIC sequence retrieved from the yeast two-hybrid screen (amino acids 126–246) is indicated by the line above the residues.
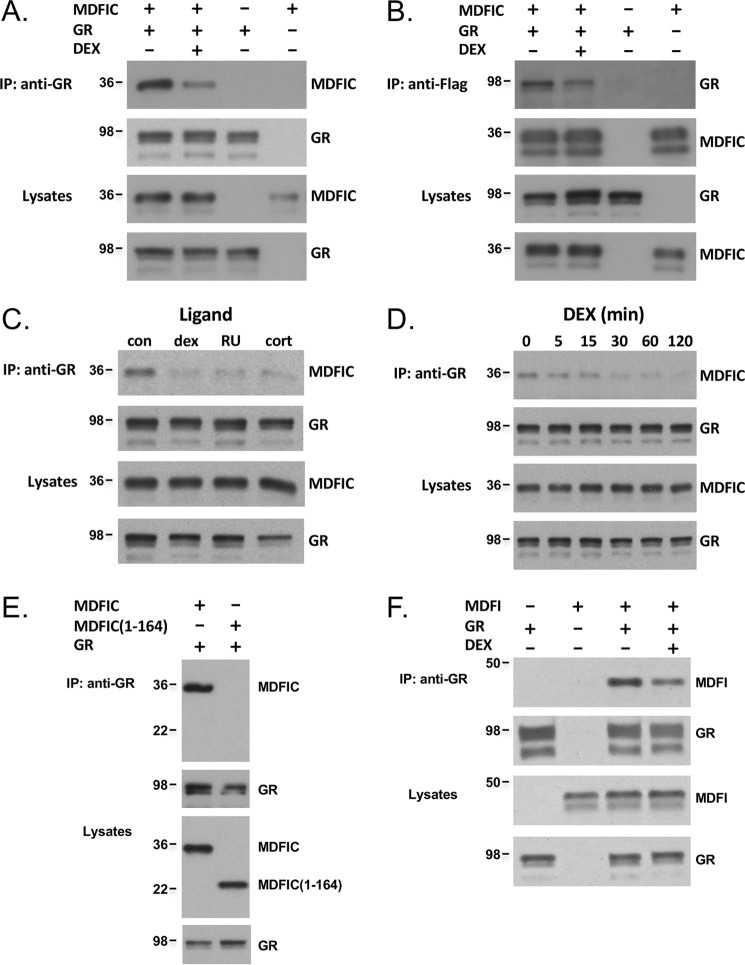
To determine whether the full-length GR and MDFIC interact in mammalian cells, we expressed GR and FLAG-MDFIC in COS-1 cells (which lack detectable levels of endogenous GR) and performed a co-immunoprecipitation assay. In the absence of glucocorticoids, MDFIC co-immunoprecipitated with GR (Fig. 2A). Treatment of cells for 1 h with the synthetic glucocorticoid dexamethasone (Dex) (100 nm) resulted in the dissociation of the complex. Reversing the order of the antibodies in the co-immunoprecipitation experiment yielded identical results; GR co-immunoprecipitated with MDFIC and the complex dissociated after Dex treatment (Fig. 2B). Dissociation of the GR-MDFIC complex was also observed with the natural glucocorticoid cortisol and the partial agonist/antagonist RU486 (Fig. 2C). A time-course was performed with 100 nm Dex to evaluate the kinetics of the GR-MDFIC dissociation. As shown in Fig. 2D, a reduction in the GR-MDFIC interaction was detected as early as 5 min after glucocorticoid addition. These data demonstrate that GR and MDFIC associate in a complex in the absence of glucocorticoids and that glucocorticoid binding to GR promotes the dissociation of the complex.
Figure 2.
GR and MDFIC form a complex in COS-1 cells that dissociates with glucocorticoid binding. A, COS-1 cells expressing GR, FLAG-MDFIC, or both GR and FLAG-MDFIC were treated for 1 h with vehicle or 100 nm Dex and processed for co-immunoprecipitation. Lysates were immunoprecipitated (IP) with an anti-GR antibody, and recovered proteins were immunoblotted for MDFIC and GR. Shown are representative immunoblots from three independent experiments. B, COS-1 cells expressing GR, FLAG-MDFIC, or both GR and FLAG-MDFIC were processed as above for co-immunoprecipitation except that the immunoprecipitation was performed with an anti-FLAG antibody. Shown are representative immunoblots from three independent experiments. C, COS-1 cells expressing both GR and FLAG-MDFIC were treated with vehicle (con), 100 nm Dex, 500 nm RU486 (RU), or 1 μm cortisol (cort) for 1 h and processed as above for co-immunoprecipitation. Shown are representative immunoblots from three independent experiments. D, COS-1 cells expressing both GR and FLAG-MDFIC were treated with 100 nm Dex for varying amounts of time and processed as above for co-immunoprecipitation. Shown are representative immunoblots from three independent experiments. E, COS-1 cells expressing both GR and FLAG-MDFIC or both GR and FLAG-MDFIC(1–164) were processed as above for co-immunoprecipitation. Shown are representative immunoblots from three independent experiments. F, COS-1 cells expressing GR, FLAG-MDFI, or both GR and FLAG-MDFI were treated for 1 h with vehicle or 100 nm Dex and processed as above for co-immunoprecipitation. Shown are representative immunoblots from three independent experiments.
The cysteine-rich carboxyl-terminal domain of MDFIC is embedded in the region of the protein retrieved from the yeast two-hybrid screen (Fig. 1B). Therefore, we examined whether this domain was necessary for the interaction of MDFIC with GR. Co-immunoprecipitation experiments were performed in COS-1 cells expressing GR and either full-length MDFIC or a truncated version missing the entire cysteine-rich carboxyl-terminal domain (MDFIC(1–164)). As shown in Fig. 2E, only the full-length MDFIC co-immunoprecipitated with GR, indicating the cysteine-rich carboxyl-terminal domain of MDFIC is necessary for its interaction with the receptor. Because the related family member MDFI also contains a homologous cysteine-rich carboxyl-terminal domain (Fig. 1B), we investigated whether MDFI interacted with GR. Similar to our findings for MDFIC, MDFI also co-immunoprecipitated with GR in untreated cells, and the addition of 100 nm Dex promoted the dissociation of the complex (Fig. 2F). These data indicate that the cysteine-rich carboxyl-terminal domain of MDFIC and MDFI mediates their interaction with GR.
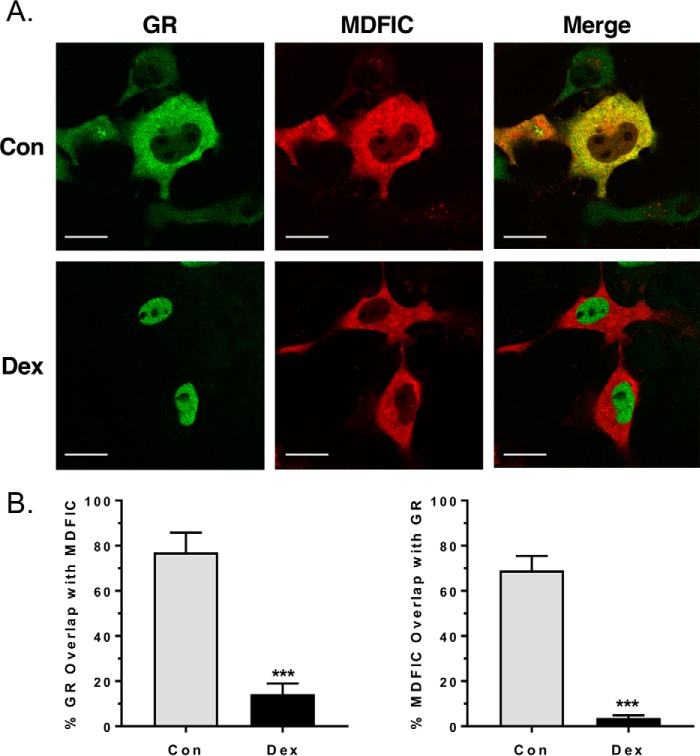
GR is distributed primarily in the cytoplasm of cells in the absence of glucocorticoids and translocates into the nucleus following glucocorticoid binding. Translocation of GR is directed by two nuclear localization signals, one of which is located at the junction of the DBD and hinge region (22, 23). To determine whether MDFIC co-localizes with GR in the cytoplasm and whether its interaction with GR in the hinge region alters receptor movement into the nucleus, we performed immunocytochemistry on COS-1 cells expressing GR and MDFIC. Both GR and MDFIC resided predominantly in the cytoplasm of control cells (Fig. 3A, upper panels). The strong overlay in their distribution, indicated by the yellow color in the merged image, is consistent with the GR-MDFIC interaction occurring in the cytoplasm of cells. Treatment with 100 nm Dex resulted in the robust translocation of GR into the nucleus and no change in the distribution of MDFIC (Fig. 3A, lower panels). Colocalization analysis revealed a 77.0% overlap of GR with MDFIC in control cells that was reduced to 14.3% in cells treated with Dex (Fig. 3B). Similarly, MDFIC exhibited a 69.0% overlap with GR in control cells that was decreased to 3.7% in response to Dex (Fig. 3B). The reduction in overlay of the two proteins after glucocorticoid treatment suggests that ligand binding triggers the dissociation of the GR-MDFIC complex. These findings are in agreement with our co-immunoprecipitation data. In addition, they suggest that the interaction of MDFIC with cytoplasmic GR does not impede its subsequent translocation into the nucleus after binding glucocorticoids.
Figure 3.
GR and MDFIC co-localize in the cytoplasm of COS-1 cells. A, COS-1 cells expressing both GR and FLAG-MDFIC were treated with vehicle (Con) or 100 nm Dex for 1 h. Cells were then fixed and processed for immunocytochemistry. Shown are representative confocal microscopic images of the distribution of GR (green) and MDFIC (red). As indicated by the yellow color in the merged image, there is a strong overlay in the distribution of GR and MDFIC in the cytoplasm of control cells. The scale bar is 20 μm. B, colocalization analysis was performed on control (n = 9) and Dex-treated cells (n = 12). Shown is the quantitation for the overlap of GR with MDFIC (left panel) and the overlap of MDFIC with GR (right panel) (Student's t test, mean ± S.E.; ***, p < 0.001).
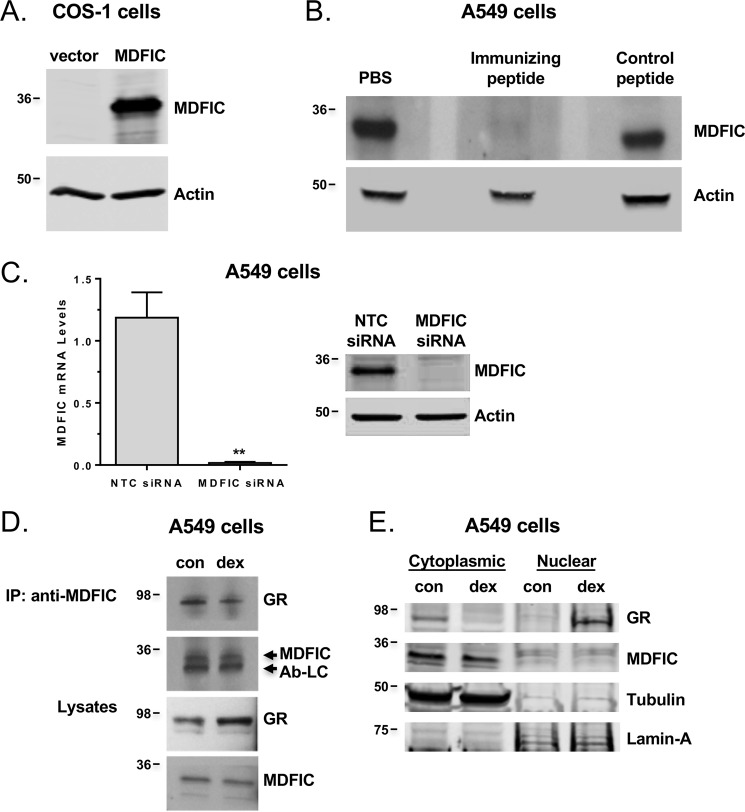
Having established that GR and MDFIC can interact in transfected COS-1 cells, we turned our attention to the endogenous GR and MDFIC. To detect endogenous MDFIC, we generated an anti-MDFIC antibody against amino acids 109–122 of human MDFIC. The anti-MDFIC antibody detected MDFIC exogenously expressed in COS-1 cells (Fig. 4A). It also detected MDFIC endogenously expressed in human A549 lung adenocarcinoma cells that are used classically to study glucocorticoid signaling (Fig. 4, B and C). The interaction was specific, as detection of MDFIC in A549 cells was prevented by preincubating the antibody with the immunizing peptide and by siRNA-mediated gene silencing of MDFIC (Fig. 4, B and C). We utilized the anti-MDFIC antibody in co-immunoprecipitation experiments to examine whether the endogenous GR and MDFIC associate in a complex. A549 cells were treated with or without 100 nm Dex for 1 h, and protein lysates were immunoprecipitated with the anti-MDFIC antibody. In the absence of glucocorticoids, GR co-immunoprecipitated with MDFIC, indicating the two endogenous proteins associate in a complex (Fig. 4D). Dex treatment resulted in the dissociation of the complex as the amount of co-immunoprecipitated GR was reduced in the glucocorticoid-treated cells (Fig. 4D). Consistent with the interaction of these two proteins occurring in the cytoplasm of cells, we found by cell fractionation studies that endogenous MDFIC and GR reside in the cytoplasm of A549 cells not exposed to glucocorticoids (Fig. 4E). After glucocorticoid treatment, GR translocated into the nucleus, whereas MDFIC remained in the cytoplasm (Fig. 4E). These data indicate that the endogenous GR and MDFIC form a complex in the cytoplasm of cells that is dissociated upon glucocorticoid binding to GR.
