Abstract
The human PAPLA1 phospholipase family is associated with hereditary spastic paraplegia (HSP), a neurodegenerative syndrome characterized by progressive spasticity and weakness of the lower limbs. Taking advantage of a new Drosophila PAPLA1 mutant, we describe here novel functions of this phospholipase family in fly development, reproduction, and energy metabolism. Loss of Drosophila PAPLA1 reduces egg hatchability, pre-adult viability, developmental speed, and impairs reproductive functions of both males and females. In addition, our work describes novel metabolic roles of PAPLA1, manifested as decreased food intake, lower energy expenditure, and reduced ATP levels of the mutants. Moreover, PAPLA1 has an important role in the glycogen metabolism, being required for expression of several regulators of carbohydrate metabolism and for glycogen storage. In contrast, global loss of PAPLA1 does not affect fat reserves in adult flies. Interestingly, several of the PAPLA1 phenotypes in fly are reminiscent of symptoms described in some HSP patients, suggesting evolutionary conserved functions of PAPLA1 family in the affected processes. Altogether, this work reveals novel physiological functions of PAPLA1, which are likely evolutionary conserved from flies to humans.
Phospholipases constitute an important class of enzymes, which serves a wide range of functions from digestive lipolysis, to membrane remodeling and cell signaling. Consistent with their fundamental functions, impaired phospholipase activity in humans is associated with numerous diseases including diabetes, cancer, neurodegenerative and neuromuscular diseases, which identifies phospholipases as important therapeutic targets1. All phospholipases catalyze the cleavage of a fatty acid from the glycerol-3-phosphate backbone of phospholipids. Depending on the fatty acid cleavage position, phospholipases are further categorized into sub-groups, with phospholipases A1 (PLA1) cleaving preferentially at the sn-1 position to generate free fatty acid and a 2-acyl-lysophospholipid. Interestingly, many PLA1 class enzymes, including Manduca sexta TGL and Drosophila melanogaster PAPLA12 have an extended substrate spectrum and next to phospholipids also hydrolyze neutral lipids such as di- and triacylglycerol (TAG)1. The PLA1 sub-family of phospholipases is evolutionary highly conserved from yeast to mammals1,3,4. Depending on their specific functions, PLA1 enzymes act either intra- or extracellularly. Whereas extracellular, secreted PLA1s typically function as digestive enzymes, intracellular PLA1s (iPLA1s) mediate membrane remodeling and trafficking, and produce lysophospholipids with signaling functions1,3,4. Genomes of invertebrates typically code for a single iPLA1, also called PAPLA1 (for Phosphatidic Acid PLA1), while mammalian iPLA1/PAPLA1 family consists of three members: DDHD1 (or PAPLA1), DDHD2 (or KIAA0725p), and the SEC23 interacting protein (SEC23IP or p125)3,4.
Mutations in DDHD15 and DDHD26 are in humans associated with hereditary spastic paraplegia (HSP), a genetically heterogeneous syndrome with progressive spasticity and weakness of lower limbs as the lead symptom7,8. The disease mechanism underlying the malfunction of the limbs is supposed to be the progressive degeneration of the pyramidal tract axon7,9. Onset of this degeneration varies; early onset is often associated with mild progression without the need for wheelchair assistance. Later disease onset leads frequently to loss of walking ability10,11. While DDHD1 mutations result in the pure, non-complicated form of HSP (type SPG28)12, DDHD2 is associated with the complex form of the disease (type SPG54)13. The SPG28 type of HSP is thus clinically characterized by weakness of the lower limbs and urinary sphincter dysfunction only, whereas the SPG54 type can be accompanied by a variety of additional neurological defects such as ataxia, mental retardation, fecal incontinence and others8,9. Apart from the neurological and locomotor symptoms, knowledge of other physiological and metabolic changes associated with HSP is rather limited. For example, the most frequent form of HSP, which is caused by mutations in the SPG11 gene, is associated with metabolic disorders such as dysphagia and mobility-independent obesity of unknown etiology14. Interestingly, dysphagia and developmental delay were also observed in some of the patients with HSP linked to DDHD2 deficiency6,15, suggesting that PAPLA1 family might have currently uncharacterized developmental and metabolic roles. However, understanding of the developmental and metabolic consequences of iPLAs deficiencies in human patients is hampered by various limitations including the low frequency of the disease, the variable penetrance and expressivity of the associated symptoms, and the confounding effects of the altered lifestyle of the patient. Yet, knowledge of the physiological effects of the iPLA1 deficiencies is important also for the disease management.
Thus, PAPLA1 mutant animal models promise valuable insights into additional roles of iPLAs in health and disease. Surprisingly, DDHD116 and SEC23IP17 mutant mice models show only minor physiological defect, i.e. male subfertility16,17. More severe phenotypes might be masked by functional redundancy among the mammalian PAPLA1 family members. Accordingly, less complex model systems that encode a single PAPLA1 gene might prove informative for a comprehensive understanding of the cellular, physiological and developmental functions of this enzyme family.
Drosophila melanogaster is a very powerful model system, which has been successfully used to study a variety of muscular, neuromuscular and neurodegenerative disorders, including HSP caused by mutations in spastin18,19,20 or KIF5A21. With respect to PAPLA1, three elegant studies proved the value of the fly model to study iPLA1- linked HSPs. Firstly, Liebl et al. showed neuromuscular junctions overgrowth caused by transposon insertion at the PAPLA1 gene region22. Later work by Schuurs-Hoeijmakers and colleagues demonstrated that RNAi-mediated knock down of PAPLA1 decreases active zones of neuromuscular junction, which causes synaptic transmission impairment similar to what occurs in the HSP patients6. Recently, Kunduri et al. proved that loss of fly PAPLA1 results in age-dependent progressive impairment of climbing ability23, reminiscent of progressive walking impairment accompanying HSP in humans7. Interestingly, this study indicated that, next to its role in the synaptic transmission, PAPLA1 might have much broader roles in fly physiology. The protein is involved in COPII transport, posttranslational modifications, and vesicular transport of G protein-coupled receptors (GPCRs). Given the plethora of signals that are transduced by GPCRs24,25, loss of PAPLA1 can be expected to severely affect fly physiology, including developmental and metabolic roles. In addition, an in vitro study of fly PAPLA1 activity has shown that this enzyme has also TAG lipase activity; and its homolog in Manduca sexta has been portrayed as the main TAG lipase in insects in general2. However, neither lipolytic, nor developmental nor metabolic roles of PAPLA1 have been addressed in vivo so far.
We thus reasoned that implementation of Drosophila as a model for human diseases caused by defective iPLA1s requires a general understanding of the physiological and metabolic consequences of PAPLA1 deficiency.
Our work indeed reveals broad physiological roles of PAPLA1 in embryonic and larval development, developmental timing, reproduction of both males and females, and in general energy metabolism. We show that impairment of energy-demanding processes such as reproduction and locomotion of PAPLA1 mutants is accompanied by reduction of food intake. Low energy intake is nevertheless antagonized by reduced basal metabolic rate. However, ATP reserves of PAPLA1 mutants remain considerably low. Interestingly, our study shows that despite the prediction of PAPLA1 coding for the main insect lipase2, deficiency for this enzyme does not increase fat storage. Nevertheless, PAPLA1 has important roles in the carbohydrate metabolism. Altogether, using Drosophila as a model system, we describe the broad developmental and metabolism-related role of this enzyme, which might be present, but due to the gene redundancy partially masked also in the mammalian family of PAPLA1 enzymes.
