ABSTRACT
Thymus-derived regulatory T (tTreg) cells are key to preventing autoimmune diseases, but the mechanisms involved in their development remain unsolved. Here, we show that the C-type lectin receptor CD69 controls tTreg cell development and peripheral Treg cell homeostasis through the regulation of BIC/microRNA 155 (miR-155) and its target, suppressor of cytokine signaling 1 (SOCS-1). Using Foxp3-mRFP/cd69+/− or Foxp3-mRFP/cd69−/− reporter mice and short hairpin RNA (shRNA)-mediated silencing and miR-155 transfection approaches, we found that CD69 deficiency impaired the signal transducer and activator of transcription 5 (STAT5) pathway in Foxp3+ cells. This results in BIC/miR-155 inhibition, increased SOCS-1 expression, and severely impaired tTreg cell development in embryos, adults, and Rag2−/− γc−/− hematopoietic chimeras reconstituted with cd69−/− stem cells. Accordingly, mirn155−/− mice have an impaired development of CD69+ tTreg cells and overexpression of the miR-155-induced CD69 pathway, suggesting that both molecules might be concomitantly activated in a positive-feedback loop. Moreover, in vitro-inducible CD25+ Treg (iTreg) cell development is inhibited in Il2rγ−/−/cd69−/− mice. Our data highlight the contribution of CD69 as a nonredundant key regulator of BIC/miR-155-dependent Treg cell development and homeostasis.
KEYWORDS: regulatory T cell development, C-type lectin, miR-155, autoimmunity, microRNA
INTRODUCTION
Regulatory T (Treg) cells are a specialized subset of lymphocytes with a dominant role in the prevention of autoimmune diseases (1). Treg cell subtypes have been classified according to their origin in the thymus, peripheral lymphoid organs, or in vitro and have been extensively characterized; however, the mechanisms that regulate their generation in the thymus remain poorly understood. Understanding how thymus-derived Treg (tTreg) cells (2) become a distinct lineage is crucial for the development of strategies to control immune responses by targeting these cells (3). A central event in tTreg cell differentiation is the induction of the transcription factor Foxp3 by early signals delivered from the T cell (TC) receptor (TCR), which results in transcriptional activation and enhanced function of the interleukin-2 (IL-2) signaling pathway (4). Among other mechanisms, Foxp3 expression is promoted by the microRNA (miRNA) 155 (miR-155) through the inhibition of SOCS-1 (suppressor of cytokine signaling 1), enhancing the activation and binding of STAT5 (signal transducer and activator of transcription 5) to the Foxp3 promoter and the Foxp3-CNS (conserved noncoding sequence) (5, 6). In a positive-feedback loop, Foxp3 increases the expression of miR-155 by binding to an intronic element of BIC, the gene encoding the miR-155 precursor transcript. Nevertheless, the mechanisms by which miRNAs impact tTreg cell differentiation and function are not fully elucidated, and the data are somewhat contradictory. For example, Dicer, a member of the RNase III complex that processes pre-miRNAs into mature miRNAs, plays a key role in tTreg cell differentiation (7) and function (8); however, the lack of Dicer is linked to enhanced miR-155 expression in MRL/lpr mice (9), suggesting that there are Dicer-independent mechanisms for miRNA regulation in Treg cells. The Treg cells of lupus-prone mice have an altered phenotype, low levels of Dicer, and a weak suppressive capacity linked to the expression of the C-type lectin receptor CD69 (9). Moreover, increased CD69 expression has been detected in activated Dicer−/− TCs, which show defective egress from lymphoid organs (10). In addition, CD4+ CD8+ thymocytes include a CD69high TCRhigh Treg cell progenitor subpopulation, indicating that CD69 expression is relevant to tTreg cell differentiation (11). We hypothesized that CD69, which contributes to the maintenance of immunological tolerance through the regulation of Treg cell function, makes a substantial contribution to Treg cell development in the thymus. The C-type lectin CD69 is expressed constitutively by a subpopulation of peripheral Treg (pTreg) cells and tTreg cells (12). Here, we report that CD69 is required for the development of Treg cells in the thymus through the promotion of STAT5 phosphorylation and the transcription of BIC/miR-155. FoxP3-mRFP/cd69−/− reporter mice have a significantly below-normal number of tTreg cells, and Treg cell differentiation was also impaired in fetal thymus organ cultures (FTOCs) of cd69−/− embryonic thymuses or wild-type embryonic thymuses treated with anti-CD69. Consistently, FoxP3+ tTreg cells are poorly generated from cd69−/− precursors in mixed bone marrow chimeras. An impairment of STAT5 phosphorylation in FoxP3-mRFP/cd69−/− tTreg cells leads to the enhanced transcription of SOCS-1 and the inhibition of miR-155-dependent tTreg cell development. CD69 thus maintains miR-155-dependent tTreg cell development through a positive-feedback regulatory mechanism, giving rise to a functional pTreg cell subset. Our results strongly support a role for CD69 as a critical receptor in the control of Treg cell development and homeostasis.
RESULTS
CD69 expression is required for development of the tTreg cell subset.
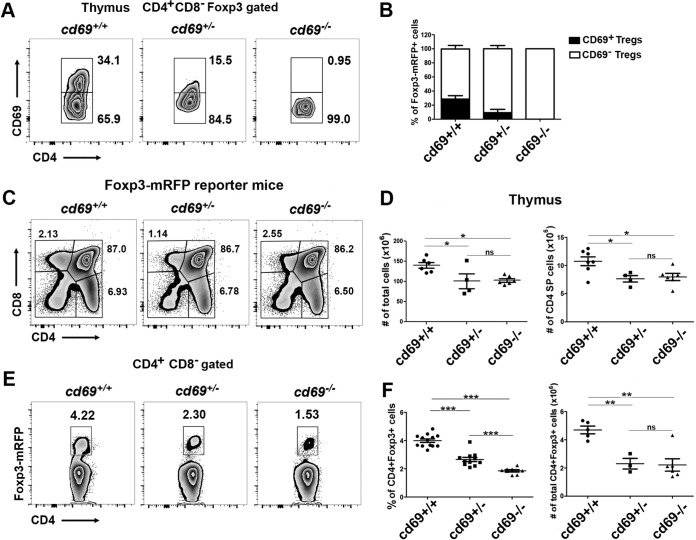
To determine whether CD69 is necessary for tTreg cell development in the thymus, we analyzed CD69 membrane expression in tTreg cells from cd69+/+, cd69+/−, and cd69−/− littermates bearing a Foxp3-mRFP reporter gene (monomeric red fluorescent protein inserted into the foxp3 locus). In agreement with previously reported data for nonreporter mice (12), about 30% of tTreg cells expressing Foxp3-mRFP in wild-type thymuses also express CD69 (Fig. 1A and B). This percentage is lower in Foxp3-mRFP/cd69+/− heterozygous mice, and this subset is absent in Foxp3-mRFP/cd69−/− mice (Fig. 1A and B). The proportions of CD4+ single-positive (SP) (CD4SP) thymocytes and the other thymocyte subsets are unaffected in cd69 heterozygous and cd69-deficient reporter littermates (Fig. 1C), but compared with Foxp3-mRFP/cd69+/+ mice, both genotypes showed a 30% lower cellularity of total and CD4SP thymocytes (Fig. 1D). These results are consistent with previously reported data showing that the overexpression of CD69 in the thymus increases the levels of SP thymocytes controlling egress to the periphery (13, 14). However, Foxp3-mRFP/cd69−/− and cd69+/− mice showed a marked reduction in the proportion of tTreg cells compared with that in cd69+/+ adult reporter mice (Fig. 1E and F), while total tTreg cell numbers were not altered in the cd69+/− and cd69−/− groups (Fig. 1F), indicating that CD69 could be playing an important role in the regulation of tTreg cell development masked by thymocyte egress defects in Foxp3-mRFP/cd69−/− mice. In addition, we found that cd69−/− adult reporter mice also showed a reduction in the proportion of pTreg cells compared with that in cd69+/+ littermates (see Fig. S1A and B in the supplemental material). These data are not consistent with data reported previously for nonreporter mice (12). To clarify the differences observed with Foxp3 reporter mice, we performed Foxp3 staining in thymuses and spleens from Foxp3-mRFP mice. The data indicate that exogenous staining with anti-Foxp3 antibodies (Abs) differs from the endogenous print of Foxp3-mRFP depending on the tissue (Fig. S2), suggesting that the use of anti-Foxp3 antibodies is not always as accurate as the use of reporter genes. In summary, CD69 could be playing a role in both tTreg cell development and pTreg cell homeostasis.
FIG 1.
CD69 expression is required for thymus-derived Treg cell homeostasis in adult mice. (A) Density plots showing CD69 expression in CD4+ CD8− Foxp3+-gated thymocytes from 8- to 12-week-old Foxp3-mRFP/cd69+/+ (wild-type), Foxp3-mRFP/cd69+/− (heterozygous), and Foxp3-mRFP/cd69−/− (deficient) reporter littermates. Numbers indicate the proportions (percentages) of gated cells. (B) Bar chart showing the percentages (±standard deviations) of CD69+ and CD69− tTreg cells within the thymus of the indicated reporter mice. (C) Flow cytometry analysis of thymocyte subsets in 8- to 10-week-old reporter littermates. The percentages of thymus-derived T cell subsets are shown. (D) Cellularity of the thymus (left) and total numbers of CD4SP cells (right) in reporter littermates. (E) Analysis of endogenous Foxp3 expression in tTreg cells in the thymuses of reporter littermates. (F) Percentages (left) and total cell numbers (right) of gated CD4+ CD8− Foxp3+ tTreg cells in adult reporter littermates. Data are from at least 7 litters with 3 to 12 littermates each. Totals of 16 Foxp3-mRFP/cd69+/+ (wild-type), 11 Foxp3-mRFP/cd69+/− (heterozygous), and 12 Foxp3-mRFP/cd69−/− (deficient) mice were analyzed. Error bars show standard deviations. Data were evaluated by ANOVA followed by Bonferroni's multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Deletion of CD69 inhibits tTreg cell differentiation in fetal thymus organ cultures.
