Abstract
Lauraceae are an important component of tropical and subtropical forests and have major ecological and economic significance. Owing to lack of clear-cut morphological differences between genera and species, this family is an ideal case for testing the efficacy of DNA barcoding in the identification and discrimination of species and genera. In this study, we evaluated five widely recommended plant DNA barcode loci matK, rbcL, trnH–psbA, ITS2 and the entire ITS region for 409 individuals representing 133 species, 12 genera from China. We tested the ability of DNA barcoding to distinguish species and as an alternative tool for correcting species misidentification. We also used the rbcL+matK+trnH–psbA+ITS loci to investigate the phylogenetic relationships of the species examined. Among the gene regions and their combinations, ITS was the most efficient for identifying species (57.5%) and genera (70%). DNA barcoding also had a positive role for correcting species misidentification (10.8%). Furthermore, based on the results of the phylogenetic analyses, Chinese Lauraceae species formed three supported monophyletic clades, with the Cryptocarya group strongly supported (PP = 1.00, BS = 100%) and the clade including the Persea group, Laureae and Cinnamomum also receiving strong support (PP = 1.00, BS = 98%), whereas the Caryodaphnopsis–Neocinnamomum received only moderate support (PP = 1.00 and BS = 85%). This study indicates that molecular barcoding can assist in screening difficult to identify families like Lauraceae, detecting errors of species identification, as well as helping to reconstruct phylogenetic relationships. DNA barcoding can thus help with large-scale biodiversity inventories and rare species conservation by improving accuracy, as well as reducing time and costs associated with species identification.
Introduction
Lauraceae in China comprise about 25 widely distributed genera (two endemic, two introduced) with 445 species (316 endemic, three introduced) [1, 2]. The family has major ecological and economic importance [3]. They play an important role in tropical and subtropical forests, often as canopy dominants [1, 4, 5–7] and also have economic significance as sources of medicine, timber, spices, nutritious fruits and perfumes [1, 8, 9]. The fruits of some species contain abundant oil and fat [1]. However, because of their biological and economic relevance, some plants have been overexploitated [10], with 109 species now listed as endangered by the China Species Red List [10].
Because it is important to know whether economic benefit estimates are stable over time [11], the accurate identification of species is important for their protection. However, the taxonomy of Lauraceae, as with other taxonomically complex groups of angiosperms (e.g., Berberis: Roy et al. [12]; Ficus: Li et al. [13]; Curcuma: Chen et al. [14]; Salix: Percy et al. [15]; Rhododendron: Yan et al. [16]), is very poorly resolved. The classification traditionally has been based mainly on morphological characters, such as phyllotaxis, perianth, inflorescence type, size of tepals, number of fertile stamens, number of locules per anther, or fate of tepals in fruit [17–20]. Chinese Lauraceae represent more than 400 species and encompass a broad range of morphological diversity [1], both reproductive and vegetative [21], of which the former are regarded as more effective for classification and identification. However, as most species are tall trees with small, inconspicuous flowers that are not easy to locate or collect in the field [3], generic delimitation within the family is problematic [17–20], particularly as many trees are sterile when sampled (expecially during biodiversity inventories) and must be identified using vegetative characters [21], making misidentifications inevitable [22]. Even with flowers and fruits, the identification and discrimination of Lauraceae taxa can be challenging for non-specialists [4, 21, 23]. So far, scant information is available regarding accurate classification and biodiversity assessment within this family, particularly in south-east Asia, so complementary methods of identification and classification are urgently needed for Lauraceae.
DNA barcoding is a technique for taxonomic identification using one or several standardized DNA regions that are universally present in the target lineages and have sufficient sequence variation to recognize species and identify individuals correctly [24–29]. In order to choose universal DNA barcodes for plants, various molecular markers have been analyzed, including the cpDNA regions, matK, rbcL and trnH–psbA [27, 28, 30, 31], as well as nuclear DNA regions such as ITS (ITS1+5.8S+ITS2) and ITS2 only [32–35]. These regions were chosen based on three main criteria: (a) universality, (b) sequence quality and (c) discriminatory power [29]. DNA barcoding has been shown to be an important tool for species identification and as a supplement to traditional morphology-based taxonomy [36, 37, 38]. Nevertheless, relatively little attention has been paid to sources of potential bias which affect species identification error rates [39], even though species-level barcodes can be prone to substantial errors related to morphological identifications [21]. Combining DNA sequences with existing morphological characters may facilitate species identification and classification [38, 40, 41], as well as broaden our understanding of phylogenetic signal within target lineages [42]. Previous studies have shown that it is not easy to reconstruct phylogenetic relationships in Lauraceae [7, 41, 43–46], so the affinities of species and generic relationships within many of the major lineages in the family are still poorly resolved [7, 18, 43, 44, 47, 48].
Accordingly, in the present study we used existing molecular barcodes: three cpDNA regions (rbcL, matK, psbA–trnH) and the nuclear marker ITS (ITS1+5.8S+ITS2), as well as the subunit ITS2, to examine the taxonomic classification and phylogeny of Lauraceae. Our main aims were to:
evaluate barcode universality in Chinese Lauraceae species;
assess DNA barcoding performance relative to species identification;
determine if these barcodes can also allow for the reconstruction of phylogenetic relationships within the Lauraceae, relative to previously recognized subdivisions and affinities.
Materials and methods
Ethics statement
Collection of these species was conducted in compliance with existing regulations for plants defined as non-commercial, as determined by local government offices. In addition, these sample collections were performed in China with the written approval from the National Forest Bureau and relevant local governments, complying with Chinese and international regulations for the collection of native plant samples.
Sampling
A total of 409 individuals of 133 species from 12 genera of Lauraceae were included in this study (S1 and S2 Tables, Supporting Information), distributed across eight provinces: Chongqing, Guangdong, Guangxi, Hainan, Hunan, Sichuan, Yunnan and Zhejiang, representing much of the diversity of this family in China. Materials for this study were collected in the field from 2002 to 2012, with 22 species represented by a single individual and 111 species represented by two to nine individuals (an average of three samples per species). The Lauraceae expert at KUN, Hsi-Wen Li, who is one of the co-authors, identified the vouchers (S1 Table) based on the reproductive or vegetative characters available. All vouchers were stored at the Herbarium of Xishuangbanna Tropical Botanical Garden (HITBC).
DNA isolation, amplification and sequencing
Total genomic DNA was extracted from silica gel-dried leaf tissue or herbarium specimens using a modified CTAB method [49]. The plastid markers rbcL, matK, trnH–psbA and nuclear markers ITS and ITS2 were amplified using multiple primers, following the suggestions of Dunning and Savolainen [50] and Yu et al. [51]. For example, four primers sets were tested for matK due to its generally poor performance of amplification and sequencing [52]. DMSO and BSA were also added to enhance the PCR performance for matK and ITS [53, 54].
PCR was performed in 20 μL reaction mixtures containing 0.2 μL of Taq polymerase (5 U), 2.0 μL of 10 × PCR buffer, 2.0 μL of 25 mM MgCl2, 2 μL of 2.5 mM dNTPs, 1 μL of 10 uM of each primer, 1 μL of DMSO, 2 μL of 1 mg/ml BSA and 2 μL template DNA. For primer combinations, PCR thermal conditions and references, see Supporting Information (S3 Table). All PCR products were sequenced at the Beijing Genomics Institute (BGI).
Sequence editing and alignment
Raw sequences were assembled and edited using Sequencher 4.14 (GeneCodes Corp., Ann Arbor, Michigan, USA) and deposited in GenBank (see S2 Table for GenBank accession numbers). Edited sequences were then aligned using Geneious 6.1.2 (Biomatters Ltd.), Clustal W [55] and MUSCLE [56], with final manual adjustment undertaken with Geneious 6.1.2 and BioEdit 7.0.9.0 [57]. All variable sites were rechecked on the original trace files for final confirmation. For the rbcL and matK markers, a global multiple sequence alignment was used. The rbcL sequences were unambiguous, due to the absence of insertions or deletions, but alignment of matK was more difficult due to the insertion of triplet codons, so the alignment results were checked visually. The trnH–psbA and ITS sequences were highly variable and very difficult to align with Geneious, so these markers were aligned several times by Clustal W and MUSCLE and then a supermatrix was created by concatenating them with the aligned sequences of the remaining markers.
Data analysis
Two widely applied methods (tree-based and similarity-based) were used to evaluate species discrimination success, following Huang et al. [22]. Five single markers and all possible combinations were applied. For the tree-based method, we used Geneious 6.1.2 to construct Neighbour-Joining (NJ) trees. For the similarity-based method, we used BLAST [58] for building local reference databases against which all sequences were then queried using the blastn program. The 22 species with only a single individual were excluded in NJ trees and BLAST (n ≥ 2) analyses. Species discrimination was considered successful only when all conspecific individuals formed a single clade supported by bootstrap values greater than 50% in the NJ tree [59], and when all individuals of the species or genus only had a top matching hit with a conspecific/congeneric individual in BLAST (the query sequence itself was excluded from the list of top hits when there were multiple individuals).
