SUMMARY
Spt5 is an essential and conserved factor that functions in transcription and co-transcriptional processes. However, many aspects of the requirement for Spt5 in transcription are poorly understood. We have analyzed the consequences of Spt5 depletion in Schizosaccharomyces pombe, using four genome-wide approaches. Our results demonstrate that Spt5 is crucial for a normal rate of RNA synthesis and distribution of RNAPII over transcription units. In the absence of Spt5, RNAPII localization changes dramatically, with reduced levels and a relative accumulation over the first ~500 bp, suggesting that Spt5 is required for transcription past a barrier. Spt5 depletion also results in widespread antisense transcription initiating within this barrier region. Deletions of this region alter the distribution of RNAPII on the sense strand, suggesting that the barrier observed after Spt5 depletion is normally a site at which Spt5 stimulates elongation. Our results reveal a global requirement for Spt5 in transcription elongation.
Keywords: Spt5, transcription elongation, antisense transcription
eTOC Blurb
Shetty et al. show that Spt5, a transcription factor conserved in all domains of life, globally regulates transcription by RNA polymerase II in vivo. Spt5 controls the level and rate of RNAPII transcription, its ability to elongate past a barrier, and the synthesis of distinct classes of antisense RNAs.

INTRODUCTION
During transcription elongation, a large number of factors dynamically associate with RNA polymerase II to facilitate transcription on a chromatin template, as well as to coordinate co-transcriptional processes (Hsin and Manley, 2012). One widely-studied and essential factor that is involved in these processes is Spt5, the only transcription factor known to be conserved in all three domains of life (Hartzog and Fu, 2013; Werner, 2012). In bacteria, the Spt5 homologue, NusG, functions as a monomer (Mooney et al., 2009), while in archaea and eukaryotes, Spt5 heterodimerizes with Spt4, a small non-essential protein, in a complex also known as DSIF (DRB-sensitivity inducing factor) (Hartzog et al., 1998; Hirtreiter et al., 2010; Schwer et al., 2009; Wada et al., 1998).
In eukaryotes, Spt5 is an integral and essential part of the RNA polymerase II (RNAPII) elongation complex. Spt5 interacts directly with both archaeal RNAP and eukaryotic RNAPII via its NusG N-terminal (NGN) and Kyprides, Ouzounis, Woese (KOW) domains (Hirtreiter et al., 2010; Martinez-Rucobo et al., 2011; Viktorovskaya et al., 2011; Yamaguchi et al., 1999) and co-localizes with elongating RNAPII across eukaryotic genomes (Mayer et al., 2010; Rahl et al., 2010). Spt5 also interacts directly with RNAPI and is likely required for rRNA synthesis (Viktorovskaya et al., 2011). Structural studies have determined that Spt5 binds over the RNAP clamp domain above the nucleic acid cleft in a manner that likely stabilizes the transcription elongation complex, enhancing its processivity (Hirtreiter et al., 2010; Klein et al., 2011; Martinez-Rucobo et al., 2011). Biochemical results have shown that the NGN domain of Spt5 also directly interacts with both the nascent RNA and the noncoding strand of the DNA template during transcription (Blythe et al., 2016; Crickard et al., 2016; Meyer et al., 2015). Interestingly, Spt5 mutants that impair interaction with the noncoding strand of the template increase arrest by RNAPII in vitro, suggesting an important function for this interaction (Crickard et al., 2016).
In addition to interactions with RNAPII, Spt5 physically recruits several factors to the elongating transcription complex, a function often dependent upon the Spt5 carboxy-terminal region (CTR; Fig. 1A), a sequence of tandem repeats of amino acids that are phosphorylated in a manner similar to the CTD of RNAPII (Hartzog and Fu, 2013). The Spt5 CTR in its unphosphorylated state aids in recruiting the mRNA capping enzyme (Doamekpor et al., 2014; Doamekpor et al., 2015; Schneider et al., 2010; Wen and Shatkin, 1999). In contrast, the phosphorylated Spt5 CTR is required for recruitment of the Paf1 complex, which plays an important role in elongation by RNAPII (Liu et al., 2009; Mbogning et al., 2013; Wier et al., 2013; Zhou et al., 2009). Furthermore, Spt5 helps to recruit the 3′ end processing factors for mRNA and snoRNA transcripts (Mayer et al., 2012; Stadelmayer et al., 2014; Yamamoto et al., 2014), and the histone deacetylase complex, Rpd3S (Drouin et al., 2010).
Figure 1.
Depletion of Spt5 using a degron allele. (A) The S. pombe spt5+ gene, diagramming the regions that encode the NGN, KOW, and CTR domains, and the system for Spt5 depletion upon addition of both thiamine (to repress transcription) and auxin (to induce protein degradation). (B) The Western blot showing the depletion of Spt5-V5-AID. The blot was probed with anti-V5 antisera. S. pombe Act1 served as a loading control. The labels 0 and 4.5 indicate the Spt5 degron strain before and 4.5 hours after the addition of thiamine and auxin respectively. The depletion condition shows the mean and standard deviation of Spt5 protein levels for three biological replicates. (C) A scatterplot comparing the spike-in normalized ChIP-seq level of Spt5 in non-depleted and depleted cells. (D) A metagene plot showing the spike-in normalized ChIP-seq levels of Spt5 over 4294 expressed genes. (E) An example of Spt5 ChIP-seq at a single gene, showing the levels of Spt5 before and after depletion at the plb1+ gene.
Genetic analysis in S. cerevisiae also provides strong support for a broad role for Spt5 during transcription. The spt5-242 mutation is suppressed by mutations in several relevant genes, including those encoding RNAPII subunits, the Paf1 complex, the H3K36 methyltransferase Set2, and members of the Rpd3S complex (Hartzog et al., 1998; Quan and Hartzog, 2010). In contrast, other spt5 alleles confer double-mutant lethality with mutations in genes encoding Paf1 complex members (Squazzo et al., 2002) as well as other transcription factors (Lindstrom and Hartzog, 2001). Mutations that delete or alter phosphorylation of the Spt5 CTR also cause multiple phenotypes, suggesting interactions with RNAPII and histone modification enzymes (Mbogning et al., 2015; Sanso et al., 2012). This combination of biochemical and genetic data strongly support the idea that Spt5 plays a central role in mediating interactions between elongating RNAPII and other chromatin regulatory proteins.
While Spt5 has been extensively studied, surprisingly little has been done to test its role as a positive transcription elongation factor genome-wide. Current evidence for a role in elongation comes from in vitro studies that showed that Spt5 reduces pausing under nucleotide-limiting conditions (Guo et al., 2000; Wada et al., 1998; Zhu et al., 2007) and in vivo studies that showed elongation defects at candidate loci when Spt4 or Spt5 is depleted by mutation or knock-down (Diamant et al., 2016a; Kramer et al., 2016; Liu et al., 2012; Mason and Struhl, 2005; Morillon et al., 2003; Quan and Hartzog, 2010; Rondon et al., 2003). Genome-wide studies of Spt5 knockdowns, done in mice, HeLa cells, and zebrafish, showed only small effects, possibly due to inefficient depletion (Diamant et al., 2016b; Komori et al., 2009; Krishnan et al., 2008; Stanlie et al., 2012). Thus, there is little known about Spt5 with respect to the nature and breadth of its requirement during transcription elongation.
To address the genome-wide role of Spt5 in transcription, we have comprehensively analyzed the effects of Spt5 depletion on transcription genome-wide in the model organism Schizosaccharomyces pombe. Using an efficient degron allele to deplete Spt5, we have demonstrated that Spt5 is globally required for normal transcription by RNAPII. Spt5 depletion leads to reduced levels of transcription, accompanied by a relative accumulation of RNAPII over the 5′ region of genes. Spt5 depletion also causes a genome-wide reduction in both transcript synthesis and steady state mRNA levels, as well as inefficient mRNA splicing. Strikingly, there is a widespread increase in convergent and divergent antisense transcription upon depletion of Spt5. Our experiments to probe the role played by convergent transcripts suggest that they do not regulate the steady state levels of sense transcripts. However, small regions required for convergent transcription do control the distribution of RNAPII over gene bodies, suggesting that these sequences serve as sites at which Spt5 stimulates elongation. Together, our studies reveal previously unknown and critical roles for Spt5 in transcription elongation.
