In this study, Ueda et al. show that paternal SSP/YDA signaling directly phosphorylates WRKY2 that in turn up-regulates transcription of the major patterning gene WOX8 in the plant zygote. Their results reveal a framework of how maternal and paternal factors are integrated in the zygote to regulate embryo patterning in plants.
Keywords: zygote asymmetry, MAPK signaling, WOX8, maternal factors
Abstract
In many plants, the asymmetric division of the zygote sets up the apical–basal axis of the embryo. Unlike animals, plant zygotes are transcriptionally active, implying that plants have evolved specific mechanisms to control transcriptional activation of patterning genes in the zygote. In Arabidopsis, two pathways have been found to regulate zygote asymmetry: YODA (YDA) mitogen-activated protein kinase (MAPK) signaling, which is potentiated by sperm-delivered mRNA of the SHORT SUSPENSOR (SSP) membrane protein, and up-regulation of the patterning gene WOX8 by the WRKY2 transcription factor. How SSP/YDA signaling is transduced into the nucleus and how these pathways are integrated have remained elusive. Here we show that paternal SSP/YDA signaling directly phosphorylates WRKY2, which in turn leads to the up-regulation of WOX8 transcription in the zygote. We further discovered the transcription factors HOMEODOMAIN GLABROUS11/12 (HDG11/12) as maternal regulators of zygote asymmetry that also directly regulate WOX8 transcription. Our results reveal a framework of how maternal and paternal factors are integrated in the zygote to regulate embryo patterning.
Unlike animal zygotes, where transcription is blocked and maternally derived mRNAs and proteins stored in the unfertilized egg drive early embryo development (Riechmann and Ephrussi 2001; Tadros and Lipshitz 2009), plant zygotes are transcriptionally active, including the transcription of embryo patterning genes (Ueda et al. 2011; Nodine and Bartel 2012). Therefore, fundamental developmental strategies that have evolved in the plant kingdom are different from those in the animal kingdom, implying the evolution of mechanisms that ensure the timely transcriptional initiation of patterning programs in the zygote.
Initiation of the main body axis is one of the first patterning steps in developing an organism from a unicellular zygote. In many plants, including Arabidopsis thaliana, Fucus (brown algae), Physcomitrella patens (moss), or rice (Brawley et al. 1977; He et al. 2007; Sato et al. 2010; Sakakibara et al. 2014), the zygote divides asymmetrically. In Arabidopsis, this division yields a small apical daughter cell that forms the embryo and a large basal daughter cell that will give rise mostly to the extraembryonic suspensor (Mansfield and Briarty 1991). The zygote expresses paralogs of the stem cell transcription factor WUSCHEL (WUS); namely, WUS HOMEOBOX2 (WOX2) and the redundant WOX8 and WOX9 proteins (also named STIMPY-LIKE and STIMPY) (Haecker et al. 2004; Wu et al. 2007; Ueda et al. 2011). After the zygotic division, WOX2 expression becomes confined to the apical cell, where it regulates embryo development, whereas expression of WOX8 becomes restricted to the basal cell. WOX8 is a master regulator of early embryogenesis, controlling suspensor development and root initiation in the basal lineage. In addition, WOX8 non-cell-autonomously regulates development of the embryo, including the expression of WOX2 and patterning of the phytohormone auxin (Breuninger et al. 2008).
Maternal parent-of-origin-effect genes have been identified that regulate endosperm development (Waters et al. 2011) or cotyledon development in mid-stage embryos (Ray et al. 1996) but, unlike in animals, none that specifically affect the zygote or early embryo patterning. Recent studies discovered several factors that are required for zygote asymmetry. The zinc finger transcription factor WRKY2 regulates symmetry breaking and unequal division of the zygote in part by directly activating transcription of the WOX8 gene (Ueda et al. 2011). In wrky2 mutants, zygotes fail to polarize after a transient symmetric stage and eventually divide in a more symmetric manner compared to wild type. The YODA (YDA) mitogen-activated protein kinase kinase kinase (MAPKKK) with the downstream MAPKs MPK3/6 is required for zygote elongation and suspensor specification (Lukowitz et al. 2004; Wang et al. 2007; Bayer et al. 2009). Activation of YDA/MPK signaling in the zygote requires the membrane-associated receptor-like kinase SHORT SUSPENSOR (SSP), whose mRNA is delivered from the sperm cell and translated in the zygote (Bayer et al. 2009). In addition, the EMBRYO-SURROUNDING FACTOR1 (ESF1) peptide and the plasma membrane receptor kinase ZYGOTE ARREST1 (ZAR1) are required for WOX8 expression and zygote development (Costa et al. 2014; Yu et al. 2016). These examples imply that zygote development depends on external cues. However, the molecular mechanisms linking external cues and transcriptional regulation in the zygote have remained elusive.
In this study, we dissected the molecular relationship between SSP/YDA–MAPK signaling and the WRKY2/WOX8 transcription module in zygote asymmetry and embryo patterning by combining molecular and biochemical approaches with genetic studies in Arabidopsis. Our results indicate that WRKY2 bridges MAPK signaling to the nucleus of the zygote and that WOX8 transcriptional control integrates paternal and novel maternal inputs during the initiation of zygote asymmetry.
Results
Both WRKY2 and SSP up-regulate WOX8 expression to polarize the zygote
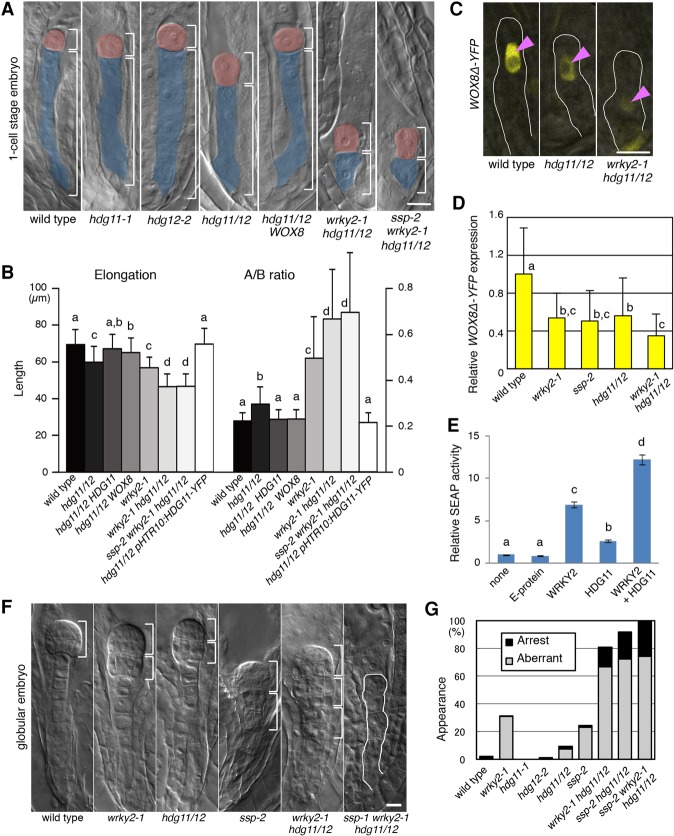
To address the relationship between the WRKY2 and SSP pathways in regulating Arabidopsis zygote asymmetry, we first compared the respective mutant phenotypes. We measured the ratio of apical to basal daughter cell lengths as an inverse proxy for the asymmetry of the zygotic division. Because no further elongation occurs at this stage (Mansfield and Briarty 1991), we measured the total length of one-cell stage embryos as a proxy for zygote elongation. Both wrky2-1 and ssp-2 display shorter zygotes and divide less asymmetrically than the wild type (Fig. 1A,B; Supplemental Table S1; Lukowitz et al. 2004; Bayer et al. 2009; Ueda et al. 2011). Notably, despite the reduction of the total length of the zygote, the length of the apical cell in these mutants is increased compared with wild type, and the basal cell is strongly reduced (Fig. 1A,B; Supplemental Table S1). This indicates that the loss of zygote asymmetry is not simply the consequence of its reduced elongation and suggests that the two processes might be independently regulated, although both are under the control of WRKY2 and SSP.