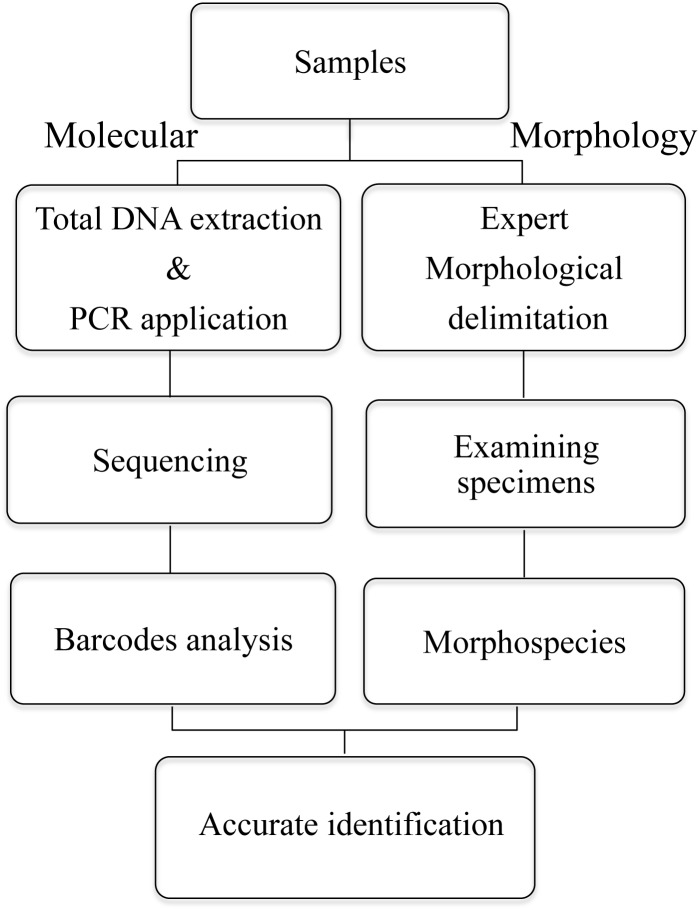
In detecting identification errors, a two-step procedure of reciprocal illumination was used. We evaluated errors in the initial morphology-based identifications combining morphology and DNA sequence data to uncover and correct mistakes in Lauraceae identification. A schematic illustration is used to show the identification process in the present study (Fig 1). Firstly, our initial morphological delimitations were identified by the Lauraceae expert and defined as morphospecies. Then we compared the specimens with herbarium specimens from HITBC, KUN and PYU. Finally, we combined DNA sequences with existing morphological characters. Potential errors were identified through examination of the NJ trees (using rbcL, matK and the combination of rbcL+matK+trnH–psbA+ITS) and BLAST. If the result indicated that the sample did not belong to an a priori assigned taxon, it was flagged as a possible error and the sample was then compared with descriptions and herbarium specimens of the species involved, using morphological characteristics in order to confirm whether an error had been made.
Fig 1. A schematic pipelines of conventional and molecular species identification analyses.
In phylogenetic analyses, combined data sets are often able to generate more resolved and better-supported phylogenies [41, 60], so this approach was also used for Lauraceae. In this study, phylogenetic analyses are inferred from sequence variation in the four-locus combination of rbcL+matK+trnH–psbA+ITS. Bayesian Inference (BI) and Maximum Parsimony (MP) phylogenetic analyses were conducted to reconstruct phylogenetic relationships using PAUP*4.0b10 [61] and MrBayes 3.1.2 [62], with gaps coded as simple indels using the program Gapcoder [63]. For the Bayesian analysis, the dataset was partitioned by markers. Modeltest 3.7 [64, 65] was used to select the best-fit evolutionary model for each partition according to the Akaike Information Criterion (AIC) [66]. The Markov chain Monte Carlo (MCMC) algorithm was run with one cold and three heated chains for 5,000,000 generations, which started from random trees and sampling one out of every 500 generations. Inspection of the log likelihood values suggested that stationarity was reached well before the first 25% implemented as default value for the burn-in and the remaining 75% were used for constructing the consensus tree with the proportion of bifurcations found in this consensus tree given as posterior probabilities (PP). MP analysis was conducted using the following heuristic search options: tree-bisection-reconnection (TBR) branch swapping, collapse of zero length branches and MulTrees on, with 1000 random taxon additions, saving 100 trees from each random sequence addition [66]. All character states were regarded as unordered and equally weighted. Bootstrap support values (BS) for internal nodes were estimated with 100 heuristic bootstrap replicates. The reliability of clades as judged by the posterior probability in Bayesian analysis was generally higher than that as judged by the bootstrap probability in MP analysis [67]. Based on known phylogenies and simulations, bootstrap values of 50% corresponding to posterior probabilities of 90% are generally considered as moderate support of true clade probabilities, and a strong relationship between bootstrap values of 70% corresponding to posterior probabilities of 95% are generally considered as strong support [68, 69]. Three species of Monimiaceae, plus Gomortega nitida Ruiz & Pav. (Gomortegaceae) were selected as outgroups, based on their sister relationship to Lauraceae in a previous study [7]. A sample of the monotypic African genus Hypodaphnis was also included, as the genus is considered to be sister to the remainder of Lauraceae [7], with ITS sequences for these five species downloaded from GenBank.
Results
Barcode universality and sequence characteristics
In total, we obtained 1474 sequences from the 409 samples, representing 133 species of 12 genera after correction. These included 381 sequences for rbcL, 381 sequences for matK, 323 for trnH–psbA, 228 for ITS2 and 161 for ITS (S2 Table). Sequence recovery success rates were very high for rbcL (92.5% of individuals, 97.7% of species, 100% of genera) and matK (92.5% of individuals, 92.5% of species, 100% of genera) with the four commonly used primers pairs matK-1RKIM-f/matK-3FKIM-r, matK-472f/matK-1248r, matK-390f/matK-1326r and matK-xf/matK-5r. Two regions showed moderate success: trnH–psbA region (78.4% of individuals, 86.5% of species, 100% of genera) and ITS2 (55.3% of individuals, 73.7% of species, 100% of genera). In contrast, the ITS region showed the lowest overall recovery rates (39.1% of individuals, 57.9% of species, 100% of genera) (see Table 1).
Table 1. Sequence recovery rates for five DNA barcodes evaluated in this study.
| Barcode regions | rbcL | matK | trnH-psbA | ITS | ITS2 |
|---|---|---|---|---|---|
| Successful individuals/sampled individuals | 381/412 (92.5%) | 381/412 (92.5%) | 323/412 (78.4%) | 161/412 (39.1%) | 228/412 (55.3%) |
| Successful species/sampled species | 130/133 (97.7%) | 123/133 (92.5%) | 115/133 (86.5%) | 77/133 (57.9%) | 98/133 (73.7%) |
| Successful genera/sampled genera | 12/12 (100%) | 12/12 (100%) | 12/12 (100%) | 12/12 (100%) | 12/12 (100%) |
For each category, the absolute number of successes is given along with the percentage relative to the total number. Successful individuals/sampled individuals; Successful species/sampled species; Successful genera/sampled genera (n ≥ 1).
Mistakes in taxonomic identification
After combining DNA sequences with existing morphological characters, various putative species were found to comprise 1–4 individuals that were divergent from the majority of individuals sequenced for their species and that were nested within other species. In these cases, a detailed reanalysis of voucher specimens combined with NJ Tree analyses and BLAST examinations was needed. The results showed that the divergent individuals had been identified incorrectly. In total, 44 individuals (10.8%) had been misidentified by the expert (Table 2, Fig 2; S1 and S2 Figs), 34 at the generic level and 10 at the species level. Following these corrections, we recognised 133 OTUs for the study. The misidentified samples and their identification after revision are listed in Table 2.
