Significance
The enzyme RNA polymerase (RNAP) transcribes DNA genetic information into RNA. Regulation of transcription occurs largely in initiation; these regulatory mechanisms must be understood. Lifetimes of transcription-capable RNAP-promoter open complexes (OCs) vary greatly, dictated largely by the DNA discriminator region, but the significance of OC lifetime for regulation was unknown. We observe that a significantly longer RNA:DNA hybrid is synthesized before RNAP escapes from long-lived λPR-promoter OCs as compared with shorter-lived T7A1 promoter OCs. We quantify the free energy needed to overcome OC stability and allow escape from the promoter and elongation of the nascent RNA, and thereby predict escape points for ribosomal (rrnB P1) and lacUV5 promoters. Longer-lived OCs produce longer abortive RNAs, which likely have specific regulatory roles.
Keywords: RNA polymerase, open complex lifetime, transcription initiation, abortive RNA, hybrid length
Abstract
To investigate roles of the discriminator and open complex (OC) lifetime in transcription initiation by Escherichia coli RNA polymerase (RNAP; α2ββ’ωσ70), we compare productive and abortive initiation rates, short RNA distributions, and OC lifetime for the λPR and T7A1 promoters and variants with exchanged discriminators, all with the same transcribed region. The discriminator determines the OC lifetime of these promoters. Permanganate reactivity of thymines reveals that strand backbones in open regions of long-lived λPR-discriminator OCs are much more tightly held than for shorter-lived T7A1-discriminator OCs. Initiation from these OCs exhibits two kinetic phases and at least two subpopulations of ternary complexes. Long RNA synthesis (constrained to be single round) occurs only in the initial phase (<10 s), at similar rates for all promoters. Less than half of OCs synthesize a full-length RNA; the majority stall after synthesizing a short RNA. Most abortive cycling occurs in the slower phase (>10 s), when stalled complexes release their short RNA and make another without escaping. In both kinetic phases, significant amounts of 8-nt and 10-nt transcripts are produced by longer-lived, λPR-discriminator OCs, whereas no RNA longer than 7 nt is produced by shorter-lived T7A1-discriminator OCs. These observations and the lack of abortive RNA in initiation from short-lived ribosomal promoter OCs are well described by a quantitative model in which ∼1.0 kcal/mol of scrunching free energy is generated per translocation step of RNA synthesis to overcome OC stability and drive escape. The different length-distributions of abortive RNAs released from OCs with different lifetimes likely play regulatory roles.
Many facets of transcription initiation by E. coli RNA polymerase (RNAP; α2ββ′ωσ70) have been elucidated, but significant questions remain about the mechanism or mechanisms by which initial transcribing complexes (ITC) with a short RNA–DNA hybrid decide to advance and escape from the promoter to enter elongation mode, or, alternately, to stall, release their short RNA, and reinitiate (abortive cycling). For RNAP to escape, its sequence-specific interactions with promoter DNA in the binary open complex (OC) must be overcome.
The open regions of promoter DNA in the binary OC are the −10 region (six residues, with specific interactions between σ2.2 and the nontemplate strand), the discriminator region (typically six to eight residues with no consensus sequence, the upstream end of which interacts with σ1.2), and the transcription start site (TSS, +1) and adjacent residue (+2), which are in the active site of RNAP (Table 1). The interactions involving and directed by the six-residue λPR discriminator make its OC long-lived and highly stable (1). A six-residue discriminator allows the OC to form without deforming (prescrunching) either open discriminator strand (2). Less extensive interactions involving and directed by the seven-residue T7A1 discriminator, together with the need to prescrunch one base of each strand, make its OC shorter lived and only moderately stable (1). The eight-residue discriminator of the ribosomal promoter rrnB P1 requires prescrunching of two residues to make an OC with the observed TSS (2); this OC is very unstable and short-lived. The instability of the rrnB P1 OC is the basis of the regulation of initiation at this promoter by concentrations of initiating nucleotides, the feedback ligand guanosine tetraphosphate (ppGpp), and protein factors such as DksA (3). A single base change (C–7G) improves the interaction of the rrnB P1 discriminator with σ1.2, shifts the TSS, greatly increases OC lifetime and stability, and eliminates regulation by these ligands (3, 4). The network of interactions directed by discriminator DNA, which determines OC lifetime, has been characterized (1). OC lifetimes vary greatly, but the impact of lifetime on initiation from stable OC was unknown.
Table 1.
Sequences of open regions and ITRs of promoter variants
| Promoter (discriminator) | −10 region | Discriminator | TSS (+1) | Transcription ITR |
| λPR(λPR) | GATAAT | GGTTGC | A | TGT AGT AAG GAG GTT CT… |
| T7A1(λPR) | GATACT | GGTTGC | A | TGT AGT AAG GAG GTT CT… |
| T7A1(T7A1) | GATACT | TACAGCC | A | TGT AGT AAG GAG GTT CT… |
| λPR(T7A1) | GATAAT | TACAGCC | A | TGT AGT AAG GAG GTT CT… |
| rrnB P1(λPR) | TATAAT | GGTTGC | A | TGT AGT AAG GAG GTT CT… |
| rrnB P1(T7A1) | TATAAT | TACAGCC | A | TGT AGT AAG GAG GTT CT… |
Promoter variant sequences are listed for the nontemplate strand. The sequence of the −10 region to approximately −60 is that of the promoter designated. The ITR used in transcription experiments (transcription ITR) is patterned after the λPR ITR with modified bases (underlined) that result in a stop at position +16 when CTP is withheld.
What drives promoter escape? Escape involves disrupting all the favorable interactions involved in forming and stabilizing the binary OC as well as –core interactions. Escape from these interactions is fundamentally driven by the favorable chemical (free) energy change of RNA synthesis, but this energy must be stored in the ITC in each step before escape. Proposed means of energy storage as the length of the RNA–DNA hybrid increases include the stresses introduced by scrunching distortions of the discriminator regions of the open strands in the cleft (2, 5, 6) and by unfavorable interactions of the RNA–DNA hybrid with the hairpin loop of 3.2 (7–10). Scrunching of the discriminator region of the template strand is proposed to be most significant for stress buildup in the initial transcribing complex (11). Here, we test the hypothesis that this accumulated stress drives promoter escape at the point where it overcomes the RNAP–promoter interactions responsible for stability and lifetime of the binary OC.
Amounts and/or rates of productive and abortive initiation have been studied using a variety of different assays and well-characterized promoters and variants including λPR, λPR′, and λPL and lacUV5, T7A1, T5N25, and rrnB P1 (12–19). In multiple-round assays, amounts of short and long RNAs are determined at a fixed time or as a function of time under conditions in which, after dissociation from the template, RNAP can rebind at the promoter and reinitiate RNA synthesis (19–21). In other cases, including the present study, the time courses of synthesis of both short and long RNA have been determined under conditions in which long RNA synthesis is constrained to be single round (16, 22–25).
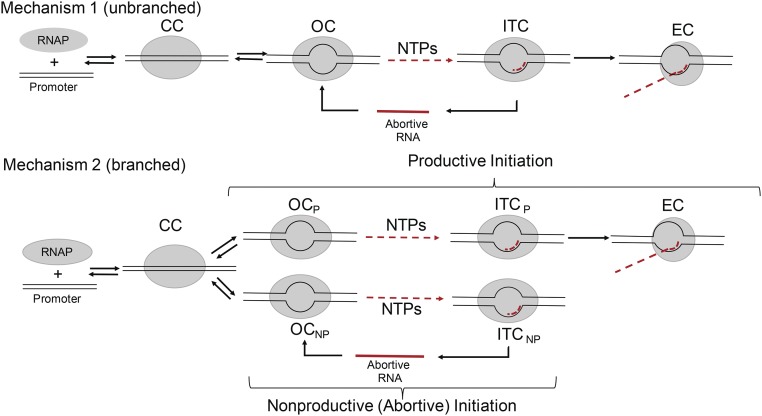
Many previous studies of initiation by Escherichia coli RNAP have been discussed using unbranched mechanism 1 (Fig. 1) (15, 26, 27), in which all binary OCs carry out both abortive and productive initiation. Short RNA–DNA hybrids (RNA lengths <6-mer) in ITCs are proposed to be unstable (12, 15). In this mechanism, dissociation of short RNAs from these hybrids is proposed to be rapid, followed by reinitiation, which can lead to promoter escape and long RNA synthesis or to abortive cycling. Multiple cycles of abortive synthesis have been proposed to occur in the process of breaking contacts between RNAP and DNA.
