Abstract
There are more than 600 receptor-like kinases (RLKs) in Arabidopsis, but due to challenges associated with the characterization of membrane proteins, only a few have known biological functions. The plant RLK FERONIA is a peptide receptor and has been implicated in plant growth regulation, but little is known about its molecular mechanism of action. To investigate the properties of this enzyme, we used a cell-free wheat germ-based expression system in which mRNA encoding FERONIA was co-expressed with mRNA encoding the membrane scaffold protein variant MSP1D1. With the addition of the lipid cardiolipin, assembly of these proteins into nanodiscs was initiated. FERONIA protein kinase activity in nanodiscs was higher than that of soluble protein and comparable with other heterologously expressed protein kinases. Truncation experiments revealed that the cytoplasmic juxtamembrane domain is necessary for maximal FERONIA activity, whereas the transmembrane domain is inhibitory. An ATP analogue that reacts with lysine residues inhibited catalytic activity and labeled four lysines; mutagenesis demonstrated that two of these, Lys-565 and Lys-663, coordinate ATP in the active site. Mass spectrometric phosphoproteomic measurements further identified phosphorylation sites that were examined using phosphomimetic mutagenesis. The results of these experiments are consistent with a model in which kinase-mediated phosphorylation within the C-terminal region is inhibitory and regulates catalytic activity. These data represent a step further toward understanding the molecular basis for the protein kinase catalytic activity of FERONIA and show promise for future characterization of eukaryotic membrane proteins.
Keywords: enzyme mutation, lipid bilayer, membrane protein, mutagenesis in vitro, receptor protein serine/threonine kinase, FERONIA, cell-free translation, nanodiscs, plant protein kinase
Introduction
The Arabidopsis receptor-like kinase (RLK)2 family currently contains more than 600 members and makes up 2.5% of the protein-coding Arabidopsis genes (1). Despite their large prevalence in the genome, only a few have known biological functions, and even fewer have been biochemically characterized. The vast majority of RLKs are single transmembrane helix-containing proteins, with a relatively conserved cytosolic Ser/Thr kinase domain, a more diverse extracellular (ecto) ligand-binding domain, and a juxtamembrane domain separating the kinase domain and the transmembrane helix. The RLKs can be subdivided into families classified by the sequence of their respective ecto domains. The largest subfamily of RLKs contains over 200 members, whose ecto domains display varying numbers of leucine-rich repeat (LRR) sequences (2). The second largest subfamily contains domains with a low degree of sequence homology to lectins, classically known sugar binding motifs, in their extracellular region (3, 4). Few RLKs have experimentally verified function; those with a known function play roles, generally, in regulating plant development, growth, or pathogen defense (5). For example, BRI1, a well characterized LRR-containing RLK, is a brassinolide receptor, which, alongside the co-receptor BAK1, regulates, among other phenotypes, plant growth (6–9). Much work has been done with the goal of determining the function of the many remaining “orphan” RLKs (i.e. those that lack known ligands). Although some progress has been made with model genetic plants, such as Arabidopsis, as with other plant proteins encoded by large gene families, the redundant functionality of RLKs arising from gene duplications and homology has complicated their functional characterization using genetic techniques (1, 10).
The family of plant RLKS known as lectin RLKs (due to the presence of an extracellular lectin-binding domain) are generally associated with stress and hormone response and development (4). The lectin RLK FERONIA has been identified as being critical for pollen tube-ovule interaction (11–13) and was thus named for the Etruscan goddess of fertility. FERONIA also controls plant growth and is involved in hormonal signaling, mechanosensing, and pathogen response (14). Recently, FERONIA was reported to be a receptor for the plant peptide hormone rapid alkalinization factor (RALF) (15).
In this study, using a quantitative mass spectrometric screen, rapid RALF-dependent changes in phosphorylation were identified in planta, on Ser residues within the FERONIA C terminus. It was further demonstrated that feronia mutants are uniquely insensitive to RALF-dependent inhibition of root growth, cytoplasmic calcium increases, and gene induction as compared with wild-type Arabidopsis. By testing mutant plants containing knock-out mutations in genes encoding dozens of other RLK protein knockouts, it was revealed that resistance to the root growth-inhibiting effect of RALF is specific to this peptide and only found in plants containing lesions in the gene encoding FERONIA, and whereas radioactively labeled RALF bound to plasma membrane in a saturable fashion, the biologically inactive (Δ2–8)RALF did not. Additional in vitro binding studies showed that the FERONIA ecto domain, produced in Escherichia coli, is capable of specifically binding to RALF, although the affinity is lower than that observed in vivo with the full-length protein (15). Given the importance of this newly described RALF receptor in plant growth and the paucity of data describing the plant RLK family, we sought to study FERONIA in vitro using an established wheat germ embryo-based cell-free protein production system.
Wheat germ embryo extract has emerged as an efficient way to synthesize proteins in vitro with greater protein yield than that obtained using other cell-free systems (16). An improved system of wheat germ-based cell-free production was first described in the early 2000s (17), which is particularly robust and capable of remaining active for more than 48 h because translation inactivation factors are removed, the machinery is eukaryotic and thus more amenable to eukaryotic protein production, and translation can occur over a wide range of temperatures (18). Additionally, cell-free translation is an open production system and thus allows co-translational addition of co-factors, binding partners, and lipids, all of which can facilitate production of a properly folded and active protein. All of these factors make wheat germ-based cell-free expression a possible platform for providing a large-scale source of the 100-kDa single-pass membrane plant receptor-like protein kinase, FERONIA.
Cell-free translation has been used to produce soluble membrane proteins by co-expressing amphiphilic helical proteins called membrane scaffold proteins (MSPs) alongside transmembrane proteins in the presence of exogenously added lipids (19, 20). In the absence of MSPs, membrane proteins are typically insoluble and cannot be purified; this co-expression technique has been shown to result in the spontaneous assembly of nanodiscs, which can then be used to biochemically characterize a membrane protein of interest within a lipid bilayer. Nanodiscs are lipid bilayers constrained by amphipathic MSPs, the majority of which in use are modifications of the human apoA1 protein (21). When combined in vitro with lipids in an appropriate ratio of lipid to MSP, MSPs can spontaneously form discs containing lipid bilayers, nanodiscs, confined by two copies of MSP, into which transmembrane domains of lipophilic proteins can insert (22). For example, the highly studied human EGFR, chemically similar to FERONIA in that it contains an ecto domain, a single-pass transmembrane domain, and a cytoplasmically oriented kinase domain, has been reconstituted into nanodiscs in vitro with significantly stabilized kinase activity (23). Lipids supplied during production determine their lipid content, and the diameter of the disc is controlled by the choice of MSP. For example, the MSP used in the experiments described below, MSP1D1, produces nanodiscs of about 9.7 nm in diameter, whereas longer variants, containing extra helices and linker regions, produce nanodiscs up to 17 nm in diameter (21). Cell-free translation of MSP1D1 in the presence of the lipid cardiolipin, followed by further translation of full-length, untagged FERONIA mRNA added into the mixture, produced highly kinase-active FERONIA in nanodiscs, the first report of a plant RLK being so produced using a cell-free system. Herein we describe the in vitro biochemical, structure/function, and regulatory characterization of the plant RLK FERONIA produced using this cell-free system.
Results
Co-expression of FERONIA with membrane scaffold protein and lipid produces monomeric FERONIA with high protein kinase activity in nanodiscs
The first attempt to produce FERONIA was made using a previously described dialysis cup cell-free translation technique (24). Wheat germ-based cell-free expression produced soluble, quantifiable amounts of FERONIA, but we observed the calculated specific activity and stability over time to be variable and generally low.
