ABSTRACT
Raccoons (Procyon lotor) are successful urban adapters and hosts to a number of zoonotic and nonzoonotic pathogens, yet little is known about their hemoplasma infections and how prevalence varies across habitat types. This study identifies hemotropic Mycoplasma species infection in raccoons from urban and undisturbed habitats and compares hemoplasma infection in sympatric urban cats (Felis catus) from the same geographic region. We collected blood from raccoons (n = 95) on an urban coastal island (n = 37) and an undisturbed coastal island (n = 58) and from sympatric urban cats (n = 39) in Georgia, USA. Based on 16S rRNA gene amplification, 62.1% (59/95) of raccoons and 17.9% (7/39) of feral cats were positive for hemoplasma. There was a greater percentage of hemoplasma-infected raccoons on the undisturbed island (79.3% [46/58]) than on the urban island (35.1% [13/37]; χ2 = 16.9, df = 1, P = 0.00004). Sequencing of the full-length 16S rRNA gene amplicons revealed six hemoplasma genotypes in raccoons, including five novel genotypes that were distinct from three known hemoplasma species identified in the sympatric cats. In addition, the hemoplasma genotypes detected in raccoons were not identified in sympatric cats or vice versa. Although all six hemoplasma genotypes were found in raccoons from urban and undisturbed islands, coinfection patterns differed between sites and among individuals, with the proportion of coinfected raccoons being greater in the undisturbed site. This study shows that raccoons are hosts for several novel hemoplasmas and that habitat type influences infection patterns.
IMPORTANCE This study provides information about novel hemoplasmas identified in raccoons (Procyon lotor), which can be used for assessments of the prevalence of these hemoplasmas in raccoon populations and for future studies on the potential pathogenic impacts of these hemoplasmas on raccoon health. Raccoons from the undisturbed habitat had a higher prevalence of hemoplasma infection than urban raccoons. There does not appear to be cross-species transmission of hemotropic mycoplasmas between urban raccoons and feral cats. Raccoons appear to be hosts for several novel hemoplasmas, and habitat type influences infection patterns.
KEYWORDS: raccoons, Procyon lotor, feral cats, Felis catus, hemoplasmas, wildlife, 16S rRNA gene, phylogenetic analysis, hemoplasma
INTRODUCTION
Urbanization is a strong ecological driver that causes significant changes in the composition of wildlife communities and in various intra- and interspecies interactions (1). Ecological responses to urbanization can lead to changes in the dynamics of wildlife-parasite interactions and in patterns of pathogen transmission through mechanisms such as loss of species diversity, changes in vector abundance, and increased exposure to invasive species and their pathogens (2). Urban-adapted wildlife frequently share anthropogenic food sources with other species and can live in close proximity to other wild and feral/domestic animals (3). Resource use overlap in urbanized environments can increase direct and indirect contact within species and facilitate cross-species transmission of pathogens between wildlife and animals, such as feral cats, while providing opportunities for pathogens to adapt to novel host species and expand their host range (2, 4, 5). In addition, smaller home range sizes, increased aggregation, and high population densities of some wild animals in urban areas (3) can further increase opportunities for contact between species and potentially increase cross-species pathogen transmission.
The raccoon (Procyon lotor) is well known for its adaptability to urbanized habitats and active interaction with domestic animals, such as cats and dogs (6). In urbanized habitats, raccoons and feral cats (Felis catus) are often highly abundant, frequently foraging in close proximity to one another on clumped anthropogenic food sources (e.g., garbage and intentional feeding). Raccoons also harbor diverse pathogens shared with or transmitted to other domestic and/or wild animals, such as canine distemper virus, parvovirus, rabies virus, and Leptospira (6). Here, we used raccoons from urban and undisturbed environments and urban feral cats as a study system to compare and investigate hemotropic Mycoplasma species (so-called genotypes here) composition, richness, coinfection patterns, and potential cross-species transmission.
Hemoplasmas (the common name for hemotropic Mycoplasma species) are facultative intracellular erythrocytic parasites without a cell wall comprising a group of noncultivable Mycoplasma species, including organisms formerly known as Haemobartonella and Eperythrozoon species, which were reclassified as Mycoplasma species based on phylogenetic analysis of their 16S rRNA gene sequences and deeper studies on cell morphological properties (7–11). Hemoplasmas are causative agents of acute or chronic infectious anemias in several mammalian species (7, 12, 13). Human infections with hemotropic mycoplasmas and potential zoonotic transmission of these organisms have also been reported (14–16). Animal infection with hemotropic mycoplasmas is usually self-limiting and well controlled by immunocompetent animals; however, the establishment of clinically inapparent chronic bacteremia is possible in stressed, immunosuppressed, immunocompromised, and immunocompetent individuals (7, 14, 17). The main hematological observation in hemoplasma-infected animals is mild or severe anemia and positive Coombs tests, but infections can occur with or without alterations in hematological parameters (13, 18). The intracellular life cycle of some hemotropic mycoplasmas may explain the chronicity of hemotropic mycoplasma infections in their natural hosts (19).
Several hemoplasma species have been reported from wild carnivores, including Darwin's foxes (Lycalopex fulvipes), black bears (Ursus thibetanus japonicus), Namibian cheetahs (Acinonyx jubatus), Iriomote cats (Prionailurus bengalensis iriomotensis), California sea lions (Zalophus californianus), Japanese badgers (Meles anakuma), raccoon dogs (Nyctereutes procyonoides viverrinus), and Asian mongooses (Herpestes javanicus) (18, 20–25). However, mycoplasma (including hemoplasma) infections of raccoons (Procyon lotor) are poorly studied (26). In 1971, Haemobartonella procyoni was described in raccoons from Maryland, USA (27). The morphology of the parasite resembled that of Mycoplasma haemomuris (formerly Haemobartonella muris), and the microorganism was found in association with the surface of host erythrocytes. The naturally infected raccoons did not have any clinical signs or hematological abnormalities, although infection prevalence was approximately 50%, and parasitemia persisted for 60 days of observation (27).
The objectives of this study were (i) to identify hemotropic mycoplasmas in raccoons and compare infection prevalence, genotype richness, and coinfection patterns in raccoons on an urban and an undisturbed barrier island, and (ii) to evaluate the possibility of cross-species transmission of hemoplasmas between urban raccoons and sympatric feral cats.
RESULTS
PCR amplification of hemoplasma sequences from blood samples.
Primary screening using the HBT-F and HBT-R primers for amplification of the 16S rRNA gene sequences demonstrated that 62.1% (59/95) of raccoons were PCR positive for hemoplasma DNA. These previously published universal primers amplified the partial 16S rRNA gene of Mycoplasma spp. and were successfully used for differentiating hemoplasma-positive and hemoplasma-negative animals. In 2012, the International Committee on Systematics of Prokaryotes (ICSP) subcommittee on the taxonomy of Mollicutes agreed the recommendation to require the full-length 16S rRNA gene sequences in papers describing new hemoplasmas (28). Based on this recommendation, we generated the full-length 16S rRNA gene sequences and used them for construction of our phylogenetic trees. To amplify of the full-length 16S rRNA genes of hemoplasmas present in blood samples of raccoons and cats, we designed new primers based on available sequences in GenBank (see Materials and Methods). Using these new primers for eight individual 16S rRNA-based PCRs (16S-PCR-1 through 16S-PCR-8), we successfully amplified the individual full-length rRNA genes of each hemoplasma present in the raccoon blood DNA samples (Table 1). Five of these eight primer pairs produced the full-length 16S rRNA amplicons from hemoplasma-positive raccoons (16S-PCR sets 1, 2, 3, 5, and 8 [Table 1]). In contrast, only three primer pairs amplified the full-length 16S rRNA amplicons from hemoplasma-positive cats (16S-PCR sets 5 to 7), i.e., the primers designed for Mycoplasma haemofelis, “Candidatus Mycoplasma turicensis,” and “Candidatus Mycoplasma haemominutum,” respectively. The 16S primer pairs 6 and 7 did not generate any amplicons from hemoplasma-positive raccoons. The 16S primer pair 5, which was designed to amplify the full-length 16S rRNA genes of both M. haemofelis and Mycoplasma haemocanis, was the only set that worked with both raccoons and cats. The 16S primer pair 4, which was designed to amplify the full-length 16S rRNA gene of Mycoplasma suis, did not generate any amplicons from either raccoons or cats.
TABLE 1.
