Abstract
Origins of DNA replication in Schizosaccharomyces pombe lack a specific consensus sequence analogous to the Saccharomyces cerevisiae autonomously replicating sequence (ARS) consensus, raising the question of how they are recognized by the replication machinery. Because all well characterized S. pombe origins are located in intergenic regions, we analyzed the sequence properties and biological activity of such regions. The AT content of intergenes is very high (≈70%), and runs of A's or T's occur with a significantly greater frequency than expected. Additionally, the two DNA strands in intergenes display compositional asymmetry that strongly correlates with the direction of transcription of flanking genes. Importantly, the sequence properties of known S. pombe origins of DNA replication are similar to those of intergenes in general. In functional studies, we assayed the in vivo origin activity of 26 intergenes in a 68-kb region of S. pombe chromosome 2. We also assayed the origin activity of sets of randomly chosen intergenes with the same length or AT content. Our data demonstrate that at least half of intergenes have potential origin activity and that the relative ability of an intergene to function as an origin is governed primarily by AT content and length. We propose a stochastic model for initiation of DNA replication in the fission yeast. In this model, the number of AT tracts in a given sequence is the major determinant of its probability of binding SpORC and serving as a replication origin. A similar model may explain some features of origins of DNA replication in metazoans.
The replicon model postulates that initiation of DNA replication takes place at specific chromosomal sequence elements (replicators) that are recognized by regulatory proteins (initiators) (1). This model was originally proposed to explain features of the replication of prokaryotic cells and viruses and has been validated in such systems by a large body of evidence (2). In eukaryotic cells, DNA replication is initiated from hundreds to thousands of different chromosomal sites in each cell cycle, raising the question of whether the replicon model provides a valid description of the initiation process. Although early genetic studies indicated that DNA replication in the budding yeast Saccharomyces cerevisiae conforms to the main features of the model, it is not yet clear that this is the case for most other eukaryotic species.
Origins of DNA replication in S. cerevisiae were identified as sequence elements that conferred the property of autonomous replication on extrachromosomal plasmids (3). Genetic analysis demonstrated that such autonomously replicating sequences (ARS) were ≈100 bp in length and contained a common 11-bp consensus sequence essential for origin activity, as well as other sequences that augmented origin activity (3–7). The characterization of S. cerevisiae ARS elements led to the identification of the yeast origin recognition complex (ScORC), the initiator protein that recognizes the ARS-consensus sequence (8). The specificity of the interaction of ScORC with origins is quite high. Single base substitutions are sufficient to abolish ScORC binding to the consensus sequence and prevent initiation of DNA replication (8, 9).
Origins of DNA replication in Schizosaccharomyces pombe differ in a number of ways from those of S. cerevisiae (10–17). They are very large (>500 bp) and extremely AT-rich. Although they often contain asymmetrically distributed clusters of A's and T's, they do not contain a highly specific consensus sequence analogous to the S. cerevisiae ARS consensus. Genetic studies have shown that S. pombe origins contain multiple redundant elements, which can be deleted or replaced by other AT-rich sequences without significantly affecting activity (14–16, 18). Most of the chromosomal origins that have been analyzed in S. pombe fire in only a minority of cell cycles, suggesting that the number of potential origins is greater than the number actually used in any given cell cycle (11, 13, 17, 19–21). These properties are not easily reconciled with the classical replicon model.
The sequencing of the S. pombe genome has made it possible to begin studying the distribution of chromosomal origins of DNA replication (22). Because almost all known S. pombe origins are located in intergenes, we studied the sequence properties and biological activity of such regions. Bioinformatic analysis revealed that the sequences of intergenes (and introns) are not random but exhibit certain underlying patterns that presumably reflect the nature of the mutational mechanisms that operate on the S. pombe genome. The sequence properties of origins of DNA replication are similar to those of intergenes in general. Functional studies of a 68-kb region of S. pombe chromosome 2 demonstrated that more than half of intergenes have potential origin activity and that the relative ability of an intergene to serve as an origin is a function of both its AT content and its length. Based on these and other data we propose that initiation of DNA replication in S. pombe (and perhaps metazoans) conforms to a stochastic model rather than the classical replicon model.
Materials and Methods
Strains and Media. The S. pombe strain TK47 (h+ ade6-M216 leu1–32) was the recipient strain for transformation assays. Cells were grown in supplemented Edinburgh minimal medium (EMM; Bio 101) at 30°C. EMM6S is EMM supplemented with 250 μg/ml adenine, 250 μg/ml uracil, 250 μg/ml leucine, 250 μg/ml lysine, 250 μg/ml histidine, and 1 mg/ml arginine. EMM-leu is EMM6S without the addition of leucine. Escherichia coli DH5α transformants were grown in Luria–Bertani medium supplemented with 100 μg/ml carbinicilin at 37°C. All solid media contained 2% agar.