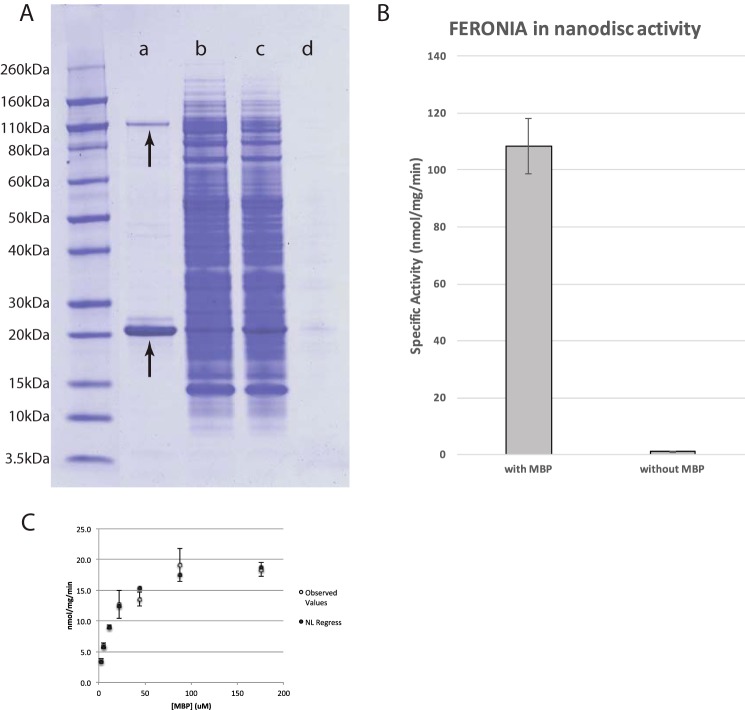
To test whether the low specific activity reflected a possible requirement for FERONIA to be embedded within a lipid bilayer, we explored the use of nanodisc technology in combination with cell-free expression (19, 25). FERONIA was co-expressed with the membrane scaffold protein MSP1D1 modified with a StrepII tag for StrepTactin purification and lipid content of 0.6 mm cardiolipin, optimized for MSP1D1 production and purification (supplemental Fig. S1). We found that non-affinity-tagged FERONIA co-expressed with StrepII-tagged MSP1D1 has significantly less background following StrepTactin purification than C-terminally His-tagged FERONIA subjected to immobilized nickel affinity purification (Fig. 1A). This observation is possibly a reflection of the lower salt StrepTactin purification conditions, which discourage nonspecific co-purification of hydrophobic wheat germ proteins and lipids. Furthermore, MSP1D1 co-expressed FERONIA exhibits significantly higher specific activity than soluble FERONIA produced without MSPs (Fig. 1B). Using FERONIA co-translated with MSPD1 and cardiolipins, respectively, Km and Vmax values of ∼12 μm and ∼19 nmol/min for MBP phosphorylation were observed (Fig. 1C).
Figure 1.
A, co-translation and purification of untagged FERONIA with nanodiscs. a, elution from streptactin-Sepharose purification; b, flow-through from streptactin-Sepharose purification of reaction; c, soluble fraction from cell-free reaction; d, insoluble fraction from cell-free reaction. Top arrow, FERONIA band; bottom arrow, MSP band. B, FERONIA co-translated with nanodiscs exhibits high specific activity with MBP as a substrate; error bars, S.D., n = 3/bar, p = 0.00004. C, kinetic parameter determination for MBP. Curve fitting reveals the Km and Vmax for MBP to be 13.7 μm and 20.2 nmol/min/mg, respectively. Error bars, S.D.; n = 3/experimental data point.
Size exclusion chromatography was performed with affinity-purified FERONIA in nanodiscs, and SDS-PAGE analysis of gel filtration fractions was performed (Fig. 2). The vast majority of FERONIA in nanodiscs elutes from the sizing column slightly after the 158-kDa standard. This observation is consistent with FERONIA (100 kDa) associating with or inserting into nanodiscs (two 23-kDa MSP1D1 molecules per disc plus an indeterminate but presumably minor mass of cardiolipin) at a stoichiometry of 1:1, although at this point, other interpretations cannot be ruled out.
Figure 2.
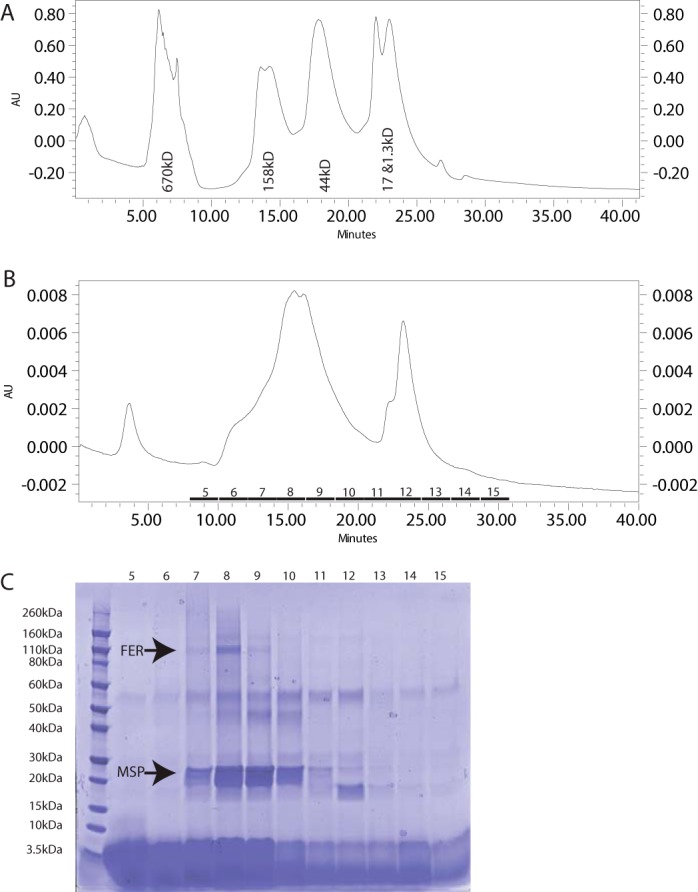
A, size exclusion chromatographic curve for standard protein mixture with labels. AU, absorbance units at 214 nm. B, size exclusion chromatographic curve for 30 μl of injection FERONIA in nanodiscs. C, SDS-PAGE of fractions labeled on the curve in B. Bands for FERONIA (FER) and MSP are labeled. Identity of the unlabeled bands is unknown.
Electron microscopy confirmed nanodisc production and aggregation observed with HPLC (supplemental Fig. S2). Due to limitations in resolution obtained in the microscope and camera used for imaging, it cannot be determined which particles within the image contain FERONIA. Rough calculations on the percentage occupancy of FERONIA:nanodisc versus empty nanodiscs by colorimetrically measuring the intensity of Coomassie-stained bands across the SDS-PAGE gel in Fig. 2, indicate that roughly 28% of discs are occupied by FERONIA.