Table 2. Original species determinations and correct species using DNA barcodes.
| Original species | Correct species |
|---|---|
| Alseodaphne andersonii J116 | Cryptocarya acutifolia J116 |
| Alseodaphne andersonii J127 | Cryptocarya acutifolia J127 |
| Alseodaphne petiolaris J470 | Cryptocarya acutifolia J470 |
| Beilschmiedia robusta G174 | Beilschmiedia purpurascens G174 |
| Beilschmiedia yunnanensis J467 | Beilschmiedia purpurascens J467 |
| Beilschmiedia yunnanensis GBOW0246 | Machilus robusta GBOW0246 |
| Beilschmiedia yunnanensis GBOW0678 | Machilus yunnanensis GBOW0678 |
| Cinnamomum chartophyllum J088 | Beilschmiedia yunnanensis J088 |
| Cinnamomum chartophyllum J193 | Beilschmiedia yunnanensis J193 |
| Cinnamomum mollifolium J677 | Beilschmiedia roxburghiana J677 |
| Cinnamomum tenuipilum J083 | Litsea acutivena J083 |
| Cryptocarya brachythyrsa J576 | Beilschmiedia brachythyrsa J576 |
| Cryptocarya calcicola L061 | Beilschmiedia purpurascens L061 |
| Cryptocarya calcicola J607 | Cryptocarya acutifolia J607 |
| Cryptocarya chinensis J386 | Beilschmiedia yunnanensis J386 |
| Cryptocarya chinensis J407 | Litsea lancilimba J407 |
| Cryptocarya densiflora GBOW0745 | Caryodaphnopsis laotica GBOW0745 |
| Cryptocarya yunnanensis J485 | Beilschmiedia yunnanensis J485 |
| Lindera latifolia CXQ09023 | Phoebe neurantha CXQ09023 |
| Lindera latifolia GBOW0930 | Machilus grijsii GBOW0930 |
| Lindera latifolia GBOW0936 | Machilus grijsii GBOW0936 |
| Litsea baviensis J227 | Litsea pierrei J227 |
| Litsea elongata G102 | Litsea salicifolia G102 |
| Litsea elongata G198 | Litsea salicifolia G198 |
| Litsea elongata J092 | Litsea acutivena J092 |
| Litsea euosma LJ2002068 | Neolitsea chuii LJ2002068 |
| Litsea glutinosa J133 | Actinodaphne henryi J133 |
| Litsea rotundifolia CXQ0069 | Cryptocary calcicola CXQ0069 |
| Machilus chuanchienensis CXQ0426 | Phoebe tavoyana CXQ0426 |
| Machilus pauhoi CXQ0080 | Litsea greenmaniana var. angustifolia CXQ0080 |
| Machilus salicina Q133 | Litsea greenmaniana var. angustifolia Q133 |
| Machilus viridis Q090 | Cinnamomum chago Q090 |
| Machilus viridis Q129 | Cinnamomum chago Q129 |
| Machilus viridis CXQ690 | Cinnamomum chago CXQ690 |
| Machilus viridis CXQ762 | Cinnamomum chago CXQ762 |
| Machilus yunnanensis LJ2002064 | Cinnamomum chago LJ2002064 |
| Machilus yunnanensis LJ2002072 | Cinnamomum chago LJ2002072 |
| Neolitsea levinei LJ2002035 | Machilus tenuipilis LJ2002035 |
| Neolitsea lunglingensis LJ0014 | Cinnamomum longepaniculatum LJ0014 |
| Neolitsea lunglingensis LJ15201 | Lindera thomsonii LJ15201 |
| Neolitsea lunglingensis LJ2002058 | Neolitsea homilantha LJ2002058 |
| Neolitsea phanerophlebia LJ2006004 | Cinnamomum camphora LJ2006004 |
| Neolitsea phanerophlebia LJ2006083 | Neolitsea chuii LJ2006083 |
| Neolitsea shingningensis CXQ0284 | Lindera fragrans CXQ0284 |
Original species determinations based on morphological characters; correct species based on NJ trees of matK, rbcL and rbcL+matK+trnH–psbA+ITS and BLAST plus re-examination of morphology.
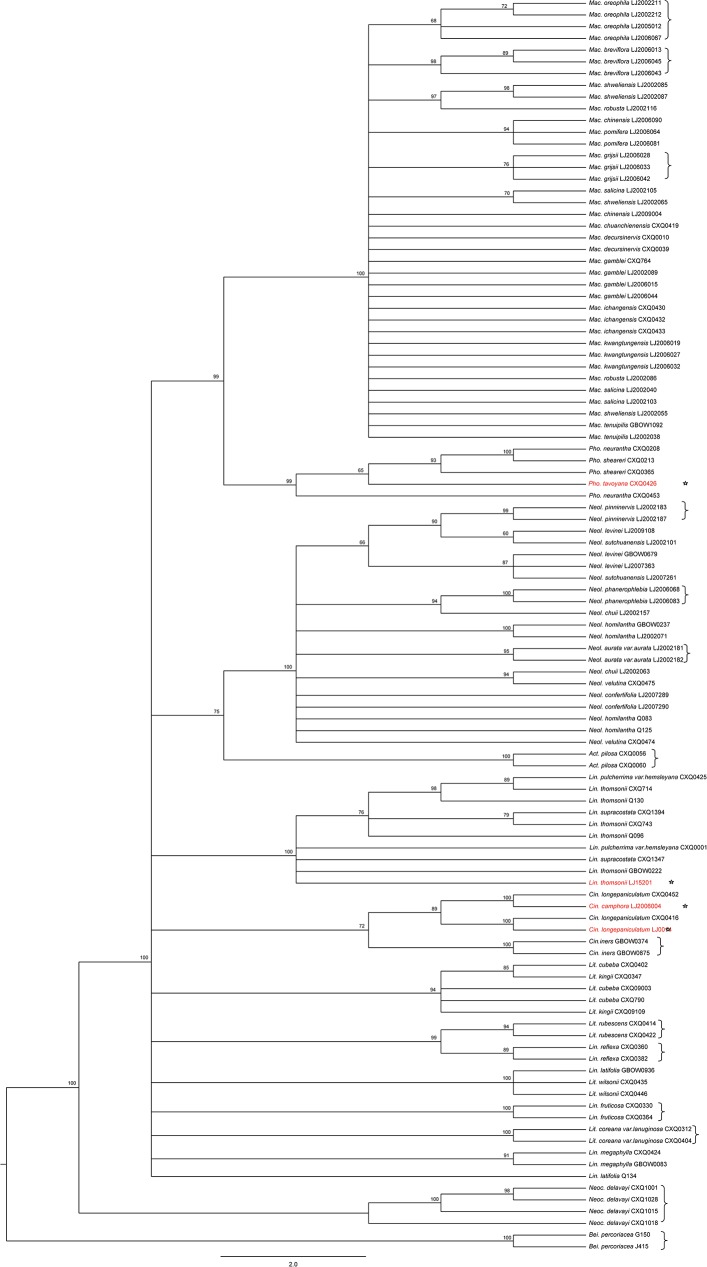
Fig 2. Species misidentification and resolution at the genus and species levels.
The NJ tree based on the combined barcodes rbcL+matK+trnH–psbA+ITS. The bootstrap values ≥ 50% are shown on the branches. The stars represent corrected individuals; brackets represent successfully identified species.
Discrimination efficiency in Lauraceae
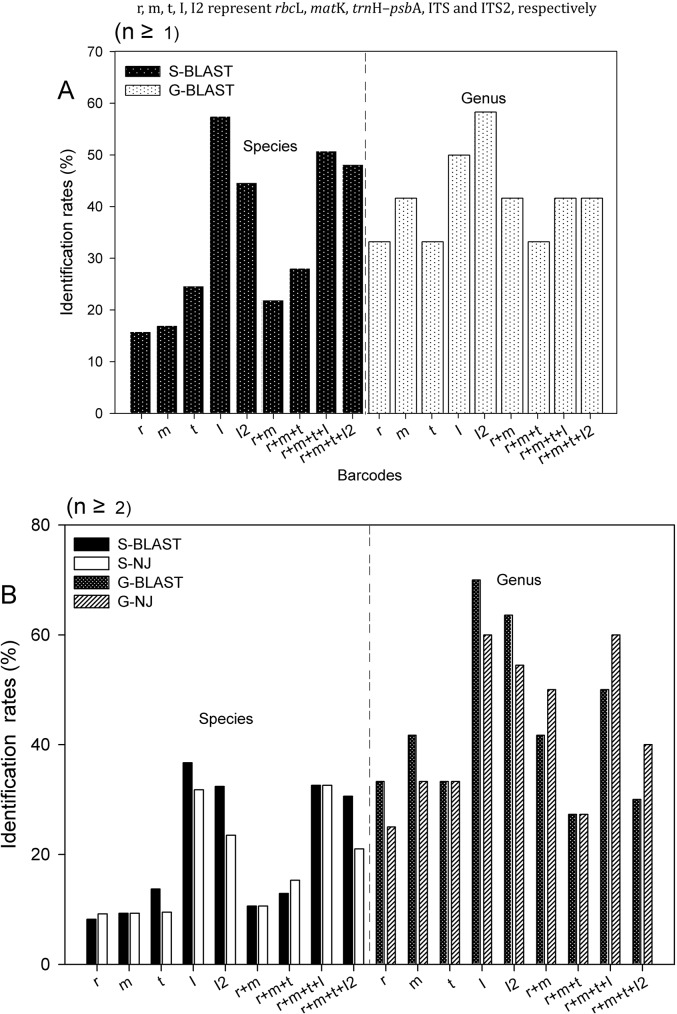
After morphological error correction, the resolution rates of species (8.2–57.5%) and genera (25–70%) were calculated, both for individual barcode sequences, as well as for various combinations (Table 1 and Fig 3). For single barcodes, ITS showed the highest discriminatory power of the five markers (Figs 3 and 4), but the discrimination rate was only 57.5% at the species level in BLAST (n ≥ 1) (see Fig 3A). At the genus level, ITS was again the most accurate (70%) in BLAST (n ≥ 2) (see Fig 3B). ITS2 showed lower sequence variation and species discrimination than ITS (see Fig 3A, 44.7% at species level; Fig 3B, 63.6% at genus level), despite its sequence recovery being more or less double that of ITS (Table 1). The discrimination rates of rbcL were the lowest (see Fig 3B, 8.2% at species level; 25% at genus level).