RESULTS
The level and distribution of RNA polymerase II are dramatically changed after depletion of Spt5
To study the essential Spt5 protein, we constructed an S. pombe strain that allows for efficient, auxin-inducible degradation of Spt5. Using this system, we were able to efficiently deplete Spt5 genome-wide, based on both ChIP-seq and western analysis, while maintaining cell viability (Figures 1 and S1). We then measured the level of RNAPII across the S. pombe genome using ChIP-seq, comparing cells before and after Spt5 depletion. We also compared wild-type cells to an spt5-ΔCTR mutant, which is deleted for sequences encoding the Spt5 carboxy-terminal region (CTR). Our spike-in normalized results showed that, after Spt5 depletion, there was a globally reduced level of RNAPII across transcribed regions (Figure 2A, Figures S2A,B). The inclusion of spike-in normalization was critical in reaching this conclusion (Figure S2B, Methods). Importantly, the distribution of RNAPII across genes also changed, with an accumulation over the first ~500 bp of genes, followed by a decreased level of RNAPII downstream (Figures 2A and S2A,B). In contrast, the level and localization of RNAPII in the spt5-ΔCTR mutant appeared similar to wild type (Figure S2C). These results show that depletion of Spt5 causes a widespread change in the level and distribution of RNAPII, suggesting an elongation defect.
Figure 2.
Spt5 is required for normal RNAPII localization. (A) A heatmap showing log2 ratios of spike-in normalized Rpb1 ChIP-seq density over 4294 expressed genes based on ChIP-seq, comparing depleted to non-depleted cells. Genes are sorted by length and aligned by their transcription start site (TSS). TSS and the cleavage/polyadenylation site (CPS) are indicated by the dotted green lines. (B) A similar heatmap based on log2 ratios of library-size-normalized NET-seq signal for the level of RNAPII on the sense strand. The NET-seq experiment was normalized by library size (see Methods). (C) A diagram depicting the calculation of the traveling ratio as the cumulative distribution function for the ratio of Rpb1 signal (in the case of ChIP-seq) or Rpb3 signal (in the case of NET-seq) over the first 500 bp of each transcript versus the last 500 bp of each transcript. (D) Traveling ratio for ChIP-seq for genes shown in 2A. Two replicates for Spt5 non-depleted and Spt5 depleted (red) are shown. (E) Traveling ratio plot for the NET-seq sense strand signal for genes shown in 2B, showing two replicates. (F) A scatterplot comparing the 3′-to-5′ shift in ChIP-seq and NET-seq signals over expressed genes after Spt5 depletion via the equation log2[ (5′T4.5/3′T4.5) / (5′T0/3′T0) ], using 500 bp bins.
To extend our ChIP-seq results, we used NET-seq, which measures the level and position of elongating RNAPII at single-nucleotide resolution in a strand-specific manner (Churchman and Weissman, 2011). Consistent with our ChIP-seq results, our NET-seq data (which was not spike-in normalized; see Methods) also showed that, after Spt5 depletion, there was a reproducible change in the distribution of transcribing RNAPII, with increased levels over the 5′ region of the sense strand relative to the levels downstream (Figure 2B). As an alternative way to view our ChIP-seq and NET-seq data, we plotted a “traveling ratio” (Figure 2C) (Rahl et al., 2010; Reppas et al., 2006) as a cumulative distribution. Both showed a reproducibly lower level of RNAPII over the 3′ ends of genes after Spt5 depletion (Figures 2D, 2E). Comparison of our ChIP-seq and NET-seq results showed a substantial overlap for the genes with this pattern (Figure 2F). Our results strongly suggest that Spt5 is required for elongation at a normal level across most genes.
To test other possible changes after Spt5 depletion, we measured the levels of the two major modifications of the C-terminal domain (CTD) of Rpb1, the largest subunit of RNAPII. Previous studies have shown that phosphorylation of serine 5 (S5P) in the CTD heptapeptide repeat occurs primarily over the 5′ end of transcription units, while the majority of phosphorylation of serine 2 (S2P) occurs downstream (Komarnitsky et al., 2000). Our results (Figure S3) show that the total level of Rpb1-S5P is unchanged after Spt5 depletion while the distribution of Rpb1-S5P is similar to the distribution of Rpb1 across the genome, with perhaps an increased elevation over the 5′ region. In contrast, the total level of Rpb1-S2P is reduced. This change in Rpb1-S2P seems likely to be a consequence of the changes in transcription caused by Spt5 depletion, rather than the cause, as complete loss of Rpb1-S2P causes less widespread transcriptional changes than what we observe after Spt5 depletion (Coudreuse et al., 2010; Schwer et al., 2014). In addition, the mRNA level for lsk1+, which encodes the major Rpb1-S2P kinase, is reduced approximately two-fold after Spt5 depletion.
Spt5 is required for a normal rate of RNA synthesis
To gain greater insight into the changes in elongation when Spt5 is depleted, we monitored cellular RNA synthesis before and after Spt5 depletion by metabolic labeling with 4tU (Miller et al., 2011) and normalization with RNA spike-in probes. The 4tU-seq results showed greatly decreased RNA synthesis rates genome-wide in the Spt5-depleted cells, consistent with a general elongation-stimulatory activity for Spt5 (Figure 3A, Figure S4). We observed a uniform decrease in RNA synthesis activity across transcription units (Figure 3B), suggesting that the overall rate of transcription is decreased when Spt5 is depleted. Taken together with our ChIP-seq and NET-seq results, these findings suggest that Spt5 is required for a normal rate of transcription by RNAPII in order to elongate past a site or barrier at a position within ~500 bp from the transcription start site (TSS), possibly a nucleosome or an Spt5-dependent transcription checkpoint (Hartzog et al., 1998; Lidschreiber et al., 2013; Viladevall et al., 2009).
Figure 3.
Spt5 is required for a normal rate of transcription genome-wide. (A) The scatter plot shows new RNA synthesis measured by 4tU-seq in Spt5-depleted versus non-depleted cells. Each point corresponds to the spike-in normalized signal from one transcript for merged replicate experiments. (B) The heatmap shows the log2 ratio of the spike-in normalized 4tU-seq signal in Spt5-depeleted versus non-depleted cells. The change is represented as described in Figure 2A. Genes are sorted by length and aligned by their TSS. The TSS and CPS are indicated by the solid black lines.
Spt5 is required for normal levels of most mRNAs and for mRNA splicing
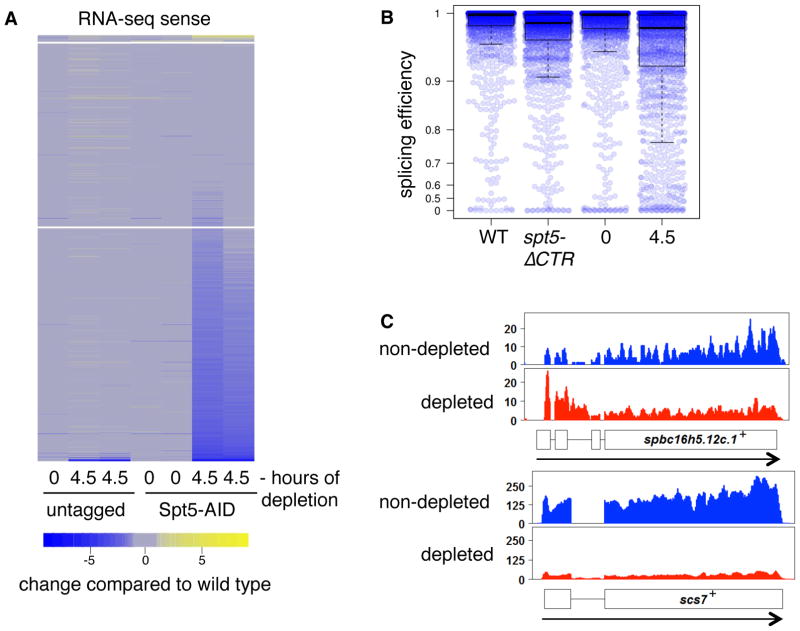
To determine the impact of these transcription defects on RNA levels and RNA processing, we performed RNA-seq. Our spike-in normalized results showed that, upon Spt5 depletion, there was a two-fold or greater reduction for over 60% of sense-strand RNAs (Figure 4A, Figure S4). Our RNA-seq data also revealed a widespread defect in splicing when Spt5 is depleted in S. pombe (Figures 4B, 4C, S4A), in agreement with previous studies in S. cerevisiae (Burckin et al., 2005; Lindstrom et al., 2003; Xiao et al., 2005). Consistent with our ChIP-seq results, there was little effect on mRNA levels and only a small effect on splicing caused by the spt5-ΔCTR mutation (Figure 4B). Our depletion studies, then, show that Spt5 controls the global levels of mRNAs and of mRNA splicing.