Figure 4.
Endogenous MDFIC expression and its interaction with GR. A, representative immunoblot from three independent experiments of COS-1 cells expressing FLAG-MDFIC or not (empty vector) using the anti-MDFIC antibody. B, representative immunoblot from two independent experiments of A549 cells using the anti-MDFIC antibody preincubated with PBS, the immunizing peptide, or a control peptide. C, A549 cells were transfected with NTC siRNA or MDFIC siRNA. The left panel shows RT-PCR analysis of MDFIC mRNA (Student's t test, mean ± S.E.; **, p < 0.01). The right panel shows representative immunoblot from three independent experiments with anti-MDFIC antibody and actin antibody. D, Endogenous GR and MDFIC associate in a complex. A549 cells were treated for 1 h with vehicle or 100 nm Dex and processed for co-immunoprecipitation (IP). Lysates were immunoprecipitated with the anti-MDFIC antibody, and recovered proteins were immunoblotted for GR and MDFIC. Nonspecific detection of the antibody light chain (Ab-LC) was observed just below the MDFIC protein. Shown are representative immunoblots from three independent experiments. con, vehicle. E, subcellular distribution of endogenous GR and MDFIC. A549 cells were treated for 1 h with vehicle or 100 nm Dex. Cytosolic and nuclear fractions were prepared, and immunoblots were performed with anti-GR, anti-MDFIC, anti-tubulin, and anti-Lamin-A antibodies. Shown are representative immunoblots from three independent experiments.
MDFIC alters the gene regulatory profile of GR
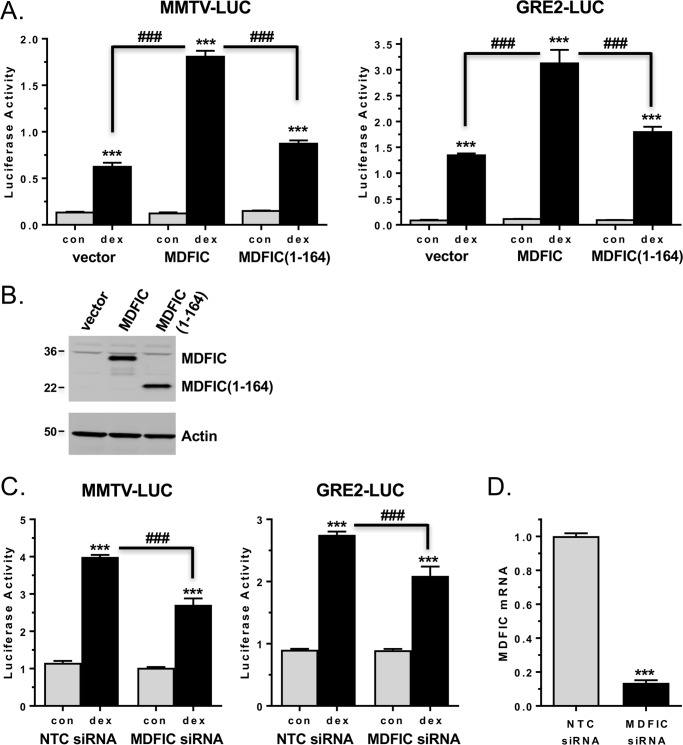
MDFIC has been characterized as a transcriptional regulator (20, 24–28); therefore, we initially investigated whether the association of MDFIC with GR altered the transcriptional activity of the receptor on glucocorticoid-responsive reporter genes. A549 cells were transfected with a luciferase reporter driven by the glucocorticoid-responsive mouse mammary tumor virus promoter (MMTV-LUC) and either empty vector or MDFIC. As shown in the left panel of Fig. 5A, an 18-h treatment with 100 nm Dex resulted in 4.7-fold induction of luciferase expression in cells receiving the empty vector. In cells transfected with MDFIC, however, glucocorticoid treatment resulted in a greater 14.8-fold induction in luciferase expression. The MMTV promoter is a complex promoter that contains binding sites not only for GR and but also for other transcription factors such as octamer binding factor 1 and nuclear factor 1. Therefore, we next tested the ability of MDFIC to alter the transcriptional activity of GR on a simple glucocorticoid-responsive promoter composed solely of 2 tandem GREs and a TATA box. A549 cells were transfected with a luciferase reporter driven by this simple glucocorticoid-responsive promoter (GRE2-LUC) and either empty vector or MDFIC. Glucocorticoid treatment resulted in a 15.6-fold increase in luciferase expression in cells receiving the empty vector (Fig. 5A, right panel). Cells transfected with MDFIC displayed an even greater 28.1-fold increase in luciferase expression in response to glucocorticoids. To determine if the observed increase in GR transactivation of these two reporter genes depended on its association with MDFIC, we utilized the MDFIC(1–164) truncation mutant, which does not interact with GR due to the absence of the cysteine-rich carboxyl-terminal domain (Fig. 2E). The enhanced transcriptional activity of GR on both these promoters was largely abolished in cells transfected with the MDFIC(1–164) truncation mutant (Fig. 5, A and B). We next examined GR transactivation of the MMTV-LUC and GRE2-LUC reporters in A549 cells depleted of endogenous MDFIC by siRNA-mediated gene silencing (Fig. 5, C and D). Loss of MDFIC resulted in a significant reduction in the ability of GR to transactivate both reporter genes after glucocorticoid treatment. These data suggest that the interaction of MDFIC with GR can alter its transcriptional activity on both complex and simple glucocorticoid-responsive promoters.
Figure 5.
MDFIC alters the transcriptional activity of GR on glucocorticoid-responsive reporter genes in A549 cells. A, A549 cells were transfected with the pMMTV-LUC reporter (left panel) or the pGRE2-LUC reporter (right panel) and either empty vector, FLAG-MDFIC, or FLAG-MDFIC(1–164). After an 18-h treatment with vehicle or 100 nm Dex, the cells were harvested, and luciferase activity was measured. Data represent the mean ± S.D. from four independent experiments performed in quadruplicate. A one-way ANOVA followed by Tukey's post hoc test was performed to determine significance. ***, p < 0.001 for Dex versus con (vehicle). ###, p < 0.001 for MDFIC Dex versus vector Dex and for MDFIC(1–164) Dex versus MDFIC Dex. B, protein lysates from A549 cells transfected as above with FLAG-MDFIC or FLAG-MDFIC(1–164) were immunoblotted with the anti-FLAG antibody (upper blot) or anti-actin antibody (lower blot). Shown is a representative immunoblot from three independent experiments. C, A549 cells were transfected with the pMMTV-LUC reporter (left panel) or the pGRE2-LUC reporter (right panel) and either NTC siRNA or MDFIC siRNA. After a 6-h treatment with vehicle or 100 nm Dex, the cells were harvested, and luciferase activity was measured. Data represent the mean ± S.D. from three independent experiments performed in quadruplicate. A one-way ANOVA followed by Tukey's post hoc test was performed to determine significance. ***, p < 0.001 for Dex versus con. ###, p < 0.001 for MDFIC siRNA Dex versus NTC siRNA Dex. D, RT-PCR analysis of MDFIC mRNA expression in A549 cells transfected as above with reporter gene and either NTC siRNA or MDFIC siRNA (Student's t test, mean ± S.E. ***, p < 0.001).
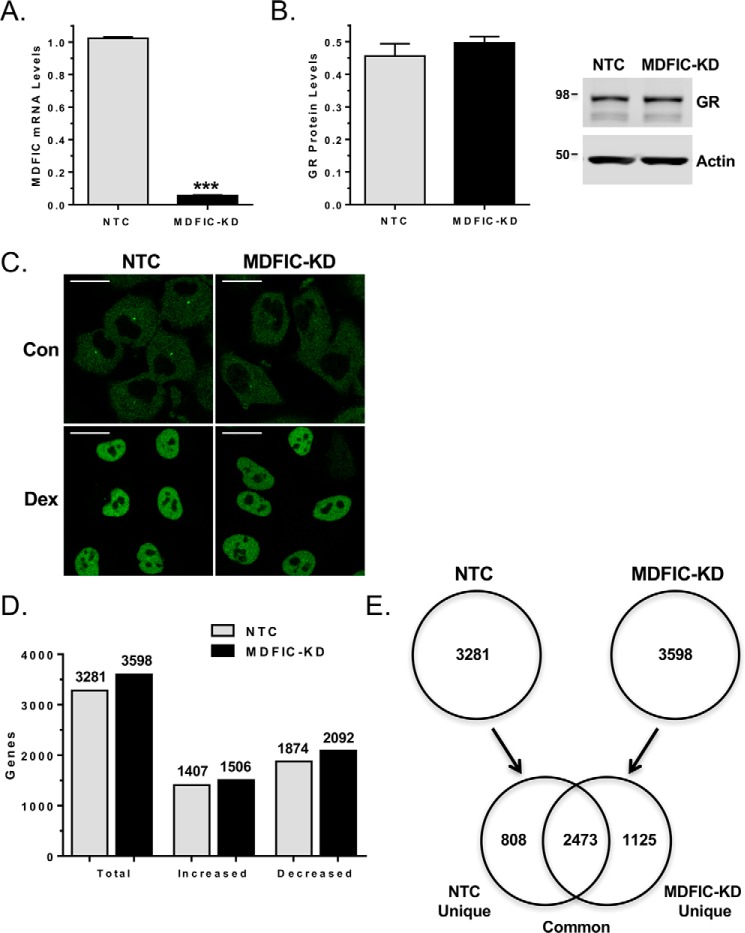
To evaluate whether the interaction of MDFIC with GR alters its regulation of endogenous genes, we performed a genome-wide microarray in A549 cells that were transfected with non-targeting control (NTC) siRNA or MDFIC siRNA (MDFIC-KD). Knockdown of MDFIC was efficient and had no effect on the expression of GR (Fig. 6, A and B). In addition, the subcellular distribution of GR was not altered by depletion of MDFIC (Fig. 6C). Treatment of cells for 6 h with 100 nm Dex resulted in the regulation of 3281 and 3598 genes in the NTC and MDFIC-KD cells, respectively (Fig. 6D). The percentage of genes induced (42.9% in NTC cells; 41.9% in MDFIC-KD cells) and repressed (57.1% in NTC cells; 58.1% in MDFIC-KD cells) by Dex was unaffected by depletion of MDFIC (Fig. 6D). A comparison of the glucocorticoid-regulated genes in the NTC and MDFIC-KD cells revealed three major groups: a group of 2473 genes (common) that were regulated by Dex independent of MDFIC, a group of 808 genes (NTC unique) that were regulated by Dex only in the presence of MDFIC, and a group of 1125 genes (MDFIC-KD unique) that were regulated by Dex only in the absence of MDFIC (Fig. 6E).
