Abstract
Oncogenic viruses are responsible for about 15% human cancers. This article explores the promise and challenges of viral proteomics in the study of the oncogenic human DNA viruses, HPV, McPyV, EBV and KSHV. These viruses have coevolved with their hosts and cause persistent infections. Each virus encodes oncoproteins that manipulate key cellular pathways to promote viral replication and evade the host immune response. Viral proteomics can identify cellular pathways perturbed by viral infection, identify cellular proteins that are crucial for viral persistence and oncogenesis, and identify important diagnostic and therapeutic targets.
Systems Virology and Viral Proteomics
Proteomics is the identification and characterization of a collection of proteins found in a specific condition or circumstance. As outlined in Fig. 1, viral proteomics can define widely different collections of proteins related to viral infection. These proteomes can range from the proteins in a virion particle, to the cellular proteins found in complex with a particular viral protein, to global changes in cellular proteins in a diseased tissue following viral infection. This article will describe some of the major advances and challenges in viral proteomics, with particular emphasis on oncogenic DNA viruses.
Fig. 1.
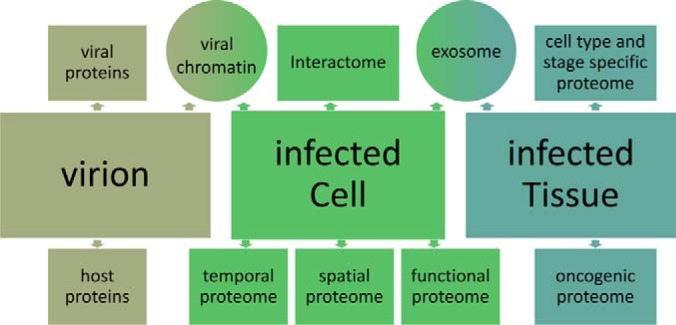
Diverse proteomic targets in the study of an oncogenic virus. Proteomes associated with virion particles, virus infected cells or virus infected tissue.
Oncogenic Viruses
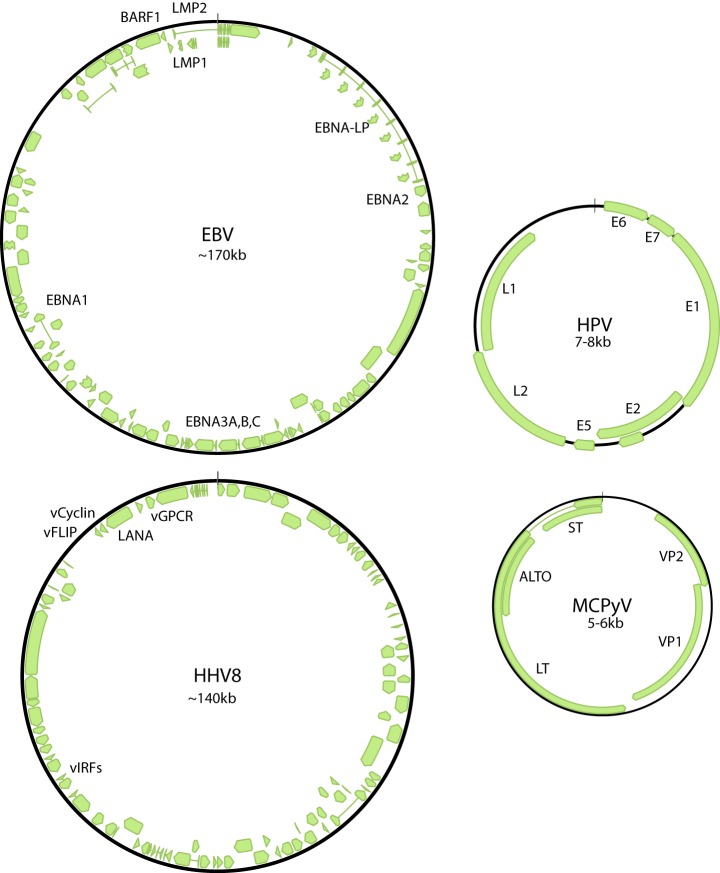
About 15% of human cancers are caused by viral infection (3). Seven human tumor viruses have been described to date: Epstein-Barr virus (EBV1 or HHV4); Kaposi's sarcoma associated virus (KSHV or HHV8); Hepatitis B and C viruses (HBV and HCV); Human T-lymphotrophic virus 1 (HTLV-1); Merkel cell polyomavirus (MCPyV); and a group of alpha Human papillomaviruses (HPVs). These are listed in Table I. The DNA viruses MCPyV, EBV, KSHV, and a subset of oncogenic alpha HPVs are direct carcinogens that encode oncogenes which are required for maintenance of the tumor phenotype (4); they will be the focus of this article. Fig. 2 shows the vast difference in genome size and coding capacity among these four oncogenic viruses.
Table I. Oncogenic DNA viruses and their oncogenes. For details see (4, 96, 97).
| Virus | Family and Genome Type | Viral Proteins contributing to Oncogenesis | Associated Cancers |
|---|---|---|---|
| A subset of human alpha Papillomaviruses, (HPV) | Papillomaviridae dsDNA genome | E6, E7, E5 | Cervical and other anogenital carcinomas; Oropharyngeal carcinoma |
| Merkel cell polyomavirus (MCPyV) | Polyomaviridae dsDNA genome | LT, ST | Merkel Cell carcinoma |
| Epstein Barr Virus (EBV or HHV4) | Herpesviridae dsDNA genome | LMP1, LMP2, BARF1, EBNA1, EBNA2, EBNA3A,B,C, EBNA-LP | Burkitt's lymphoma; Nasopharyngeal carcinoma; Gastric cancer Hodgkin's and Non-Hodgkin's lymphoma Post-transplant lymphoproliferative disorder |
| Kaposi's sarcoma herpesvirus (KSHV or HHV8) | Herpesviridae dsDNA genome | vFLIP, vCyclin, LANA, vGPCR, vIRF-1 | Kaposi's sarcoma Primary Effusion lymphoma Multicentric Castleman's disease |
| Hepatitis B virus (HBV) | Hepadnaviridae ssDNA/dsDNA genome | HBx | Hepatocellular carcinoma |
| Hepatitis C virus (HCV) | Flaviviridae +ssRNA genome | Core, NS3, NS5a | Hepatocellular carcinoma |
| Human T cell lymphotrophic virus (HTLV-1) | Retroviridae +ssRNA genome | Tax | Adult T cell leukemia |
Fig. 2.
Oncogenic viraL genomes. Shown are the circular dsDNA genomes of the four oncogenic viruses, EBV, KSHV, McPyV, and HPV. Coding regions are shown in green and the approximate positions of regions encoding viral proteins associated with oncogenesis (listed in Table I) are indicated. For the small HPV and MCPyV genomes, additional genes are indicated.