Construction of Plasmids. Plasmid pRS305 (23) was used as vector for all constructions. Sequence data were downloaded from the Sanger Institute (ftp://ftp.sanger.ac.uk/pub/yeast/pombe/Chromosome_contigs). Twenty-six intergenic regions in 68 kb starting at base pair 2,326,561 (the promoter region of gene SPBC1921.01C) of virtual chromosome 2 (September 5, 2002, release of the S. pombe Genome Project at the Sanger Institute, www.sanger.ac.uk/Projects/S_pombe) were amplified by PCR techniques with oligonucleotide primers containing either a XmaI or SacII site. The PCR products were digested with XmaI and SacII (New England Biolabs) and ligated to pRS305 plasmids that had been digested with the same restriction enzymes. Other intergenes and genes tested in this study were cloned by the same method.
Dimeric inserts were generated by a three-fragment ligation. The three fragments were as follows: (i) the large fragment from pRS305 digested with XmaI and SacII, (ii) the PCR product of the intergene cleaved at the 5′ terminus with XbaI or SpeI and at the 3′ terminus with SacII, and (iii) the same PCR product cleaved at the 5′ terminus with XmaI and at the 3′ terminus with XbaI or SpeI. XbaI and SpeI have compatible overhangs and can be joined by ligation. All constructed plasmids were confirmed by sequencing the vector-insert junctions and by restriction digestion as appropriate.
ars Activity Assays. Transformation of S. pombe by electroporation was carried out as described in ref. 14. TK47 was grown in EMM6S with shaking at 30°C and harvested at an OD600 of 0.5–0.6 by centrifugation. Cells then were washed once with ice-cold water and once with 1 M ice-cold sorbitol and then resuspended in 1 M sorbitol at 1 × 109 cells per ml. Then, 1 μl of plasmid DNA at a concentration of 100 ng/μl was mixed with 40 μl of cell suspension. After incubation on ice for 5 min, the mixture was transferred to an ice-cold 0.2-cm cuvette (Bio-Rad) immediately before electroporation. Cells were pulsed at 2.0 kV, 200 Ω, and 25 μF by using a Bio-Rad gene pulser. Observed time constants varied between 4.4 and 4.5 ms. Immediately after the pulse, 0.9 ml of ice-cold 1 M sorbitol was added to the cuvette. Aliquots of the transformation were plated onto EMM-leu for selection. Colonies were counted after 5 and 10 days of incubation at 30°C and normalized against pRC20, a plasmid containing ars1 (14). Vector pRS305 was used as a negative control.
Software. Programs for analysis of the S. pombe genome were written in the c language and compiled for execution on a Macintosh G3 processor. The genome data were obtained from the Sanger Institute S. pombe Genome Project (www.sanger.ac.uk/Projects/S_pombe).
Results
Sequence Properties of Intergenes and ars Elements in the S. pombe Genome. To date, approximately 12 S. pombe ars elements have been characterized. The completion of the S. pombe genome sequence has made it possible to determine the chromosomal locations of these elements and to ask whether their sequences have unique features. All of the well characterized ars elements are located in the regions between coding sequences (“intergenic” DNA). There are 4,978 annotated intergenes and 5,006 annotated coding sequences (“genes”) in the S. pombe genome (virtual chromosomes, September 5, 2002, release of the S. pombe Genome Project at the Sanger Institute). The difference between the numbers of intergenes and genes is due to 22 pairs of overlapping genes, 2 pairs of immediately adjacent genes, alternative start codons in 1 gene, and 5 gaps in the sequence.
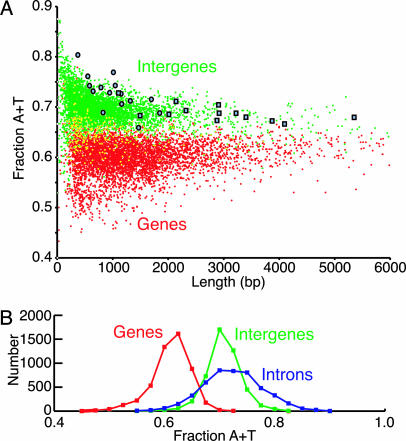
Bioinformatic analysis of the S. pombe genome sequence revealed that the sequences of intergenes are not random but exhibit some underlying patterns. The AT contents of intergenes are much greater than those of genes and, as shown in Fig. 1, the base compositions of intergenes and genes do not overlap significantly. The average intergene is 976 bp in length with an AT content of 69.4%, whereas the average gene is 1,423 bp in length with an AT content of 60.1%. S. pombe introns have compositions similar to those of intergenes. The average intron is 83 bp in length and has an AT content of 71.2% (Fig. 1B).
Fig. 1.
Sequence properties of genes, intergenes, introns, and ars elements in the S. pombe genome. (A) Length–composition diagram of genes (red points), intergenes (green points), known ars elements (blue circles), and the intergenes that contain known ars elements (blue squares). Yellow points represent the superimposition of gene and intergene points. A small number of genes and intergenes with lengths >6,000 bp are not shown in the diagram. (B) Distributions of AT contents of genes (red), intergenes (green), and introns (blue).
