Abstract
Hepatitis B virus (HBV) is an etiological agent of viral hepatitis, which may lead to cirrhosis, and hepatocellular carcinoma. Current treatment strategies have not shown promising effect to date but various complications such as, drug toxicity-resistance have been reported. Study on newly discovered compounds, with minimal side effects, as specific HBV inhibitors is a fundamental subject introducing new biologic drugs. Here, we aimed to, by prediction, estimate interactions of HBF-0259 as a non-toxic anti-HBV compound on inhibiting the HBV through either interaction with the viral entry or HBsAg secreting factors using In Silico procedure. Molecular docking was performed by Hex 8.0.0 software to predict the interaction energy (Etot) between HBF-0259 and known cellular factors involved in HBV entry and HBsAg secreting factors. Hex 8.0.0 also employed to create protein–protein complexes. These interactions were then used to analyze the binding site of HBF-0259 within the assumed receptors by MGLTools software. Finally, the amino acid sequences involved in this interaction were aligned for any conservancy. Here, we showed that HBF-0259 Etot with CypA (−545.41 kcal/mol) and SCCA1 (499.68 kcal/mol), involved in HBsAg secretion and HBV integration, respectively, was higher than other interactions. Furthermore, HBF-0259 predicted interaction energy was even higher than those of CypA inhibitors. In addition, we claim that preS1 and/or preS2 regions within HBsAg are not suitable targets for HBF-0259. HBF-0259 has higher interaction energy with CypA and SCCA1, even more than other known receptors, co-receptors, viral ligands, and secretory factors. HBF-0259 could be introduced as potent anti-viral compound in which CypA and or SCCA1, as previously shown, are involved.
Keywords: HBF-0259, HBV, Interaction, HBsAg secretion, Cyclophilin A, Molecular docking
Introduction
Hepatitis B Virus (HBV) is a causative agent of viral hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) with a relatively high prevalence all around the world [32]. As the untreated chronic infections may result in HCC, the treatment is crucial. The majority of current treatment strategies have shown to be helpless in complete removal of the pathogenic agent. Several FDA-approved common agents have shown no promising efficacy upon restricting HBV. Moreover, drug resistance infections and drug toxicities have been reported in long period applications as well as the cost burden [6].
A well-established mechanism in the treatment of viral pathogens is to block the binding of virus to the receptor(s) and consequently prevent the penetration [29]. HBsAg is a HBV product in chronic infections which plays an important role in pathogenesis of HBV and modulation of host immune responses [2]. HBV is translated into three major surface antigens including Large HBsAg (LHBsAg), Medium (MHBsAG) and Small (SHBsAg) according to the position of start codon within the open reading frame. Pre-S1 is a highly conserved domain within LHBsAg which plays a key role in recognizing and binding to the specific receptor molecules on the host cells receptors [19, 28]. In addition to preS1, preS2 domain of LHBS is involved in the interaction with known HBV co-receptors such as Fibronectin (FN) and Ferritin Light Chain (FTL). The viral ligands (preS1 and preS2) and cellular receptors and co-receptors such as Squamous Cell Carcinoma Antigen 1 (SCCA1) and FTL complex facilitate the penetration of viral particles into the cell [8, 41]. Over expression of SCCA1 is believed to increase the binding capability of virus to the host cells. However, FTL has a potential role for this approach [8, 18]. ASGPR and its co-receptor, FN, and NTCP are also other proposed HBV receptor/co-receptors, which could target by drug compounds [31, 40].
Novel specific receptor blockers could result in a noticeable decrease in the secretion of HBsAg and the pathogenicity of HBV. HBF-0259, (7-(2-Chloro-6-fluorophenyl)-5-(4-chlorophenyl)-4, 5, 6, 7-tetrahydro-tetrazolo [1, 5-a] pyrimidine), could act as an inhibitor of HBsAg secretion which is proved to be efficient in certain in vitro studies [4]. Moreover, the triazole derivates of this compound have similar anti-HBV effects [38, 39]. Anti-HBV potential of this non-toxic compound are hypothesized to be result of HBsAg inhibition through direct interaction with the viral or host molecules [4, 38, 42]. Cyclophilin A (CypA), Annexin II, and Ras-Associated Protein 7 (Rab7) are known cellular components involved in HBsAg secretion (Table 1). In order to have a precise perspective over the underlying molecular mechanism involved in HBsAg secretion inhibition, cellular components involved in this process should be analyzed separately and also in a network.