One common feature of oncogenic viruses is that they cause very persistent infection of the host and must evade immune detection for very long periods of time (4). The oncogenic herpesviruses have large genomes that encode many proteins to facilitate the viral life cycle, but also to escape immune detection. In contrast, the oncogenic polyoma and papillomaviruses have very limited coding capacity (see Fig. 2) and rely almost completely on interactions with host proteins to fulfill the same functions. Remarkably, these divergent oncogenic viruses target many of the same cellular proteins and pathways to facilitate viral replication and this has provided great insight into the study of oncogenesis.
Advances in Proteomic Studies of Oncogenic Viruses
SV40 and adenovirus were the first intensely studied oncogenic viruses and they were instrumental in the discovery of the major cellular tumor suppressors, p53 and pRb. Although SV40 and adenoviruses do not cause tumors in their natural hosts, they form tumors in rodents (5, 6) and this is dependent on SV40 Large Tag and adenovirus E1A and E1B. Pioneering studies by several laboratories in the 1970s and 80s showed that these viral oncogenes bound to the cellular proteins p53 and pRb, and this binding correlated with their ability to transform cells (7–9). The associated cellular proteins were first noted in co-immunoprecipitates with viral proteins. These early studies relied heavily on highly specific antibodies against the viral tumor antigens and the host interacting proteins (7, 8, 10). Partial peptide mapping and protein sequencing techniques were also used to compare and identify proteins (8, 11), although it was some time before the functions of the host proteins were revealed. There was great excitement when follow up studies showed that the viral oncogenes HPV E6 and E7, SV40 large Tag, and adenovirus E1A and E1B all bound and inactivated pRb and p53 (12–16).
Thirty years later, under the same principle of the commonality of tumor virus targets, Rozenblatt-Rosen and colleagues undertook an impressive systems virology analysis of cellular pathways perturbed by the human oncogenic viruses EBV, HPV, adenoviruses, and polyomaviruses (17). The authors proposed the Variome to Virome hypothesis that stated that a comparison of cellular pathways perturbed by these viruses should facilitate the identification of driver versus passenger mutations in human tumors. This study identified additional cellular partners of the viral oncoproteins by both high-throughput yeast-two hybrid analyses, and tandem affinity purification (TAP) combined with LC-MS/MS (liquid chromatography tandem mass spectrometry). This was further integrated with a microarray-based transcriptome analysis that compared how each viral oncoprotein perturbed cellular networks. The authors concluded that viral oncogenes and cancer-associated mutations in the host genome converged on common cellular pathways. These studies highlight the remarkable progress that has been made in systems virology in the last few decades.
Early comparative proteomic studies of oncogenic viruses used 2D gel electrophoresis to identify differential protein expression in virally infected cells or tumors, and later, differentially expressed proteins could often be identified by mass spectrometry techniques. The yeast two hybrid technique, and related mammalian two hybrid techniques, have been extensively used to identify interacting cellular factors of oncogenic viruses (17–21). However, MS based techniques are advancing rapidly, and are used widely, and so studies based on this technology are the focus of this chapter.
Approaches used in Oncogenic Viral Proteomics
There are many different ways to study the viral associated proteome. Sophisticated virion purification methods can provide a highly enriched sample to study protein content and post-translational modifications. In infected cells, viral and cellular proteins can be defined with a global and unbiased shotgun approach in which all viral and cellular proteins can be identified at a specific time of infection, an intracellular location, or associated with a specific activity (e.g. replication). In a complementary approach, viral interactomes can be defined by determining all interactions among viral and cellular proteins using highly specific protein complex purification techniques. In practice, many of these approaches can be combined to yield important information about viral infection. These approaches are outlined in Fig. 1.
Proteomics of Virion Particles
Proteomics can define the proteins contained in virion particles. The components of the large, enveloped EBV and KHSV virions have been analyzed by LC-MS/MS, revealing 24–34 viral proteins in the viral capsid, tegument, and envelopes, as well as several host proteins (22, 23). In the EBV study, purified virions were further fractionated into envelope, tegument, and capsid containing components (verified by electron microscopy) to further define the structure of the virion (22). Treatment of the virions with deglycosylases helped identify the highly glycosylated proteins associated with the viral envelope (22). Mass spectrometry techniques can also reveal post-translational modifications of virion proteins; Lind et al. identified phosphoproteins in the adenovirus type 2 virion using LC MS/MS techniques but, despite the highly purified nature of virion particles, they had to employ several additional strategies such as TiO2 enrichment and alternative digestion strategies to identify virion associated phosphoproteins (24). These studies provide important insight into the viral life cycle strategy as virion associated proteins are often required to evade the host intrinsic immune system as well as to initiate the immediate-early viral transcriptional process.
In comparison, the small nonenveloped polyoma and papilloma virions are very simple with just one major and one minor capsid protein (25, 26). One unique, and relatively unstudied, feature of these small DNA viruses is that their genomes are packaged in host nucleosomes (27, 28). The activity of chromatin is highly modified by extensive post-translational modifications (PTMs) of histones, which could greatly determine the efficacy of the early stages of viral infection. Early studies on polyomaviruses showed that virion associated histones are highly acetylated (27); this has been confirmed, and many additional modifications identified, by Fang and colleagues who mapped extensive post-translational modifications in the histones of the polyomavirus BKPyV virion particles and minichromosome using triton-acetic acid-urea (TAU)-polyacrylamide gel electrophoresis separation followed by nanoflow LC-MS/MS analysis (29). Notably, the authors found that N-terminal acetylation of histone H2A occurred only on the viral genome packaged in virions, and not of those in infected cells (29). However, as yet, these studies have not provided functional insight into these modifications. However, it would be surprising if the polyoma and papillomavirus life cycle strategies did not take advantage of epigenetic modulation of virion DNA to facilitate viral infection.
Infected Cells: Viral Chromatin
Global profiling of chromatin modifications by mass spectrometry is challenging, in part because most modifications are located in the highly basic N-terminal tails of the histones. The Garcia laboratory has developed a very detailed workflow that includes propionic anhydride derivatization of lysine residue side chains before trypsin digestion to circumvent this problem. An additional propionylation step targets the N terminus of the digested peptides to help identify these very short peptides by nano LC-MS/MS (30). The workflow also incorporates a TiO2 binding step to enrich for phosphopeptides. Garcia and colleagues used this method for dynamic global profiling of adenovirus (30) and cytomegalovirus (31) histone PTMs during infection. Mass spectrometry based techniques can provide detailed and unbiased information about global histone post-translational modifications in a virus. However, the availability of highly specific antibodies directed against individual histone modifications enables relatively easy purification of modified chromatin (ChIP) and subsequent identification of the specific sequence of bound DNA by PCR or DNA sequencing technologies (32). At this point, the easy accessibility of ChIP techniques, and the sequence-specific information obtained, make them the method of choice.