Figure 6.
MDFIC alters the GR transcriptome in A549 cells. A549 cells were transfected with non-targeting control siRNA (NTC) or MDFIC siRNA (MDFIC-KD). A, RT-PCR analysis of MDFIC mRNA expression in NTC and MDFIC-KD cells (Student's t test, mean ± S.E. ***, p < 0.001). B, GR protein levels were evaluated by immunoblot analysis in NTC and MDFIC-KD cells. Quantitation of GR normalized to actin (mean ± S.E. from three independent experiments) is presented in the left panel, and a representative immunoblot is presented in the right panel. C, NTC and MDFIC-KD cells were treated with vehicle (Con) or 100 nm Dex for 1 h and processed for immunocytochemistry. Representative confocal microscopic images depict the distribution of GR. The scale bar is 20 μm. D, microarray analysis was performed on RNA isolated from NTC and MDFIC-KD cells treated with vehicle or 100 nm Dex for 6 h. Shown are the total number of genes regulated by Dex in NTC cells and MDFIC-KD cells and the number of genes with increased or decreased expression. Differentially expressed genes were determined using an error-weighted ANOVA and Benjamini-Hochberg False Discovery Rate multiple test correction with a p value of p < 0.01. E, genes regulated by Dex in NTC cells were compared with genes regulated by Dex in the MDFIC-KD cells. The Venn diagram depicts Dex-regulated genes that are unique to the NTC cells, common to both the NTC and MDFIC-KD cells, and unique to the MDFIC-KD cells.
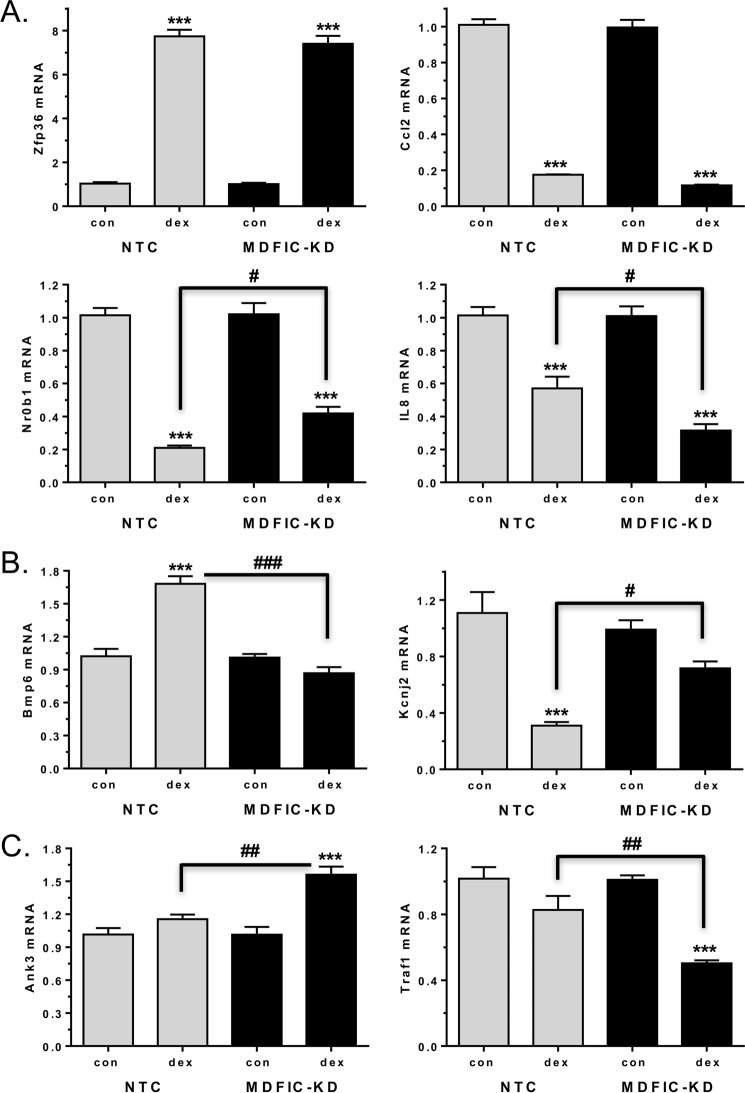
For validation of the microarray, we performed RT-PCR on an independent set of NTC and MDFIC-KD samples and evaluated the expression of genes belonging to each of the three groups. Members of the common group include zinc finger protein 36 (Zfp36) and C-C motif chemokine ligand 2 (Ccl2), and these two genes were induced and repressed, respectively, to a similar extent by Dex in the NTC and MDFIC-KD cells (Fig. 7A, upper panels). Other members of the common group exhibited differences in the magnitude of the observed glucocorticoid regulation (Fig. 7A, lower panels). Dex treatment resulted in a 79.3% repression of the DAX-1 nuclear receptor (Nr0b1) in NTC cells but a smaller, 58.9%, repression in the MDFIC-KD cells. On the other hand, administration of Dex resulted in a 43.7% repression of interleukin-8 (IL-8; Cxcl8 gene) in the NTC cells but a greater, 68.8%, repression in the MDFIC-KD cells. Members of the NTC unique group include the bone morphogenetic protein 6 (Bmp6) and potassium channel, inwardly rectifying subfamily J, member 2 (Kcnj2) genes (Fig. 7B). In the presence of MDFIC, glucocorticoid treatment resulted in the induction of Bmp6 and the repression of Kcnj2. However, in cells lacking MDFIC, the glucocorticoid-dependent regulation of these two genes was largely abolished. Members of the MDFIC-KD unique group include the ankyrin-3 (Ank3) and tumor necrosis factor (TNF) receptor-associated factor 1 (Traf1) genes (Fig. 7C). These two genes were not regulated by glucocorticoids in cells expressing MDFIC. However, in the MDFIC-KD cells, GR gained the ability to induce Ank3 and to repress Traf1 after glucocorticoid exposure. Collectively, these data demonstrate that the interaction of GR with MDFIC in the cytoplasm of cells alters the global gene regulatory profile of GR such that GR gains the ability to regulate some genes but loses the ability to regulate others.
Figure 7.
GR differentially regulates genes in A549 cells depending on the presence or absence of MDFIC. Total RNA was isolated from A549 cells transfected with NTC siRNA or MDFIC siRNA (MDFIC-KD) and treated with vehicle or 100 nm Dex for 6 h. A, genes commonly regulated by GR in the NTC and MDFIC-KD cells. The expression of zinc finger protein 36 (Zfp36), Ccl2, Nr0b1, and IL-8 was measured by RT-PCR. B, genes uniquely regulated by GR in the NTC cells. The expression of Bmp6 and KcnJ2 was measured by RT-PCR. C, genes uniquely regulated by GR in the MDFIC-KD cells. The expression of Ank3 and Traf1 was measured by RT-PCR. Data are plotted as -fold change and represent the mean ± S.E. from 3–6 independent experiments. A one-way ANOVA followed by Tukey's post hoc test was performed to determine significance. ***, p < 0.001 for Dex versus vehicle (con). #, p < 0.05; ##, p < 0.01; ###, p < 0.001 for MDFIC-KD Dex versus NTC Dex.
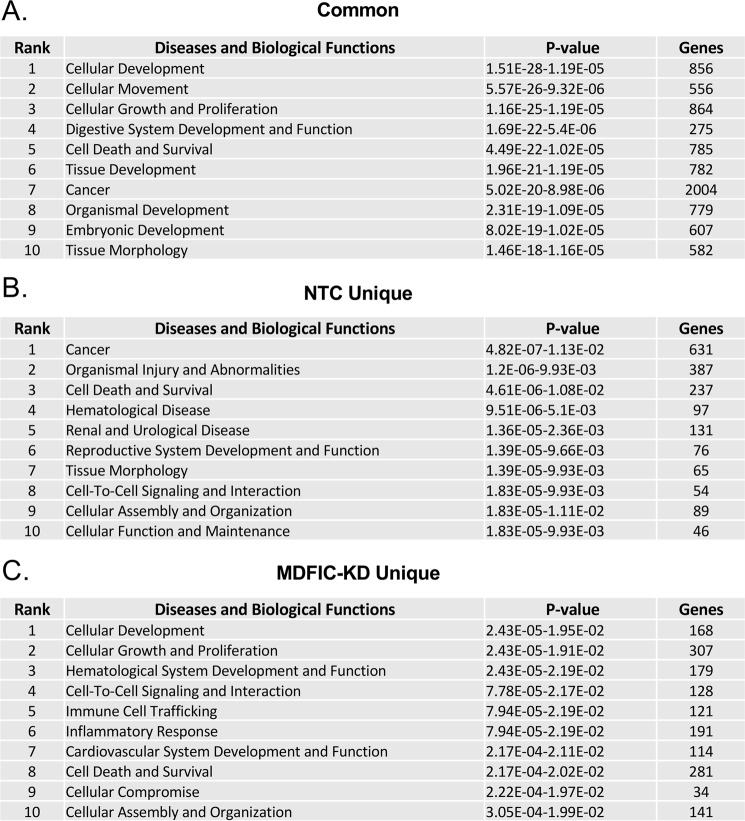
We analyzed the common, NTC unique, and MDFIC-KD unique gene sets using literature-based Ingenuity Pathway Analysis software to gain insight into the diseases and biological functions most significantly associated with the glucocorticoid regulated genes. Shown in Fig. 8 are the top 10 annotations for each gene group. Remarkably, only one annotation, Cell Death and Survival, was shared across all 3 sets of genes. Among the annotations displaying the greatest divergence between the NTC unique and MDFIC-KD unique gene sets were Immune Cell Trafficking and Inflammatory Response. This is of particular interest given the widespread clinical use of glucocorticoids to suppress the immune system and inhibit inflammation (29). Immune Cell Trafficking and Inflammatory Response were strongly associated with the MDFIC-KD unique gene set (rank = 5 and 6, respectively) but very poorly associated with the NTC unique gene set (rank = 50 and 51, respectively). Loss of MDFIC not only resulted in different genes becoming regulated by glucocorticoids but also an expansion (∼2.5-fold) in the total number of regulated genes associated with these two annotations. For example, a total of 44 genes associated with Immune Cell Trafficking were regulated by Dex only in the presence of MDFIC, whereas 121 genes were regulated by Dex only in the absence of MDFIC (supplemental Fig. S1). Similarly, a total of 79 genes associated with Inflammatory Response were regulated by Dex only in the presence of MDFIC, whereas 191 genes were regulated by Dex only in the absence of MDFIC (supplemental Fig. S2).
Figure 8.
Distinct biological functions are associated with the MDFIC-dependent alteration in the GR transcriptome in A549 cells. The set of genes commonly regulated by glucocorticoids in the NTC and MDFIC-KD cells (2473 total genes), uniquely regulated by glucocorticoids in the NTC cells (808 total genes), and uniquely regulated by glucocorticoids in the MDFIC-KD cells (1125 total genes) were analyzed using literature-based Ingenuity Pathway Analysis software. Shown are the top 10 diseases and biological functions most significantly associated with the common (A), NTC unique (B), and MDFIC-KD unique (C) genes.