Fig. 1.
Proposed mechanisms of transcription initiation. In unbranched mechanism 1, abortive synthesis is proposed to occur on the pathway to promoter escape. Branched mechanism 2 proposes two classes of initiating complexes: productive complexes (subscripted P) that escape from the promoter without releasing any short RNA, and nonproductive (abortive) complexes (subscripted NP) that cannot escape and only synthesize and release short RNA. CC, closed complex; EC, escaped (elongation) complex; ITC, initial transcribing complex; OC, open complex; R, RNAP.
An alternative, branched mechanism of initiation (mechanism 2 of Fig. 1) was proposed based on kinetic studies at the T7A1 and λPR promoters under conditions in which long RNA synthesis is single round (25). Only a minority (∼20%) of OCs were found to be capable of promoter escape and productive initiation (10, 16, 25, 28). This subpopulation of complexes escapes and synthesizes a long RNA without any short (abortive) RNA release. Indeed, in initiation from poly dAT, the rate of synthesis and release of 3-mer RNA (the shortest, presumably least stably held RNA) is much slower than the rate of synthesis of full-length RNA (23). Other OCs (termed “moribund OCs”) synthesize, slowly release, and resynthesize short RNA in an abortive cycle without escaping to make a long RNA (24, 25). In mechanism 2, abortive cycling is not on the pathway to productive initiation. Studies that have been interpreted using mechanism 1 do not exclude mechanism 2.
To address the above energetic and mechanistic questions regarding initiation, we compare initiation rates, RNA transcript distributions, and properties of binary OCs (lifetime, thymine reactivity) for a set of four promoters with different combinations of T7A1 and λPR upstream recognition sequences and discriminator sequences and lengths (the 7-bp T7A1 discriminator and the 6-bp λPR discriminator), all with the same TSS and with λPR-based initial transcribed regions (ITRs) (Table 1). We also characterize two phases of initiation (initial and slow phases) and two subsets of initiating complexes (productive and nonproductive) at these widely studied promoters and their discriminator variants. We obtain substantial kinetic–mechanistic evidence for initiation mechanism 2 and for the role of the discriminator sequence in determining the RNA–DNA hybrid length at which promoter escape occurs. A free-energy analysis of promoter escape is applied to quantify the opposing roles of binary OC stability and stress build-up from scrunching. We conclude that these two factors determine the RNA–DNA hybrid length for promoter escape and the length distribution of the short RNAs produced in abortive initiation.
Results
WT and Hybrid Promoters Investigated.
λPR, T7A1, and rrnB P1 promoters were synthesized with the WT promoter sequence from −60 to +1, including the UP element and the −35, spacer, −10, and discriminator regions (SI Appendix, Table S1). For this study of the effects of the discriminator region on properties of the OC including lifetime, MnO4− reactivity, and transcription initiation and hybrid promoters were created by the interchange of discriminator regions as described in Materials and Methods. All WT and hybrid promoters are designated as promoter (discriminator); for example, λPR(T7A1) is the λPR promoter variant with the T7A1 discriminator replacing the λPR discriminator. Sequences of the open regions (−10 element, discriminator, start site region) and ITR of the promoter fragments used in this research are given in Table 1. All promoters have the same start site (+1), base (template strand T), and the same ITR patterned after λPR, with the indicated sequence changes to stop transcription after synthesis of a 16-mer RNA when only ATP, GTP, and UTP are provided. In the absence of CTP, a unique TSS is ensured even for OCs of promoters with prescrunched discriminators, because CTP would be required to initiate transcription upstream of +1.
Lifetimes of Stable OCs with λPR and T7A1 Discriminators.
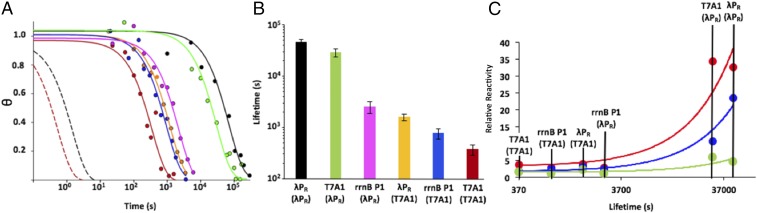
The kinetics of irreversible dissociation of 32P-labeled OCs of λPR, T7A1, and rrnB P1 (ribosomal) promoters with λPR or T7A1 discriminators, determined by the filter-binding assay after the addition of unlabeled λPR + UP promoter DNA as competitor, are shown in Fig. 2A in which the fraction of OCs remaining (θ) is plotted vs. log t (time). Because the range of dissociation rates is so wide, the log time scale is needed to compare the different promoters. The kinetics of dissociation are first-order. Analysis yields dissociation rate constants (kd) for the overall process of reversibly converting the stable OC to the initial OC and subsequent DNA closing. DNA closing is irreversible because closed complexes dissociate rapidly and free RNAP is trapped by the unlabeled competitor.
Fig. 2.
Lifetimes and permanganate reactivities of thymines of OCs formed by discriminator variants. (A) The fraction (θ) of OC remaining at 37 °C as a function of time (log scale) after the addition of an inert competitor for λPR(λPR) (black), T7A1(λPR) (green), rrnBP1(λPR) (magenta), λPR(T7A1) (orange), rrnBP1(T7A1) (blue), and T7A1(T7A1) (red) promoters. For comparison, dissociation kinetics are shown for initial (unstable) OC intermediates at λPR and T7A1 promoters [black and red dashed lines, respectively (29, 60)]. (B) Comparison of OC lifetimes [1/kd on a log scale) (SI Appendix, Table S4) of discriminator variants investigated here. (C) Plot of permanganate reactivities (SI Appendix, Figs. S2 and S4) vs. OC lifetime for the template-strand Ts present in all promoter variants (−12/−11, red; −10/−9, blue; +1, green). Reactivities are normalized by that of +1 T of rrnBP1(T7A1), the least reactive of these Ts. All results are reported as the average of multiple replications ± standard error.
The lifetimes (τ) of stable OCs (τ = 1/kd) at these promoters, shown in the bar graph of Fig. 2B (see also SI Appendix, Table S4), span a range of 120-fold [from 12.6 h for λPR (λPR) to 6.2 min for T7A1(T7A1)]. Fig. 2B shows that the rank order of lifetimes is λPR (λPR) > T7A1(λPR) >> rrnB P1 (λPR) > λPR (T7A1) > rrnB P1(T7A1) > T7A1(T7A1). T7A1 and λPR promoters with the λPR discriminator are much longer lived than their counterparts with the T7A1 discriminator, and all three promoters with the λPR discriminator are longer lived than even the longest lived variant with the T7A1 discriminator.
Comparison of lifetimes of λPR (λPR) and T7A1(T7A1) with λPR(T7A1) and T7A1(λPR) reveals that the discriminator is the major determinant of the lifetime differences among these four promoters. Looked at another way, the effects of changing the upstream promoter sequence in the context of any of these discriminators are much smaller than the effects of changing the discriminator in the context of any of these upstream promoter sequences. The lifetime of λPR (λPR) is 120-fold greater than that of T7A1(T7A1). Conversion of T7A1(T7A1) to T7A1(λPR) increases OC lifetime ∼80-fold, to within a factor of two of that of λPR (λPR). Conversion of λPR (λPR) to the λPR(T7A1) hybrid reduces OC lifetime by ∼30-fold, only approximately fourfold above that of T7A1(T7A1).
The lifetime of the rrnB P1(rrnB P1) OC under these conditions could not be determined because it is too unstable to populate, but it has been estimated to be no more than ∼1 s (1). This characteristic short lifetime, previously shown to result from its discriminator (3), is transferrable. We find that the λPR(rrnB P1) promoter also does not form a stable OC. On the other hand, replacement of the rrnB P1(rrnB P1) discriminator by either the T7A1 or λPR discriminator increases OC stability greatly. Conversion to rrnB P1(T7A1) increases lifetime to ∼770 s (an ∼770-fold increase), which exceeds the lifetime of T7A1(T7A1). The fold increase in OC lifetime from converting rrnB P1(rrnB P1) to rrnB P1(λPR) is similar to that observed for converting T7A1(T7A1) to T7A1(λPR), although the lifetime of rrnB P1(λPR) is only a small fraction (6%) of that of λPR(λPR).