Figure 1.
WRKY2 and SSP/MAPK signaling regulate zygote elongation and asymmetric division by square brackets. WOX8 expression. (A) One-cell stage embryos of the indicated genotypes. The apical (red) and basal (blue) cells are color-coded, and their lengths are marked by square brackets. (WOX8) pEASE:WOX8-YFP. (B) The total length of one-cell stage embryos (sum of apical and basal cell lengths), denoted as elongation, and the ratio of apical divided by basal cell lengths, denoted as A/B ratio, are shown. (WRKY2) pWRKY2:WRKY2-YFP; [WRKY2(Ala)] pWRKY2:WRKY2(Ala)-YFP; [WRKY2(Asp)] pWRKY2:WRKY2(Asp)-YFP. (C) WOX8Δ-YFP expression in zygotes of the indicated genotypes. The zygotes are outlined, and the nuclei are indicated by arrowheads. (D) Relative WOX8Δ-YFP signal intensities in the zygote. Wild type is set as 1. Bars, 10 µm. Error bars represent SD. n ≥ 80 (B); n ≥ 22 (D). Letters on columns indicate significantly associated categories. P < 0.05 by the Tukey-Kramer test.
To test whether WRKY2 and SSP genetically interact, we first analyzed double mutants. We found that defective zygote asymmetry in the wrky2-1 mutant is not significantly enhanced by ssp-2, suggesting a single pathway (Fig. 1A,B; Supplemental Table S1). In contrast, the effects on zygote elongation are stronger in the double mutant than in each single mutant (Fig. 1A,B; Supplemental Table S1). Because WRKY2 regulates the zygote and the extraembryonic suspensor through direct transcriptional activation of the patterning gene WOX8 (Ueda et al. 2011), we tested whether SSP might also act through WOX8 in this process by using the WOX8Δ-YFP reporter gene, which faithfully mimics the endogenous WOX8 mRNA pattern (Breuninger et al. 2008; Ueda et al. 2011). We found that both ssp-2 and wrky2-1 zygotes display a similarly strong reduction in WOX8Δ-YFP expression compared with wild type (Fig. 1C,D). Notably, WOX8Δ-YFP expression in the wrky2-1 ssp-2 double mutant is not significantly altered compared with either single mutant (Fig. 1D), supporting the hypothesis that SSP and WRKY2 act in a single pathway regulating WOX8. Moreover, restoring expression of WOX8 in the zygote through the pEASE:WOX8-YFP transgene suppresses all ssp-2 zygote defects (ssp-2 WOX8 in Fig. 1A,B; Supplemental Table S1), similar to the previously reported suppression of wrky2 zygote defects by the same transgene (Ueda et al. 2011).
Together, these results suggest that WRKY2 and SSP activities act in one genetic pathway required for the asymmetric division of the zygote through positively regulating WOX8 expression, whereas they appear to act at least in part independently in zygote elongation.
WRKY2 binds to MPK3/6 and is activated by phosphorylation
Previous studies showed that, in the zygote, SSP acts through the YDA MAPKKK and the MAPKs MPK3 and MPK6 (Wang et al. 2007; Musielak and Bayer 2014). Therefore, one possible way in which WRKY2 and SSP could interact is that SSP/YDA–MAPK signaling targets the WRKY2 protein. In yeast two-hybrid assays, we found that WRKY2 binds to MPK3 and MPK6 but not to MAPK MPK9 as a negative control (Fig. 2A) and confirmed this interaction by bimolecular fluorescence complementation (BiFC) in Arabidopsis protoplasts (Supplemental Fig. S1A).
Figure 2.
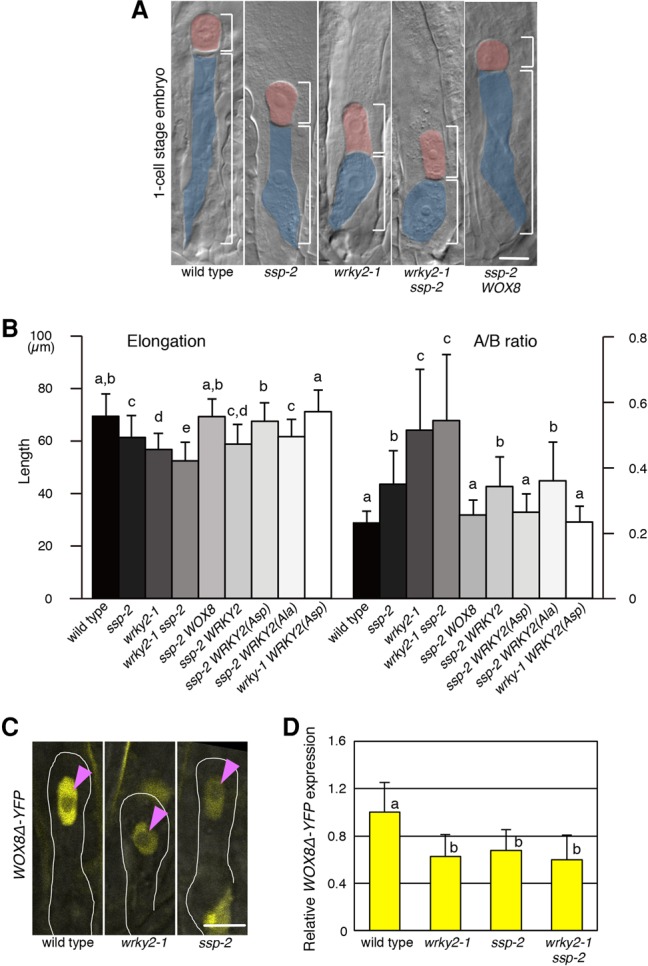
MPK3/6 phosphorylate WRKY2 to promote zygote asymmetry via transcriptional activation of WOX8. (A) Yeast two-hybrid assay of interactions between WRKY2 and MPK3 or MPK6. Panels show serial decimal dilutions of yeast on media with (permissive) and without (restrictive) histidine. Arrows mark growth on the restrictive medium. WOX8 and MPK9 were used as negative controls for WRKY2 and MPK3/6, respectively. (B) In vitro phosphorylation of WRKY2. (+) The presence of constitutively active MKK4 and MKK5 (MKK4/5DD) or MPK3; (−) the absence of constitutively active MKK4 and MKK5 (MKK4/5DD) or MPK3. WRKY2-GST (wt) and WRKY2(Ala)-GST (Ala) expressed in Escherichia coli show two main products, possibly due to partial degradation. An image of the entire gel is shown in Supplemental Figure S1C. (C) In vivo phosphorylation of WRKY2-GFP. Arabidopsis protoplasts were cotransformed with MPK3 and MKK4DD. The five serine residues of the serine–proline motifs (SP cluster) were replaced with alanine in WRKY2(Ala)-GFP. WRKY2-GFP, but not WRKY2(Ala)-GFP, shows a mobility shift due to phosphorylation that disappears after incubation (+) with λ phosphatase (λPP). (D) Luciferase (LUC) assay of the indicated effector proteins on the WOX8 cisB element in protoplasts. Expression of control without effector (none) is set as 1. Unmodified WRKY2, phospho-blocked WRKY2(Ala), and phospho-mimic WRKY2(Asp) variants were used. (WRKY3) Negative control. The characters on the graph indicate the significantly associated categories. P < 0.05 by the Tukey-Kramer test. (E) Relative intensity of WOX8Δ-YFP signals in the zygote and the egg cell. Egg cell is set as 1. (***) P < 0.001, Student's t-test. Error bars represent SD. n ≥ 3 (D); n ≥ 30 (E).