Results
Generation of PAPLA1-deficient flies by CRISPR/Cas9-mediated genome engineering
To analyze the biological roles of PAPLA1 in the Drosophila model system, we first created mutants deficient for all of the functional domains that are conserved in the mammalian iPLA1s, i.e. the DDHD and WWE domain, and the GHSLG sequence, which matches the canonical lipase consensus sequence GxSxG (Fig. 1a). Since the genomic region of fly PAPLA1 is exceptionally large and codes for five additional genes (Fig. 1b), we specifically targeted the genome region that is unique to PAPLA1. The corresponding deletion was triggered by simultaneous mutagenesis of exon 2 and exon 7 (Fig. 1b) by co-expression of two gRNAs and a source of CAS9 in the male germline according to Kondo and Ueda26. This approach led to two independent deletion alleles, which were termed PAPLA11 and PAPLA12 (Fig. 1b). Both deletions represent functional null alleles. These deletion mutants were backcrossed into a common genetic background for nine generations prior to the analysis (for details see Materials and Methods).
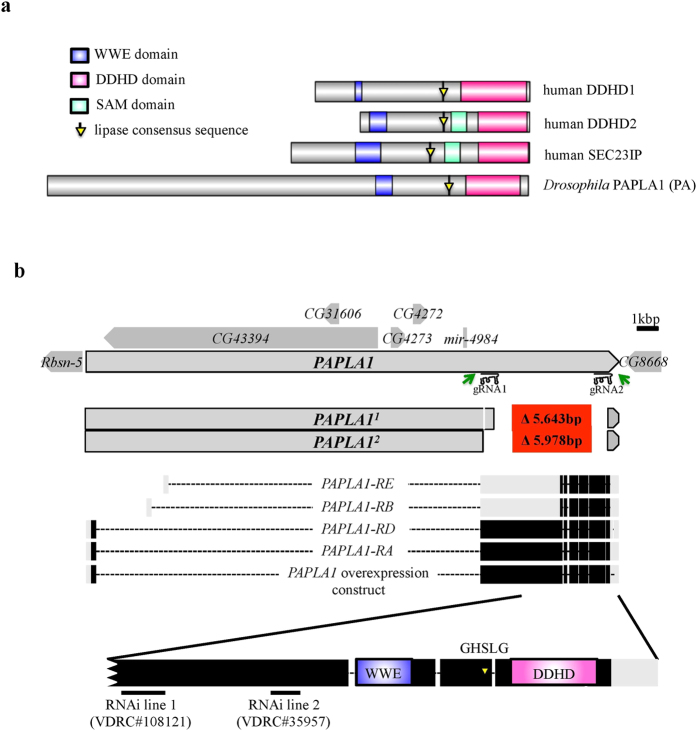
Figure 1. Structure and functional domains of the human and Drosophila PAPLA1 members, molecular identity of the fly PAPLA1 mutants, and schematic representation of other genetic tools used to study the gene functions.
(a) Human and fly PAPLA1 members and their functional domains. (b) The Drosophila PAPLA1 locus at chr. 2 L is flanked by the genes Rbsn-5 and CG8668 and encompasses additional five genes (CG43394, CG31606, CG4272, CG4273 and mir-4984; dark grey). FlyBase (FlyBase release r6.06) annotates four PAPLA1 transcript isoforms (RA to RD; light grey = non-coding exons; black = coding exons) PAPLA11 and PAPLA12 deletion mutants were induced by CRISPR/Cas9-assisted genome editing with gRNAs 1 and 2 flanking all main PAPLA1 domains shared by all protein isoforms. The PAPLA11 allele has a short 6 nt deletion (pos. 2 L:8140678–8140673), and a large 5.643 bp deletion (pos. 2 L: 8140324–813468), with two additional nucleotides (AT) in-between the breakpoints. PAPLA12 represents a 5.978 kbp deletion (pos. 2 L: 8140659–8134682), with a short ectopic sequence (CCATCCA) between the breakpoints (REFSEQ:NT_033779; FlyBase release r6.06). Sequence of the other genes encoded by the PAPLA1 locus remains intact in both mutant alleles. Green arrows indicate the binding sites of primers that were used for genotyping. To study the PAPLA1 gain-of function, we created UAS-PAPLA1 overexpression line, allowing overexpression of the full-length PAPLA1 identical to the RA transcript. Black bars depict the targeting positions of the two RNAi lines used in this study.
PAPLA1 is necessary for pre-adult development
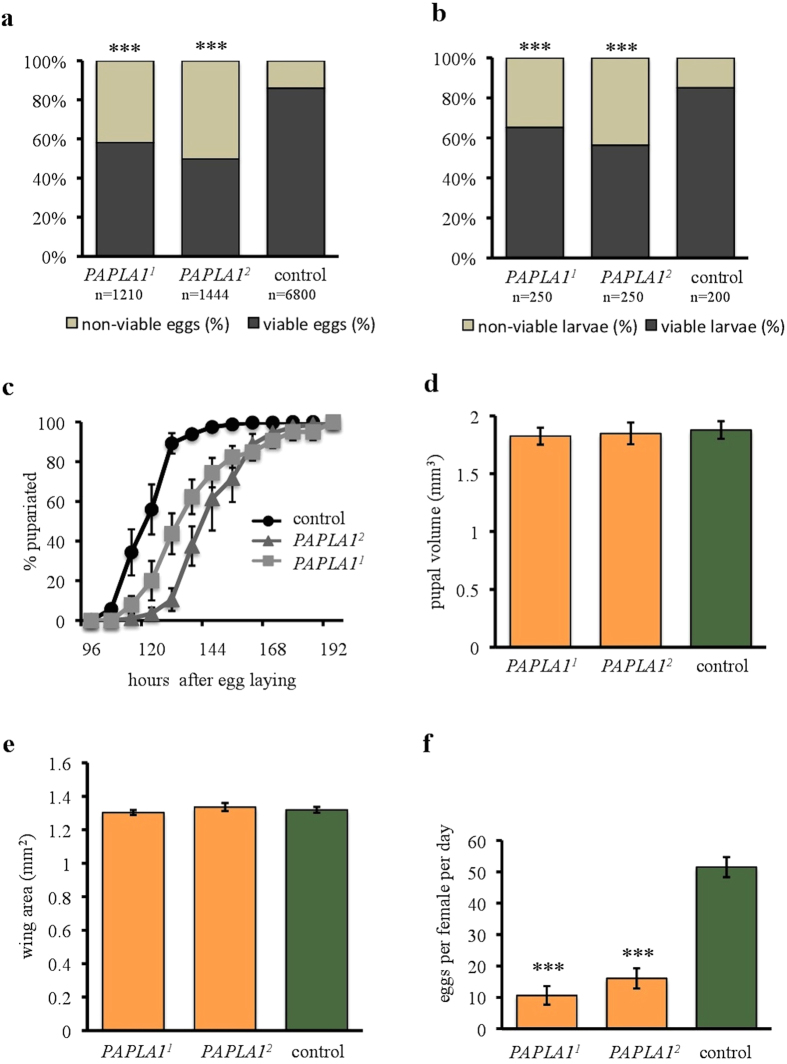
Firstly, we tested the potential developmental roles of PAPLA1. PAPLA11 and PAPLA12 mutations resulted in reduced hatchability, measured as proportion of eggs that developed into the 1st instar larvae (Fig. 2a). Viability of the mutants, estimated as the proportion of 1st instar larvae that successfully developed into adulthood, was reduced, too (Fig. 2b). Moreover, developmental speed of PAPLA1 mutants, estimated as the time from the egg deposition until the puparium formation, was considerably reduced (Fig. 2c). Altogether, ontogenetic defects of the tested mutants imply that the PAPLA1 family has important developmental functions.