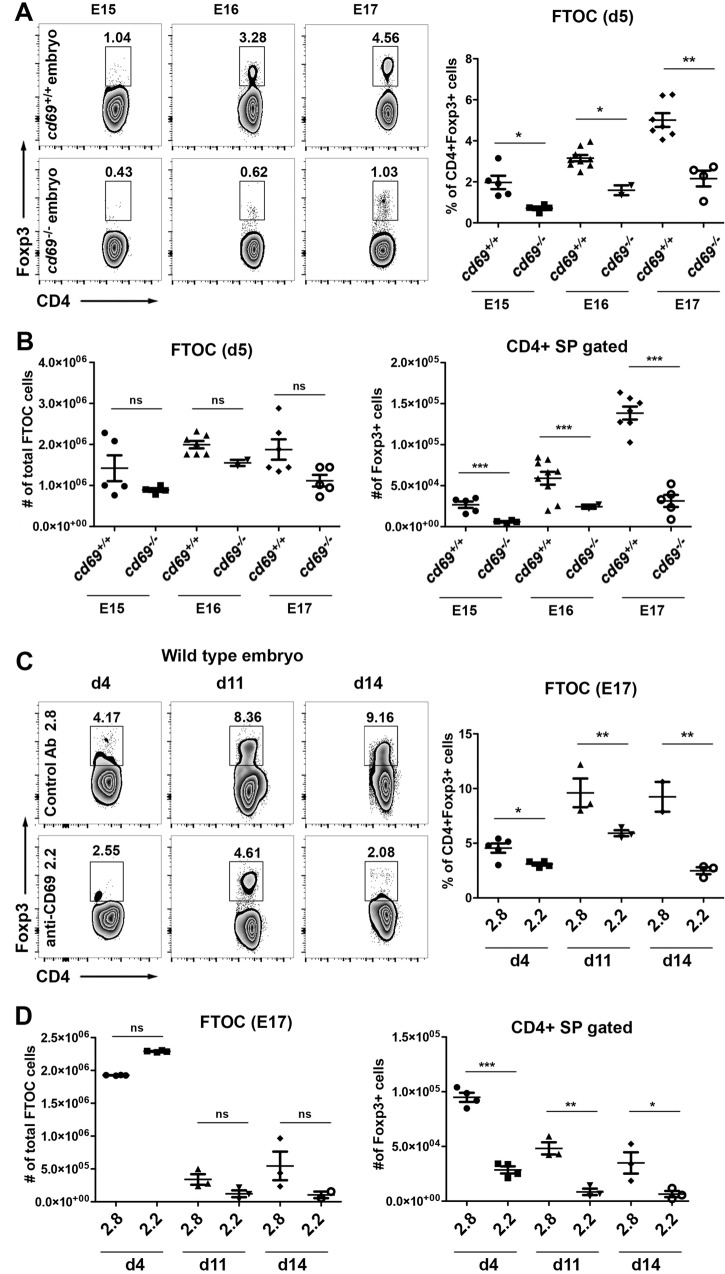
To determine if cd69 deficiency leads to decreased tTreg cell development, independently of the thymic maturation state or the sphingosine 1-phosphate receptor 1 (S1P1)-induced thymocyte egress capacity, we performed an FTOC assay on thymuses from 15- to 17-day-old mouse embryos (embryonic day 15 [E15] to E17) and analyzed total CD4SP thymocytes and tTreg cell differentiation over 5 days of culture. Compared with cd69+/+ FTOCs, cd69−/− E15 to E17 FTOCs displayed marked reductions in the proportions and absolute cell numbers of Foxp3+ tTreg cells, with insignificant changes in total cell numbers (Fig. 2A and B), indicating that CD69 is required during tTreg cell differentiation at early stages of development. To confirm these results, we treated E15 FTOCs with an anti-CD69 monoclonal antibody (MAb) (2.2), which downregulates CD69 expression and hence blocks downstream signaling (15), and monitored Treg cell development over 14 days of culture. Consistent with the cd69−/− FTOC data, throughout the culture period, anti-CD69-treated FTOCs showed notably lower proportions and cell numbers of Foxp3+ tTreg cells than did FTOCs treated with the isotype control antibody (2.8) (Fig. 2C and D), whereas total FTOC cell numbers were unaltered by either treatment (Fig. 2D). These findings are consistent with previously reported evidence indicating that immature activated CD69+ thymocytes are the precursors of intrathymic Treg cells in humans and mice (11, 16).
FIG 2.
tTreg cell differentiation in fetal thymus organ culture requires CD69 expression. (A) Representative density plots of 5-day FTOCs from cd69+/+ and cd69−/− embryos in the C57BL/6 background. Embryonic thymuses were removed from 15- to 17-day-old embryos, and the percentages of tTreg development in the lobes were determined by FACS analysis. (B) Cellularity of fetal thymus lobes (left) and total cell numbers of CD4+ Foxp3+ cells (right) from cd69+/+ and cd69−/− embryos. (C) FTOCs from wild-type 17-day-old embryos (E17) were maintained for up to 14 days in culture in the presence of anti-CD69 monoclonal antibody (2.2) or the isotype control antibody (2.8). Density plots shows the percentages of tTreg cells on days 4, 11, and 14 after culture. (D) Cellularity of fetal thymus lobes (left) and total numbers of CD4+ Foxp3+ cells (right) under each condition. Totals of 31 and 36 embryos from five cd69+/+ and four cd69−/− females, respectively, were analyzed. The 2 lobes from each fetal thymus were analyzed separately. Error bars show standard deviations. Values were calculated relative to data for cd69+/+ control lobes from four independent FTOC assays. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test).
Defective tTreg and pTreg cell generation from cd69−/− progenitors is a cell-autonomous defect.
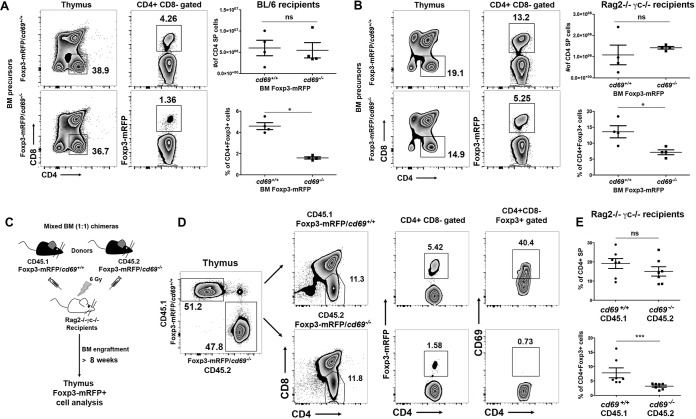
To further explore the role of CD69 in tTreg cell differentiation, we transferred bone marrow (BM) hematopoietic stem cells from Foxp3-mRFP/cd69+/+ or Foxp3-mRFP/cd69−/− littermates into lethally γ-irradiated C57BL/6 recipients (Fig. 3A). Twelve weeks after reconstitution, percentages and numbers of CD4+ Foxp3+ Treg cells derived from cd69−/− BM precursors were markedly lower in the thymus (Fig. 3A) and blood (see Fig. S3 in the supplemental material) than in those derived from cd69+/+ precursors, indicating an impaired Treg cell regeneration capacity of cd69−/− BM hematopoietic stem cells. Moreover, we analyzed the potential of these precursors to differentiate into tTreg cells in sublethally irradiated Rag2−/− γc−/− recipients, which lack lymphoid cells (Fig. 3B). Because Rag2−/− γc−/− recipient mice lack NK cells, we depleted donor BM precursors of T cells before transplantation to avoid graft-versus-host disease (17). As described above, cd69−/− BM precursors had the lowest tTreg cell regeneration potential, even though in both systems, there were no differences in CD4SP cell numbers between thymuses of chimeric mice from cd69+/+ and those from cd69−/− BM precursors (Fig. 3A and B). These results suggest that the differences observed in the percentages of CD4+ Foxp3+ tTreg cells in the thymus are due to an impaired differentiation of this cell subset and not to defective thymocyte egress (Fig. 3A and B).
FIG 3.
CD69+ hematopoietic stem cells are more prone to developing tTreg cells after reconstitution. (A and B) Eight- to twelve-week-old C57BL/6 (A) or Rag2−/− γc−/− (B) recipient mice received two or one split dose of 6.5 Gy gamma radiation, respectively, and were i.v. injected with bone marrow cells from Foxp3-mRFP/cd69+/+ or Foxp3-mRFP/cd69−/− littermates. (C) In mixed chimeras, irradiated Rag2−/− γc−/− recipients were transplanted with a mixture of CD45.1-Foxp3-mRFP/cd69+/+ or CD45.2-Foxp3-mRFP/cd69−/− bone marrow precursors at a ratio of 1:1. (D) After at least 10 weeks, the contributions of the different donor bone marrow precursors to tTreg cell development and CD69 expression in tTreg cells were determined by FACS analysis. (E) Percentages of gated CD4+ SP cells and CD4+ CD8− Foxp3+ tTreg cells within CD45.1 or CD45.2 donors in the thymus. All data are representative of results from at least 3 independent experiments with at least 3 recipient mice per group or 6 recipient mice for mixed chimeras. Error bars show standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test).