Infected Cells: Temporal Studies
Temporal studies can reveal global changes in the viral and cellular proteome at different stages of infection. These studies can characterize the cellular and viral proteome at different stages of infection, or can define the viral interactome for one or more viral proteins. To date, most temporal studies have examined the nononcogenic herpesviruses, HSV1 (herpes simplex virus) and HCMV (human cytomagalovirus). For the oncogenic viruses, it is more difficult to produce large amounts of viral particles, to infect cells synchronously, and to complete the viral life cycle in a short time frame. Furthermore, most early gene products are expressed at very low levels. Oncogenic viruses establish persistent infections and the late stages of infection must be induced by manipulation of the host cell (e.g. differentiation). Quasivirus particles (recircularized viral genomes packaged in a cell line overexpressing the capsid proteins) can be used to generate papillomavirus and polyomavirus particles to study the early stages of infection, and in theory epitope tagged versions of viral proteins could be packaged in these recombinant particles to facilitate their detection and localization (33, 34). More efficient methods are also being established to induce the late stages of infection.
Infected Cells: Spatial Studies
Throughout the course of infection, there can be dramatic changes in cellular organelles as well as the formation of viral replication and assembly factories. Subcellular fractionation and analysis of the protein components in these intracellular structures can provide great insight into the infectious process and reveal ways in which viruses manipulate cellular organization. Baquero-Pérez and Whitehouse took advantage of the fact that KSHV replication and transcription centers are associated with the nuclear envelope and this allowed them to purify, and then identify cellular factors enriched here using LC-MS/MS (35). These studies revealed that the molecular chaperone hsp70 was crucial for the formation of these compartments (35). Bartee et al. used SILAC (stable isotope labeling with amino acids in cell culture) and 2D-LC-MS/MS to compare the proteomics of plasma, golgi, and endoplasmic reticulum membranes of cells in the presence or absence of the KSHV K5 protein, which was known to downregulate MHC class I molecules on the surface of cells (36). Using this approach, they identified three additional immunomodulatory proteins that were underrepresented in the presence of K5 (36). Similar approaches with other oncogenic viruses should reveal a wealth of additional information about how these viruses manipulate cellular function and organization.
Infected Cells: Exosomes
Exosomes are small membrane bound vesicles that are secreted from cells into bodily fluids, and are thought to regulate the cellular microenvironment, particularly when secreted from tumor cells. Exosomes by their very nature contain an easily purified source of proteins. LC-MS/MS analyses of exosomes secreted from EBV and KSHV infected cells have shown that they contain complex mixtures of proteins that are dramatically modulated by viral infection (37). Another study examined the protein content of exosomes secreted from MCpyV positive and negative Merkel cell carcinoma cell lines; using LC-MS/MS, proteins involved in cellular motility and oncogenesis were identified (38). There is strong interest in defining the contents of the exosomes in the quest for tumor biomarkers because they can be isolated noninvasively from body fluids such as saliva.
Infected Cells: Functional Studies
Activity based protein profiling (ABBP) uses highly specific probes that consist of a reactive warhead (that creates an irreversible bond between probe and enzyme), a tag that specifically binds to the catalytic sites of the targeted enzymes, and a reporter that allows their detection or purification (39, 40). Comparative ABPP can compare the activity of a class of enzymes in the presence or absence of viral infection. For example, this approach was used to show that both EBV and HPV induced oncogenesis correlates with up-regulation of a series of deubiquitinating enzymes (41, 42). To date, there are about 12 different classes of enzymes that can be targeted by ABPP chemistries (43). They hold great promise for viral proteomics, as well as for identifying and optimizing anti-viral therapeutics that can bind to the active site of viral or host enzymes (39).
Advances in click chemistry have also allowed the development of small, cell permeable activity based probes that are highly active in the intracellular environment (44). Cortez and colleagues developed a technique to identify proteins at replication forks; it is named Isolation of Proteins on Nascent DNA or iPOND, coupled with mass spectrometry (45). This technique uses the nucleoside analog 5-ethynyl-2-deoxyuridine (EdU), which is incorporated into nascently replicated DNA, and click chemistry, to define the replisome. Dembowski and DeLuca used this technique to identify both host and viral factors located in replication centers of HSV1 (46), and it is likely that this technique will prove useful for the study of oncogenic viral replication. Many viruses manipulate the DNA damage and repair response to replicate their own DNA (47, 48) and the iPOND technique could help determine which of these factors are utilized at the replication fork (46).
Infected Cells: Viral Interactomes
Shotgun proteomics can classify the global proteome at specific stages or locations of infection in an unbiased fashion. However, almost all biological processes and pathways function through protein-protein interactions (PPIs) and so many targeted approaches have been developed that can be used to identify viral-viral and viral-host interactomes (49). These approaches have been especially fruitful in the study of oncogenic viruses, which have a long-term association with the host, and many key cellular regulatory proteins have been identified because they are targeted by oncogenic viral proteins.
High throughput protein complementation assays such as the yeast two hybrid assay (Y2H) can identify binary interactions between proteins (50) and a number of oncogenic virus interactomes have been developed using this or derivative techniques. For example, extensive and comparative interactomes have been identified for the E2, E6, and E7 proteins from different HPV types using both yeast and mammalian complementation assays (51, 52). The Y2H approach was also used to define a global viral-host interaction network of all KSHV proteins (53), and for EBV (21). These studies showed that viral networks tended to appear as single, highly coupled modules (53).
In the last few years, the combination of affinity purification and mass spectrometry (AP-MS) has dominated the viral proteomics field. Viral protein complexes can be affinity purified and interacting proteins rapidly identified by LC-MS/MS techniques. These viral proteins can be expressed alone (fused to high affinity tags) or expressed from the viral genome in the context of an infection. Fluorescent tags enable the location of the viral protein to be monitored in living cells and correlated with protein interactions at various times post infection (54). For example, White et al. identified and compared the interaction partners of a series of E6 proteins from different HPVs using AP-tandem MS (55). By comparing E6 proteins from sixteen different HPVs, the authors could distinguish protein partners of the alpha-HPV E6 proteins (the ubiquitin ligase E6AP) from those of the cutaneous beta-HPV E6 proteins (MAML and associated Notch proteins) (55). Proteomic profiling of EBV EBNA1 by AP-LC-MS/MS defined protein interactions in EBV-associated cancers in both latent and lytic infection (56). Similar studies using KSHV LANA, showed that the RFC complex interacted with LANA and is important for viral replication (57). Another study defined cellular proteins that interacted with LANA, and determined which of these interactions were mediated through the LANA SUMO Interacting motif (SIM) (58).
BioID is a relatively new method that enables the identification of both proximal and interacting proteins in living cells (59, 60). The target protein is fused to a biotin ligase that, in the presence of excess biotin, will biotinylate adjacent proteins. The biotinylated proteins are purified by affinity methods and identified by standard LC-MS/MS techniques. This method is particularly useful in identifying transient interacting partners, or protein complexes that are difficult to extract intact from cells. Although not yet used for an oncogenic viral protein, Ortiz et al. used BioID to identify both viral and cellular interacting partners of the HCMV tegument protein pUL103 (61). Of note, they identified the ESCRT-associated protein, ALIX as a binding partner and discovered a previously unidentified ALIX binding domain in pUL103 (61).
Viral interactomes do not necessarily require that a viral protein be the bait. Si et al. used the KSHV terminal repeat DNA as an affinity ligand and identified 123 bound proteins, mostly present in KSHV infected cells, using LCQ-MS (62).