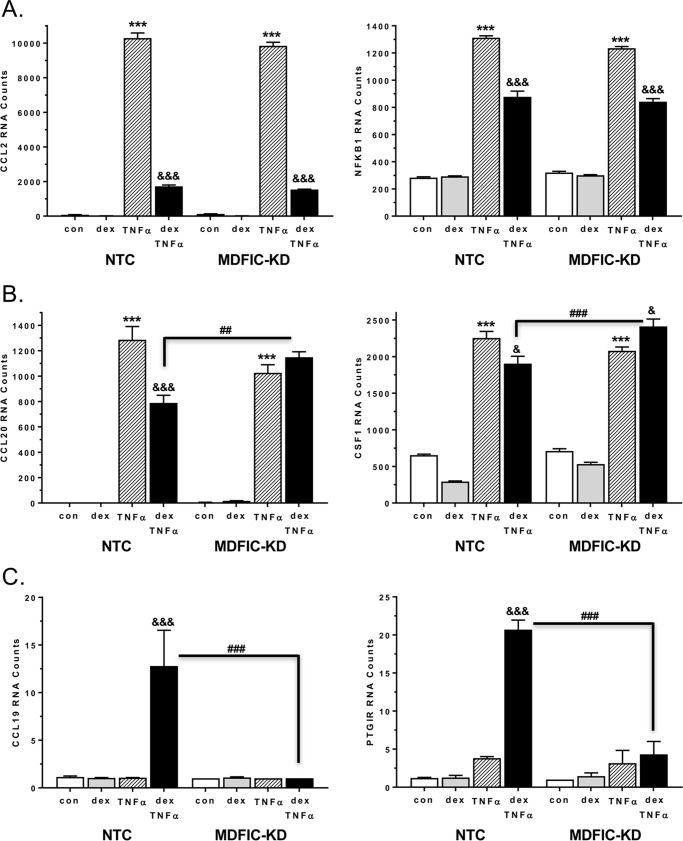
The gene enrichment predictions suggest that glucocorticoids differentially affect the inflammatory response depending on the association of GR with MDFIC. We tested this directly by evaluating glucocorticoid-mediated antagonism of TNFα-induced pro-inflammatory gene changes in A549 cells transfected with NTC siRNA or MDFIC siRNA. For this experiment we used nanostring technology, which provides direct measurement of mRNA expression without the synthesis of cDNA or amplification of transcripts. Cells were treated with vehicle, 100 nm Dex, 10 ng/ml TNFα, or both 100 nm Dex and 10 ng/ml TNFα for 6 h. Many of the inflammatory genes induced by TNFα, such as Ccl2 and nuclear factor κB subunit 1 (Nfkb1), were antagonized by co-administration of glucocorticoids, and the extent of this inhibition was unaffected by MDFIC (Fig. 9A). However, the ability of glucocorticoids to antagonize the TNFα-mediated up-regulation of other genes, including C-C motif chemokine ligand 20 (Ccl20) and colony stimulating factor 1 (Csf1), was abolished in cells depleted of MDFIC (Fig. 9B). In fact, co-treatment of MDFIC-KD cells with Dex and TNFα resulted in a significant up-regulation of Csf1 mRNA compared with TNFα treatment alone. Although glucocorticoids and TNFα are generally considered to act in an opposing manner, recent reports have identified genes that are co-regulated by these molecules (30). We observed a similar phenomenon as co-administration of Dex and TNFα robustly stimulated the expression of C-C motif chemokine 19 (Ccl19) and prostaglandin I2 receptor (Ptgir) mRNA in NTC cells (Fig. 9C). Strikingly, the up-regulation of these two genes was eliminated in cells lacking MDFIC. These data provide evidence that the interaction of MDFIC with GR may play an important functional role fine-tuning glucocorticoid regulation of the inflammatory response.
Figure 9.
MDFIC alters glucocorticoid regulation of the inflammatory response in A549 cells. Nanostring analysis was performed on RNA isolated from A549 cells transfected with NTC siRNA or MDFIC siRNA (MDFIC-KD) and treated for 6 h with vehicle, 100 nm Dex, 10 ng/ml TNFα, or both 100 nm Dex and 10 ng/ml TNFα. A, RNA counts for the Ccl2 and nuclear factor κB subunit 1 (Nfkb1) genes. B, RNA counts for the Ccl20 and Csf1 genes. C, RNA counts for the C-C motif chemokine 19 (Ccl19) and prostaglandin I2 receptor (Ptgir) genes. Data shown are the raw RNA counts normalized to six housekeeping genes as described under “Experimental Procedures” and represent the mean ± S.E. from three independent experiments. A one-way ANOVA followed by Tukey's post hoc test was performed to determine significance. ***, p < 0.001 for TNFα versus con (vehicle). &, p < 0.05; &&&, p < 0.001 for Dex+TNFα versus TNFα. ##, p < 0.01. ###, p < 0.001 for MDFIC-KD Dex+TNFα versus NTC Dex+TNFα.
MDFIC alters the phosphorylation status of GR
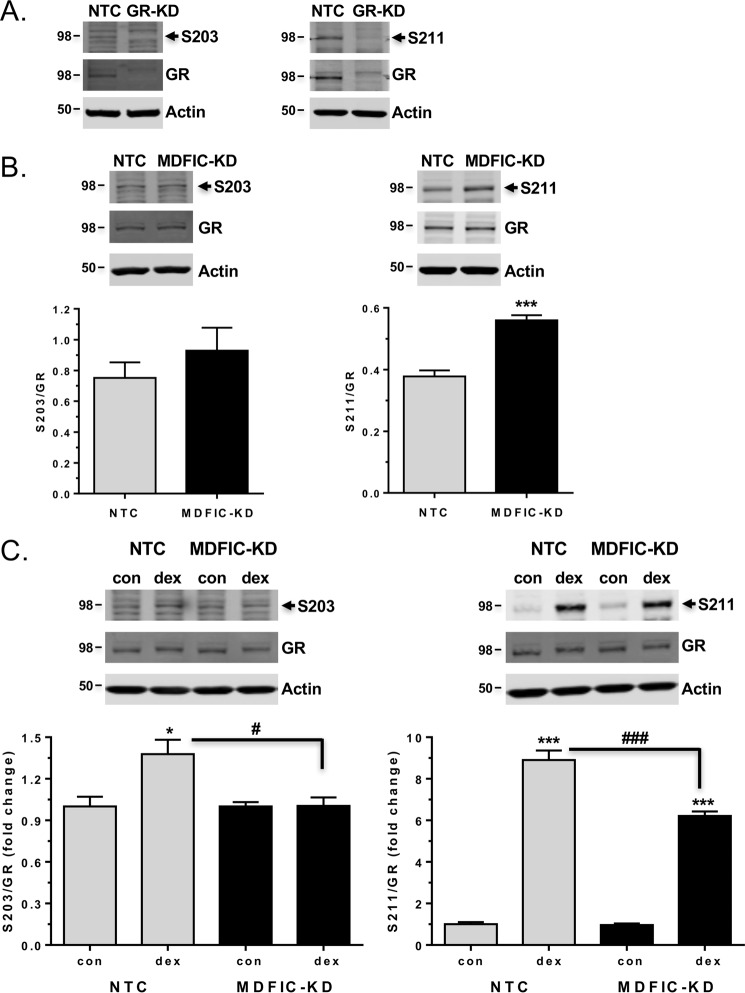
GR is phosphorylated on multiple serine residues (see the Fig. 1A schematic) in both an agonist-independent and agonist-dependent manner, and these phosphorylation events have been shown to influence the transcriptional activity of GR in a gene-specific fashion (31, 32). Among the well studied GR phosphorylation sites are serine 203 (Ser-203) and serine 211 (Ser-211). Ser-203 and Ser-211 exhibit a basal level of phosphorylation and become hyper-phosphorylated after glucocorticoid treatment (31, 32). We hypothesized that the interaction of MDFIC with GR in the cytoplasm of A549 cells might modulate the phosphorylation status of the receptor and thereby contribute to the observed diversity in GR signaling. To explore this possibility, we examined GR phosphorylation at Ser-203 and Ser-211 in NTC and MDFIC-KD cells using phospho-specific antibodies. Specificity of the antibodies for phosphorylated GR was confirmed by siRNA-mediated knockdown of the receptor (Fig. 10A). In untreated cells, knockdown of MDFIC did not alter the basal level of phosphorylation measured for Ser-203 but did lead to a significant 1.5-fold increase in the basal phosphorylation of Ser-211 (Fig. 10B). Treatment of cells for 1 h with 100 nm Dex resulted in a 1.4-fold induction of Ser-203 phosphorylation and an 8.9-fold induction of Ser-211 phosphorylation (Fig. 10C). However, in cells depleted of MDFIC, glucocorticoid-dependent phosphorylation of Ser-203 was abolished, and the induction of Ser-211 phosphorylation was attenuated (Fig. 10C). These data suggest that the association of MDFIC with GR can modulate the phosphorylation status of the receptor both in the absence and presence of glucocorticoids and thereby alter the GR transcriptome and glucocorticoid response.
Figure 10.
MDFIC alters the basal and agonist-induced phosphorylation state of GR in A549 cells. A, specificity of anti-GR phospho-specific antibodies. A549 cells were transfected with NTC siRNA or GR siRNA (GR-KD). Representative immunoblot from two independent experiments of GR protein expression in NTC and GR-KD cells using the anti-GR(Ser-203) antibody (left panel) and the anti-GR(Ser-211) antibody (right panel). B, A549 cells were transfected with NTC siRNA or MDFIC siRNA (MDFIC-KD). Cells were harvested, and GR phosphorylation was evaluated by immunoblot using phospho-specific antibodies to Ser-203 (left panel) and Ser-211 (right panel). Shown are representative immunoblots and quantitation of phosphorylated GR to total GR. Data represent the mean ± S.E. from six independent experiments (Student's t test, mean ± S.E. ***, p < 0.001). C, A549 cells were transfected with NTC siRNA or MDFIC siRNA (MDFIC-KD). After a 1-h treatment with vehicle or 100 nm Dex, cells were harvested, and GR phosphorylation was evaluated by immunoblot using phospho-specific antibodies to Ser-203 (left panel) and Ser-211 (right panel). Shown are representative immunoblots and quantitation of phosphorylated GR to total GR. Data are plotted as -fold change and represent the mean ± S.E. from six independent experiments. A one-way ANOVA followed by Tukey's post hoc test was performed to determine significance. *, p < 0.05; ***, p < 0.001 for Dex versus Con (vehicle) #, p < 0.05. ###, p < 0.001 for MDFIC-KD Dex versus NTC Dex.
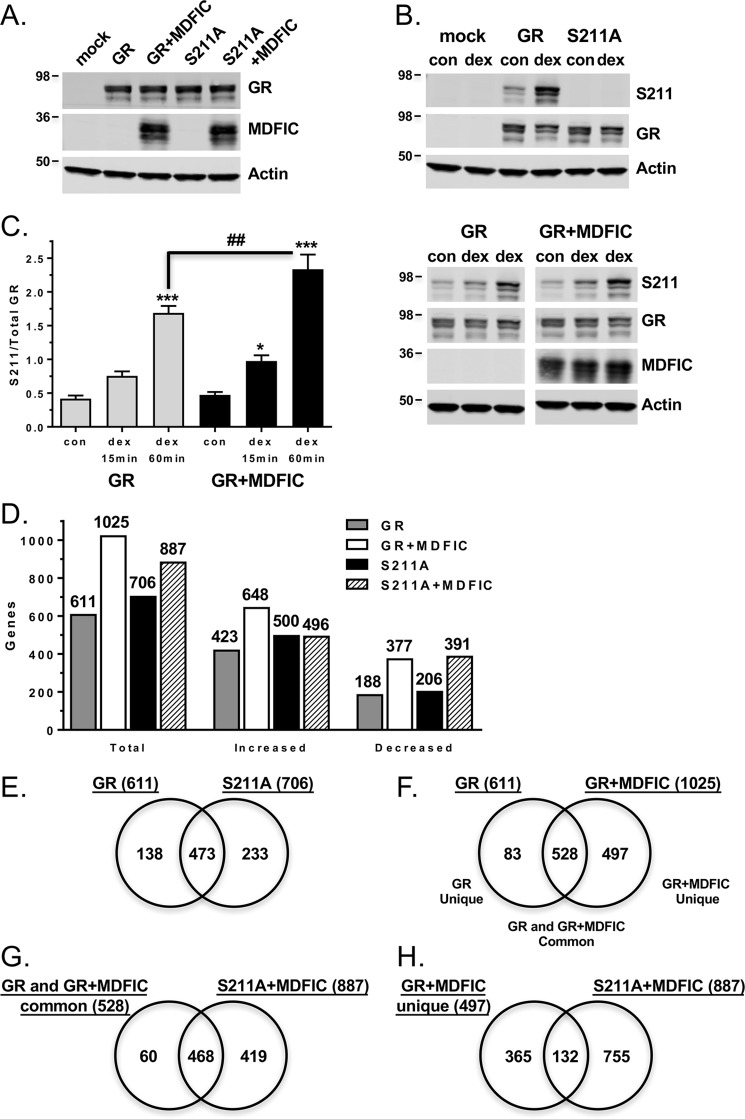
To test whether MDFIC-mediated changes in GR phosphorylation are responsible for the alterations in the receptor transcriptome, we compared the ability of MDFIC to modulate the gene regulatory profile of wild-type GR with that of a GR mutant that cannot be phosphorylated on Ser-211 (S211A). Microarrays were performed on COS-1 cells that were transfected with GR alone, GR and MDFIC, S211A alone, or S211A and MDFIC and then treated with vehicle or 100 nm Dex for 6 h. S211A was expressed at similar levels to GR both in the absence and presence of overexpressed MDFIC (Fig. 11A), and the mutant receptor was not phosphorylated under basal conditions or in response to glucocorticoids (Fig. 11B). In addition, whereas knockdown of MDFIC attenuated glucocorticoid-dependent Ser-211 phosphorylation in A549 cells (Fig. 10C), overexpression of MDFIC increased glucocorticoid-dependent Ser-211 phosphorylation in COS-1 cells (Fig. 11C). Results from the microarray revealed that 611 genes were regulated by GR and 706 genes were regulated by the S211A mutant after Dex treatment (Fig. 11D, supplemental Tables S1 and S2). The percentage of genes induced (69.2% in GR cells; 70.8% S211A cells) and repressed (30.8% in GR cells; 29.2% in S211A cells) by glucocorticoids was unaffected by the loss of Ser-211 phosphorylation. However, consistent with previous reports (33), loss of Ser-211 phosphorylation altered the gene regulatory profile of GR (Fig. 11E). A comparison of the Dex-regulated genes showed that 138 genes require Ser-211 phosphorylation for regulation and 233 genes require its absence. The majority of the GR transcriptome (473/611 genes or 77.4%) was regulated independently of Ser-211 phosphorylation.