Figure 4.
Spt5 is required for normal steady-state levels of mRNA and mRNA splicing. (A) The heatmap shows the log2 fold change of spike-in normalized RNA-seq signal for sense-strand transcripts. Each column depicts the signal for the indicated strain compared to one replicate for a wild-type untreated strain. Genes are arranged in rows and placed in three different bins, demarcated by the horizontal gaps, based on whether their mRNA level is increased greater than two-fold (yellow gradient), changed less than two-fold (gray), or decreased greater than two-fold (blue gradient) in the average of the Spt5-depleted samples. The lanes labeled “untagged” have an spt5+ wild-type allele and the strains labeled “Spt5-AID” have the depletion construct. Two replicates are shown for each strain except for the untagged T0 sample, as the other untagged T0 sample is the denominator for all columns. (B) The box plot depicts the distribution of splicing efficiencies (spliced/total reads at 5′ splice junctions) in the indicated strains from merged RNA-seq data from two biological replicates for each strain. The bars represent the median, 25%, and 75% quartile ranges. A value of 0 indicates no splicing and a value of 1 represents complete splicing. The dots represent the values for the individual measurements. (C) Single-gene profiles illustrate the increased RNA-seq signal over introns before and after Spt5 depletion. The exons are shown as boxes and the introns as lines connecting the exons.
Widespread antisense transcription occurs after Spt5 depletion
In addition to dramatic effects on sense-strand transcription, our analysis demonstrated a surprising role for Spt5 in the regulation of antisense transcription. RNA-seq analysis showed that over 60% of all genes had a two-fold or greater elevated level of antisense transcripts after Spt5 depletion (Figure 5A) and that these changes are localized at the 5′ regions of genes (Figure 5B). In the spt5-ΔCTR mutant, there was a small increase in antisense levels that was much less prominent than after Spt5 depletion (Figure S4B). Antisense transcription near the 5′ ends of genes has been previously observed in both wild-type S. cerevisiae (Kim et al., 2012) and mammalian cells (Lavender et al., 2016; Mayer et al., 2015), albeit occurring at a smaller percentage of genes. Our results suggest that in those cases Spt5 limits the level of antisense transcription. We note that our 4tU-seq results showed that antisense transcription, similar to sense transcription, generally occurs at a lower level despite of producing new species of RNAs (Figure S4C).
Figure 5.
Spt5 represses convergent and divergent antisense transcription. (A) The heatmap shows the log2 fold change of spike-in normalized RNA-seq signal for transcripts on the antisense strand. Strains and details are as described in the legend to Figure 4A. (B) The heatmap shows the log2 ratio of spike-in normalized RNA-seq signal for the antisense strand over transcribed regions in Spt5-depleted versus non-depleted cells. (C) A Northern blot probed to detect both the asn1+ sense and convergent antisense transcripts. (D) RNA-seq and (E) NET-seq heatmaps show the antisense signal in the region between −500 bp to +500bp (with respect to the sense TSS) in Spt5-depleted cells compared to non-depleted cells. Shown are 1568 (RNA-seq) and 1336 (NET-seq) genes, selected based on criteria described in Methods. The green dotted line indicates the TSS. Genes with antisense transcription were sorted into three categories, shown from top to bottom: divergent only, both divergent and convergent, and convergent only.
To verify our RNA-seq results, we also assayed for antisense RNAs at two genes by Northern analysis. In these experiments we included several S. pombe mutants to test whether the derepression of antisense transcription occurs via one of the known defects that occur when S. pombe Spt5 is mutant or depleted, including reduced recruitment of the Paf1 complex (Mbogning et al., 2013) and loss of H3K4 trimethylation (Mbogning et al., 2015; Sanso et al., 2012) (Figure S5A). We also included an spt6-1 mutant, which has greatly elevated levels of antisense transcripts (DeGennaro et al., 2013), and an rrp6 mutant, to test if antisense levels are controlled by the nuclear exosome (Fox and Mosley, 2016). Our results show that for the two genes tested, rif1+ and asn1+, we detected the 5′ antisense RNAs only when Spt5 is depleted but not in any of the other mutant conditions (Figures 5C, S5B). Importantly, the depletion phenotype is complemented when spt5+ is expressed from a plasmid. Therefore, the widespread repression of 5′ antisense RNAs in wild-type S. pombe is dependent upon Spt5, but not other known Spt5-dependent events.
The location of the antisense transcription that occurs after Spt5 depletion suggested that it might include two classes of antisense transcripts – convergent transcripts, initiating within the gene, and divergent transcripts, initiating upstream of the transcription start site of the gene. Notably, divergent transcription, a feature of both S. cerevisiae (Neil et al., 2009) and mammalian cell transcription (Core et al., 2008), has not been detectable in S. pombe (Booth et al., 2016). To characterize antisense transcription in greater detail, we screened both our RNA-seq and NET-seq datasets for transcripts enriched over two regions: TSS to +500 for convergent transcripts and −500 to TSS for divergent transcripts. In this analysis, we only included genes whose transcripts did not overlap with adjacent transcripts (Eser et al., 2016). Our results revealed that, specifically after Spt5 depletion, there are distinct sets of genes enriched for one or both of those classes of transcripts (Figures 5D and 5E). From these results we conclude that Spt5 normally represses both convergent and divergent antisense transcription in S. pombe. We note that the number of genes with divergent transcription was greater when measured by NET-seq than when measured by RNA-seq (Figures 5D and 5E). As NET-seq measures synthesis, while RNA-seq measures steady-state levels, this suggests that many divergent transcripts in S. pombe are unstable, as previously shown in S. cerevisiae (Neil et al., 2009) and human cells (Preker et al., 2008).
Evidence for a site of action for Spt5 in stimulating transcription elongation
The unprecedented widespread nature of convergent antisense transcription after Spt5 depletion motivated us to characterize its expression and to test for possible regulatory roles. To do this, we chose the asn1+ and rif1+ genes, where Spt5 depletion causes both an increased level of convergent antisense transcripts and a decreased level of sense transcripts. For each gene, we mapped the antisense TSS by 5′ RACE and then constructed a 51 bp deletion that removed the antisense TSS plus 50 bp upstream (Figures 6A and S6A). In both deletion mutants, antisense transcription was undetectable after Spt5 depletion (Figures 6B and S6B), demonstrating that the deleted sequences are required for convergent antisense transcription. We measured the effect of each deletion and found that the corresponding sense RNA levels were still decreased after Spt5 depletion (Figures 6B, 6C, S6B and S7). This result suggests that antisense transcription at these two genes does not control steady-state sense transcript levels; however, our results do not rule out a role for antisense transcription at other genes or in controlling the kinetics of expression changes, as has been seen for other antisense RNAs (Kim et al., 2012; Lenstra et al., 2015).
Figure 6.
Analysis of convergent antisense promoters. (A) A diagram of the asn1+ gene showing the locations of the sense and antisense transcripts. The exons are shown as boxes and the intron as a line connecting the exons. The small gray box indicates the location of the 51 bp region deleted, including the TSS and upstream sequences for the antisense transcript, creating the asn1-Δ1 allele. The black bars labeled A and B indicate the regions tested for the RNAPII-ChIP analysis shown in panel D. (B) A Northern blot was probed with DNA that detects both the asn1+ sense and convergent antisense transcripts. (C) RT-qPCR analysis was performed to measure asn1+ sense RNA levels, normalized to adg1+ levels, for the strains indicated. Shown are the mean and standard deviation for three biological replicates. (D) ChIP-qPCR shows the enrichment of RNAPII at the asn1+ locus. The ChIP/input signal at asn1+ was normalized to the ChIP/input signal at the spiked in S. cerevisiae ADH1 gene. Shown are the mean and standard deviation from six to eight biological replicates. The p-values were calculated using Student’s t test.
To test if antisense transcription might control RNAPII localization across transcription units, we also measured the level of RNAPII by ChIP at the asn1+ and rif1+ genes, in both the presence and the absence of the antisense promoter sequences. Consistent with our ChIP-seq and NET-seq data, for both asn1+ and rif1+ (Figure S7), we observed a significant decrease in RNAPII levels at the 3′ end of the wild-type gene after Spt5 depletion (Figures 6D, and S6D). However, deletion of the antisense promoter virtually eliminated this dependence of RNAPII levels upon Spt5 (Figures 6D and S7D). Unexpectedly, the major phenotype caused by the deletions occurred when Spt5 was present, a state when antisense transcription is not detectable. These results suggest that the sequence defined by the promoter deletions functions as a site required for Spt5 to stimulate transcription elongation.