Accessibility of Thymines in OCs with λPR and T7A1 Discriminators.
Gel lanes in SI Appendix, Fig. S2 show permanganate (MnO4−) footprints (34) of template-strand thymines (T) in OCs formed by λPR, T7A1, and rrnB P1 promoter variants. All experiments were performed at the same MnO4− dose and conditions. Relative MnO4− reactivities of T bases in −10 and discriminator regions and at the +1 start site are quantified by phosphorimager analysis as described in Materials and Methods, using a common block of downstream background bands in the ITR (+5 to +24; outlined in red in SI Appendix, Fig. S2) as normalization. Differences in the −10 region and discriminator sequence result in a range of three to six template-strand T residues. MnO4− reactivities of nontemplate-strand thymines of λPR and T7A1 promoters and discriminator variants are shown in SI Appendix, Fig. S3.
Together, these studies reveal that the open region extends from −11 to +2 for promoters with the λPR discriminator and from −12 to +2 for promoters with the T7A1 discriminator. All template-strand T bases in these open regions are MnO4− reactive, but nontemplate-strand T bases at −7 and −10 of λPR(λPR) and T7A1(λPR) and nontemplate-strand T bases at −8T and −11T of T7A1(T7A1) and λPR(T7A1), are unreactive. The lack of permanganate reactivity for −7T(λPR)/−8T(T7A1) is expected; this highly conserved T is bound in a pocket of σ2.2 (11, 37, 38). Although no similar binding pocket is evident for −10T(λPR)/−11T(T7A1), this base is as fully protected from attack by permanganate as −7T(λPR)/−8T(T7A1).
All T bases in the open region of the template strand are permanganate-reactive, but reactivity varies widely. Relative reactivities of all template strand Ts are compared in the bar graph of SI Appendix, Fig. S4. All reactivities are expressed relative to that of the start site +1 T of rrnB P1(T7A1), which reproducibly is the least reactive T. From SI Appendix, Figs. S3 and S4, two general trends are observed. (i) T bases in the −10 region of each promoter are more reactive than those in the discriminator region and the start site +1 T. Differences in reactivity between the −10 region and downstream regions are most pronounced for T7A1(λPR) and λPR (λPR). (ii) At all positions, the T bases of T7A1(λPR) and λPR(λPR) promoters are much (three- to 10-fold) more reactive and hence more solvent-accessible than their counterparts in OCs formed with the other four promoters.
T7A1(λPR) and λPR(λPR) promoter OCs are much longer lived than those at the other four promoters (Fig. 2B; also see SI Appendix, Table S4). Fig. 2C compares MnO4− reactivity and OC lifetimes for this set of promoters and reinforces previous observations that T bases in the open region of long-lived, stable OCs are more accessible to attack by MnO4− than T bases in unstable OCs because tight binding of the strand backbone results in increased unstacking of T bases (1, 35).
Initiation from OCs with λPR and T7A1 Discriminators.
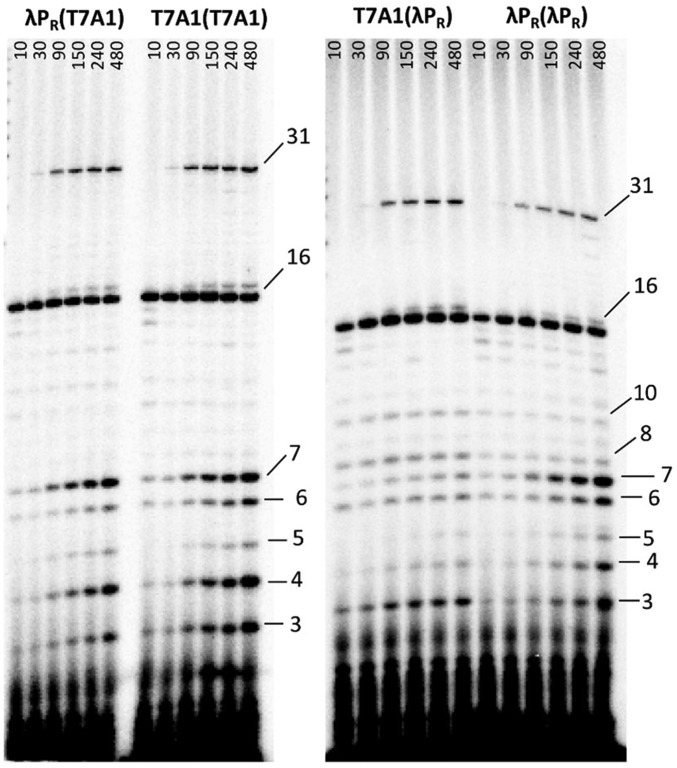
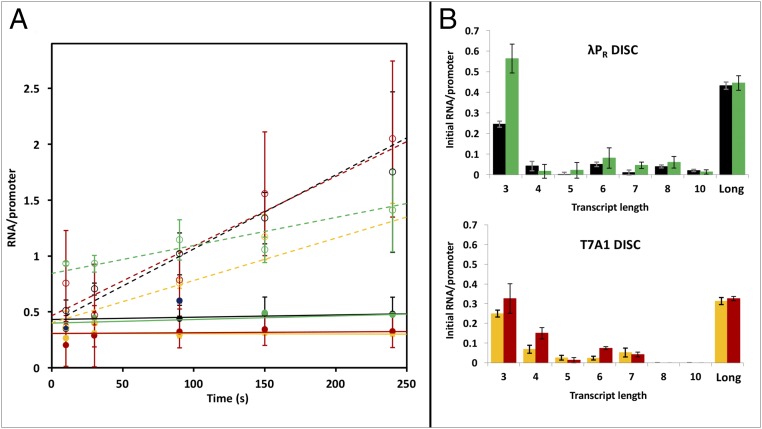
Transcription assays in which long RNA synthesis is single-round were performed with λPR(λPR), T7A1(T7A1), λPR(T7A1), and T7A1(λPR) promoters at 37 °C to examine the roles of these discriminator regions in determining (i) the fraction of RNAP-promoter OCs capable of promoter escape, (ii) the time courses of synthesis of short RNA products and of promoter escape, and (iii) the population distributions of short vs. long RNAs at different times. Representative phosphorimager scans of transcription gels as a function of time after the addition of NTP to preformed OCs with these four promoters are shown in Fig. 3. Total amounts of short (3–10 nt) and long (>10 nt) RNA synthesized per OC as a function of time are plotted in Fig. 4A. From this plot, two kinetic phases of RNA synthesis are observed. An initial (burst) phase of rapid RNA synthesis, complete by 10 s after NTP addition, is followed by a much slower, linear (steady-state) phase extending at least to 480 s. From the initial phase (t = 0 intercept) values for long and short RNA synthesis at each promoter, we observe that approximately one RNA is rapidly synthesized per OC in this initial phase [0.7 0.2 (λPR(T7A1), 0.8 0.2 (λPR(λPR), 0.9 0.2 T7A1(T7A1), and 1.3 0.4 (T7A1(λPR)]. Individual amounts of each RNA length synthesized in this initial (fast) phase are shown on the bar graphs in Fig. 4B.
Fig. 3.
Time courses of synthesis of short (3- to 10-mer) and long (16- or 31-mer) RNAs from λPR(T7A1), T7A1(T7A1), T7A1(λPR), and λPR(λPR) promoter(discriminator) constructs. RNA products are identified by size and were quantified from acrylamide gels over the time course (10–480 s) of transcription initiation from each promoter after the addition of ATP, GTP, and limiting UTP (+ α-32P-UTP).
Fig. 4.