Fig 3. Species resolution at the genus and species levels for single regions and combinations.
Results based on BLAST and Neighbor-Joining Tree analyses of the samples (A: n ≥ 1; B: n ≥ 2).
Fig 4. Species resolution success at the genus and species levels for ITS.
Result based on Neighbor-Joining Tree analysis (n ≥ 2). The bootstrap values ≥ 50% are shown on the branches. Brackets represent successfully identified species.
Among the marker combinations (rbcL+matK, rbcL+matK+trnH–psbA, rbcL+matK+trnH–psbA+ITS and rbcL+matK+trnH–psbA+ITS2), rbcL+matK+trnH–psbA+ITS showed the highest discriminatory power, with discrimination rates of 50.8% at the species level in BLAST (n ≥ 1) (Fig 3A) and 60% at the genus level in NJ Tree (n ≥ 2) (Fig 3B), whereas rbcL+matK+trnH–psbA+ITS2 showed lower discrimination rates (48.2% and 40%) at the same level (Fig 3). In contrast, the combination of rbcL+matK showed quite low discrimination rates (10.6%) at species level in both BLAST and NJ Tree (n ≥ 2) (see Fig 3B), but discrimination rate were relatively high (50%) at the genus level in NJ Tree (n ≥ 2) (see Fig 3B). Overall, the tree-based method (NJ Tree) and the similarity-based method (BLAST) provided unsatisfactory discrimination rates.
Relationships within Lauraceae
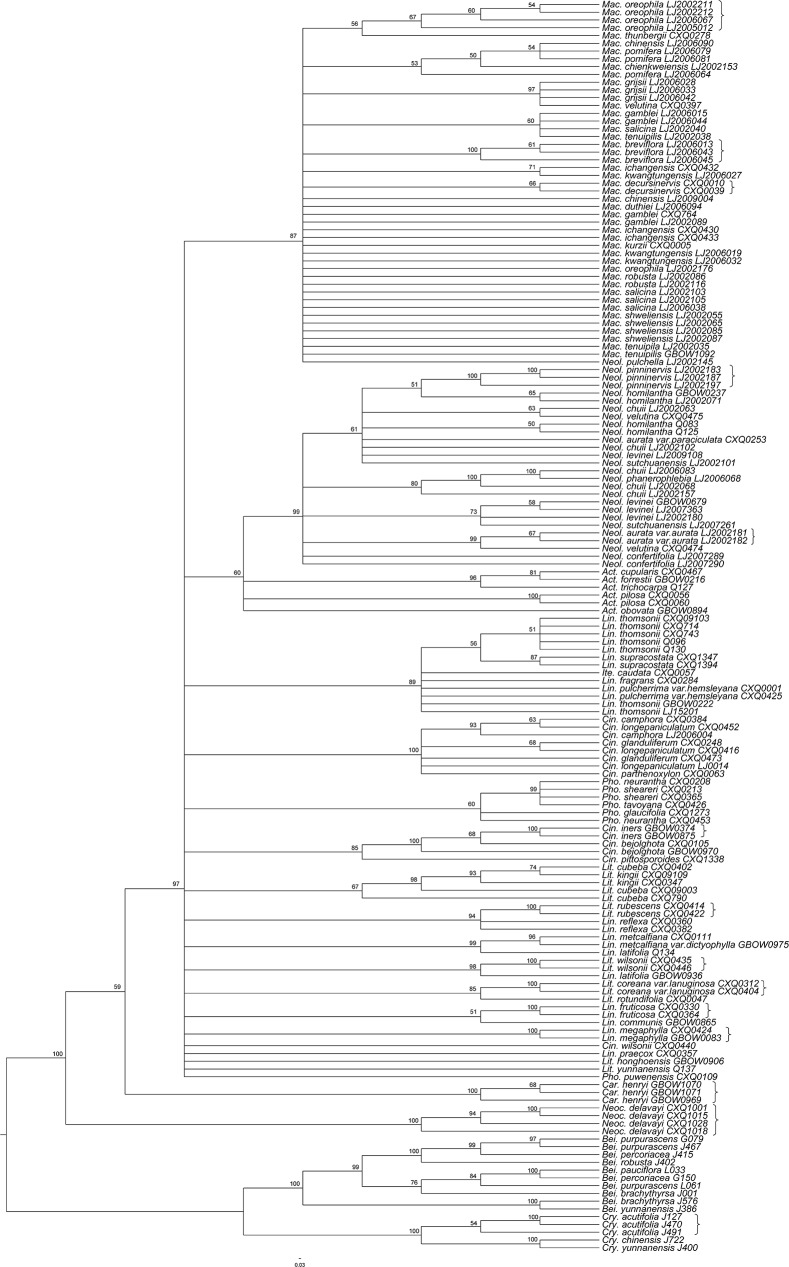
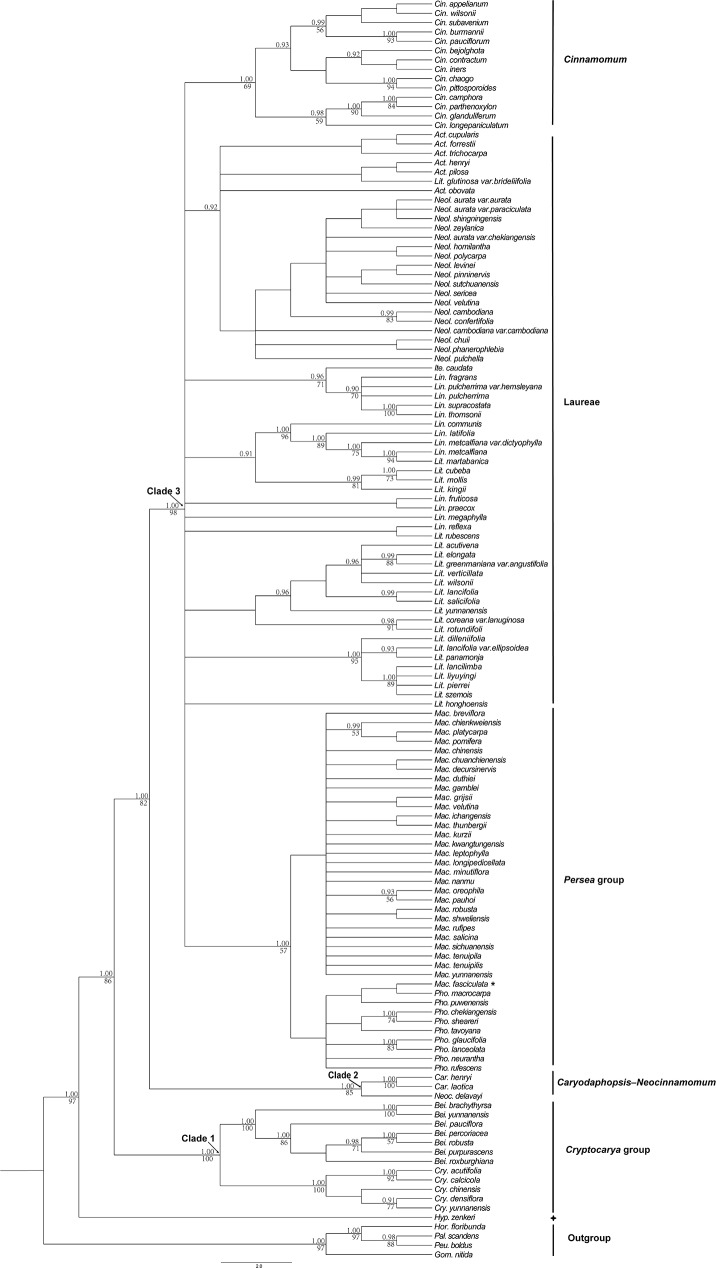
Phylogenetic relationships among 133 species of Lauraceae representing 12 of the 25 genera were analysed after correction. The four DNA markers, rbcL, matK, trnH–psbA and ITS (ITS1+5.8S+ITS2) produced 665, 746, 508 and 845 bp aligned positions respectively, yielding 64, 50, 74, and 149 informative sites and were best fitted to the TVM (Transversional model) +I+G, TIM (Transitional model) +G, K81uf (Two transversion-parameters model 1 unequal frecuencies) +I+G and TrN (Tamura-Nei) +I+G substitution models respectively. As the consensus trees obtained from the BI and MP analyses were almost identical in their topologies, only the Bayesian consensus tree based on rbcL+matK+trnH–psbA+ITS with PP (Posterior Probabilities) and BS values (Bootstrap Support values) is presented here (Fig 5). The Bayesian tree contains three principal Lauraceae clades, with Hypodaphnis strongly supported in BI and MP analyses as their sister group. Clade 1 (PP = 1.00, BS = 100%) includes members of the Cryptocarya group; Caryodaphnopsis and Neocinnamomum form Clade 2 (PP = 1.00, BS = 85%); the remainder, representing the Persea group, Laureae and Cinnamomum formed Clade 3 (PP = 1.00, BS = 98%). In the Clade 3, there is some support for a clade including the representatives of the Persea group (PP = 1.00, BS = 57%) and moderate support for a clade including all members of Cinnamomum investigated here (PP = 1.00, BS = 69%). All the remaining samples, including members of Neolitsea, Actinodaphne, Litsea, Lindera and Iteadaphne belong to the Laureae, which do not form a clade in our analysis, but rather a large polytomy of eight clades, plus Cinnamomum and the Persea group. The outgroup samples, three species of Monimiaceae plus Gomortegaceae also form a monophyletic lineage with strong support (PP = 1.00, BS = 97%).