Finally, to definitely rule out that the differences observed are due to differential egress between cd69+/+ and cd69−/− thymocytes (Fig. 1D), we generated mixed BM chimeric mice by reconstituting sublethally irradiated Rag2−/− γc−/− mice with a 1:1 mixture of wild-type (B6SJL) CD45.1 and cd69−/− CD45.2 BM hematopoietic stem cells from either Foxp3 reporter (Fig. 3C) or nonreporter (see Fig. S4A in the supplemental material) mice. Thymuses, spleens, lymph nodes, and blood were harvested starting from 8 to 10 weeks after transfer. CD4+ Foxp3+ Treg cells generated from cd69+/+ and cd69−/− precursors were analyzed separately (Fig. 3D; Fig. S4B and S5). We detected a marked difference in the frequencies of CD4+ Foxp3+ tTreg cells and pTreg cells originating from the two precursors in both models, with a lower proportion of Treg cells being derived from cd69−/− CD45.2 CD4+ SP precursors than from cd69+/+ CD45.1 precursors (Fig. 3D and E; Fig. S4C and S5A to C); this occurred even though CD4+ SP cells originated from both precursors in equal proportions in the thymuses (Fig. 3E), spleens (Fig. S5A), and lymph nodes (Fig. S5B). These data indicate that cd69+/+ BM hematopoietic stem cells are necessary for the generation of CD4+ Foxp3+ tTreg cells and, subsequently, pTreg cell homeostasis. Our data are consistent with the finding that Treg cell precursors in the human thymus form part of the CD69+ thymocyte cell subset (11).
CD69 deficiency impairs STAT5 signaling and BIC/miR-155-dependent tTreg cell differentiation.
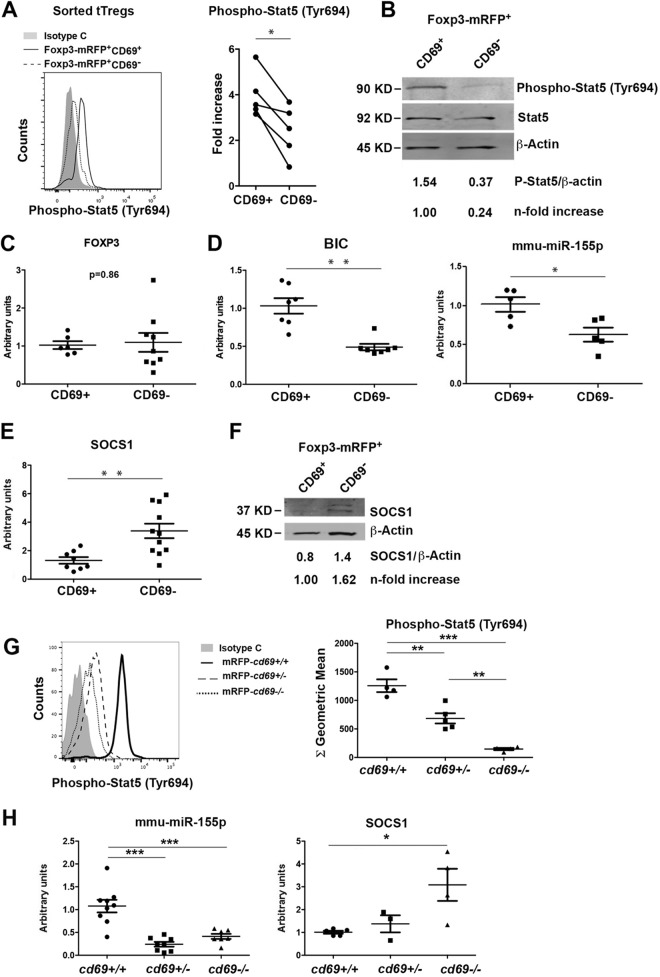
To investigate the mechanism of CD69-modulated tTreg cell development, we examined the STAT5 pathway that stimulates the foxp3 promoter, inducing tTreg cell development (4). Sorted Foxp3-mRFP+-CD69+ and -CD69− Treg cells from wild-type reporter mice (see Fig. S6 in the supplemental material) were analyzed by intracellular staining and Western blotting. The analyses showed diminished STAT5 phosphorylation in sorted CD69− tTreg cells in steady state (Fig. 4A and B), indicating that CD69 expression maintains STAT5 bystander activation of tTreg cells within the thymus. The analysis of sorted pTreg cells in the spleen confirmed diminished STAT5 phosphorylation in secondary lymphoid organs (Fig. S7A). Although we detected no differences in Foxp3 activation or expression between CD69-expressing and nonexpressing tTreg cells (Fig. 1E and 4C) or pTreg cells (12), the transcriptional activation of bic was abrogated in CD69− tTreg cells, and consequently, miR-155 expression was inhibited in these cells (Fig. 4D) and pTreg cells (Fig. S7B). It has been reported that miR-155 inhibits the expression of SOCS-1, supporting Foxp3+ tTreg cell development (6). Importantly, expression levels of both the socs-1 gene and protein were upregulated in CD69− tTreg cells (Fig. 4E and F) and pTreg cells (Fig. S7B), which had very low levels of miR-155. Moreover, we analyzed the STAT5 pathway in sorted tTreg cells from cd69+/+, cd69+/−, and cd69−/− Foxp3-mRFP reporter mice. STAT5 phosphorylation is partially inhibited in cd69+/− compared to cd69−/− tTreg cells, which almost abrogated the pathway (Fig. 4G). Thus, cd69+/− and cd69−/− tTreg cells have very low levels of miR-155 compared to those of cd69+/+ cells. Accordingly, the socs-1 gene is modestly and strongly upregulated in cd69+/− and cd69−/− tTreg cells, respectively (Fig. 4H). Our data suggest that the loss of at least one cd69 allele modifies at least in part the expression of the receptor on the membrane (Fig. 1A and B) but is sufficient to fully prevent the activation of the STAT5 pathway, miR-155 transcription, SOCS-1 inhibition, and the proper differentiation of tTreg cells.
FIG 4.
Expression of miR-155 and target proteins in CD69-deficient and -proficient Treg cells. (A, left) Representative histogram showing the levels of STAT5 phosphorylation determined by FACS analysis in sorted CD69+ or CD69− tTreg cells. (Right) Levels of STAT5 phosphorylation shown as fold differences compared with isotype control-treated cells. Lines link measurements of CD69+ and CD69− tTreg cells from the same mouse. (B) Representative Western blot showing the levels of STAT5 phosphorylation in tTreg cells sorted as described above for panel A. Phosphorylation levels are normalized to STAT5 and β-actin total protein levels. (C to E) qPCR analysis of the relative expression levels of Foxp3 (C), the BIC promoter, mmu-miR-155 (D), and socs-1 (E) in CD69+ and CD69− tTreg cells. Expression was normalized to the levels in CD69+ tTreg cells. (F) Representative Western blot of SOCS-1 protein expression in sorted CD69+ and CD69− Foxp3-mRFP+ tTreg cells. SOCS-1 levels are normalized to mean β-actin levels from of at least 4 independent sortings. (G, left) Representative histogram showing the levels of STAT5 phosphorylation in sorted tTreg cells from cd69+/+, cd69+/−, and cd69−/− Foxp3 reporter mice. (Right) Quantification of STAT5 phosphorylation levels shown as geometric mean fluorescence intensities. (H) mmu-miR-155 and socs-1 transcriptional levels analyzed by qPCR in tTreg cells from cd69+/+, cd69+/−, and cd69−/− Foxp3 reporter mice. All data are derived from at least 5 independent sortings/experiments (3 animals per sorting). For panels A to E, data were analyzed by a t test, except for Western blot analyses, for which representative gels are shown. Error bars show standard deviations. **, P < 0.01; ***, P < 0.001 (Student's t test). For panels G and H, data were analyzed by ANOVA followed by Bonferroni's multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
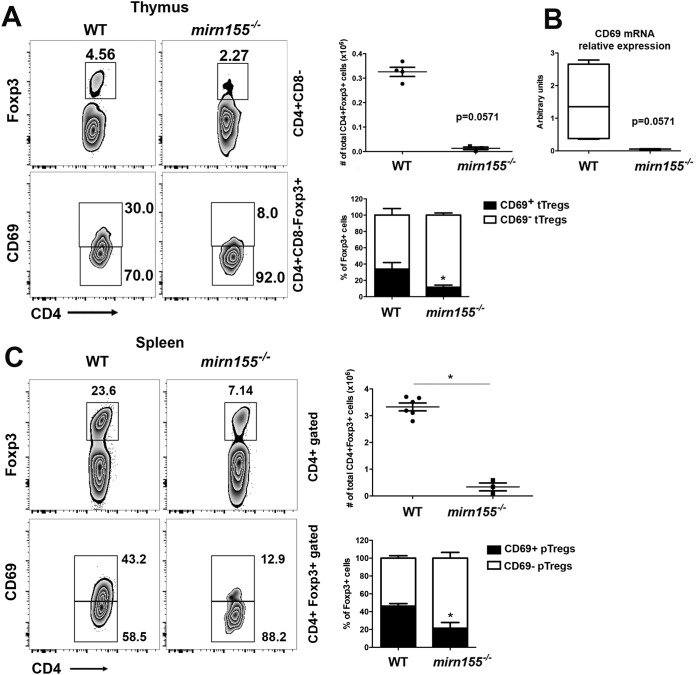
The overexpression of SOCS-1 regulates STAT5 signaling, reducing the proportions of tTreg cells in cd69−/− mice to levels similar to those in mirn155−/− mice (6). We analyzed the CD69+/CD69− ratio within tTreg and pTreg cells from mirn155−/− mice (Fig. 5A and C). Consistent with data from previous work, mirn155−/− mice display impaired numbers of tTreg cells and pTreg cells as well as an important reduction in the development of CD69+ Treg cells in both thymuses (Fig. 5A) and spleens (Fig. 5C). Interestingly, cd69 gene expression was almost abrogated in the thymuses of mirn155−/− mice (Fig. 5B), suggesting that cd69 and mirn155 could have common regulation pathways.
FIG 5.