The juxtamembrane domain is required for full kinase activity when FERONIA is translated without MSP1D1 and cardiolipin
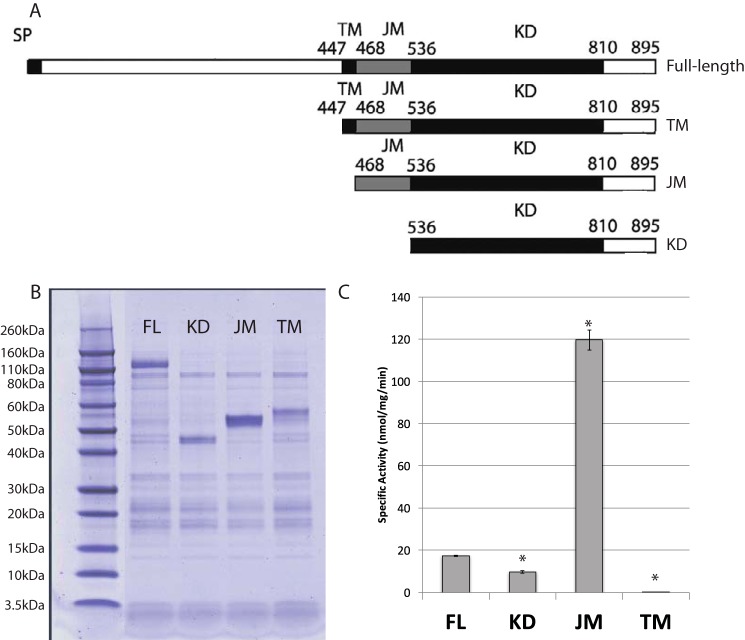
To determine which domains are required for kinase activity, three FERONIA mutants were made containing different sequence truncations: 1) KD, containing only the kinase domain plus C terminus; 2) JM, containing the C-terminal domain plus kinase domain plus the juxtamembrane domain; and 3) TM, containing the C-terminal domain, the kinase domain, the juxtamembrane domain, and the transmembrane domain (Fig. 3A). To provide a consistent comparison without the complicating factor of membrane insertion, these mutants were produced as C-terminally His-tagged proteins without nanodisc co-expression, because the KD and JM truncations lack transmembrane helices and would presumably not associate with lipids and thus be unable to insert into nanodiscs.
Figure 3.
A, truncation mutant design. Numbers, specific amino acid delineations between domains. B, c-His truncation mutants are soluble and can be expressed and purified with nickel-Sepharose resin. C, specific activity of truncation mutants. Error bars, S.D.; n = 2/bar. *, statistical significance (p ≤ 0.005).
We observed that the JM domain is necessary for full kinase activity in vitro when translated in the absence of MSPs and cardiolipin (Fig. 3C), a result consistent with data reported for other receptor protein kinases (26). In contrast, full-length FERONIA and the truncations TM and KD had significantly reduced activity compared with the JM truncation (Fig. 3C). Although these results indicate important roles for the TM and ecto domains in regulating the kinase catalytic activity, we cannot rule out other more trivial interpretations, such as TM-mediated aggregation that may be occurring in the absence of cardiolipin and causing steric inhibition of kinase activity.
ATP analogue DB-acyl-ATP inhibits kinase activity and labels lysines involved in ATP coordination
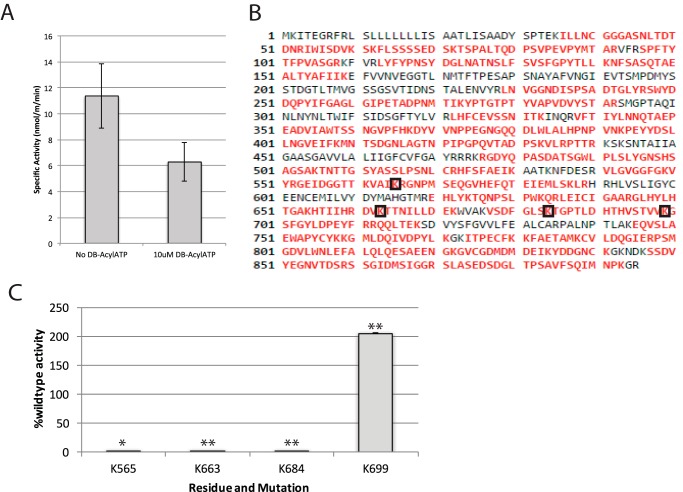
DB-acyl-ATP binds covalently and efficiently to lysine residues within kinase active sites, a property that has been used to enrich proteome samples for kinases and increase kinase sampling in mass spectrometry experiments (27). We tested the ability of DB-acyl-ATP to inhibit the kinase activity of FERONIA when FERONIA was made with MSP1D1 and cardiolipin. DB-acyl-ATP inhibited kinase activity in wild-type FERONIA (Fig. 4A). Using tandem mass spectrometry, DB-acyl-ATP labeling was identified on Lys-565, Lys-663, Lys-684, and Lys-699, all within the kinase domain, (Fig. 4B). These results were promising and consistent with important literature, which has shown that FERONIA K565R cannot autophosphorylate (13).
Figure 4.
A, inhibition of specific activity with DB-acyl-ATP treatment. Error bars, S.D., n = 2, p = 0.13. B, sequence diagram of FERONIA. Red type, areas sequenced in MS experiment; black type, unidentified. Black boxes, lysines modified with DB-acyl-ATP. C, specific activity of FERONIA Lys → Arg mutants co-translated with nanodiscs. Specific activity displayed is normalized to percentage of wild-type activity. Error bars, S.D.; n = 2/bar. Asterisks, statistical significance (*, p ≤ 0.05; **, p ≤ 0.01).
We next singly mutated each of these Lys residues to Arg and translated these variants in the presence of MSP1D1 and cardiolipin to experimentally evaluate whether any of these residues were required for FERONIA protein kinase activity (Table 1 and Fig. 4C). All of the Lys-mutated FERONIA variant genes produced 100-kDa proteins except for K684R, which generated a truncated, 60-kDa protein (supplemental Fig. S3). When the in vitro kinase activity of K565R, K663R, K684R, and K699R was tested (Fig. 4C), both K565R and K663R exhibited significantly reduced kinase activity, suggesting that these two Lys residues are important for ATP coordination, as the DB-acyl-ATP experiment had suggested. K684R also had significantly reduced kinase activity, but whether this is a result of truncation or Lys → Arg mutagenesis has yet to be determined. Conversely, K699R appears to be more active than wild-type FERONIA by 2-fold, suggesting that, unlike the other Lys residues, Lys-699 may destabilize ATP turnover or binding, such that Lys-699 mutation yields a more active kinase. Taken together, the DB-acyl-ATP labeling and activity evaluation of FERONIA Lys mutants implicate residues Lys-663 and Lys-565 in ATP coordination.
TABLE 1.
List of FERONIA mutants described herein, their domain, and justification for their choice
| Residue | Mutation | Domain | Reasoning |
|---|---|---|---|
| Tyr-495 | Phe | Juxtamembrane | Phosphoproteomic screen |
| Lys-565 | Arg | Kinase | DB-acyl-ATP screen |
| Lys-663 | Arg | Kinase | DB-acyl-ATP screen |
| Lys-684 | Arg | Kinase | DB-acyl-ATP screen |
| Thr-696 | Ala/Asp | Kinase | Phosphoproteomic screen |
| Lys-699 | Arg | Kinase | DB-acyl-ATP screen |
| Ser-701 | Ala/Asp | Kinase | Phosphoproteomic screen |
| Ser-871 | Ala/Asp | C terminus | Phosphoproteomic screen |
| Ser-877 | Ala/Asp | C terminus | Phosphoproteomic screen |
Phosphomimetic mutation suggests that C-terminal phosphorylation is regulatory
FERONIA amino acids that were identified in phosphoproteomic data sets were chosen for Ser/Thr → Ala/Asp mutagenesis (Table 1) (28, 29). A single Tyr residue within the JM domain was also chosen for mutagenic analysis, because JM domain phosphorylation is involved in regulation of other plant RLKs (30). Because a common regulatory mechanism for receptor kinases is phosphorylation on the C terminus (31), and previous in vivo experiments involving RALF-dependent changes in FERONIA implicated this domain as a potential regulatory domain (15), we chose Ser-871 and Ser-877 within the C terminus as well as Thr-696 and Ser-701 within the kinase domain for analysis (Table 1 and supplemental Fig. S3).