Figure 2. Developmental and reproduction-related roles of PAPLA1.
(a) PAPLA1 deficiency reduces hatchability (egg to 1st larval instar survival). Fischer’s exact test: P < 0.001. (b) PAPLA1 deficiency reduces viability (survival from the 1st larval instar to adulthood). Larvae that did not give rise to adult flies are denoted as non-viable. Fischer’s exact test: P < 0.001. (c) PAPLA1 deficiency reduces developmental speed. Shown are mean % of pupariated flies at the given time point ± SEM. Mann Whitney test to compare median developmental times: P < 0.001 for both PAPLA1 mutants. (d) PAPLA1 deficient flies develop into pupae with standard volume. Two-tailed Student’s t–test: P > 0.05. (e) PAPLA1 deficiency does not affect body size of adults (measured as wing area). Two-tailed Student’s t–test: P > 0.05 for both assays. (f) PAPLA1 deficient flies have lowered egg production. Two-tailed Student’s t–test: P < 0.001.
PAPLA1 has reproduction-associated functions in males and females
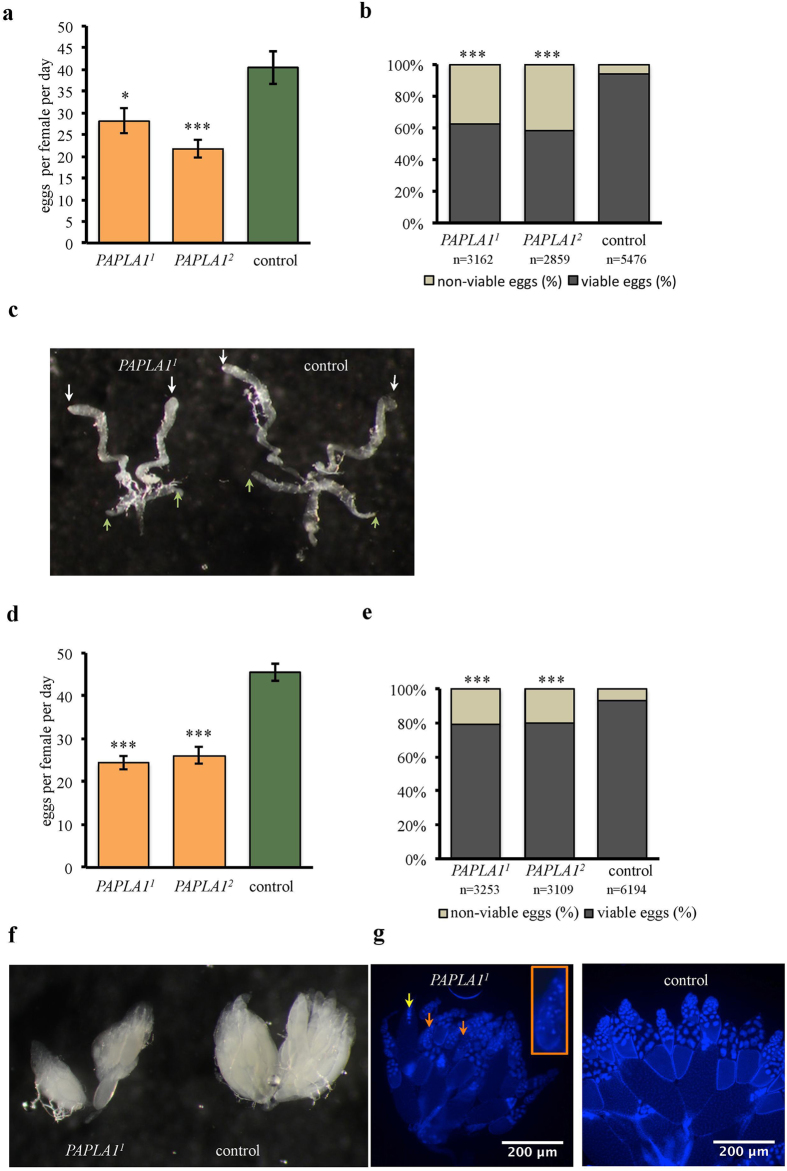
Despite the extended developmental time, PAPLA11 and PAPLA12 mutant flies reached normal size, documented by standard volume of pupae (Fig. 2d) and wing area of adults (Fig. 2e). Interestingly, egg production of homozygous PAPLA1-deficient parents was dramatically reduced (Fig. 2f), indicating important reproduction-related functions of this gene. To differentiate between the male- and female-specific requirements of PAPLA1 in reproduction, we first mated PAPLA1-deficient males with virgin PAPLA1+ tester females. These tester females laid considerably less eggs (Fig. 3a) than tester females mated with control males, indicating that the gene has a male-specific role in inducing female ovulation. In line with the previously described requirement of PAPLA1 for spermatogenesis23, hatchability of eggs laid by the tester females mated to PAPLA1 mutants was significantly decreased, too (Fig. 3b). Males induce ovulation in females by proteins that are produced in the male accessory glands, and deposited into females during copulation27. Consistent with the reduced capacity to induce ovulation when mated to tester females, PAPLA11 and PAPLA12 mutant males had smaller accessory glands (Fig. 3c). In addition, testes of the PAPLA1 deficient males were smaller as well (Fig. 3c), as reported previously23, suggesting a general role of PAPLA1 in the development of male reproductive tract.
Figure 3. PAPLA1 has important reproduction-related roles in both males and females.
(a) PAPLA1 function in males is necessary to stimulate normal egg laying in tester females. Two-tailed Student’s t–test: PAPLA11 vs. control P < 0.05, PAPLA12 vs. control P < 0.001. (b) PAPLA1 deficient males are subfertile. Sub-fertility of PAPLA1 deficient males shown by low hatchability of eggs produced upon mating with tester females. Tested is the ratio between the viable and non-viable eggs produced by single pair crosses upon mating with tester females. Fischer’s exact test: P < 0.001. (c) Hypotrophy of male reproductive organs in PAPLA1 mutants; note the smaller size of testes (white arrows) and of accessory glands (yellow arrows). (d–g) PAPLA1 is required for female reproduction. (d) PAPLA1 mutant females have reduced fecundity when mated with tester males. Two-tailed Student’s t–test: P < 0.001. (e) PAPLA1 deficiency in females decreases egg hatchability. Fischer’s exact test: P < 0.001. (f) PAPLA1 deficient females have smaller ovaries with reduced number of vitellogenic stage 10 chambers (see also g). (g) Note the occasional degeneration of egg chambers at the mid-oogenesis checkpoint (orange arrows, detail of the degenerating chamber in orange-lined rectangular). Stage 14 egg chambers occasionally contain nurse cell nuclei (yellow arrow).