Table 1.
HBV ligands and their cognate receptors and co-receptors with PDB IDs
| Viral ligands (ref) | Receptors (ref)/HBsAg secreting factorsa (ref) | PDB ID (ref) |
|---|---|---|
| Pre-S1 [8, 31] | ASGPRCRD [41] and SCCA1 [8] | 1DV8 [17] and 2ZV6 [43] |
| Pre-S2 [3, 8] | FTL [8] and FNIII1 [41] | 2FG4 [35] and 2HA1 [34] |
| Pre-S1 [20] | NTCP [40] | UA |
| HBsAg [13] | Annexin II [13] | 1W7B (UA) |
| HBsAg [10] | Rab7 [10] | 1YHN [36] |
| HBsAg [26] | Cyclophilin A [26] | 1BCK [11] |
ASGPRCRD Asialoglycoprotein carbohydrate recognition domain, SCCA1 human Squamous cell carcinoma antigen 1, FTL Ferritin light chain, FNIII 1 Fibronectin III class 1, ref Reference, PDB Protein DataBank, UA unavailable
aHBsAg secretory factors are Annexin II, Rab7, and Cyclophilin A
In this study, in order to predict the mechanism(s) of action by which HBF-0259 decreases HBsAg secretion, we predict the interaction of HBF-0259 with suggested HBV receptors, preS regions of HBsAg (preS121–47 and preS21–11) and cellular HBsAg secreting factors by computational approaches (Table 1).
Materials and methods
Preparation of HBV cellular receptors and co-receptors, HBF-0259, CypA and the inhibitors
ACD/ChemSketch software (ACDLABS 12.0, Toronto, Canada) was used to design and refine the tautomer and 3D structure of HBF-0259. The structures of four CypA inhibitors, Alisporivir, NIM811, SCY635, and Sanglifehrin A were obtained in SDF format from ChemSpider database [23]. As shown in Table 1, the sequences and validated structures of CypA, Annexin II, and Rab7 as well as relative HBV receptors were obtained from protein databank (PDB) [1]. The crystallographic structure of molecules were cleared for the presence of water atoms and other unnecessary extra chains using UCSF Chimera 1.8.1 [25] and the results were saved in PDB format as separate files. The amino acid sequences of PreS121–47 and PreS21–11 domains were obtained from UniProtKB Swiss-Prot database [33] (UniProt Entry: P03141). These sequences were then used for De Novo prediction of 3D structures in Pep-Fold server [16]. The amino acid sequence of NTCP was also obtained from UniProtKB Swiss-Prot database (UniProt Entry: Q12908) and used in for homology based 3D structure prediction RaptorX server [24]. The predicted structures were finally validated by Ramachandran plot in RAMPAGE server (Fig. 1) [14].
Fig. 1.
Ramachandran plot validation of predicted structures; a 100 % NTCP84-165 residues are in favoured (97.5 %) and allowed (2.5 %) zones. b preS21-11 residues were 100 % present in favoured area, c preS121-47 residues with 95.8 % occurrences in favor
Docking software and parameters
Standalone software Hex 8.0.0 (Team Orpailleur, Nancy, France) was used to dock HBF0259 ligand with predicted HBV receptors. We also used Hex 8.0.0 in docking of HBF-0259 together with any of Alisporivir, NIM811, SCY635 and Sanglifehrin A compounds in correlation with secretory molecules. Since ASGPR/FNIII1 and SCCA1/FTL complexes are involved in binding to HBV [8, 41], the HBF-0259 binding energy of these complexes were evaluated. Hex 8.0.0 was also used to form the protein–protein complexes in the presence or absence of HBF0259 and with PreS1 and PreS2 binding domains. Correlation Type and FFT Mode software parameters were changed to Shape + Electro + DARS and 5D, respectively. CypA, Annexin II, and Rab7 molecules were considered as receptors, while HBF-0259 and CypA inhibitors were investigated as ligands.