CD69+ Treg cell development is impaired in the thymus and spleen of mirn155−/− mice. (A and C) Density plots showing CD4+ CD8− Foxp3+ tTreg cells and CD69 expression in gated CD4+ CD8− Foxp3+ thymocytes (A) or splenocytes (C) from wild-type (WT) or mirn155−/− mice. Numbers indicate the proportions (percentages) of gated cells. Bar charts show total cell numbers of gated CD4+ CD8− Foxp3+ tTreg cells (top) and percentages (±standard deviations) of CD69+ and CD69− tTreg cells (bottom) within thymuses from wild-type or mirn155−/− mice. (B) Relative expression levels of cd69 in thymocytes from wild-type or mirn155−/− mice analyzed by qPCR. All data are derived from 5 wild-type mice and 3 mirn155−/− mice. Data were analyzed by a t test. Error bars show standard deviations. *, P < 0.05 (Student's t test).
In agreement, we found that cd69+/− and cd69−/− thymic precursors are less able to differentiate toward tTreg cells than are CD69-proficient precursors in the same mice (Fig. 1F and 3E). These data thus strongly suggest that the maintenance of miR-155 expression in tTreg cells is dependent on CD69-induced STAT5 phosphorylation, reflecting a unique property of CD69 in the development of tTreg cells.
Signaling of both IL-2 receptor γ (IL-2Rγ) and CD69 is required for the development of in vitro-inducible CD25+ Treg cells.
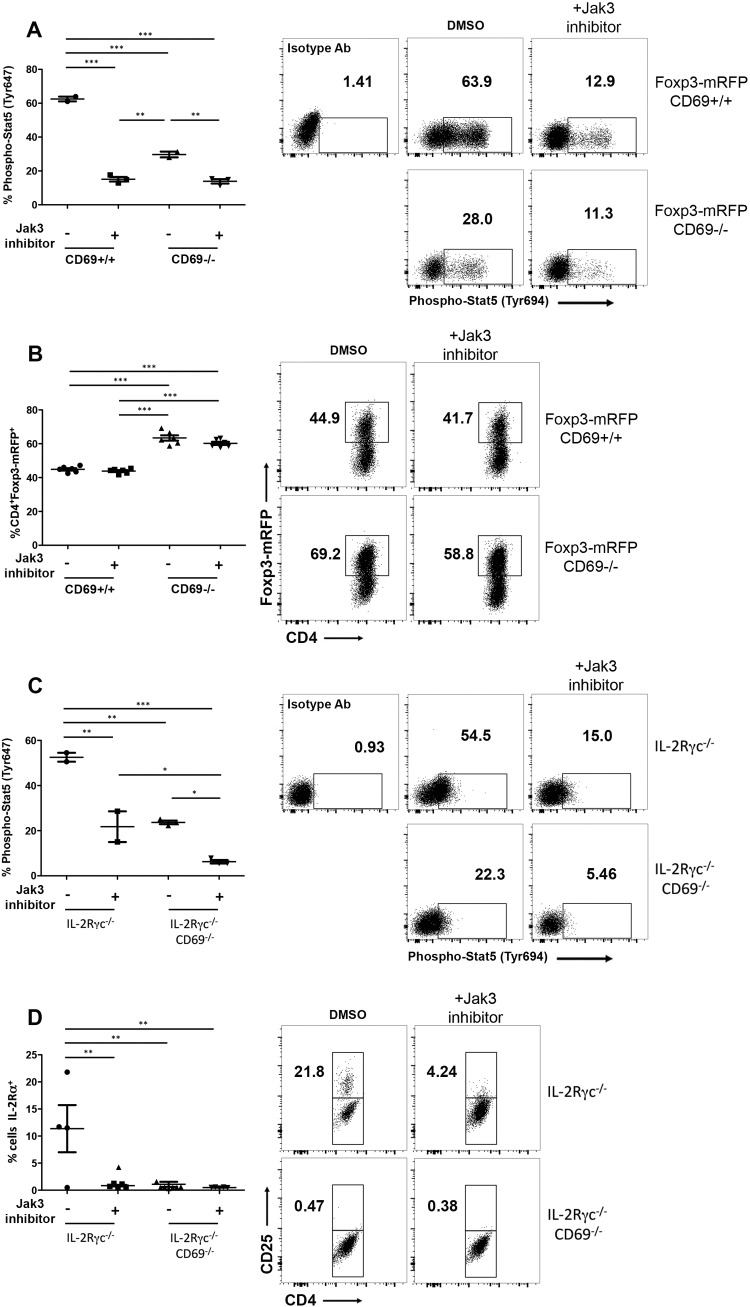
To further explore the nonredundant role of CD69 in the development of iTreg cells, we analyzed the levels of Foxp3 in the absence of Jak3-STAT5 signaling. We cultured naive CD4 T cells under Treg-skewed conditions with transforming growth factor β (TGF-β) plus IL-2 in the presence of antigen-presenting cells. The use of Jak3 chemical inhibitors decreased STAT5 phosphorylation in cd69+/+ iTreg cells to cd69−/− iTreg cell levels (Fig. 6A); however, the percentages of Foxp3-mRFP+ cells are comparable for both genotypes, even high in cd69−/− Treg cell cultures and independently of Jak-STAT5 inhibition (Fig. 6B), indicating that the Jak3-STAT5 signaling pathway is not required for Foxp3 expression of inducible Treg cells, corroborating the above-described data for tTreg cells (Fig. 4C).
FIG 6.
CD69 expression rescues iTreg cell differentiation in the absence of the IL-2Rγc/Foxp3 signaling pathway. (A) Naive CD4+ T cells from Foxp3-mRFP/cd69+/+ or Foxp3-mRFP/cd69−/− littermates were cultured for 72 h under Treg-skewed conditions and treated with a chemical Jak3 inhibitor or an equal concentration of dimethyl sulfoxide (DMSO) for the last 9 h. The percentages of phospho-STAT5+ cells and the levels of STAT5 phosphorylation determined by FACS analysis and compared to an isotype Ab are shown. (B) Quantification of reporter Foxp3-mRFP+ cells treated as described above for panel A. (C) Naive CD4+ T cells from Il2rγ−/−/cd69−/− and Il2rγ−/− mice were cultured as described above for panel A, and the percentages of phospho-STAT5+ cells and the levels of STAT5 phosphorylation were determined by FACS analysis. (D) Quantification of CD25+ Treg cells by FACS analysis. Data are from two independent experiments (n = 3 for each genotype). Error bars show standard deviations. Data were evaluated by ANOVA followed by Bonferroni's multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
It has been described that Foxp3 expression is dependent on IL-2Rγc; thus, Il2rγ−/− mice had no detectable Foxp3+ cells in thymus or spleen (18). However, the expression of CD25+ Treg cells is detectable in thymus and spleen of these mice (18). We aimed to address the role of CD69 in the development of CD25+ iTreg cells in the absence of IL-2Rγ/Foxp3 signaling pathways. For this purpose, we generated Il2rγ−/−/cd69−/− double-knockout mice. We analyzed the levels of CD25+ iTreg cells after induction with TGF-β plus IL-2 in the presence of Jak3 inhibitors in cells from Il2rγ−/− mice compared to Il2rγ−/−/cd69−/− mice. Jak3 inhibition decreased STAT5 phosphorylation in Il2rγ−/− iTreg cells to the levels in Il2rγ−/−/cd69−/− Treg cells (Fig. 6C). Interestingly, the differentiation of CD25+ iTreg cells is completely abolished in both Il2rγ−/−/cd69−/− iTreg cells and Il2rγ−/− iTreg cells plus Jak3 inhibitors (Fig. 6D). These data indicate that in the absence of the IL-2Rγ/Foxp3 pathway, CD69-induced Jak3-STAT5 activation is pivotal for the development of CD25+ iTreg cells.
It has been proposed that miR-155 could regulate different cell type functions depending on the biological context, and miR-155-mediated SOCS-1 repression regulates the competitive fitness of Treg cells (19). We analyzed the expression of mir-155, socs-1, T-bet, and Eomes in order to investigate if other miR-155 target genes are affected in iTreg cell differentiation in the absence of Jak3-STAT5 signaling pathway activation through CD69. We observed a diminished expression of miR-155 in cd69−/− compared to cd69+/+ iTreg cells (see Fig. S8A in the supplemental material), as in ex vivo CD69− thymus-derived Treg cells (Fig. 4D and G). However, Jak3 inhibition does not contribute to miR-155 inhibition (Fig. S8A), suggesting that other signaling pathways could contribute to miR-155 regulation in iTreg cells. Moreover, socs-1 expression is strongly induced in cd69−/− iTreg cells compared to cd69+/+ iTreg cells (Fig. S8B), but the expression of other miR-155 target genes such as T-bet and Eomes is not (Fig. S8C). Interestingly, Jak3 inhibits the expression of socs-1, T-bet, and Eomes in the absence of CD69 (Fig. S8B and C), supporting the hypothesis that other CD69-dependent mechanisms could be involved in the regulation of these target genes. Altogether, these data suggest that CD69 controls socs-1 expression and Treg cell differentiation through miR-155 regulation, although other molecules could be involved in this process.
Expression levels of miR-155 and CD69 are coregulated in a positive-feedback loop.