Infected Tissues: Differentiation Dependent Viral Life Styles and Oncogenesis
Shotgun proteomics of virally infected tissues and associated cancers allows the unbiased identification of proteins and has the potential to discover novel therapeutic targets, pathogenic virus signatures, or biomarkers (63). For example, Malik et al. provide a comprehensive review of candidate biomarkers, discovered by a wide range of proteomic techniques, in tissues and saliva from individuals with oral squamous cell carcinoma (64). However, the proteome of undissected tissue biopsies can only give a general overview of protein content, as tumors and infected tissues contain a heterogeneous mixture of cells and stroma. Identification of protein differences between different and specific cell populations requires a combination of highly sophisticated microdissection techniques and ultrasensitive proteomics analysis.
The oncogenic DNA viruses form a long-term, complex relationship with the host. For example, EBV can infect both oral epithelial cells and B-lymphocytes, and viral infection is thought to transition between both cell types for the life of the host (65). HPVs infect the basal cells of a stratified epithelium and establish a relatively quiescent, but long term, infection in these proliferating cells; high levels of viral DNA synthesis, transcription and protein expression are switched on as the infected cells differentiate and traverse to the surface of the epithelium (66). In HPV-associated oncogenesis, the infected cells acquire characteristics that enable them to resist differentiation signals and proliferate continuously throughout the full thickness of the epithelium (66). Further, genetic changes promote invasion through the basement membrane (66). In each of these scenarios, different cells within the tissue are supporting different stages of viral infection or oncogenesis. The spatial distribution of infections and cancer progression in tissue make the technique of laser capture microdissection (LCM) ideally suited to these situations as it allows biological materials (DNA, RNA, proteins) to be directly isolated from cells of interest within a tissue (67).
Most studies to date have used LCM to study the progressive stages of virus-associated cancer. Some of the earliest studies used LCM, 2D gel electrophoresis and mass spectrometry to identify protein changes among normal, premalignant, and cervical cancer tissues (68), or oral squamous cell carcinoma (69). The latter study was able to detect differences in the proteomes of HPV positive and HPV negative cancers (69). LCM followed by mass spectrometry can measure cell type specific protein expression in the tumor microenvironment, and this approach has shown that the stroma of EBV associated nasopharyngeal carcinomas overexpress periostin compared with normal stromal tissue, and this correlates with clinical stage and outcome (70). An important study used LCM and proteomic analysis to demonstrate that the CD21 EBV receptor was only expressed on tonsil epithelial cells (71). A quantitative proteomic analysis of Merkel Cell carcinoma indicated that the MAPK, PI3K/Akt/mTOR, Wnt, and apoptosis signaling pathways were involved in the development of the tumor (72). As mass spectrometry techniques become more sensitive, studies such as these will be able to use proteomics to gain direct information about infection and disease states.
Viral Proteomics in the Clinical Forum
There is intense interest in the use of proteomics to diagnose disease (63, 73). As described above, proteomics is being used to identify biomarkers, pathogenic viral signatures, and cellular pathways that are dysregulated by viruses. However, an important goal is to identify highly specific biomarkers that enable clinicians to diagnose, stage and predict the outcome of different disease states, and to develop highly sensitive and accurate proteomic methods to detect these markers (74).
Of particular interest for oncogenic viruses are biomarkers that could be used for early detection in bodily fluids such as saliva. The Pap smear enables early histological detection of cervical abnormalities in HPV associated cancers, but there is no equivalent screen for abnormalities in the oropharynx or nasopharynx, the sites of HPV and EBV associated cancers. Noninvasive screening for biomarkers, or molecular signatures, in the salivary proteome could allow early detection of these diseases. However, some of the techniques employed have not been reproducible (75) and at this point, very few biomarkers have been successful in the clinical arena (76).
Comparative Proteomics: Evolutionary Proteomics
Comparative proteomics can be very useful in dissecting protein-protein interactions important for viral-mediated oncogenesis. These studies can either compare the interactome of very closely related viral types that have different oncogenic potential (such as the papillomaviruses), or compare the interactome of oncogenes from different viruses to identify common interactions and pathways.
The papillomaviruses are particularly well suited for phylogenetically driven comparative proteomics. There are over 200 different types of HPV and they all infect similar cell types (keratinocytes), yet have different disease outcomes. Comparative proteomic studies can identify pathogenic viral signatures and help define the key features that make a viral protein oncogenic, as well as identify structural and molecular differences that could be key therapeutic targets. In contrast, polyomaviruses are more diverse (77), their infections are often asymptomatic, and they have multiple tissue tropisms. There have been several studies using AP-MS to compare the interactome of papillomavirus E6, E7 and E2 proteins from distinct phylogenetic groups (17, 55, 78–80), and these have identified key differences in the interactome of oncogenic and nononcogenic viruses.
Another approach is to analyze the interactome of different oncogenic viruses to identify common targets (17). As described above, the DNA tumor virus oncogenes were instrumental in identifying p53 and pRb pathways as crucial targets for viral-mediated oncogenesis (17). In the systematic study already described above, by comparing the interactomes (defined by TAP-MS and Y2H) and transcriptomes of HPV, polyomavirus, adenovirus and EBV, Rozenblatt-Rosen and colleagues identified that Notch signaling was targeted by all DNA tumor viruses (17).
Technical Advances, Considerations and Challenges
Advances
Virology is an ideal discipline for the use and development of proteomic techniques. Uninfected cells provide a robust negative control, and spatial and temporal changes in the both viral and host proteomes during infection can be tracked, often in concert with live cell microscopy (54, 81). Viruses can often be easily manipulated to express tagged viral proteins that are expressed in the context of an infection, and well characterized viral mutations can help unravel the connection between interacting protein partners and function. A comparison of interacting partners among closely related viruses can determine which interactions are required for different disease outcomes and can identify novel therapeutic targets.
Both yeast two hybrid (Y2H) and MS-based approaches have been used in the study of oncogenic viruses and each has advantages and disadvantages (82). However, advances in both the sensitivity and high-throughput capabilities of quantitative MS-based proteomics are likely to make it the method of choice. Advances in instrumentation and technologies will not only improve the precision, sensitivity and speed of MS-based analysis but “plug and play” MS systems will make protein identification an accessible and routine technique for many laboratories not expert in protein chemistry (83).
Powerful techniques such as chemical cross-linking coupled with mass spectrometry (CXMS) can elucidate the higher order of protein macromolecular complexes (84, 85) and Chait and colleagues envisage the eventual development of a “multiscale molecular microscope.” This would entail the use of chemical crosslinkers in vivo to stabilize complexes, the rapid isolation and identification of proteins using quantitative AP-MS, and further chemical crosslinking ex vivo to map at high resolution the spatial proximities of subunits within a complex (84).
Advances in both CRISPR and chromatin immunoprecipitation technologies have led to CRISPR-based Chromatin Affinity Purification with Mass Spectrometry (CRISPR-ChAP-MS), which can define the epiproteome (86). In this technique, a catalytically inactive, Cas9 protein and complementary gRNA can target, and be used to purify, a specific genomic region of interest along with associated proteins. Such approaches should prove very fruitful in the study of oncogenic viruses, which are dependent on, and take advantage of, host epigenetic mechanisms.