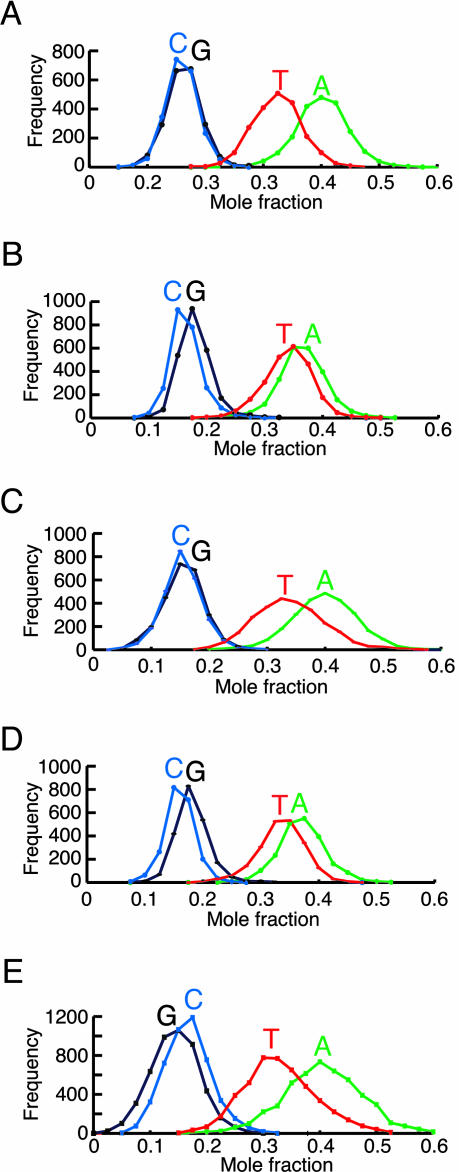
The foregoing data indicate that noncoding sequences have drifted toward high AT contents during the evolution of S. pombe. If the mutational and selection pressures accounting for this trend affected each strand equally, we would expect a symmetric base composition, i.e., that A = T and G = C for each of the two strands of an intergene. However, as shown in Fig. 2, this is not the case. We observed large-scale compositional asymmetries in intergenes that are strongly correlated with the direction of transcription of adjacent genes. In the analyses shown in Fig. 2 A–D, we divided each intergene in half and determined the composition of each DNA strand. We define the template strand as the strand that serves as the template for transcription of the gene adjacent to each half-intergene. All classes of intergenes exhibit significant compositional asymmetry with A > T on the template strand. The asymmetry is most pronounced for half-intergenes adjacent to the 3′ ends of genes (Fig. 2 A and C), where the template strands have an average A content of 38.8 ± 0.1% and an average T content of 32.0 ± 0.1%. The template strands of half-intergenes adjacent to the 5′ ends of genes (i.e., promoter proximal) have an average A content of 35.6 ± 0.1% and an average T content of 32.6 ± 0.1%. The association of the compositional asymmetry with the direction of transcription was confirmed by analysis of introns (Fig. 2E). The template strands of introns have an average A content of 39.4 ± 0.1% and an average T content of 31.8 ± 0.1%.
Fig. 2.
Asymmetric base composition of the two strands of S. pombe intergenes. Each intergene was divided in half. For each half, the composition of the strand that serves as template for transcription of the adjacent gene was determined. (A) Convergent half-intergenes. The data for the template strands of the two halves were combined. (B) Divergent half-intergenes. The data for the template strands of the two halves were combined. (C and D) Unidirectional half-intergenes. The data for the 3′ (C) and 5′ (D) halves of the template strands of unidirectional intergenes were combined. (E) Composition of the template strands of introns within coding sequences.
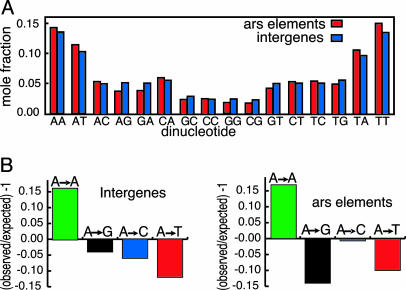
The compositions and lengths of previously published S. pombe ars elements and the intergenes that contain them are plotted on the length–composition diagram of Fig. 1 A. These elements are somewhat longer and more AT-rich than the average S. pombe intergene. It is not possible to determine at this point whether this pattern reflects the basic properties of S. pombe origins of DNA replication or whether it is the result of ascertainment bias, because this collection of ars elements was not selected at random. More detailed analysis revealed that frequencies of dinucleotides (Fig. 3A) and trinucleotides (data not shown) in ars elements are similar to those of intergenes in general. We also compared the frequency of runs of A's and T's in ars elements and intergenes (Fig. 3B). For this purpose, we determined the frequency with which an A residue was followed by A, G, C, or T in each ars or intergene. The resulting frequencies were compared with the frequencies expected if the sequences were random. The data indicate that the probability that an A residue will be followed by another A residue is significantly greater than expected on a random basis, whereas the probabilities that an A residue will be followed by G, C, or T is less than expected on a random basis. Significantly, the same general pattern was observed for both ars elements and intergenes. Thus, the data in Figs. 1, 2, 3 indicate that the sequence properties of ars elements are quite similar to those of intergenes in general. It previously had been reported that S. pombe ars elements contain AT-rich sequences that exhibit local compositional asymmetry of A and T residues and are enriched in runs of A's and T's. Our analysis shows that the same is true of intergenes in general. These observations led us to speculate that a high proportion of S. pombe intergenes might be capable of functioning as origins of DNA replication.