Positioning the binding sites of HBF0259 HBV receptors and secreting factors
The results of each receptor, co-receptor, and viral ligand docking with HBF-0259 were analyzed by MGLTools 1.5.6 software (The Scripps Research Institute, California, USA) for determining amino acids in HBF-0259 binding site. The interaction analysis between HBF-0259 and possible HBV receptors in the setting of MGLTools were done according to VDW scaling factor 1 Å and the amino acid position(s) of all binding sites were determined and investigated in UniProt (30) regarding the molecular domains. The binding sites for HBF0259 and CypA to Cyclophilin A inhibitors were evaluated in the same way. Catalytic Site Atlas (CSA) 2.0 and Allosteric ASBench (ASD/ASBench) databases were also mined for anycatalytic sites, esteric and orthosteric residues within macromolecules [7, 9].
Alignment
Amino acids resulted from binding site analyses were used for ClustalW alignment by CLS Sequence Viewer version 7.5 (QIAGEN Company, USA) software. Briefly, the amino acid sequences of the ligand binding sites were imported to the CLS software manually. Graphic maps of aligned amino acids were drawn using the ClustalW tool and analyzed for the presence of any consensus sequence.
Results
Assessment of HBF-0259 interactions with HBV receptors
As shown in Table 2, the HBF-0259 compound has the greatest interaction energy (total energy) to SCCA1 (−499.68 kcal/mol) in comparison to all other receptors and co-receptors. The HBF-0259 Etot to PreS121–47 (−130.11 kcal/mol) and PreS21–11 (−288.22 kcal/mol) were also lower comparing to other predicted receptors. These results indicate that HBF-0259 interacts more strongly with SCCA1 and could be introduced as a more effective receptor blocker with probable negative influence on recognition, binding and penetration of HBV to the host cell with therapeutic benefits. No hits were found in CSA and ASD/ASBench databases for any of these molecules.
Table 2.
ClustalW alignment of HBF-0259 binding sites and docking total energy
| HBV receptors, co-receptors and ligands | Amino acid residues within interaction site of HBF-0259 | Total Energy (Kcal/mol) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SCCA1 | – | – | – | S | L | G | L | F | V | H | Q | F | V | −499.68 |
| FNIII1 | – | – | – | W | R | P | R | W | K | Y | – | – | – | −352.89 |
| preS2 | – | – | – | – | M | Q | S | T | H | – | – | – | – | −288.22 |
| NTCP84–165 | F | I | L | V | I | G | T | A | I | – | – | – | – | −260.26 |
| FTL | – | – | – | – | Q | K | K | A | K | – | – | – | – | −215.78 |
| ASGPRCRD | – | – | – | – | S | G | N | V | T | Y | R | – | – | −130.72 |
| preS1 | – | – | – | P | S | N | N | W | P | – | – | – | – | −130.11 |
Among different HBsAg secreting molecules, HBF-0259 interact CypA with higher interaction energy
The interaction energy of HBF-0259 to CypA (−545.41 kcal/mol) was obviously higher than Annexin II (−355.10) and Rab7 (−381.99 kcal/mol). We also compared interaction energy of four other CypA inhibitors with HBF-0259 (Table 3). The energy of Alisporivir, NIM811, SCY635, and Sanglifehrin A were very close to each other with average lower than that of HBF-0259 (−436.12 ± 3.75 kcal/mol). For HbsAg secreting molecules no entries were found in CSA and ASD/ASBench databases, but Rab7 Leu67 in its catalytic site.
Table 3.
Interaction energy of HBF-0259 to HBsAg secreting molecules and its energy to CypA in comparison with other four CypA inhibitors
| Molecule | HBF-0259 Interaction Energy (Kcal/mol) |
|---|---|
| CypA | −545.41 |
| Annexin II | −355.10 |
| Rab7 | −381.99 |
| CypA inhibitors | Interaction Energy to CypA (Kcal/mol) |
|---|---|
| HBF-0259 | −545.41 |
| Alisporivir | −439.93 |
| NIM811 | −438.75 |
| SCY635 | −432.40 |
| Sanglifehrin A | −433.41 |
HBF-0259 interaction sites within HBV receptors, co-receptors, and viral ligands
The HBF0259 receptor binding sites of interest were studied for the type of amino acids. Amino acids involved in interaction site of HBF-0259 were analyzed for conservancies among target molecules including, SCCA1, FNIII1, preS21–11, NTCP84–165, FTL, ASGPRCRD, preS121–47. The results showed no conserved sequences in any of mentioned receptors (Table 2).