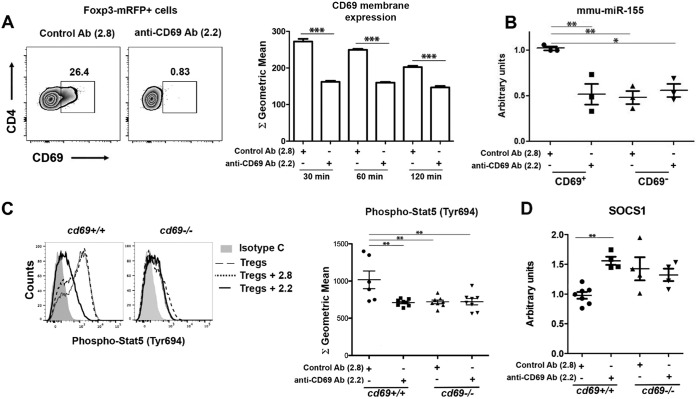
CD69 and BIC/miR-155 promoter sequences have two putative STAT5 binding elements upstream of the TATA box and AP-1 element (20) (see Fig. S9 in the supplemental material). Moreover, the transcription factor AP-1, highly induced after TCR stimulation, regulates the activation of both promoters (20, 21), suggesting that both promoters might be concomitantly activated, in a positive-feedback loop, by the same TCR/CD3-triggered pathway (Fig. S9). To test this hypothesis, we next investigated whether CD69 downstream signaling regulates miR-155 expression in tTreg cells. Sorted Foxp3+ tTreg cells from Foxp3-mRFP/cd69+/+ mice, expressing CD69 at steady state, were incubated with anti-CD69 antibody (2.2), which downregulates CD69 membrane expression and dampens its signaling (22) (Fig. 7A). As described above, we observed strong CD69 dampening on the membrane compared with that in cells incubated with control mouse IgG1 MAb (2.8) (Fig. 7A). Quantitative PCR (qPCR) analysis revealed decreased miR-155 expression in 2.2-treated CD69+ tTreg cells (Fig. 7B), to levels comparable to those in CD69− or cd69−/− tTreg cells (Fig. 7B and 4D and H). Moreover, CD69 blockade with 2.2 Abs impairs STAT5 phosphorylation (Fig. 7C) and prevents SOCS-1 inhibition (Fig. 7D), meaning that CD69 expression is necessary for the miR-155-dependent inhibition of SOCS-1 and the bona fide formation of tTreg cells.
FIG 7.
CD69 downstream signaling regulates miR-155, STAT5, and socs-1 expression in Treg cells. (A, left) Representative plots of CD69 expression in sorted mouse Foxp3-mRFP/cd69+/+ tTreg cells treated with anti-CD69 Ab 2.2 or the 2.8 isotype control antibody. (Right) CD69 expression after Ab treatment determined by FACS analysis. Bars correspond to the means ± standard deviations of data from one representative experiment of four. (B) qPCR analysis of mmu-miR-155 expression in sorted CD69+ or CD69− Foxp3-mRFP+ tTreg cells after Ab treatment. Results are normalized by sno135 snRNA expression, and expression is relative to that in 2.8-treated CD69+ cells. (C, left) Representative histogram showing the levels of STAT5 phosphorylation in iTreg cells from cd69+/+ or cd69−/− reporter mice treated with anti-CD69 Ab 2.2 or the 2.8 isotype control Ab. (Right) Quantification of STAT5 phosphorylation levels shown as geometric mean fluorescence intensities. (D) socs-1 transcriptional levels analyzed by qPCR. Data shown in panels A and B are derived from 3 independent sortings/experiments (3 animals per sorting), and data shown in panels C and D are iTreg cells differentiated from at least 4 mice per group. Data were analyzed by 1-way ANOVA and Bonferroni's posttest (B). CD69 expression after Ab treatment was analyzed by a t test (A). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test).
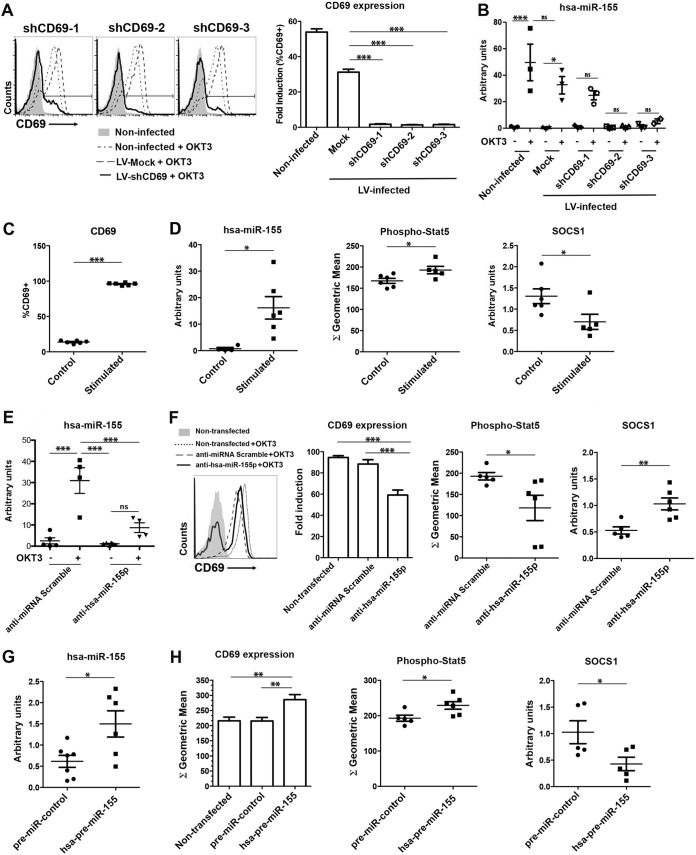
To verify whether these findings could be extended to human cells, activated CD4+ CD25+ peripheral blood lymphocytes (PBLs) were infected with lentiviruses (LVs) carrying different short hairpin RNA (shRNA) sequences targeting CD69 (shCD69-1 to -3). Endogenous levels of membrane CD69 and hsa-miR-155 were analyzed by fluorescence-activated cell sorter (FACS) analysis and qPCR, respectively (Fig. 8A and B). LV infection of PBLs with three shCD69 sequences fully inhibited CD69 expression compared to mock LV infection (Fig. 8A), inducing the loss of hsa-miR-155 transcription (Fig. 8B). Our data indicate that human CD69 and hsa-miR-155 are regulated together as in mouse cells. In parallel, we induced the expression of CD69 in vitro (Fig. 8C) to corroborate that the STAT5 pathway and hsa-miR-155 are activated together with the receptor, whereas SOCS-1 is inhibited (Fig. 8D).
FIG 8.
Coregulation of CD69 and miR-155 expression in human Treg cells. (A, left) Representative histograms of CD69 expression after LV infection with 3 different shCD69 sequences (shCD691 to -3) or an shRNA control sequence, stimulated with human anti-CD3 Abs (OKT3 clone) or not stimulated. (Right) Fold CD69 induction relative to values for nonstimulated cells. (B) qPCR analysis of hsa-miR-155 expression in human CD4+ T cells after LV infection. (C and D) Human PBLs were stimulated with PMA-ionomycin for 4 h or not stimulated, and the percentages of CD69+ cells (C) and phospho-STAT5 (D) were determined by FACS analysis. (D) hsa-miR-155 and human socs-1 gene expression levels were analyzed by qPCR. (E) Human PBLs were transfected with anti-hsa-miR-155-5p or an anti-miRNA scramble control, and hsa-miR-155 expression was analyzed by qPCR. (F) Representative histograms and quantification of CD69 expression, STAT5 phosphorylation, and human socs-1 transcription levels in CD4+ PBLs treated as described above for panel E. (G) Human PBLs were transfected with hsa-pre-miR-155-5p or the pre-miRNA control, and hsa-miR-155 expression was analyzed by qPCR. (H) CD69 expression, STAT5 phosphorylation, and human socs-1 transcription in CD4+ PBLs treated as described above for panel G. Results from miRNA qPCRs are normalized to sno135 snRNA expression levels. All data are means ± standard deviations of results from at least 3 independent donors for a total of 10 donors. Data were analyzed by 1-way ANOVA and Bonferroni's posttest or by a t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To test this mechanism functionally, we performed loss- and gain-of-function assays by transfecting human Treg cells with anti-hsa-miR-155 or hsa-pre-miR-155. First, we transfected control and anti-CD3 (OKT3)-stimulated CD4+ CD25+ human PBLs with anti-hsa-miR-155-5p or scrambled anti-miRNA (Fig. 8E). CD69 expression in activated PBLs dropped dramatically after the inhibition of hsa-miR-155 (Fig. 8F). Moreover, STAT5 activation was reduced, and in agreement, socs-1 gene expression was enhanced, indicating that miR-155 blockade regulates the CD69 signaling pathway. In contrast, the overexpression of hsa-miR-155 in CD69− Treg cells (Fig. 8G) revealed significant increases in the expression of CD69, STAT5 activation, and socs-1 inhibition (Fig. 8H). Thus, the reciprocal modulation of the C-type lectin and miR-155 in a positive-feedback loop could be pivotal to maintaining tTreg cell fitness and pTreg cell homeostasis.
DISCUSSION
In this study, we have shown that the C-type lectin CD69 plays a key role in the development and homeostasis of Treg cells. Using a combined genetic model of Foxp3 reporter and cd69 knockout mice and genetic inhibition approaches, we unequivocally demonstrate that the activation of the CD69 pathway promotes STAT5 phosphorylation, BIC/miR-155 expression, and SOCS-1 inhibition. The role of CD69 as a negative regulator of the immune system has remained a controversial issue during the last years (23). However, very recent studies by independent groups show that CD69 plays a crucial role in the suppressor function of mouse and human Treg cells as well as in the generation of in vitro-induced Treg cells (12, 16, 24–26). Nevertheless, the specific role of the C-type lectin in the development of Treg cells in the thymus remains elusive.