Fig. 3.
Intergenes and ars elements have similar sequence characteristics. (A) Dinucleotide frequencies of intergenes (blue) and ars elements (red). (B) Observed vs. expected frequencies of dinucleotides with A in the first position. The frequencies of the nucleotides A (green), G (black), C (blue), and T (red) after an A residue was determined for S. pombe intergenes (Left) and known ars elements (Right). The frequencies expected for random sequences of the same composition were calculated. The differences between the expected and observed frequencies were normalized to the expected frequencies.
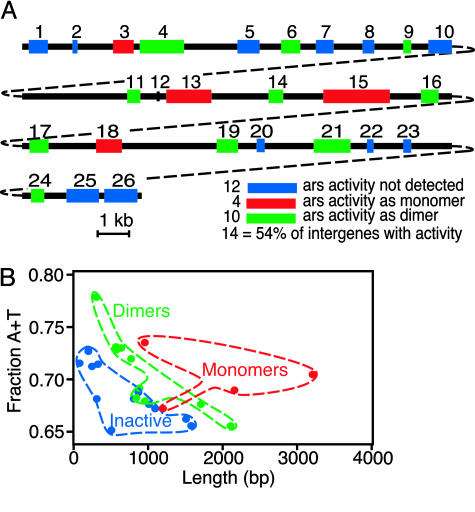
Biological Activity of S. pombe Intergenes. To test the ability of S. pombe intergenes to function as origins of DNA replication, we studied a randomly chosen 68-kb contig in chromosome 2 that contained 26 intergenes. Each intergene was cloned into a plasmid that lacked a functional origin of DNA replication and tested for ars activity by the standard assay (see Materials and Methods). The ability to support the autonomous replication of a plasmid is a relatively stringent assay for origin activity, because the long-term maintenance of the plasmid requires that it replicate approximately once each cell cycle. Most S. pombe chromosomal origins fire considerably less frequently than once per cell cycle. We observed that 4 of the 26 intergenes had ars activity by this assay (Fig. 4A and Table 1, which is published as supporting information on the PNAS web site). It seemed possible that additional intergenes might be capable of functioning as origins of DNA replication but might have efficiencies below the threshold of detection of the ars assay. To detect less efficient origins, we constructed head-to-tail dimers of the remaining 22 intergenes and tested them for ars activity by the same assay. The rationale for this approach was that dimerization might increase the origin activity of an intergene by a factor of 2 by doubling the likelihood that it was recognized by the replication machinery. We found that 10 additional intergenes exhibited ars activity under these conditions (Fig. 4A and Table 1). Thus, more than half of the S. pombe intergenes tested (54%) had the potential to function as origins of DNA replication. The frequency of potential replication origins that we observed (one ars element per 4.9 kb on average) is much higher than the previous estimations of one ars element per 20 kb (10, 21) or 55 kb (20).
Fig. 4.
ars activity of S. pombe intergenes. (A) We tested 26 consecutive intergenes in a 68-kb contig in chromosome 2 for ars activity in the standard assay. Intergenes are represented as boxes and marked with numbers. Red, intergenes active as monomers; green, intergenes active as dimers; blue, intergenes with no detectable activity. The activity of each intergene is given in Table 1. (B) ars activity of intergenes as a function of length and composition. The 26 intergenes are divided into three groups according to whether they are active as monomers or dimers or are inactive. The color scheme is as in A.
Origin Activity of Intergenes Is a Function of Length and AT Content. Based on their biological activity, the 26 intergenes fell into three classes: (i) those that are active as monomers, (ii) those that are active as dimers, or (iii) those that are not active in the ars assay. Interestingly, the three classes largely clustered in distinct regions of the length–composition diagram (Fig. 4B). Intergenes that were active as monomers generally clustered toward the upper right portion of the diagram (higher AT contents and longer sequences), whereas intergenes that were inactive in the ars assay clustered toward the lower left (lower AT contents and shorter sequences). The intergenes that were active as dimers clustered at intermediate positions. The shorter sequences tended to have higher AT contents, whereas the longer sequences tended to have lower AT contents. Although there were some exceptions to these correlations, the overall pattern was strikingly nonrandom (Fig. 4B). These data strongly suggest that the relative origin activity of an intergene is a function of both length and composition and are consistent with the hypothesis that the ability of an intergene to function as an origin simply depends on the cumulative number of AT tracts that it contains.