DISCUSSION
Our studies have revealed several previously unknown requirements for the essential factor, Spt5, in transcription in vivo. We have learned from both ChIP-seq and NET-seq that Spt5 is required for a normal distribution of RNAPII across transcription units. When Spt5 is depleted, RNAPII levels are reduced and accumulated over the 5′ regions of most genes. In addition, our 4tU-seq data suggests that transcription occurs at a lower rate after Spt5 depletion. Taken together, these results suggest that Spt5 is required to enable RNAPII to transcribe at a sufficient rate to elongate past a barrier. This model is consistent with our RNA-seq results that demonstrate that most mRNAs are present at reduced levels after Spt5 depletion. Our studies have also revealed that Spt5 normally represses two classes of antisense transcripts: convergent, initiating downstream of the TSS, and divergent, initiating upstream of the TSS. While previous studies have shown that the Spt5 CTR is important for the Spt5-dependent recruitment of several other factors, our results suggest that the CTR does not play a prominent role in the transcriptional roles for Spt5 studied here, nor does the recruitment of those factors, including the Paf1 complex. Together, our results provide strong evidence that Spt5 plays a critical and broad role in controlling transcription.
The change in the distribution of RNAPII that we observed in S. pombe after Spt5 depletion is distinct from previously-studied promoter proximal pausing in metazoans. After Spt5 depletion, the accumulation occurs over a much-greater distance from the TSS, approximately 500 bp, than that of promoter-proximal pausing, which occurs at approximately 20–60 bp from the TSS (Kwak and Lis, 2013). The accumulation that occurs in S. pombe suggests that when Spt5 is depleted, RNAPII is unable to elongate past a barrier. One obvious possibility for such a barrier is a nucleosome, as suggested by previous studies of spt5 mutants in S. cerevisiae (Hartzog et al., 1998). Although nucleosome positions have been mapped in S. pombe (Lantermann et al., 2010; Soriano et al., 2013), our data are not yet sufficient to resolve the position of the barrier with respect to that of nucleosomes, nor do we yet know whether Spt5 plays any role in controlling nucleosome position. Spt5 may also, directly or indirectly, help to clear a possible checkpoint during elongation by the recruitment of other factors. A comprehensive analysis to identify the factors recruited by Spt5 during elongation will be important in understanding such putative transcriptional checkpoints.
A recent study of an S. pombe spt4Δ mutant (Booth et al., 2016) showed two interesting results relevant to our studies. First, their results showed a similar change in transcriptional profiles at approximately 25% of those where we observed changes after Spt5 depletion. Second, their results, using PRO-seq, suggested that S. pombe, like metazoan cells, has promoter-proximal pausing at a subset of genes in wild type, but not in the spt4Δ mutant. We have examined our results for the S. pombe genes that were reported to have promoter-proximal pausing and we were unable to detect this event, suggesting that our ChIP-seq and NET-seq data are not sufficiently sensitive. However, given that loss of Spt5 causes stronger phenotypes than loss of Spt4, it seems likely that Spt5 is required for this type of promoter-proximal pausing in S. pombe.
Our results with respect to the effect of Spt5 on mRNA levels and on the distribution of RNAPII across transcription units stand in contrast to earlier studies that examined the effect of knockdown of Spt5 in either zebrafish or mammalian cells (Diamant et al., 2016b; Komori et al., 2009; Krishnan et al., 2008; Stanlie et al., 2012). In those studies, mRNA levels were changed for only a small percentage of genes, in contrast to our finding that over 60% of mRNAs have a decreased level of two-fold or greater. Our inclusion of spike-in normalization is likely one reason why we observed global effects for Spt5 depletion. In addition, two previous studies in mammalian cells concluded that Spt5 depletion results in an increased level of RNAPII across gene bodies (Brannan et al., 2012; Rahl et al., 2010), also in contrast to our results. The differences between those studies and ours are likely due to the strong requirement for Spt5 in widespread promoter-proximal pausing in mammalian cells (Rahl et al., 2010), as well as our inclusion of spike-in controls.
Our study reveals that divergent antisense transcription, a common feature in many eukaryotes, is widespread in S. pombe. Although the function of such divergent transcription remains elusive, our data have revealed that it is repressed by Spt5 and possibly is unstable, in agreement with previous observations of divergent transcription (Neil et al., 2009; Preker et al., 2008). These characteristics might explain why S. pombe divergent transcription has gone undetected by previous studies (Booth et al., 2016).
Our results have also identified widespread convergent antisense transcription after Spt5 depletion. This class of transcription has been previously found in a small number of cases in S. cerevisiae (Kim et al., 2012) as well as in mammalian cells (Lavender et al., 2016; Mayer et al., 2015) where it has been detected in up to 25% of genes. Conceivably, the elevation in antisense transcription is an indirect effect of releasing elevated levels of RNAPII after Spt5 depletion. While we currently cannot rule out this possibility, we think it is unlikely given the evidence that the antisense promoter region serves functions both before and after Spt5 depletion. While our results showed that antisense transcription did not control the steady-state level of sense transcripts at the two genes tested, previous studies have suggested that antisense transcription might have a more nuanced function, possibly in regulating the kinetics of sense-strand expression during transitions in growth conditions (Kim et al., 2012; Lenstra et al., 2015; Xu et al., 2011) or in the recruitment of particular histone modifications or transcription complexes (Lavender et al., 2016; Margaritis et al., 2012).
Our deletion analysis of the convergent antisense promoter regions of two genes, asn1+ and rif1+, suggested two functions for these sequences. As expected, they are required for convergent antisense transcription after Spt5 depletion. Unexpectedly, our results also revealed a second possible role for these regions under conditions when Spt5 is present at normal levels: controlling the level of RNAPII across each gene. As it has been established that Spt5 is recruited just downstream of the site of transcription initiation (Lidschreiber et al., 2013; Mayer et al., 2010) (Fig. 1D), our results suggest a model in which sites downstream from Spt5 recruitment are required for Spt5 to stimulate elongation (Figure 7). This model fits with Spt5 having a role similar to that of NusG in E. coli (Hartzog and Kaplan, 2011; Ray-Soni et al., 2016; Werner, 2012). Although both antisense promoter deletions behaved similarly, we cannot yet rule out that the decreased level of RNAPII downstream is caused by some other mechanism that destabilizes the elongation complex. The mechanism by which these sites control elongation is an important issue for future study.
Figure 7.
A model for the role of Spt5 during transcription elongation. Previous studies have shown that Spt5 is recruited to elongating RNAPII shortly after initiation. We propose a model in which there is a site ~500 bp downstream of the TSS, where RNAPII is converted, in an Spt5-dependent fashion, to a form that is more capable of transcription elongation. In the absence of either Spt5 (middle diagram) or the Spt5 stimulatory site (bottom diagram), RNAPII is impaired in elongation, resulting in accumulation of RNAPII over the 5′ region and a lower level of RNAPII downstream. In the case of Spt5 depletion there is also activation of antisense transcription. This model for Spt5 function is similar to those previously proposed for the function of NusG, an Spt5 orthologue, in prokaryotes.
In conclusion, our studies have shown a strong and broad requirement for Spt5 during transcription elongation. In addition, they have provided evidence for a positive site for stimulation of transcription by Spt5, helping to unify the function of Spt5 and NusG between eukaryotes and prokaryotes. Our results have also raised questions that can be addressed in future studies, including the mechanism by which Spt5 stimulates elongation and the possible role for convergent antisense transcription.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Correspondence and requests for materials should be addressed to Fred Winston (winston@genetics.med.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All yeast strains are listed in Table S1. S. pombe strains were grown in EMM complete, YES, or PMGSC media (DeGennaro et al., 2013) and grown at 30°C, unless otherwise indicated. Strains with epitope tags and deletions were generated by homologous recombination of DNA fragments synthesized by PCR. The Spt5-degron strain, FWP486, was generated by first fusing the 3′ end of the endogenous spt5+ gene in strain FWP484 to a sequence encoding the V5-IAA17 tag, which was amplified from a plasmid containing the V5-KanMx cassette. Then the 5′ end of spt5+ was fused to the nmt1-81x promoter, amplified from a plasmid with the hph-MX6-nmt181xpr cassette. The spt5-ΔCTR strain, FWP488, was generated by transformation of a restriction fragment from a plasmid with spt5-ΔCTR-ura4+ allele with flanking homologous sequences for transformation. The truncation in Spt5 was confirmed by Sanger sequencing and by western blotting using an antibody that recognizes both full length and Spt5-ΔCTR. The pSpt5-pr450 plasmid was generated by cloning Spt5 with 450 base pairs of sequence upstream of Spt5 into the vector pFY20 (Wahls and Davidson, 2008). The 51 bp deletions in the antisense promoters were made in the asn1+(−195 to −145 from ATG) and rif1+(+84 to +135 from ATG) genes and encompassed the mapped antisense start sites (see below). The putative antisense promoters were deleted using two-step gene replacement. Northern blots and qPCR were performed as described previously (DeGennaro et al., 2013). Spt5 depletion experiments were performed by adding NAA (0.5mM in DMSO (Kanke et al., 2011), napthaleneacetic acid, (Sigma) and thiamine (100nM, thiamine hydrochloride (Tommasino and Maundrell, 1991), Sigma), to cells grown to a density of 107cells/ml (O.D.600 ~ 0.5). Cultures were incubated with shaking at 30°C for 4.5 hours. Viability assays were performed by pelleting 1 ml of cells at the indicated time points, washing the cells twice with 1ml of water, and plating dilutions on EMM complete medium. Colony counts were performed after 4–5 days of growth at 32°C.