The WRKY2 protein contains a cluster of five serine–proline motifs (SP cluster) as putative MPK phosphorylation sites at the N terminus that is in close proximity to predicted MPK-docking sites (Supplemental Fig. S1B; Eulgem et al. 2000; Sharrocks et al. 2000; Ishihama and Yoshioka 2012). To investigate whether MPK3 can phosphorylate the WRKY2 protein, we used an in vitro assay with proteins synthesized in Escherichia coli. We obtained two main WRKY2 products of slightly different sizes that are phosphorylated only in the presence of both MPK3 and the constitutively active MKK4/5DD that activates MPK3 (Fig. 2B; Supplemental Fig. S1C,D). To analyze whether the SP cluster is important for in vitro WRKY2 phosphorylation, we exchanged the serine residues of the five SP motifs within the cluster with alanines to block phosphorylation [WRKY2(Ala)] (Fig. 2B; Supplemental Fig. S1C). This resulted in a reduced phosphorylation of WRKY2(Ala) protein by MPK3 (Ala in Fig. 2B; Supplemental Fig. S1C), indicating that, in vitro, MPK3 targets at least some of the exchanged SP sites within the SP cluster of WRKY2. Nevertheless, the remaining detectable phosphorylation of WRKY2(Ala) in this assay suggests additional phosphorylation sites outside the SP cluster. Mass spectrometric analysis of Arabidopsis WRKY2 transiently expressed in tobacco (Nicotiana benthamiana) leaves suggests that phosphorylation occurs on several SP sites—including Ser116 and Ser120 within the SP cluster—and several other Ser and Thr residues (Supplemental Figs. S1B, S2). We confirmed that WRKY2 protein is phosphorylated also in Arabidopsis protoplasts using a phospho-retardation assay (Fig. 2C). Importantly, WRKY2(Ala) does not show any phosphorylation in protoplasts (Fig. 2C), suggesting that, in this experimental system, the SP cluster is the main phosphorylation target site of WRKY2. Therefore, we focused our functional analysis on the WRKY2(Ala) variant.
To assess whether phosphorylation of the SP cluster is crucial for the transcriptional activity of WRKY2, we investigated the WRKY2 sequence variants using a luciferase (LUC) transcription assay in Arabidopsis protoplasts (Fig. 2D). We found that unmodified WRKY2 or the constitutively active phospho-mimic version WRKY2(Asp), where aspartates replace all serine residues of the SP sites in the SP cluster, significantly activates transcription from a WOX8 intron fragment containing the WRKY2-binding site (cisB) (Fig. 2D; Supplemental Fig. S3A). In contrast, the phospho-blocked WRKY2(Ala) or WRKY3 as negative control does not (Fig. 2D). Thus, phosphorylation of the WRKY2 SP cluster is essential for transcriptional activation of the WOX8 regulatory region in protoplasts.
To investigate whether WRKY2 phosphorylation by the SSP/MAPK cascade is crucial for the regulation of zygote asymmetry, we analyzed whether the WRKY2 sequence variants can rescue the ssp-2 zygote defects. We used a functional WRKY2-YFP protein as a backbone, enabling us to select transgenic plants with similar expression levels (data not shown). We found that expression of the phospho-mimic WRKY2(Asp)-YFP from the endogenous WRKY2 promoter not only fully rescues the wrky2 zygote but also suppresses the elongation and asymmetry defects of ssp-2 zygotes (Fig. 1B; Supplemental Table S1). In contrast, expression of unmodified WRKY2-YFP or the phospho-blocked WRKY2(Ala)-YFP does not rescue the ssp-2 zygote defects (Fig. 1B; Supplemental Table S1).
Because SSP activity is derived from the sperm-delivered mRNA (Bayer et al. 2009), we tested whether WOX8 transcription increases after fertilization. We found that WOX8 expression levels in the wild-type zygote is increased about twofold compared with the egg cell (Fig. 2E) but is low in wrky2-1 and ssp-2 zygotes (Fig. 1D).
Together, these findings suggest that the phosphorylation of the SP cluster of WRKY2 triggered by sperm-delivered SSP causes up-regulation of WOX8 expression after fertilization and that this step is crucial for zygote elongation and asymmetry.
HOMEODOMAIN GLABROUS11 (HDG11) and HDG12 are direct regulators of WOX8 transcription
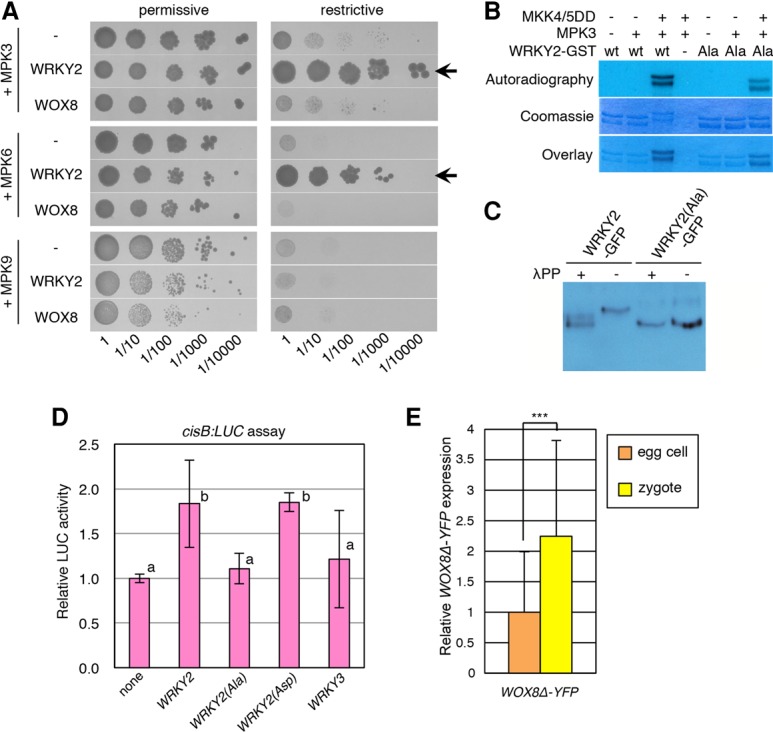
In addition to the WRKY2-binding site in cisB, the second WOX8 intron contains the 85-base-pair (bp)-long cisC element (Supplemental Fig. S3A) that is also sufficient for the correct spatial and temporal WOX8 expression pattern during embryogenesis (Ueda et al. 2011). Using a yeast one-hybrid approach (see the Supplemental Material), we identified the class IV homeodomain–leucine zipper transcription factor family (HD-ZIP IV) protein HDG11 (At1g73360) as a cisC-binding protein (Fig. 3A). In contrast, HDG11 does not bind to a mutated cisC sequence where the core sequence of the predicted HD-ZIP IV-binding motif TAAA was changed (cisCm in Fig. 3A; Supplemental Fig. S3A; Nakamura et al. 2006) or to the unrelated cisB motif (Fig. 3A). We confirmed that HDG11 activates transcription from cisC in Arabidopsis protoplasts (Supplemental Fig. S3B). A similar activation was found with the closely related HDG12 (At1g17920) but not with HDG6/FLOWERING WAGENINGEN (FWA) as a negative control (Supplemental Fig. S3B; Kinoshita et al. 2004).
Figure 3.
HDG11 binds to the WOX8 cisC fragment and is expressed in the female gametophyte, the zygote, and the embryo. (A) Yeast one-hybrid assay of interactions between HDG11 and the indicated WOX8 cis-regulatory fragments. (B) Two-photon microscope images showing pHDG11:YFP expression in ovules containing a mature embryo sac with the egg cell (female gametophyte) or the zygote. Autofluorescence is shown in magenta. Arrowheads point to the nucleus of the egg cell (ec), synergid cell (sy), central cell (cc), and zygote (zyg). (C) pHDG11:YFP expression during embryogenesis at the indicated stages. (Inset) Enhanced YFP signal without differential interference contrast image. Bars, 10 µm.