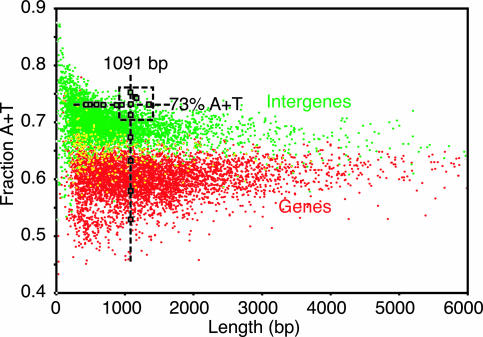
To further explore the correlation between activity and length or composition, we studied randomly chosen intergenes and genes with the same length (≈1,090 bp) or the same composition (73% AT) (Fig. 5). We observed that the intergenes with the same length were active in the ars assay when their AT contents were >70% but were inactive when their AT content was <70%. Similarly, we found that the intergenes with the same AT content were active in the ars assay when their length was greater than ≈900 bp (Fig. 5). Significantly, the average S. pombe intergene is 976 bp and has an AT content of 69.4%. These data support the hypothesis that the ability of an intergene to function as an origin of DNA replication does not depend on its precise nucleotide sequence but only on its length and AT content.
Fig. 5.
ars activity of intergenes is a function of length and AT content. Intergenes (green dots) and genes (red dots) are shown as in Fig. 1. Two series of randomly chosen intergenes (green squares) and genes (red squares) were tested for ars activity in the standard assay as follows: (i) intergenes or genes with similar lengths (1,090–1,092 bp) but different AT contents (vertical line), and (ii) intergenes with similar AT contents (73%) but different lengths (horizontal line). All fragments with ars activity are enclosed in the box delimited by dashed lines.
Discussion
Our data indicate that the sequences of intergenes in S. pombe are not random but exhibit certain patterns. These patterns are most likely the result of mutational biases, although it is possible that selection plays a role as well. S. pombe intergenes exhibit three major characteristics. First, the AT contents of intergenes are much higher than those of coding sequences. The drift toward higher AT contents has proceeded to the point that there is little compositional overlap between genes and intergenes. Deamination of cytosine residues (C → T) is probably the major contributor to this drift (24, 25). Second, the sequences of S. pombe intergenes exhibit systematic compositional asymmetry of A and T residues on the two DNA strands. This asymmetry is clearly correlated with the local direction of transcription with A residues significantly more frequent than T residues on the strand that serves as the transcriptional template. The asymmetry is detectable throughout the lengths of the intergenes but is more pronounced on the downstream (3′) side of the coding sequences (Fig. 2 A and B), presumably because transcription often (but not always) terminates within intergenes (26–30). A similar strand-specific compositional asymmetry associated with transcription has been observed in introns and in the third position of codons of S. pombe, S. cerevisiae, Arabidopsis thaliana, mouse, and human genes (31). The origin of transcription-mediated, strand-specific compositional bias in S. pombe intergenes is not clear, but studies of a similar phenomenon in bacteria have uncovered two plausible mechanisms. (i) The rate of cytosine deamination (C → T) is higher on the coding strand than the template strand, probably because the former is transiently exposed in a single-stranded state, whereas the latter is protected by the nascent RNA and the polymerase (32, 33). (ii) The rates of repair of mutagenic lesions and possibly base mismatches is higher on the template strand than the coding strand due to the operation of transcription-coupled repair (33–36). Although differential rates of cytosine deamination on the template and coding strands may play a major role in generating the observed asymmetry of A and T residues in S. pombe intergenes, such a mechanism cannot entirely explain our observations because it would result in asymmetry of G and C residues of a similar magnitude. Our data indicate that the compositional asymmetry of G and C residues in intergenes is small and largely confined to the region immediately upstream of coding sequences where there is a slight accumulation of purines in the template strand (A > T and G > C) (Fig. 2 A and B). Thus, there must be additional strand-specific mutational biases, perhaps arising as a consequence of transcription-coupled repair. The third characteristic of S. pombe intergenes is the presence of runs of A or T residues at a higher frequency than expected if the sequence were completely random. The accumulation of runs over time is likely due to DNA polymerase slippage during DNA replication (37).
Studies in several laboratories of S. pombe origins of DNA replication have failed to uncover a common consensus sequence analogous to that found in all S. cerevisiae origins. However, several groups have observed that S. pombe origins contain multiple stretches of asymmetric A or T residues (14, 15, 18, 38, 39), some of which have been shown to contribute to origin activity. The results presented here indicate that the sequence properties previously ascribed to origins are actually characteristic of intergenes in general and are not specific to origins. In fact, the only features that appear to distinguish known S. pombe ars elements from the bulk of intergenes are average AT content and length. Our analysis of a 68-kb contig of chromosome 2 demonstrated that intergenes capable of supporting autonomous plasmid replication in vivo are generally long and AT-rich. We also found that many intergenes with shorter lengths and lower AT contents were active in the ars assay after dimerization. Strikingly, the shorter members of this class of intergenes generally had high AT contents, whereas the longer members had low AT contents. Additional studies showed that randomly chosen intergenes with a constant length exhibited ars activity when their AT contents exceeded a threshold value and that randomly chosen intergenes with a constant AT content exhibited ars activity when their lengths exceeded a threshold value. The observation that both length and average AT content contribute to the ability of an intergene to function as an origin suggests that the total AT content is a major determining factor. The density of AT base pairs is important as well because large increases in length are required to compensate for small reductions in AT content (Fig. 4B). These findings are generally consistent with previous observations that S. pombe genomic segments 0.5–1 kb in length with AT contents >72% have a high probability of functioning as origins in vivo, as determined by two-dimensional electrophoretic analysis of replication intermediates (17).