A major issue that has limited this study has been the key role of CD69 in the egress of lymphocytes from lymphoid organs and in particular from the thymus to the periphery (13, 14, 27–29). Although thymic positive and negative T cell selection processes are unaffected by CD69 deficiency (30), CD69 controls the egress of mature T cells into the periphery via corticomedullary blood vessels, through the negative regulation of S1P1 receptors (27, 28), making it not an easy task to study its role in the development of Treg cells in the thymus. With the help of Foxp3 reporter mice, we have performed studies of tTreg cell differentiation in FTOC and in mixed chimeric mice to avoid the effects derived from the different migratory potentials of CD69+ and CD69− cells. We demonstrate that the expression of the C-type lectin CD69 is pivotal for the development of tTreg cells, as they are virtually absent in FTOCs from cd69−/− or anti-CD69-treated embryonic thymuses or in mixed bone marrow chimeras from cd69−/− precursors. In both systems, total numbers of cells within the thymus do not change, whereas tTreg cell proportions originating from CD69− precursors are consistently diminished, demonstrating unequivocally that this effect is not due to a different migratory behavior.
We have found that proportions of Foxp3+ pTreg cells are also diminished after analysis of spleen and lymph nodes from adult Foxp3-mRFP/cd69−/− reporter mice compared to cd69+/+ and cd69+/− littermates. In addition, CD69-deficient pTreg cells have a defective suppressive function (12). Thus, defects observed in CD69-deficient precursors affect both tTreg cell development and pTreg cell homeostasis, strongly indicating that CD69-proficient precursors give rise to the CD69+ functionally active pTreg cell subset. In this regard, two different genetic approaches in mice and a recent study in humans indicate that CD69 expression in pTreg cells is required to maintain immunological tolerance. CD69 deficiency in mice compromises T cell-induced colitis and the establishment of oral tolerance after antigen challenge in vivo (24), and CD69+ pTreg cells are essential for the prevention of asthmatic reactions to harmless antigens (12). Furthermore, a subset of CD69+ Treg cells in the blood of healthy human donors seems to have a relevant immune-regulatory role (25).
The C-type lectin CD69 interacts with Jak3/STAT5 proteins independently of the IL-2 pathway, thus inhibiting Th17 responses (31) and controlling the suppressor potential of pTreg cells (12). STAT5 phosphorylation stimulates the foxp3 promoter, inducing tTreg cell development (4), and Foxp3 binds to an intron within the promoter region of the miR-155 host gene bic in Treg cells (32). Both mirn155−/− and bic−/− mice have below-normal numbers of Foxp3+ Treg cells in thymuses and secondary lymphoid organs, indicating an essential role for miR-155 in the development of Foxp3+ Treg cells (5, 6). We have explored if this pathway could be the responsible for the defects observed in Treg cell development in cd69-deficient mice, finding a strong inhibition of STAT5 phosphorylation in freshly isolated Foxp3-mRFP+-CD69− compared to Foxp3-mRFP+-CD69+ tTreg cells. Moreover, the transcriptional level of bic/miR-155 is reduced in Foxp3-mRFP+-CD69− Treg cells, and consequently, its target SOCS-1 is upregulated at both the mRNA and protein levels. In a mouse model of SOCS-1 overexpression, negative regulation of STAT5 signaling reduces the proportion of Foxp3+ thymocytes to levels similar to those seen in mirn155−/− mice (6). miR-155 inhibits SOCS-1 expression, enhancing Foxp3+ tTreg cell development (6). Our data demonstrate that CD69 expression enhanced BIC/miR-155 transcription, inhibited SOCS-1, and therefore maintained Treg cell differentiation and the fitness of Treg cells. However, IL-2R signaling also activates the Jak/STAT5 pathway in Treg cells; specifically, Foxp3 expression is dependent on IL-2Rγc signaling, as Il2rγ−/− mice have no detectable Foxp3+ cells, although a small proportion of CD25+ Treg cells is still detectable in these mice (18). Our study shows that the differentiation of CD25+ iTreg cells is inhibited in Il2rγ−/− cultures plus Jak3 inhibitors or Il2rγ−/−/cd69−/− mice, indicating that Jak3-STAT5 signaling pathway activation through CD69 is essential for the development of Treg cells.
CD69 does not appear as an miR-155 target in the PicTar, Targetscan, or miRanda miRNA target prediction database, and there are no miR-155 target sequences in the CD69 3′ untranslated region (UTR) (33). However, several studies have shown a correlation between Dicer, a member of the RNase III complex that processes pre-miRNAs into mature miRNAs, miR-155 regulation, and CD69 expression. Treg cells from MRL/lpr mice are Dicer insufficient and yet overexpress miR-155 and show increased CD69 expression (9), suggesting that there are Dicer-alternative mechanisms for miRNA regulation. In another study, Dicer−/− TCs showed increased CD69 expression after TCR stimulation and, consequently, defective egress from lymphoid organs (10). As described above for CD69, Dicer plays a key role in tTreg cell differentiation (7) and Treg cell function (8). In this regard, CD69 is expressed in lymphocytes early after TCR/CD3 stimulation (34), and its cytoplasmic tail interacts with Jak3/STAT5 molecules (35), triggering this pathway in pTreg cells (12) and tTreg cells and therefore inhibiting SOCS-1 transcription and protein expression. Similarly, TCR-induced IL-2 signaling triggers STAT5 signaling and enhances Foxp3-dependent miR-155 expression, limiting SOCS-1 expression and promoting Treg cell homeostasis (6). Recent data show that microRNAs could regulate different cell type functions modulating different target genes, depending on the biological context (19). We analyzed the expression of miR-155 and SOCS-1 in the absence of Jak3-STAT5 signaling pathway activation through CD69 in the differentiation of iTreg cells. miR-155 expression is inhibited and SOCS-1 is upregulated in cd69−/− compared to cd69+/+ iTreg cells; however, Jak3 inhibition does not contribute to miR-155 dampening, suggesting that other microRNAs and/or target genes could be involved. Interestingly, the STAT5 binding elements of the BIC/miR-155 and CD69 promoter sequences are similar, with each element containing two putative STAT binding elements upstream of the TATA box and AP-1 element (20). Moreover, the transcription factor AP-1, which is highly induced after TCR stimulation, regulates the activation of both promoters (20, 21). This suggests that both promoters might be concomitantly activated, in a positive-feedback loop, by the same TCR/CD3-triggered pathway.
Our present study shows that Foxp3-RFP/cd69−/− reporter mice have a dramatically reduced tTreg cell population in adult thymuses. Moreover, tTreg cells are unable to develop properly in FTOCs from cd69−/− or anti-CD69-treated embryonic thymuses or in mixed bone marrow chimeras from cd69−/− precursors. The in vitro data confirm that the phosphorylation of STAT5 is abrogated in CD69-deficient tTreg cells and results in the inhibition of the BIC/miR-155 pathway, increased SOCS-1 expression, and impaired tTreg cell development. Our previous studies show that the suppressor function of Treg cells is compromised in cd69-deficient mice (12), indicating that CD69 is a key molecule in the development of Foxp3+ CD69+ Treg cells in the thymus that will give rise to the functionally active subset of Treg cells in the periphery. Therefore, we postulated that the C-type lectin CD69 is a pivotal molecule for the maintenance of immune homeostasis in health and disease.
MATERIALS AND METHODS
Mice.
cd69−/− mice were generated in the 129/Sv background as described previously (31) and backcrossed onto the C57BL/6 strain for at least 12 generations. C57BL/6.Ly5.1 mice (CD45.1+) were purchased from The Jackson Laboratory (B6.SJL-Ptprca Pepcb/BoyJ, stock number 002014). Rag2−/− γc−/− (Rag2/Il2rg) mice were provided by the laboratory of M. L. Toribio (Centro de Biología Molecular, CSIC, Spain) and were intercrossed with C57BL/6 mice to generate Il2rγ−/− mice, which were subsequently intercrossed with cd69−/− mice to generate Il2rγ−/−/cd69−/− mice. FoxP3-mRFP reporter mice (FIR mice; C57BL/6 background) were generated and provided by the Flavell laboratory (Yale University School of Medicine, New Haven, CT) (36) and were intercrossed with cd69−/− mice to generate Foxp3-mRFP/cd69+/+ wild-type, Foxp3-mRFP/cd69+/− heterozygous, and Foxp3-mRFP/cd69−/− CD69-deficient littermates. Animals were housed and used under specific-pathogen-free (SPF) conditions at the Centro Nacional de Investigaciones Cardiovasculares (CNIC) animal facility. mirn155−/− mice were provided by R. Nakagawa (The Francis Crick Institute, London, United Kingdom). All animal procedures were approved by the ethics committee of the Comunidad Autónoma de Madrid and conducted in accordance with the institutional guidelines that comply with European Institutes of Health directives (37).
Intracellular staining and FACS analysis.
Single-cell suspensions were obtained from adult or fetal thymuses and incubated in FACS buffer (phosphate-buffered saline [PBS], 0.5% bovine serum albumin [BSA], 1 μM EDTA, 0.1% NaN3) with fluorochrome-conjugated mouse-specific antibodies against CD4, CD8, CD69, CD45.1, and CD45.2. All antibodies were purchased from BD Biosciences. For Foxp3 intracellular staining, we used the Foxp3 staining kit (eBioscience). CD69+- and CD69−-Foxp3-mRFP+ tTreg cells were sorted from Foxp3-mRFP/cd69+/+ thymuses by using a FACSAria III instrument (BD Biosciences). For intracellular STAT5 staining, sorted tTreg cells were fixed with 0.2% paraformaldehyde and permeabilized with 90% methanol, and cells were incubated with anti-phospho-STAT5 (Tyr694) (Cell Signaling), an Alexa Fluor 647-IgG1 isotype control, and Alexa Fluor 647–anti-phospho-STAT5 (pY694) (Becton Dickinson). Human PBLs were obtained after Ficoll separation from buffy coats and maintained in RPMI medium supplemented with 10% fetal calf serum (FCS), 20 mM HEPES, l-glutamine, antibiotics, nonessential amino acids, sodium pyruvate, and β-mercaptoethanol. Treated PBLs were incubated with fluorochrome-conjugated human-specific antibodies against CD4, CD25, and CD69 (BD Biosciences) and Foxp3 (Miltenyi Biotec). Cells were analyzed in an LSRFortessa flow cytometer (BD Biosciences) equipped with four lasers (405, 488, 561, and 640 nm), and the data were processed with FlowJo v10.0.4 (TreeStar).