These results show that HDG11/12 bind to the cisC regulatory site of WOX8 in yeast and activate transcription from cisC in Arabidopsis protoplasts.
HDG11/12 regulate zygote development through WOX8
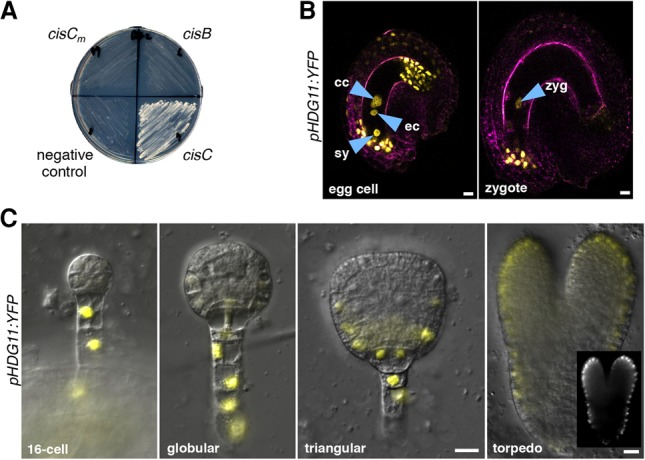
To investigate the role of HDG11/12 genes in embryogenesis, we first analyzed their expression patterns in the female gametophyte and during embryogenesis. We detected expression of a pHDG11:YFP reporter in the egg cell, synergid cells, and central cell of the embryo sac and in cells at the micropylar and antipodal side of the integuments (Fig. 3B). After fertilization, pHDG11:YFP expression remains high in the zygote and the integument at the micropyle but ceases in the endosperm (Fig. 3B). In preglobular and globular embryos, expression is restricted to the suspensor (Fig. 3C), similar to WOX8 and WRKY2 expression (Ueda et al. 2011). At the triangular embryo stage, pHDG11:YFP expression becomes gradually visible in the epidermis (Fig. 3C). Expression of a functional HDG11-YFP fusion protein from the endogenous promoter fully rescues the hdg11-1 hdg12-2 double mutant (hdg11/12 HDG11 in Fig. 4B; Supplemental Table S1) and is expressed similarly to the transcriptional reporter, albeit weaker, in the embryo (Supplemental Fig. S4A), indicating that the observed expression pattern is sufficient for gene function. The expression pattern of a pHDG12:YFP reporter gene is strikingly similar to that of pHDG11:YFP (Supplemental Fig. S4B).
Figure 4.
HDG11/12 regulate zygote asymmetry and embryo patterning. (A) One-cell stage embryos of the indicated genotypes. The apical (red) and basal (blue) cells are color-coded, and their lengths are marked by square brackets. (hdg11/12) hdg11-1 hdg12-2 double mutant; (WOX8) pEASE:WOX8-YFP. (B) Elongation and the A/B ratio of one-cell stage embryos of the indicated genotypes. (HDG11) pHDG11:HDG11-YFP. (C) WOX8Δ-YFP expression in zygotes. The zygote is outlined, and arrowheads indicate the nuclei. (D) Relative WOX8Δ-YFP signal intensities in the zygote. Wild type is set as 1. (E) Secreted alkaline phosphatase (SEAP) assay of the indicated effector proteins on the reporter with the WOX8 intron fragment containing cisB and cisC. Mammalian E protein was used as a negative control. (F) Early globular stage embryos of the indicated genotypes. The sphere-like structures are marked by square brackets, and the arrested embryo is outlined. (G) Frequency of mutant embryo phenotypes. (Aberrant) Embryo whose cell division pattern is different from wild type; (arrest) embryo arrested with <10 cells. Bars, 10 µm. Error bars represent SD. n ≥ 80 (B); n ≥ 28 (D); n ≥ 4 (E); n ≥ 238 (G). Wild type and wrky2-1 in B correspond to Figure 1B because these measurements were performed in one experiment. The characters on graphs indicate the significantly associated categories. P < 0.05 by the Tukey-Kramer test in B, D, and E.
We then analyzed the phenotypes of the strong loss-of-function T-DNA insertion mutants hdg11-1 and hdg12-2 (Nakamura et al. 2006; Khosla et al. 2014). In both single mutants, the zygotes are indistinguishable from wild type, whereas zygotes of the hdg11-1 hdg12-2 double mutant do not fully elongate and divide more symmetrically than wild type (Figs. 4A, 5A; Supplemental Table S1). Furthermore, double-mutant zygotes show a strong reduction of WOX8Δ-YFP expression (Fig. 4C,D) similar to wrky2-1 and ssp-2 zygotes (Fig. 4D). Importantly, the asymmetry defects of hdg11-1 hdg12-2 zygotes are completely suppressed and elongation defects are partially suppressed by restoring WOX8 expression through the pEASE:WOX8-YFP transgene (Fig. 4A,B; Supplemental Table S1), again similar to wrky2-1 and ssp-2 (Fig. 1A,B; Ueda et al. 2011). Thus, HDG11/12 directly activate WOX8 transcription, which in turn affects zygote elongation and asymmetric division.
Figure 5.
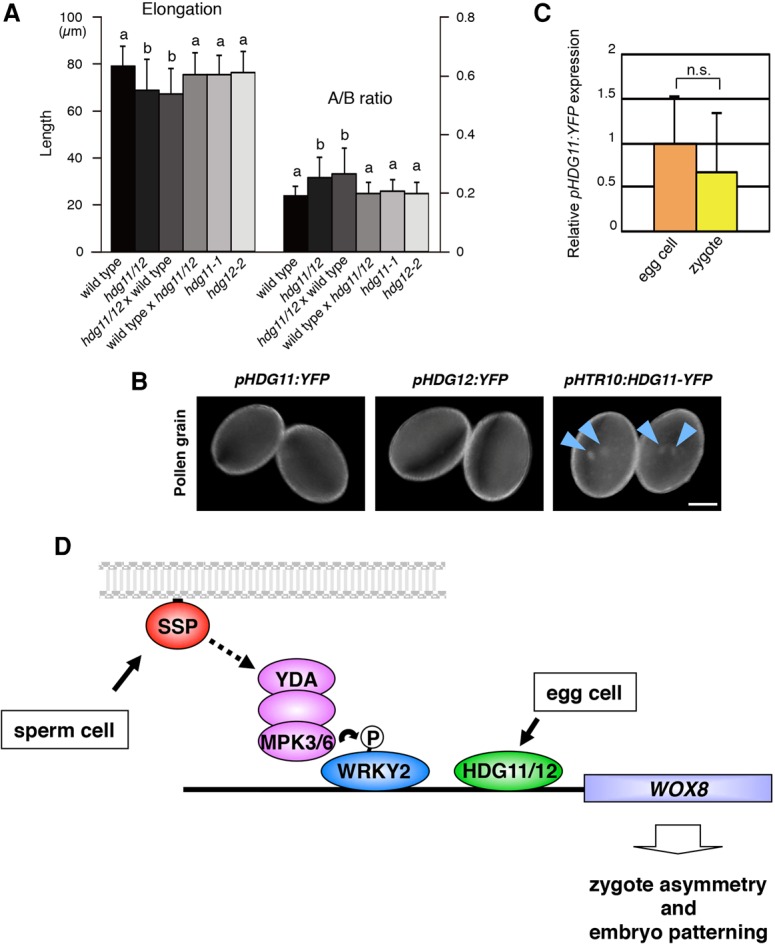
HDG11/12 act as maternal parent-of-origin factors in zygote asymmetry. (A) Elongation and the A/B ratio of one-cell stage embryos of the indicated genotypes. Crosses are denoted as female × male. (hdg11/12) hdg11-1 hdg12-2. (B) Expression of the indicated reporter genes in pollen grains. Arrowheads point to the sperm cell nuclei. (C) Relative intensity of pHDG11:YFP signals in the zygote and the egg cell. Egg cell is set as 1. (D) Model of the regulation of zygote asymmetry. Sperm-derived SSP triggers the YDA–MPK3/6 cascade in the zygote to phosphorylate WRKY2. WRKY2 and maternally derived HDG11/12 directly bind to the WOX8 intron and activate its transcription to regulate elongation and asymmetry of the zygote and embryo patterning. Bar, 10 µm. Error bars represent SD. n ≥ 80 (A); n ≥ 19 (C). The characters on columns indicate the significantly associated categories. P < 0.05 by the Tukey-Kramer test. (n.s) Not significant (Student's t-test).