Enrichment, Extraction, and Fractionation
All proteomic approaches require proteins to be efficiently and reproducibly extracted from their original biological materials. However, approaches that require isolation of subcellular compartments or protein complexes must balance efficient extraction of the target with accurate retention of low affinity protein-protein interactions within complexes. A number of extraction techniques have been developed to overcome this and one example is cryogenic cell lysis in which frozen cell pellets or tissues are ground to a powder in a ball mill before complex purification (87). Cross-linking agents such as formaldehyde can be used to stabilize interactions and allow more stringent washing conditions in affinity approaches and this approach is used in the iPOND technique that isolates proteins bound to nascent replication forks (45, 46). The technique of Tandem Affinity Purification (TAP) uses proteins with two epitope tags and protein complexes are purified sequentially using these tags. This approach can remove contaminants but low-affinity, transient interactions are likely to be lost (80, 82). A related challenge is the identification and elimination of false positive and false negative interactions identified by AP methods. Techniques such as I-DIRT (Isotopic Differentiation of Interactions as Random or Targeted) can define false interactions (88) and online data repositories can help identify common contaminants (89). Trinkle-Mulcahy and colleagues have defined “the bead proteome,” a comprehensive list of proteins that bind to the Sepharose, agarose and magnetic beads used for affinity purification (90). These issues are discussed in detail by White and Howley (82).
Computational Requirements and Challenges
Many of the proteomic studies described here generate enormous datasets and processing and interpretation of the data can be computationally intense. Proteomic data is often a small part of a larger systems virology analysis and robust computational methods are needed to integrate and compare these datasets (17). A recent ambitious study used MS-based proteomics, live cell microscopy and organelle fractionation to analyze spatial changes in the HCMV and host proteome at different times of infection (81). This required intensive machine learning to classify temporal changes in subcellular localization during infection (81).
As described below, it is crucial to share proteomic data but there also needs to be efficient ways to compare proteomic data (both from MS-based and alternative approaches) across different studies, and to integrate these data with that obtained from other systems virology data sets. Many virology research groups can undertake sophisticated proteomic experiments, often in concert with expert mass spectrometry facilities, but they often struggle to find appropriate bioinformatics support to interpret and analyze the data.
Databases: Shared Knowledge and Resources
It is important that open access resources are available to provide reliable bioinformatic information about each virus family to assist in the development of proteomic analyses, and it is also crucial that large proteomic datasets are disseminated and shared to allow others to mine the data using their own tools and expertise. Viral-host interaction databases are also invaluable. A compendium of different resources related to viroinformatics has recently been compiled by Sharma and colleagues (91).
Bioinformatic databases and online resources exist that are specific for each oncogenic virus, and these have often been developed by investigators determined to provide curated and organized datasets to the community. Examples are the Papillomavirus Episteme, or PaVE (https://pave.niaid.nih.gov/) (92, 93), PyVE for polyomaviruses (https://home.ccr.cancer.gov/lco/PyVE.asp), the EBV Portal (https://ebv.wistar.upenn.edu/EBVportal/), and ViPR for gammaherpesviruses (https://www.viprbrc.org/brc/home.spg?decorator=herpes).
There are also highly valuable resources that help in the interpretation of proteomic data and design of proteomic studies. Highly popular is The Crapome, which is a repository that contains lists of contaminants often found in AP-MS data (89) and COMPASS, an open-source proteomic software pipeline (94). The Pandey lab have developed http://www.silac.org/to assist in the design of quantitative proteomics using SILAC.
It is common, and often required, that genomic sequencing and transcriptome data are deposited in open access online repositories. This is less common for proteomic data but the ProteomeXchange consortium has developed a resource to assist in the submission and access of proteomic datasets (http://www.proteomexchange.org/). As yet, the ProteomeXchange does not contain many virus-related datasets, but this should improve with time. Additional resources have been reviewed by Perez-Riverol and colleagues (95).
There are a number of online databases that contain curated virus-virus and virus-host interactions. Examples are VirHostNet (VirHostNet http://virhostnet.prabi.fr/), which contains viral-host interaction data derived from the literature for the oncogenic adenoviruses, polyomaviruses, papillomaviruses, and γherpes viruses and VirusMentha (http://virusmentha.uniroma2.it/).
Concluding Remarks
Despite the enormous advances that have been made in viral proteomics over the last few decades, our knowledge of global viral-host interactions is still somewhat rudimentary. Technology is advancing at a rapid pace, and imaginative new techniques are being developed frequently. However, as described here, there are still many technical challenges. Most proteomic experiments are high throughput and discovery-based, and cellular proteins and pathways that are perturbed by viral infection need careful validation. Rigor and reproducibility are especially important because of the complex and diverse nature of proteomic studies. Proteomic studies can generate vast amounts of data and highly organized collaborative efforts and computer resources are necessary to analyze, integrate, manage, and make this data publically available. Hanash (2011) describes some of the collaborative efforts and resources required to fullfill the full potential of proteomics in human disease (74).
Footnotes
Author contributions: A.A.M. analyzed data; A.A.M. wrote the paper.
* Alison McBride's research is supported by the Intramural Research Program of the NIAID, NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
1 The abbreviations used are:
- EBV
- Epstein-Barr virus
- KSHV
- Kaposi's sarcoma-associated virus
- HBV
- hepatitis B virus
- HCV
- hepatitis C virus
- HTLV
- human T-lymphotrophic virus
- MCPyV
- Merkel cell polyoma virus
- HPV
- human papilloma virus
- PTM
- post-translational modification.