Fig 5. Bayesian consensus tree based on rbcL+matK+trnH–psbA+ITS.
Bayesian posterior probabilities (≥ 0.9) / Bootstrap support values (≥ 50%) are shown above the branches. Abbreviations: Act. = Actinodaphne, Bei. = Beilschmiedia, Cin. = Cinnamomum, Car. = Caryodaphnopsis, Cry. = Cryptocarya, Ite. = Iteadaphne, Lin. = Lindera, Lit. = Litsea, Mac. = Machilus, Neoc. = Neocinnamomum, Neol. = Neolitsea, Pho. = Phoebe, Hyp. = Hypodaphnis, Hor. = Hortonia, Pal. = Palmeria, Peu. = Peumus, Gom. = Gomortega.
Discussion
Universality of DNA barcodes
Primer universality is an important criterion for a useful DNA barcode [27]. In this regard, the core barcodes (rbcL and matK) for Lauraceae plants had the best performance in PCR amplification and sequencing among the five regions (successfully amplifying and sequencing 92.5% individuals), consistent with a previous study [70]. Compared to the above core barcodes, ITS had a relatively low sequencing success rate of 39.1%, because of the lack of universal primers (either published or with potential development by using current information) and poor success by using existing primers [25]. The poor success by using existing primers is probably due largely to the problem of secondary structure formation resulting in poor quality sequence data, multiple copy numbers, etc. [29, 32, 33, 71, 72]. Thus, this region is probably unsuitable as a universal barcode, although it may be useful in particular cases.
Detecting identification mistakes
Characters such as phyllotaxis, perianth, inflorescence type, size of tepals, or fate of tepals in fruit have been used to delimit the species of Lauraceae [1, 3, 5, 41]. Among these characters, there are some polymorphic characters considered useful at the between-genus level, while they are rarely present together on a specimen when sampled. In Cryptocarya, the fruit completely enclosed in the accrescent receptacular is a remarkable character distinguishing it from other genera; however, only some species were flowering when sampled. Hence, Beilschmiedia purpurascens L061 was wrongly recognized as Cryptocarya calcicola (Table 2). Likewise, the persistent and spreading to reflexed tepals in the fruit of Machilus are important morphological characters for generic delimitation from the closely related genus Phoebe, in which tepals are leathery to woody, conspicuously thickened and clasping the base of the fruit [41]. These characters are also obviously different, but some of these species were also only flowering when sampled, resulting in identification errors, such as Phoebe tavoyana CXQ0426 (Table 2). There are also some morphological identification errors due to scant information about the species. For example, Cinnamomum chago B.S. Sun et H.L. Zhao [73], which had not been included in Flora of China, where if the expert had seen the topotype prior to this study (which has an axillary panicle and short perianth tube), identification errors may not have happened. Furthermore, some genera, such as Lindera, Litsea, Neolitsea and Actinodaphne, which form the Laureae, are really not well defined. All the above factors hampered the accurate identification of Lauraceae. Although each sample in the current study is represented by a voucher that was compared to a reference collection, some species often cannot be distinguished in the absence of complete flowering and fruiting material.
DNA barcoding can act as a tool for detecting errors in species identifications [23]. The tree-based and similarity-based approaches using DNA barcoding in combination with morphology are thus very useful to address identification mistakes based only on morphology [22, 74–77]. Examination of the initially misidentified samples showed that misidentifications were most likely to occur when the samples were only flowering or fruiting and their morphological characters and geographical distributions were similar. Once morphology-based errors listed above were taken into account, mistakes in individual identifications were then only detectable through DNA sequencing.
Revision of morphological identifications based mainly on the core barcodes, or the combination of rbcL+matK+trnH–psbA+ITS, supplemented by BLAST analyses, determined that 10.8% individuals had been misidentified a priori based on morphology (Table 2). This error rate is higher than those reported for some other studies (5.6–10.5%, Archaux et al. [78]; 7.4%, Scott & Hallam [79]; 6.8–7.6%, Dexter et al. [17]; 9.9%, Huang et al. [22]), suggesting that the Lauraceae require careful interpretation of the characters used for specific and generic definition. In particular, accurate recognition of Lauraceae would be very useful because it is the most diverse family in China and is known to be taxonomically problematic.
Evaluation of DNA barcodes for Lauraceae
Our study gives a reliable assessment of barcoding efficacy in the family Lauraceae based on a large sample size, comparable to the results of studies for other diverse angiosperm groups (e.g., Percy et al.: 77 species of Salix [15]; Edwards et al.: 82 species of Aspalathus [80]; Yu et al.: 88 species of Pedicularis [81]). An ideal DNA barcode must combine conserved regions for universal primer design, which show high rates of PCR amplification and sequencing [28] and should also provide a high rate of success for species discrimination and identification [25, 30, 82].
In the present study, the five barcodes performed differently for all samples (Table 1 and Fig 3) and out of all regions tested, ITS performed best, showing the greatest level of species discrimination. However, other studies have described inherent difficulties with this marker [29, 32, 33, 71, 72] and some researchers have advocated using ITS2 alone as a replacement for ITS because it is easier to amplify and sequence this subset of the marker [32, 33]. In contrast, ITS2 showed lower sequence variation and species identification ability than ITS in our study, even though its sequence recovery rate is about two times that of ITS, but we did not observe the other difficulties usually associated with ITS as a barcode marker, so the marker appears to have potential for Lauraceae as long as the low sequencing success rate can be addressed.
ITS was proposed as a DNA barcode for seed plants because of its high species identification ability [25, 33] and in this study ITS provided the highest species resolution, agreeing with the results of recent studies in other plant groups (e.g., Poaceae: Cai et al. [83]; Schisandraceae: Zhang et al. [84]; Orchidaceae: Li et al. [85]). The other four barcoding regions investigated here (rbcL, matK, trnH–psbA and ITS2 alone) have all been proposed as core or supplementary regions for plant barcoding [25, 28, 29,32, 82, 86], but in our study they exhibited low species-level resolution and only Cryptocarya and Beilschmiedia were distinguished clearly from the other genera. This suggests that ITS is the best candidate for Lauraceae when using a single barcode.
Combining DNA barcodes is generally considered to improve species identification [28, 33, 87, 88] and in this study, the discrimination rates of the combinations varied from 10.6% to 32.6% with rbcL+matK < rbcL+matK+trnH–psbA < rbcL+matK+trnH–psbA+ITS2 < rbcL+matK+trnH–psbA+ITS at the species level (Fig 3). However, we can see that the discrimination rates of rbcL+matK are higher than those of rbcL+matK+trnH–psbA and rbcL+matK+trnH–psbA+ITS2 at the genus level. The utility of a marker is not only affected by its discriminatory power, but also by its rate of sequence recovery (Figs 2–5).