Because all zygote phenotypes of the hdg11-1 hdg12-2 double mutant are strikingly similar to the mutants of the SSP/WRKY2 pathway and because all factors appear to act through the regulation of WOX8 transcription, we asked how these pathways genetically interact. At the zygote stage, we found that the wrky2-1 hdg11-1 hdg12-2 triple mutant and the ssp-2 wrky2-1 hdg11-1 hdg12-2 quadruple mutant display stronger elongation and asymmetry defects than either wrky2-1 or ssp-2 single mutants or the hdg11-1 hdg12-2 double mutant (Fig. 4A,B; Supplemental Table S1). In a complementary experiment, we confirmed additive transcriptional activation by WRKY2 and HDG11/12 from a WOX8 intron fragment encompassing cisB and cisC (Supplemental Fig. S3A) using a transcriptional assay in human embryonic kidney 293 (HEK-293T) cells (Fig. 4E).
During subsequent embryo development, we observed aberrant cell division patterns in ssp-2 and wkry2-1 single mutants and the hdg11-1 hdg12-2 double mutant but not hdg11-1 or hdg12-2 single mutants (Fig. 4F,G). These abnormal embryos generated spherical structures reminiscent of secondary proembryos (Fig. 4F) that lack WOX8Δ-YFP expression (Supplemental Fig. S5A). The defects are strongly enhanced in the wrky2-1 hdg11-1 hdg12-2 and ssp-2 hdg11-1 hdg12-2 triple and ssp-2 wrky2-1 hdg11-1 hdg12-2 quadruple mutants, with the strongest defects being arrest of embryo development with <10 cells formed (Fig. 4F,G).
In summary, we conclude that HDG11/HDG12 and SSP/WRKY2 activities together are required to promote WOX8 expression and regulate zygote asymmetric division and early embryo patterning.
HDG11/12 are maternal effect factors promoting zygote polarization
Because WRKY2 activity in the zygote is under control of the sperm-supplied factor SSP, we examined whether HDG11/12 also have a parent-of-origin effect. Fertilization of hdg11-1 hdg12-2 egg cells with wild-type pollen does not suppress zygote defects (Fig. 5A; Supplemental Table S1). In contrast, the reciprocal cross of wild-type egg cells with hdg11-1 hdg12-2 pollen does suppress all zygote defects (Fig. 5A; Supplemental Table S1). This indicates that the maternal HDG11/12 gene copies are sufficient for normal zygote development, whereas the paternal copies are not. In contrast to the zygote defects of the hdg11-1 hdg12-2 double mutant, pollination with wild-type pollen did complement later embryo defects at the globular stage (Supplemental Fig. S5E).
To investigate why the paternal HDG11/12 gene copies are unable to complement the mutant zygote defects, we analyzed their expression before and after fertilization. In self-pollinated plants, pHDG11:YFP is expressed at similar levels in the egg cell and the zygote (Fig. 5C). In the sperm cells, expression of pHDG11:YFP and pHDG12:YFP reporters is undetectable (Fig. 5B). After fertilization, paternally provided pHDG11:YFP, pHDG12:YFP, and pHDG11:HDG11-YFP reporters remain unexpressed in the young (short) zygotes (Supplemental Fig. S5B) when zygote polarization occurs (Kimata et al. 2016). However, expression of pHDG11:YFP, pHDG11:HDG11-YFP, and pHDG12:YFP becomes detectable in the mature elongated zygotes, and, during subsequent embryo stages, expression patterns were identical to those in self-pollinated embryos (Supplemental Fig. S5B–D; cf. Fig. 3C and Supplemental Fig. S4A,B). Unlike wild-type pollen, pollination of hdg11-1 hdg12-2 mother plants with pollen carrying the pHTR10:HDG11-YFP transgene (Fig. 5B), which expresses the functional HDG11-YFP fusion protein in sperm cells (Ingouff et al. 2007), fully complements all mutant zygote defects (Fig. 4B; Supplemental Table S1).
Discussion
Embryogenesis requires the initiation of specific patterning programs in the zygote. Because plant zygotes, unlike their animal counterparts, are transcriptionally active (Autran et al. 2011; Nodine and Bartel 2012; Del Toro-De Leon et al. 2014), control mechanisms must have evolved to ensure transcriptional regulation of patterning genes. Our results reveal that a collaboration of paternal and maternal factors in the zygote underlies the transcriptional up-regulation of the WOX8 patterning gene in controlling zygote asymmetry.
WRKY2 bridges paternally potentiated MAPK signaling to transcriptional regulation in the zygote
MAPK signaling cascades are highly conserved intracellular modules that connect external signals to transcriptional regulation in plants and animals. The YDA–MAPK pathway has been studied intensively in stomata development, where it connects peptide signals from neighboring cells to the regulation of cell fate through phosphorylation of the SPEECHLESS transcription factor (Lampard et al. 2008). In the zygote, YDA–MAPK signaling is required for the development of the zygote and the basal descendants (Lukowitz et al. 2004). Our finding that the activator of WOX8 transcription, WRKY2, is a direct target of SSP/YDA/MPK3/6 phosphorylation connects external signals to the transcriptional output of the zygote (Fig. 5D). Importantly, because SSP mRNA is delivered by the sperm and is a limiting factor of YDA signaling, the transcriptional up-regulation of WOX8 in the zygote is under paternal control and thus requires fertilization. What are the signals activating this process and how are they perceived? The membrane-associated IRAK/Pelle kinase SSP, required for YDA/MPK3/6 activation, is thought to function in assembly of the receptor complex (based on the function of mammalian homologs) rather than as receptor itself (Bayer et al. 2009). A recent study shows that the membrane receptor kinase ZAR1 is required for normal WRKY2 and WOX8 expression levels in the zygote and interacts physically with SSP in pull-down assays (Yu et al. 2016), but whether ZAR1 might provide the receptor for YDA/MAPK signaling still needs to be addressed. The recently discovered ESF1 peptides that are produced mainly by the central cell and the endosperm (nutrient tissue that surrounds the embryo) promote WOX8 expression and zygote asymmetry (Costa et al. 2014). However, ESF1 and SSP appear to act synergistically, suggesting that ESF1 might function in parallel to a putative SSP/receptor complex and not through it. Moreover, ESF1 mRNAs are also detectable in early embryonic transcriptome data (Bayer et al. 2016). Thus, the receptor and the signal causing WRKY2 phosphorylation through the YDA–MAPK pathway in the zygote remain to be determined.
Because WRKY proteins have been shown to be targets of MAPK signaling in pathogen defense, our findings support the concept that the genetic repertoire involved in fertilization is similar to the one active in pathogen defense (Okuda et al. 2009; Kessler et al. 2010), which has been interpreted as an evolutionary relationship of plant responses to the invasions of pollen tubes or pathogens (Dresselhaus and Marton 2009).
Maternal control of HDG11/12
Only very few cases of maternal parent-of-origin mutants are known to affect plant embryo development. As one example, expression of the microRNA processing factor DICER-LIKE1 in the sporophytic mother plant is required for normal cotyledon formation at mid-embryo stages (Ray et al. 1996). However, unlike in animals, where maternal control of early embryo patterning is well documented (Riechmann and Ephrussi 2001; Tadros and Lipshitz 2009), maternal effects on earlier embryo stages in plants have not been reported previously.