METHOD DETAILS
Western blotting and antibodies
Extracts for western blotting were prepared using trichloroacetic acid and sodium hydroxide (Knop et al., 1999). The primary antibodies used were: anti-Rpb1 (8WG16, Covance), anti-H3 (ab1791; Abcam), anti-H3K4me3 (ab8580; Abcam), anti-β-actin (ab8224; Abcam) and anti-V5 (Invitrogen, R960). The Spt5-FL antibody against S. pombe Spt5 was provided by Dr. Beate Schwer and Dr. Stewart Shuman. Anti-mouse and anti-rabbit IR-dye-coupled antibodies from LiCor Biosciences were used as secondary antibodies. Band intensities on blots were quantified with the Image Studio Lite software from LiCor biosciences and normalized to the loading control as described in the text.
Northerns blotting and RNA-seq library preparation
RNA was prepared from S. pombe and Northerns were done as previously described (Ausubel et al., 1991). Probes were made using the oligos listed in Table S2. RNA-seq was done on Spt5-degron tagged strains as well as untagged controls, with and without auxin added. For RNA-seq libraries, 200 μg of total RNA from each strain was spiked-in with 10 μl of 1:100 diluted ERCC control RNA (Invitrogen). Then, poly(A)+ RNA was enriched by two rounds of Dynabeads Oligo (dT)25 (Invitrogen) purification, followed by alkaline fragmentation and size selection of fragments 40–70 nucleotides. Size-selected RNA was dephosphorylated and ligated to an RNA linker at the 3′ end. First-strand cDNA was synthesized using a primer with sequence complementary to the 3′-linker and an additional adapter sequence, using Superscript III reverse transcriptase (Invitrogen). cDNA was circularized using CircLigase (Epicentre). The library was amplified with 8–14 cycles of PCR, using primers specific for the adaptor sequence and the products were gel purified. Library size distributions and concentrations were determined using an Agilent Bioanalyzer. All strains were done in duplicate. RNA-seq libraries were sequenced on the Illumina HiSeq platform at the Tufts University Core Facility.
ChIP and ChIP-seq
ChIP-seq and ChIP-qPCR were performed as described previously (DeGennaro et al., 2013) with the following modifications. For the spike-in control, S. cerevisiae chromatin was added to S. pombe chromatin before immunoprecipitation with the relevant antibodies. S. cerevisiae chromatin corresponding to 2.5% (for early ChIP-seq) or 10% (for late ChIP-seq and ChIP-qPCR) of S. pombe chromatin, as estimated by Bradford assay (Bio-Rad), was used. For ChIP of Rpb1S2P and Rpb1S5P, phosphatase inhibitors (PhoSTOP, Roche) were added to the lysis buffer along with protease inhibitors (cOMPLETE tablet, Roche). Oligos for ChIP are listed in Table S2. ChIP-seq library prep was either as described previously (DeGennaro et al., 2013) or using the GeneRead DNA library prep kit (Qiagen) for end repair, A tailing and adapter ligation followed by two rounds of cleanup with 0.7 X SPRI beads (Beckman Coulter) for size selection. DNA was then amplified using an appropriate number of PCR cycles determined using a titration curve. The amplified DNA was subjected to two rounds of cleanup with 0.7 X SPRI beads. The DNA libraries were then processed as described previously (DeGennaro et al., 2013). All strains were done in duplicate. ChIP-seq libraries were sequenced on the Illumina HiSeq platform at the Tufts University or the NextSeq platform at the Harvard Bauer Core Facility.
NET-seq
NET-seq was performed as previously described (Churchman and Weissman, 2012) with a few modifications to the lysis and immunoprecipitation steps. These modifications gave a better and more reproducible solubilization of RNAPII in S. pombe samples. Approximately 80% of total RNAPII was solubilized reproducibly using this method. Briefly, one liter cultures of S. pombe at 107 cells/ml were spiked-in with S. cerevisiae (10% by cell number), and the cells were collected by filtration. One of the libraries used for our analysis (Spt5-AID 0 hr II e1) did not have S. cerevisiae as spike-in, as explained in the NET-seq computational analysis section below. Cells were flash frozen by forcing the cells through a syringe into liquid nitrogen. The frozen cells were mixed with 4 ml of frozen (by dropping 50 μl drops into liquid nitrogen) lysis-buffer without MnCl2 and protease inhibitors and lysed in a mixer-mill using 8 cycles at 15 Hz (Lysis buffer: 20mM HEPES, 110mM KOAc, 0.5% Triton X-100, 0.1% Tween 20, 10mM MnCl2, 1X protease inhibitors Roche EDTA-free). This lysate was thawed on ice and 1 ml lysis buffer with 50mM MnCl2 and 5x protease inhibitor, was added to the lysate along with 660 μl of DNAseI (RQ1, Promega) and incubated on ice for 30 minutes. Solubilized RNAPII was then immunoprecipitated along with the associated RNA using FLAG M2 beads (SIGMA), and eluted using 3X-FLAG peptide (SIGMA). RNA was extracted from the eluate using Qiagen miRNEasy kit and subjected to library prep as previously described (Churchman and Weissman, 2011) except that a modified linker-1 with a randomized hexamer was used for some libraries. All strains were done in duplicate. Libraries were sequenced on a HiSeq platform at either the Biopolymers Facility at Harvard Medical School or the Harvard Bauer Core Facility.
4tU-seq
The 4tU-seq experiments and analyses were performed as previously described (Eser et al., 2016; Miller et al., 2011). The Spt5 depletion strain was grown as described earlier and cultures were labeled with 4tU for 10 minutes at 0 and 4.5 hours after the addition of thiamine and auxin. RNA spike-ins were added to cell pellets at the first step of RNA purification (Schwalb et al., 2016). The amount of spike-ins was adjusted to the cell number for each sample (120 ng of spike-in mix for 2.5 × 10^8 cells for all samples). Sequencing libraries were prepared according to the manufacturer’s recommendations using the Ovation Universal RNA-Seq System (NuGen). Libraries were sequenced on an Illumina HiSeq 2500 at LAFUGA, Ludwig-Maximilians-University of Munich.
Transcription start site mapping
Ten μg of total RNA was purified from Spt5-depleted cells, and was verified by Northern for expression of the antisense transcript at rif1+. Then, the RNA was dephosphorylated (calf intestinal phosphatase, NEB). The 5′ cap from the mRNA was then cleaved using cap-clip enzyme (CELLSCRIPT). The 5′ phosphate group thus generated was then ligated to an RNA adapter using T4 RNA ligase (NEB). The adapter-ligated RNA was reverse transcribed with Superscript III reverse transcriptase (Thermo Fisher Scientific) with a primer complementary to the desired antisense transcript, and the cDNA was used for PCR with primers nested within the adapter and cDNA. The PCR product was gel purified, cloned into pGEM-T vector (Promega) and five clones per antisense transcript were sequenced. The 5′ end of the antisense transcripts were as follows: for asn1+, −195 with respect to the sense ATG and +213 with respect to the sense TSS; for rif1+, +87 with respect to the sense ATG and +221 with respect to the sense TSS.