REFERENCES
- 1. Law G. L., Korth M. J., Benecke A. G., and Katze M. G. (2013) Systems virology: host-directed approaches to viral pathogenesis and drug targeting. Nat. Rev. 11, 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aderem A., Adkins J. N., Ansong C., Galagan J., Kaiser S., Korth M. J., Law G. L., McDermott J. G., Proll S. C., Rosenberger C., Schoolnik G., and Katze M. G. (2011) A systems biology approach to infectious disease research: innovating the pathogen-host research paradigm. mBio 2, e00325-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Plummer M., de Martel C., Vignat J., Ferlay J., Bray F., and Franceschi S. (2016) Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob. Health 4, Se609–S616 [DOI] [PubMed] [Google Scholar]
- 4. Moore P. S., and Chang Y. (2010) Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer 10, 878–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trentin J. J., Yabe Y., and Taylor G. (1962) The quest for human cancer viruses. Science 137, 835–841 [DOI] [PubMed] [Google Scholar]
- 6. Eddy B. E., Borman G. S., Grubbs G. E., and Young R. D. (1962) Identification of the oncogenic substance in rhesus monkey kidney cell cultures as simian virus 40. Virology 17, 65–75 [DOI] [PubMed] [Google Scholar]
- 7. Lane D. P., and Crawford L. V. (1979) T-Antigen Is Bound to a Host Protein in Sv40-Transformed Cells. Nature 278, 261–263 [DOI] [PubMed] [Google Scholar]
- 8. Linzer D. I., and Levine A. J. (1979) Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17, 43–52 [DOI] [PubMed] [Google Scholar]
- 9. DeCaprio J. A. (2009) How the Rb tumor suppressor structure and function was revealed by the study of Adenovirus and SV40. Virology 384, 274–284 [DOI] [PubMed] [Google Scholar]
- 10. Gurney E. G., Harrison R. O., and Fenno J. (1980) Monoclonal-Antibodies against Simian Virus-40 T-Antigens - Evidence for Distinct Subclasses of Large T-Antigen and for Similarities among Non-Viral T-Antigens. J. Virol. 34, 752–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leppard K., Totty N., Waterfield M., Harlow E., Jenkins J., and Crawford L. (1983) Purification and partial amino acid sequence analysis of the cellular tumour antigen, p53, from mouse SV40-transformed cells. EMBO J. 2, 1993–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dyson N., Howley P. M., Munger K., and Harlow E. (1989) The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243, 934–937 [DOI] [PubMed] [Google Scholar]
- 13. Dyson N., Buchkovich K., Whyte P., and Harlow E. (1989) The cellular 107K protein that binds to adenovirus E1A also associates with the large T antigens of SV40 and JC virus. Cell 58, 249–255 [DOI] [PubMed] [Google Scholar]
- 14. Werness B. A., Levine A. J., and Howley P. M. (1990) Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248, 76–79 [DOI] [PubMed] [Google Scholar]
- 15. Sarnow P., Ho Y. S., Williams J., and Levine A. J. (1982) Adenovirus E1b-58kd Tumor-Antigen and Sv40 Large Tumor-Antigen Are Physically Associated with the Same 54 Kd Cellular Protein in Transformed-Cells. Cell 28, 387–394 [DOI] [PubMed] [Google Scholar]
- 16. DeCaprio J. A., Ludlow J. W., Figge J., Shew J. Y., Huang C. M., Lee W. H., Marsilio E., Paucha E., and Livingston D. M. (1988) SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell 54, 275–283 [DOI] [PubMed] [Google Scholar]
- 17. Rozenblatt-Rosen O., Deo R. C., Padi M., Adelmant G., Calderwood M. A., Rolland T., Grace M., Dricot A., Askenazi M., Tavares M., Pevzner S. J., Abderazzaq F., Byrdsong D., Carvunis A. R., Chen A. A., Cheng J. W., Correll M., Duarte M., Fan C. Y., Feltkamp M. C., Ficarro S. B., Franchi R., Garg B. K., Gulbahce N., Hao T., Holthaus A. M., James R., Korkhin A., Litovchick L., Mar J. C., Pak T. R., Rabello S., Rubio R., Shen Y., Singh S., Spangle J. M., Tasan M., Wanamaker S., Webber J. T., Roecklein-Canfield J., Johannsen E., Barabasi A. L., Beroukhim R., Kieff E., Cusick M. E., Hill D. E., Munger K., Marto J. A., Quackenbush J., Roth F. P., DeCaprio J. A., and Vidal M. (2012) Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487, 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muller M., Jacob Y., Jones L., Weiss A., Brino L., Chantier T., Lotteau V., Favre M., and Demeret C. (2012) Large scale genotype comparison of human papillomavirus E2-host interaction networks provides new insights for e2 molecular functions. Plos Pathog. 8, e1002761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fertey J., Ammermann I., Winkler M., Stoger R., Iftner T., and Stubenrauch F. (2010) Interaction of the papillomavirus E8–E2C protein with the cellular CHD6 protein contributes to transcriptional repression. J. Virol. 84, 9505–9515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fossum E., Friedel C. C., Rajagopala S. V., Titz B., Baiker A., Schmidt T., Kraus T., Stellberger T., Rutenberg C., Suthram S., Bandyopadhyay S., Rose D., von Brunn A., Uhlmann M., Zeretzke C., Dong Y. A., Boulet H., Koegl M., Bailer S. M., Koszinowski U., Ideker T., Uetz P., Zimmer R., and Haas J. (2009) Evolutionarily conserved herpesviral protein interaction networks. Plos Pathog. 5, e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Calderwood M. A., Venkatesan K., Xing L., Chase M. R., Vazquez A., Holthaus A. M., Ewence A. E., Li N., Hirozane-Kishikawa T., Hill D. E., Vidal M., Kieff E., and Johannsen E. (2007) Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U.S.A. 104, 7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johannsen E., Luftig M., Chase M. R., Weicksel S., Cahir-McFarland E., Illanes D., Sarracino D., and Kieff E. (2004) Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U.S.A. 101, 16286–16291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu F. X., Chong J. M., Wu L., and Yuan Y. (2005) Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79, 800–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lind S. B., Artemenko K. A., Elfineh L., Zhao Y. H., Bergquist J., and Pettersson U. (2012) The phosphoproteome of the adenovirus type 2 virion. Virology 433, 253–261 [DOI] [PubMed] [Google Scholar]
- 25. Schowalter R. M., and Buck C. B. (2013) The Merkel cell polyomavirus minor capsid protein. Plos Pathog. 9, e1003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Buck C. B., Cheng N., Thompson C. D., Lowy D. R., Steven A. C., Schiller J. T., and Trus B. L. (2008) Arrangement of L2 within the papillomavirus capsid. J. Virol. 82, 5190–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schaffhausen B. S., and Benjamin T. L. (1976) Deficiency in histone acetylation in nontransforming host range mutants of polyoma virus. Proc. Natl. Acad. Sci. U.S.A. 73, 1092–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Favre M., Breitburd F., Croissant O., and Orth G. (1977) Chromatin-like structures obtained after alkaline disruption of bovine and human papillomaviruses. J. Virol. 21, 1205–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fang C. Y., Shen C. H., Wang M., Chen P. L., Chan M. W., Hsu P. H., and Chang D. (2015) Global profiling of histone modifications in the polyomavirus BK virion minichromosome. Virology 483, 1–12 [DOI] [PubMed] [Google Scholar]
- 30. Kulej K., Avgousti D. C., Weitzman M. D., and Garcia B. A. (2015) Characterization of histone post-translational modifications during virus infection using mass spectrometry-based proteomics. Methods 90, 8–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O'Connor C. M., DiMaggio P. A., Shenk T., and Garcia B. A. (2014) Quantitative proteomic discovery of dynamic epigenome changes that control human cytomegalovirus (HCMV) infection. Mol. Cell. Proteomics 13, 2399–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gunther T., Theiss J. M., Fischer N., and Grundhoff A. (2016) Investigation of Viral and Host Chromatin by ChIP-PCR or ChIP-Seq Analysis. Current Protocols Microbiol. 40, 1E.10.1–21 [DOI] [PubMed] [Google Scholar]