Indeed, the phosphomimetic mutations S871D and S877D on the C terminus inhibited in vitro kinase activity, whereas the S871A and S877A mutation increased activity by 3-fold when compared with wild-type FERONIA (Fig. 5), supporting the hypothesis that phosphorylation in the C-terminal domain may inhibit kinase activity in vivo as part of FERONIA regulation. In contrast, mutation of two phosphate acceptor sites within the kinase domain, Thr-696 and Ser-701, to either Asp or Ala, significantly decreased the kinase activity of FERONIA (Fig. 5), suggesting that these residues are either important for maintaining an active kinase conformation or have some necessary role in catalysis. The Y495F mutation, located within the JM domain, resulted in a minor increase in kinase activity (Fig. 5).
Figure 5.
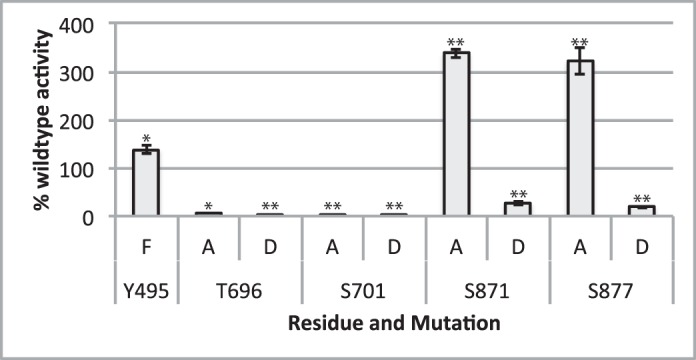
Specific activity of FERONIA mutant variants co-translated with nanodiscs. Specific activity displayed is normalized to percentage of wild-type activity. Error bars, S.D.; n = 2/bar. Asterisks, statistical significance (*, p ≤ 0.05; **, p ≤ 0.01).
Altogether, these observations suggest that C-terminal FERONIA phosphorylation may be involved in regulation of the kinase via inhibition in vivo, whereas Ser and Thr residues within the kinase domain may be important for structural stability of the protein. Tyr-495 phosphorylation may also have some role in inhibition, although indirect structural changes resulting from Tyr to Phe mutation cannot be ruled out.
Discussion
Co-expression of FERONIA in the presence of MSP1D1 and cardiolipin greatly increased in vitro FERONIA specific activity compared with expression without added lipid and MSP mRNA (Fig. 1). Nanodiscs are particularly suited for biochemical characterization of membrane protein kinases such as FERONIA because, unlike for a liposome, there is no directionality of FERONIA insertion into the lipid bilayer and thus no potential problems due to substrate accessibility on the two sides of the bilayer (i.e. there is no inside and outside to a nanodisc). It has been demonstrated in previous studies that nanodiscs are a suitable environment for such proteins. For example, a 2008 study with purified epidermal growth factor receptor (EGFR) demonstrated that reconstitution into nanodiscs greatly increased the length of time the kinase was active when stored at 4 °C compared with EGFR embedded in dodecyl maltoside detergent micelles (23). Additionally, the cell-free translation system utilized in our studies has several unique advantages over the more widely used nanodisc assembly protocols that utilize detergent-solubilized membrane proteins and lipids (21, 23). First, co-expression of FERONIA and MSP1D1 circumvents the need to produce and purify MSP and FERONIA separately before nanodisc assembly. Second, producing and purifying FERONIA-containing nanodiscs using StrepII-tagged MSP1D1 yields FERONIA lacking an affinity tag. Affinity tags on either the N or C terminus interrupt the native sequence of a protein and thus may interfere with kinase activity involving C-terminal regulation and/or overall protein conformation. Finally, this system allows for solubilization and membrane-directed folding of FERONIA in the absence of detergents that may perturb enzyme stability, folding, or functionality. We found that FERONIA co-expressed with nanodiscs exhibits high specific activity, similar to previously published observations with EGFR-containing nanodiscs after activation with EGF (23).
Size exclusion chromatography and electron microscopy revealed that cell-free synthesis and StrepTactin affinity purification resulted in a heterogeneous mixture of both empty nanodiscs and nanodiscs containing FERONIA (Fig. 2 and supplemental Fig. S2). Most of the FERONIA in nanodiscs eluted slightly later than a standard 158-kDa protein, leading to the simplest interpretation that most of the FERONIA produced is associated with discs at a ratio of 1:1, as monomeric FERONIA. The mass ratio of MSP1D1/FERONIA in this size exclusion fraction was calculated to be 1.25:1, which is consistent with a mixture of monomeric and dimeric FERONIA within each nanodisc, because two molecules of MSP1D1 are needed to form each nanodisc. In the calculation of molecular weight by size exclusion chromatography, it is assumed that the Stokes radius is proportional to mass of protein, and thus the simplest explanation from the data is that FERONIA exists mainly as a monomer in nanodiscs. However, it is possible that the Stokes radius of the dimeric and monomeric FERONIA nanodiscs may be very similar. Computational analysis revealed that FERONIA, along with other members of its subfamily, contains the transmembrane motif (A/G)XXX(A/G) (32), suggesting that the kinase may indeed dimerize under certain conditions (supplemental Table S1). Further experimentation is needed to conclusively determine the number of FERONIA molecules in each disc.
The electron microscopy data (supplemental Fig. S2) demonstrated that FERONIA co-expressed with nanodiscs may be a good candidate for further microscopy and, possibly, for high-resolution cryo-electron microscopy experiments aimed at determining three-dimensional structure with single amino acid resolution. Currently, the technology for visualizing nanodisc-embedded protein via cryo-EM is still in its infancy, although prospects for ultimate success are promising (33, 34).
FERONIA truncations demonstrated the necessity of the JM domain for kinase activity when translated without MSP1D1 and cardiolipin. Although the KD construct alone appears minimally active, the KD in addition to the JM domain is the most active version of the protein (Fig. 3). Truncations that also contained the TM-produced protein were much less active than the JM construct, as was full-length protein. The lack of MSP1D1 and added cardiolipin may cause aggregation within the samples containing a TM domain, perhaps inhibiting kinase activity. The JM domain of EGFR has been studied and shown to be necessary for both the dimerization and activation of EGFR (26). Similarly, the plant brassinosteroid receptor kinase BRI1 requires its juxtamembrane domain for kinase domain activation (35). Consistent with these previously published reports, our data suggest that the JM domain of FERONIA may be necessary for kinase activity as well. Whether the JM domain plays a role in dimerization, membrane interaction, or structural integrity is unknown. Interestingly, it has also been reported that EGFR dimerization and activation occurs via the transmembrane helices as well, although the EGF-induced rotational orientation of the JM domain and, by extension, the TM domain is important for stabilization of EGFR dimers (36). Thus, the lack of an MSP1D1-constrained lipid bilayer for the TM domain to insert into may cause the inhibition in activity that we observed.