The similarity of HBF-0259 binding site to CypA in comparison with Alisporivir and NIM811
Among three cellular secreting molecules, Annexin II, Rab7, and CypA, HBF-0259 had the highest Etot for CypA. Therefore, CypA was selected to study the HBF-0259 binding site in comparison to the other four CypA inhibitors including Alisporivir, NIM811, SCY635 and Sanglifehrin A. As shown in Table 4, HBF-0259 have common binding site with Alisporivir in amino acids R, F, and with NIM811 in R, H, and G. Sanglifehrin A and SCY635 had no similar sequences neither with each other nor other inhibitors, but Alisporivir and NIM811 had five analogous amino acids in their binding sites including, H, R, N, G, and T.
Table 4.
Interaction site of HBF-0259 and four known CypA inhibitors within CypA
| Compounds | AAs presented in interaction site within CypA | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HBF-0259 | R | F | M | H | F1 | K | G | |||||||||||
| Alisporivir | Q | H | R | I | F | N | G | T | A | N | G | Q | L | H | R | N | ||
| NIM811 | H | R | T | R | H | N | G | T | K | A | N | G | ||||||
| Sanglifehrin A | P | P | T | A | K | E | R | G | S | R | ||||||||
| SCY635 | F | R | R | T | G | E | K | D1 | Q | L | E | |||||||
HBF-0259 binds to HBV Receptor/Co-receptor Complexes with lower energies than receptors or co-receptors alone
The AAs on the HBF-0259 binding site within SCCA1/FTL complex were S, K, L, E, K, and T, which interact with −173.05 kcal/mol. The HBF-0259 binding site within NTCP84–165 was similar to preS121–47 and the AAs contained in this contact were S, F, Q, V, W, K, G, and S with 209.65 kcal/mol interaction energy. The AAs in HBF-0259 binding site for ASGPR/FNIII1 complex were K, P, G, T, E, P, Q, and Y157 which could makes −291.60 kcal/mol.
None of the mentioned amino acids were individually identical to receptors and co-receptors. The binding energy of these complexes showed a dramatic decrease comparing to docking results of HBF-0259 with each receptor alone.
HBF-0259 could reduce affinity of the receptor-co-receptor interactions which is necessary for HBV cell entry
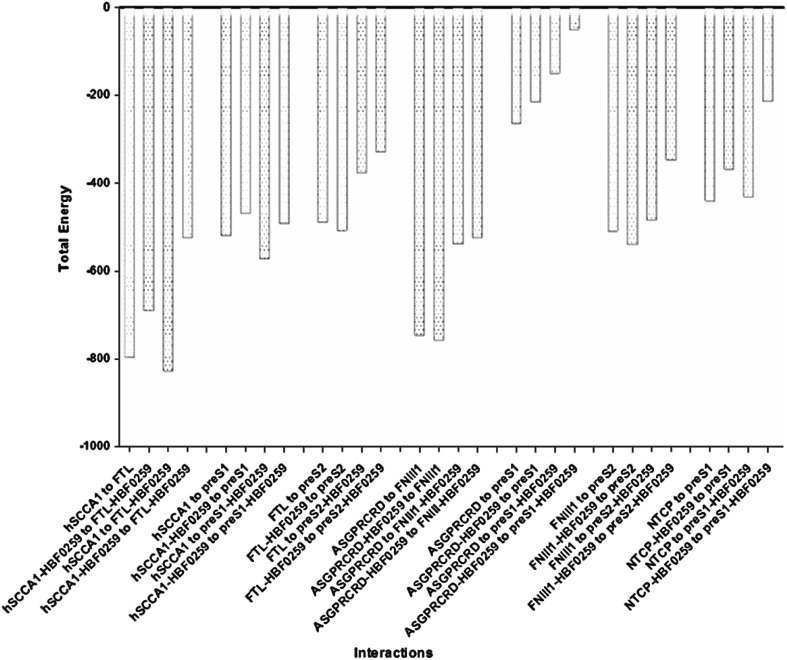
Different potential states of HBF-0259 binding to SCCA1/FTL and ASGPRCRD/FNII1 protein complexes as receptors for viral ligands were studied. Etot of receptors and co-receptors, as well as ligand interactions in the presence and/or absence of HBF-0259 were obtained (Table 5). In the presence of HBF-0259 the Interaction energy of FTL and PreS121–47 to SCCA1 is lower. Similarly, when PreS121–47 and/or FTL are bound to HBF-0259 the total energy is reduced. The interaction energy of pre-S121–47 to ASGPRCRD in the presence of HBF-0259 was reduced either (Table 5, Fig. 2). With existence of HBF-0259, the PreS121–47 interaction energy to NTCP84–165 was clearly lower.