Species delimitation in Lauraceae is often complicated by a lack of unique qualitative morphological characters that can be used to define them. DNA barcode data can therefore provide useful additional information for evaluation of observed morphological diversity [89]. Efficient species identification is also important for customs and other authorities to prevent the illegal export and commercial use of protected or rare species [90]. Thus, it is suggested here that using ITS as single barcode, or a combination of barcode markers that included ITS, would be the most suitable approach for barcoding in Lauraceae.
Relationships among major clades
The BI and MP analyses provided relatively good phylogenetic resolution for Lauraceae at both generic and intrageneric levels (Fig 5), especially in basal lineages, with the Cryptocarya group, the Caryodaphnopsis–Neocinnamomum group and the Persea group plus Laureae and Cinnamomum corresponding to our Clades 1, 2 and 3 respectively. Within the Cryptocarya group, which is basal within Lauraceae [7, 47], Cryptocarya is sister to the non-cupulate clade of Beilschmiedia. Cryptocarya has a deeply urceolate floral hypanthium that develops into a deep cupule enclosing the drupe at maturity, except for a small terminal orifice [7, 46], but Beilschmiedia lacks these characters; a synapomorphy that separates Beilschmiedia and related genera (Endiandra and Syndiclis) from the rest of the Cryptocarya group.
Caryodaphnopsis and Neocinnamomum are associated in the present study and have been found previously to have a relatively close relationship [47, 91, 92]. They share triplinerved venation and four-locular anthers with the loculi arranged in a shallow arc [7], sometimes two-locular in Caryodaphnopsis, or in a horizontal row, such as in Neocinnamomum delavayi (Lecomte) H. Liu.
The remaining clade (the Persea group, Laureae and Cinnamomum) with Machilus and Phoebe as subsets of the Persea group received moderate support, agreeing with the studies of Chanderbali et al. [7], Li et al. [41] and Rohwer et al. [44]. However, as with these earlier studies, there was poor resolution for species relationships within Machilus and its presently accepted sections and subsections (e.g. Li et al. [93]) are still questionable. Nevertheless, the present study does suggest that M. fasciculata H. W. Li belongs in Phoebe. Cinnamomum was divided into two clades corresponding to sect. Camphora Meissn. and sect. Cinnamomum [63], reflecting morphological traits such as leaf arrangement, leaf venation pattern, presence or absence of perulate buds or domatia.
The remaining sampled Laureae were poorly resolved, even though a close relationship between Actinodaphne, Lindera, Litsea and Neolitsea has been recognized in almost all Lauraceae classifications [7]. All of these genera are dioecious and most have umbellate inflorescences subtended by involucral bracts [7], but further character evolution study is needed to determine if these features actually represent synplesiomorphies. This suggests that although multilocus molecular markers still do not give well-resolved phylogenies for all Lauraceae, DNA barcoding is nevertheless useful for resolving phylogenetic relationships at the generic or species level within some groups in the family.
Conclusions
The barcodes used here produced positive results for correcting species identification errors and reconstructing phylogenetic relationships of Lauraceae, even though identification rates were not high. Furthermore, because DNA barcoding plays an important role in the conservation of rare species and for forest crime prosecutions, we advocate the use of DNA barcodes, in combination with other techniques, in order to develop adequate management strategies for the long term conservation of Lauraceae. In particular, barcodes such as ITS show promise for large-scale biodiversity assessment and inventory, particularly for tropical tree species, where the use of a single barcode could significantly reduce the time and costs involved with species identification. However, our study also indicates the critical need for additional data from both more taxa and more sequence regions to help resolve issues in Lauraceae taxonomy and conservation, as there is clearly no simple one-size-fits-all barcoding solution for the family.
Supporting information
(XLSX)
(DOCX)
(DOCX)
Erroneous identifications which recognized based on rbcL marker at the genus and species levels are marked by stars.
(TIFF)
Erroneous identifications which recognized based on matK marker at the genus and species levels are marked by stars.
(TIFF)
Acknowledgments
The authors would like to thank the Center for Integrative Conservation. We are grateful to the Plant Phylogenetics & Conservation Group members for previous data accumulation. We also thank Richard T. Corlett, Jens G. Rohwer, Yong Yang and two anonymous reviewers for their comments on this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Natural Science Foundation of China (31370245, 31500454) and a Ministry of Science and Technology (MOST) Grant (2011FY120200-4, 2012FY110400), as well as Southeast Asia Biodiversity Research Institute, Chinese Academy of Science (Y4ZK111B01).
References
- 1.Li HW, Li J, Huang PH, Wei FN, Cui HB, van der Werff H. Lauraceae. In: Wu ZY, Raven PH editors. Flora of China, Calycanthaceae–Schisandraceae. Vol.7 Beijing, and St. Louis, Missouri: Science Press and Missouri Botanical Garden Press; 2008. pp. 102–254. [Google Scholar]
- 2.Yang Y, Liu B. Species catalogue of Lauraceae in China: problems and perspectives. Biod Sci. 2015; 23(2): 232–236. [Google Scholar]
- 3.van der Werff H, Richter HG. Toward an improved classification of Lauraceae. Ann Missouri Bot Gard. 1996; 83: 409–418. [Google Scholar]
- 4.Van der Werff H. An annotated key to the genera of Lauraceae in the Flora Malesiana Region. Blumea. 2001; 46: 125–140. [Google Scholar]
- 5.Rohwer JG. Lauraceae In: Kubitzki K, Rohwer JG, Brittrich V editors. The families and genera of vascular plants. Vol.2 Berlin: Springer-Verlag; 1993. pp. 366–391. [Google Scholar]
- 6.Mabry CM, Hamburg SP, Lin TC, Horng FW, King HB, et al. Typhoon disturbance and stand-level damage patterns at a subtropical forest in Taiwan. Biotropica. 1998; 30: 238–250. [Google Scholar]
- 7.Chanderbali AS, van der Werff HS, Renner S. Phylogeny and historical biogeography of Lauraceae: evidence from the chloroplast and nuclear genomes. Ann Missouri Bot Gard. 2001; 88: 104–134. [Google Scholar]
- 8.Loi DT. Medicinal plants and medicinal taste of vietnam. Science and Technological Publishing House, Hanoi: 1996. [Google Scholar]
- 9.Bolson M, Smidt ED, Brotto ML, Silva-Pereira V. ITS and trnH–psbA as efficient DNA barcodes to identify threatened commercial woody angiosperms from southern Brazilian Atlantic rainforests. PLoS ONE. 2015; 10: e0143049 doi: 10.1371/journal.pone.0143049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S, Xie Y. China species red list. Beijing: Higher Education Press; 2004. pp. 330–334. [Google Scholar]
- 11.Lew DK, Wallmo K. Temporal stability of stated preferences for endangered species protection from choice experiments. Ecol Econ. 2017; 131: 87–97. [Google Scholar]
- 12.Roy S, Tyagi A, Shukla V, Kumar A, Singh UM, Chaudhary LB, et al. Universal plant DNA barcode loci may not work in complex groups: A case study with Indian Berberis species. PLoS ONE. 2010; 5: e13674 doi: 10.1371/journal.pone.0013674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li HQ, Chen JY, Wang S, Xiong SZ. Evaluation of six candidate DNA barcoding loci in Ficus (Moraceae) of China. Mol Ecol Resour. 2012; 12: 783–790. doi: 10.1111/j.1755-0998.2012.03147.x [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Zhao JT, Erickson DL, Xia NH, Kress WJ. Testing DNA barcodes in closely related species of Curcuma (Zingiberaceae) from Myanmar and China. Mol Ecol Resour. 2015; 15: 337–348. doi: 10.1111/1755-0998.12319 [DOI] [PubMed] [Google Scholar]