The conclusion that the efficiency of S. pombe origin activity depends largely on total AT content and is relatively indifferent to the precise nucleotide sequence is supported by both biochemical and genetic data. Biochemical studies have shown that the S. pombe origin recognition complex is targeted to origins by the N-terminal domain of SpOrc4, which contains nine AT-hook motifs (40–43). The AT-hook motif binds to short AT tracts of 4–6 nt in length in the minor groove of DNA (44). Proteins that contain multiple AT-hook motifs, like HMGA [HMGI(Y)], can bind to multiple AT tracts in close proximity (45). In this case, the overall affinity of the protein for the site depends on the spacing of the AT tracts and, to a lesser extent, on the length of the AT tracts. For example, in the case of HMGA, which has three AT hooks and can bind with high affinity to three adjacent AT tracts, the optimum spacing between the tracts is in the range of 4–8 nt. When the spacing becomes significantly larger (e.g., 12 nt), a single HMGA molecule no longer can span all three sites, and the binding affinity is much lower (45). Because SpOrc4 has nine AT hooks, it would be expected that there would be many possible ways that it could interact with a sequence containing multiple AT tracts (e.g., an intergene). The affinity of each possible binding mode would be determined largely by the number of AT hooks engaged, which would in turn depend on the spacing of the AT tracts. Thus, the overall affinity of SpOrc4 for such a sequence would be a complex function of the number of possible binding modes and the binding affinity of each mode and would be expected to increase with AT content and length. Consistent with these ideas, it has been demonstrated experimentally that SpOrc4 has significant affinity for multiple sites within a single ars element (41–43, 46). Moreover, genetic studies have shown that sequences capable of functioning as origins contain multiple redundant AT-rich elements that contribute to activity (14–16, 18, 38). It has been shown in some cases that such AT-rich elements can be replaced by different AT-rich sequences without significant loss of activity (15).
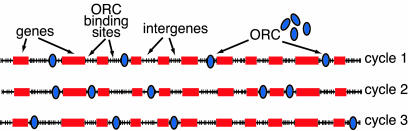
Thus, the picture of S. pombe origins that emerges from these considerations is quite different from the classical replicon model, which postulates that the initiator protein binds a limited number of sites with high specificity (1). Instead, the initiation of S. pombe DNA replication is likely to be a much more stochastic process. In the stochastic model shown in Fig. 6, we suggest that a typical AT-rich intergene contains many potential SpOrc4 binding sites with widely varying affinities because it contains many short AT tracts with different spacings. Because there are 5,000 intergenes in the S. pombe genome and each contains multiple potential SpOrc4 binding sites, the number of potential binding sites would be expected to greatly exceed the number of SpORC molecules in the cell. It follows that the available SpORC will distribute over different binding sites during each successive cell cycle, thus explaining the observation that nearly all S. pombe origins that have been studied to date fire in only a minority of cell cycles. (e.g., see refs. 11, 13, 17, and 19–21) It is clear that the potential origins in some intergenes function in a greater fraction of cell cycles than others (this paper and ref. 17). The stochastic model suggests that long AT-rich intergenes function with higher efficiency simply because they contain more AT tracts and are more likely to have multiple tracts with the appropriate spacing for high-affinity SpOrc4 binding. However, it is important to emphasize that the efficiency with which intergenes function as origins is likely to be a broad continuum and that intergenes with lower AT contents and thus fewer SpOrc4 binding sites likely will contribute significantly to the duplication of the fission yeast genome. The potential origins in such intergenes will fire in fewer cell cycles, but because there are many of them in the genome they will likely account for a significant fraction of the origins that fire in any given cell cycle. It may be quite difficult to detect these weaker origins with the currently available methods such as two-dimensional gel electrophoresis or strand-abundance assays, so such methods will likely underestimate significantly the number of functional origins in the S. pombe genome. In our experiments, we only were able to detect potential origin activity in some intergenes by dimerizing them. We suggest that these intergenes contain origins that are likely to function in vivo but at a somewhat lower efficiency than those in the intergenes that functioned as monomers in our ars assays. Finally, it is important to remember that not all potential SpOrc4 binding sites in the genome may be accessible to SpORC because of the constraints of chromatin structure. Further work will be required to determine whether and how such constraints might affect the distribution of origins of DNA replication during each S. pombe cell cycle.