To test the potential role of PAPLA1 in female reproduction, we crossed virgin PAPLA1 deficient females with PAPLA1+ tester males. Oviposition of the mutant females was considerably reduced (Fig. 3d). Moreover, hatchability of eggs laid by the PAPLA11 and PAPLA12 mutant females was mildly but significantly reduced (Fig. 3e), suggesting that PAPLA1 regulates egg production also qualitatively. Ovaries of PAPLA1 females appeared normal without any severe malformations, just smaller due to the reduced number of vitellogenic egg chambers (Fig. 3f,g), suggesting reduced speed of egg production. We also noted increased degeneration of egg chambers at the mid-oogenesis stage (Fig. 3g, orange arrows, for detail see also the enlarged degenerating chamber in the orange rectangle), which is a hallmark of nutritional stress28, and occasional persistence of nurse cell nuclei after stage 13 of oogenesis (Fig. 3g, yellow arrow).
Altogether, our work revealed novel roles of PAPLA1 in Drosophila reproduction; in addition to its known role in spermatogenesis23, male PAPLA1 is required also for proper development of accessory glands, and for male ability to induce female ovulation upon mating. In female reproduction, PAPLA1 is required for both oogenesis rate and viability of the produced egg.
PAPLA1 controls general energy homeostasis
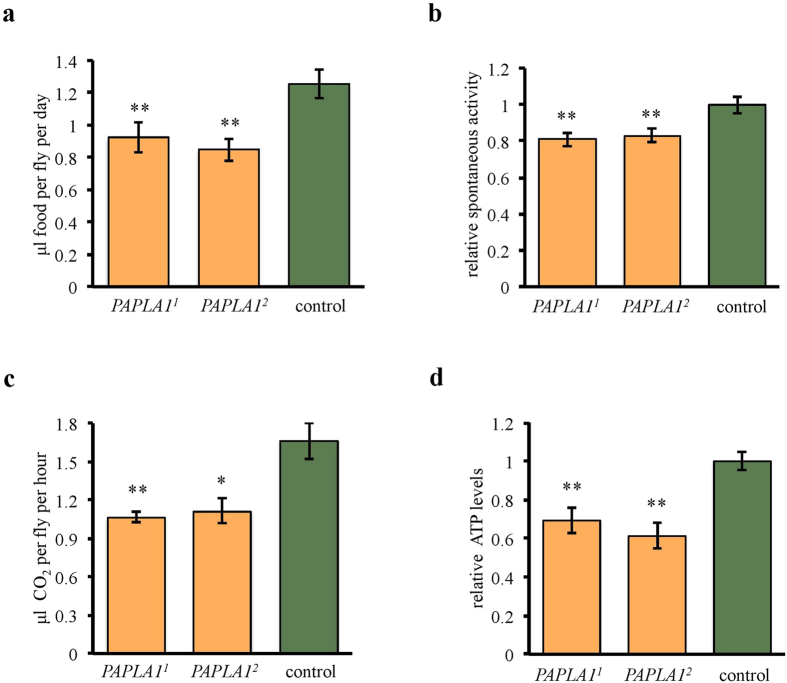
Since reproduction is an energetically demanding process, we hypothesized that the reduced reproductive success of PAPLA1 mutants might be at least partially caused by a general impairment of energy metabolism. Moreover, some patients with the DDHD2–associated spastic paraplegia have dysregulated food intake6,14, also suggesting defects of energy balance in the absence of PAPLA1 enzymes. Thus, we aimed to test whether PAPLA1 deficiency affects energy intake in flies. Monitoring the food intake by capillary feeding assay revealed significant reduction in daily food intake in both PAPLA11 and PAPLA12 mutants (Fig. 4a). Next, we measured activity-dependent energy expenditure, indicated by spontaneous locomotion. Spontaneous walking, monitored over a week period, was reduced in the mutants, too (Fig. 4b). Consistently, the CO2 production, which is a measure of the metabolic rate, was also decreased upon PAPLA1 loss-of-function (Fig. 4c). In addition, the lower metabolic rate was accompanied by considerably decreased level of ATP (Fig. 4d).
Figure 4. PAPLA1 deficiency reduces both energy intake and energy expenditure.
(a) PAPLA11 and PAPLA12 mutants have reduced food intake. (b) Spontaneous locomotor activity of PAPLA11 and PAPLA12 mutants is reduced. (c) Metabolic rate (estimated as CO2 production) of PAPLA11 and PAPLA12 mutants is reduced. (d) ATP levels of PAPLA11 and PAPLA12 mutants are reduced. All panels: Two-tailed Student’s t–test: *P < 0.05; **P < 0.01.
Altogether, our data show that PAPLA1 deficiency reduces both food intake and energy expenditure, decreasing energy turnover in general. Reduced metabolic capacity of the PAPLA1 mutants thus likely contributes to the observed impairment of energetically demanding processes such as reproduction and locomotion.
Differential effects of PAPLA1 on body fat storage
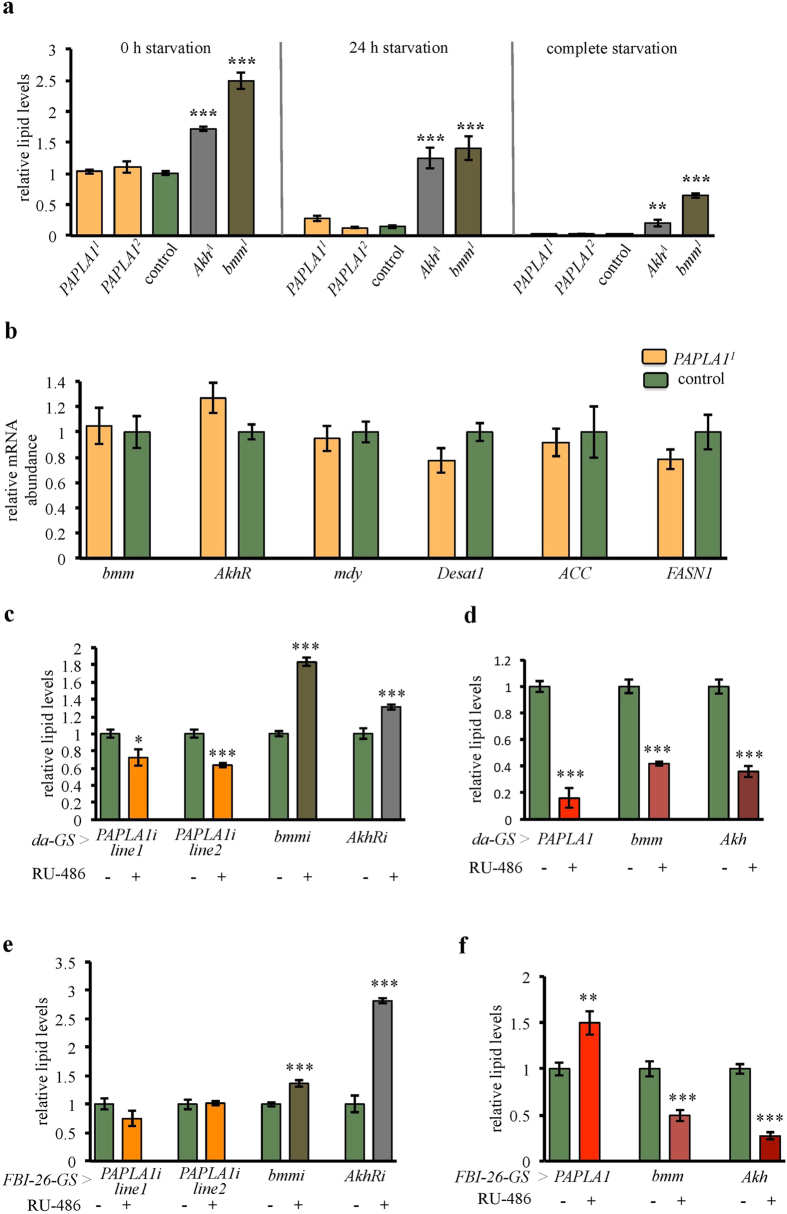
The above-described defects in the energy metabolism, and the prediction that PAPLA1 encodes the main insect TAG lipase2 prompted us to address a potential role of the gene in the energy reserves. In order to test the potential roles of PAPLA1 in the fat storage, we compared the PAPLA1 mutants to the lipolysis-defective bmm1 29 and AkhA 30 mutants. These mutants are obese and display incomplete storage lipids mobilization under starvation29,30. To exclude genetic background effects on the fat storage, these mutants were backcrossed into the same genetic background as the PAPLA1 mutants. In contrast to bmm1 and AkhA flies, fat storage in PAPLA11 and PAPLA12 mutants was normal, and they completely mobilized their lipid reserves upon starvation (Fig. 5a). To exclude that a potential role of PAPLA1 in body fat storage is masked by compensatory changes in PAPLA1 mutants, we tested the expression levels of several key regulators of fat storage that are known to be transcriptionally regulated. Consistent with their normal fat storage, PAPLA1 mutants had normal mRNA levels of the key lipid catabolism genes bmm and Akh receptor (AkhR) (Fig. 5b). Similarly, expression levels of the lipogenesis genes midway (mdy; encoding Drosophila diacylglycerol acyltransferase 1), Desaturase 1 (Desat1), Acetyl-CoA carboxylase (ACC), and Fatty acid synthase 1 (FASN1) were unaffected in PAPLA1 loss-of-function mutants (Fig. 5b).