- 33. McKinney C., Kim M. J., Chen D., and McBride A. A. (2016) Brd4 activates early viral transcription upon HPV18 infection of primary keratinocytes. mBio in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Doorslaer K., Porter S., McKinney C., Stepp W. H., and Mcbride A. A. (2016) Novel recombinant papillomavirus genomes expressing selectable genes. Sci Rep-Uk in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baquero-Perez B., and Whitehouse A. (2015) Hsp70 Isoforms Are Essential for the Formation of Kaposi's Sarcoma-Associated Herpesvirus Replication and Transcription Compartments. Plos Pathog. 11, e1005274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bartee E., McCormack A., and Fruh K. (2006) Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. Plos Pathog. 2, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meckes D. G. Jr, Gunawardena H. P., Dekroon R. M., Heaton P. R., Edwards R. H., Ozgur S., Griffith J. D., Damania B., and Raab-Traub N. (2013) Modulation of B-cell exosome proteins by gamma herpesvirus infection. Proc. Natl. Acad. Sci. U.S.A. 110, SE2925–SE2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Konstantinell A., Bruun J. A., Olsen R., Aspar A., Skalko-Basnet N., Sveinbjornsson B., and Moens U. (2016) Secretomic analysis of extracellular vesicles originating from polyomavirus-negative and polyomavirus-positive Merkel cell carcinoma cell lines. Proteomics 16, 2587–2591 [DOI] [PubMed] [Google Scholar]
- 39. Blais D. R., Nasheri N., McKay C. S., Legault M. C., and Pezacki J. P. (2012) Activity-based protein profiling of host-virus interactions. Trends Biotechnol. 30, 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Strmiskova M., Desrochers G. F., Shaw T. A., Powdrill M. H., Lafreniere M. A., and Pezacki J. P. (2016) Chemical methods for probing virus–host proteomic interactions. ACS Infectious Diseases in press [DOI] [PubMed] [Google Scholar]
- 41. Ovaa H., Kessler B. M., Rolen U., Galardy P. J., Ploegh H. L., and Masucci M. G. (2004) Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc. Natl. Acad. Sci. U.S.A. 101, 2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rolen U., Kobzeva V., Gasparjan N., Ovaa H., Winberg G., Kisseljov F., and Masucci M. G. (2006) Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol. Carcinogenesis 45, 260–269 [DOI] [PubMed] [Google Scholar]
- 43. Cravatt B. F., Wright A. T., and Kozarich J. W. (2008) Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Ann. Rev. Biochem. 77, 383–414 [DOI] [PubMed] [Google Scholar]
- 44. Kolb H. C., Finn M. G., and Sharpless K. B. (2001) Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem. Int. Ed. Engl. 40, 2004–2021 [DOI] [PubMed] [Google Scholar]
- 45. Sirbu B. M., McDonald W. H., Dungrawala H., Badu-Nkansah A., Kavanaugh G. M., Chen Y., Tabb D. L., and Cortez D. (2013) Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J. Biol. Chem. 288, 31458–31467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dembowski J. A., and DeLuca N. A. (2015) Selective recruitment of nuclear factors to productively replicating herpes simplex virus genomes. Plos Pathog. 11, e1004939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sakakibara N., Chen D., and McBride A. A. (2013) Papillomaviruses use recombination-dependent replication to vegetatively amplify their genomes in differentiated cells. Plos Pathog. 9, e1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chaurushiya M. S., and Weitzman M. D. (2009) Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amst) 8, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Snider J., Kotlyar M., Saraon P., Yao Z., Jurisica I., and Stagljar I. (2015) Fundamentals of protein interaction network mapping. Mol. Syst. Biol. 11, 848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fields S., and Song O. (1989) A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 [DOI] [PubMed] [Google Scholar]
- 51. Neveu G., Cassonnet P., Vidalain P. O., Rolloy C., Mendoza J., Jones L., Tangy F., Muller M., Demeret C., Tafforeau L., Lotteau V., Rabourdin-Combe C., Trave G., Dricot A., Hill D. E., Vidal M., Favre M., and Jacob Y. (2012) Comparative analysis of virus-host interactomes with a mammalian high-throughput protein complementation assay based on Gaussia princeps luciferase. Methods 58, 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Muller M., and Demeret C. (2012) The HPV E2-Host Protein-Protein Interactions: A Complex Hijacking of the Cellular Network. Open Virol. J. 6, 173–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Uetz P., Dong Y. A., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S. V., Roupelieva M., Rose D., Fossum E., and Haas J. (2006) Herpesviral protein networks and their interaction with the human proteome. Science 311, 239–242 [DOI] [PubMed] [Google Scholar]
- 54. Moorman N. J., Sharon-Friling R., Shenk T., and Cristea I. M. (2010) A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol. Cell. Proteomics 9, 851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. White E. A., Kramer R. E., Tan M. J., Hayes S. D., Harper J. W., and Howley P. M. (2012) Comprehensive analysis of host cellular interactions with human papillomavirus E6 proteins identifies new E6 binding partners and reflects viral diversity J. Virol. 86, 13174–13186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Malik-Soni N., and Frappier L. (2012) Proteomic profiling of EBNA1-host protein interactions in latent and lytic Epstein-Barr virus infections. J. Virol. 86, 6999–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun Q., Tsurimoto T., Juillard F., Li L., Li S., De Leon Vazquez E., Chen S., and Kaye K. (2014) Kaposi's sarcoma-associated herpesvirus LANA recruits the DNA polymerase clamp loader to mediate efficient replication and virus persistence. Proc. Natl. Acad. Sci. U.S.A. 111, 11816–11821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gan J., Wang C., Jin Y., Guo Y., Xu F., Zhu Q., Ding L., Shang H., Wang J., Wei F., Cai Q., and Robertson E. S. (2015) Proteomic profiling identifies the SIM-associated complex of KSHV-encoded LANA. Proteomics 15, 2023–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roux K. J., Kim D. I., Raida M., and Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Varnaite R., and MacNeill S. A. (2016) Meet the neighbors: Mapping local protein interactomes by proximity-dependent labeling with BioID. Proteomics 16, 2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ortiz D. A., Glassbrook J. E., and Pellett P. E. (2016) Protein-protein interactions suggest novel activities of human cytomegalovirus tegument protein pUL103. J. Virol. 90, 7798–7810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Si H., Verma S. C., and Robertson E. S. (2006) Proteomic analysis of the Kaposi's sarcoma-associated herpesvirus terminal repeat element binding proteins J. Virol. 80, 9017–9030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hanash S. (2003) Disease proteomics. Nature 422, 226–232 [DOI] [PubMed] [Google Scholar]
- 64. Malik U. U., Zarina S., and Pennington S. R. (2016) Oral squamous cell carcinoma: Key clinical questions, biomarker discovery, and the role of proteomics. Arch. Oral Biol. 63, 53–65 [DOI] [PubMed] [Google Scholar]
- 65. Chesnokova L. S., and Hutt-Fletcher L. M. (2014) Epstein-Barr virus infection mechanisms. Chin. J. Cancer 33, 545–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McBride A. A. (2016) Oncogenic human papillomaviruses. Philosophical transactions of the Royal Society of London. Series B, Biological sciences [Google Scholar]