Fetal thymus organ culture.
Uteri were removed from female mice at the indicated gestational time points, and the embryos were placed into a petri dish with fresh cold PBS for the extraction of thymuses. To place the fetal thymus lobes in culture, we placed 0.8-μm nitrocellulose membrane filters (Millipore) on top of 12- to 7-mm Gelfoam sponges embedded in prewarmed Iscove´s modified Dulbecco´s medium (IMDM) (supplemented with 10% FCS, l-glutamine, antibiotics, and β-mercaptoethanol). FTOCs were maintained for 4 to 14 days, with medium being replaced every 3 days. An anti-CD69 monoclonal antibody (2.2) or the isotype control antibody (2.8) was added (50 μg/ml) to the culture medium as indicated and replaced every 3 days. At the end of the culture period, single-cell suspensions were prepared from the lobes, and cells were counted and analyzed by FACS analysis.
Western blotting.
Lysates of sorted CD69+- and CD69−-Foxp3-mRFP+ tTreg cells were prepared in PD buffer (40 mM Tris HCl [pH 8.0], 0.5 M NaCl, 6 mM EDTA, 6 mM EGTA, 0.1% NP-40) containing a protease inhibitor cocktail (Complete Mini; Roche). Proteins (20 μg) were size separated on 12% SDS-polyacrylamide gels and transferred onto Trans-Blot nitrocellulose membranes (Bio-Rad). Primary antibodies for immunoblotting were as follows: anti-β-actin, anti-SOCS-1, and anti-STAT5 (Santa Cruz) and anti-phospho-STAT5 (Cell Signaling). Quantitative assessment of protein expression was performed with the Odyssey scanner and analyzed with Image Studio Lite v4.0 Western blot analysis software (Li-Cor).
In vitro differentiation of Treg cells.
Inducible Treg cells were differentiated from Foxp3-mRFP/cd69+/+, Foxp3-mRFP/cd69−/−, Il2rγ−/−/cd69−/−, and Il2rγ−/− mice. Naive CD4 T cells from these mice were isolated and cocultured for 72 h with irradiated antigen-presenting cells in the presence of plate-bound anti-CD3 (2 μg/ml) and soluble anti-CD28 (2 μg/ml) plus recombinant TGF-β1 (10 ng/ml) and IL-2 (2 ng/ml). The last 9 h, the cells were incubated with or without Jak3 inhibitor I (catalog number CAS 202475-60-3; Calbiochem) (10 μg/ml). For experiments with inhibitor antibodies, after differentiation, Treg cells were cultured for 4 h with anti-2.2 Ab or the 2.8 isotype control Ab.
RNA extraction and gene expression analysis.
RNA and microRNA were extracted from 2 × 104 to 6 × 104 sorted mouse tTreg cells or 106 human PBLs with the miRNeasy minikit (Qiagen), followed by DNase treatment with the Turbo DNase-free kit (Ambion). For analysis of SOCS-1, Foxp3, and BIC transcripts, reverse transcription was performed by using the High Capacity cDNA reverse transcription kit (Applied Biosystems). SOCS-1 and Foxp3 gene expression levels were analyzed by real-time PCR using SYBR green PCR mix (Applied Biosystems). Mouse and human Gapdh genes were used as the endogenous controls. The following primers were used to amplify murine genes: forward (F) primer 5′-CTGCGGTTCTATTGGGGAC-3′ and reverse (R) primer 5′-AAAAGGCAGTCGAAGGTCTCG-3′ for socs-1, F primer 5′-CACCCAGGAAAGACAGCAACC-3′ and R primer 5′-GCAAGAGCTCTTGTCCATTGA-3′ for Foxp3, F primer 5′-CCCTTGGGCTGTGTTAATAGTG-3′ and R primer 5′-AACTTCTCGTACAAGCCTGGG-3′for cd69, and F primer 5′-TGAAGCAGGCATCTGAGGG-3′ and R primer 5′-CGAAGGTGGAAGAGTGGGAG-3′ for Gapdh. The following primers were used to amplify human genes: F primer 5′-TTTTCGCCCTTAGCGTGAAGA-3′ and R primer 5′-GAGGCAGTCGAAGCTCTCG-3′ for socs-1 and F primer 5′-AATGGACTGGTCGTGGAG-3′ and R primer 5′-CCCTCCAGGGGATCGTTTG-3′ for gapdh. BIC gene expression was analyzed by real-time PCR using TaqMan universal PCR master mix and specific TaqMan probe and primers for bic (catalog numbers Mm01716204-m1 and Hs01374570-m1; Applied Biosystems). The expression of microRNA was analyzed by using a TaqMan microRNA reverse transcription kit, individual TaqMan microRNA assays for mmu-miR-155-5p (catalog number 002571) and hsa-miR-155-5p (catalog number 002287), and TaqMan universal PCR master mix (Applied Biosystems). sno135 snRNA (catalog number 001230) was used as the endogenous control. Real-time quantitative PCR analysis was performed with an ABI Prism 7900HT 384 thermal cycler (Applied Biosystems). The relative gene expression level was determined by using the method.
Chimeric mice.
Eight- to twelve-week-old Rag2−/− γc−/− recipient mice were irradiated with one split dose of 6.5 Gy gamma radiation, whereas C57BL/6 recipients were irradiated with two split 6.5-Gy doses. The mice were intravenously (i.v.) injected with bone marrow cells from Foxp3-mRFP/cd69+/+ or Foxp3-mRFP/cd69−/− littermates. In mixed chimeras, irradiated Rag2−/− γc−/− recipients were transplanted with a mixture of CD45.1 cd69+/+ or CD45.2 cd69−/− bone marrow precursors from nonreporter or reporter Foxp3-mRFP+ cells, at a ratio of 1:1. After at least 10 weeks, the contribution of the different donor bone marrow precursors to the tTreg cell subset was determined by FACS analysis.
Transient transfection.
PBLs (106) were transiently transfected for 4 h with 50 pM anti-miR-155 (catalog number AM12601; Ambion) by using Lipotransfectin (Niborlab) according to the manufacturer's instructions. As a negative control, a random anti-miRNA sequence control (negative control 1, catalog number AM1701; Ambion) was included in the assay mixture. Transfected cells were stimulated with plate-bound anti-CD3 antibody (OKT3; 3 μg/ml) for 24 h. When indicated, PBLs (0.5 × 106) were transfected for 7 h with 50 pM pre-miR-hsa-miR-155 or a pre-miRNA negative control (Ambion) by using Lipofectamine RNA iMAX (Invitrogen). Transfected cells were stimulated with phorbol myristate acetate (PMA) during 4 h with 50 ng/ml PMA and 750 ng/ml ionomycin (P+I). After stimulation, the levels of CD69 and phospho-STAT5 were monitored by flow cytometry, and transcriptional levels of hsa-miR-155 and socs-1 were monitored by qPCR.
Plasmids.
The pLKO lentiviral plasmids containing shCD69 sequences were obtained from Sigma-Aldrich (catalog numbers TRCN0000057693, TRCN0000057694, and TRCN0000057695), and the pLKO lentiviral control plasmid is a pLKO empty vector from Sigma-Aldrich (catalog number SHC001). The shCD69 sequences used were as follows (5′ to 3′): CCGGGCATGGAATGTGAGAAGAATTCTCGAGAATTCTTCTCACATTCCATGCTTTTTG for shCD69-1, CCGGAGGCCAATACACATTCTCAATCTCGAGATTGAGAATGTGTATTGGCCTTTTTTG for shCD69-2, and CCGGGTGGTCAAATGGCAAAGAATTCTCGAGAATTCTTTGCCATTTGACCACTTTTTG for shCD69-3.
LV production, titration, and infection.
HEK-293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) (Sigma-Aldrich) and l-glutamine plus antibiotics. HEK-293 cells were transiently transfected by the calcium phosphate method with 3 HIV-derived plasmids and the vesicular stomatitis virus (VSV)-pseudotyped LV system (provided by F. Sánchez-Madrid, Hospital de la Princesa, Spain) to obtain LVs expressing the shCD69 sequences. The supernatant containing LV particles was collected 48 h after the removal of the calcium phosphate precipitate and ultracentrifuged for 2 h (Optima L-100 XP ultracentrifuge; Beckman). LVs were collected by adding cold PBS and were titrated by qPCR. PBLs isolated from healthy donors were infected with LV particles (multiplicity of infection [MOI] of 10) for 5 h. Subsequently, virus-containing medium was replaced with fresh complete RPMI medium supplemented with 10% FBS. After 12 h, infected cells were selected with puromycin for 48 h. Selected cells were stimulated with plate-bound anti-CD3 antibody (OKT3; 3 μg/ml) for 24 h. After stimulation, the levels of CD69 were monitored by flow cytometry, and levels of miR-155 were monitored by TaqMan qPCR.
Statistical analysis.
Experiments were performed according to a randomized complete block design (treatments and different time points have been taken into account) or a fully randomized design. To determine significant differences, P values were calculated by Student's t test as appropriate, and differences were considered significant at P values of <0.05. Means from more than two experimental groups were compared by 1-way analysis of variance (ANOVA). To account for multiple comparisons, the Tukey test was used to compared selected pairs of means, and the Bonferroni posttest was used to compare all pairs of means. All statistical analyses were carried out with Prism v5 (GraphPad Software). Each experiment was repeated at least three times, unless otherwise indicated in the figure legends.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Bartlett for editorial assistance and Richard A. Flavell (Yale University, New Haven, CT) for kindly providing the foxp3-mRFP reporter mice.