ChIP-seq computational analysis
Read mapping for the initial Rpb1 ChIP-seq was performed with Bowtie2 (Langmead and Salzberg, 2012) successively to the S. pombe and then the S. cerevisiae genomes using default parameters. Alignment rates were 73–95% for fission yeast and 12–68% for budding yeast. Mapped reads for fission yeast IP samples ranged from 4.5 million to 8.2 million, while those for budding yeast IP samples ranged from 150,000 to 700,000. Binding density was produced using SPP software. Profiles were normalized to library size (reads-per-million; RPM) (Bonhoure et al., 2014; Nagalakshmi et al., 2008). To test for global changes in Pol II ChIP levels, a spike-in normalization method was applied (Orlando et al., 2014). This method compares the total number of reads mapping to each organism per sample. This S. cerevisiae/S. pombe ratio was low across input samples but consistently higher specifically among Spt5-depleted cells, indicating a real effect. A second mapping using Bowtie and a unique mapping constraint (“-m 1”) to a concatenated fission+budding yeast “genome” was also performed and corroborated this result. The second ChIP-seq experiment (Rpb1, S2P, S5P, Spt5) was mapped this way, using Bowtie. Unique read alignment rates were above 85% for all samples. Total read counts mapping from IP samples to the fission yeast genome ranged from 4.1 to 16.7 million and 340,000 to 6 million to the budding yeast genome (the latter was a Spt5-depletion Spt5 IP sample, for which half the reads were from the budding yeast spike-in, as expected).
RNA-seq computational analysis
All RNA-seq experiments were performed in duplicate. The total number of reads ranged from 11 to 21 million. After adapter trimming, reads were aligned using TopHat2 (Kim et al., 2013) with minimum and maximum intron lengths 20 and 5000, respectively (“-i 20 -I 5000”). Alignment rates were 75–83%, with approximately 60% of reads mapping uniquely to the transcriptome, 17% multiply to transcriptome, 3% uniquely to the rest of genome, and 0.2% multiply to the rest of genome, with little variability across samples. Per-gene RPKM values were calculated with Cuffdiff (Trapnell et al., 2010). Spike-in normalization factors were obtained by calculating all pairwise linear model slopes for ERCC spike-ins, then deconvoluting to a vector of normalization factors. The adjusted RPKM values were linearly transformed by these scaling factors. Spike-in normalized data showed a change of two-fold or more in RNA levels for approximately 62% of genes after Spt5 depletion, whereas the same data normalized to library size, without spike-in normalization, showed a change in only 24% of genes.
Computational analysis of intron retention levels
Combining replicates, using the 5′ end of each junction, and keeping only reads that extend at least 4 bp upstream and downstream of the junction, we counted the number properly spliced as defined in the annotation (“spliced”) and those that are not spliced at all (“unspliced”). Any other reads are discarded. For the boxplots, per-junction splicing efficiency = (spliced) / (spliced + unspliced). The y-axis scale is transformed to allow easier visualization of conditional differences. Individual splicing efficiencies per junction, per sample are plotted behind boxplots. Lines above indicate pairwise significant difference via Kolmogorov–Smirnov test (α=0.05).
NET-seq computational analysis
For NET-seq libraries, there were 32–71 million reads per sample. The adapter was removed with CutAdapt (Martin, 2011), and low-quality reads were removed with Prinseq (Lite, version 0.20.2, parameters “-no_qual_header -min_len 7 -min_qual_mean 20 -trim_right 1 -trim_ns_right 1 -trim_qual_right 20 -trim_qual_type min -trim_qual_window 1 -trim_qual_step 1”) (Schmieder and Edwards, 2011). Molecular barcodes were removed from FASTQ files with a custom script. Other RNAs, including rRNAs, snRNAs, and snoRNAs were removed by mapping via TopHat2 (Kim et al., 2013) to a genome file containing just those sequences from both S. pombe and S. cerevisiae, allowing multiple mappings. Approximately 75–88% of reads were removed at this step, and 95% of these aligned multiply. RT mispriming, which occurs when reverse transcriptase primes within a template rather than at the end, was observed and filtered out by mapping all reads prior to hexameric barcode removal. Those that mapped were assumed to have lost the barcode in a mispriming event and were filtered out downstream. Approximately 9–12% of libraries mapped without barcode removal versus 80–90% after barcode removal. Alignment was performed with Tophat 2 (Kim et al., 2013) to a concatenated FASTA composed of both S. pombe and S. cerevisiae genomes, allowing only unique mapping. PCR duplicates were removed by comparing all reads that start and end at the same position and contain the same barcode. Surprisingly, this showed that 90% of reads in Spt5-AID T0 replicate I were from PCR duplication (vs 10–23% in the others). Thus removal of PCR duplicates is critical for QC and downstream analysis. Because removal of these duplicates reduced the sample to < 1M reads, we focused mainly on the other replicate and included a T0 sample from an earlier NET-seq experiment. This earlier experiment did not include the 5′ barcode, but quantification and removal of reads that show the same chromosome, strand, start, end, and sequence, as well as high correlation (r=0.96) with the remaining T0 replicate (Figure S2), indicated that it was of normal quality. This earlier T0 replicate also did not include a spike-in control. Because spike-ins were used for the later samples, we performed an analysis similar to that for ChIP-seq (Orlando et al., 2014), comparing the ratio of reads mapping uniquely to S. pombe vs S. cerevisiae. For these samples, 10% budding yeast cells were spiked in, although the variation in ratio of budding/fission yeast reads was 4.1–5.5% for all samples, not correlating between replicate pairs. This suggested that there was no significant difference between T0 and T4.5 Spt5-depletion cells for NET-seq. Therefore, spike-in normalization was not included for the NET-seq data analysis.
Multigene heatmaps
Multigene heatmaps were produced by binning profiles every 10 bp, sorting genes by length, then aligning all at the 5′ TSS. Flanking regions 500 bp upstream and downstream are also shown. Genes longer than 4 kb are allowed to run off the right side of the plot; this occurs for 216 (6.6%) of 4294 genes that are protein-coding and >10 RPKM in the average of NET-seq T0 samples. For pairwise comparison of profiles from sample A and B, the red-blue colors depict the ratio log2(B / A), where B and A are library- or spike-normalized values per bin. In the heatmaps depicting TSS+/−500 bp, genes were sorted into bins based on whether they showed signal increase upstream of TSS, downstream of TSS, or both. For these heatmaps, we chose non-overlapping (over TSS+/−500) genes longer than 700 bp that showed a 2-fold or greater increase in the −500 to +500 region; therefore, genes with no increase are not depicted.
Metagene plots
Metagene profiles present averages for genes longer than 1000 bp and which have expression above a threshold of 10 RPKM over full genes in the average of NET-seq T0 replicates. Replicates were merged via averaging at the stage of normalized genomic profiles. The dark lines in the metagene profiles present arithmetic mean values at each 10 bp bin across genes that are long enough to have signal at the given position, aligned at the transcription start site. Shaded areas represent 95% confidence intervals calculated at each gene position from a t-distribution with degrees of freedom representing the number of genes of at least that length.
Traveling ratio plots
“Traveling ratio”-type plots for ChIP-seq and NET-seq were produced by quantifying signal in a 5′ bin and 3′ bin for all protein-coding genes greater than a minimum length (ChIP-seq: 1 kb; NET-seq: 1050 bp), using the gene set with expression above a threshold of 10 RPKM over full genes in the average of NET-seq T0 replicates. For ChIP-seq, the 5′ bin used was TSS+1:+500, while for NET-seq, the 5′ bin was TSS+50:+550 because NET-seq does not reliably show 5′ signal due to the minimal requirement for transcription before mapping a read and taking the 3′ read position as the single-base position of RNAPII. There were 3716 genes in the ChIP-seq plot and 3630 genes in the NET-seq plot. Ratios of 5′/3′ bins were sorted before plotting against percentile.
Determining overlapping genes
Because the S. pombe genome is very gene-dense, quantification of antisense transcripts near and beyond 5′ TSSs is easily confounded by accidental inclusion of genes in which an adjacent opposite-strand sense transcript overlaps the antisense region in question. Additionally, not all annotated transcripts are expressed in all conditions, and overlap with unexpressed transcripts should not be reason for removal. We chose to filter on a gene set that included all protein-coding genes plus non-coding transcripts that were expressed above 1 RPKM in the average of our NET-seq T0 samples. For different analyses, we filtered out genes with protein-coding or expressed transcripts overlapping TSS−500 to TSS+500, TSS+1000, or CPS+500, depending on the region in question.
Novel annotation set
Recent studies have shown that the annotations in PomBase may be inaccurate at 5′ and 3′ ends (Booth et al., 2016; Eser et al., 2016; Li et al., 2015). As we were interested in the distribution of novel antisense reads relative to gene TSSs, we used the annotations of Eser et al. (Eser et al., 2016), including all those protein-coding annotations that differed from the PomBase 5′ or 3′ ends, except in a small number of cases where annotations were not included; in those cases we used the PomBase annotations.