Figure 5. PAPLA1 deficiency does not increase fat storage nor affects lipid mobilization.
(a) PAPLA1 mutants store normal fat reserves and are able to mobilize them completely. Note the difference to the AkhA and bmm1 mutants. Lipid content was determined prior to starvation, after 24 h of starvation, and after complete starvation. Lipid levels were normalized to the protein levels at the given time point, and standardized to the relative lipid levels of control flies prior to starvation. (b) Consistent with normal body fat storage, expression of lipolysis (bmm, AkhR) and lipogenesis (mdy, Desat1, ACC, FASN1) genes in PAPLA1 mutants is normal. (c) Adulthood-specific ubiquitous knockdown of PAPLA1 driven by daughterless-GeneSwitch (da-GS) decreases fat storage. Note the contrast to the corresponding knockdown of lipolytic genes bmm and AkhR. (d) Adulthood-specific overexpression of wild type PAPLA1 reduces lipid levels, similarly as overexpression of lipolytic genes bmm and AkhR. (e) Adulthood-specific knockdown of PAPLA1 in the fat body driven by the FBI-26-GeneSwitch (FBI-26-GS) driver does not affect fat storage. Two independent RNAi lines were used. Note the contrast to the obesogenic effects of the corresponding knockdown of the lipolytic genes bmm and AkhR. (f) Adulthood-specific overexpression of PAPLA1 in the fat body considerably increases fat storage. Note the contrast to the lean phenotype triggered by overexpression of Akh or bmm. All panels: Two-tailed Student’s t–test: *P < 0.05; **P < 0.01; ***P < 0.001.
Taken together, PAPLA1 mutants are able to adapt to the lower ATP levels and energy flux without compromising their fat storage. Next we investigated fat storage in flies with partial knock-down of PAPLA1. We used two independent RNAi lines, both of which significantly reduced PAPLA1 mRNA levels (Supplementary Fig. S1), and induced their expression in adult flies using the ubiquitous daughterless-GeneSwitch (da-GS) driver31. Adult-specific PAPLA1 knockdown reduced fat storage, opposite to the obese phenotype caused by the corresponding manipulation of the lipid catabolism genes AkhR and bmm32 (Fig. 5c). Nevertheless, da-GS-driven overexpression of wild type form of PAPLA1 decreased fat content (Fig. 5d), similar to overexpression of Akh32 or bmm29. Yet the low fat content triggered by this manipulation might have been contributed also by the toxicity of the manipulation, as around 50% of flies died prior to the analysis. Next, we tested whether the observed leanness in response to the ubiquitous PAPLA1 RNAi is due to an organ-specific function of the gene in the fat body or rather a secondary consequence of functional impairment of PAPLA1 in non-adipose tissues. Therefore, we knocked down PAPLA1 exclusively in the fat body, using the FBI-26-GeneSwitch driver (FBI-26-GS)33. Fat body-specific RNAi targeting of AkhR or bmm phenocopies the obesity of the respective null mutants (Fig. 5e). In contrast, impairment of PAPLA1 function in the fat body did not affect the body fat content neither in response to gene knockdown during adulthood (FBI-26-GS; Fig. 5e), nor when driven throughout the development (Lpp-GAL4; Supplementary Fig. S2). Interestingly, PAPLA1 overexpression in the fat body even increased the fat reserves, contrasting with the lean phenotype caused by overexpression of AkhR or bmm (Fig. 5f).
Taken together, PAPLA1 deletion mutants exhibit normal fat storage and mobilization. Yet, conditional gene knockdown experiments revealed a role of PAPLA1 in the fat storage control, which might be compensated in the null mutant situation. Importantly, none of the tested modulations of PAPLA1 mimiced the fat storage changes caused by the corresponding manipulations of central lipid mobilization genes.