DB-acyl-ATP labeling identified Lys residues involved in ATP coordination in the hypothesized active site of FERONIA (Fig. 4). Both K565R and K663R mutations effectively produced kinase-inactive FERONIA, suggesting that these two Lys residues are essential for ATP coordination and are located within the active site, given that they were also labeled with DB-acyl-ATP. K565R FERONIA, produced in E. coli, has been shown to lose the ability to autophosphorylate (37), and sequence alignments have shown that Lys-565 is conserved among most of the plant RLK family (37, 38); however, there are no data suggesting conservation of the other Lys residues that were labeled in this study with DB-acyl-ATP. In vivo, K565R is only able to partially complement the loss of mechanosensing induced by feronia mutation (39) and can complement the unfertilized ovule phenotype observed in feronia mutants (38), consistent with the hypothesis that FERONIA has many roles within a plant, some of which may be independent of protein kinase functionality. Interestingly, it has recently been shown that RIPK, a receptor-like cytoplasmic kinase, interacts with FERONIA and can phosphorylate K565R in vitro (40), suggesting that FERONIA phosphorylation observed in vivo may not be limited to autophosphorylation.
The K699R mutation, in contrast to K565R and K663R, produced FERONIA that was more active than wild type, suggesting that it is not involved in ATP coordination but may be involved in maintaining structural integrity via salt bridge formation. Thus, by replacing a Lys with Arg (which is predicted to allow salt bridge formation) this mutation could produce a more active conformational state. K684R mutation leads to a truncated protein product in the cell-free system (supplemental Fig. S3), and this product demonstrated significantly reduced kinase activity; however, the possibility that this reduction in activity is a result of a lack of functionality caused by the truncation rather than the K684R mutation has not been tested. Taken together, these results suggest that Lys-565 and Lys-663 are located within the active site and play a role in ATP coordination, whereas Lys-699 does not share this function.
Mutants S871A and S877A were much more active than wild-type FERONIA, whereas phosphomimetic S871D and S877D mutants were much less active than wild-type FERONIA (Fig. 5), suggesting that these sites are both important for FERONIA phosphoregulation at the C terminus. Such regulation may be similar to EGFR, in which autophosphorylation of the C-terminal region ultimately inhibits kinase activity (31). However, EGFR autophosphorylates mainly on Tyr residues within the C terminus as a means of regulation (41); full kinase activity of EGFR is dependent upon EGF stimulation (42); there are only 2 Tyr residues within the FERONIA C terminus; there is not strong evidence for C-terminal Tyr phosphorylation; and EGFR is an animal Tyr kinase rather than a Ser/Thr protein kinase, like FERONIA. Thus, a more direct comparison may be BRI1, the plant brassinosteroid receptor. Interestingly, BRI1 autophosphorylation occurs sequentially, with Ser and Thr phosphorylation preceding Tyr autophosphorylation (43). Similar to what we observed with FERONIA, C-terminal phosphorylation is inhibitory to BRI1 kinase activity (35).
Both Thr-696 and Ser-701 Ala and Asp mutants caused significant loss of kinase activity compared with the wild-type enzyme, suggesting that the native residues in these positions are important for conformational stability and/or activity. Similar results have been observed for both BRI1 and BAK1, the RLK co-receptor of BRI1 (30). In vitro kinase activity was significantly decreased when Ser residues in their kinase domains were mutated to either Ala, Glu, or Asp. Although BRI1 has been shown to exhibit inhibitory autophosphorylation within its kinase domain, it is possible that the FERONIA phosphorylation identified in planta may not serve this purpose, because in vitro mutagenesis showed that both Ala and Asp substitution significantly decreased activity. It should be noted, however, that whereas the phosphorylation of some residues may be inhibitory, some may be activating, and there may be an effect of cumulative phosphorylation on residues over time that results in more controlled adjustment of FERONIA activity. Phosphorylation in vivo with some of these mutations has been tested. Ser-695, Thr-696, and Ser-701 were mutated singly and cumulatively to Ala and Asp residues and tested for the ability to complement lack of ovule fertilization in the feronia background, demonstrating that single, double, and triple A mutations could fully complement the phenotype, whereas triple D could not (38). Taken together with the complementation data for K565R, this suggests that FERONIA kinase functionality is not necessary for ovule formation, although it may play a role in mechanosensing or other pathways in which FERONIA is involved.
JM domain phosphorylation has been shown to be important for plant receptor kinase signaling. For example, transphosphorylation of BRI1 within the JM domain by co-receptor RLK BAK1 is necessary for full in vitro kinase activity, and loss of these phosphorylation events reduces brassinosteroid signaling capacity in vivo (30). The Y495F mutation within the FERONIA juxtamembrane domain resulted in a slight increase in kinase activity. There are several possible interpretations of this result. Phosphorylation at Tyr-495 may be inhibitory; removal of the hydroxyl group from the benzene ring of Tyr-495 may modify the kinase's conformation; or altered charge distribution near the kinase domain may be altering activity, resulting in a slightly more active form. Nevertheless, Tyr-495 phosphorylation is not necessary for maximal kinase activity of FERONIA produced in vitro with this cell-free translation system.
Taken together, these data suggest a model in which FERONIA kinase activity is dependent on the JM domain and can be inhibited at least in part by C-terminal phosphorylation (Fig. 6). The active site contains Lys-565 and Lys-663, whereas Lys-699 may be in close proximity to the active site but may not be directly involved in ATP coordination. Although in vivo models invoke dimerization as a means of activation (35, 36), the data obtained, taken at their simplest, suggest that the majority of FERONIA in nanodiscs produced here is monomeric, although other interpretations cannot be ruled out.
Figure 6.
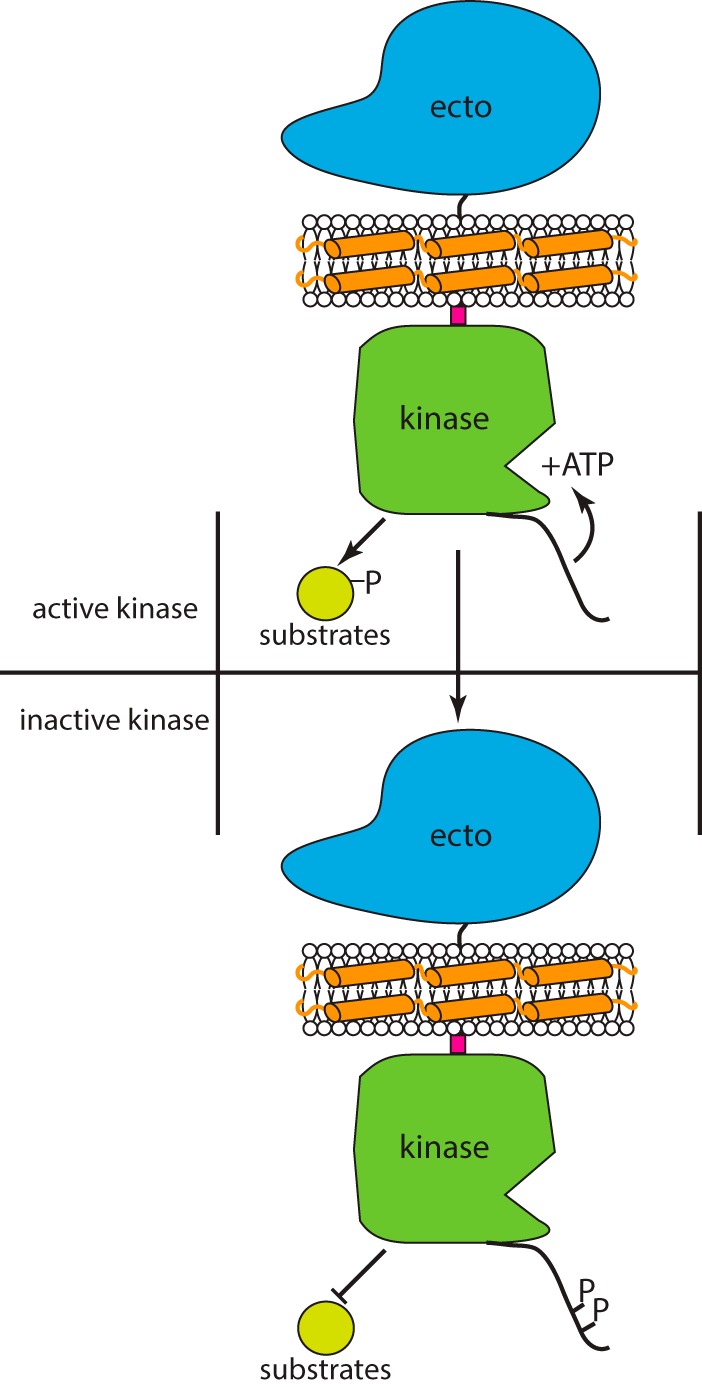
Current model for regulatory mechanism of FERONIA. FERONIA can phosphorylate substrates, the juxtamembrane domain (pink) is necessary for kinase activity, and Ser-871 and Ser-877 may be phosphorylated to inhibit kinase activity.