Hemotropic mycoplasmas detected in raccoons
| GenBank accession no.a | Sample ID | Hemoplasma genotype detected in raccoons | Primer set used for full-length 16S rRNA amplification (see Table S1) | Sequence homology to other hemoplasmas (GenBank accession no.) |
|---|---|---|---|---|
| KC920443 | PRLO50 | 3 | 16S-PCR-1 | 96% to “Candidatus Mycoplasma erythrodidelphis” (AF178676) |
| KC920441 | PRLO49 | |||
| KC920447 | PRLO72 | |||
| KC920446 | PRLO84 | |||
| KC920445 | PRLO62 | |||
| KC920444 | PRLO57 | |||
| KC920442 | PRLO42 | |||
| KC920448 | PRLO53 | |||
| KC920440 | PRLO25 | |||
| KC920439 | PRLO96 | |||
| KF743729 | PRLO102 | 2 | 16S-PCR-3 | 92–93% to “Candidatus Mycoplasma haemozalophi” (GU905012) and “Candidatus Mycoplasma haemolamae” (AF306346) |
| KF743724 | PRLO88 | |||
| KF743727 | PRLO92 | |||
| KF743722 | PRLO86 | |||
| KF743713 | PRLO56 | |||
| KC936280 | PRLO87 | |||
| KF743717 | PRLO65 | |||
| KF743726 | PRLO91 | 4 | 16S-PCR-2 | 86–88% to raccoon hemoplasma genotypes 2 and 3, and to “Candidatus Mycoplasma haemominutum,” M. wenyonii, and M. ovis |
| KF743711 | PRLO46 | |||
| KF743728 | PRLO101 | |||
| KF743721 | PRLO84 | |||
| KF743719 | PRLO77 | |||
| KF743718 | PRLO74 | |||
| KF743716 | PRLO64 | |||
| KF743706 | PRLO24 | 5 | 16S-PCR-5 | 96–97% to M. haemocanis (AY529641) (including M. haemocanis detected in Japanese raccoon dog [AB848714] and M. haemocanis/M. haemofelis-like sp. detected in Japanese black bear [AB725596]), to M. haemofelis (AF548631), and to raccoon hemoplasma genotype 1 |
| KF743734 | PRLO_56HC | |||
| KF743715 | PRLO62 | |||
| KF743736 | PRLO_103HC | |||
| KF743704 | PRLO14 | |||
| KF743710 | PRLO44 | 6 | 16S-PCR-8 | 91–92% to raccoon hemoplasma genotype 5, and to “Candidatus Mycoplasma haemobos” (EF460765) |
| KF743733 | PRLO_55HB | |||
| KF743707 | PRLO33 | |||
| KF743731 | PRLO104 | |||
| KF743720 | PRLO80 | |||
| KF743714 | PRLO59 | 1 (M. haemocanis/M. haemofelis-like sp.) | 16S-PCR-5 | 99% to M. haemocanis (AY529641) and M. haemofelis (AF548631) |
| KF743709 | PRLO40 | |||
| KF743723 | PRLO87 | |||
| KF743735 | PRLO_92HC | |||
| KF743732 | PRLO_43SC | |||
| KF743705 | PRLO21 | |||
| KF743712 | PRLO55 | |||
| KF743725 | PRLO90 | |||
| KF743708 | PRLO38 | |||
| KF743730 | PRLO103 |
The reference sequence for each hemoplasma genotype is indicated in bold. The numbering of raccoon hemoplasma genotypes (1 to 6) is based on their sequential discovery.
Based on our full-length 16S rRNA PCR analyses, only 54.7% of raccoons (versus 62.1% with the HBT-F and HBT-R primers) were hemoplasma positive. The difference (54.7% versus 62.1%) between our new primers and HBT-F/R primers can be attributed to (i) the difference in the length of amplicons (1,400 to 1,460 bp versus 595 to 620 bp, respectively), (ii) the possible difference in hemoplasma DNA load in blood among the raccoons, which could affect the efficiency of amplification of the full-length 16S rRNA sequences, (iii) the degree of optimization of our PCR conditions for these new primers, and/or (iv) a combination of these factors.
Six hemotropic mycoplasma sequences (genotypes) with partial identity with the 16S rRNA gene sequences of known hemoplasma sequences (available in GenBank) were detected in raccoon populations (Table 1; see also Table S2 in the supplemental material). Except for two M. haemocanis/M. haemofelis-like raccoon hemoplasmas (called raccoon hemoplasma genotypes 1 and 5), which had 96 to 97% nucleotide sequence identity with each other in their 16S rRNA genes, the other hemoplasma sequences (genotypes 2 to 4 and 6) detected in raccoons demonstrated lower levels of genetic similarity (≤86 to 96%) among these genotypes and to other known hemoplasma species (Tables 1 and S2).
Raccoon hemoplasma genotypes 1 and 5 were amplified from independent raccoon blood samples using the primers designed to amplify the full-length 16S rRNA gene of M. haemocanis/M. haemofelis. Genotype 5 (GenBank accession no. KF743706) had only 96 to 97% nucleotide sequence identity with M. haemocanis/M. haemofelis and genotype 1, whereas genotype 1 (GenBank accession no. KF743705) had 99% nucleotide similarity to M. haemocanis/M. haemofelis (Tables 1 and S2). When amplification from the rpoB and gyrB genes was attempted on all the raccoon samples positive for these two M. haemocanis/M. haemofelis-like hemoplasma genotypes (PCR-A to -D primer sets for rpoB [PCR-A to -C] and PCR-D set for gyrB [Table S1]), amplification from the rpoB gene was unsuccessful for both genotypes of M. haemocanis/M. haemofelis-like raccoon hemoplasmas, and amplification from the gyrB gene was possible only for genotype 1 and not for genotype 5. Sequence analysis of the amplified gyrB genes (GenBank accession no. KF743740 to KF743744) and their deduced protein sequences demonstrated low identity (73 to 75% nucleotide and 87 to 88% amino acid) with both M. haemocanis and M. haemofelis. Thus, these M. haemocanis/M. haemofelis-like spp. (genotypes 1 and 5) detected in raccoon blood samples were unlikely to belong to the known species M. haemocanis or M. haemofelis but were closely related to them phylogenetically. All attempts to amplify rpoB gene sequences from the other raccoon hemoplasma-positive samples using previously published primers designed for rpoB of Mycoplasma spp. (29) failed to yield amplicons.
In feral cats on St. Simons island, M. haemofelis (n = 1), “Ca. Mycoplasma haemominutum” (n = 5), and “Ca. Mycoplasma turicensis” (n = 1) were identified by the full-length 16S rRNA gene amplification and sequencing; however, these species were not detected in raccoons. The presence of M. haemofelis and “Ca. Mycoplasma haemominutum” in cat blood was also confirmed by amplification and sequencing of their partial rpoB genes (GenBank accession no. KF743746 to KF743751).
Phylogenetic analysis of the 16S rRNA genes.
We used sequences of the 16S rRNA genes to determine the phylogenetic relatedness of the hemoplasmas (genotypes) detected in raccoons with those of other known hemotropic Mycoplasma species available in GenBank (Fig. 1). No chimeras were detected from all 16S rRNA gene sequences generated in this study. The dendrogram in Fig. 1 shows the inferred phylogenetic position of the hemoplasma sequences identified in raccoons among known hemotropic Mycoplasma species. The interspecies similarity of the 16S rRNA genes among the species in the phylogenetic tree was also assessed (Table S2). The 16S rRNA-based phylogenetic analysis and the interspecies similarity data showed that the raccoon hemoplasma genotypes were phylogenetically related to known hemoplasma species, i.e., M. haemocanis, M. haemofelis, “Candidatus Mycoplasma haemobos,” “Candidatus Mycoplasma wenyonii,” Mycoplasma ovis, “Candidatus Mycoplasma haemocervae,” “Candidatus Mycoplasma erythrocervae,” “Candidatus Mycoplasma erythrodidelphis,” “Candidatus Mycoplasma haemolamae,” “Candidatus Mycoplasma haemozalophi,” and “Candidatus Mycoplasma kahanei” (see Fig. 1). However, based on the low levels (i.e., ≤97%) of sequence identity (29, 30) for five of the six raccoon hemoplasma genotypes (genotypes 2 to 6), we believe these five genotypes represent novel hemoplasma genotypes or putatively new hemoplasma species (see Fig. 1 and Table 1) not yet described in other animal species.
FIG 1.