PAPLA1 regulates glycogen storage
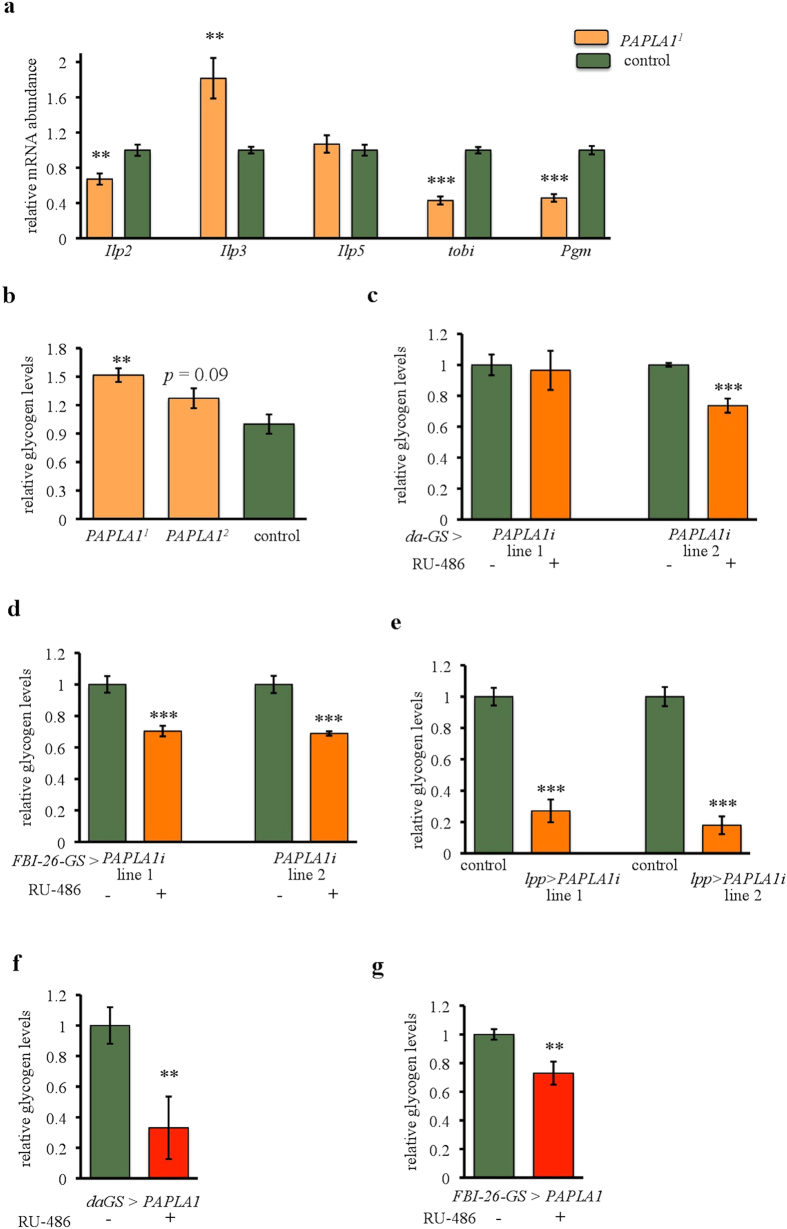
While fat is the calorically most important energy reserve, carbohydrates stored in the form of glycogen constitute the second pillar of energy storage in flies. Carbohydrate metabolism is regulated primarily by the insulin/insulin-like pathway34,35. Hence, we firstly tested the expression of fly Insulin-like peptides (Ilps). Expression of Ilp2 was reduced and Ilp3 was up-regulated, while Ilp5 remained unchanged in the PAPLA1 mutants (Fig. 6a). However, downstream targets of insulin-like signaling, such as Thor and l(2)efl, were not changed (Supplementary Fig. S3). Nevertheless, consistent with low levels of Ilp3, expression of target of brain insulin (tobi) was reduced in PAPLA1 mutants (Fig. 6a). The tobi gene codes for an evolutionary conserved α-glucosidase, which is positively regulated by insulin signaling36. This enzyme is involved in glycogen catabolism, cleaving the polysaccharide and releasing free glucose molecules. Similarly to tobi, expression of phosphoglucose mutase (Pgm), another enzyme acting downstream of glycogen cleavage, was also reduced in the absence of PAPLA1 (Fig. 6a). This suggests lowered glycogen catabolism in PAPLA1 mutants. Consistently, glycogen levels of PAPLA1 mutants were indeed increased (Fig. 6b).
Figure 6. PAPLA1 regulates glycogen storage and controls the expression of carbohydrate metabolism regulators.
(a) PAPLA1 mutants have mis-regulated expression of genes involved in the carbohydrate metabolism. Expression of Ilp2 is reduced, whereas expression of Ilp3 increased. Expression of genes encoding enzymes involved in glycogen cleavage, tobi and Pgm, is reduced. (b) PAPLA1 mutants have increased glycogen storage. (c) Ubiquitous, adulthood-specific knockdown of PAPLA1 by daughterless-GeneSwitch (da-GS) decreases glycogen storage when the stronger RNAi line is used (i.e. line 2; see also Supplementary Fig. S1). The weaker RNAi line (line 1) has no significant effect on glycogen storage. (d) Fat body targeted, adulthood-specific PAPLA1 knockdown by FBI-26-GeneSwitch (FBI-26-GS) reduces glycogen content. (e) Fat body targeted PAPLA1 RNAi driven by Lpp-Gal4 dramatically reduces glycogen content. (f) Adulthood-specific overexpression of PAPLA1 driven ubiquitously from the daughterless-GeneSwitch (da-GS) decreases glycogen storage. (g) Adulthood-specific overexpression of PAPLA1 in the fat body, driven by the FBI-26-GeneSwitch (FBI-26-GS), decreases glycogen storage. All panels: Two-tailed Student’s t–test: **P < 0.01; ***P < 0.001.
Considering the developmental and tissue-specific effects of PAPLA1 function on fat storage (Fig. 5), we next tested the effect of conditional PAPLA1 gene knockdown on glycogen reserves. Opposite to the PAPLA1 null phenotype, adulthood-specific ubiquitous PAPLA1 RNAi decreased glycogen reserves (Fig. 6c), and so did the fat body-specific knock down, irrespective whether conducted during adulthood only (Fig. 6d) or throughout the whole ontogenesis (Fig. 6e). Similar to RNAi, overexpression of the wild type isoform of PAPLA1 also decreased glycogen content when driven ubiquitously (Fig. 6f) or specifically in the fat body (Fig. 6g). Thus, both low and high levels of PAPLA1 in the fat body affect glycogen storage.
Collectively, our experiments provide the first evidence that an enzyme from the PAPLA1 family is required for proper carbohydrate metabolism. The impact of PAPLA1 function on carbohydrate but also on fat storage depends on the timing/strength of manipulation of the gene, suggesting existence of compensating mechanisms that underlie the energy homeostasis function of PAPLA1 in the fly.
Discussion
Given their roles in fundamental cellular processes such as membrane dynamics and trafficking, PAPLA1 enzymes are expected to serve a wide spectrum of physiological functions1,3,4. However, analysis of these roles in mammals might be complicated by redundancy among the multiple mammalian enzymes. Consistently, mice models lacking a single PAPLA1 family gene result in a surprisingly subtle phenotype4,16,17. This study aimed to investigate PAPLA1 functions using the Drosophila model, which has only a single member of this family, and is thus free of any confounding effects of redundancy that might occur in the mammals. Taking advantage of this model system we show that PAPLA1 is required for a broad range of functions, from early development to energy storage. Interestingly, PAPLA1 deficiency is not completely lethal in flies, suggesting that other mechanisms might partially compensate for its function. PAPLA1 mutants develop into normally sized adults, nevertheless, they have severely reduced metabolism, with lowered food intake and energy expenditure. This metabolic impairment likely contributes to the decreased spontaneous movement and to the lower reproductive success observed in these mutants. Subfertility of the PAPLA11 deficient flies correlates well with the spermatogenesis defects of mutants in its mammalian homologs Sec23IP4,17 and Ddhd116. This phenotype can be partially attributed to the previously reported spermatid individualization defects in flies lacking PAPLA1 function23, but also to the here-reported hypotrophy of accessory glands. Remarkably, the small size of both the testes and the accessory glands in PAPLA1 mutant flies is reminiscent of the hypogonadism that was described for the spastic paraplegia caused by deficiency in the phospholipase PNPLA637. In Drosophila, male fertility depends on spermatogenesis, but also on products of the accessory glands called sex peptides, which are transferred to females during copulation27. These peptides are necessary to induce ovulation and behavioral changes in females upon mating, maximizing thus the reproductive success of the male27. We speculate that loss of PAPLA1 interferes with sex-peptide function, as PAPLA11 and PAPLA12 males fail to stimulate oviposition when mated to females with normal PAPLA1 function. Thus, PAPLA1 deficiency may reduce male reproductive success in two ways – by impairing the production of sperm and by failure to induce standard ovulation in the mated females. Notably, whereas Kunduri et al. reported complete sterility of their PAPLA1 knockout males23, we observed severe subfertility, but not complete loss of the reproductive capacity of PAPLA11 and PAPLA12 mutant males. This suggests that the penetrance of the phenotype is likely modulated by genetic background and/or on environmental conditions. Intriguingly, these factors might also be responsible for the variability of symptoms in human patients suffering from PAPLA1 family-linked HSP.