Our results that WOX8 transcription and zygote asymmetry are directly regulated by maternally, but not paternally, derived HDG11/12 activity show that there is maternal control of early embryogenesis also in plants. What is the basis of this maternal parent-of-origin effect? Our data suggest that the paternal HDG11/12 gene copies are not expressed in the sperm cells and during early zygote stages and thus are unable to drive zygote polarization but are expressed and thus able to complement hdg11/12 mutant defects thereafter. This interpretation is in line with recent findings that zygote polarization, such as cytoskeleton rearrangement and nuclear migration, is evident already a few hours after fertilization, whereas the zygotic division takes place ∼20 h after fertilization (Gooh et al. 2015; Kimata et al. 2016). Silencing of parental gene copies is common in animals and plants and has been suggested for several paralogs of HDG11/12 (Gehring et al. 2009, 2011). In this view, HDG11/12 might be similar to a recently reported group of genes that display transient maternal parent-of-origin effects due to delayed transcription of the paternal gene copy (Del Toro-De Leon et al. 2014).
Two mechanisms, not mutually exclusive, can explain the maternal parent-of-origin effect of HDG11/12 in zygote asymmetry. First, the maternal gene copies could be transcribed in the zygote and then regulate its development. The fact that transcription initiated in the zygote can regulate its own asymmetric division has been shown by the ability to restore WOX8 expression and zygote polarity through a sperm-delivered WRKY2 gene copy (Ueda et al. 2011). Second, the zygote could have inherited HDG11/12 mRNA or protein from the egg cell, similar to animal zygotes. This mechanism has not been described yet in plants, but because HDG11 expression is detected already in the egg cell, it is conceivable that at least some of the egg cell HDG11/12 reservoir could be passed on to the zygote.
A complex network regulates zygote asymmetry and embryo patterning
Comparison of the different mutant combinations indicates that zygote asymmetry requires a complex regulatory network with redundant and synergistic activities. For example, the wrky2 ssp double mutant displays a more severe reduction in zygote elongation compared with the wrky2 single mutant, suggesting that SSP affects additional factors functionally redundant with WRKY2. One potential candidate might be WRKY34, which, in pollen development, is phosphorylated by MPK3/6, acts redundantly to WRKY2 (Guan et al. 2014), and is also expressed at levels similar to that of WRKY2 in the zygote (Xiang et al. 2011). Nevertheless, despite the redundancy within the regulatory network, the complete loss of SSP/WRKY2 and HDG11/12 activities cannot be compensated for, as indicated by the strong defects and the early developmental arrest of ssp wrky2 hdg11/12 quadruple-mutant embryos. Because WRKY2, HDG11/12, and WOX8 are also expressed after the division of the zygote, these defects could be either the consequence of the defective zygote or due to additional functions of these genes in specifying later events in embryo development. One example for the latter possibility is the expression of HDG11/12 in the protoderm at the triangular stage, where they redundantly regulate epidermis development with their paralogs, ATML1 and PDF2 (Lu et al. 1996; Abe et al. 2003; Khosla et al. 2014; Ogawa et al. 2015).
WOX8 transcriptional regulation as an integrator for maternal and paternal cues into embryogenesis
The absence of either WRKY2 activation or HDG11/12 in the zygote similarly reduces WOX8 transcription and causes asymmetry defects, indicating that both inputs must be present to reach the WOX8 transcription levels necessary for zygote development (Fig. 5D). Because HDG11 expression levels are unchanged between the egg cell and zygote, it appears plausible that the up-regulation of WOX8 after fertilization might be caused mainly by the sperm-triggered phosphorylation of WRKY2.
Thus, regulation of WOX8 transcription serves as an integrator of different regulatory inputs from the egg cell and the sperm. One possible advantage of transcriptional control of WOX8 through both parents could be to ensure that the WOX8 expression level necessary for zygote asymmetry is faithfully reached after fertilization. Comparison of a large set of publicly available transcriptome data and our own expression studies (data not shown) indicates that in early embryos and the suspensor, WOX8 expression is indeed higher than in all other tissues, where it is usually at the detection limit. Paradoxically, however, some of these other tissues coexpress HDG11/12, WRKY2, and MPK3/6, raising the question of why WOX8 is not up-regulated there. A plausible hypothesis is that up-regulation of WOX8 transcription is limited to the zygote by the combination of HDG11/12 transcription factors, WRKY2 phosphorylation, and a transcriptionally competent chromatin. Because plant zygotes are deeply embedded in maternal tissue and cannot be isolated in sufficient amounts, future development of sensitive chromatin analysis methods will be necessary to test this hypothesis.
The parental conflict theory proposed for plants and mammals holds that, for embryo nourishment, the two parental genomes act antagonistically (Haig and Westoby 1989; Moore and Haig 1991). Whereas paternal genes favor nutrient supply to their own offspring, maternal genes favor the distribution of resources among all offspring. Our results—that cues from both parents conjointly regulate zygote asymmetry through WOX8 transcription—suggest that during early embryo patterning, a parental cooperation model applies.
Materials and methods
Growth conditions and strains
Plants were grown on soil under continuous light at 18°C–20°C. WOX8Δ-YFP and pEASE:WOX8-YFP were described previously (Ueda et al. 2011). The Arabidopsis lines generated in this study are described in Supplemental Table S2.
Microscopy
Nomarski microscopy analysis was performed after clearing seeds (1:8:3 glycerin:chloral hydrate:water) with a Zeiss Imager A1 or Axioskop2 microscope. For fluorescent analysis, the seeds were placed in 10% glycerin. A two-photon microscope (Zeiss LSM780) was used to observe female gametophytes and zygotes, and intensity of nuclear YFP signal was measured by using ImageJ (http://rsb.info.nih.gov/ij) software. For BiFC assays, an Olympus IX71 microscope was used. Cell lengths of one-cell stage embryos were measured with AxioVision (Zeiss) or ImageJ software.
Yeast assays
The yeast one-hybrid screen was performed as described (Lopato et al. 2006) using the synthetic library REGIA (Regulatory Gene Initiative in Arabidopsis), which contains ∼1500 Arabidopsis transcription factors (Paz-Ares 2002). The yeast two-hybrid assay was performed according to the manufacturer's guidebook (Clontech). For all constructs, full-length cDNAs were cloned into pGADT7 for prey or Clontech pGBKT7 for bait. The detailed methods are described in the Supplemental Material.
BiFC assay
BiFC constructs were created as described previously (Taoka et al. 2011). The 35S CaMV promoter was used to drive the full-length genes fused to the VENUS-C-terminal fragment for bait or the VENUS-N-terminal fragment for prey. For transient expression analysis, 5 µg each of bait, prey, and tagRFP (transformation marker; Evrogen) plasmids were simultaneously introduced into 100 µL of protoplast suspension, generated from the Arabidopsis suspension culture “deep cells” by PEG-mediated transformation (Walter et al. 2004; Saito et al. 2011).
After overnight incubation, YFP-positive (representing interaction) cells were counted among the cells expressing tagRFP (representing transformed cells).
LUC assay
The effector containing transcription factors and the reporter plasmids, including firefly LUC, were prepared as described previously (Yamaguchi et al. 2010). As the effectors, full-length cDNAs were placed after the 35S CaMV promoter. For the reporter plasmids, the quadruple repeat of the 84-bp-long cisB fragment or the 85-bp-long cisC fragment was connected to a 35S minimal promoter, LUC, and a NOS terminator (Ueda et al. 2011). A plasmid containing the Renilla LUC gene driven by the 35S CaMV promoter was used as an internal control. Transient expression in Arabidopsis suspension culture “deep cells” was performed as for the BiFC assays (see above). LUC activity was assayed using the dual-LUC reporter assay system (Promega) and a Mithras LB940 reader (Berthold Technologies). To assess the efficiency of protoplast transformation, the data were used for analysis only when the firefly and Renilla LUC values both exceeded 1000.