Although some of the molecular details and requirements for FERONIA kinase functionality have been revealed, many questions remain. Models of receptor kinase functionality suggest that in vivo, co-receptors may be required for full signaling capacity (44). Thus, a model invoking a FERONIA co-receptor would open many avenues of study. In this context, it is worth noting a few putative co-receptors. The previously published phosphoproteomic data set indeed shows a second RLK in the same subfamily as FERONIA, coined ERULUS, whose phosphorylation was decreased in response to RALF, in contrast to FERONIA (15). Additionally, a recent report presents strong genetic evidence that LLG1, a glycosylphosphatidylinositol-anchored membrane protein, is required for in vivo RALF-dependent phenotypes and demonstrates that llg1 knockouts are phenotypically similar to feronia knockouts (45). Another glycosylphosphatidylinositol-anchored membrane protein, LORELEI, LLG1, or both may be required for full localization and thus signaling capacity for FERONIA, in planta (45). Finally, the aforementioned RIPK cytoplasmic phosphorylates FERONIA, potentially more strongly with RALF treatment (40). Given all of these putative co-receptors, and that the plant RLK BRI1 also functions in planta via a co-receptor, BAK1 (7), a future model of FERONIA phosphosignaling may include a co-receptor, although the mechanistic details of such have yet to be determined.
Further experiments will explore in greater detail how RALF binding at the ecto domain alters kinase conformation and activity. In the in vitro expression system utilized herein, we were unable to observe RALF-dependent changes in kinase activity. Although cell-free wheat germ co-expression with MSP1D1 and cardiolipin proved successful at producing highly kinase-active FERONIA, this in vitro system nevertheless lacks glycosylating enzymes or other components that may be responsible for post-translational FERONIA ecto domain modification as observed in our laboratory (data not shown) and/or a necessary RALF co-receptor, possibly LLG1, LORELAI, or RIPK, for FERONIA to attain the proper conformation to correctly perceive RALF. Thus, the lack of RALF-dependent changes in kinase activity may reflect the fact that the ecto domain is not conformationally active, although whether this is due to the lack of a co-receptor, a lack of coupling of RALF binding to an ultimate change in in vitro kinase activity, or some combination of the two is unknown.
First used in 2008 to incorporate bacteriorhodopsin into nanodiscs without the need for detergents (19), MSP co-expression of membrane proteins is also promising for in vitro experiments with plant membrane proteins (46). Our results with the plant RLK FERONIA, produced using cell-free translation coupled with nanodisc production, demonstrate that this system provides sufficient quantities of the protein for biochemical and structural studies as well as a facile means of testing structure-function predictions using molecular genetic tools.
Experimental procedures
FERONIA and MSP1D1 constructs
FERONIA:His6 was cloned from previously described plasmids into the pEU omega cell-free wheat germ expression vector containing ampicillin resistance using Gibson assembly (47) and transformed/maintained into One Shot TOP10 cells using the manufacturer's protocol (Invitrogen). Primers are listed in supplemental Table S2. The MSP1D1 variant used was cloned using Gibson assembly with a C-terminal StrepII tag added. FERONIA without any tag and truncations of FERONIA:His6 were cloned from the aforementioned FERONIA pEU vector using Gibson assembly. All sequences were verified using BigDye Sanger sequencing. FERONIA mutagenesis and truncations were performed using QuikChange II or QuikChange Lightning kits according to the manufacturer's protocol (Agilent Technologies).
Cell-free production of full-length and truncated FERONIA constructs
Cell-free expression plasmids were isolated from E. coli using spin miniprep kits (Qiagen) according to the manufacturer's protocol. Final elution was made into 40 μl of 18-megaohm H2O. Cell-free dialysis cup translation was performed following the published protocol (24), up to the “affinity purification” section. No detergent was used. Translations were fractionated by centrifugation for 5 min at room temperature and 17,000 × g. Soluble fractions were subjected to immobilized metal affinity chromatography using nickel-Sepharose HP resin (GE Healthcare) and eluted with 500 mm imidazole in the absence of reducing agents. Elutions were split into 10-μl aliquots and flash-frozen and then stored at −80 °C for future use.
Nanodisc production/co-expression
Translations were performed as described above using a reaction buffer supplemented with 0.6 mm cardiolipin (Avanti Polar Lipids) in place of listed detergents or added liposomes. This was the only exogenous lipid supplied to the translation mix. Before the addition of FERONIA, MSP1D1 RNA was initially added and translated at room temperature for 24 h, followed by the addition of FERONIA RNA to the translation reaction and replenishment of the dialysis buffer with amino acids. The reaction was allowed to translate for an additional 24 h at room temperature. Protein purification was performed using the same protocol as above but using StrepTactin Sepharose high-performance resin (GE Healthcare) to bind the StrepII tag on the MSP rather than nickel Sepharose. 2.5 mm desthiobiotin was used for elution in the absence of reducing agents.
SDS-polyacrylamide gel quantification of cell-free products
1 μl from the insoluble and soluble fractions and 4 μl from the flow-through and elution fractions were sampled for SDS-PAGE analysis. To enable quantification of bands specifically for FERONIA, a standard mixture containing BSA was prepared in 18-megaohm water and electrophoresed to create a standard curve, including 0.05-, 0.1-, 0.25-, and 0.5-μg loads. The intensity of the BSA bands was determined from the gel image using ImageJ, plotted as a standard curve in Excel, and used to quantify the FERONIA concentration based on band intensity.
Size exclusion chromatography
Sizing chromatography was performed on StrepTactin-purified FERONIA/MSP1D1-SII/cardiolipin co-translations and a standard mixture of proteins (Bio-Rad, catalogue no. 151-1901) using a Yarra 3u SEC-4000 column with dimensions 300 × 7.8 mm (Phenomenex) in 15 mm NaCl, 5 mm Na2HPO4, and 5 mm NaH2PO4 with a flow rate of 0.5 ml/min. Fractions were collected every 2 min and dried with vacuum centrifugation. Dried samples were resolubilized in 20 μl of 1% SDS, mixed 1:1 with Laemmli sample buffer, and analyzed using SDS-PAGE.