Fig. 6.
Stochastic model for the initiation of S. pombe DNA replication. A segment of S. pombe chromosomal DNA is shown with many potential AT-rich SpORC binding sites (tick marks) in intergenes. The stochastic model differs from the classic replicon model in the following ways. (i) There are no highly specific replicator sequences. SpORC binds simple sequences (AT tracts) that are very common in the genome. (ii) There are many more potential SpORC binding sites than SpORC molecules. (iii) The distribution of SpORC over the potential sites is quasi-random, depending largely on the local density of AT tracts. Accessibility of sites may be affected by chromatin organization. (iv) Different sets of initiation sites are used in each cell cycle because the ratio of available binding sites to SpORC molecules is high. This feature of the model accounts for the observation that the utilization of S. pombe“origins” is very inefficient.
Several lines of evidence suggest that the stochastic model may be relevant to initiation of DNA replication in mammalian cells. In several cases initiation has been shown to occur with relatively low efficiency at many different sites in intergenic regions of the genome (47, 48). Recent work suggests that human ORC binds preferentially to AT-rich sequences but otherwise has little sequence specificity (49). Moreover, recombinant human ORC can direct initiation of DNA replication on any DNA molecule in a cell-free replication system (49). Although these observations are clearly consistent with a stochastic model, there are other instances where initiation of mammalian DNA replication appears to be localized to relatively small regions of the chromosome, indicating that ORC binding and/or initiation are not completely random in mammalian cells (50). Such regions are generally intergenic and often AT-rich. As in the case of the AT-rich intergenes of S. pombe, these regions simply may contain a high density of potential ORC binding sites, or, alternatively, the localization of initiation may depend on specific features of chromatin structure.
Supplementary Material
Acknowledgments
We thank Pamela Simancek and Deborah Tien for technical assistance and the other members of the Kelly laboratory for stimulating discussions. This work was supported by National Institutes of Health Grants CA40414 and GM50806.
Author contributions: J.D., R.-Y.C., and T.J.K. designed research; J.D. and T.J.K. performed research; J.D., R.-Y.C., and T.J.K. contributed new reagents/analytic tools; J.D. and T.J.K. analyzed data; and J.D. and T.J.K. wrote the paper.
Abbreviations: ARS, autonomously replicating sequence; ORC, origin recognition complex.
References
- 1.Jacob, F. & Brenner, S. (1963) Comptes Rendus Hebdomadaires Seances Acad. Sci. 256, 298-300. [PubMed] [Google Scholar]
- 2.Kornberg, A. & Baker, T. A. (1992) DNA Replication (Freeman, New York).
- 3.Newlon, C. S. & Theis, J. F. (1993) Curr. Opin. Genet. Dev. 3, 752-758. [DOI] [PubMed] [Google Scholar]
- 4.Yabuki, N., Terashima, H. & Kitada, K. (2002) Genes Cells 7, 781-789. [DOI] [PubMed] [Google Scholar]
- 5.Wyrick, J. J., Aparicio, J. G., Chen, T., Barnett, J. D., Jennings, E. G., Young, R. A., Bell, S. P. & Aparicio, O. M. (2001) Science 294, 2357-2360. [DOI] [PubMed] [Google Scholar]
- 6.Raghuraman, M. K., Winzeler, E. A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D. J., Davis, R. W., Brewer, B. J. & Fangman, W. L. (2001) Science 294, 115-121. [DOI] [PubMed] [Google Scholar]
- 7.Breier, A. M., Chatterji, S. & Cozzarelli, N. R. (March 4, 2004) Genome Biol. 5, R22. Available at http://genomebiology.com/2004/5/4/R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell, S. P. & Stillman, B. (1992) Nature 357, 128-134. [DOI] [PubMed] [Google Scholar]
- 9.Van Houten, J. V. & Newlon, C. S. (1990) Mol. Cell. Biol. 10, 3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maundrell, K., Hutchison, A. & Shall, S. (1988) EMBO J. 7, 2203-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubey, D. D., Zhu, J., Carlson, D. L., Sharma, K. & Huberman, J. A. (1994) EMBO J. 13, 3638-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston, L. H. & Barker, D. G. (1987) Mol. Gen. Genet. 207, 161-164. [DOI] [PubMed] [Google Scholar]