Figure 11.
MDFIC modulation of the GR transcriptome is impaired in COS-1 cells expressing a phosphorylation-defective receptor. A, COS-1 cells were transfected with GR alone, GR and MDFIC, S211A alone, or S211A and MDFIC. GR, S211A, and MDFIC levels were evaluated by immunoblot analysis. Shown is a representative immunoblot from three independent experiments. B, COS-1 cells were transfected with GR alone or S211A alone. Basal and Dex-dependent phosphorylation of GR and the S211A receptor mutant were evaluated by immunoblot analysis using the anti-GR(Ser-211) antibody. Shown is representative immunoblot from three independent experiments. C, COS-1 cells were transfected with GR alone or GR and MDFIC. After treatment with vehicle or 100 nm Dex for the indicated times, the cells were harvested, and GR phosphorylation was evaluated by immunoblot using the anti-GR(Ser-211) antibody. Shown are representative immunoblots and quantitation of phosphorylated GR to total GR. Data represent the mean ± S.E. from four independent experiments. A one-way ANOVA followed by Tukey's post hoc test was performed to determine significance. *, p < 0.05; ***, p < 0.001 for Dex versus Con (vehicle). ##, p < 0.01 for GR+MDFIC Dex versus GR Dex. D, microarray analysis was performed on RNA isolated from COS-1 cells transfected with GR alone, GR and MDFIC, S211A alone, or S211A and MDFIC. Cells were treated with vehicle or 100 nm Dex for 6 h. Shown are the total number of genes regulated by Dex and the number of genes with increased or decreased expression. Differentially expressed genes were determined using an error-weighted ANOVA and Benjamini-Hochberg False Discovery Rate multiple test correction with a p value of p < 0.01. E, genes regulated by Dex in GR cells were compared with genes regulated by Dex in the S211A cells using a Venn diagram. F, genes regulated by Dex in GR cells were compared with genes regulated by Dex in the GR+MDFIC cells using a Venn diagram. G, genes regulated by Dex-activated GR independent of MDFIC (GR and GR+MDFIC common genes shown in panel F) were compared with genes regulated by Dex in S211A+MDFIC cells using a Venn diagram. H, genes regulated by Dex-activated GR only in the presence of MDFIC (GR+MDFIC unique genes shown in panel F) were compared with genes regulated by Dex in S211A+MDFIC cells using a Venn diagram.
When co-expressed with MDFIC, GR regulated a greater number of genes (1025) than observed in cells expressing GR alone (611), reflecting a 1.7-fold expansion in the GR transcriptome (Fig. 11D and supplemental Table S3). Interestingly, a smaller 1.25-fold increase in the number of regulated genes was observed for the S211A mutant co-expressed with MDFIC (706 genes for S211A cells; 887 genes for S211A+MDFIC cells) (Fig. 11D and supplemental Table S4), suggesting a deficiency in the ability of MDFIC to modulate the activity of the phosphorylation-defective receptor. A comparison of the Dex-regulated genes in the GR cells and GR+MDFIC cells showed that GR lost the ability to regulate 83 genes (GR unique) but gained the ability to regulate 497 new genes (GR+MDFIC unique) in the presence of MDFIC (Fig. 11F). The regulation of 528 genes (GR and GR+MDFIC common) occurred independently of MDFIC (Fig. 11F). We next compared the 528 genes regulated by Dex in both the GR and GR+MDFIC cells with the 887 genes regulated by Dex in S211A+MDFIC cells (Fig. 11G). Nearly 90% of the genes (468/528) regulated by GR independently of MDFIC were also regulated by the S211A mutant. Markedly different results were found, however, when we compared the 497 new genes uniquely regulated by GR only in the presence of MDFIC with the 887 genes regulated by Dex in the S211A+MDFIC cells (Fig. 11H). Only 26.6% of these genes (132/497) were regulated by the S211A mutant. These findings demonstrate a major loss in the ability of MDFIC to modulate the GR transcriptome when Ser-211 phosphorylation is prevented, suggesting that MDFIC-mediated changes in Ser-211 phosphorylation underlie many of its effects on GR signaling.
Glucocorticoids negatively regulate MDFIC gene expression
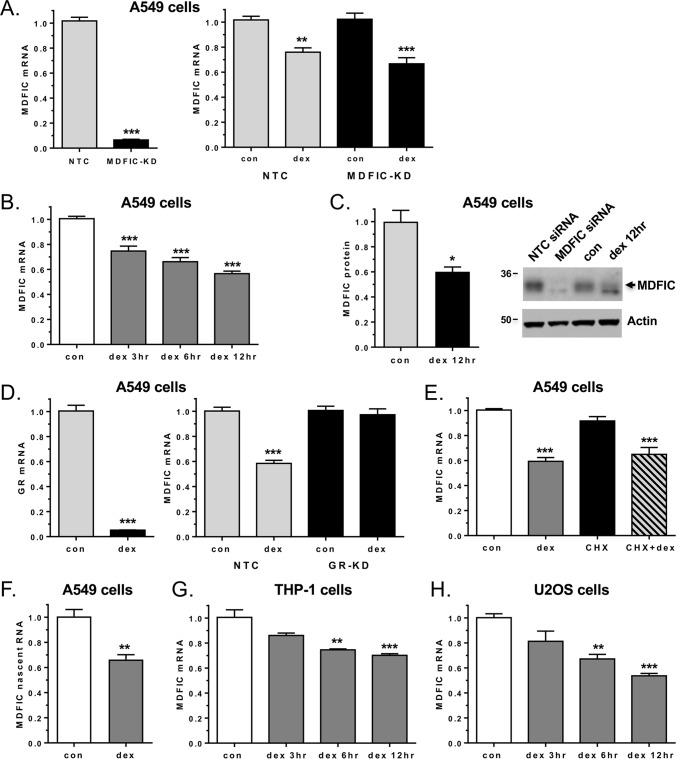
The interaction of MDFIC with GR has a profound influence on the receptor transcriptome. Therefore, factors that control MDFIC expression will shape the cellular response to glucocorticoids. A search of the genes identified in the A549 microarray to be regulated by glucocorticoids in both the presence and absence of MDFIC uncovered the MDFIC gene itself as a target for glucocorticoid-dependent repression. We confirmed the negative regulation of MDFIC by glucocorticoids in an independent set of NTC and MDFIC-KD samples (Fig. 12A). The magnitude of the repression was time-dependent: a 25.7%, 34.2%, and 43.7% decrease in MDFIC mRNA was observed after a 3-, 6-, and 12-h treatment of A549 cells with 100 nm Dex, respectively (Fig. 12B). The glucocorticoid-mediated decrease in MDFIC mRNA also led to a 39.9% reduction in MDFIC protein levels after a 12-h Dex treatment (Fig. 12C). To define the molecular mechanism underlying the repression, we first examined whether GR was required for the regulatory event. The Dex-dependent decrease in MDFIC gene expression was completely abolished in A549 cells depleted of GR by siRNA (Fig. 12D). We next investigated whether MDFIC was a primary or secondary target of glucocorticoid regulation by using the protein synthesis inhibitor cycloheximide. In the presence of cycloheximide, Dex administration still resulted in a significant repression of MDFIC, suggesting MDFIC is a direct target of GR regulation (Fig. 12E). Furthermore, we used nascent RNA primers targeting intronic sequences of MDFIC to examine whether GR altered the transcription of the MDFIC gene. After a 3-h exposure to glucocorticoids, a 34.1% reduction in MDFIC nascent RNA was observed (Fig. 12F). Finally, we examined whether the glucocorticoid-dependent regulation of MDFIC occurred in other human cell types. As shown in Fig. 12, G and H, Dex treatment resulted in a time-dependent reduction of MDFIC in both human THP-1 monocytes and human U2OS osteosarcoma cells. Collectively, these data indicate that glucocorticoids act through GR to directly repress the expression of MDFIC in multiple cell types.
Figure 12.
GR directly represses MDFIC gene expression in multiple human cell types. A, total RNA was isolated from A549 cells transfected with NTC siRNA or MDFIC siRNA (MDFIC-KD) and treated with vehicle or 100 nm Dex for 6 h. The left panel shows MDFIC knockdown as measured by RT-PCR. The right panel shows glucocorticoid-dependent regulation of MDFIC mRNA in NTC and MDFIC-KD cells as measured by RT-PCR and plotted as -fold change. B, A549 cells were treated with 100 nm Dex for the indicated times, and MDFIC mRNA was measured by RT-PCR. con, vehicle. C, A549 cells were treated with 100 nm Dex for 12 h, and MDFIC levels were evaluated by immunoblot. The left panel shows quantitation of MDFIC normalized to actin (mean ± S.E. from three independent experiments). The right panel shows a representative immunoblot. Protein lysates from A549 cells transfected with NTC siRNA or MDFIC siRNA were included as controls. D, total RNA was isolated from A549 cells transfected with NTC siRNA or GR siRNA (GR-KD) and treated with vehicle or 100 nm Dex for 6 h. The left panel shows GR knockdown as measured by RT-PCR. The right panel shows glucocorticoid-dependent regulation of MDFIC mRNA in NTC and GR-KD cells as measured by RT-PCR and plotted as -fold change. E, A549 cells pretreated for 1 h with vehicle or 10 μg/ml of cycloheximide (CHX) were exposed to 100 nm Dex for 6 h. MDFIC mRNA levels were analyzed by RT-PCR. F, A549 cells were treated with 100 nm Dex for 3 h, and the level of MDFIC nascent RNA was analyzed by RT-PCR. G, THP-1 monocytes were treated with 100 nm Dex for indicated times, and MDFIC mRNA was measured by RT-PCR. H, U2OS osteosarcoma cells stably expressing GR were treated with 100 nm Dex for the indicated times, and MDFIC mRNA was measured by RT-PCR. Data represent the mean ± S.E. from 3–7 independent experiments. Student's t test or a one-way ANOVA followed by Tukey's post hoc test was performed to determine significance. *, p < 0.05; **, p < 0.01; ***, p < 0.001 for Dex versus Con.
Discussion
Elucidating the mechanisms by which glucocorticoids generate cell type- and tissue-specific effects is an area of intense investigation because of the widespread clinical use of these steroids, the adverse effects resulting from sustained elevations in glucocorticoids, and the development of glucocorticoid resistance which limits the therapeutic benefit. Many factors appear to shape the cellular response to glucocorticoids, including the nature and concentration of the GR agonist, the expression level of GR, the repertoire of various splicing and translational GR isoforms, post-translational modifications of the receptor, the availability of specific co-regulators, and the chromatin landscape. In this report we have identified a novel protein-protein interaction between unliganded GR and MDFIC that occurs in the cytoplasm of cells. This interaction is dissociated in the presence of glucocorticoids but has a profound impact on the ensuing transcriptional and cellular responses elicited by activated GR. The association of MDFIC with GR alters both the pattern and magnitude of receptor phosphorylation, which contributes to the modulated GR transcriptome. Cross-talk between GR and MDFIC is bi-directional, as we show that glucocorticoids operate in a negative feedback loop to directly repress MDFIC gene expression. These findings identify MDFIC as a new binding partner for cytoplasmic GR that contributes to the heterogeneity and tissue specificity of glucocorticoid action.