- 15.Percy DM, Argus GW, Cronk QC, Fazekas AJ, Kesanakurti PR, Burgess KS, et al. Understanding the spectacular failure of DNA barcoding in willows (Salix): Does this result from a trans-specific selective sweep? Mol Ecol. 2014; 23: 4737–4756. doi: 10.1111/mec.12837 [DOI] [PubMed] [Google Scholar]
- 16.Yan LJ, Liu J, Möller M, Zhang L, Zhang XM, et al. DNA barcoding of Rhododendron (Ericaceae), the largest Chinese plant genus in biodiversity hotspots of the Himalaya-Hengduan Mountains. Mol Ecol Resour. 2015; 15: 932–944. doi: 10.1111/1755-0998.12353 [DOI] [PubMed] [Google Scholar]
- 17.Li J, Christophel DC, Conran JG, Li HW. Phylogenetic relationships within the 'core' Laureae (Litsea complex, Lauraceae) inferred from sequences of the chloroplast gene matK and nuclear ribosomal DNA ITS regions. Plant Syst Evol. 2004; 246: 19–34. [Google Scholar]
- 18.Kostermans AJGH. Lauraceae. Pengumuan Balai Besar Penjelidikan Kehutanan Indonesia. 1957; 57: 1–64. [Google Scholar]
- 19.Hutchinson J. The genera of flowering plants. Vol.1 Oxford: Clarendon Press; 1964. [Google Scholar]
- 20.Richter HG. Anatomie des sekundären Xylems und der Rinde der Lauraceae Vol.5 Sonderbande des Naturwiss. Vereins Hamburg Press; 1981. [Google Scholar]
- 21.Dexter KG, Pennington TD, Cunningham CW. Using DNA to assess errors in tropical tree identifications: How often are ecologists wrong and when does it matter? Ecol Monographs. 2010; 80: 267–286. [Google Scholar]
- 22.Huang XC, Ci XQ, Conran JG, Jie L. Application of DNA barcodes in Asian tropical trees—a case study from Xishuangbanna nature reserve, southwest China. PLoS ONE. 2015; 10(6): e0129295 doi: 10.1371/journal.pone.0129295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez MA, Baraloto C, Engel J, Mori SA, Pétronelli P, Riéra B, et al. Identification of Amazonian trees with DNA barcodes. PLoS ONE. 2009; 4: e7483 doi: 10.1371/journal.pone.0007483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hebert PDN, Cywinska A, Ball SL, DeWaard JR. Biological identifications through DNA barcodes. Proc Roy Soc B Bio. 2003; 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci U S A. 2005; 102: 8369–8374. doi: 10.1073/pnas.0503123102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chase MW, Cowan RS, Hollingsworth PM, Van Den Berg C, Madriñán S, Petersen G, et al. A proposal for a standardised protocol to barcode all land plants. Taxon. 2007; 56(2): 295–299. [Google Scholar]
- 27.Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the noncoding trnH–psbA spacer region. PLoS ONE. 2007; 2: e508 doi: 10.1371/journal.pone.0000508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.CBOL Plant Working Group. A DNA barcode for land plants. Proc Natl Acad Sci U S A. 2009; 106: 12794–12797. doi: 10.1073/pnas.0905845106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollingsworth PM. Refining the DNA barcode for land plants. Proc Natl Acad Sci U S A. 2011; 108 (49): 19451–19452. doi: 10.1073/pnas.1116812108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahaye R, Van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, et al. DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci U S A. 2008; 105: 2923–2928. doi: 10.1073/pnas.0709936105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollingsworth ML, Andra CA, Forrest LL, Richardson J, Pennington R, Long D, et al. Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour. 2009; 9: 439–457. doi: 10.1111/j.1755-0998.2008.02439.x [DOI] [PubMed] [Google Scholar]
- 32.Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE. 2010; 5(1): e8613 doi: 10.1371/journal.pone.0008613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci U S A. 2011; 108: 19641–19646. doi: 10.1073/pnas.1104551108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang CY, Wang FY, Yan HF, Hao G, Hu CM, Ge XJ, et al. Testing DNA barcoding in closely related groups of Lysimachia L. (Myrsinaceae). Mol Ecol Resour. 2012; 12(1): 98–108. doi: 10.1111/j.1755-0998.2011.03076.x [DOI] [PubMed] [Google Scholar]
- 35.Aubriot X, Lowry PP II, Cruaud C, Couloux A, Haevermans T. DNA barcoding in biodiversity hotspot: potential value for the identification of Malagasy Euphorbia L. listed in CITES appendices I and II. Mol Ecol Resour. 2013; 13: 57–65. doi: 10.1111/1755-0998.12028 [DOI] [PubMed] [Google Scholar]
- 36.Hebert PD, Gregory TR. The promise of DNA barcoding for taxonomy. Syst Biol. 2005; 54: 852–859. doi: 10.1080/10635150500354886 [DOI] [PubMed] [Google Scholar]
- 37.Packer L, Gibbs J, Sheffield C, Hanner R. DNA barcoding and the mediocrity of morphology. Mol Ecol Resour. 2009; 9: 42–50. doi: 10.1111/j.1755-0998.2009.02631.x [DOI] [PubMed] [Google Scholar]
- 38.Wang FH, Lu JM, Wen J, Ebihara A, Li DZ. Applying DNA barcodes to identify closely related species of ferns: A case study of the Chinese Adiantum (Pteridaceae). PLoS ONE. 2016; 11(9): e0160611 doi: 10.1371/journal.pone.0160611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hull JM, Fish AM, Keane JJ, Mori SR, Sacks BN, Hull AC. Estimation of species identification error: implications for raptor migration counts and trend estimation. J Wildlife Manage. 2010; 74: 1326–1334. [Google Scholar]
- 40.Desalle R. Species discovery versus species identification in DNA barcoding efforts: response to Rubinoff. Conserv Biol. 2006; 20: 1545–1547. doi: 10.1111/j.1523-1739.2006.00543.x [DOI] [PubMed] [Google Scholar]
- 41.Li L, Li J, Rohwer JG, van der Werff H, Wang ZH, Li HW. Molecular phylogenetic analysis of the Persea group (Lauraceae) and its biogeographic implications on the evolution of tropical and subtropical Amphi–Pacific disjunctions. Am J Bot. 2011; 98: 1520–1536. doi: 10.3732/ajb.1100006 [DOI] [PubMed] [Google Scholar]
- 42.Hajibabaei M, Singer GA, Hebert PD, Hickey DA. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 2007; 23: 167–172. doi: 10.1016/j.tig.2007.02.001 [DOI] [PubMed] [Google Scholar]
- 43.Rohwer JG. Toward a phylogenetic classification of the Lauraceae: Evidence from matK sequences. Syst Bot. 2000; 25: 60–71. [Google Scholar]
- 44.Rohwer JG, Li J, Rudolph B, Schmidt SA, van der Werff H, Li HW. Is Persea (Lauraceae) monophyletic? Evidence from nuclear ribosomal ITS sequences. Taxon. 2009; 58: 1153–1167. [Google Scholar]
- 45.Rohwer JG, Rudolph B. Jumping genera: The phylogenetic positions of Cassytha, Hypodaphnis, and Neocinnamomum (Lauraceae) based on different analyses of trnK intron sequences. Ann Missouri Bot Gard. 2005; 92: 153–178. [Google Scholar]
- 46.Fijridiyanto IA, Murakami N. Phylogeny of Litsea and related genera (Laureae-Lauraceae) based on analysis of rpb2 gene sequences. J Plant Res. 2009; 122: 283–298. doi: 10.1007/s10265-009-0218-8 [DOI] [PubMed] [Google Scholar]
- 47.Li J, Conran JG, Christophel DC, Li ZM, Li L, Li HW. Phylogenetic relationships of the Litsea complex and core Laureae (Lauraceae) using ITS and ETS sequences and morphology. Ann Missouri Bot Gard. 2008; 95: 580–599. [Google Scholar]
- 48.Rohwer JG, de Moraes PLR, Rudolph B, van der Werff H. A phylogenetic analysis of the Cryptocarya group (Lauraceae), and relationships of Dahlgrenodendron, Sinopora, Triadodaphne, and Yasunia. Phytotaxa. 2014; 158: 111–132. [Google Scholar]
- 49.Doyle JJ, Doyle JS. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987; 19: 11–15. [Google Scholar]
- 50.Dunning LT, Savolainen V. Broad-scale amplification of matK for DNA barcoding plants, a technical note. Bot J Lin Soc. 2010; 164: 1–9. [Google Scholar]
- 51.Yu J, Xue JH, Zhou SL. New universal matK primers for DNA barcoding angiosperms. J Syst Evol. 2011; 49: 176–181. [Google Scholar]
- 52.Fazekas AJ, Kesanakurti PR, Burgess KS, Percy DM, Graham SW, Barrett SCH, et al. Are plant species inherently harder to discriminate than animal species using DNA barcoding markers? Mol Ecol Resour. 2009; 9: 130–139. doi: 10.1111/j.1755-0998.2009.02652.x [DOI] [PubMed] [Google Scholar]
- 53.Ralser M, Querfurth R, Warnatz HJ, Lehrach H, Yaspo ML, et al. An efficient and economic enhancer mix for PCR. Biochem Bioph Res Co. 2006; 347: 747–751. [DOI] [PubMed] [Google Scholar]