- 13.Caddle, M. S. & Calos, M. P. (1994) Mol. Cell. Biol. 14, 1796-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clyne, R. K. & Kelly, T. J. (1995) EMBO J. 14, 6348-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okuno, Y., Satoh, H., Sekiguchi, M. & Masukata, H. (1999) Mol. Cell. Biol. 19, 6699-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, S. M. & Huberman, J. A. (1998) Mol. Cell. Biol. 18, 7294-7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segurado, M., de Luis, A. & Antequera, F. (2003) EMBO Rep. 4, 1048-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, S. M., Zhang, D. Y. & Huberman, J. A. (2001) BMC Mol Biol., 10.1186/1471–2199-2–1. [DOI] [PMC free article] [PubMed]
- 19.Smith, J. G., Caddle, M. S., Bulboaca, G. H., Wohlgemuth, J. G., Baum, M., Clarke, L. & Calos, M. P. (1995) Mol. Cell. Biol. 15, 5165-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wohlgemuth, J. G., Bulboaca, G. H., Moghadam, M., Caddle, M. S. & Calos, M. P. (1994) Mol. Biol. Cell 5, 839-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okuno, Y., Okazaki, T. & Masukata, H. (1997) Nucleic Acids Res. 25, 530-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood, V., Gwilliam, R., Rajandream, M. A., Lyne, M., Lyne, R., Stewart, A., Sgouros, J., Peat, N., Hayles, J., Baker, S., et al. (2002) Nature 415, 871-880. [DOI] [PubMed] [Google Scholar]
- 23.Sikorski, R. S. & Hieter, P. (1989) Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fryxell, K. J. & Zuckerkandl, E. (2000) Mol. Biol. Evol. 17, 1371-1383. [DOI] [PubMed] [Google Scholar]
- 25.Sueoka, N. (2002) Gene 300, 141-154. [DOI] [PubMed] [Google Scholar]
- 26.Munoz, M. J., Daga, R. R., Garzon, A., Thode, G. & Jimenez, J. (2002) Mol. Genet. Genomics 267, 792-796. [DOI] [PubMed] [Google Scholar]
- 27.Humphrey, T., Birse, C. E. & Proudfoot, N. J. (1994) EMBO J. 13, 2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birse, C. E., Lee, B. A., Hansen, K. & Proudfoot, N. J. (1997) EMBO J. 16, 3633-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patrikakis, M., Izant, J. G. & Atkins, D. (1996) Curr. Genet. 30, 151-158. [DOI] [PubMed] [Google Scholar]
- 30.Hansen, K., Birse, C. E. & Proudfoot, N. J. (1998) EMBO J. 17, 3066-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu, D. K., Lin, K. & Zhang, D. Y. (2003) J. Mol. Evol. 57, 325-334. [DOI] [PubMed] [Google Scholar]
- 32.Beletskii, A. & Bhagwat, A. S. (1996) Proc. Natl. Acad. Sci. USA 93, 13919-13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francino, M. P. & Ochman, H. (2001) Mol. Biol. Evol. 18, 1147-1150. [DOI] [PubMed] [Google Scholar]
- 34.Oller, A. R., Fijalkowska, I. J., Dunn, R. L. & Schaaper, R. M. (1992) Proc. Natl. Acad. Sci. USA 89, 11036-11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanawalt, P. C. (1995) Mutat. Res. 336, 101-113. [DOI] [PubMed] [Google Scholar]
- 36.Green, P., Ewing, B., Miller, W., Thomas, P. J. & Green, E. D. (2003) Nat. Genet. 33, 514-517. [DOI] [PubMed] [Google Scholar]
- 37.Tautz, D. & Schlotterer (1994) Curr. Opin. Genet. Dev. 4, 832-837. [DOI] [PubMed] [Google Scholar]
- 38.Dubey, D. D., Kim, S. M., Todorov, I. T. & Huberman, J. A. (1996) Curr. Biol. 6, 467-473. [DOI] [PubMed] [Google Scholar]
- 39.Clyne, R. K. & Kelly, T. J. (1997) Methods 13, 221-233. [DOI] [PubMed] [Google Scholar]
- 40.Chuang, R. Y. & Kelly, T. J. (1999) Proc. Natl. Acad. Sci. USA 96, 2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuang, R. Y., Chretien, L., Dai, J. & Kelly, T. J. (2002) J. Biol. Chem. 277, 16920-16927. [DOI] [PubMed] [Google Scholar]
- 42.Lee, J. K., Moon, K. Y., Jiang, Y. & Hurwitz, J. (2001) Proc. Natl. Acad. Sci. USA 98, 13589-13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong, D. & DePamphilis, M. L. (2001) Mol. Cell. Biol. 21, 8095-8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reeves, R. (2001) Gene 277, 63-81. [DOI] [PubMed] [Google Scholar]
- 45.Maher, J. F. & Nathans, D. (1996) Proc. Natl. Acad. Sci. USA 93, 6716-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi, T., Ohara, E., Nishitani, H. & Masukata, H. (2003) EMBO J. 22, 964-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dijkwel, P. A., Vaughn, J. P. & Hamlin, J. L. (1994) Nucleic Acids Res. 22, 4989-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dijkwel, P. A., Mesner, L. D., Levenson, V. V., d'Anna, J. & Hamlin, J. L. (2000) Exp. Cell Res. 256, 150-157. [DOI] [PubMed] [Google Scholar]
- 49.Vashee, S., Cvetic, C., Lu, W., Simancek, P., Kelly, T. J. & Walter, J. C. (2003) Genes Dev. 17, 1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilbert, D. M. (2001) Science 294, 96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.