[32P]ATP kinase reactions
Kinase assay conditions and calculations were adapted from two publications (48, 49). Kinase used was always 1 μl of the affinity-purified translation mix. Total reaction volume was 17 μl, reactions were performed in duplicate or triplicate as noted in the figure legends, and each experiment was repeated. Myelin basic protein (Sigma-Aldrich) was dissolved in kinase assay buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 100 μm EGTA, 10 mm MgCl2, 1.0% glycerol) and supplied at 500 μm final concentration. For the reactions performed for Km/Vmax measurements, the concentrations of substrate used are noted in Fig. 2, and ∼120 μm non-radioactive ATP and 1 μCi of radioactive ATP (γ-phosphate, 32P, PerkinElmer Life Sciences) were added to each reaction. To stop the reaction and remove excess ATP, 15 μl of 17 μl was spotted onto a single 1.5 × 1.5-cm square of P81-phosphocellulose paper (Millipore) and immediately immersed into 0.4% phosphoric acid. Squares were washed twice more for 5 min in 0.4% phosphoric acid and once in acetone and then allowed to air-dry. Squares were put into liquid scintillation vials, and counts of ambient ionizing (Cerenkov) radiation were measured for 1 min. Additionally, two 1-μl aliquots of ATP mix were measured to determine specific activity for 1 nmol of ATP. The calculation for specific activity is as follows: (((raw cpm measurement − blank value)/specific activity/nmol of ATP) × spotting factor × dilution factor/reaction time)/concentration in mg/ml, where the spotting factor is, in this case, 1.15, to normalize for 15/17 μl of reaction spotted to P81 square, and the dilution factor extrapolates kinase activity to 1 ml, followed by conversion to mg of FERONIA. This yields specific activity units of nmol/mg/min and is normalized to the quantified amount of FERONIA and reaction time. Thus, FERONIA mutants that may have differing expression levels are normalized across all experiments. Reaction n values are listed in the figure legends, and all T-testing was performed in Microsoft Excel using two-tailed, homoscedastic testing.
DB-acyl-ATP labeling and MS analysis
FERONIA was mixed with DB-acyl-ATP at 10 μm and allowed to sit at room temperature for 20 min. A kinase assay was then performed to verify inhibition of kinase activity. A second aliquot of FERONIA was used for MS analysis. Protein was precipitated with 6% cold saturated TCA on ice for 30 min, washed with acetone, and solubilized in 8 m urea. Samples were reduced for 45 min with 5 mm DTT at 42 °C and then alkylated for 45 min with 15 mm iodoacetimide at room temperature in the dark. Samples were diluted to 1 m urea with 25 mm ammonium bicarbonate, and 0.5 μg of trypsin (Promega) and 0.5 μg of LysC (Wako) were added. Samples were incubated overnight at 37 °C. Following digestion, samples were desalted and concentrated using OMIX 100-μl C18 tips (Agilent Technologies) according to the manufacturer's protocol, dried to completion with a vacuum centrifuge, and resolubilized in LC/MS grade 0.1% formic acid (Fisher). 1 μl of resolubilized samples was injected onto an LTQ Orbitrap Elite (Thermo Fisher). Conditions were as described previously (15), using an 80-min chromatographic gradient. Data were processed using the Mascot search algorithm, with the following parameters: trypsin protease digestion with two missed cleavages allowed, a precursor ion tolerance of 20 ppm, and a fragment ion tolerance of 0.6 Da. Cysteine carbamidomethylation was set as a fixed modification, and methionine oxidation and asparagine/glutamine deamidation were set as variable modifications. The DB-acyl-ATP variable modification was added to the Mascot list of modifications and defined as +68 atomic mass units on lysines. Raw data have been uploaded to and are available via the Chorus project.
Author contributions
B. B. M., M. H., and M. R. S. designed the experiments and wrote the paper. M. H. supplied unpublished phosphoproteomic data. R. L. W. helped to clone and provide plasmid constructs. S. M., E. T. B., and B. G. F. helped to establish cell-free protein production and optimize nanodisc co-translation.
Supplementary Material
Acknowledgments
We thank Heather Burch for help with experiments, both on the bench and in discussion, and the Alessandro Senes Laboratory for computational prediction of transmembrane dimerization motifs.
This work was supported by National Science Foundation Molecular and Cellular Biosciences (MCB) Grant 0929395 (to M. R. S.), NIGMS (National Institutes of Health) Protein Structure Initiative Grant U54 GM094584 (to B. G. F.), and the National Human Genome Research Institute-University of Wisconsin Genomic Sciences Training Program (NIH Grant 5T32HG002760) (to B. B. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S3.
- RLK
- receptor-like kinase
- DB-acyl-ATP
- desthiobiotinylated acyl-ATP
- ecto
- extracellular
- LRR
- leucine-rich repeat
- RALF
- rapid alkalinization factor
- MSP
- membrane scaffold protein
- EGFR
- EGF receptor
- RIKP
- RPM1-induced protein kinase.
References
- 1. Shiu S.-H., and Bleecker A. B. (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 98, 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Torii K. U. (2004) Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int. Rev. Cytol. 234, 1–46 [DOI] [PubMed] [Google Scholar]
- 3. Shiu S.-H., and Bleecker A. B. (2001) Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE 2001, re22. [DOI] [PubMed] [Google Scholar]
- 4. Vaid N., Macovei A., and Tuteja N. (2013) Knights in action: lectin receptor-like kinases in plant development and stress responses. Mol. Plant 6, 1405–1418 [DOI] [PubMed] [Google Scholar]
- 5. Afzal A. J., Wood A. J., and Lightfoot D. A. (2008) Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol. Plant Microbe Interact. 21, 507–517 [DOI] [PubMed] [Google Scholar]
- 6. Wang Z.-Y., Seto H., Fujioka S., Yoshida S., and Chory J. (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383 [DOI] [PubMed] [Google Scholar]
- 7. Li J., Wen J., Lease K. A., Doke J. T., Tax F. E., and Walker J. C. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222 [DOI] [PubMed] [Google Scholar]
- 8. He K., Xu S., and Li J. (2013) BAK1 directly regulates brassinosteroid perception and BRI1 activation. J. Integr. Plant Biol. 55, 1264–1270 [DOI] [PubMed] [Google Scholar]
- 9. Sun Y., Han Z., Tang J., Hu Z., Chai C., Zhou B., and Chai J. (2013) Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 23, 1326–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shiu S.-H., and Bleecker A. B. (2003) Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132, 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huck N., Moore J. M., Federer M., and Grossniklaus U. (2003) The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130, 2149–2159 [DOI] [PubMed] [Google Scholar]
- 12. Rotman N., Rozier F., Boavida L., Dumas C., Berger F., and Faure J.-E. (2003) Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr. Biol. 13, 432–436 [DOI] [PubMed] [Google Scholar]
- 13. McCormick S. (2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317, 606–607 [DOI] [PubMed] [Google Scholar]
- 14. Li C., Wu H.-M., and Cheung A. Y. (2016) FERONIA and her pals: functions and mechanisms. Plant Physiol. 171, 2379–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haruta M., Sabat G., Stecker K., Minkoff B. B., and Sussman M. R. (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sawasaki T., Ogasawara T., Morishita R., and Endo Y. (2002) A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl. Acad. Sci. 99, 14652–14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Madin K., Sawasaki T., Ogasawara T., and Endo Y. (2000) A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: plants apparently contain a suicide system directed at ribosomes. Proc. Natl. Acad. Sci. U.S.A. 97, 559–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Madono M., Sawasaki T., Morishita R., and Endo Y. (2011) Wheat germ cell-free protein production system for post-genomic research. New Biotechnol. 28, 211–217 [DOI] [PubMed] [Google Scholar]