Next to the known role in male reproduction, our work establishes for the first time the role of a PAPLA1 member in female reproduction. Interestingly, the decrease in oogenesis with occasional degeneration of egg chambers at the nutritionally controlled mid-oogenesis checkpoint is reminiscent of the same phenotypes triggered in wild type flies by nutritional shortage28. Since the PAPLA1 mutation reduces general metabolism and ATP levels, these metabolic defects likely also contribute to the reduced reproduction of the mutants.
Deficiency for PAPLA1 causes a complex metabolic phenotype suggestive of low metabolic flux; hypophagia, low basal metabolism and reduced spontaneous locomotion. The primary cause of this metabolic reduction is not known; however, several lines of evidence suggest that these defects might be triggered by impaired mitochondrial function. Recent work showed that Yor022c, the yeast homolog of PAPLA1, regulates mitochondrial dynamics and is thus required for the proper ATP production38. Moreover, impaired spermatogenesis of PAPLA1-deficient mice is also associated with reduced mitochondrial function16. Finally, the DDHD1-linked HSP in some human patients is also associated with lowered mitochondrial function39, decreased oxidative metabolism, and reduced ATP levels5. Our corresponding findings in mutant flies suggest, that PAPLA1 enzymes have a role in oxidative metabolism and ATP production that is evolutionarily conserved from yeast to human. Moreover, mitochondrial dysfunction and low basal metabolic rate might be serious, if not the principal cause of the reduced reproductive capacity and spontaneous movement of PAPLA1 mutants.
Interestingly, fat stores in PAPLA1 mutant flies are normal, suggesting that the reduced metabolic rate antagonizes the low food intake. On the other hand, normal fat storage of the fly mutants is surprising in view of an in vitro study showing that PAPLA1 encodes the main fat body TAG lipase of the tobacco hornworm Manduca sexta, and that a truncated Drosophila melanogaster PAPLA1 has TAG lipase activity as well2. Our work emphasizes the importance of genetic manipulations in investigations of the physiological roles of a gene, as neither PAPLA1 deficiency nor conditional PAPLA1 gene knockdown increases lipid stores in flies, whereas adipose tissue-specific PAPLA1 over-expression results in obesity. However, while PAPLA1 deficiency does not affect global fat storage, we cannot exclude that the TAG lipase activity of PAPLA1 is required in certain tissues that do not contribute significantly to the total body fat storage. Such a role has been described for DDHD2 in the nervous system of mice40. Absence of PAPLA1-associated lipid storage phenotype is also surprising in view of PAPLA1 acting in the COPII-mediated vesicular transport23. Components of the COPII machinery have been previously identified as regulators of Drosophila fat storage in genetic screens41,42. Moreover, PAPLA1 is required for post-translational processing and transport of GPCRs23, which mediate metabolic signaling via a broad spectrum of ligands including peptide hormones. As insect neuropeptides regulate numerous processes ranging from feeding to energy mobilization25, impaired GPCR signaling might underlie the hypophagia and reduced metabolic rate of the PAPLA1 mutants.
To the best of our knowledge, our work provides the first evidence that an iPLA1 family enzyme is involved in glycogen storage. Interestingly, the effect of PAPLA1 on carbohydrate storage depends on the range of the manipulation. Complete gene knockout in the null mutants increases glycogen storage, whereas ubiquitous or fat body-specific knockdown by RNAi in adult flies only has the opposite effect. We assume that PAPLA1 deficiency activates compensatory mechanisms that counteract the mutation, but are not activated by partial, RNAi-mediated gene knockdown. Mechanism of genetic compensation, which ameliorates the effect of complete null, but not knockdown by RNAi, was described in zebrafish43 and recently reported44 and discussed45 also for Drosophila. However, it is also possible that the lack of stronger phenotypes in the PAPLA1 mutants might simply result from the very general and broad role of PAPLA1 in GPCRs transport. Given the plethora of ligands with metabolic functions that signal via GPCRs, it is possible that PAPLA1 deficiency simultaneously affects antagonistically acting pathways, and the effect of impairment thus averages out. Nevertheless, temporally and/or spatially restricted PAPLA1 gene knockdown might affect only part of the signaling network, shifting thus the signaling balance, and disclosing the role of the gene in the energy homeostasis.
Altogether, this study reveals novel, unexpected functions of PAPLA1, including roles in development and developmental timing, female reproduction, as well as metabolic functions such as regulation of food intake, metabolic rate, ATP production, and carbohydrate storage. These functions might be evolutionary conserved, but difficult to disentangle in mammalian models with potential functional redundancy among the PAPLA1 enzymes. Importantly, similar defects as we observed in the PAPLA1 mutants might contribute to the complications accompanying HSP in humans. Of particular interest are the developmental delay and the dysphagia, which were also observed in the DDHD2-linked HSPs6,15. This suggests that requirement for PAPLA1 in developmental progression and food intake is evolutionary conserved from flies to humans. Whether the additional here-described roles of PAPLA1 in development, reproduction and metabolism are conserved in humans awaits clinical correlation studies. For many years, genetic causes of HSPs were studied only by linkage analysis. Yet times are changing thanks to the almost routine use of next-generation sequencing in clinical studies, which led to numerous reports of PAPLA1-family linked HSPs over the last years13,46,47, and to the discovery of PAPLA1-failure being involved also in some of the previously-reported linkage studies6. Nevertheless, even though clinical studies identify the disease-associated genes, their functional analysis is still dependent on the basic research in the model organisms. The case of the PAPLA1 family-associated HSPs demonstrates that Drosophila can be even more informative than mammalian animal models, as only the fly mutant recapitulates some of the HSP-linked phenotypes like walking defects and impairment of the neuromuscular junctions6,16,23.
Material and Methods
Fly stocks and fly husbandry
Flies were reared at 25 °C in 12 h light – 12 h dark cycle under controlled density conditions. Details on the rearing conditions and diets are available in the Supplementary Information. List of the fly stocks generated or used in this study is available in the Supplementary Table S1.
Creation of the PAPLA1 gRNAs transgenic line
PAPLA1 gRNA target sites were identified using the target prediction tool available at www.shigen.nig.ac.jp/fly/nigfly/cas9. The PAPLA1-targeting vector was based on pBFv.U6.2.B26. The vector carries two gRNAs, which target PAPLA1 exon 2 and 7 sequences, separated by 5978 bp (Fig. 1b). This vector was used for transgenesis by custom embryo injection (BestGene Inc.) to generate a stable line co-expressing these two gRNAs. For details on the vector and gRNA line construction see the Supplementary Information.