Table 5.
Total energy (Etot)a of receptor, co-receptor and ligand interactions in the presence and or absence of HBF-0259
| Pre-S1 | Pre-S1-HBF-0259 | Pre-S2 | Pre-S2/HBF-0259 | FTL | FTL/HBF-0259 | FNIII1 | FNIII1/HBF-0259 | |
|---|---|---|---|---|---|---|---|---|
| SCCA1 | −519.28 | −571.27 | NP | NP | −795.72 | −826.65 | NP | NP |
| SCCA1/HBF-0259 | −469.01 | −491.54 | NP | NP | −690.32 | −523.80 | NP | NP |
| FTL | NP | NP | −488.28 | −376.38 | NP | NP | NP | NP |
| FTL/HBF-0259 | NP | NP | −507.62 | −329.18 | NP | NP | NP | NP |
| ASGPRCRD | −263.58 | −149.60 | NP | NP | NP | NP | −745.57 | −537.86 |
| ASGPRCRD/HBF-0259 | −214.15 | −49.93 | NP | NP | NP | NP | −756.92 | −523.80 |
| FNIII1 | NP | NP | −509.19 | −484.10 | NP | NP | NP | NP |
| FNIII1/HBF-0259 | NP | NP | −539.81 | −346.72 | NP | NP | NP | NP |
| NTCP84–165 | −439.82 | −431.13 | NP | NP | NP | NP | NP | NP |
| NTCP-HBF0259 | −368.16 | −213.44 | NP | NP | NP | NP | NP | NP |
The slashes (/) show the complex. For example SCCA1/HBF-0259 implicate that SCCA1 is bound to HBF-0259
aHex 8.0.0 Total Energy (Kcal/mol)
NP not possible
Fig. 2.
Viral ligands interaction energy variation to their cognates receptors and co-receptors in the both present and absent of HBF-0259
Discussion
Current treatment strategies for HBV, as the most important causative agent of viral hepatitis, cirrhosis, and HCC have not meet in complete removal of the virus. Recent studies on virus cell counterparts and HBsAg secretory factors could make us capable to get advantages. To achieve this goal, we performed molecular docking and computational simulation for predicting mechanism(s) of action of this recently developed lead-like compound, HBF-0259. Among known cellular receptors involved in virus integration, SCCA1 and its cellular cognate, FTL, are reported to be overexpressed in HBV infection. Moreover, the interaction of these receptors with viral ligands PreS1 and PreS2 facilitates virus entry [8, 37]. HBF-0259 was presumed to have an inhibitory effect over the virus entry by blocking SCCA1 and FTL receptor and co-receptor. For this purpose, molecular docking was performed and the interaction energy of each receptor and co-receptor in conjunction with HBF-0259 was calculated.
The interaction energy between HBF-0259 and SCCA1 was high (−499.68 kcal/mol) among all other cellular receptors. The interaction energy of HBF-0259 to PreS121–47 and PreS21–11 were lower comparing to other predicted receptors. HBF-0259 obviously interacts more strongly with SCCA1. The powerful interaction of a ligand to a receptor is compulsory in designing effective receptor blockers. The common binding site in SCCA1 for HBF-0259 and FTL (in SCCA1/FTL complex) could explain energy reduction from −499.68 to −173.05 kcal/mol (Table 3; Fig. 2). On the other hand, according to the lower interaction energy of HBF-0259 to PreS121–47 and FTL, also with SCCA1 higher affinity (in FTL/SCCA1 complex), PreS121–47 and FTL are probably not HBF-0259 targets. Regarding other HBF-0259 binding sites, the interaction energy of HBF-0259 was lower for FNIII1 than ASGPRCRD and FTL. Interaction energy of FNIII1-HBF-0259 complex to ASGPRCRD shows significant decrease in comparison to the state of HBF-0259 absence (Table 2). The docking results of HBF-0259 to NTCP84–165 [12] also showed minimal interaction energy. Reduction of this interaction energy in the presence of HBF-0259 might be a result of the same binding sites within the receptors. Therefore, HBF-0259 could be introduced as an effective receptor blocker with probable negative influence on recognition, binding and penetration of HBV to the host cell with therapeutic benefits by blocking SCCA1.