- 54.Lister DL, Bower MA, Howe CJ, Jones MK. Extraction and amplification of nuclear DNA from herbarium specimens of emmer wheat: a method for assessing DNA preservation by maximum amplicon length recovery. Taxon. 2008; 57: 254–258. [Google Scholar]
- 55.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32: 1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 1999; 41: 95–98. [Google Scholar]
- 58.Blaxter M, Mann J, Chapman T, Thomas F, Whitton C, Floyd R, et al. Defining operational taxonomic units using DNA barcode data. Philos Trans Roy Soc B Bio. 2005; 360: 1935–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J, Provan J, Gao LM, Li DZ. Sampling strategy and potential utility of indels for DNA barcoding of closely related plant species: A case study in Taxus. Int J Mol Sci. 2012; 13: 8740–8751. doi: 10.3390/ijms13078740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L, Li J, Conran JG, Li XW, Li HW. Phylogeny of Neolitsea (Lauraceae) inferred from Bayesian analysis of nrDNA ITS and ETS sequences. Plant Syst Evol. 2007; 269: 203–221. [Google Scholar]
- 61.Swofford DL. PAUP*: Phylogenetic analysis using parsimony (* and other methods), version 4.0b10. Sunderland: Sinauer; 2003. [Google Scholar]
- 62.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001; 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 63.Young ND, Healy J. GapCoder automates the use of indel characters in phylogenetic analysis. Bioinformatics. 2003; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998; 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 65.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004; 53: 793–808. doi: 10.1080/10635150490522304 [DOI] [PubMed] [Google Scholar]
- 66.Huang JF, Li L, van der Werff H, Li HW, Rohwer JG, Crayn DM. Origins and evolution of cinnamon and camphor: A phylogenetic and historical biogeographical analysis of the Cinnamomum group (Lauraceae). Mol Phylogenet Evol. 2016; 96: 33–44. doi: 10.1016/j.ympev.2015.12.007 [DOI] [PubMed] [Google Scholar]
- 67.Suzuki Y, Glazko GV, Nei M. Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proc Natl Acad Sci U S A. 2002; 99(25): 16138–16143. doi: 10.1073/pnas.212646199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zharkikh A, Li WH. Statistical properties of bootstrap estimation of phylogenetic variability from nucleotide sequences: I. Four taxa with a molecular clock. Mol Biol Evol. 1992; 9:1119–1147. [DOI] [PubMed] [Google Scholar]
- 69.Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing condence in phylogenetic analysis. Syst Biol. 1993; 42:182–192. [Google Scholar]
- 70.Li FW, Kuo LY, Rothfels CJ, Ebihara A, Chiou WL, Windham MD, et al. rbcL and matK earn two thumbs up as the core DNA barcode for ferns. PLoS ONE. 2011; 6(10): e26597 doi: 10.1371/journal.pone.0026597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Starr JR, Naczi RF, Chouinard BN. Plant DNA barcodes and species resolution in sedges (Carex, Cyperaceae). Mol Ecol Resour. 2009; 9: 151–163. doi: 10.1111/j.1755-0998.2009.02640.x [DOI] [PubMed] [Google Scholar]
- 72.Xiao LQ, Moller M, Zhu H. High nrDNA ITS polymorphism in the ancient extant seed plant Cycas: Incomplete concerted evolution and the origin of pseudogenes. Mol Phylogenet Evol. 2010; 55: 168–177. doi: 10.1016/j.ympev.2009.11.020 [DOI] [PubMed] [Google Scholar]
- 73.Sun BS, Zhao HL. A new species of Cinnamomum from Yunnan. J Yunnan Univ. 1991; 13: 93–94. [Google Scholar]
- 74.Ferri G, Alu M, Corradini B, Beduschi G. Forensic botany: species identification of botanical trace evidence using a multigene barcoding approach. Int J Legal Med. 2009; 123: 395–401. doi: 10.1007/s00414-009-0356-5 [DOI] [PubMed] [Google Scholar]
- 75.Yassin A, Markow TA, Narechania A, O'Grady PM, DeSalle R. The genus Drosophila as a model for testing tree- and character-based methods of species identification using DNA barcoding. Mol Phylogenet Evol. 2010; 57: 509–517. doi: 10.1016/j.ympev.2010.08.020 [DOI] [PubMed] [Google Scholar]
- 76.Piredda R, Simeone MC, Attimonelli M, Bellarosa R, Schirone B. Prospects of barcoding the Italian wild dendroflora: oaks reveal severe limitations to tracking species identity. Mol Ecol Resour. 2011; 11: 72–83. doi: 10.1111/j.1755-0998.2010.02900.x [DOI] [PubMed] [Google Scholar]
- 77.Oihana R, Benjamín JGM, Katerina V, Martínez‐Ortí A, Madeira MJ. Species delimitation for cryptic species complexes: Case study of Pyramidula (Gastropoda, Pulmonata). Zool Scr. 2016. [Google Scholar]
- 78.Archaux F, Gosselin F, Berges L, Chevalier R. Effects of sampling time, species richness and observer on the exhaustiveness of plant censuses. J Veg Sci. 2006; 17: 299–306. [Google Scholar]
- 79.Scott WA, Hallam CJ. Assessing species misidentification rates through quality assurance of vegetation monitoring. Plant Ecol. 2003; 165: 101–115. [Google Scholar]
- 80.Edwards D, Horn A, Taylor D, Savolain V, Hawkins JA. DNA barcoding of a large genus, Aspalathus L. (Fabaceae). Taxon. 2008; 57: 1317–1327. [Google Scholar]
- 81.Yu WB, Huang PH, Ree RH, Liu ML, Li DZ, Wang H. DNA barcoding of Pedicularis L. (Orobanchaceae): Evaluating four universal barcode loci in a large and hemiparasitic genus. J Syst Evol. 2011; 49: 425–437. [Google Scholar]
- 82.Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS ONE. 2011; 6(5): e19254 doi: 10.1371/journal.pone.0019254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cai ZM, Zhang YX, Zhang LN, Gao LM, Li DZ. Testing four candidate barcoding markers in temperate woody bamboos (Poaceae: Bambusoideae). J Syst Evol. 2012; 50: 527–539. [Google Scholar]
- 84.Zhang J, Chen M, Dong XY, Lin RZ, Fan JH, Chen Z. Evaluation of four commonly used DNA barcoding loci for Chinese medicinal plants of the family Schisandraceae. PLoS ONE. 2015; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li YL, Tong Y, Xing FW. DNA barcoding evaluation and its taxonomic implications in the recently evolved genus Oberonia Lindl. (Orchidaceae) in China. Front Plant Sci. 2016; 7: 1791 doi: 10.3389/fpls.2016.01791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tripathi AM, Tyagi A, Kumar A, Singh A, Singh S, Chaudhary LB, et al. The internal transcribed spacer (ITS) region and trnH–psbA are suitable candidate loci for DNA barcoding of tropical tree species of India. PLoS ONE. 2013; 8: e57934 doi: 10.1371/journal.pone.0057934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, et al. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE. 2008; 3: e2802 doi: 10.1371/journal.pone.0002802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang JB, Wang YP, Moller M, Gao LM, Wu D. Applying plant DNA barcodes to identify species of Parnassia (Parnassiaceae). Mol Ecol Resour. 2012; 12: 267–275. doi: 10.1111/j.1755-0998.2011.03095.x [DOI] [PubMed] [Google Scholar]
- 89.Muellner AN, Schaefer H, Lahaye R. Evaluation of candidate DNA barcoding loci for economically important timber species of the mahogany family (Meliaceae). Mol Ecol Resour. 2011; 11: 450–460. doi: 10.1111/j.1755-0998.2011.02984.x [DOI] [PubMed] [Google Scholar]
- 90.Newmaster S, Fazekas A, Steeves R, Janovec J. Testing candidate plant barcode regions in the Myristicaceae. Mol Ecol Resour. 2008; 8: 480–490. doi: 10.1111/j.1471-8286.2007.02002.x [DOI] [PubMed] [Google Scholar]
- 91.Wang ZH, Li J, Conran JG, Li HW. Phylogeny of the Southeast Asian endemic genus Neocinnamomum H. Liu (Lauraceae). Plant Syst Evol. 2010; 290: 173–184. [Google Scholar]
- 92.Li L, Madriñán S, Li J. Phylogeny and biogeography of Caryodaphnopsis (Lauraceae) inferred from low-copy nuclear gene and ITS sequences. Taxon. 2016; 65: 433–443. [Google Scholar]
- 93.Li HW, Pai PY, Lee SK. Wei FN, Wei YT, Yang YC, et al. Lauraceae In: Li H-W editor. Flora Reipublicae Popularis Sinicae. Vol.31 Beijing: Science Press; 1984. pp.1–463. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(DOCX)
(DOCX)
Erroneous identifications which recognized based on rbcL marker at the genus and species levels are marked by stars.
(TIFF)
Erroneous identifications which recognized based on matK marker at the genus and species levels are marked by stars.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.