Secreted alkaline phosphatase (SEAP) assay
For the effectors, a triple de novo assembly was designed by generating three PCR-amplified linear DNA fragments: coding sequence (CDS) of WRKY2 or HDG11, VP16 with a nuclear localization signal (NLS) sequence, and the SV40 promoter-driven mammalian expression vector. Each pair of adjacent fragments shared 32 bp of overlapping sequence. For the reporter, four times the repeats of the second intron of WOX8 and pAP1(PMA)-SEAP that encodes the SEAP reporter gene (Clontech, catalog no. 631907) were amplified and assembled following the AQUA cloning procedure as described previously (Beyer et al. 2015). A plasmid containing a mammalian E protein driven by SV40 promoter was used as a negative control. HEK-293T cells were maintained in DMEM (Cell Culture Technologies) supplemented with 10% fetal bovine serum (PAN Biotech, catalog no. P30-3602, batch no. P101003TC), 100 U/mL penicillin, and 0.1 mg/mL streptomycin (PAN Biotech). Seventy-five-thousand cells were seeded per well of a 24-well plate 1 d prior to transfection. Cells were transfected with the plasmids (0.75 µg of DNA in total) using polyethylenimine. After 24 h of incubation, SEAP reporter activity was determined as described previously (Schlatter et al. 2002).
Plasmid construction for transgenic plants
Details of cloning procedures are in the Supplemental Material, and all constructs for transgenic plants are listed in Supplemental Table S2.
In vivo phosphorylation assay
Protoplasts were isolated as described previously (Wu et al. 2009). Protoplasts were transfected with plasmids carrying 35S:WRKY2-GFP or 35S:WRKY2(Ala)-GFP, 35S:MKK4DD, and 35S:MPK3 and incubated for 16 h at room temperature. The protoplasts were resuspended in Tris buffer (50 mM Tris-HCl at pH 7.5, 0.15 M NaCl) containing 1% Igepal CA-630 (Sigma-Aldrich) and sonicated three times for 15 sec. All buffers were supplemented with 2× Complete protease inhibitor cocktail (Roche), 1 mM PMSF, and phosphatase inhibitors (25 mM sodium fluoride, 1 mM sodium orthovanadate, 50 mM β–glycerophosphate, 10 mM sodium pyrophosphate). The protoplast solution was diluted to a final concentration of 0.2% Igepal CA-630. The debris was separated by centrifugation, and the supernatant was incubated with anti-GFP microbeads (Miltenyi Biotec) on a rotating wheel for 15 min. The lysate was applied on a column in the magnetic field (Miltenyi Biotech) and washed with Tris buffer containing 0.1% Igepal CA-630. WRKY2-GFP or WRKY2(Ala)-GFP was eluted in HEPES buffer by removing the columns from the magnetic field. The eluate was supplemented with λ phosphate buffer (New England Biolabs) and 1 mM MnCl2 and treated with λ protein phosphatase (New England Biolabs) for 5 min at 30°C. Proteins were separated on 7.5% SDS-PAGE supplemented with 50 μM Phos tag and 100 μM MnCl2 and visualized with anti-GFP antibody (Abcam, ab290) and anti-rat-HRP antibody (Thermo).
In vitro phosphorylation assay
Recombinant proteins for the in vitro phosphorylation assay were expressed from the pGEX4T1 vector in E. coli BL21 (DE3) pLysS cells and purified as described in the manufacturer's instructions (GE Healthcare). Protein expression was induced by adding IPTG to a final concentration of 0.5 mM and incubation for 3 h at 28°C. The culture was pelleted, resuspended in lysis buffer, and sonicated three times for 30 sec with 50% pulses. The GST-tagged proteins were purified using GraviTrap columns according to the manufacturer's instructions (GE Healthcare). MPK3 was activated by incubation in kinase buffer (20 mM HEPES at pH 7.5, 10 mM MgCl2, 1 mM DTT, 25 μM ATP) in the presence of MKK4DD and MKK5DD for 2 h at room temperature. Phosphorylation of WRKY2 and MBP was tested by incubation of WRKY2 or MBP protein with activated MPK3 in kinase buffer supplemented with 1 μCi of ATP-γ32P per reaction for 30 min and autoradiography. Uninduced E. coli lysate was processed in parallel and tested for background signals in the phosphorylation assay.
Mass spectrometry and data analysis
N. benthamiana leaves were infiltrated with p35S:WRKY2-GFP, p35S:MKK4DD-p35S:MPK6, and p35S:MKK5DD-p35S:MPK3 as described previously (Leuzinger et al. 2013). The leaf samples were harvested, and the phospho-enrichment experiment was performed as described previously (Vu et al. 2016). Detailed methods are in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank Taku Takahashi for providing various materials and information of HDG11 and HDG12, including hdg11-1and hdg12-2 seeds, and Tomomi Yamada and Hanae Tsuchiya for technical support. We thank Julia Baaske, Patrick Gonschorek, Wilfried Weber, and Matias Zurbriggen for help with SEAP assays. We thank Wolfgang Lukowitz for ssp-2 seeds and helpful discussion, Nobuaki Ishihama and Hirofumi Yoshioka for prediction of WRKY2 phosphorylation sites, Javier Paz-Ares for providing the yeast library, Hiroyuki Tsuji and Ko Shimamoto for BiFC vectors, Yoko Ikeda and Tetsu Kinoshita for the FWA construct, Hitoshi Endo and Taku Demura for help with LUC assays, and Daisuke Kurihara and Yoshikatsu Sato for help with measurement of fluorescent intensity. We gratefully acknowledge financial support from Grants-in-Aid for Scientific Research (KAKENHI) on Innovative Areas (nos. JP24113514, JP15H05955, JP15H05962, JP17H05838, and JP26113710), Grants-in-Aid for Young Scientists (B, nos. JP24770045 and JP26840093), a Grant-in-Aid for Challenging Exploratory Research (no. JP16K14753), the Sumitomo Foundation, the Kato Memorial Bioscience Foundation, and Exploratory Research for Advanced Technology, Japan Science and Tech Agency (JST; no. JP25-J-J4216) to M. Ueda; a Marie Curie fellowship (275988) to E.A.; Ministry of Education, Culture, Sports, Science and Technology KAKENHI (no. JP22119009), Japan Society for the Promotion of Science KAKENHI (no. JP26291061), and CREST, JST to M. Umeda; Grants-in-Aid for Scientific Research on Innovative Areas (nos. JP16H06465, JP16H06464, and JP16K21727) to T.H.; and the Deutsche Forschungsgemeinschaft to T.L. (ERA-NET, SIREN, and LA606/4). This work was supported by the World Premier International Research Center Initiative of the Institute of Transformative Bio-Molecules (WPI-ITbM) of Nagoya University and Japan Advanced Plant Science Network.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.292409.116.