4tU-seq computational analysis
Paired-end 50 base reads with additional 6 base reads of barcodes were obtained in replicates for all samples. Reads were demultiplexed and mapped with STAR 2.3.0 (Dobin and Gingeras, 2015) to the concatenated S.pombe genome and external spike-in sequences with maximum one mismatch (--outFilterMismatchNmax 2) allowing for only one mapping position (--outFilterMultimapScoreRange 0). SAM files were filtered using Samtools (Li et al., 2009; Schwalb et al., 2016) for alignments with MAPQ of at least 7 (-q 7) and only proper pairs (-f99, -f149, -f84, -f163) were selected. This resulted in 44–95 million reads per sample. Further data processing was carried out using the R/Bioconductor environment. Samples were normalized using external spike-ins as previously described (Schwalb et al., 2016).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification and statistical tests employed for each experiment are described in the figure legends or in the methods section.
DATA AND SOFTWARE AVAILABILITY
The raw sequencing data reported in this paper has been deposited at the NCBI Gene Expression Ominbus, accession number GSE85182. Other data are available at Mendeley (http://dx.doi.org/10.17632/v5jy3367rs.3).
Supplementary Material
Highlights.
Upon depletion of Spt5 the level and rate of RNA polymerase II is globally reduced.
RNAPII distribution suggests an elongation barrier in the absence of Spt5.
Spt5 represses both divergent and convergent antisense transcription.
The convergent antisense promoter may function as an elongation stimulation site.
Acknowledgments
We thank Stirling Churchman and Olga Viktorovskaya for helpful comments on the manuscript. We also thank Julia di Iulio and Steve Doris for helpful discussions, and Rajaraman Gopalakrishnan for help with the figures. We are also grateful to Beate Schwer and Steward Shuman for anti-Spt5 antisera and S. pombe strains, to Danesh Moazed for S. pombe strains, and to the McCarroll lab for use of their Bioanalyzer. S.K. was supported by the Boston-area Research Training Program in Biomedical Informatics (T15LM007092). C.D. was supported by a DFG Fellowship through the Graduate School of Quantitative Biosciences Munich (QBM). P.C. was supported by the Deutsche Forschungsgemeinschaft (SFB860, SPP1935), the European Research Council Advanced Grant TRANSREGULON (grant agreement No 693023), and the Volkswagen Foundation. F.W. was supported by the William F. Milton Fund, Harvard Medical School, and NIH grant GM032967.
Footnotes
Supplemental information includes seven figures and two tables and can be found with this article online at …
AUTHOR CONTRIBUTIONS
A.S. performed the ChIP-seq, NET-seq, and RNA-seq experiments. S.P.K., B.H.A., and P.J.P. performed and interpreted the computational analysis for the ChIP-seq, NET-seq, and RNA-seq experiments. C.D., K.C.M., and P.C. performed the 4tU-seq experiments and computational analysis, and D.S. performed the Northerns and RT-qPCR splicing analysis. F.W. and A.S. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl KE. Current protocols in molecular biology. Green Publishing Associates and Wiley-Interscience; New York, NY: 1991. [Google Scholar]
- Blythe AJ, Yazar-Klosinski B, Webster MW, Chen E, Vandevenne M, Bendak K, Mackay JP, Hartzog GA, Vrielink A. The yeast transcription elongation factor Spt4/5 is a sequence-specific RNA binding protein. Protein Sci. 2016 doi: 10.1002/pro.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhoure N, Bounova G, Bernasconi D, Praz V, Lammers F, Canella D, Willis IM, Herr W, Hernandez N, Delorenzi M, et al. Quantifying ChIP-seq data: a spiking method providing an internal reference for sample-to-sample normalization. Genome Res. 2014;24:1157–1168. doi: 10.1101/gr.168260.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth GT, Wang IX, Cheung VG, Lis JT. Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast. Genome Res. 2016 doi: 10.1101/gr.204578.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Native elongating transcript sequencing (NET-seq) Curr Protoc Mol Biol. 2012;Chapter 4(Unit 4 14):11–17. doi: 10.1002/0471142727.mb0414s98. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudreuse D, van Bakel H, Dewez M, Soutourina J, Parnell T, Vandenhaute J, Cairns B, Werner M, Hermand D. A gene-specific requirement of RNA polymerase II CTD phosphorylation for sexual differentiation in S. pombe. Curr Biol. 2010;20:1053–1064. doi: 10.1016/j.cub.2010.04.054. [DOI] [PubMed] [Google Scholar]
- Crickard JB, Fu J, Reese JC. Biochemical Analysis of Yeast Suppressor of Ty 4/5 (Spt4/5) Reveals the Importance of Nucleic Acid Interactions in the Prevention of RNA Polymerase II Arrest. J Biol Chem. 2016;291:9853–9870. doi: 10.1074/jbc.M116.716001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGennaro CM, Alver BH, Marguerat S, Stepanova E, Davis CP, Bahler J, Park PJ, Winston F. Spt6 regulates intragenic and antisense transcription, nucleosome positioning, and histone modifications genome-wide in fission yeast. Mol Cell Biol. 2013;33:4779–4792. doi: 10.1128/MCB.01068-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamant G, Bahat A, Dikstein R. The elongation factor Spt5 facilitates transcription initiation for rapid induction of inflammatory-response genes. Nat Commun. 2016a;7:11547. doi: 10.1038/ncomms11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamant G, Eisenbaum T, Leshkowitz D, Dikstein R. Analysis of Subcellular RNA Fractions Revealed a Transcription-Independent Effect of Tumor Necrosis Factor Alpha on Splicing, Mediated by Spt5. Mol Cell Biol. 2016b;36:1342–1353. doi: 10.1128/MCB.01117-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doamekpor SK, Sanchez AM, Schwer B, Shuman S, Lima CD. How an mRNA capping enzyme reads distinct RNA polymerase II and Spt5 CTD phosphorylation codes. Genes Dev. 2014;28:1323–1336. doi: 10.1101/gad.242768.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doamekpor SK, Schwer B, Sanchez AM, Shuman S, Lima CD. Fission yeast RNA triphosphatase reads an Spt5 CTD code. RNA. 2015;21:113–123. doi: 10.1261/rna.048181.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Gingeras TR. Mapping RNA-seq Reads with STAR. Curr Protoc Bioinformatics. 2015;51:11 14 11–19. doi: 10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 2010;6:e1001173. doi: 10.1371/journal.pgen.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser P, Wachutka L, Maier KC, Demel C, Boroni M, Iyer S, Cramer P, Gagneur J. Determinants of RNA metabolism in the Schizosaccharomyces pombe genome. Mol Syst Biol. 2016;12:857. doi: 10.15252/msb.20156526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MJ, Mosley AL. Rrp6: Integrated roles in nuclear RNA metabolism and transcription termination. Wiley Interdiscip Rev RNA. 2016;7:91–104. doi: 10.1002/wrna.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Yamaguchi Y, Schilbach S, Wada T, Lee J, Goddard A, French D, Handa H, Rosenthal A. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature. 2000;408:366–369. doi: 10.1038/35042590. [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Fu J. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta. 2013;1829:105–115. doi: 10.1016/j.bbagrm.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Kaplan CD. Competing for the clamp: promoting RNA polymerase processivity and managing the transition from initiation to elongation. Mol Cell. 2011;43:161–163. doi: 10.1016/j.molcel.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirtreiter A, Damsma GE, Cheung AC, Klose D, Grohmann D, Vojnic E, Martin AC, Cramer P, Werner F. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010;38:4040–4051. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- Kanke M, Nishimura K, Kanemaki M, Kakimoto T, Takahashi TS, Nakagawa T, Masukata H. Auxin-inducible protein depletion system in fission yeast. BMC Cell Biol. 2011;12:8. doi: 10.1186/1471-2121-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Munster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci U S A. 2011;108:546–550. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Inukai N, Yamada T, Yamaguchi Y, Handa H. Role of human transcription elongation factor DSIF in the suppression of senescence and apoptosis. Genes Cells. 2009;14:343–354. doi: 10.1111/j.1365-2443.2008.01273.x. [DOI] [PubMed] [Google Scholar]