The HBF-0259 mechanism of effect on HBsAg secretion inhibition is still unknown [4, 42]. For this, we performed docking on known cellular factors involved in HBsAg and HBV particle secretion including CypA, Annexin II and Rab7 [10, 13, 26]. Our results showed that the interaction energy of HBF-0259 to CypA (−545.41 kcal/mol) was obviously higher than Annexin II and Rab7 (Table 3). CypA is a peptidyl- proline cis to trans isomerase in the cells and is believed to have a role in the pathogenesis of some diseases [5, 21] which make it a potent molecule for therapeutic approaches [22]. Tian et al. [30] demonstrated the synergic role of HBV in increasing cellular CypA and its action in HBsAg secretion. As mentioned previously [21], CypA has a key role in life cycle and pathogenesis of several viruses, such as Human Immunodeficiency virus type 1(HIV-1), Hepatitis C virus (HCV), and HBV. It could be suppressed by CypA inhibitors (Alisporivir, NIM811), which their anti-HCV activities studied previously [15, 27]. According to the comparable interaction between these inhibitors and HBF-0259 to CypA (in both positions and interaction energies, Tables 3 and 4), HBF-0259 could be introduced as a new candidate for further investigation in antiviral therapies.
CypA and SCCA1 molecules probably are targeted by HBF-0259, followed by consequent inhibition of HBsAg. Studies with animal models or siRNA interfering and in vitro experiments with live viruses could be so useful for investigating HBF-0259 mechanism of action more deeply.
Acknowledgments
This work has supported by the School of Medicine, Golestan University of Medical Sciences, Gorgan, Iran.
Contributor Information
Alireza Mohebbi, Phone: 00989354674593.
Saeed Mohammadi, Phone: 00989155087104.
Ali Memarian, Phone: 00981732422644, Email: alimemarian@goums.ac.ir.
References
- 1.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertoletti A, Gehring AJ. The immune response during hepatitis B virus infection. J Gen Virol. 2006;87(Pt 6):1439–1449. doi: 10.1099/vir.0.81920-0. [DOI] [PubMed] [Google Scholar]
- 3.Budkowska A, Bedossa P, Groh F, Louise A, Pillot J. Fibronectin of human liver sinusoids binds hepatitis B virus: identification by an anti-idiotypic antibody bearing the internal image of the pre-S2 domain. J Virol. 1995;69(2):840–848. doi: 10.1128/jvi.69.2.840-848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dougherty AM, Guo H, Westby G, Liu Y, Simsek E, Guo JT, et al. A substituted tetrahydro-tetrazolo-pyrimidine is a specific and novel inhibitor of hepatitis B virus surface antigen secretion. Antimicrob Agents Chemother. 2007;51(12):4427–4437. doi: 10.1128/AAC.00541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer G, Bang H, Mech C. Determination of enzymatic catalysis for the cis-trans-isomerization of peptide binding in proline-containing peptides. Biomed Biochim Acta. 1984;43(10):1101–1111. [PubMed] [Google Scholar]
- 6.Fletcher SP, Delaney WE. New therapeutic targets and drugs for the treatment of chronic hepatitis B. Semin Liver Dis. 2013;33(2):130–137. doi: 10.1055/s-0033-1345713. [DOI] [PubMed] [Google Scholar]
- 7.Furnham N, Holliday GL, de Beer TA, Jacobsen JO, Pearson WR, Thornton JM. The Catalytic Site Atlas 2.0: cataloging catalytic sites and residues identified in enzymes. Nucleic Acids Res. 2014;42(Database issue):D485–D489. doi: 10.1093/nar/gkt1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao Z, Zheng L, Kluwe L, Huang W. Ferritin light chain and squamous cell carcinoma antigen 1 are coreceptors for cellular attachment and entry of hepatitis B virus. Int J Nanomed. 2012;7:827–834. doi: 10.2147/IJN.S27803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Z, Mou L, Shen Q, Lu S, Li C, Liu X, et al. ASD v2.0: updated content and novel features focusing on allosteric regulation. Nucleic Acids Research. 