MDFIC belongs to a small family of proteins that possess a unique cysteine rich carboxyl-terminal domain. The physiological function of MDFIC remains incompletely understood in large measure because mice with a disrupted MDFIC gene have not yet been described. In loss of function studies performed in Xenopus, MDFIC was found to be necessary for development (34). Embryos depleted of MDFIC were missing head structures, neural tube, notochord, and paraxial mesoderm, and these effects were attributed to the loss of MDFIC-dependent repression of the transcription factor T cell factor 3 (TCF-3). In mammalian cells MDFIC has been shown to interact with a variety of different proteins that directly or indirectly modulate transcription, including axin, cyclin T1, cyclin T2, lymphocyte enhancer factor 1 (LEF-1), and the viral transactivators human immunodeficiency virus type 1 (HIV-1) Tat and human T-cell leukemia virus type 1 (HTLV-1) Tax (24, 26, 27, 35). The association of MDFIC with these proteins is mediated by the cysteine-rich carboxyl-terminal domain and results in an altered transcriptional output. For this reason, MDFIC is generally described as a transcriptional regulator. Although these early studies laid a foundation for our understanding of MDFIC, they were limited by their heavy reliance on reporter genes and overexpression of MDFIC. Our current work, performed on endogenous MDFIC and on a genome-wide scale, reveals an expanded role for MDFIC as a gene expression regulator. We find that MDFIC is required for GR to regulate the expression of 808 unique genes in A549 cells. Unexpectedly, we also discovered that GR gains the ability to regulate 1125 unique genes in the absence of MDFIC. These findings suggest that the interaction of MDFIC with GR is required not only for the transcriptional activity of GR on one set of genes but also for the silencing of GR activity on a completely different set of genes.
Transcriptional regulators of GR typically associate with the receptor in the nucleus of cells after ligand activation. The GR-MDFIC interaction is unique because it occurs in the cytoplasm of cells and is dissociated after binding of glucocorticoids. MDFIC has been shown to inhibit the activity of other nuclear proteins by sequestering them in the cytoplasm, preventing their DNA binding, and/or promoting their degradation (24, 35, 36). In A549 cells depleted of MDFIC, we did not detect alterations in the expression level or cellular distribution of GR. However, we did observe changes in both the pattern and magnitude of GR phosphorylation. Knockdown of MDFIC increased the basal level of Ser-211 phosphorylation, and the glucocorticoid-dependent phosphorylation of Ser-203 and Ser-211 was impaired. In COS-1 cells, overexpression of MDFIC augmented GR phosphorylation at Ser-211 after glucocorticoid treatment. Microarray data from COS-1 cells expressing wild-type GR or the phosphorylation-defective mutant S211A revealed an important role for Ser-211 phosphorylation in the ability of MDFIC to modulate the GR transcriptome. Not all MDFIC-sensitive gene changes depended on Ser-211 phosphorylation, however, suggesting Ser-203 phosphorylation may also contribute to the regulatory activity of MDFIC on receptor signaling.
These findings suggest that the interaction of MDFIC with GR can influence receptor conformation, making it more or less favored as a substrate for kinase and/or phosphatase activity. The resultant perturbations in GR phosphorylation would then lead to distinct gene transcription programs and cellular responses to glucocorticoids. One of the primary functions of the site-specific phosphorylation is to regulate the transcriptional activity of GR. Via changes in cofactor recruitment and chromatin occupancy, alterations in phosphorylation have been shown to dramatically change the genomic gene regulatory profile of GR and result in the activation of distinct signaling pathways (33, 37–39). GR is also subject to a variety of other post-translational modifications, including sumoylation and acetylation (1). Sumoylation of GR has been shown recently to modulate the chromatin occupancy of GR and the ensuing transcriptional and cellular response to glucocorticoids (40). Whether the association of MDFIC with GR can influence other receptor post-translational modifications is currently unknown but is an important question for future studies.
The ability of MDFIC to alter the GR transcriptome suggests that the expression level of MDFIC will have a major impact on cellular responsiveness to glucocorticoids. Multiple studies have reported that MDFIC is expressed in a tissue- and cell type-specific manner. MDFIC is expressed in human spleen, thymus, prostate, uterus, and small intestine but is not detected in testis and colon (20). MDFIC is also found in primary human immune cells, but its expression varies considerably among immune cell subsets with NK cells (CD56+) and monocytes (CD14+) having high levels, CD4+ T-cells and CD8+ T-cells displaying intermediate levels, and B-cells (CD19+) expressing low levels (41). Many transformed cell lines also exhibit differential expression of MDFIC (27, 41). These cell type-specific expression patterns of MDFIC may contribute to the diverse actions of glucocorticoids that are observed in different tissues.
Factors that induce or repress MDFIC gene expression within a given cell type will also have an important influence on GR signaling. In the current study we made the novel discovery that glucocorticoids themselves directly repress MDFIC gene expression in a negative feedback loop. This repression requires GR, is direct, occurs at the level of transcription, and takes place in multiple cell types. By feeding back on MDFIC, glucocorticoids can limit the changes in GR signaling mediated by MDFIC. The only previously described regulator of MDFIC gene expression is the pro-inflammatory cytokine interleukin 2 (IL-2), which has been shown to induce the expression of MDFIC in a variety of hematopoietic cell lines and in primary human immune cells (41). That anti-inflammatory glucocorticoids and pro-inflammatory IL-2 regulate MDFIC gene expression in an opposing manner suggests that MDFIC might have an important function in the immune system. In support of this conclusion, two of the biological functions most strongly affected by depletion of MDFIC in A549 cells were Immune Cell Trafficking and Inflammatory Response. In addition, the ability of glucocorticoids to antagonize TNFα-mediated up-regulation of certain inflammatory genes was abolished in A549 cells lacking MDFIC. Therefore, the interaction of MDFIC with GR may play an important role modulating the anti-inflammatory actions of glucocorticoids.
In our study we also demonstrate that GR interacts with the related family member MDFI in a manner analogous to MDFIC. A549 cells express MDFI, but its expression is low compared with other cell types (27). Analysis of 12 different cell types revealed that cells with high levels of MDFIC have low levels of MDFI and, conversely, cells with low levels of MDFIC have high levels of MDFI (27). MDFI has been reported to play an important role in development. Via its conserved cysteine-rich carboxyl-terminal domain, MDFI binds and represses the MyoD family of transcription factors that are necessary for myogenesis (42). Additionally, disruption of MDFI in C57BL/6 mice results in embryonic lethality due to the loss of MDFI-dependent repression of the transcription factor MASH2, which leads to placental defects (43). MDFI knock-out mice generated in the 129Sv strain survive but exhibit abnormal skeletal development (43). MDFI interacts with many of the same proteins that associate with MDFIC and has both redundant and unique roles on their activity (26, 27, 35). The distinctive outcomes are not unexpected given that the two proteins show little homology outside the conserved cysteine-rich carboxyl-terminal domain. Glucocorticoids play a key role in the maturation of many organs including the lung, liver, heart, gastrointestinal tract, and adrenal gland (44–47). They have also been shown to regulate myoblast differentiation and proliferation and affect muscle development (48). It will be important for future studies to examine the influence MDFI has on the GR transcriptome and to investigate whether MDFI contributes to the tissue-specific patterns of gene regulation elicited by glucocorticoids during development. In addition, because MDFI is co-expressed with MDFIC in various cell types (27), it will be informative to evaluate GR signaling and stress hormone responses in cells devoid of both MDFI and MDFIC.
In summary, we provide evidence of cross-talk between GR and MDFIC. MDFIC can bind GR, alter its phosphorylation status, and redirect its global gene regulatory profile. GR, in turn, can directly repress MDFIC gene expression and limit its ability to modulate glucocorticoid responses. As the chief mediator of homeostasis under conditions of physiological and pathological stress, GR is subject to multiple inputs that can regulate its activity. Deciphering the molecular mechanisms underlying the diversity in GR signaling is critical for the development of new glucocorticoids and/or treatment regimens with improved risk/benefit ratios.
Experimental procedures
Reagents
Dexamethasone, RU486, and cortisol were purchased from Steraloids (Newport, RI). The rabbit anti-MDFIC antibody 2075 was produced using a peptide (CIHHGAKHGSADNRK) synthesized by AnaSpec (San Jose, CA), and the antisera was produced by Covance (Denver, PA).
Yeast two-hybrid assay
ProNet technologies automated yeast two-hybrid screening was performed by Myriad Genetics as previously described (49). A DNA sequence encoding amino acids 466–546 of human GR was used as bait. Human brain, spleen, pooled breast cancer/prostate cancer, and pooled liver/small intestine/adipose cDNA libraries were used to screen for prey proteins. Isolated bait and prey plasmids were co-transformed into yeast for confirmation of interactions by liquid β-galactosidase assays. DNA sequencing was used to determine the identity of the prey.
Cell culture
A549, COS-1, and U2OS cells (ATCC) were maintained in Dulbecco's modified Eagle's medium F-12 (DMEM) supplemented with 10% heat-inactivated FBS, 50 units/ml penicillin, 50 μg/ml streptomycin, and 2 mm l-glutamine. U2OS cells stably expressing the human GR have been previously described (50). THP-1 monocytes were maintained in RPMI medium supplemented with 10% heat-inactivated FBS, 50 μm β-mercaptoethanol, 25 mm HEPES (pH 7.0), and 100 units/ml penicillin/streptomycin. Before glucocorticoid treatment, cells were cultured overnight in medium supplemented with 10% charcoal-stripped FBS.
Plasmids
MDFIC was cloned from a human spleen cDNA library (Ambion) into the cloning vector pCR2.1-TOPO (Invitrogen) using the primers 5′-GCCACCATGTCCGGCGCGGGCGAAGC-3′ and 5′-TTATGAAGGAAAACAAATTCCACAGC-3′. The FLAG epitope was added to the amino terminus of MDFIC using the primers 5′-GTTAAGCTTGCCACCATGGACTACAAGGACGATGACGACAAGTCCGGCGCGGGCGAAGCCCTC-3′ and 5′-GTTTCTAGATTATGAAGGAAAACAAATTCCACAGC-3′ and subcloned into the expression vector pcDNA3.1zeo+ (Invitrogen) using HindIII/XbaI. MDFIC(1–164), a truncated version of MDFIC without the cysteine-rich carboxyl-terminal domain, was generated in pcDNA3.1zeo+ using the primers 5′-GTTAAGCTTGCCACCATGGACTACAAGGACGATGACGACAAGTCCGGCGCGGGCGAAGCCCTC-3′ and 5′-GTTTCTAGAAGATCTTTATTCAGGTGAAGAGCCTGTCTTTTG-3′. MDFI was cloned from a human skeletal muscle cDNA library (Ambion) into the cloning vector pCR2.1-TOPO using the primers 5′-GTTCAGAAGCTTGCCACCATGTACCAGGTGAGCGGCCAGCGC-3′ and 5′-GTTCACTCTAGACTAGGAGGAGAAGCAGAGCCCACAGC-3′. The FLAG epitope was added to the amino terminus of MDFI using the primers 5′-GTTCAGAAGCTTGCCACCATGGACTACAAGGACGATGACGACAAGTACCAGGTGAGCGGCCAGCGC-3′ and 5′-GTTCACTCTAGACTAGGAGGAGAAGCAGAGCCCACAGC-3′ and subcloned into the expression vector pcDNA3.1zeo+ (Invitrogen) using HindIII/XbaI. The GR phosphorylation-defective mutant S211A was generated by site-directed mutagenesis (AGT to GCT) using the QuikChange kit (Stratagene). The sequence of all constructs was confirmed by DNA sequencing.