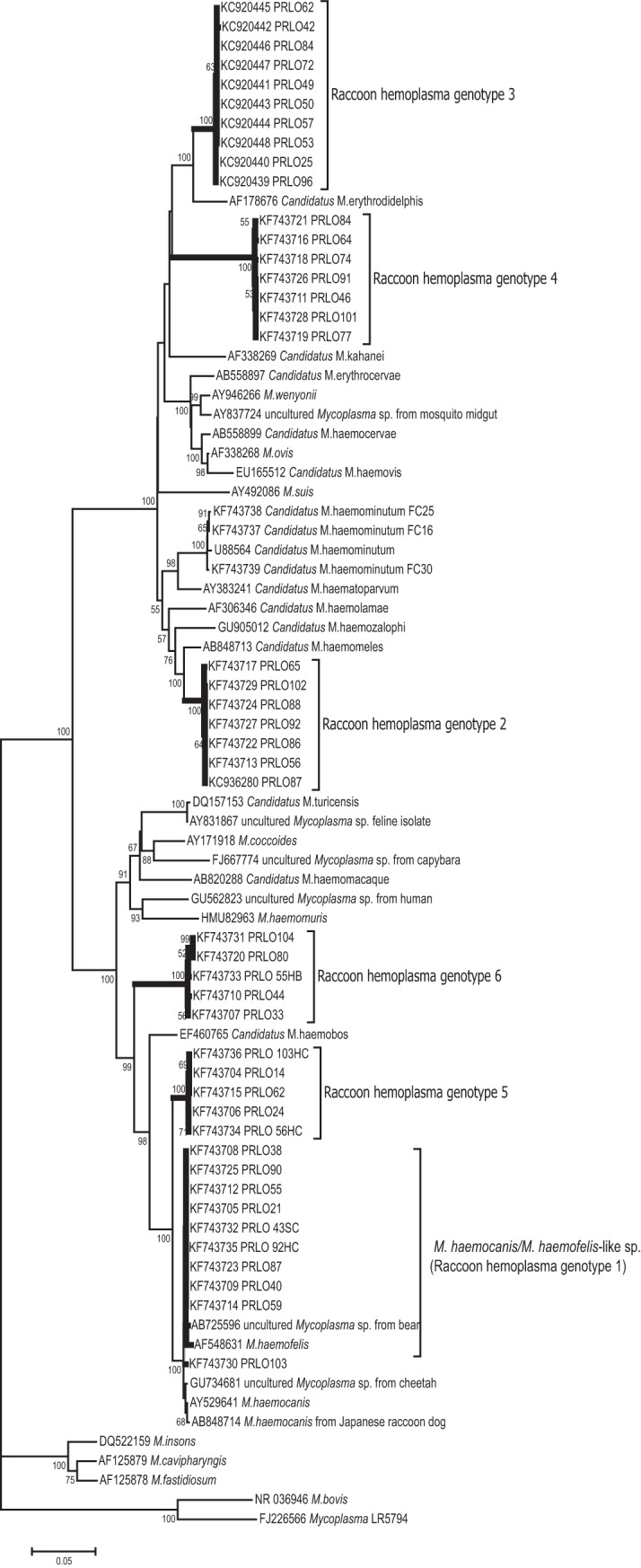
Dendrogram showing phylogenetic relationships based on nucleotide sequence data for the 16S rRNA gene among the hemoplasma genotypes detected in raccoons (Procyon lotor) with other hemotropic Mycoplasma spp., and three nonhemotropic phylogenetically closely related Mycoplasma spp. (M. insons, M. fastidiosum, and M. cavipharyngis) and two not phylogenetically closely related Mycoplasma species. The trees were constructed by the minimum evolution method in the MEGA 6 package. Accession numbers are shown to the left of each organism name; strain or isolate names are also shown.
The presence of regions of the low interspecies sequence similarity in the 16S rRNA genes among these five novel raccoon hemoplasma genotypes (genotypes 2 to 6) and the 16S rRNA genes of other hemoplasmas available in GenBank allowed us to design the species-specific PCR primers (Fig. S3 to S7 show detailed sequence comparisons) that can be used for qualitative detection of each hemoplasma genotype. In the current study, we used these species-specific 16S rRNA primers to demonstrate selective amplification of each hemoplasma genotype in raccoon samples that were coinfected with different hemoplasma genotypes. No cross-amplification between raccoon hemoplasma genotypes or false-positive or false-negative amplifications were observed for these species-specific primers when used in PCR assays on DNA from all hemoplasma-positive and hemoplasma-negative raccoon blood DNA samples.
Prevalence of hemoplasmas in the studied animals in urban versus undisturbed ecosystems and coinfection.
The proportion of raccoons infected with hemotropic Mycoplasma spp. was 62.1% (95% confidence interval [CI], 51.5, 71.7%; n = 95), with overall proportions of 35.1% (95% CI, 20.7, 52.6%; n = 37) on St. Simons Island (developed habitat) and 79.3% (95% CI, 66.3, 88.4%; n = 58) on St. Catherines Island (undisturbed habitat). Hemoplasma infection prevalence was 17.9% (95% CI, 8.1, 34.1%; n = 39) in feral cats from St. Simons. Overall, infection rates with hemoplasma were 44.4% (95% CI, 28.3, 61.7%; n = 36) in female raccoons and 72.9% (95% CI, 59.5, 83.3%; n = 59) in male raccoons.
In univariate analyses, habitat and sex were significantly associated with hemoplasma infection in raccoons (habitat type, χ2 = 16.9, df = 1, P = 0.00004; sex, χ2 = 6.52, df = 1, P = 0.01); hemoplasma infection prevalence was greater in male raccoons and on the undisturbed island. We also found a significant association between hemoplasma infection and body mass (Mann-Whitney U test, W = 732, P = 0.01), although habitat type and body mass were not correlated (Mann-Whitney U test, W = 927.5, P = 0.27). We found no significant association between host species (raccoon or feral cat) and hemoplasma infection on the urbanized island (χ2 = 2.89, df = 1, P = 0.09). The best-fit generalized linear model (GLM) associated with raccoon hemoplasma infection included weight, habitat, and the interaction between weight and habitat (Table S3). When the weight × urbanized habitat interaction was accounted for, the urbanized habitat was less likely to be associated with hemoplasma infection (odds ratio [OR], −0.34, P = 0.02) than the undisturbed habitat. Specifically, heavier raccoons had greater odds of hemoplasma infection on the undisturbed island (OR, 1.3) but had lower odds of infection on the urban island (OR, 0.21) (Fig. S1).
Coinfection of raccoons with multiple hemoplasma genotypes was observed at both sampling sites: undisturbed habitat coinfection prevalence was 87% (40/46), and urban habitat coinfection prevalence was 53.8% (7/13). Raccoons from the undisturbed habitat had a significantly higher number of individuals coinfected with more than one hemoplasma genotype than those from the urbanized habitat (χ2 = 52.989, df = 1, P < 0.00001). The ratio of single infection to coinfection for the urban habitat was 1/1.2 and was 1/5.7 for the undisturbed habitat. Hemoplasma genotype richness and infection patterns in raccoons are shown in Table 2. At the population level, there was no difference in overall hemoplasma genotype richness (both sites had a total richness of 6 hemoplasma genotypes identified). However, at the individual level, raccoons on St. Simons Island (urban) had lower hemoplasma genotype richness than those on St. Catherines Island (undisturbed) (Kruskal-Wallis test, χ2 = 24.03, df = 1, P < 0.0001; GLM, z = −5.88, df = 1, P < 0.0001).
TABLE 2.
Hemoplasma genotypes identified on developed (urban) and protected (undisturbed) islands
| Hemoplasma genotype detected in raccoons | Urban |
Undisturbed |
Total |
|||
|---|---|---|---|---|---|---|
| % (n = 13) | 95% CI | % (n = 46) | 95% CI | % (n = 59) | 95% CI | |
| 5 | 15.4 | 2.7, 46.3 | 58.7 | 43.2, 72.7 | 49.2 | 36.0, 62.4 |
| 1 (M. haemocanis/M. haemofelis-like sp.) | 61.5 | 32.37, 84.9 | 41.3 | 27.3, 56.7 | 45.8 | 32.9, 59.2 |
| 2 | 7.7 | 0.4, 3.79 | 56.5 | 41.2, 70.8 | 45.8 | 32.9, 59.2 |
| 3 | 30.8 | 10.4, 61.1 | 60.9 | 45.4, 74.5 | 54.2 | 40.8, 67.1 |
| 6 | 46.2 | 20.4, 73.9 | 32.6 | 20.0, 48.1 | 35.6 | 23.9, 49.2 |
| 4 | 23.1 | 6.16, 54.0 | 43.5 | 29.2, 58.8 | 39.0 | 26.8, 52.6 |
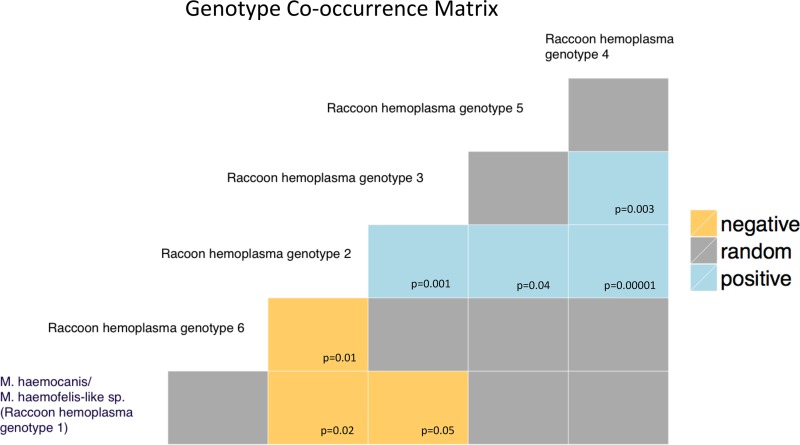
The mean number of hemoplasma genotypes identified per infected raccoon was 1.85 (range, 1 to 4 genotypes) on the urbanized island and 2.93 (range, 1 to 6 genotypes) on the undisturbed island. There was a slight positive but nonsignificant correlation between hemoplasma genotype richness and individual raccoon body weight (Spearman rank correlation, rho = 0.25, P = 0.06). Although raccoons at both sites had coinfections ranging from 2 to 4 hemoplasma genotypes, coinfection with 5 to 6 genotypes was seen only in raccoons from the undisturbed site (Fig. S2). We also observed varied composition of hemoplasma genotypes among raccoons; coinfections with two to three hemoplasma genotypes were relatively evenly distributed. An association plot (Fig. 2) showed positive associations for coinfection for raccoon hemoplasma genotypes 2 and 3, 2 and 4, 2 and 5, and 3 and 4. Negative associations of coinfection were seen between genotypes 2 and 6, as well as M. haemocanis/M. haemofelis-like sp. (genotype 1) and genotype 2, and between M. haemocanis/M. haemofelis-like sp. (genotype 1) and genotype 3.