Synthesis of short and long RNA in two phases of transcription initiation. (A) Short (≤10-mer) and long (>10-mer) RNA transcripts synthesized per OC are plotted vs. time for λPR(λPR) (black), T7A1(λPR) (green), λPR(T7A1) (orange), and T7A1(T7A1) (red) promoters. Plotted points are the averages of two to four experiments (± standard error) like that shown in Fig. 3. (B) The bar graphs show intercepts from extrapolation of the linear (steady-state) regions of A (and individual RNA lengths in SI Appendix, Fig. S6). These intercepts represent the number of short (dashed lines, open circles in A) and long (solid lines, filled circles in A) RNA synthesized in the initial phase (t <10 s) of transcription initiation. Determinations at 480 s fall on these lines and are used in the fits but are omitted to show the results to 240 s more clearly. Color coding in B is the same as in A. DISC, discriminator.
Long RNA Synthesis in the Initial Kinetic Phase of Initiation.
Strikingly, Fig. 4A reveals that all promoter escape and long RNA synthesis occurs rapidly in the initial kinetic phase. Long RNA is synthesized by ∼40–45% of the OCs at the two promoters with the λPR discriminator and by 30–35% of the OCs at the two promoters with the T7A1 discriminator. The rest of the OC population stalls after rapid synthesis of a short RNA.
The Major Short RNA That Accumulates Initially Is the 3-mer.
Amounts of short RNAs that are rapidly synthesized and accumulate in the initial phase of initiation from these four promoters, obtained from extrapolation of linear plots analogous to Fig. 4A (SI Appendix, Fig. S6) to t = 0, are compared in the bar graphs of Fig. 4B. For all four promoters, the major short RNA that accumulates initially is the 3-mer. The fraction of OCs that stall after synthesis of a 3-mer in the initial phase is similar to the fraction of OCs that successfully synthesize a full-length RNA. Initial levels of accumulation of most other short RNAs are small by comparison. Because the amount of long RNA does not increase after the initial phase (Fig. 4A), it is unlikely that many, if any, of the short RNAs that accumulate initially are intermediates in full-length RNA synthesis.
Discriminator Effects on the Initial Pattern of Short RNA Accumulation.
Fig. 4B reveals a major and very significant discriminator-specific difference in the initial distributions of short RNA. Significant levels of 8-mer and 10-mer RNAs accumulate in initiation from the more stable OCs at promoters with the λPR discriminator, whereas no short RNA longer than a 7-mer accumulates in initiation from the two less stable OCs at promoters with the T7A1 discriminator.
Abortive RNA Synthesis in the Slow Kinetic Phase of Initiation.
Fig. 4A reveals that no net synthesis of long RNA occurs in the slower kinetic phase of initiation (t >10 s) for any of the four promoters investigated. Some conversion of 16-mer to 31-mer (the next C-stop) is observed in this phase (Fig. 3 and SI Appendix, Fig. S5), at a rate consistent with misincorporation of NTP when a subset of NTPs are present (39).
Short RNA synthesis in the second kinetic phase of initiation is reiterative (abortive) and is much slower than in the initial phase. The bar graphs in Fig. 5 compare RNA synthesis rates in this phase for the subclass of OCs that are engaged in abortive synthesis at the four promoters. Individual abortive synthesis rates range from 0.1–2.8 RNAs per abortively synthesizing OC per 1,000 s. Overall abortive rates range from six to seven RNAs per abortively synthesizing OC per 1,000 s for λPR(λPR) and T7A1(T7A1) to four and 2.5 RNA per abortively synthesizing OC per 1,000 s for λPR(T7A1) and T7A1(λPR), respectively. In all cases, the rates are much slower than the rates of synthesis of the first short RNA at these complexes in the initial phase (one RNA synthesized per OC in less than 10 s). This profound difference in the kinetics of short RNA synthesis in the two phases indicates that the release of short RNA from the hybrid determines the rate of subsequent rounds of abortive RNA synthesis.
Fig. 5.
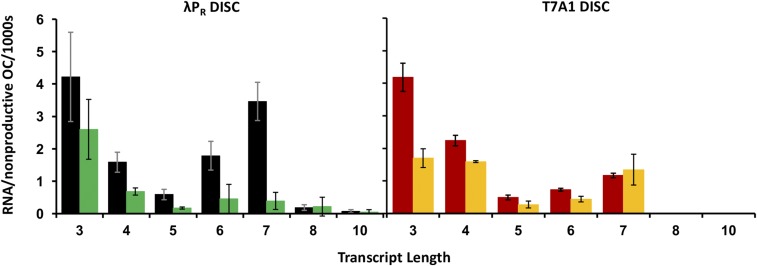
Rates of synthesis of short RNAs per nonproductive (i.e., abortively initiating) OC in the slower phase of transcription initiation. Bar graphs summarize steady-state synthesis rates for all short RNAs synthesized in the linear (slow, abortive) phase from promoters with λPR (Left) and T7A1 (Right) discriminators (DISC). Rates are the averages obtained from best-fit slopes of the linear (steady-state) regions of two to four kinetics experiments ± standard error (see example in SI Appendix, Fig. S6 and Table S6) and are expressed as the number of RNAs synthesized per nonproductive promoter OC per 1,000 s. Color-coding is the same as in Fig. 4.
Discriminator Effects on the Slow-Phase Pattern of Abortive RNAs.
A comparison of the panels of Fig. 5 reveals the same discriminator-specific difference in the distributions of short RNAs that accumulate in the slow phase of initiation as observed in the initial phase. Significant amounts of 8-mer and 10-mer RNAs accumulate in both the initial and slow kinetic phases of initiation at the two promoters with the λPR discriminator, whereas no RNA longer than a 7-mer accumulates in initiation at the two promoters with the T7A1 discriminator.
For the shorter (3-mer to 7-mer) abortive RNAs synthesized at all four promoters, the rates of synthesis show qualitatively similar variations with RNA length. From 3-mer to 5-mer, the abortive synthesis rate decreases strongly, as might be expected from the increasing stability of the hybrid as its length increases (15). But, the abortive rate then increases for the 6-mer. For all but T7A1(λPR), the abortive rate for the 7-mer exceeds that of the 6-mer, and for λPR(λPR) the abortive rate for the 7-mer is comparable to that of the 3-mer.
Shift in Length Distribution of Abortive RNAs in the Slow Phase.
A comparison of the distributions of RNA length shown in Fig. 4B with the rates shown in Fig. 5 reveals a shift to longer RNAs in the slow phase compared with the initial phase. Although the 3-mer remains a major product of abortive synthesis for all four promoter OCs, the fraction of 3-mers is reduced significantly, and fractions of most larger RNAs increase in the slow phase. Figs. 4B and 5 show that the details of these patterns are quite promoter-specific, as are the details of the shifts in pattern from the initial phase to the slow phase. It is noteworthy that this shift in the population distribution to longer RNAs in the slow phase is not accompanied by any promoter escape and long RNA synthesis.
Discussion
Discriminator Effects on OC Lifetime and Conformation.
Discriminator sequence and length dictate a series of in-cleft and downstream conformational changes and interactions that convert the initial open intermediate to a stable OC (1, 40). Rates of closing the initial unstable OC are found to be similar for the different promoters and RNAP variants studied to date (1), and therefore the extent of stabilization of the initial open intermediate largely determines OC lifetime. These lifetimes span at least five orders of magnitude.
Because the WT rrnB P1(rrnB P1) promoter forms a stable OC only on negatively supercoiled DNA or with high concentrations of the two initiating NTPs present (41–43), its lifetime on linear DNA is not well known; under the conditions investigated here, it is estimated to be ≤1 s, by analogy with the lifetimes of unstable initial OCs at the T7A1(T7A1) and λPR(λPR) promoters (Fig. 2A, red and black dashed lines) (1, 44). The OC formed at the hybrid λPR (rrnB P1) promoter is also unstable under the conditions investigated here.
Based on this analysis and the lifetime data in Fig. 2, we propose that the T7A1(λPR) hybrid promoter forms an OC structurally similar to that of λPR(λPR). By contrast, promoters containing the T7A1 discriminator are structurally similar to the T7A1 WT OC, in which conformational changes within RNAP and DNA do not stabilize the OC to the same extent as λPR(λPR) (1, 29). We also propose that both the rrnB P1 discriminator and an additional upstream feature that distinguishes rrnB P1(rrnB P1) from T7A1(T7A1) and λPR(λPR) may be responsible for the instability and short lifetime of its OC. A likely candidate is the difference in spacer length between the −35 and −10 regions [16 bp for rrnB P1(rrnB P1) vs.17 bp for T7A1(T7A1) and λPR(λPR)]. Changes in spacer length affect OC formation kinetics and transcription levels in vitro (45, 46) and could explain why introduction of the λPR discriminator has a similar fold effect on lifetime in the context of rrnB P1 and T7A1 but why the resulting promoter lifetime is comparable to that of λPR(λPR) only in the context of T7A1.