RNA isolation and quantitative RT-PCR analysis
Total RNA was harvested from A549 cells using the RNeasy Mini kit and RNase-Free DNase kit (Qiagen). Individual mRNA abundance was determined using a TaqMan one-step RT-PCR procedure on the 7900HT sequence detection system (Applied Biosystems), and all primer/probe sets were from Applied Biosystems. For analysis of MDFIC nascent RNA, primer sequences were designed to amplify a region spanning an exon-intron boundary and thereby detect only unprocessed, newly expressed transcripts. The primer sequences were as follows: forward primer, 5′-ACAGCCCAGGGTGAGTG-3′ (exon 2/intron2); probe, 5′-TGCAAGTGGCAAGCTTCTTTGTCA-3′; reverse primer, 5′-TCTGTGCACTTGTGAGCAAAC-3′ (intron 2). Relative expression values for each gene were calculated using the ΔΔCt analysis method and the house-keeping gene peptidylprolyl isomerase B, which was unaffected by glucocorticoid treatment.
Co-immunoprecipitation assay
COS-1 cells were transfected with GR alone, FLAG-MDFIC alone, FLAG-MDFIC(1–164) alone, FLAG-MDFI alone, both GR and FLAG-MDFIC, both GR and FLAG-MDFIC(1–164), or both GR and FLAG MDFI. Forty-eight hours after transfection the cells were treated with vehicle or Dex for 1 h. Cells were harvested in cold PBS and resuspended in TENT lysis buffer (20 mm Tris-HCl (pH 7.5), 2 mm EDTA, 150 mm NaCl, 0.5% Triton X-100) containing protease inhibitors. After incubating with rotation for 1 h at 4 °C, the samples were centrifuged, and supernatant was removed for protein quantification. Equivalent amounts of protein were immunoprecipitated overnight with anti-FLAG antibody (Sigma, #F3165), anti-GR antibody 41 (BD Biosciences, #611227), or anti-GR antibody 57 (51). The following day protein A/G-agarose beads were added for a 90-min incubation at 4 °C. The beads were washed with cold TENT lysis buffer, resuspended in sample buffer containing 5% β-mercaptoethanol, and heated for 6 min at 95 °C. For co-immunoprecipitation of endogenous GR-MDFIC complexes, A549 cells were treated with vehicle or Dex for 1 h. Cells were washed with cold PBS, harvested in TENT lysis buffer containing protease inhibitors, Dounce-homogenized on ice, and processed as above. Immunoprecipitations were performed overnight with the anti-MDFIC 2075 antibody. Recovered proteins were resolved on Tris-glycine gels, transferred to nitrocellulose, and immunoblotted as described below.
Immunoblot analysis
Cells were washed once with cold PBS, lysed in SDS sample buffer (Invitrogen) supplemented with β-mercaptoethanol, sonicated on ice, and boiled for 6 min. Total protein was determined using the Pierce 660-nm protein assay with an ionic detergent compatibility reagent (Thermo Scientific). Cell fractionation experiments were performed using the Nuclear/Cytosol Fractionation kit (Biovision). For analysis of glucocorticoid regulation of MDFIC expression, A549 cells were harvested using radioimmune precipitation assay buffer. Equivalent amounts of protein were separated on 4–20% Tris-glycine gels. Proteins were then transferred to nitrocellulose membranes and probed overnight with the following antibodies: rabbit anti-GR antibody 57 or mouse anti-GR antibody 59 (51), anti-GR antibody D8H2 (Cell Signaling, #3660S), anti-FLAG antibody (Sigma, #F3165), anti-FLAG antibody (Sigma, #F7425), anti-MDFIC antibody 2075, anti-GR(Ser-203) antibody (Abcam,#ab195703), anti-GR(Ser-211) antibody (Cell Signaling, #4161S), and/or anti-actin antibody (Millipore, #MAB1501). After washing, blots were incubated with goat anti-rabbit Alexa Fluor 680-conjugated secondary antibody (Life Technologies, #A21109) and/or goat anti-mouse IRDye800-conjugated secondary antibody (LI-COR Biosciences, #926–32210) and developed in the linear dynamic range using the LI-COR Odyssey imaging system. For some experiments, blots were incubated with HRP-linked secondary antibodies and developed using enhanced chemiluminescence (GE Healthcare).
Immunocytochemistry
COS-1 cells were transfected with GR and FLAG-MDFIC, and A549 cells were transfected with NTC siRNA or MDFIC siRNA. The cells were plated in 35-mm glass-bottom dishes (MatTek, Ashland, MA). The next day, cells were treated with vehicle or 100 nm Dex for 1 h. Cells were then washed with cold PBS, fixed for 30 min at room temperature with 4% paraformaldehyde, and processed as previously described (52). The anti-GR antibody 57 (51), anti-GR antibody D8H2 (Cell Signaling, #3660S), and/or the anti-FLAG antibody (Sigma, #F3165) were incubated with cells overnight at 4 °C. The next day, goat anti-mouse Alexa Fluor 594 and/or goat anti-rabbit Alexa Fluor 488 secondary antibodies were incubated with the cells for 1 h at room temperature. A Zeiss laser-scanning confocal microscope (LSM 510 or LSM710; Carl Zeiss, Thornwood, NY) with a Plan-Apochromat 63×/1.4 oil objective was used to analyze the cells. Images were collected sequentially using dual excitation (488 nm from argon laser, 543 nm from HeNe laser) and emission filter sets (band pass, 500–530; long pass, 560 nm). Colocalization analysis in COS-1 cells was performed with ImageJ (National Institutes of Health) using the JACoP Plugin (Institut Curie, France) (53). The Manders colocalization coefficient was computed from each image using a pixel intensity threshold of 75 for both channels.
Luciferase assays
A549 cells were transfected in six-well plates using Transit-LT1 (Mirrus) with a firefly luciferase reporter (pMMTV-LUC or pGRE2-LUC), Renilla luciferase reporter pGL3-hRL (50), and either empty vector, MDFIC, or MDFIC(1–164). For knockdown experiments, A549 cells were transfected with the reporters above and either NTC siRNA or MDFIC siRNA using Dharmafect Duo (Thermo Scientific). The glucocorticoid-responsive firefly luciferase reporters, pMMTV-LUC and pGRE2-LUC, have been described previously (52). Cells were harvested the day after transfection and re-plated at equal densities in a 48-well plate. Vehicle or 100 nm Dex was added to the cells for an overnight (∼18 h) incubation. For cells transfected with siRNA, vehicle or 100 nm Dex was added for a 6-h incubation. Cells were then lysed in passive lysis buffer, and luciferase activity was measured using the Dual Luciferase Reporter Assay (Promega). Firefly luciferase values divided by Renilla luciferase values are reported as luciferase activity.
RNA interference assays
NTC siRNA, MDFIC SMARTpool siRNA, and GR SMARTpool siRNA were purchased from Thermo Scientific (Lafayette, CO). A549 cells were transfected with 60 nm concentrations of each siRNA using Dharmafect1 transfection reagent (Thermo Scientific) according to the manufacturer's instructions. Cells were harvested the day after transfection and re-plated at appropriate tissue culture densities. All experiments were performed 72 h post transfection.
Microarray analysis
A549 cells were transfected with NTC siRNA or MDFIC siRNA. Seventy-two hours post-transfection, cells were stimulated with either vehicle or 100 nm Dex for 6 h. Total RNA from three biological replicates of NTC and MDFIC siRNA-treated cells was harvested as described above. COS-1 cells were transfected with GR alone, GR and MDFIC, S211A alone, or S211A and MDFIC. Forty-eight hours post-transfection, cells were stimulated with either vehicle or 100 nm Dex for 6 h. Total RNA from three biological replicates of the transfected COS-1 cells was harvested as described above. Gene expression profiles were analyzed using Agilent Human Whole Genome 4-by-44 multiplex format oligonucleotide arrays (catalogue number 014850, Agilent technologies, Santa Clara, CA) according to the Agilent one-color microarray-based gene expression analysis protocol. To identify differentially expressed probes, an analysis of variance (ANOVA) was used to determine if there was a statistical difference between the means of the different groups. In addition, an error-weighted ANOVA and Benjamini-Hochberg False Discovery Rate multiple test correction, with a p value of p < 0.01, was performed using Rosetta Resolver (A549 data) or OmicSoft Array Studio software (COS-1 data) to reduce the numbers of false positives. Finally, the statistically significant probes were analyzed by Ingenuity Pathway Analysis software (Ingenuity Systems) for functional analysis. Gene enrichment p values (p < 0.05) for biological functions were determined using Fischer's exact test. The microarray data presented in this publication were deposited in the NCBI Gene Expression Omnibus (www.ncbi.nlm.nih.gov) (54) and are accessible through Gene Expression Omnibus series accession numbers GSE86115, GSE93899, and GSE93900.
Nanostring analysis
A549 cells were transfected with NTC siRNA or MDFIC siRNA. Seventy-two hours post-transfection, cells were stimulated with either vehicle, 100 nm Dex, 10 ng/ml TNFα, or 100 nm Dex and 10 ng/ml TNFα for 6 h. Total RNA from three biological replicates of NTC and MDFIC knockdown cells was harvested as described above. Gene expression was examined utilizing the human inflammation code set (Nanostring Technologies) that measures 249 endogenous genes and 6 housekeeping genes. RNA expression was quantified on the nCounter Digital Analyzer according to the manufacturer's protocol (Nanostring Technologies). Raw and normalized counts were generated with nSolver (v3.0)TM software. Data were normalized utilizing the manufacturer's positive and negative experimental control probes as well as all six housekeeping genes. To identify significant differences in RNA expression, an ANOVA was performed with post-hoc Benjamini-Hochberg FDR (false discovery rate)-corrected p values (p < 0.01).
Statistical analysis
Student's t test or one-way ANOVA with Tukey's post hoc analysis were used to determine whether differences between groups were statistically significant. The statistical analyses were performed using GraphPad Prism software.
Author contributions
R. H. O., J. M. B., and J. A. C. designed and analyzed the experiments. R. H. O. and J. M. B. performed the experiments. R. H. O., J. M. B., and J. A. C. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Christine M. Jewell (Signal Transduction Laboratory, NIEHS, National Institutes of Health) for supplying the GR phosphorylation mutant S211A expression vector. We thank Jeff Tucker of the Fluorescence Microscopy and Imaging Center (NIEHS) for assistance with confocal microscopy and quantitation of the GR and MDFIC colocalization. We thank Dr. Kevin Gerrish and Rick D. Fannin of the Molecular Genomics core laboratory (NIEHS) for assistance with microarray and nanostring analyses.
This work was supported by the Intramural Research Program of the NIEHS, National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The microarray data presented in this publication have been deposited in the NCBI Gene Expression Omnibus (www.ncbi.nlm.nih.gov) and are accessible through Gene Expression Omnibus series accession numbers GSE86115, GSE93899, and GSE93900.
This article contains supplemental Figs. 1 and 2 and Tables 1–4.
- GR
- glucocorticoid receptor
- MDFIC
- MyoD family inhibitor domain-containing protein
- MDFI
- MyoD family inhibitor isoform 1
- NTD
- amino-terminal transactivation domain
- DBD
- DNA binding domain
- LBD
- carboxyl-terminal ligand binding domain
- GRE
- glucocorticoid-responsive element
- Dex
- dexamethasone
- MMTV
- mouse mammary tumor virus
- NTC
- non-targeting control
- KD
- knockdown
- Ccl2
- C-C motif chemokine ligand 2
- Bmp6
- bone morphogenetic protein 6
- Kcnj2
- potassium channel, inwardly rectifying subfamily J, member 2
- Ank3
- ankyrin-3
- Traf1
- TNF receptor-associated factor 1
- Csf1
- colony stimulating factor 1
- ANOVA
- analysis of variance.