FIG 2.
Species cooccurrence matrix showing patterns of coinfection among the hemoplasma genotypes detected in raccoons. Results of the genotype cooccurrence matrix represent the probability (P value) of the cooccurrence observed being greater or less than that expected due to chance. Only significant P values (≤0.05) are shown.
DISCUSSION
The genus Mycoplasma currently comprises 20 hemotropic species (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=2093), and except for the well-established species M. haemocanis, M. haemofelis, and M. haemomuris, all other 17 hemoplasmas have the provisional taxonomic status “Candidatus,” as they are incompletely described prokaryotes (29–31). Hemoplasmas infect different hosts and are able attach to, and sometimes intracellularly invade, their erythrocytes (7, 10, 19). Hemoplasmas have not been cultured in vitro, and the detection of hemotropic mycoplasmas using Romanowsky-Giemsa- and/or acridine orange-stained blood smears in combination with the PCR amplification of target hemoplasma genes is common laboratory practice for diagnosing these infections in animals and humans (7, 10, 13, 14, 32–34). However, the examination of stained blood films for hemoplasmas (not performed in our study) has low sensitivity and specificity, and the absence of hemoplasma-like bodies on blood film often does not correlate with the PCR results. Hemoplasma concentrations in the blood of animals may also fluctuate during the course of a hemoplasma infection, and the low concentrations may not be detected by microscopy (35), especially in immunocompetent chronically hemoplasma-infected animals. Therefore, microscopic blood film evaluation is occasionally omitted in multiple published studies on the investigation of hemoplasma infections in domestic or wild animals (20, 21, 36).
The reported prevalence of hemoplasma-infected animals in a variety of species ranges from 0.5 to 56.7% (13, 18, 20, 34, 37). In this study, the PCR screening using the HBT-F and HBT-R primers revealed that 62.1% of raccoons were hemoplasma infected. The use of these published universal primers allowed us to successfully identify hemoplasma-positive animals; however, the direct sequencing (i.e., without cloning into a plasmid vector prior to the sequencing) of PCR products from animals coinfected with different mycoplasma genotypes was complicated or impossible due to the presence of the mixed PCR amplicons generated by these universal primers. The primers amplify the partial 16S rRNA of 595 to 620 bp in size, and the difference in 25 nucleotides (nt) was indistinguishable on electrophoresis in a 1% agarose gel. Thus, if the primers amplified the 16S fragment from animals coinfected with different mycoplasma genotypes, it was impossible to discriminate them on a gel and to obtain a clear sequence of them using the same HBT-F/R primers. To amplify all hemoplasmas (genotypes), we designed and used new primers that allowed us to amplify all genotypes separately and produce clear sequences from direct DNA sequencing of PCR amplicons. The new primers were able to amplify full-length 16S rRNA gene sequences of these hemoplasmas in raccoons in compliance with the recommendation of the subcommittee on the taxonomy of Mollicutes that in 2012 agreed to require the full-length 16S rRNA gene sequences in papers describing new hemoplasmas (28). The use of full-length 16S rRNA gene sequences for further phylogenetic analysis may be especially necessary to distinguish between closely related bacterial species, and thus the sequencing of the full-length 16S rRNA gene is desirable and usually required when describing new species (38–40).
After the full-length 16S rRNA sequences for raccoon hemoplasmas were determined, HBT-F/R primers were retrospectively analyzed for their matching to the hemoplasma sequences (against all 6 detected genotypes). The reverse primer (HBT-R) matched all genotypes with 100% identity. The forward primer (HBT-F) matched genotypes 1, 5, and 6 with 100% identity; however, one mismatch (in bold) was present in this region for all sequences of genotype 3 (HBT-F primer sequence, ATACGGCCCATATTCCTACG, versus the sequence of genotype 3, ATATGGCCCATATTCCTACG). Two mismatches (in bold) were present in this region for all sequences of genotype 4 (HBT-F primer sequence, ATACGGCCCATATTCCTACG, versus the sequence in genotype 4, ATATGGCCCATATCCCTACG). Despite the presence of these mismatches between the forward primer and the 16S rRNA sequences of genotypes 3 and 4, we did not observe any negative impact on the results of our qualitative PCR with these universal primers. However, for future studies of hemoplasma infections in raccoons using HBT-F/R primers, we recommend introducing an ambiguous base (Y = C/T) at positions 4 and 14 of HBT-F, which may improve the sensitivity and yield of amplification of target hemoplasmas, especially in tested samples with low DNA concentrations.
New primers for amplification of the full-length 16S rRNA genes allowed us to detect six hemoplasmas (genotypes) in raccoons with partial identity to the 16S rRNA gene sequences of known hemoplasma species. Based on low levels (i.e., ≤97%) of sequence similarity of these hemoplasma genotypes to other described hemoplasmas and the mammalian host in which these genotypes were detected, we believe that five of these six genotypes represent novel hemoplasma genotypes or putatively novel hemoplasma species not yet described in other animal species.
Except for two raccoon hemoplasma genotypes, 1 and 5 (M. haemocanis/M. haemofelis-like), which had 96 to 97% nucleotide sequence identity with each other in their 16S rRNA genes, the other genotypes, 2 to 4 and 6, demonstrated genetic similarity of ≤86 to 96% among these genotypes and to other known hemoplasma species. Only the 16S rRNA gene sequences of genotype 1 demonstrated 99% sequence identity to the 16S rRNA gene of M. haemocanis and M. haemofelis. M. haemocanis and M. haemofelis by themselves also have 99% sequence identity with each other in their 16S rRNA genes and are indistinguishable by the 16S rRNA gene analysis, without the sequencing of additional housekeeping genes, e.g., rpoB, gyrB, and others. To deeply identify genotype 1 as either M. haemocanis or M. haemofelis, we performed amplification of two housekeeping genes (rpoB and gyrB). The amplification from the rpoB gene was unsuccessful using different M. haemocanis/M. haemofelis rpoB-specific primers, and only the gyrB gene was amplified for genotype 1; however, nucleotide sequence analysis of the gyrB gene and the deduced protein sequences demonstrated low similarities to both M. haemocanis and M. haemofelis. Thus, based on our results, we decided that the genotype 1 detected in raccoons is unlikely to belong to the known species M. haemocanis or M. haemofelis but were closely related to them phylogenetically.
Phylogenetic studies of the 16S rRNA gene of closely related Mycoplasma species propose to use the arbitrary interspecies sequence similarity value of ≤97% as a minimum level indicating a separate genetically distant species (29, 30, 41). Data based on the expanded analysis of the 16S rRNA gene sequences of the species within the family Mycoplasmataceae generally support this proposition (29). Nevertheless, at least 20 pairs of closely related well-established Mycoplasma species with 16S rRNA gene similarity greater than 97% demonstrated serological, genetic, and ecological features that defined them as individual species, despite the high percentage of similarity of their 16S rRNA genes (for details, see reference 29). Thus, the 16S rRNA sequence identity of any new isolate of ≥98 to 99% may not be a clear indication that the Mycoplasma species is the same or different. Similar examples exist and are well known for some other closely related species with identical or nearly identical 16S rRNA sequences (42), e.g., Bacillus and Listeria species (43, 44).
Although about 62.1% of the raccoons in thus study were infected with hemoplasmas, they appeared normal upon physical examination. The pathogenicity of the detected hemoplasmas for the raccoons is unknown and should be investigated in the future. From a taxonomic point of view, similar to cases of Eperythrozoon teganodes in cattle (28) and Haemobartonella spp. in horses (33), the genetic relationship between the previously studied hemoplasma-like species Haemobartonella procyoni in raccoons of Maryland (27) and the hemoplasmas detected in raccoons this study is unknown, because there is no known H. procyoni genetic material in any national or international collection of microorganisms.
The three species of hemoplasma identified in feral cats on St. Simons Island have been previously detected in blood samples from cats throughout the world, and the overall proportion of infected cats (17.9%) was within the reported prevalence in cats seen worldwide (16, 45–51). The most common hemoplasma species found in cats in our study, “Ca. Mycoplasma haemominutum,” also appears to be the most common hemoplasma species in cats across many different studies (50, 51). Our study detected no evidence of cross-species hemoplasma transmission between feral cats and raccoons, despite the close proximity of feral cats and raccoons on St. Simons Island.