Structural analysis revealed that the strands of a six-base discriminator are bound without scrunching and that the additional bases of longer discriminators are accommodated by prescrunching of the discriminator so that interactions of the −10 region are the same for promoters with different discriminator lengths (47). Hence positions −12 to −8 of promoters with the T7A1 discriminator are expected to be positioned in the upstream cleft similarly to positions −11 to −7 of promoters with the λPR discriminator. This expectation is confirmed by the permanganate reactivity data of SI Appendix, Fig. S3 that show complete protection of −7T and −10T on the nontemplate strand of promoters with the λPR discriminator and protection of −8T and −11T of the nontemplate strand of promoters with the T7A1 discriminators. The relative reactivity, and therefore the accessibility of the template-strand +1 T is similar for all six promoters, providing further indication that the template strand is similarly positioned in the active site for transcription. Hence there is prescrunching of one base of the seven-base T7A1 discriminator region in the OC.
Subpopulations of Productive and Abortive Initiation Complexes.
Although all OCs initiate RNA synthesis, only 30–50% these OCs go on to escape and make a full-length RNA. Almost all full-length RNA synthesis occurs in the initial phase of initiation. The 50–70% of complexes that stall in the initial phase make short RNAs ranging in size from 3-mer to 7-mer or 10-mer, depending on their discriminator. Why different initiating complexes stall at different RNA lengths is unclear. What is clear is that this subpopulation of stalled initiating complexes is unable to elongate and escape. Even after the release of the initially synthesized RNA, these complexes do not recover the ability to initiate synthesis productively. Instead they somehow are programmed to make only short RNA.
Previous ensemble and recent single-molecule studies of transcription initiation at the λPR, T7A1, and lacUV5 promoters also observed multiple subpopulations of initiating complexes (Fig. 1, mechanism 2) (10, 12, 14, 24, 25, 48, 49). The subpopulation that stalls after producing a short RNA and that cannot elongate the RNA and escape was originally termed “moribund” (25, 50).
Moribund complexes are not the result of RNAP heterogeneity (i.e., active vs. inactive). OCs prepared using RNAP purified from productive complexes gave a similar distribution of productively initiating and moribund complexes according to mechanism 2 (25). This finding indicates that the heterogeneity in initiation is a property of the OC or ITC and not of the RNAP itself.
Taken together, these results and ours reported here show that abortive synthesis is not in any sense a precursor to promoter escape but rather is the consequence of a subpopulation of initiating complexes that are unable to elongate further and escape. This conclusion is obscured in assays in which long RNA synthesis occurs in multiple rounds, because such multiround assays cannot distinguish between mechanisms 1 and 2. Our findings support initiation mechanism 2, in which different subpopulations of OCs are responsible for synthesis of full-length and abortive RNA, and are inconsistent with mechanism 1. The factors that dictate which OCs can transcribe productively and which cannot are unknown. For the promoters and conditions investigated here, only a small number of cycles (fewer than three) of abortive initiation per promoter are observed in a 480-s assay.
Hybrid Length for Promoter Escape and Its Correlation with OC Lifetime.
Escape of RNAP from the T7A1 promoter with its natural discriminator and ITR was found to occur in the step in which the RNA is elongated from 7 to 8 nt (12, 13), and a 7-mer RNA is the longest short RNA synthesized from this WT T7A1 promoter (19). We find that the distribution of short RNAs ends with the 7-mer for both promoters with the T7A1 discriminator [T7A1(T7A1) and λPR (T7A1)] in both phases of initiation of RNA synthesis.
Escape of RNAP from the WT λPR promoter with its own discriminator and ITR (and also WT lacUV5) was found to occur at a hybrid length between 8 and 14 nt (13, 15). Consistent with this observation, we find that the distribution of short RNAs extends to 8-mers and 10-mers for both promoters with the λPR discriminator [λPR(λPR) and T7A1(λPR)] in both phases of initiation of RNA synthesis. In multiround assays with the WT λPR promoter the length distribution of short RNAs also ends at 8-mer and 10-mer (19). Particularly in the slow phase, when little additional promoter escape occurs, these RNAs cannot be intermediates in the synthesis of full-length RNA. Their presence indicates that RNAs of these lengths can be synthesized without triggering promoter escape.
The endpoint of the short RNA distribution is the most significant difference between the initiation patterns of the OCs of promoters with T7A1 and λPR discriminators. For these promoter variants, the discriminator, and not the upstream promoter sequence, determines the RNA hybrid length for escape of RNAP. The discriminator region also determines the OC lifetime (1/kd; see above). For the promoters studied, the hybrid length for promoter escape increases with increasing OC lifetime (Fig. 6; also see SI Appendix, Table S7) (51). A quantitative interpretation of this result in terms of scrunching is provided in the following section.
Fig. 6.
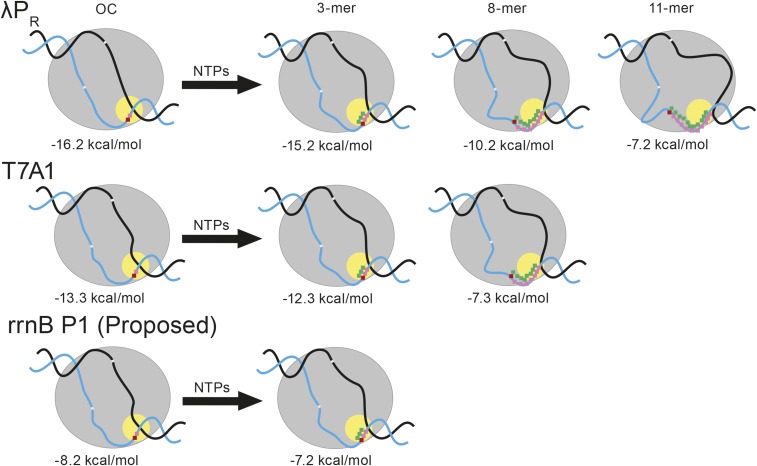
Scrunching in the binary OC and in steps of initiation leading to escape from λPR(λPR), T7A1(T7A1), and rrnB P1(rrnB P1) promoters. Models of binary OC at λPR(λPR) (no prescrunching), T7A1 (one prescrunched base), and rrnB P1 (two prescrunched bases) and their stabilities are shown at left. The template strand +1 (red) and +2 (pink) bases in the active site (yellow circle) are shown. The −10 and discriminator regions of the template strand in the λPR(λPR) binary OC are also indicated. Translocation in RNA synthesis moves the RNA (green)–DNA (pink) hybrid into the cleft, increasing the scrunching of the discriminator strands (and the scrunching of the downstream region of nontemplate strand). The helical curve of the hybrid is indicated; this region is thought not to be scrunched (see SI Appendix). The ITC for each promoter is shown after synthesis of (i) 3-mer RNA [proposed escape point of rrnB P1(rrnB P1)], (ii) 8-mer RNA [proposed escape point of T7A1(T7A1)], and (iii) 11-mer RNA [proposed escape point of λPR(λPR)]. Effects of increased scrunching on stability are indicated below each ITC.
Scrunching Free Energy Offsets OC Stability at the Escape Point.
Scrunching (compacting) of regions of the open strands and other stresses develop as the length of the RNA–DNA hybrid grows after initiation. These stresses allow the ITC to store some of the excess free energy [−3 kcal/mol (39)] of hydrolysis of the high-energy NTP phosphate bond and PPi release not expended in RNA synthesis. Here, we test the proposal that scrunching and other stresses reduce the stability of the ITC relative to the OC and thereby generate the driving force for promoter escape (5, 6).
The stability of the binary OC is quantified by the standard free-energy change for forming it from reactants (R, P): ∆GoR+P→OC =−RTlnKobs −RTln(ka/kd), where ka is the overall RNAP-promoter forward rate constant for OC formation and kd is the dissociation rate constant. The promoters selected for study here all have similar ka, but differ widely in kd (see SI Appendix, Table S7). Values of ∆GoR+P→OC for the λPR, T7A1, and lacUV5 promoters and estimates of ∆GoR+P→OC for rrnBP1 are listed in SI Appendix, Table S7. The stability difference ∆∆GoR+P→OC between OCs formed at T7A1 and λPR promoters is ∼2.9 kcal/mol at 37 °C.