References
- Abe M, Katsumata H, Komeda Y, Takahashi T. 2003. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130: 635–643. [DOI] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M, Grob S, Steimer A, Barann M, Klostermeier UC, Leblanc O, et al. 2011. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719. [DOI] [PubMed] [Google Scholar]
- Bayer M, Nawy T, Giglione C, Galli M, Meinnel T, Lukowitz W. 2009. Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488. [DOI] [PubMed] [Google Scholar]
- Bayer M, Slane D, Jurgens G. 2016. Early plant embryogenesis-dark ages or dark matter? Curr Opin Plant Biol 35: 30–36. [DOI] [PubMed] [Google Scholar]
- Beyer HM, Gonschorek P, Samodelov SL, Meier M, Weber W, Zurbriggen MD. 2015. AQUA cloning: a versatile and simple enzyme-free cloning approach. PLoS One 10: e0137652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley SH, Quatrano RS, Wetherbee R. 1977. Fine-structural studies of the gametes and embryo of Fucus vesiculosus L. (Phaeophyta). III. Cytokinesis and the multicellular embryo. J Cell Sci 24: 275–294. [DOI] [PubMed] [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T. 2008. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell 14: 867–876. [DOI] [PubMed] [Google Scholar]
- Costa LM, Marshall E, Tesfaye M, Silverstein KA, Mori M, Umetsu Y, Otterbach SL, Papareddy R, Dickinson HG, Boutiller K, et al. 2014. Central cell-derived peptides regulate early embryo patterning in flowering plants. Science 344: 168–172. [DOI] [PubMed] [Google Scholar]
- Del Toro-De Leon G, Garcia-Aguilar M, Gillmor CS. 2014. Non-equivalent contributions of maternal and paternal genomes to early plant embryogenesis. Nature 514: 624–627. [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Marton ML. 2009. Micropylar pollen tube guidance and burst: adapted from defense mechanisms? Curr Opin Plant Biol 12: 773–780. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. 2000. The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. 2009. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Missirian V, Henikoff S. 2011. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One 6: e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooh K, Ueda M, Aruga K, Park J, Arata H, Higashiyama T, Kurihara D. 2015. Live-cell imaging and optical manipulation of Arabidopsis early embryogenesis. Dev Cell 34: 242–251. [DOI] [PubMed] [Google Scholar]
- Guan Y, Meng X, Khanna R, LaMontagne E, Liu Y, Zhang S. 2014. Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet 10: e1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A, Groß-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. 2004. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668. [DOI] [PubMed] [Google Scholar]
- Haig D, Westoby M. 1989. Parent-specific gene expression and the triploid endosperm. Am Nat 134: 147–155. [Google Scholar]
- He YC, He YQ, Qu LH, Sun MX, Yang HY. 2007. Tobacco zygotic embryogenesis in vitro: the original cell wall of the zygote is essential for maintenance of cell polarity, the apical–basal axis and typical suspensor formation. Plant J 49: 515–527. [DOI] [PubMed] [Google Scholar]
- Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F. 2007. Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol 17: 1032–1037. [DOI] [PubMed] [Google Scholar]
- Ishihama N, Yoshioka H. 2012. Post-translational regulation of WRKY transcription factors in plant immunity. Curr Opin Plant Biol 15: 431–437. [DOI] [PubMed] [Google Scholar]
- Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U. 2010. Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971. [DOI] [PubMed] [Google Scholar]
- Khosla A, Paper JM, Boehler AP, Bradley AM, Neumann TR, Schrick K. 2014. HD-Zip proteins GL2 and HDG11 have redundant functions in Arabidopsis trichomes, and GL2 activates a positive feedback loop via MYB23. Plant Cell 26: 2184–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Higaki T, Kawashima T, Kurihara D, Sato Y, Yamada T, Hasezawa S, Berger F, Higashiyama T, Ueda M. 2016. Cytoskeleton dynamics control the first asymmetric cell division in Arabidopsis zygote. Proc Natl Acad Sci 113: 14157–14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. 2004. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523. [DOI] [PubMed] [Google Scholar]
- Lampard GR, Macalister CA, Bergmann DC. 2008. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Leuzinger K, Dent M, Hurtado J, Stahnke J, Lai H, Zhou X, Chen Q. 2013. Efficient agroinfiltration of plants for high-level transient expression of recombinant proteins. J Vis Exp 10.3791/50521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato S, Bazanova N, Morran S, Milligan AS, Shirley N, Langridge P. 2006. Isolation of plant transcription factors using a modified yeast one-hybrid system. Plant Methods 2: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Porat R, Nadeau JA, O'Neill SD. 1996. Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8: 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Roeder A, Parmenter D, Somerville C. 2004. A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119. [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. 1991. Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot 69: 461–476. [Google Scholar]
- Moore T, Haig D. 1991. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet 7: 45–49. [DOI] [PubMed] [Google Scholar]
- Musielak TJ, Bayer M. 2014. YODA signalling in the early Arabidopsis embryo. Biochem Soc Trans 42: 408–412. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T. 2006. Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol 141: 1363–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP. 2012. Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482: 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa E, Yamada Y, Sezaki N, Kosaka S, Kondo H, Kamata N, Abe M, Komeda Y, Takahashi T. 2015. ATML1 and PDF2 Play a redundant and essential role in Arabidopsis embryo development. Plant Cell Physiol 56: 1183–1192. [DOI] [PubMed] [Google Scholar]
- Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D, et al. 2009. Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361. [DOI] [PubMed] [Google Scholar]
- Paz-Ares J. 2002. REGIA, an EU project on functional genomics of transcription factors from Arabidopsis thaliana. Comp Funct Genomics 3: 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Golden T, Ray A. 1996. Maternal effects of the short integument mutation on embryo development in Arabidopsis. Dev Biol 180: 365–369. [DOI] [PubMed] [Google Scholar]
- Riechmann V, Ephrussi A. 2001. Axis formation during Drosophila oogenesis. Curr Opin Genet Dev 11: 374–383. [DOI] [PubMed] [Google Scholar]
- Saito C, Uemura T, Awai C, Tominaga M, Ebine K, Ito J, Ueda T, Abe H, Morita MT, Tasaka M, et al. 2011. The occurrence of ‘bulbs,’ a complex configuration of the vacuolar membrane, is affected by mutations of vacuolar SNARE and phospholipase in Arabidopsis. Plant J 68: 64–73. [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Reisewitz P, Aoyama T, Friedrich T, Ando S, Sato Y, Tamada Y, Nishiyama T, Hiwatashi Y, Kurata T, et al. 2014. WOX13-like genes are required for reprogramming of leaf and protoplast cells into stem cells in the moss Physcomitrella patens. Development 141: 1660–1670. [DOI] [PubMed] [Google Scholar]
- Sato A, Toyooka K, Okamoto T. 2010. Asymmetric cell division of rice zygotes located in embryo sac and produced by in vitro fertilization. Sex Plant Reprod 23: 211–217. [DOI] [PubMed] [Google Scholar]
- Schlatter S, Rimann M, Kelm J, Fussenegger M. 2002. SAMY, a novel mammalian reporter gene derived from Bacillus stearothermophilus α-amylase. Gene 282: 19–31. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD, Yang SH, Galanis A. 2000. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem Sci 25: 448–453. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, et al. 2011. 14–3–3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335. [DOI] [PubMed] [Google Scholar]
- Ueda M, Zhang Z, Laux T. 2011. Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell 20: 264–270. [DOI] [PubMed] [Google Scholar]
- Vu LD, Stes E, Van Bel M, Nelissen H, Maddelein D, Inze D, Coppens F, Martens L, Gevaert K, De Smet I. 2016. Up-to-date workflow for plant (phospho)proteomics identifies differential drought-responsive phosphorylation events in maize leaves. J Proteome Res 15: 4304–4317. [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. 2007. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AJ, Makarevitch I, Eichten SR, Swanson-Wagner RA, Yeh CT, Xu W, Schnable PS, Vaughn MW, Gehring M, Springer NM. 2011. Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23: 4221–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chory J, Weigel D. 2007. Combinations of WOX activities regulate tissue proliferation during Arabidopsis embryonic development. Dev Biol 309: 306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. 2009. Tape-Arabidopsis Sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D, Venglat P, Tibiche C, Yang H, Risseeuw E, Cao Y, Babic V, Cloutier M, Keller W, Wang E, et al. 2011. Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiol 156: 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H, Demura T. 2010. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22: 1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TY, Shi DQ, Jia PF, Tang J, Li HJ, Liu J, Yang WC. 2016. The Arabidopsis receptor kinase ZAR1 is required for zygote asymmetric division and its daughter cell fate. PLoS Genet 12: e1005933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.