- 19. Cappuccio J. A., Blanchette C. D., Sulchek T. A., Arroyo E. S., Kralj J. M., Hinz A. K., Kuhn E. A., Chromy B. A., Segelke B. W., Rothschild K. J., Fletcher J. E., Katzen F., Peterson T. C., Kudlicki W. A., Bench G., et al. (2008) Cell-free co-expression of functional membrane proteins and apolipoprotein, forming soluble nanolipoprotein particles. Mol. Cell Proteomics 7, 2246–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cappuccio J. A., Hinz A. K., Kuhn E. A., Fletcher J. E., Arroyo E. S., Henderson P. T., Blanchette C. D., Walsworth V. L., Corzett M. H., Law R. J., Pesavento J. B., Segelke B. W., Sulchek T. A., Chromy B. A., Katzen F., et al. (2009) Cell-free expression for nanolipoprotein particles: building a high-throughput membrane protein solubility platform. Methods Mol. Biol. 498, 273–296 [DOI] [PubMed] [Google Scholar]
- 21. Ritchie T. K., Grinkova Y. V., Bayburt T. H., Denisov I. G., Zolnerciks J. K., Atkins W. M., and Sligar S. G. (2009) Chapter eleven: reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bayburt T. H., and Sligar S. G. (2010) Membrane protein assembly into nanodiscs. FEBS Lett. 584, 1721–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mi L.-Z., Grey M. J., Nishida N., Walz T., Lu C., and Springer T. A. (2008) Functional and structural stability of the epidermal growth factor receptor in detergent micelles and phospholipid nanodiscs. Biochemistry 47, 10314–10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Makino S., Beebe E. T., Markley J. L., and Fox B. G. (2014) Cell-free protein synthesis for functional and structural studies. Methods Mol. Biol. 1091, 161–178 [DOI] [PubMed] [Google Scholar]
- 25. Katzen F., Fletcher J. E., Yang J.-P., Kang D., Peterson T. C., Cappuccio J. A., Blanchette C. D., Sulchek T., Chromy B. A., and Hoeprich P. D., Coleman M. A., and Kudlicki W. (2008) Insertion of membrane proteins into discoidal membranes using a cell-free protein expression approach. J. Proteome Res. 7, 3535–3542 [DOI] [PubMed] [Google Scholar]
- 26. Red Brewer M., Choi S. H., Alvarado D., Moravcevic K., Pozzi A., Lemmon M. A., and Carpenter G. (2009) The juxtamembrane region of the EGF receptor functions as an activation domain. Mol. Cell 34, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Villamor J. G., Kaschani F., Colby T., Oeljeklaus J., Zhao D., Kaiser M., Patricelli M. P., and van der Hoorn R. A. (2013) Profiling protein kinases and other ATP binding proteins in Arabidopsis using acyl-ATP probes. Mol. Cell Proteomics 12, 2481–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nühse T. S., Stensballe A., Jensen O. N., and Peck S. C. (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16, 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mayank P., Grossman J., Wuest S., Boisson-Dernier A., Roschitzki B., Nanni P., Nühse T., and Grossniklaus U. (2012) Characterization of the phosphoproteome of mature Arabidopsis pollen. Plant J. 72, 89–101 [DOI] [PubMed] [Google Scholar]
- 30. Wang X., Kota U., He K., Blackburn K., Li J., Goshe M. B., Huber S. C., and Clouse S. D. (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15, 220–235 [DOI] [PubMed] [Google Scholar]
- 31. Kovacs E., Das R., Wang Q., Collier T. S., Cantor A., Huang Y., Wong K., Mirza A., Barros T., and Grob P., Jura N., Bose R., and Kuriyan J. (2015) Analysis of the role of the C-terminal tail in the regulation of the epidermal growth factor receptor. Mol. Cell. Biol. 35, 3083–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mueller B. K., Subramaniam S., and Senes A. (2014) A frequent, GxxxG-mediated, transmembrane association motif is optimized for the formation of interhelical Cα–H hydrogen bonds. Proc. Natl. Acad. Sci. U.S.A. 111, E888–E895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gogol E. P., Akkaladevi N., Szerszen L., Mukherjee S., Chollet-Hinton L., Katayama H., Pentelute B. L., Collier R. J., and Fisher M. T. (2013) Three dimensional structure of the anthrax toxin translocon-lethal factor complex by cryo-electron microscopy. Protein Sci. 22, 586–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frauenfeld J., Gumbart J., Sluis E. O., Funes S., Gartmann M., Beatrix B., Mielke T., Berninghausen O., Becker T., Schulten K., and Beckmann R. (2011) Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18, 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang X., Li X., Meisenhelder J., Hunter T., Yoshida S., Asami T., and Chory J. (2005) Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 8, 855–865 [DOI] [PubMed] [Google Scholar]
- 36. Moriki T., Maruyama H., and Maruyama I. N. (2001) Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J. Mol. Biol. 311, 1011–1026 [DOI] [PubMed] [Google Scholar]
- 37. Escobar-Restrepo J. M., Huck N., Kessler S., Gagliardini V., Gheyselinck J., Yang W. C., and Grossniklaus U. (2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317, 656–660 [DOI] [PubMed] [Google Scholar]
- 38. Kessler S. A., Lindner H., Jones D. S., and Grossniklaus U. (2015) Functional analysis of related CrRLK1L receptor-like kinases in pollen tube reception. EMBO Rep. 16, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shih H.-W., Miller N. D., Dai C., Spalding E. P., and Monshausen G. B. (2014) The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr. Biol. 24, 1887–1892 [DOI] [PubMed] [Google Scholar]
- 40. Du C., Li X., Chen J., Chen W., Li B., Li C., Wang L., Li J., Zhao X., and Lin J., Liu X., Luan S., and Yu F. (2016) Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, E8326–E8334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Downward J., Parker P., and Waterfield M. (1984) Autophosphorylation sites on the epidermal growth factor receptor. Nature 311, 483–485 [DOI] [PubMed] [Google Scholar]
- 42. Schlessinger J. (2002) Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110, 669–672 [DOI] [PubMed] [Google Scholar]
- 43. Oh M.-H., Bender K. W., Kim S. Y., Wu X., Lee S., Nou I.-S., Zielinski R. E., Clouse S. D., and Huber S. C. (2015) Functional analysis of the BRI1 receptor kinase by Thr-for-Ser substitution in a regulatory autophosphorylation site. Front. Plant Sci. 6, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Santiago J., Henzler C., and Hothorn M. (2013) Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341, 889–892 [DOI] [PubMed] [Google Scholar]
- 45. Li C., Yeh F.-L., Cheung A. Y., Duan Q., Kita D., Liu M.-C., Maman J., Luu E. J., Wu B. W., and Gates L. (2015) Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. Elife 4, e06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li B., Makino S., Beebe E. T., Urano D., Aceti D. J., Misenheimer T. M., Peters J., Fox B. G., and Jones A. M. (2016) Cell-free translation and purification of Arabidopsis thaliana regulator of G signaling 1 protein. Protein Expr. Purif. 126, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gibson D. G. (2011) Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hastie C. J., McLauchlan H. J., and Cohen P. (2006) Assay of protein kinases using radiolabeled ATP: a protocol. Nat. Protoc. 1, 968–971 [DOI] [PubMed] [Google Scholar]
- 49. Peck S. C. (2006) Analysis of protein phosphorylation: methods and strategies for studying kinases and substrates. Plant J. 45, 512–522 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.