Modeling scrunching as uniform bending of elastic rods (SI Appendix, Fig. S8) and applying elastic deformation analysis (SI Appendix, Free Energy Analysis of Scrunching of the Open Strands of Promoter DNA), we find that the free-energy change from scrunching the single-stranded regions of the template strand (discriminator region) and nontemplate strand (discriminator region and downstream region) is a linear function of the RNA–DNA hybrid length (NH, expressed in base pairs). At 37 °C, for a representative value of the stiffness of the open strands (persistence length of 14 Å, in the accepted range for ssDNA) and an axial distance between ssDNA residues of 4 Å (also in the accepted range and consistent with structural information for transcription initiation complexes), we predict Goscr = 1.0 NH (in kilocalories per mole). Given the different escape points for λPR and T7A1 at hybrid lengths of 11 bp and 8 bp, respectively, the difference in scrunching free energies at the different escape points would be ∆Goscr,ESC = 1.0∆NH,ESC = 3 kcal/mol, which would quantitatively compensate the stability difference between the binary OCs, ∆∆GoR+P→OC = 2.9 kcal/mol. Scrunching of the strands in the conversion of the binary OC to the ITC at the point of escape therefore is capable of offsetting the stability of the initial OC and driving escape. Fig. 6 shows our quantitative proposal for how scrunching reduces initial OC stability and drives promoter escape. At the different escape points, scrunching has reduced the originally different binary OC stabilities of λPR and T7A1 to a common value of −7.2 to −7.3 kcal/mol.
From this analysis, we propose that RNA–DNA hybrid lengths at escape (NH,ESC) can be predicted for other promoters from the stability of their binary OCs. Only a few test cases are available. For example, the stability of the lacUV5 OC (SI Appendix, Table S7) is found to be between that of λPR and T7A1. Hence we predict that NH,ESC, should be between 11 bp and 8 bp. Reported values are 9–10 bp (15). For a heteroduplex variant of the lac promoter, for which ∆GoR+P→OC must be much larger in magnitude, the point of escape appears to be shifted to a substantially larger RNA–DNA hybrid length (10), as expected from the above analysis.
The situation for the rrnB P1 promoter is interesting. On linear DNA at 37 °C, the binary rrnB P1 OC is short-lived and unstable, and productive initiation occurs without abortive synthesis (47). Qualitatively, comparison of this initiation behavior with that observed for stable OCs formed at the λPR, T7A1, and lacUV5 promoters and variants leads us to propose that promoter escape occurs at a short RNA–DNA hybrid length. Quantitative analysis supports this proposal; the estimated stability of the rrnB P1 OC (−8.2 ± 1 kcal/mol) is ∼8 kcal/mol less than that of λPR, resulting in a predicted NH,ESC of 3 ± 1 bp. The lack of abortive synthesis of a 3-mer at rrnB P1 supports the proposal that escape occurs in the initial translocation step (Fig. 6), which reduces the stability of the initiating complex to approximately −7.2 kcal/mol by the scrunching that occurs in this step, as shown in Fig. 6.
In Vivo Implications of OC Lifetime: Abortive RNA Lengths.
Why have promoter OCs evolved to have a 105-fold range of lifetimes? The thermodynamic instability and very short lifetime of the ribosomal rrnB P1 promoter OCs (at the lower end of this range of lifetimes) are the key to its regulation by initiating nucleotides, ppGpp, and DksA (3). These factors act on the unstable OCs, subsequent to initial binding and formation of the initial closed complex. Mutations in this promoter that increase its lifetime largely eliminate this regulation (3). Other rRNA and tRNA promoter OCs also are unstable and are regulated by the same ligands as rrnB P1(52-54). Presumably, initiation from these OCs also proceeds without abortive RNA synthesis. These regulatory mechanisms appear to be applicable only with short-lived, unstable OCs (3).
The very large differences in lifetime between different stable OCs, illustrated here by T7A1 and λPR discriminator variants, could in principle have evolved to regulate initiation directly by regulating the escape rate. This does not appear to be the case; escape from all these promoters occurs similarly rapidly (<10 s). Instead, we find that the primary difference between promoter OCs with very different lifetimes is in the range of the lengths of the abortive RNAs produced by the fraction of nonproductive ITC at the promoter. It is possible that late-acting E. coli regulatory factors analogous to bacteriophage T7gp2 (55, 56) exist that interact selectively with the different downstream structures of OCs with different lifetimes (1, 29) to regulate initiation, but none have been discovered. We therefore propose that the longer abortive RNAs produced by long-lived, stable promoter OCs play regulatory roles.
Very short RNA oligomers (2-mers to 4-mers) replace the initiating NTP, serve as primers of RNA synthesis, and play regulatory roles in vivo, shifting expression levels from promoters as much as 40-fold in either direction (57, 58). Longer oligomers also are capable of participating in initiation (e.g., an 8-mer DNA complementary to the template-strand discriminator region with an RNA base and triphosphate at the downstream end) (59). Length distributions and relative amounts of longer RNA oligomers produced in abortive initiation are promoter-specific, and relative amounts differ in the pre–steady-state and steady-state phases, as shown here (Figs. 4 and 5). These longer abortive RNAs may serve to communicate with other long-lived promoter OCs and ITCs. In vitro initiation kinetics and in vivo GFP expression studies using these and other promoter/discriminator combinations are in progress to test this hypothesis.
Materials and Methods
Reagents and General Procedures.
Reagents and solvents used in preparing buffers were the highest grade available and were used as received. dNTPs and NTPs (Thermo Fisher Scientific) used in PCR and transcription reactions were 99% pure and were used as received. All enzymes used in PCR reactions were purchased from New England Biolabs and were used according to the manufacturer’s protocols. All buffers and reagents were made using 18 MΩ water purified using an ultrafiltration system and filtered before use. Stocks of heparin (50 mg/mL), DTT (0.1 M), and BSA (50 mg/mL) were filtered and stored at −20 °C before use. Buffers and solutions used in filter-binding assays and permanganate footprinting are the same as used previously (SI Appendix) (1, 29). The transcription buffer (TB) was 40 mM Tris base (pH 8.0), 5 mM MgCl2, 1 mM DTT, 0.1 mg/mL BSA, and 60 mM KCl. The initiation solution (IS) for transcription assays was 1 mM GTP, 1 mM ATP, 200 µM UTP, 7 µCi α-32P-UTP (87.5 nM), and 0.25 mg/mL heparin in TB. The quench solution (QS) for transcription assays was 8 M urea, 0.015 M EDTA, 0.05% (wt/vol) xylene cyanol, and 0.05% (wt/vol) bromophenol blue in Tris/borate/EDTA buffer and was diluted 1:1 with the sample during quenching.
Overexpression and Purification of RNAP.
Overexpression and purification of RNA polymerase core enzyme was performed as previously described using E. coli BL21(DE3) transformed with pVS10 (30). Activities of different purified stocks of RNAP in OC formation [determined by filter binding (31)] ranged from 50 to 90%; all concentrations reported here are active concentrations.
Preparation of Promoter DNAs.
WT and variant promoter DNA templates were prepared by annealing two oligonucleotides with a 13-bp overlap filled in using Taq polymerase. The resulting template was extended with short primers (HTOP and HBOT) and was amplified by PCR to prepare ∼124-bp promoter fragments (extending from approximately −82 to +42). Sequences of the key regions of promoter fragments are given in Table 1, and the sequences of primers and templates used in this study are given in SI Appendix, Table S1. For filter-binding assays and permanganate footprinting experiments, HTOP or HBOT was kinase-labeled using T4 polynucleotide kinase and γ-[32P]ATP before mixing with the template DNA for extension and amplification (32).
Nitrocellulose Filter-Binding Dissociation Assays.