- 67. Bonner R. F., Emmert-Buck M., Cole K., Pohida T., Chuaqui R., Goldstein S., and Liotta L. A. (1997) Laser capture microdissection: molecular analysis of tissue. Science 278, 1481, 1483. [DOI] [PubMed] [Google Scholar]
- 68. Arnouk H., Merkley M. A., Podolsky R. H., Stoppler H., Santos C., Alvarez M., Mariategui J., Ferris D., Lee J. R., and Dynan W. S. (2009) Characterization of molecular markers indicative of cervical cancer progression. Proteomics Clin. Appl. 3, 516–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Melle C., Ernst G., Winkler R., Schimmel B., Klussmann J. P., Wittekindt C., Guntinas-Lichius O., and von Eggeling F. (2009) Proteomic analysis of human papillomavirus-related oral squamous cell carcinoma: identification of thioredoxin and epidermal-fatty acid binding protein as upregulated protein markers in microdissected tumor tissue. Proteomics 9, 2193–2201 [DOI] [PubMed] [Google Scholar]
- 70. Li M., Li C., Li D., Xie Y., Shi J., Li G., Guan Y., Li M., Zhang P., Peng F., Xiao Z., and Chen Z. (2012) Periostin, a stroma-associated protein, correlates with tumor invasiveness and progression in nasopharyngeal carcinoma. Clin. Exp. Metastasis 29, 865–877 [DOI] [PubMed] [Google Scholar]
- 71. Jiang R., Gu X., Nathan C. O., and Hutt-Fletcher L. (2008) Laser-capture microdissection of oropharyngeal epithelium indicates restriction of Epstein-Barr virus receptor/CD21 mRNA to tonsil epithelial cells. J. Oral Pathol. 37, 626–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shao Q., Byrum S. D., Moreland L. E., Mackintosh S. G., Kannan A., Lin Z., Morgan M., Stack B. C. Jr, Cornelius L. A., Tackett A. J., and Gao L. (2013) A proteomic study of human merkel cell carcinoma. J. Proteomics Bioinformatics 6, 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Amaya M., Baer A., Voss K., Campbell C., Mueller C., Bailey C., Kehn-Hall K., Petricoin E 3rd, Narayanan A. (2014) Proteomic strategies for the discovery of novel diagnostic and therapeutic targets for infectious diseases. Pathog. Dis. 71, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hanash S. (2011) Progress in mining the human proteome for disease applications. Omics 15, 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Baggerly K. A., Morris J. S., and Coombes K. R. (2004) Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics 20, 777–785 [DOI] [PubMed] [Google Scholar]
- 76. Crutchfield C. A., Thomas S. N., Sokoll L. J., and Chan D. W. (2016) Advances in mass spectrometry-based clinical biomarker discovery. Clin. Proteomics 13, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Buck C. B., Van Doorslaer K., Peretti A., Geoghegan E. M., Tisza M. J., An P., Katz J. P., Pipas J. M., McBride A. A., Camus A. C., McDermott A. J., Dill J. A., Delwart E., Ng T. F., Farkas K., Austin C., Kraberger S., Davison W., Pastrana D. V., and Varsani A. (2016) The ancient evolutionary history of polyomaviruses. Plos Pathog. 12, e1005574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tan M. J., White E. A., Sowa M. E., Harper J. W., Aster J. C., and Howley P. M. (2012) Cutaneous beta-human papillomavirus E6 proteins bind Mastermind-like coactivators and repress Notch signaling. Proc. Natl. Acad. Sci. U.S.A. 109, SE1473–SE1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. White E. A., Sowa M. E., Tan M. J., Jeudy S., Hayes S. D., Santha S., Munger K., Harper J. W., and Howley P. M. (2012) Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc. Natl. Acad. Sci. U.S.A. 109, SE260–SE267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jang M. K., Anderson D. E., van Doorslaer K., and McBride A. A. (2015) A proteomic approach to discover and compare interacting partners of papillomavirus E2 proteins from diverse phylogenetic groups. Proteomics 15, 2038–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jean Beltran P. M., Mathias R. A., and Cristea I. M. (2016) A portrait of the human organelle proteome in space and time during cytomegalovirus infection. Cell Syst. 3, 361–373 e366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. White E. A., and Howley P. M. (2013) Proteomic approaches to the study of papillomavirus-host interactions. Virology 435, 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Maldonado-Torres M., Lopez-Hernandez J. F., Jimenez-Sandoval P., and Winkler R. (2014) 'Plug and Play' assembly of a low-temperature plasma ionization mass spectrometry imaging (LTP-MSI) system. J. Proteomics 102, 60–65 [DOI] [PubMed] [Google Scholar]
- 84. Chait B. T., Cadene M., Olinares P. D., Rout M. P., and Shi Y. (2016) Revealing Higher Order Protein Structure Using Mass Spectrometry. J. Am. Soc. Mass Spectrom. 27, 952–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tran B. Q., Goodlett D. R., and Goo Y. A. (2016) Advances in protein complex analysis by chemical cross-linking coupled with mass spectrometry (CXMS) and bioinformatics. Biochim. Biophys. Acta 1864, 123–129 [DOI] [PubMed] [Google Scholar]
- 86. Waldrip Z. J., Byrum S. D., Storey A. J., Gao J., Byrd A. K., Mackintosh S. G., Wahls W. P., Taverna S. D., Raney K. D., and Tackett A. J. (2014) A CRISPR-based approach for proteomic analysis of a single genomic locus. Epigenetics 9, 1207–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lum K. K., and Cristea I. M. (2016) Proteomic approaches to uncovering virus-host protein interactions during the progression of viral infection. Expert Rev. Proteomics 13, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tackett A. J., DeGrasse J. A., Sekedat M. D., Oeffinger M., Rout M. P., and Chait B. T. (2005) I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J. Proteome Res. 4, 1752–1756 [DOI] [PubMed] [Google Scholar]
- 89. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z. Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A. C., and Nesvizhskii A. I. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., and Lamond A. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J.Cell Biol. 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sharma D., Priyadarshini P., and Vrati S. (2015) Unraveling the web of viroinformatics: computational tools and databases in virus research. J. Virol. 89, 1489–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Van Doorslaer K., Tan Q., Xirasagar S., Bandaru S., Gopalan V., Mohamoud Y., Huyen Y., and McBride A. A. (2013) The Papillomavirus Episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Res. 41, SD571–SD578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. van Doorslaer K., Li Z., Xirasagar S., Maes P., Kaminsky D., Liou D., Sun Q., Kaur R., Huyen Y., and Mcbride A. A. (2016) The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res. in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wenger C. D., Phanstiel D. H., Lee M. V., Bailey D. J., and Coon J. J. (2011) COMPASS: a suite of pre- and post-search proteomics software tools for OMSSA. Proteomics 11, 1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Perez-Riverol Y., Alpi E., Wang R., Hermjakob H., and Vizcaino J. A. (2015) Making proteomics data accessible and reusable: Current state of proteomics databases and repositories. Proteomics 15, 930–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mesri E. A., Feitelson M. A., and Munger K. (2014) Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe 15, 266–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mesri E. A., Cesarman E., and Boshoff C. (2010) Kaposi's sarcoma and its associated herpesvirus. Nat. Rev. Cancer 10, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]