Interactions between pathogen prevalence, diversity, and anthropogenic disturbance, such as urbanization, can be positive, negative, or neutral, depending on the type of environmental change and how it affects abundance, density, and/or contact within and between host species, other coinfecting pathogens, and environmental influences on host immunity and pathogen susceptibility (52–54). The proportion of hemoplasma-infected raccoons was greater on the undisturbed than on the urbanized island, and the proportion of hemoplasma-infected raccoons in both locations was higher than in urban cats. If hemoplasma transmission is population density dependent, an increase in hemoplasma infection in urbanized habitats is expected (55, 56). We did not see evidence of this, because trapping success on the urban island (0.41 animals/trap night) was higher than on the protected island (0.24 animals/trap night). However, we did not perform mark-recapture studies for density estimation. Regardless, in urbanized areas, raccoons may be more likely to enter traps due to greater habituation and differing food preferences. Alternatively, hemoplasma transmission may be frequency dependent, due to bites, scratches, licking (50), and potential vector-borne (flea/tick) transmission. Assuming that hemoplasma infection can be vector-borne in wild raccoons, as has been found in cats (10, 46), habitat-related differences in microclimate could influence hemoplasma transmission. The prevalence of tick-borne diseases and tick infestation rates in host species is often higher in natural habitats than in urban environments (57, 58), related to insufficient host diversity to maintain complex tick life cycles in urbanized areas (59).
The best-fit GLM showed a positive relationship between heavier animals and hemoplasma infection in undisturbed habitats, whereas heavier animals in urban habitats had lower odds of infection. Although associations between sex and hemoplasma infection were male biased and marginally significant in univariate analyses, sex was not a significant predictor of hemoplasma infection in our best-fit GLMs. The positive association between body weight and hemoplasma infection in the undisturbed habitat could be related to host age; although we did not determine the age of captured raccoons, larger individuals were likely to be older and may have had increased exposure to ectoparasites (if arthropod-borne transmission is a dominant mode of transmission) or higher ectoparasite infestation. The observation of a positive relationship between weight and hemoplasma positivity in the undisturbed environment might suggest that heavier animals may have greater contact with other infected animals or vectors. For the opposite result in an urban habitat, supplemental feeding (garbage and deliberate feral animal feeding by residents) on St. Simons Island might have led to higher tolerance by raccoons toward conspecific animals, which may reduce aggressive (60) behavior, reducing the likelihood of fighting and hemoplasma transmission through infected blood or saliva. Higher food availability or quality in urban habitats (if raccoons were being fed nutritionally rich supplements, such as cat food) could allow heavier raccoons to mount a more successful immune defense against hemoplasma (61). Additional routes of transmission for hemoplasmas, such as transplacental and transmammary transmission (32), may also influence differences in hemoplasma prevalence, particularly if one population has a different population age structure.
Differences in population management of raccoons between islands may influence the prevalence of hemoplasma infection. Although there are no published data available on the relative population densities of raccoons on these two islands, raccoons are routinely culled on St. Catherines Island to protect damage to sea turtle nests. Culling may inadvertently increase the transmission of pathogens with frequency-dependent transmission in part by increasing the birth rate, leading to an increase in susceptible individuals in a population (62). However, further study is required to thoroughly understand the epidemiology and mode of hemotropic mycoplasmas in raccoons in order to pinpoint the causes for differences in infection rates between undisturbed and disturbed habitats.
Coinfection with multiple hemoplasmas has been described in humans (16), domestic animals (45, 63, 64), and wildlife (36). However, it remains unknown why patterns of hemoplasma coinfection vary among raccoons and habitats. For instance, negative cooccurrence of some hemoplasmas (Fig. 2) may be due to cross-immunity, ecological interference, or differing contact networks (65) between raccoons in different habitat types. One possible explanation for our results is that the protected island offers a greater opportunity for within-species and cross-species hemoplasma transmission due to contacts with a more diverse host community.
Horizontal transmission of hemotropic mycoplasmas in species other than raccoons has been hypothesized to be potentially associated with blood-feeding arthropod vectors, as well as direct transmission via infected blood (e.g., aggressive interactions and injuries related to animal-to-animal contact, and contact with blood) (10, 66–68). All these transmission routes may account for the widespread occurrence of hemoplasmas in the studied raccoon populations and require additional investigation.
To conclude, this study identified novel hemoplasma genotypes in raccoons and provided new molecular tools to detect these species. We identified six hemoplasma genotypes in raccoons that were phylogenetically related to hemoplasma species previously reported in other mammalian hosts (see Fig. 1). Five of these six hemoplasmas appear to be novel hemoplasma genotypes (i.e., never previously reported or deposited in GenBank). Future studies should (i) explore the probability of cross-species transmission with additional samples from various sympatric host species and (ii) evaluate the pathogenicity of hemoplasmas in raccoons, particularly using hematological and immunological assays. The potential mechanism of intra- and interspecies transmission of hemoplasmas and the drivers of its species composition in sympatric host species remain to be elucidated.
Conclusion.
This study provides information about novel hemoplasmas in raccoons (Procyon lotor), which can be used for assessments of the prevalence of these hemoplasmas in raccoon populations. Raccoons from the undisturbed habitat had higher hemoplasma infection rates than raccoons in a rural habitat. There does not appear to be cross-species transmission of hemotropic mycoplasmas between urban raccoons and feral cats.
MATERIALS AND METHODS
Field sites and study populations.
Raccoons were live-trapped on two Georgia coast barrier islands, St. Simons Island (31°9′40″N 81°23′13W) and St. Catherines Island (31°37′50″N 81°9′36.5W), which is approximately 50 km north of St. Simons Island. St. Simons Island has complex ecosystems, including ocean beach, salt marsh, maritime forest, and freshwater slough. In addition, St. Simons Island is one of the most urbanized Georgia Barrier Islands, with a large resident human population and rapidly increasing residential developments (69). St. Catherines Island also has diverse habitats, including marsh, deciduous and evergreen forest, palmetto scrub, and open savannah (70). St. Catherines is a protected barrier island for scientific research, with no human development, residential areas, or domestic animals. Adult feral cats were live-trapped on St. Simons Island as a part of spay/neuter program and physically examined by local veterinarians; no information on the sex of these feral cats was recorded.
Sample collection.
Raccoons (n = 95) were trapped in the spring and summer of 2012 along trapping transects using 20 Tomahawk traps (Tomahawk Live Trap Company, Tomahawk, WI, USA). Raccoons were anesthetized by intramuscular injection of ketamine (20 mg/kg of body weight; Aveco Co., Fort Dodge, IA, USA) mixed with xylazine (4 mg/kg; Mobay Corp., Animal Health Division, Shawnee, KS, USA). The body weight of each animal was measured, and their general physical health was evaluated by local veterinarians. Approximately 3 ml of blood was collected from all animals (raccoons and cats) by jugular venipuncture into vacuum tubes containing anticoagulant (EDTA) tubes. Blood samples from feral urban cats (n = 39) were collected by local veterinarians as part of the physical examination for the St. Simons Island spay/neuter program. Whole-blood samples in the EDTA tubes were stored at −20°C until laboratory analysis. In total, we collected 37 and 58 raccoon blood samples from St. Simons Island and St. Catherines Island, respectively. Institutional Animal Care and Use Committee (A2011 03-042-Y2-A2) and Georgia Department of Natural Resources wildlife permits (29-WBH-12-100) were obtained before sampling (71).
DNA extraction, PCR amplification, and sequencing of amplicons.
Total DNA was extracted from 200 μl of blood from each individual using the DNeasy blood and tissue kit (Qiagen, Valencia, CA, USA) or the Quick-gDNA MiniPrep kit (Zymo Research Corporation, Orange, CA, USA) according to the manufacturers' protocols and with standard clinical PCR laboratory precautions to avoid cross-contamination. DNA samples were stored at −80°C until use.
The primary screening for the presence of hemoplasmas was performed by PCR using previously published HBT-F and HBT-R universal primers for amplification of the partial 16S rRNA hemoplasma genes (37). These primers amplify the 16S rRNA gene region from positions 313 to 332 to positions 889 to 908 based on the 16S rRNA gene reference sequence of M. haemofelis (accession no. AF178677) (37, 72). Based on our in silico PCR analysis (72) of these universal primers against the different mycoplasma 16S rRNA gene sequences available in the GenBank database, it was demonstrated that depending on the target Mycoplasma spp., these primers produce PCR fragments with sizes of 595 to 620 bp. In addition, these universal primers were successfully used for amplification of the partial 16S rRNA hemoplasma genes in a few published studies (73–75).
A second aliquot of whole blood from each of the hemoplasma-positive samples was used to isolate additional DNA for amplification of the full-length 16S rRNA gene, the RNA polymerase beta-subunit gene (rpoB), and the DNA gyrase subunit B gene (gyrB) using PCR primers designed in this study (Table S1). Eight primer pairs were designed to amplify the full-length 16S rRNA genes, three primer sets were designed to amplify part of rpoB, and one primer set was designed to amplify part of gyrB based on sequences of other known hemoplasma species available at GenBank (Table S1, 16S-PCR-1 through 16S-PCR-8, and PCR-A through PCR-D).
The 16S rRNA amplicons produced were directly sequenced (with and without cloning into a plasmid vector) by Macrogen, and the rpoB and the gyrB amplicons were sequenced directly without cloning. Prior to sequencing, PCR amplicons were purified by electrophoresis using 1.5% agarose gels and extracted with the QIAquick gel extraction kit (Qiagen). Amplicons were sequenced with the same primers used for PCR amplification and then with internal (walking) primers when needed. Cloned amplicons were produced as described elsewhere (29), and 15 to 20 clones of the 16S rRNA gene PCR products of each amplicon were sequenced and analyzed.