Dissociation kinetic assays were performed as previously described (29, 33). RNAP-promoter OCs were formed by incubating 2–6 nM RNAP for 1 h in binding buffer (BB) (see SI Appendix, General Procedures for composition) at 37 °C with γ-32P–radiolabeled promoter DNA (∼36,000 cpm). Irreversible dissociation was initiated by the addition of a 10-fold molar excess of a rapid- and tight-binding, unlabeled λPR promotor variant (λPR + UP) (29). Samples were filtered over nitrocellulose filters on a vacuum manifold, rinsed with wash buffer, and quantified using a Packard 1600TR Liquid Scintillation Counter. All filter-binding data were collected in triplicate and averaged.
Permanganate Footprinting.
OCs were formed by incubating 10–50 nM RNAP with enough radiolabeled promoter DNA to yield 70,000 counts (∼1 nM) for 1 h at 37 °C. Samples were challenged with 20 µg/mL heparin, reacted with 0.4 mM KMnO4 for 10 s, and quenched in a solution containing 1 M β-mercaptoethanol and 7.5 M NH4OAc (34). Promoter DNA was cleaved at sites of MnO4− reaction with 1 M piperidine for 30 min at 90 °C, and fragments were separated on an 8% acrylamide gel with negative control and (A + G) sequencing lanes (34–36). MnO4− gels were imaged and analyzed using a GE Typhoon FLA 9000 phosphorimager and GE ImageQuant software.
Single-Round Transcription Assays.
Promoter DNAs (10 nM final concentration) with the engineered λPR ITR (see SI Appendix, Table S1 for sequences) were incubated with a twofold excess of active RNAP for 1 h at 37 °C to form OCs. Transcription was initiated by the addition of IS to obtain final concentrations of 200 μM ATP and GTP, 10 μM UTP, plus 17.5 nM α-32P-UTP. Samples were quenched at the desired time points (10–480 s) with 15 mM EDTA loading dye containing 8 M urea, xylene cyanol, and bromophenol blue and were run on a 20% polyacrylamide gel to separate RNA products. Phosphorimaging screens were exposed to the gel for 18 h and then were imaged.
To quantify the mole amount of RNA in a gel band for each time point in an initiation experiment, a standard line with a concentration series of α-32P-UTP was run briefly on a 20% polyacrylamide gel and imaged. Areas of α-32P-UTP standard bands were determined with GE ImageQuant software and were plotted vs. the number of moles of α-32P-UTP loaded. The resulting linear calibration was used to obtain the mole amount of α-32P-UTP in each short and long RNA transcript from its gel band area. To calculate the mole amount of each RNA transcript, each mole amount was divided by the probability of incorporating an α-32P-UTP at each transcript length. Probabilities of α-32P-UTP incorporation were calculated using a binomial density function for transcripts containing one or more α-32P-UTP sites. The total moles of RNA in a gel band were divided by the volume loaded to obtain the RNA concentration in nanomoles, multiplied by the quench dye dilution factor, and divided by the OC concentration (10 nM) to obtain the number of RNA molecules synthesized per OC (RNA/OC) at that time. Plots of RNA/OC vs. time (as in SI Appendix, Fig. S6) were fitted linearly. Intercepts and slopes from two to four independent experiments were averaged to use in further analyses. The calibration curve, linear fit, and calculations are given in the figures, tables, and text of the SI Appendix. Steady-state rates of short RNA synthesis in the slow phase of initiation obtained from the slopes of these plots (RNA per OC per second) were renormalized by dividing by the fraction of OCs engaged in abortive initiation. These normalization fractions [5.7 nM λPR(λPR), 5.5 nM T7A1(λPR), 6.7 nM T7A1(T7A1), and 6.9 nM λPR(T7A1)] were obtained for each promoter/discriminator complex from the fraction of OCs that made a long RNA in the initial phase (see Fig. 4B and SI Appendix, Table S6).
Supplementary Material
Acknowledgments
We thank Robin Davies and Laura Vanderploeg for assistance in generating high-quality figures for this article and the editor and reviewers for helpful comments on the manuscript. This work was supported by NIH Grants GM118100 (previously GM103061) (to M.T.R.) and GM122303-01 (to K.L.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618675114/-/DCSupplemental.
References
- 1.Ruff EF, et al. E. coli polymerase determinants of open complex lifetime and structure. J Mol Biol. 2015;427:2435–2450. doi: 10.1016/j.jmb.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winkelman JT, et al. Crosslink mapping at amino acid-base resolution reveals the path of scrunched DNA in initial transcribing complexes. Mol Cell. 2015;59:768–780. doi: 10.1016/j.molcel.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haugen SP, et al. rRNA promoter regulation by nonoptimal binding of σ region 1.2: An additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 4.Winkelman JT, et al. Multiplexed protein-DNA cross-linking: Scrunching in transcription start site selection. Science. 2016;351:1090–1093. doi: 10.1126/science.aad6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapanidis AN, et al. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samanta S, Martin CT. Insights into the mechanism of initial transcription in Escherichia coli RNA polymerase. J Biol Chem. 2013;288:31993–32003. doi: 10.1074/jbc.M113.497669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pupov D, Kuzin I, Bass I, Kulbachinskiy A. Distinct functions of the RNA polymerase σ subunit region 3.2 in RNA priming and promoter escape. Nucleic Acids Res. 2014;42:4494–4504. doi: 10.1093/nar/gkt1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulbachinskiy A, Mustaev A. Region 3.2 of the σ subunit contributes to the binding of the 3′-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J Biol Chem. 2006;281:18273–18276. doi: 10.1074/jbc.C600060200. [DOI] [PubMed] [Google Scholar]
- 10.Duchi D, et al. RNA polymerase pausing during initial transcription. Mol Cell. 2016;63:939–950. doi: 10.1016/j.molcel.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuo Y, Steitz TA. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol Cell. 2015;58:534–540. doi: 10.1016/j.molcel.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metzger W, Schickor P, Meier T, Werel W, Heumann H. Nucleation of RNA chain formation by Escherichia coli DNA-dependent RNA polymerase. J Mol Biol. 1993;232:35–49. doi: 10.1006/jmbi.1993.1368. [DOI] [PubMed] [Google Scholar]
- 13.Stackhouse TM, Telesnitsky AP, Meares CF. Release of the σ subunit from Escherichia coli RNA polymerase transcription complexes is dependent on the promoter sequence. Biochemistry. 1989;28:7781–7788. doi: 10.1021/bi00445a038. [DOI] [PubMed] [Google Scholar]
- 14.Johnston DE, McClure WR. Abortive initiation of in vitro RNA synthesis on bacteriophage λ DNA. In: Losick R, Chamberlin MJ, editors. RNA Polymerase. Cold Spring Harbor Laboratory; Cold Spring Harbor: 1976. pp. 413–428. [Google Scholar]
- 15.Straney DC, Crothers DM. A stressed intermediate in the formation of stably initiated RNA chains at the Escherichia coli lac UV5 promoter. J Mol Biol. 1987;193:267–278. doi: 10.1016/0022-2836(87)90218-x. [DOI] [PubMed] [Google Scholar]
- 16.Munson LM, Reznikoff WS. Abortive initiation and long ribonucleic acid synthesis. Biochemistry. 1981;20:2081–2085. doi: 10.1021/bi00511a003. [DOI] [PubMed] [Google Scholar]
- 17.Gralla JD, Carpousis AJ, Stefano JE. Productive and abortive initiation of transcription in vitro at the lac UV5 promoter. Biochemistry. 1980;19:5864–5869. doi: 10.1021/bi00566a031. [DOI] [PubMed] [Google Scholar]
- 18.Levin JR, Krummel B, Chamberlin MJ. Isolation and properties of transcribing ternary complexes of Escherichia coli RNA polymerase positioned at a single template base. J Mol Biol. 1987;196:85–100. doi: 10.1016/0022-2836(87)90512-2. [DOI] [PubMed] [Google Scholar]
- 19.Vo NV, Hsu LM, Kane CM, Chamberlin MJ. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 3. Influences of individual DNA elements within the promoter recognition region on abortive initiation and promoter escape. Biochemistry. 2003;42:3798–3811. doi: 10.1021/bi026962v. [DOI] [PubMed] [Google Scholar]