2014;42(Database issue):D510–D516. doi: 10.1093/nar/gkt1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue J, Krueger EW, Chen J, Cao H, Ninomiya M, McNiven MA. HBV secretion is regulated through the activation of endocytic and autophagic compartments mediated by Rab7 stimulation. J Cell Sci. 2015;128(9):1696–1706. doi: 10.1242/jcs.158097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kallen J, Mikol V, Taylor P, Walkinshaw MD. X-ray structures and analysis of 11 cyclosporin derivatives complexed with cyclophilin A. J Mol Biol. 1998;283(2):435–449. doi: 10.1006/jmbi.1998.2108. [DOI] [PubMed] [Google Scholar]
- 12.Konig A, Doring B, Mohr C, Geipel A, Geyer J, Glebe D. Kinetics of the bile acid transporter and hepatitis B virus receptor Na+/taurocholate cotransporting polypeptide (NTCP) in hepatocytes. J Hepatol. 2014;61(4):867–875. doi: 10.1016/j.jhep.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Gao P. Modulation of hepatitis B surface antigen secretion by annexin II expressed in hepatitis B virusproducing hepatoma cells. Mol Med Rep. 2014;10(6):3113–3117. doi: 10.3892/mmr.2014.2602. [DOI] [PubMed] [Google Scholar]
- 14.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, et al. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 15.Ma S, Boerner JE, TiongYip C, Weidmann B, Ryder NS, Cooreman MP, et al. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob Agents Chemother. 2006;50(9):2976–2982. doi: 10.1128/AAC.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maupetit J, Derreumaux P, Tuffery P. PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res. 2009;37(Web Server issue):W498–W503. doi: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier M, Bider MD, Malashkevich VN, Spiess M, Burkhard P. Crystal structure of the carbohydrate recognition domain of the H1 subunit of the asialoglycoprotein receptor. J Mol Biol. 2000;300(4):857–865. doi: 10.1006/jmbi.2000.3853. [DOI] [PubMed] [Google Scholar]
- 18.Moore PL, Ong S, Harrison TJ. Squamous cell carcinoma antigen 1-mediated binding of hepatitis B virus to hepatocytes does not involve the hepatic serpin clearance system. J Biol Chem. 2003;278(47):46709–46717. doi: 10.1074/jbc.M302842200. [DOI] [PubMed] [Google Scholar]
- 19.Neurath AR, Kent SB, Strick N, Parker K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell. 1986;46(3):429–436. doi: 10.1016/0092-8674(86)90663-X. [DOI] [PubMed] [Google Scholar]
- 20.Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146(4):1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 21.Nigro P, Pompilio G, Capogrossi MC. Cyclophilin A: a key player for human disease. Cell Death Dis. 2013;4:e888. doi: 10.1038/cddis.2013.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obchoei S, Wongkhan S, Wongkham C, Li M, Yao Q, Chen C. Cyclophilin A: potential functions and therapeutic target for human cancer. Med Sci Monit Int Med J Exp Clin Res. 2009;15(11):RA221–RA232. [PubMed] [Google Scholar]
- 23.Pence HE, Williams A. ChemSpider: an online chemical information resource. J Chem Educ. 2010;87(11):1123–1124. doi: 10.1021/ed100697w. [DOI] [Google Scholar]
- 24.Peng J, Xu J. RaptorX: exploiting structure information for protein alignment by statistical inference. Proteins. 2011;79(Suppl 10):161–171. doi: 10.1002/prot.23175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Phillips S, Chokshi S, Chatterji U, Riva A, Bobardt M, Williams R, et al. Alisporivir inhibition of hepatocyte cyclophilins reduces HBV replication and hepatitis B surface antigen production. Gastroenterology. 2015;148(2):403–414. doi: 10.1053/j.gastro.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quarato G, D’Aprile A, Gavillet B, Vuagniaux G, Moradpour D, Capitanio N, et al. The cyclophilin inhibitor alisporivir prevents hepatitis C virus-mediated mitochondrial dysfunction. Hepatology. 2012;55(5):1333–1343. doi: 10.1002/hep.25514. [DOI] [PubMed] [Google Scholar]