- Kramer NJ, Carlomagno Y, Zhang YJ, Almeida S, Cook CN, Gendron TF, Prudencio M, Van Blitterswijk M, Belzil V, Couthouis J, et al. Spt4 selectively regulates the expression of C9orf72 sense and antisense mutant transcripts. Science. 2016;353:708–712. doi: 10.1126/science.aaf7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan K, Salomonis N, Guo S. Identification of Spt5 target genes in zebrafish development reveals its dual activity in vivo. PLoS One. 2008;3:e3621. doi: 10.1371/journal.pone.0003621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:251–257. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- Lavender CA, Cannady KR, Hoffman JA, Trotter KW, Gilchrist DA, Bennett BD, Burkholder AB, Burd CJ, Fargo DC, Archer TK. Downstream Antisense Transcription Predicts Genomic Features That Define the Specific Chromatin Environment at Mammalian Promoters. PLoS Genet. 2016;12:e1006224. doi: 10.1371/journal.pgen.1006224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra TL, Coulon A, Chow CC, Larson DR. Single-Molecule Imaging Reveals a Switch between Spurious and Functional ncRNA Transcription. Mol Cell. 2015;60:597–610. doi: 10.1016/j.molcel.2015.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Hou J, Bai L, Hu C, Tong P, Kang Y, Zhao X, Shao Z. Genome-wide analysis of core promoter structures in Schizosaccharomyces pombe with DeepCAGE. RNA Biol. 2015;12:525–537. doi: 10.1080/15476286.2015.1022704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidschreiber M, Leike K, Cramer P. Cap completion and C-terminal repeat domain kinase recruitment underlie the initiation-elongation transition of RNA polymerase II. Mol Cell Biol. 2013;33:3805–3816. doi: 10.1128/MCB.00361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL, Hartzog GA. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics. 2001;159:487–497. doi: 10.1093/genetics/159.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR, 3rd, Hartzog GA. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CR, Chang CR, Chern Y, Wang TH, Hsieh WC, Shen WC, Chang CY, Chu IC, Deng N, Cohen SN, et al. Spt4 is selectively required for transcription of extended trinucleotide repeats. Cell. 2012;148:690–701. doi: 10.1016/j.cell.2011.12.032. [DOI] [PubMed] [Google Scholar]
- Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol. 2009;29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritis T, Oreal V, Brabers N, Maestroni L, Vitaliano-Prunier A, Benschop JJ, van Hooff S, van Leenen D, Dargemont C, Geli V, et al. Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLoS Genet. 2012;8:e1002952. doi: 10.1371/journal.pgen.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011:17. [Google Scholar]
- Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015;161:541–554. doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- Mayer A, Schreieck A, Lidschreiber M, Leike K, Martin DE, Cramer P. The spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol Cell Biol. 2012;32:1321–1331. doi: 10.1128/MCB.06310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbogning J, Nagy S, Page V, Schwer B, Shuman S, Fisher RP, Tanny JC. The PAF complex and Prf1/Rtf1 delineate distinct Cdk9-dependent pathways regulating transcription elongation in fission yeast. PLoS Genet. 2013;9:e1004029. doi: 10.1371/journal.pgen.1004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbogning J, Page V, Burston J, Schwenger E, Fisher RP, Schwer B, Shuman S, Tanny JC. Functional interaction of Rpb1 and Spt5 C-terminal domains in co-transcriptional histone modification. Nucleic Acids Res. 2015;43:9766–9775. doi: 10.1093/nar/gkv837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer PA, Li S, Zhang M, Yamada K, Takagi Y, Hartzog GA, Fu J. Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5. Mol Cell Biol. 2015;35:3354–3369. doi: 10.1128/MCB.00520-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Schwalb B, Maier K, Schulz D, Dumcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dolken L, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol. 2011;7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol. 2009;391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, O’Sullivan J, Kent N, Proudfoot N, Mellor J. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell. 2003;115:425–435. doi: 10.1016/s0092-8674(03)00880-8. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER, Fritz CC, Bradner JE, Guenther MG. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep. 2014;9:1163–1170. doi: 10.1016/j.celrep.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Quan TK, Hartzog GA. Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4-Spt5 in transcription. Genetics. 2010;184:321–334. doi: 10.1534/genetics.109.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Soni A, Bellecourt MJ, Landick R. Mechanisms of Bacterial Transcription Termination: All Good Things Must End. Annu Rev Biochem. 2016;85:319–347. doi: 10.1146/annurev-biochem-060815-014844. [DOI] [PubMed] [Google Scholar]
- Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Rondon AG, Garcia-Rubio M, Gonzalez-Barrera S, Aguilera A. Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J. 2003;22:612–620. doi: 10.1093/emboj/cdg047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanso M, Lee KM, Viladevall L, Jacques PE, Page V, Nagy S, Racine A, St Amour CV, Zhang C, Shokat KM, et al. A positive feedback loop links opposing functions of P-TEFb/Cdk9 and histone H2B ubiquitylation to regulate transcript elongation in fission yeast. PLoS Genet. 2012;8:e1002822. doi: 10.1371/journal.pgen.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Pei Y, Shuman S, Schwer B. Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol Cell Biol. 2010;30:2353–2364. doi: 10.1128/MCB.00116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B, Michel M, Zacher B, Fruhauf K, Demel C, Tresch A, Gagneur J, Cramer P. TT-seq maps the human transient transcriptome. Science. 2016;352:1225–1228. doi: 10.1126/science.aad9841. [DOI] [PubMed] [Google Scholar]
- Schwer B, Bitton DA, Sanchez AM, Bahler J, Shuman S. Individual letters of the RNA polymerase II CTD code govern distinct gene expression programs in fission yeast. Proc Natl Acad Sci U S A. 2014;111:4185–4190. doi: 10.1073/pnas.1321842111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Schneider S, Pei Y, Aronova A, Shuman S. Characterization of the Schizosaccharomyces pombe Spt5-Spt4 complex. RNA. 2009;15:1241–1250. doi: 10.1261/rna.1572709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano I, Quintales L, Antequera F. Clustered regulatory elements at nucleosome-depleted regions punctuate a constant nucleosomal landscape in Schizosaccharomyces pombe. BMC Genomics. 2013;14:813. doi: 10.1186/1471-2164-14-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadelmayer B, Micas G, Gamot A, Martin P, Malirat N, Koval S, Raffel R, Sobhian B, Severac D, Rialle S, et al. Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat Commun. 2014;5:5531. doi: 10.1038/ncomms6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanlie A, Begum NA, Akiyama H, Honjo T. The DSIF subunits Spt4 and Spt5 have distinct roles at various phases of immunoglobulin class switch recombination. PLoS Genet. 2012;8:e1002675. doi: 10.1371/journal.pgen.1002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasino M, Maundrell K. Uptake of thiamine by Schizosaccharomyces pombe and its effect as a transcriptional regulator of thiamine-sensitive genes. Curr Genet. 1991;20:63–66. doi: 10.1007/BF00312766. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viktorovskaya OV, Appling FD, Schneider DA. Yeast transcription elongation factor Spt5 associates with RNA polymerase I and RNA polymerase II directly. J Biol Chem. 2011;286:18825–18833. doi: 10.1074/jbc.M110.202119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladevall L, St Amour CV, Rosebrock A, Schneider S, Zhang C, Allen JJ, Shokat KM, Schwer B, Leatherwood JK, Fisher RP. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol Cell. 2009;33:738–751. doi: 10.1016/j.molcel.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP, Davidson MK. Low-copy episomal vector pFY20 and high-saturation coverage genomic libraries for the fission yeast Schizosaccharomyces pombe. Yeast. 2008;25:643–650. doi: 10.1002/yea.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417:13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier AD, Mayekar MK, Heroux A, Arndt KM, VanDemark AP. Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc Natl Acad Sci U S A. 2013;110:17290–17295. doi: 10.1073/pnas.1314754110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Yang YH, Burckin TA, Shiue L, Hartzog GA, Segal MR. Analysis of a splice array experiment elucidates roles of chromatin elongation factor Spt4-5 in splicing. PLoS Comput Biol. 2005;1:e39. doi: 10.1371/journal.pcbi.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Clauder-Munster S, Smolik M, Huber W, Steinmetz LM. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol. 2011;7:468. doi: 10.1038/msb.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- Yamamoto J, Hagiwara Y, Chiba K, Isobe T, Narita T, Handa H, Yamaguchi Y. DSIF and NELF interact with Integrator to specify the correct post-transcriptional fate of snRNA genes. Nat Commun. 2014;5:4263. doi: 10.1038/ncomms5263. [DOI] [PubMed] [Google Scholar]
- Zhou K, Kuo WH, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci U S A. 2009;106:6956–6961. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Wada T, Okabe S, Taneda T, Yamaguchi Y, Handa H. DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation. Nucleic Acids Res. 2007;35:4064–4075. doi: 10.1093/nar/gkm430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.