When the full-length 16S rRNA gene sequences of the hemoplasma genotypes of raccoons were determined, species-specific 16S rRNA primers to selectively amplify each hemoplasma genotype identified in raccoons were designed (see Table S1, 16S-PCR-9 through 16S-PCR-13). We used these species-specific 16S rRNA primers to selectively identify each hemoplasma genotype in blood samples from raccoons coinfected with different hemoplasma genotypes. The selectivity of these primers for each hemoplasma genotype was demonstrated by gel electrophoresis (i.e., the presence of a single amplicon band) and direct sequencing of amplicons.
The amplification mixture for all PCRs (for direct sequencing without cloning) contained 5 μl of 10× HotStarTaq PCR buffer, 1.5 mM MgCl2, 200 mM dinucleoside triphosphate (dNTP) mixture, 1 mM each primer, and 2.5 U of HotStarTaq Plus DNA polymerase (Qiagen) in a final volume of 50 μl, including 3 μl of DNA template. The Vent DNA polymerase kit (New England BioLabs), which contains high-fidelity thermophilic Vent DNA polymerase, was used for the amplification of PCR products for subsequent cloning and sequencing using plasmid DNA. The absence of PCR inhibitors in isolated blood DNAs was confirmed by PCR amplification of the Procyon lotor mitochondrial gene for 16S rRNA, as an extraction positive control (76) (with primers F1-Animal and R1-Animal) on each sample and negative (no DNA added) PCR control were run for each PCR assay. The DNA of M. haemocanis and “Ca. Mycoplasma haemominutum” was used as a positive control for the PCRs with the primers PCR-A through PCR-D.
All PCRs in this study were conducted under the following conditions: a polymerase activation step at 94°C for 5 min (or 15 min for HotStarTaq only), 40 cycles of 95°C for 30 s, 60°C for 60 s, and 72°C for 60 s, and a final extension at 72°C for 10 min. PCR products were detected by electrophoresis through 1% Tris-acetate-EDTA (TAE)–agarose gels containing ethidium bromide concentrations, followed by UV visualization.
Phylogenetic analysis.
The 16S rRNA sequences determined in this study were compared to those available in the GenBank database using procedures, algorithms, and methods for phylogenetic tree inference, as described elsewhere (18, 29). To avoid the potential presence for chimeric sequences or PCR-derived variants in the data set, all hemoplasma 16S rRNA PCR products for phylogenetic analyses were directly amplified from blood DNA samples of raccoons and cats with two different DNA polymerases (HotStarTaq and Vent) and were directly sequenced without cloning (77, 78). All gene sequences prior to the downstream phylogenetic analysis were subjected to the chimeric sequence analysis using DECIPHER (79) and UCHIME (80). All sequences are deposited in GenBank and are publicly available.
Ecological data analysis.
All statistical analyses were performed using R (http://cran.r-project.org) (81). A descriptive analysis of the hemoplasma infection status of animals collected in each habitat type was performed. For univariate analyses, we used Pearson's chi-square tests to compare the frequency of hemoplasma infection in raccoons in urban and undisturbed environments, raccoon sex and hemoplasma infection, raccoon hemoplasma coinfection and habitat type, and hemoplasma infection in urban cats and raccoons. Additional univariate analyses included a Mann-Whitney U test to evaluate differences between habitat type and raccoon body mass, hemoplasma infection and body mass, and habitat type and body mass. A Kruskal-Wallis test and a generalized linear model (GLM) with Poisson errors were used to evaluate associations between hemoplasma species richness in individual raccoons and body weight. We used a global GLM with individual hemoplasma infection status (positive or negative) as a response variable with a binomial error structure to identify factors that may affect mycoplasma prevalence (82). Sex, habitat type, and body weight were included as explanatory variables alongside biologically meaningful interactions between covariates. Using the R package AICcmodavg, we applied a stepwise algorithm and calculated Akaike's information criteria corrected for small sample sizes (AICc) to determine which set of covariates provided the best fit to the data. A species cooccurrence matrix was calculated to evaluate if coinfecting putative hemoplasma species were negatively, randomly, or positively associated with one another within each host using the R package cooccur (83).
Accession number(s).
All DNA sequences from this study were deposited in GenBank under the accession numbers KF743704 to KF743751, KC920439 to KC920448, and KC936280.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this research was provided by St. Catherines Island Foundation, Inc. through the American Museum of Natural History and the Odum School of Ecology, UGA, through Vanessa Ezenwa and Andrew Park. H. Danaceau was supported by grant 9T35OD010433-06 from the Office of the Director, a component of the National Institutes of Health (NIH). The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
We thank Joanne Messick, Andreas Santos, and Michael Yabsley for advice, Mark Heth, Sea Island Hospital of St. Simons Island, for providing samples, and Veronica Greco, Royce Hayes, and on-site staff of St. Catherines Island for technical advice and help during the fieldwork on the island. We also thank Daniel Becker for critical review of the manuscript and three anonymous reviewers for providing comments on an earlier draft of the manuscript.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00211-17.
REFERENCES
- 1.Shochat E, Warren PS, Faeth SH, McIntyre NE, Hope D. 2006. From patterns to emerging processes in mechanistic urban ecology. Trends Ecol Evol 21:186–191. doi: 10.1016/j.tree.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Bradley CA, Altizer S. 2007. Urbanization and the ecology of wildlife diseases. Trends Ecol Evol 22:95–102. doi: 10.1016/j.tree.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gehrt SD, Riley SPD, Cypher BL. 2010. Urban carnivores: ecology, conflict, and conservation. Johns Hopkins University Press, Baltimore, MD. [Google Scholar]
- 4.Wright AN, Gompper ME. 2005. Altered parasite assemblages in raccoons in response to manipulated resource availability. Oecologia 144:148–156. doi: 10.1007/s00442-005-0018-3. [DOI] [PubMed] [Google Scholar]
- 5.Becker DJ, Streicker DG, Altizer S. 2015. Linking anthropogenic resources to wildlife-pathogen dynamics: a review and meta-analysis. Ecol Lett 18:483–495. doi: 10.1111/ele.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadidian J, Prange S, Rosatte R, Riley SPD, Gehrt SD. 2010. Raccoons (Procyon lotor), p 35–47. In Gehrt SD, Riley SPD, Cypher BL (ed), Urban carnivores: ecology, conflict, and conservation. Johns Hopkins Press, Baltimore, MD. [Google Scholar]
- 7.Messick JB. 2004. Hemotrophic mycoplasmas (hemoplasmas): a review and new insights into pathogenic potential. Vet Clin Pathol 33:2–13. doi: 10.1111/j.1939-165X.2004.tb00342.x. [DOI] [PubMed] [Google Scholar]
- 8.Neimark H, Kocan KM. 1997. The cell wall-less rickettsia Eperythrozoon wenyonii is a mycoplasma. FEMS Microbiol Lett 156:287–291. doi: 10.1111/j.1574-6968.1997.tb12742.x. [DOI] [PubMed] [Google Scholar]
- 9.Neimark H, Peters W, Robinson BL, Stewart LB. 2005. Phylogenetic analysis and description of Eperythrozoon coccoides, proposal to transfer to the genus Mycoplasma as Mycoplasma coccoides comb. nov. and request for an opinion. Int J Syst Evol Microbiol 55:1385–1391. doi: 10.1099/ijs.0.63437-0. [DOI] [PubMed] [Google Scholar]
- 10.Willi B, Boretti FS, Tasker S, Meli ML, Wengi N, Reusch CE, Lutz H, Hofmann-Lehmann R. 2007. From Haemobartonella to hemoplasma: molecular methods provide new insights. Vet Microbiol 125:197–209. doi: 10.1016/j.vetmic.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Uilenberg G, Thiaucourt F, Jongejan F. 2006. Mycoplasma and Eperythrozoon (Mycoplasmataceae). Comments on a recent paper. Int J Syst Evol Microbiol 56:13–14. doi: 10.1099/ijs.0.63998-0. [DOI] [PubMed] [Google Scholar]