- 28.Schadler S, Hildt E. HBV life cycle: entry and morphogenesis. Viruses. 2009;1(2):185–209. doi: 10.3390/v1020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teissier E, Penin F, Pecheur EI. Targeting cell entry of enveloped viruses as an antiviral strategy. Molecules (Basel, Switzerland) 2011;16(1):221–250. doi: 10.3390/molecules16010221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian X, Zhao C, Zhu H, She W, Zhang J, Liu J, et al. Hepatitis B virus (HBV) surface antigen interacts with and promotes cyclophilin a secretion: possible link to pathogenesis of HBV infection. J Virol. 2010;84(7):3373–3381. doi: 10.1128/JVI.02555-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Treichel U, zum Buschenfelde KHM, Stockert RJ, Poralla T, Gerken G. The asialoglycoprotein receptor mediates hepatic binding and uptake of natural hepatitis B virus particles derived from viraemic carriers. J Gen Virol. 1994;75(Pt 11):3021–3029. doi: 10.1099/0022-1317-75-11-3021. [DOI] [PubMed] [Google Scholar]
- 32.Trepo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384(9959):2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 33.UniProt C. The Universal Protein Resource (UniProt) 2009. Nucleic acids research. 2009;37(Database issue):D169–74. doi:10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed]
- 34.Vakonakis I, Staunton D, Rooney LM, Campbell ID. Interdomain association in fibronectin: insight into cryptic sites and fibrillogenesis. EMBO J. 2007;26(10):2575–2583. doi: 10.1038/sj.emboj.7601694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Li C, Ellenburg M, Soistman E, Ruble J, Wright B, et al. Structure of human ferritin L chain. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 7):800–806. doi: 10.1107/S0907444906018294. [DOI] [PubMed] [Google Scholar]
- 36.Wu M, Wang T, Loh E, Hong W, Song H. Structural basis for recruitment of RILP by small GTPase Rab7. EMBO J. 2005;24(8):1491–1501. doi: 10.1038/sj.emboj.7600643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia HB, Chen ZY, Chen XG. Overexpression of hepatitis B virus-binding protein, squamous cell carcinoma antigen 1, extends retention of hepatitis B virus in mouse liver. Acta Biochim Biophys Sin. 2006;38(7):484–491. doi: 10.1111/j.1745-7270.2006.00190.x. [DOI] [PubMed] [Google Scholar]
- 38.Xu YB, Yang L, Wang GF, Tong XK, Wang YJ, Yu Y, et al. Benzimidazole derivative, BM601, a novel inhibitor of hepatitis B virus and HBsAg secretion. Antivir Res. 2014;107:6–15. doi: 10.1016/j.antiviral.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Xun YH, Zhang YJ, Pan QC, Mao RC, Qin YL, Liu HY, et al. Metformin inhibits hepatitis B virus protein production and replication in human hepatoma cells. J Viral Hepat. 2014;21(8):597–603. doi: 10.1111/jvh.12187. [DOI] [PubMed] [Google Scholar]
- 40.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Wang F, Tian L, Su J, Zhu X, Lin L, et al. Fibronectin and asialoglyprotein receptor mediate hepatitis B surface antigen binding to the cell surface. Arch Virol. 2010;155(6):881–888. doi: 10.1007/s00705-010-0657-5. [DOI] [PubMed] [Google Scholar]
- 42.Yu W, Goddard C, Clearfield E, Mills C, Xiao T, Guo H, et al. Design, synthesis, and biological evaluation of triazolo-pyrimidine derivatives as novel inhibitors of hepatitis B virus surface antigen (HBsAg) secretion. J Med Chem. 2011;54(16):5660–5670. doi: 10.1021/jm200696v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng B, Matoba Y, Kumagai T, Katagiri C, Hibino T, Sugiyama M. Crystal structure of SCCA1 and insight about the interaction with JNK1. Biochem Biophys Res Commun. 2009;380(1):143–147. doi: 10.1016/j.bbrc.2009.01.057. [DOI] [PubMed] [Google Scholar]