This study was funded by a grant from the Spanish Ministry of Economy and Competitiveness (SAF2013-44857-R to M.L.T.), grant INDISNET 01592006 from the Comunidad de Madrid to P.M. and F.S.-M., and grants from the Instituto de Salud Carlos III (PI-FIS-2016-9488 to P.M.), the CIBER de Enfermedades Cardiovasculares to F.S.-M. and P.M., and the Fundació La Marató TV3 (20152330 31) to P.M. and F.S.-M. R.S.-D. was funded by a predoctoral fellowship from the Comunidad de Madrid, S.L. was funded by a contract from the RETICS Enfermedades Cardiovasculares (Instituto de Salud Carlos III), and K.T. is cofunded by the European Union Marie Curie Program (COFUND CNIC IPP). This research has been cofinanced by FEDER. The CNIC is supported by the Ministry of Economy, Industry, and Competitiveness (MINECO) and the Pro CNIC Foundation and is a Severo Ochoa Center of Excellence (MINECO award SEV-2015-0505).
R.S.-D. and R.B.-D. performed research and analyzed the data; S.L., K.T., H.D.L.F., B.L.-P., and E.M.-G. performed research; R.N. contributed with mirn155−/− mice; F.S.-M. and M.L.T. designed research and analyzed the data; and P.M. designed research, collected and analyzed the data, and wrote the paper.
We declare that we have no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00341-16.
REFERENCES
- 1.Campbell DJ, Koch MA. 2011. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat Rev Immunol 11:119–130. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbas AK, Benoist C, Bluestone JA, Campbell DJ, Ghosh S, Hori S, Jiang S, Kuchroo VK, Mathis D, Roncarolo MG, Rudensky A, Sakaguchi S, Shevach EM, Vignali DA, Ziegler SF. 2013. Regulatory T cells: recommendations to simplify the nomenclature. Nat Immunol 14:307–308. doi: 10.1038/ni.2554. [DOI] [PubMed] [Google Scholar]
- 3.Josefowicz SZ, Lu LF, Rudensky AY. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burchill MA, Yang J, Vang KB, Moon JJ, Chu HH, Lio CW, Vegoe AL, Hsieh CS, Jenkins MK, Farrar MA. 2008. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity 28:112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. 2009. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol 182:2578–2582. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- 6.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. 2009. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. 2006. A role for Dicer in immune regulation. J Exp Med 203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liston A, Lu LF, O'Carroll D, Tarakhovsky A, Rudensky AY. 2008. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med 205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Divekar AA, Dubey S, Gangalum PR, Singh RR. 2011. Dicer insufficiency and microRNA-155 overexpression in lupus regulatory T cells: an apparent paradox in the setting of an inflammatory milieu. J Immunol 186:924–930. doi: 10.4049/jimmunol.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang N, Bevan MJ. 2010. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci U S A 107:21629–21634. doi: 10.1073/pnas.1016299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin-Gayo E, Sierra-Filardi E, Corbi AL, Toribio ML. 2010. Plasmacytoid dendritic cells resident in human thymus drive natural Treg cell development. Blood 115:5366–5375. doi: 10.1182/blood-2009-10-248260. [DOI] [PubMed] [Google Scholar]
- 12.Cortes JR, Sanchez-Diaz R, Bovolenta ER, Barreiro O, Lasarte S, Matesanz-Marin A, Toribio ML, Sanchez-Madrid F, Martin P. 2014. Maintenance of immune tolerance by Foxp3+ regulatory T cells requires CD69 expression. J Autoimmun 55:51–62. doi: 10.1016/j.jaut.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng C, Woodside KJ, Vance BA, El-Khoury D, Canelles M, Lee J, Gress R, Fowlkes BJ, Shores EW, Love PE. 2002. A potential role for CD69 in thymocyte emigration. Int Immunol 14:535–544. doi: 10.1093/intimm/dxf020. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama T, Kasprowicz DJ, Yamashita M, Schubert LA, Gillard G, Kimura M, Didierlaurent A, Koseki H, Ziegler SF. 2002. The generation of mature, single-positive thymocytes in vivo is dysregulated by CD69 blockade or overexpression. J Immunol 168:87–94. doi: 10.4049/jimmunol.168.1.87. [DOI] [PubMed] [Google Scholar]
- 15.Lamana A, Sancho D, Cruz-Adalia A, del Hoyo GM, Herrera AM, Feria M, Diaz-Gonzalez F, Gomez M, Sanchez-Madrid F. 2006. The role of CD69 in acute neutrophil-mediated inflammation. Eur J Immunol 36:2632–2638. doi: 10.1002/eji.200636355. [DOI] [PubMed] [Google Scholar]
- 16.Wirnsberger G, Mair F, Klein L. 2009. Regulatory T cell differentiation of thymocytes does not require a dedicated antigen-presenting cell but is under T cell-intrinsic developmental control. Proc Natl Acad Sci U S A 106:10278–10283. doi: 10.1073/pnas.0901877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noval Rivas M, Hazzan M, Weatherly K, Gaudray F, Salmon I, Braun MY. 2010. NK cell regulation of CD4 T cell-mediated graft-versus-host disease. J Immunol 184:6790–6798. doi: 10.4049/jimmunol.0902598. [DOI] [PubMed] [Google Scholar]
- 18.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. 2005. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol 6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 19.Lu LF, Gasteiger G, Yu IS, Chaudhry A, Hsin JP, Lu Y, Bos PD, Lin LL, Zawislak CL, Cho S, Sun JC, Leslie CS, Lin SW, Rudensky AY. 2015. A single miRNA-mRNA interaction affects the immune response in a context- and cell-type-specific manner. Immunity 43:52–64. doi: 10.1016/j.immuni.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin Q, Wang X, McBride J, Fewell C, Flemington E. 2008. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J Biol Chem 283:2654–2662. doi: 10.1074/jbc.M708218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castellanos MC, Munoz C, Montoya MC, Lara-Pezzi E, Lopez-Cabrera M, de Landazuri MO. 1997. Expression of the leukocyte early activation antigen CD69 is regulated by the transcription factor AP-1. J Immunol 159:5463–5473. [PubMed] [Google Scholar]
- 22.Esplugues E, Sancho D, Vega-Ramos J, Martinez C, Syrbe U, Hamann A, Engel P, Sanchez-Madrid F, Lauzurica P. 2003. Enhanced antitumor immunity in mice deficient in CD69. J Exp Med 197:1093–1106. doi: 10.1084/jem.20021337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez-Amaro R, Cortes JR, Sanchez-Madrid F, Martin P. 2013. Is CD69 an effective brake to control inflammatory diseases? Trends Mol Med 19:625–632. doi: 10.1016/j.molmed.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radulovic K, Manta C, Rossini V, Holzmann K, Kestler HA, Wegenka UM, Nakayama T, Niess JH. 2012. CD69 regulates type I IFN-induced tolerogenic signals to mucosal CD4 T cells that attenuate their colitogenic potential. J Immunol 188:2001–2013. doi: 10.4049/jimmunol.1100765. [DOI] [PubMed] [Google Scholar]
- 25.Vitales-Noyola M, Doniz-Padilla L, Alvarez-Quiroga C, Monsivais-Urenda A, Portillo-Salazar H, Gonzalez-Amaro R. 2015. Quantitative and functional analysis of CD69(+) NKG2D(+) T regulatory cells in healthy subjects. Hum Immunol 76:511–518. doi: 10.1016/j.humimm.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Lin CR, Wei TW, Tsai HY, Wu YT, Wu PY, Chen ST. 2015. Glycosylation-dependent interaction between CD69 and S100A8/S100A9 complex is required for regulatory T-cell differentiation. FASEB J 29:5006–5017. doi: 10.1096/fj.15-273987. [DOI] [PubMed] [Google Scholar]
- 27.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. 2004. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 28.Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL, Cyster JG, Matloubian M. 2006. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 29.Weinreich MA, Hogquist KA. 2008. Thymic emigration: when and how T cells leave home. J Immunol 181:2265–2270. doi: 10.4049/jimmunol.181.4.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lauzurica P, Sancho D, Torres M, Albella B, Marazuela M, Merino T, Bueren JA, Martinez AC, Sanchez-Madrid F. 2000. Phenotypic and functional characteristics of hematopoietic cell lineages in CD69-deficient mice. Blood 95:2312–2320. [PubMed] [Google Scholar]
- 31.Martin P, Gomez M, Lamana A, Cruz-Adalia A, Ramirez-Huesca M, Ursa MA, Yanez-Mo M, Sanchez-Madrid F. 2010. CD69 association with Jak3/Stat5 proteins regulates Th17 cell differentiation. Mol Cell Biol 30:4877–4889. doi: 10.1128/MCB.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. 2007. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 33.Ziegler SF, Levin SD, Johnson L, Copeland NG, Gilbert DJ, Jenkins NA, Baker E, Sutherland GR, Feldhaus AL, Ramsdell F. 1994. The mouse CD69 gene. Structure, expression, and mapping to the NK gene complex. J Immunol 152:1228–1236. [PubMed] [Google Scholar]
- 34.Testi R, Phillips JH, Lanier LL. 1989. T cell activation via Leu-23 (CD69). J Immunol 143:1123–1128. [PubMed] [Google Scholar]
- 35.Martin P, Sanchez-Madrid F. 2011. CD69: an unexpected regulator of TH17 cell-driven inflammatory responses. Sci Signal 4:pe14. doi: 10.1126/scisignal.2001825. [DOI] [PubMed] [Google Scholar]
- 36.Wan YY, Flavell RA. 2005. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc Natl Acad Sci U S A 102:5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.European Institutes of Health. 2010. Directive 2010/63/EU of the European Parliament and the Council on the Protection of Animals Used for Scientific Purposes. Official J Eur Union 53:33–79. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.