- 12.Sykes JE. 2010. Feline hemotropic mycoplasmas. Vet Clin North Am Small Anim Pract 40:1157–1170. doi: 10.1016/j.cvsm.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Dieckmann SM, Hoelzle K, Dieckmann MP, Straube I, Hofmann-Lehmann R, Hoelzle LE. 2012. Occurrence of hemotrophic mycoplasmas in horses with correlation to hematological findings. Vet Microbiol 160:43–52. doi: 10.1016/j.vetmic.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 14.dos Santos AP, dos Santos RP, Biondo AW, Dora JM, Goldani LZ, de Oliveira ST, de Sa Guimaraes AM, Timenetsky J, de Morais HA, Gonzalez FH, Messick JB. 2008. Hemoplasma infection in HIV-positive patient, Brazil. Emerg Infect Dis 14:1922–1924. doi: 10.3201/eid1412.080964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steer JA, Tasker S, Barker EN, Jensen J, Mitchell J, Stocki T, Chalker VJ, Hamon M. 2011. A novel hemotropic Mycoplasma (hemoplasma) in a patient with hemolytic anemia and pyrexia. Clin Infect Dis 53:e147–. doi: 10.1093/cid/cir666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sykes JE, Lindsay LL, Maggi RG, Breitschwerdt EB. 2010. Human coinfection with Bartonella henselae and two hemotropic mycoplasma variants resembling Mycoplasma ovis. J Clin Microbiol 48:3782–3785. doi: 10.1128/JCM.01029-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reynolds CA, Lappin MR. 2007. “Candidatus Mycoplasma haemominutum” infections in 21 client-owned cats. J Am Anim Hosp Assoc 43:249–257. doi: 10.5326/0430249. [DOI] [PubMed] [Google Scholar]
- 18.Volokhov DV, Norris T, Rios C, Davidson MK, Messick JB, Gulland FM, Chizhikov VE. 2011. Novel hemotrophic mycoplasma identified in naturally infected California sea lions (Zalophus californianus). Vet Microbiol 149:262–268. doi: 10.1016/j.vetmic.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 19.Groebel K, Hoelzle K, Wittenbrink MM, Ziegler U, Hoelzle LE. 2009. Mycoplasma suis invades porcine erythrocytes. Infect Immun 77:576–584. doi: 10.1128/IAI.00773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabello J, Altet L, Napolitano C, Sastre N, Hidalgo E, Davila JA, Millan J. 2013. Survey of infectious agents in the endangered Darwin's fox (Lycalopex fulvipes): high prevalence and diversity of hemotrophic mycoplasmas. Vet Microbiol 167:448–454. doi: 10.1016/j.vetmic.2013.09.034. [DOI] [PubMed] [Google Scholar]
- 21.Iso T, Suzuki J, Sasaoka F, Sashida H, Watanabe Y, Fujihara M, Nagai K, Harasawa R. 2013. Hemotropic mycoplasma infection in wild black bears (Ursus thibetanus japonicus). Vet Microbiol 163:184–189. doi: 10.1016/j.vetmic.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Krengel A, Meli ML, Cattori V, Wachter B, Willi B, Thalwitzer S, Melzheimer J, Hofer H, Lutz H, Hofmann-Lehmann R. 2013. First evidence of hemoplasma infection in free-ranging Namibian cheetahs (Acinonyx jubatus). Vet Microbiol 162:972–976. doi: 10.1016/j.vetmic.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Hirata M, Tateno M, Sakuma M, Nakanishi N, Izawa M, Asari Y, Okamura M, Shimokawa Miyama T, Setoguchi A, Endo Y. 2012. An epidemiological survey of hemoplasma infection in Iriomote cats (Prionailurus bengalensis iriomotensis). J Vet Med Sci 74:1531–1537. doi: 10.1292/jvms.12-0094. [DOI] [PubMed] [Google Scholar]
- 24.Harasawa R, Orusa R, Giangaspero M. 2014. Molecular evidence for hemotropic mycoplasma infection in a Japanese badger (Meles meles anakuma) and a raccoon dog (Nyctereutes procyonoides viverrinus). J Wildl Dis 50:412–415. doi: 10.7589/2013-09-229. [DOI] [PubMed] [Google Scholar]
- 25.Sharifiyazdi H, Nazifi S, Shirzad Aski H, Shayegh H. 2014. Molecular characterization and phylogenetic analysis of the causative agent of hemoplasma infection in small Indian mongoose (Herpestes javanicus). Comp Immunol Microbiol Infect Dis 37:243–247. doi: 10.1016/j.cimid.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Pinard CL, Brightman AH, Yeary TJ, Everson TD, Cox LK, Chengappa MM, Davidson HJ. 2002. Normal conjunctival flora in the North American opossum (Didelphis virginiana) and raccoon (Procyon lotor). J Wildl Dis 38:851–855. doi: 10.7589/0090-3558-38.4.851. [DOI] [PubMed] [Google Scholar]
- 27.Frerichs WM, Holbrook AA. 1971. Haemobartonella procyoni sp. n. in the raccoon, Procyon lotor. J Parasitol 57:1309–1310. doi: 10.2307/3277988. [DOI] [PubMed] [Google Scholar]
- 28.Firrao G, Brown DR. 2013. International Committee on Systematics of Prokaryotes. Subcommittee on the taxonomy of Mollicutes: minutes of the meetings, July 15th and 19th 2012, Toulouse, France. Int J Syst Evol Microbiol 63:2361–2364. [DOI] [PubMed] [Google Scholar]
- 29.Volokhov DV, Simonyan V, Davidson MK, Chizhikov VE. 2012. RNA polymerase beta subunit (rpoB) gene and the 16S-23S rRNA intergenic transcribed spacer region (ITS) as complementary molecular markers in addition to the 16S rRNA gene for phylogenetic analysis and identification of the species of the family Mycoplasmataceae. Mol Phylogenet Evol 62:515–528. doi: 10.1016/j.ympev.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Brown DR, Whitcomb RF, Bradbury JM. 2007. Revised minimal standards for description of new species of the class Mollicutes (division Tenericutes). Int J Syst Evol Microbiol 57:2703–2719. doi: 10.1099/ijs.0.64722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray RG, Stackebrandt E. 1995. Taxonomic note: implementation of the provisional status Candidatus for incompletely described prokaryotes. Int J Syst Bacteriol 45:186–187. doi: 10.1099/00207713-45-1-186. [DOI] [PubMed] [Google Scholar]
- 32.Almy FS, Ladd SM, Sponenberg DP, Crisman MV, Messick JB. 2006. Mycoplasma haemolamae infection in a 4-day-old cria: support for in utero transmission by use of a polymerase chain reaction assay. Can Vet J 47:229–233. [PMC free article] [PubMed] [Google Scholar]
- 33.Dieckmann SM, Winkler M, Groebel K, Dieckmann MP, Hofmann-Lehmann R, Hoelzle K, Wittenbrink MM, Hoelzle LE. 2010. Haemotrophic Mycoplasma infection in horses. Vet Microbiol 145:351–353. doi: 10.1016/j.vetmic.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Hu Z, Yin J, Shen K, Kang W, Chen Q. 2009. Outbreaks of hemotrophic mycoplasma infections in China. Emerg Infect Dis 15:1139–1140. doi: 10.3201/eid1507.090174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson KA, do Nascimento NC, Bauer AE, Weng HY, Hammac GK, Messick JB. 2016. Detection of hemoplasma infection of goats by use of a quantitative polymerase chain reaction assay and risk factor analysis for infection. Am J Vet Res 77:881–888. [DOI] [PubMed] [Google Scholar]
- 36.Maggi RG, Chitwood MC, Kennedy-Stoskopf S, DePerno CS. 2013. Novel hemotropic Mycoplasma species in white-tailed deer (Odocoileus virginianus). Comp Immunol Microbiol Infect Dis 36:607–611. doi: 10.1016/j.cimid.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Criado-Fornelio A, Martinez-Marcos A, Buling-Sarana A, Barba-Carretero JC. 2003. Presence of Mycoplasma haemofelis, Mycoplasma haemominutum and piroplasmids in cats from southern Europe: a molecular study. Vet Microbiol 93:307–317. doi: 10.1016/S0378-1135(03)00044-0. [DOI] [PubMed] [Google Scholar]
- 38.Clarridge JE., III 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev 17:840–862. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franzén O, Hu JZ, Bao XL, Itzkowitz SH, Peter I, Bashir A. 2015. Improved OTU-picking using long-read 16S rRNA gene amplicon sequencing and generic hierarchical clustering. Microbiome 3:1–14. doi: 10.1186/s40168-014-0066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schloss PD. 2010. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput Biol 6:e1000844. doi: 10.1371/journal.pcbi.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersson B, Tully JG, Bolske G, Johansson KE. 2000. Updated phylogenetic description of the Mycoplasma hominis cluster (Weisburg et al. 1989) based on 16S rDNA sequences. Int J Syst Evol Microbiol 50:291–301. doi: 10.1099/00207713-50-1-291. [DOI] [PubMed] [Google Scholar]
- 42.Rosselló-Mora R, Amann R. 2001. The species concept for prokaryotes. FEMS Microbiol Rev 25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 43.Fox GE, Wisotzkey JD, Jurtshuk P Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol 42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 44.Sallen B, Rajoharison A, Desvarenne S, Quinn F, Mabilat C. 1996. Comparative analysis of 16S and 23S rRNA sequences of Listeria species. Int J Syst Bacteriol 46:669–674. doi: 10.1099/00207713-46-3-669. [DOI] [PubMed] [Google Scholar]