- 20.Hsu LM, et al. Initial transcribed sequence mutations specifically affect promoter escape properties. Biochemistry. 2006;45:8841–8854. doi: 10.1021/bi060247u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang G-Q, Roy R, Bandwar RP, Ha T, Patel SS. Real-time observation of the transition from transcription initiation to elongation of the RNA polymerase. Proc Natl Acad Sci USA. 2009;106:22175–22180. doi: 10.1073/pnas.0906979106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimamoto N, Wu C-W. Mechanism of ribonucleic acid chain initiation. 2. A real time analysis of initiation by the rapid kinetic technique. Biochemistry. 1980;19:849–856. doi: 10.1021/bi00546a004. [DOI] [PubMed] [Google Scholar]
- 23.Shimamoto N, Wu C-W. Mechanism of ribonucleic acid chain initiation. 1. A non-steady-state study of ribonucleic acid synthesis without enzyme turnover. Biochemistry. 1980;19:842–848. doi: 10.1021/bi00546a003. [DOI] [PubMed] [Google Scholar]
- 24.Kubori T, Shimamoto N. A branched pathway in the early stage of transcription by Escherichia coli RNA polymerase. J Mol Biol. 1996;256:449–457. doi: 10.1006/jmbi.1996.0100. [DOI] [PubMed] [Google Scholar]
- 25.Susa M, Kubori T, Shimamoto N. A pathway branching in transcription initiation in Escherichia coli. Mol Microbiol. 2006;59:1807–1817. doi: 10.1111/j.1365-2958.2006.05058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu LM. Promoter clearance and escape in prokaryotes. Biochim Biophys Acta. 2002;1577:191–207. doi: 10.1016/s0167-4781(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 27.Carpousis AJ, Gralla JD. Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry. 1980;19:3245–3253. doi: 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- 28.Ko J, Heyduk T. Kinetics of promoter escape by bacterial RNA polymerase: Effects of promoter contacts and transcription bubble collapse. Biochem J. 2014;463:135–144. doi: 10.1042/BJ20140179. [DOI] [PubMed] [Google Scholar]
- 29.Drennan A, et al. Key roles of the downstream mobile jaw of Escherichia coli RNA polymerase in transcription initiation. Biochemistry. 2012;51:9447–9459. doi: 10.1021/bi301260u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svetlov V, Artsimovitch I. Purification of bacterial RNA polymerase: Tools and protocols. Methods Mol Biol. 2015;1276:13–29. doi: 10.1007/978-1-4939-2392-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roe J-H, Burgess RR, Record MT., Jr Kinetics and mechanism of the interaction of Escherichia coli RNA polymerase with the λ PR promoter. J Mol Biol. 1984;176:495–522. doi: 10.1016/0022-2836(84)90174-8. [DOI] [PubMed] [Google Scholar]
- 32.Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual. 4th Ed Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2012. [Google Scholar]
- 33.Ruff EF, Kontur WS, Record MT., Jr . Using solutes and kinetics to probe large conformational changes in the steps of transcription initiation. In: Artsimovitch I, Stanangelo TJ, editors. Bacterial Transcriptional Control: Methods and Protocols. Springer; New York: 2015. pp. 241–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasse-Dwight S, Gralla JD. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989;264:8074–8081. [PubMed] [Google Scholar]
- 35.Gries TJ, Kontur WS, Capp MW, Saecker RM, Record MTJ., Jr One-step DNA melting in the RNA polymerase cleft opens the initiation bubble to form an unstable open complex. Proc Natl Acad Sci USA. 2010;107:10418–10423. doi: 10.1073/pnas.1000967107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Craig ML, et al. DNA footprints of the two kinetically significant intermediates in formation of an RNA polymerase-promoter open complex: Evidence that interactions with start site and downstream DNA induce sequential conformational changes in polymerase and DNA. J Mol Biol. 1998;283:741–756. doi: 10.1006/jmbi.1998.2129. [DOI] [PubMed] [Google Scholar]
- 37.Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli σ70 promoters. Nucleic Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell. 2011;147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erie DA, Yager TD, von Hippel PH. The single-nucleotide addition cycle in transcription: A biophysical and biochemical perspective. Annu Rev Biophys Biomol Struct. 1992;21:379–415. doi: 10.1146/annurev.bb.21.060192.002115. [DOI] [PubMed] [Google Scholar]
- 40.Ruff EF, Record MT, Jr, Artsimovitch I. Initial events in bacterial transcription initiation. Biomolecules. 2015;5:1035–1062. doi: 10.3390/biom5021035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gourse RL. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaal T, Bartlett MS, Ross W, Turnbough CL, Jr, Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 43.Leirmo S, Gourse RL. Factor-independent activation of Escherichia coli rRNA transcription. I. Kinetic analysis of the roles of the upstream activator region and supercoiling on transcription of the rrnB P1 promoter in vitro. J Mol Biol. 1991;220:555–568. doi: 10.1016/0022-2836(91)90100-k. [DOI] [PubMed] [Google Scholar]
- 44.Kontur WS, Saecker RM, Capp MW, Record MT., Jr Late steps in the formation of E. coli RNA polymerase-λ P R promoter open complexes: Characterization of conformational changes by rapid [perturbant] upshift experiments. J Mol Biol. 2008;376:1034–1047. doi: 10.1016/j.jmb.2007.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aoyama T, et al. Essential structure of E. coli promoter: Effect of spacer length between the two consensus sequences on promoter function. Nucleic Acids Res. 1983;11:5855–5864. doi: 10.1093/nar/11.17.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stefano JE, Gralla JD. Spacer mutations in the lac ps promoter. Proc Natl Acad Sci USA. 1982;79:1069–1072. doi: 10.1073/pnas.79.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winkelman JT, Chandrangsu P, Ross W, Gourse RL. Open complex scrunching before nucleotide addition accounts for the unusual transcription start site of E. coli ribosomal RNA promoters. Proc Natl Acad Sci USA. 2016;113:E1787–E1795. doi: 10.1073/pnas.1522159113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krummel B, Chamberlin MJ. RNA chain initiation by Escherichia coli RNA polymerase. Structural transitions of the enzyme in early ternary complexes. Biochemistry. 1989;28:7829–7842. doi: 10.1021/bi00445a045. [DOI] [PubMed] [Google Scholar]
- 49.Roberts CW, Roberts JW. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 50.Sen R, Nagai H, Shimamoto N. Polymerase arrest at the lambdaP(R) promoter during transcription initiation. J Biol Chem. 2000;275:10899–10904. doi: 10.1074/jbc.275.15.10899. [DOI] [PubMed] [Google Scholar]
- 51.Lerner E, et al. Backtracked and paused transcription initiation intermediate of Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 2016;113:E6562–E6571. doi: 10.1073/pnas.1605038113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maeda M, Shimada T, Ishihama A. Strength and regulation of seven rRNA promoters in E. coli. PLoS One. 2015;10:e0144697. doi: 10.1371/journal.pone.0144697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Travers AA. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J Bacteriol. 1980;141:973–976. doi: 10.1128/jb.141.2.973-976.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 55.Mekler V, Minakhin L, Sheppard C, Wigneshweraraj S, Severinov K. Molecular mechanism of transcription inhibition by phage T7 gp2 protein. J Mol Biol. 2011;413:1016–1027. doi: 10.1016/j.jmb.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bae B, et al. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of σ70 domain 1.1. Proc Natl Acad Sci USA. 2013;110:19772–19777. doi: 10.1073/pnas.1314576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Druzhinin SY, et al. A conserved pattern of primer-dependent transcription initiation in E. coli and V. cholerae revealed by 5′ RNA-seq. PLoS Genet. 2015;11:e1005348. doi: 10.1371/journal.pgen.1005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nickels BE. A new way to start: NanoRNA-mediated priming of transcription initiation. Transcription. 2012;3:300–304. doi: 10.4161/trns.21903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grachev MA, et al. Oligonucleotides complementary to a promoter over the region -8...+2 as transcription primers for E. coli RNA polymerase. Nucleic Acids Res. 1984;12:8509–8524. doi: 10.1093/nar/12.22.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kontur WS, Capp MW, Gries TJ, Saecker RM, Record MTJ., Jr Probing DNA binding, DNA opening, and assembly of a downstream clamp/jaw in Escherichia coli RNA polymerase-lambdaP(R) promoter complexes using salt and the physiological anion glutamate. Biochemistry. 2010;49:4361–4373. doi: 10.1021/bi100092a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.