References
- 1. Oakley R. H., and Cidlowski J. A. (2013) The biology of the glucocorticoid receptor: new signaling mechanisms in health and disease. J. Allergy Clin. Immunol. 132, 1033–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sapolsky R. M., Romero L. M., and Munck A. U. (2000) How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr. Rev. 21, 55–89 [DOI] [PubMed] [Google Scholar]
- 3. Liggins G. C. (1994) The role of cortisol in preparing the fetus for birth. Reprod. Fertil. Dev. 6, 141–150 [DOI] [PubMed] [Google Scholar]
- 4. Miracle X., Di Renzo G. C., Stark A., Fanaroff A., Carbonell-Estrany X., Saling E., and Coordinators Of World Associatin of Perinatal Medicine Prematurity Working Group (2008) Guideline for the use of antenatal corticosteroids for fetal maturation. J. Perinat. Med. 36, 191–196 [DOI] [PubMed] [Google Scholar]
- 5. Evans R. M. (1988) The steroid and thyroid hormone receptor superfamily. Science 240, 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grad I., and Picard D. (2007) The glucocorticoid responses are shaped by molecular chaperones. Mol. Cell. Endocrinol. 275, 2–12 [DOI] [PubMed] [Google Scholar]
- 7. Pratt W. B., and Toft D. O. (1997) Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 18, 306–360 [DOI] [PubMed] [Google Scholar]
- 8. Freedman N. D., and Yamamoto K. R. (2004) Importin 7 and importin α/importin β are nuclear import receptors for the glucocorticoid receptor. Mol. Biol. Cell 15, 2276–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gorovits R., Ben-Dror I., Fox L. E., Westphal H. M., and Vardimon L. (1994) Developmental changes in the expression and compartmentalization of the glucocorticoid receptor in embryonic retina. Proc. Natl. Acad. Sci. U.S.A. 91, 4786–4790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hsu S. C., and DeFranco D. B. (1995) Selectivity of cell cycle regulation of glucocorticoid receptor function. J. Biol. Chem. 270, 3359–3364 [DOI] [PubMed] [Google Scholar]
- 11. Kino T., De Martino M. U., Charmandari E., Mirani M., and Chrousos G. P. (2003) Tissue glucocorticoid resistance/hypersensitivity syndromes. J. Steroid Biochem. Mol. Biol. 85, 457–467 [DOI] [PubMed] [Google Scholar]
- 12. Lamberts S. W., Huizenga A. T., de Lange P., de Jong F. H., and Koper J. W. (1996) Clinical aspects of glucocorticoid sensitivity. Steroids 61, 157–160 [DOI] [PubMed] [Google Scholar]
- 13. Lu N. Z., Collins J. B., Grissom S. F., and Cidlowski J. A. (2007) Selective regulation of bone cell apoptosis by translational isoforms of the glucocorticoid receptor. Mol. Cell. Biol. 27, 7143–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ren R., Oakley R. H., Cruz-Topete D., and Cidlowski J. A. (2012) Dual Role for Glucocorticoids in Cardiomyocyte Hypertrophy and Apoptosis. Endocrinology 153, 5346–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scoltock A. B., Heimlich G., and Cidlowski J. A. (2007) Glucocorticoids inhibit the apoptotic actions of UV-C but not Fas ligand in hepatoma cells: direct evidence for a critical role of Bcl-xL. Cell Death Differ. 14, 840–850 [DOI] [PubMed] [Google Scholar]
- 16. Barnes P. J. (2013) Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 131, 636–645 [DOI] [PubMed] [Google Scholar]
- 17. Yang N., Ray D. W., and Matthews L. C. (2012) Current concepts in glucocorticoid resistance. Steroids 77, 1041–1049 [DOI] [PubMed] [Google Scholar]
- 18. John S., Sabo P. J., Thurman R. E., Sung M. H., Biddie S. C., Johnson T. A., Hager G. L., and Stamatoyannopoulos J. A. (2011) Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43, 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uhlenhaut N. H., Barish G. D., Yu R. T., Downes M., Karunasiri M., Liddle C., Schwalie P., Hübner N., and Evans R. M. (2013) Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol Cell 49, 158–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thébault S., Gachon F., Lemasson I., Devaux C., and Mesnard J. M. (2000) Molecular cloning of a novel human I-mfa domain-containing protein that differently regulates human T-cell leukemia virus type I and HIV-1 expression. J. Biol. Chem. 275, 4848–4857 [DOI] [PubMed] [Google Scholar]
- 21. Thébault S., and Mesnard J. M. (2001) How the sequestration of a protein interferes with its mechanism of action: example of a new family of proteins characterized by a particular cysteine-rich carboxyl-terminal domain involved in gene expression regulation. Curr. Protein Pept. Sci. 2, 155–167 [DOI] [PubMed] [Google Scholar]
- 22. Picard D., and Yamamoto K. R. (1987) Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 6, 3333–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Savory J. G., Hsu B., Laquian I. R., Giffin W., Reich T., Haché R. J., and Lefebvre Y. A. (1999) Discrimination between NL1- and NL2-mediated nuclear localization of the glucocorticoid receptor. Mol. Cell. Biol. 19, 1025–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gautier V. W., Sheehy N., Duffy M., Hashimoto K., and Hall W. W. (2005) Direct interaction of the human I-mfa domain-containing protein, HIC, with HIV-1 Tat results in cytoplasmic sequestration and control of Tat activity. Proc. Natl. Acad. Sci. U.S.A. 102, 16362–16367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kusano S., and Eizuru Y. (2010) Human I-mfa domain proteins specifically interact with KSHV LANA and affect its regulation of Wnt signaling-dependent transcription. Biochem. Biophys. Res. Commun. 396, 608–613 [DOI] [PubMed] [Google Scholar]
- 26. Kusano S., and Raab-Traub N. (2002) I-mfa domain proteins interact with Axin and affect its regulation of the Wnt and c-Jun N-terminal kinase signaling pathways. Mol. Cell. Biol. 22, 6393–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Q., Young T. M., Mathews M. B., and Pe'ery T. (2007) Developmental regulators containing the I-mfa domain interact with T cyclins and Tat and modulate transcription. J. Mol. Biol. 367, 630–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Young T. M., Wang Q., Pe'ery T., and Mathews M. B. (2003) The human I-mfa domain-containing protein, HIC, interacts with cyclin T1 and modulates P-TEFb-dependent transcription. Mol. Cell. Biol. 23, 6373–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rhen T., and Cidlowski J. A. (2005) Antiinflammatory action of glucocorticoids: new mechanisms for old drugs. N. Engl. J. Med. 353, 1711–1723 [DOI] [PubMed] [Google Scholar]
- 30. Lannan E. A., Galliher-Beckley A. J., Scoltock A. B., and Cidlowski J. A. (2012) Proinflammatory actions of glucocorticoids: glucocorticoids and TNFα coregulate gene expression in vitro and in vivo. Endocrinology 153, 3701–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beck I. M., Vanden Berghe W., Vermeulen L., Yamamoto K. R., Haegeman G., and De Bosscher K. (2009) Crosstalk in inflammation: the interplay of glucocorticoid receptor-based mechanisms and kinases and phosphatases. Endocr. Rev. 30, 830–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Galliher-Beckley A. J., and Cidlowski J. A. (2009) Emerging roles of glucocorticoid receptor phosphorylation in modulating glucocorticoid hormone action in health and disease. IUBMB Life 61, 979–986 [DOI] [PubMed] [Google Scholar]
- 33. Peffer M. E., Chandran U. R., Luthra S., Volonte D., Galbiati F., Garabedian M. J., Monaghan A. P., and DeFranco D. B. (2014) Caveolin-1 regulates genomic action of the glucocorticoid receptor in neural stem cells. Mol. Cell. Biol. 34, 2611–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Snider L., and Tapscott S. J. (2005) XIC is required for Siamois activity and dorsoanterior development. Mol. Cell. Biol. 25, 5061–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kusano S., Yoshimitsu M., Hachiman M., and Ikeda M. (2015) I-mfa domain proteins specifically interact with HTLV-1 Tax and repress its transactivating functions. Virology 486, 219–227 [DOI] [PubMed] [Google Scholar]
- 36. Snider L., Thirlwell H., Miller J. R., Moon R. T., Groudine M., and Tapscott S. J. (2001) Inhibition of Tcf3 binding by I-mfa domain proteins. Mol. Cell. Biol. 21, 1866–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Galliher-Beckley A. J., Williams J. G., and Cidlowski J. A. (2011) Ligand-independent phosphorylation of the glucocorticoid receptor integrates cellular stress pathways with nuclear receptor signaling. Mol. Cell. Biol. 31, 4663–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Galliher-Beckley A. J., Williams J. G., Collins J. B., and Cidlowski J. A. (2008) Glycogen synthase kinase 3β-mediated serine phosphorylation of the human glucocorticoid receptor redirects gene expression profiles. Mol. Cell. Biol. 28, 7309–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lambert W. M., Xu C. F., Neubert T. A., Chao M. V., Garabedian M. J., and Jeanneteau F. D. (2013) Brain-derived neurotrophic factor signaling rewrites the glucocorticoid transcriptome via glucocorticoid receptor phosphorylation. Mol. Cell. Biol. 33, 3700–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paakinaho V., Kaikkonen S., Makkonen H., Benes V., and Palvimo J. J. (2014) SUMOylation regulates the chromatin occupancy and anti-proliferative gene programs of glucocorticoid receptor. Nucleic Acids Res. 42, 1575–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gu L., Dean J., Oliveira A. L., Sheehy N., Hall W. W., and Gautier V. W. (2009) Expression profile and differential regulation of the Human I-mfa domain-containing protein (HIC) gene in immune cells. Immunol. Lett. 123, 179–184 [DOI] [PubMed] [Google Scholar]
- 42. Chen C. M., Kraut N., Groudine M., and Weintraub H. (1996) I-mf, a novel myogenic repressor, interacts with members of the MyoD family. Cell 86, 731–741 [DOI] [PubMed] [Google Scholar]
- 43. Kraut N., Snider L., Chen C. M., Tapscott S. J., and Groudine M. (1998) Requirement of the mouse I-mfa gene for placental development and skeletal patterning. EMBO J. 17, 6276–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Abel M. H., Bass F. G., Krane E. J., Thomas A. L., and Liggins G. C. (1978) Pituitary stalk-section and some of its effects on endocrine function in the fetal lamb. Q. J. Exp. Physiol. 63, 211–219 [DOI] [PubMed] [Google Scholar]
- 45. Cole T. J., Blendy J. A., Monaghan A. P., Krieglstein K., Schmid W., Aguzzi A., Fantuzzi G., Hummler E., Unsicker K., and Schütz G. (1995) Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 9, 1608–1621 [DOI] [PubMed] [Google Scholar]
- 46. Liggins G. C. (1976) Adrenocortical-related maturational events in the fetus. Am. J. Obstet. Gynecol. 126, 931–941 [DOI] [PubMed] [Google Scholar]
- 47. Rog-Zielinska E. A., Thomson A., Kenyon C. J., Brownstein D. G., Moran C. M., Szumska D., Michailidou Z., Richardson J., Owen E., Watt A., Morrison H., Forrester L. M., Bhattacharya S., Holmes M. C., and Chapman K. E. (2013) Glucocorticoid receptor is required for foetal heart maturation. Hum. Mol. Genet. 22, 3269–3282 [DOI] [PubMed] [Google Scholar]
- 48. te Pas M. F., de Jong P. R., and Verburg F. J. (2000) Glucocorticoid inhibition of C2C12 proliferation rate and differentiation capacity in relation to mRNA levels of the MRF gene family. Mol. Biol. Rep. 27, 87–98 [DOI] [PubMed] [Google Scholar]
- 49. Garrus J. E., von Schwedler U. K., Pornillos O. W., Morham S. G., Zavitz K. H., Wang H. E., Wettstein D. A., Stray K. M., Côté M., Rich R. L., Myszka D. G., and Sundquist W. I. (2001) Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107, 55–65 [DOI] [PubMed] [Google Scholar]
- 50. Lu N. Z., and Cidlowski J. A. (2005) Translational regulatory mechanisms generate N-terminal glucocorticoid receptor isoforms with unique transcriptional target genes. Mol. Cell 18, 331–342 [DOI] [PubMed] [Google Scholar]
- 51. Cidlowski J. A., Bellingham D. L., Powell-Oliver F. E., Lubahn D. B., and Sar M. (1990) Novel antipeptide antibodies to the human glucocorticoid receptor: recognition of multiple receptor forms in vitro and distinct localization of cytoplasmic and nuclear receptors. Mol. Endocrinol. 4, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 52. Zhou J., Oakley R. H., and Cidlowski J. A. (2008) DAX-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X-chromosome, gene 1) selectively inhibits transactivation but not transrepression mediated by the glucocorticoid receptor in a LXXLL-dependent manner. Mol. Endocrinol. 22, 1521–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bolte S., and Cordelières F. P. (2006) A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 [DOI] [PubMed] [Google Scholar]
- 54. Edgar R., Domrachev M., and Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.