- 45.Willi B, Tasker S, Boretti FS, Doherr MG, Cattori V, Meli ML, Lobetti RG, Malik R, Reusch CE, Lutz H, Hofmann-Lehmann R. 2006. Phylogenetic analysis of “Candidatus Mycoplasma turicensis” isolates from pet cats in the United Kingdom, Australia, and South Africa, with analysis of risk factors for infection. J Clin Microbiol 44:4430–4435. doi: 10.1128/JCM.00987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willi B, Filoni C, Catao-Dias JL, Cattori V, Meli ML, Vargas A, Martinez F, Roelke ME, Ryser-Degiorgis MP, Leutenegger CM, Lutz H, Hofmann-Lehmann R. 2007. Worldwide occurrence of feline hemoplasma infections in wild felid species. J Clin Microbiol 45:1159–1166. doi: 10.1128/JCM.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tasker S, Helps CR, Day MJ, Harbour DA, Shaw SE, Harrus S, Baneth G, Lobetti RG, Malik R, Beaufils JP, Belford CR, Gruffydd-Jones TJ. 2003. Phylogenetic analysis of hemoplasma species: an international study. J Clin Microbiol 41:3877–3880. doi: 10.1128/JCM.41.8.3877-3880.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willi B, Boretti FS, Cattori V, Tasker S, Meli ML, Reusch C, Lutz H, Hofmann-Lehmann R. 2005. Identification, molecular characterization, and experimental transmission of a new hemoplasma isolate from a cat with hemolytic anemia in Switzerland. J Clin Microbiol 43:2581–2585. doi: 10.1128/JCM.43.6.2581-2585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lobetti RG, Tasker S. 2004. Diagnosis of feline haemoplasma infection using a real-time PCR assay. J S Afr Vet Assoc 75:94–99. [DOI] [PubMed] [Google Scholar]
- 50.Sykes JE. 2010. Feline hemotropic mycoplasmas. J Vet Emerg Crit Care (San Antonio) 20:62–69. doi: 10.1111/j.1476-4431.2009.00491.x. [DOI] [PubMed] [Google Scholar]
- 51.Duarte A, Marques V, Correia JH, Neto I, Braz BS, Rodrigues C, Martins T, Rosado R, Ferreira JP, Santos-Reis M, Tavares L. 2015. Molecular detection of haemotropic Mycoplasma species in urban and rural cats from Portugal. J Feline Med Surg 17:516–522. doi: 10.1177/1098612X14550172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poulin R. 2007. Evolutionary ecology of parasites, 2nd ed Princeton University Press, Princeton, NJ. [Google Scholar]
- 53.Budria A, Candolin U. 2013. How does human-induced environmental change influence host-parasite interactions? Parasitology 141:462–474. doi: 10.1017/S0031182013001881. [DOI] [PubMed] [Google Scholar]
- 54.Lafferty KD. 1997. Environmental parasitology: what can parasites tell us about human impacts on the environment? Parasitol Today 13:251–255. doi: 10.1016/S0169-4758(97)01072-7. [DOI] [PubMed] [Google Scholar]
- 55.Miller R, Kaneene JB, Fitzgerald SD, Schmitt SM. 2003. Evaluation of the influence of supplemental feeding of white-tailed deer (Odocoileus virginianus) on the prevalence of bovine tuberculosis in the Michigan wild deer population. J Wildl Dis 39:84–95. doi: 10.7589/0090-3558-39.1.84. [DOI] [PubMed] [Google Scholar]
- 56.Storm DJ, Samuel M, Rolley RE, Shelton P, Keuler NS, Richards BJ, Van Deelen TR. 2013. Deer density and disease prevalence influence transmission of chronic wasting disease in white-tailed deer. Ecosphere 4:1–14. [Google Scholar]
- 57.Evans KL, Gaston KJ, Sharp SP, McGowan A, Simeoni M, Hatchwell BJ. 2009. Effects of urbanisation on disease prevalence and age structure in blackbird Turdus merula populations. Oikos 118:774–782. doi: 10.1111/j.1600-0706.2008.17226.x. [DOI] [Google Scholar]
- 58.Geue D, Partecke J. 2008. Reduced parasite infestation in urban Eurasian blackbirds (Turdus merula): a factor favoring urbanization? Can J Zool 86:1419–1425. doi: 10.1139/Z08-129. [DOI] [Google Scholar]
- 59.Gregoire A, Faivre B, Heeb P, Cezilly F. 2002. A comparison of infestation patterns by Ixodes ticks in urban and rural populations of the common blackbird Turdus merula. Ibis 144:640–645. doi: 10.1046/j.1474-919X.2002.00102.x. [DOI] [Google Scholar]
- 60.Prange S, Gehrt S, Wiggers E. 2004. Influences of anthropogenic resources on raccoon (Procyon lotor) movements and spatial distribution. J Mammal 85:483–490. doi: 10.1644/BOS-121. [DOI] [Google Scholar]
- 61.Becker DJ, Hall RJ. 2014. Too much of a good thing: resource provisioning alters infectious disease dynamics in wildlife. Biol Lett 10:pii:20140309. doi: 10.1098/rsbl.2014.0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choisy M, Rohani P. 2006. Harvesting can increase severity of wildlife disease epidemics. Proc Biol Sci 273:2025–2034. doi: 10.1098/rspb.2006.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tagawa M, Takeuchi T, Fujisawa T, Konno Y, Yamamoto S, Matsumoto K, Yokoyama N, Inokuma H. 2012. A clinical case of severe anemia in a sheep coinfected with Mycoplasma ovis and “Candidatus Mycoplasma haemovis” in Hokkaido, Japan. J Vet Med Sci 74:99–102. doi: 10.1292/jvms.11-0296. [DOI] [PubMed] [Google Scholar]
- 64.de Bortoli CP, Andre MR, Seki MC, Pinto AA, Machado SDTZ, Machado RZ. 2012. Detection of Hemoplasma and Bartonella species and co-infection with retroviruses in cats subjected to a spaying/neutering program in Jaboticabal, SP, Brazil. Rev Bras Parasitol Vet 21:219–223. doi: 10.1590/S1984-29612012000300008. [DOI] [PubMed] [Google Scholar]
- 65.Hirsch BT, Prange S, Hauver SA, Gehrt SD. 2013. Raccoon social networks and the potential for disease transmission. PLoS One 8:e75830. doi: 10.1371/journal.pone.0075830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Valle SDF, Messick JB, Dos Santos AP, Kreutz LC, Duda NC, Machado G, Corbellini LG, Biondo AW, Gonzalez FH. 2014. Identification, occurrence and clinical findings of canine hemoplasmas in southern Brazil. Comp Immunol Microbiol Infect Dis 37:259–265. doi: 10.1016/j.cimid.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 67.Willi B, Boretti FS, Baumgartner C, Tasker S, Wenger B, Cattori V, Meli ML, Reusch CE, Lutz H, Hofmann-Lehmann R. 2006. Prevalence, risk factor analysis, and follow-up of infections caused by three feline hemoplasma species in cats in Switzerland. J Clin Microbiol 44:961–969. doi: 10.1128/JCM.44.3.961-969.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grazziotin AL, Santos AP, Guimaraes AM, Mohamed A, Cubas ZS, de Oliveira MJ, dos Santos LC, de Moraes W, Vieira RF, Donatti L, de Barros Filho IR, Biondo AW, Messick JB. 2011. Mycoplasma ovis in captive cervids: prevalence, molecular characterization and phylogeny. Vet Microbiol 152:415–419. doi: 10.1016/j.vetmic.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 69.Schoettle HT. A naturalist's guide to St. Simons Island. Watermarks Publisher, Darien, GA. [Google Scholar]
- 70.Thomas DH. 1978. The anthropology of St. Catherines Island. American Museum of Natural History, New York, NY. [Google Scholar]
- 71.Hwang J, Gottdenker NL. 2013. Bartonella species in raccoons and feral cats, Georgia, USA. Emerg Infect Dis 19:1167–1168. doi: 10.3201/eid1907.130010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Christen R. 2008. Identifications of pathogens–a bioinformatic point of view. Curr Opin Biotechnol 19:266–273. doi: 10.1016/j.copbio.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 73.Barker EN, Langton DA, Helps CR, Brown G, Malik R, Shaw SE, Tasker S. 2012. Haemoparasites of free-roaming dogs associated with several remote Aboriginal communities in Australia. BMC Vet Res 8:55. doi: 10.1186/1746-6148-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raimundo JM, Guimaraes A, Botelho CF, Peixoto MP, Pires MS, Machado CH, Santos HA, Massard CL, Andre MR, Machado RZ, Baldani CD. 2016. Hematological changes associated with hemoplasma infection in cats in Rio de Janeiro, Brazil. Rev Bras Parasitol Vet 25:441–449. doi: 10.1590/s1984-29612016086. [DOI] [PubMed] [Google Scholar]
- 75.Abd Rani PA, Irwin PJ, Coleman GT, Gatne M, Traub RJ. 2011. A survey of canine tick-borne diseases in India. Parasit Vectors 4:141. doi: 10.1186/1756-3305-4-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Volokhov DV, Kong H, George J, Anderson C, Chizhikov VE. 2008. Biological enrichment of Mycoplasma agents by cocultivation with permissive cell cultures. Appl Environ Microbiol 74:5383–5391. doi: 10.1128/AEM.00720-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hugenholtz P, Huber T. 2003. Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int J Syst Evol Microbiol 53:289–293. doi: 10.1099/ijs.0.02441-0. [DOI] [PubMed] [Google Scholar]
- 79.Wright ES, Yilmaz LS, Noguera DR. 2012. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78:717–725. doi: 10.1128/AEM.06516-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.R Development Core Team. 2010. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 82.Crawley MJ. 2013. The R book, 2nd ed John Wiley and Sons, Ltd., New York, NY. [Google Scholar]
- 83.Griffith DM, Veech JA, Marsh CJ. 2016. cooccur: probabilistic species co-occurrence analysis in R. J Stat Softw 69:17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.