CRISPR/Cas9-mediated mutagenesis of the PAPLA1 gene
The PAPLA11 and PAPLA2 mutants were generated by CRISPR/Cas9-mediated genomic engineering according to Kondo and Ueda26. Fly line expressing the two PAPLA1 gRNAs was crossed to a line that expressed nos > CAS9, to generate F1 founder males, which co-expressed CAS9 and PAPLA1 gRNAs in the germline. Male progeny of these founders was propagated and screened for the desired large deletions in the PAPLA1 region. The resulting PAPLA11 and PAPLA12 alleles were molecularly characterized and backcrossed into the control genetic background. Details on the mutagenesis, mutation detection and backcrossing are available in the Supplementary Information.
Generation of the UAS-PAPLA1 over-expression line
Full-length cDNA representing the PAPLA1-RA transcript isoform (clone LD21067) was obtained from BDGP48,49. The cDNA was excised from the pBluescript_SK(-) backbone and ligated into the pUAST attB vector. The resulting plasmid was used for custom transgenesis by embryo injection (BestGene Inc.), to generate the stock w1118; +/+; UAS-PAPLA1 / TM3 Ser1. Details on the construction of the plasmid and overexpression line are available in the Supplementary Information.
Hatchability measurement
Hatchability was determined as the ratio between the viable and non-viable eggs (i.e. eggs that did not develop into the 1st instar larvae). Eggs were collected from single male-single female crosses of homozygotes; ≥15 parental pairs were used per genotype. Pairs were placed on a standard food supplemented with active yeast, allowed to mate and lay eggs, and flipped to fresh food vials daily. Hatchability was determined daily, approximately 30 h after the end of oviposition. Eggs laid over one week period were analyzed. In total, at minimum 1210 eggs were tested per genotype.
Viability measurement (larva to adult survival)
Flies were allowed to mate and lay eggs on Petri dishes with standard food supplemented with active yeast. 1st instar larvae were collected from the dishes into vials with standard food, at a density of 50 larvae per 68 ml vial. At least four replicates with 50 larvae each were tested per genotype.
Developmental rate measurement (time to pupariation)
1st instar larvae were collected as for the viability measurement. At least six replicates of around 50 larvae were tested per genotype. Number of pupariated animals was checked every eight hours, until the last larva entered metamorphosis.
Determination of pupal volume
Pupal volume was measured using a standard approach50; photographs of pupae were taken using stereomicroscope Zeiss Discovery. V8 with Canon EOS-5D Mark II digital camera attached to it and analyzed by ImageJ. Volume was calculated according to the formula 4/3π(L/2)(l/2)2 (L = length; l = diameter). At least 16 pupae were measured per genotype.
Determination of body size based on wing area
Body size was determined based on the wing area51 with modifications as described previously30. Left wings of 20 males were analyzed per genotype.
Fecundity assay of PAPLA1 deficient parents
Homozygous PAPLA11 and PAPLA12 virgin females and w1118 control virgin females were collected, and single male-single virgin female intercrosses among the same genotypes were put on standard food supplemented with active yeast. Food was exchanged daily. Daily egg scores were counted over one week. Per genotype, ≥15 couples were tested.
Testing the male-specific requirements of PAPLA1 in reproduction
Homozygous PAPLA11, PAPLA12 and control virgin males were crossed individually to virgin females of w1118 tester stock. Fecundity and hatchability was determined as described above.
Testing the female-specific requirements of PAPLA1 in reproduction
Homozygous PAPLA11, PAPLA12 and control virgin females were crossed individually to males of the genetically matched w1118 tester stock (RKF1084). Fecundity and hatchability were determined as described above.
Imaging of reproductive organs
Male and female reproductive tracts were dissected in PBS, and imaged directly, or after fixation in 4% paraformaldehyde followed by staining with Hoechst 33342 according to a standard protocol. Bright-field images of non-fixed reproductive tracts were taken using stereomicroscope Zeiss Discovery. V8 with Canon EOS-5D Mark II digital camera attached to it. Hoechst 33342-stained samples were imaged using Zeiss Axiophot microscope with Zeiss AxioCam HRC.
Metabolic rate measurements
Metabolic rate was estimated by respirometry according to Yatsenko et al.52, with experimental details and modification described previosly32. At least five replicates of five flies were tested per genotype.
ATP measurements
ATP levels were determined by bioluminescence assay using the ATP determination kit (Molecular Probes A22066), as described53. Five to six replicates of five flies each were tested per genotype.
Determination of spontaneous locomotion
Spontaneous locomotion was tested using the Drosophila Activity monitoring 2 system (TriKinetics), as described previously30. The assay started with 1-day-old flies, and locomotion was monitored over a one-week period. 32 males were tested per each genotype.
Capillary feeding assay
Food intake measurement was based on the capillary feeding described by Ja et al.54, with experimental details and modification as described previously32. Daily food intake of 20–23 individual males per genotype was monitored over three days, starting one day after eclosion.
Measurement of lipid, glycogen and protein levels
Lipid, glycogen and protein levels were measured by colorimetric assays from the same starting homogenate as described previously32. Four to six replicates of five flies each were tested for each genetic manipulation. Lipid and glycogen content was standardized to the protein levels, and the values were normalized to the values of the genetically matched controls. To ensure direct comparison of body fat storage, bmm1 and AkhA mutants have been backcrossed into the same genetic background as the PAPLA1 mutants prior to analysis.
Testing the changes in the fat reserves upon starvation
One-week old males were collected for colorimetric lipid and protein measurements before starvation (0 h starvation), after starvation on 0.6% agarose medium for 24 h, and after complete starvation to death, respectively. Each replicate consisted of five flies and 4–6 replicates were tested per genotype. The lipid content was standardized to protein levels of the given replicate, and the obtained values were normalized to the 0 h starvation value of the control flies.
Quantitative real-time PCR
Total RNA was extracted using the Zymo Research Quick-RNA™ MiniPrep kit. Three biological replicates, each consisting of 10 flies were used. cDNA synthesis, qRT-PCR and analyses were done as described in details previously32. Information on the primers is available in the Supplement.
GeneSwitch-mediated genetic manipulations
The GeneSwitch was induced at the age of three days after eclosion, when flies were transferred on standard food supplemented with 200 μM RU-486 (in the figures referred to as RU-486+), or on standard food without RU-486 (in the figures referred as RU-486−). Details on the RU-486 treatment are available in the Supplementary Information.
Illustration of proteins and their functional domains
Illustrations were done by the IBS Illustrator for Biosequences (http://www.ibs.biocuckoo.org). Protein sequences were obtained from NCBI (https://www.ncbi.nlm.nih.gov/protein), and motifs/domains were found using MOTIF Search (http://www.genome.jp/tools/motif).
Statistical analyses
Nominal variables were analyzed by the Fischer’s exact test (using GraphPad). Measurement variables were analyzed either by the two-tailed Student’s t-test (using Excel) or, when data distribution was not normal, by the Mann-Whitney U test (PAST55). Statistical significance of the differences between the samples is indicated by asteriks (*P < 0.05, **P < 0.01, ***P < 0.001). Plotted are means ± SEM.
Additional Information
How to cite this article: Gáliková, M. et al. Spastic paraplegia-linked phospholipase PAPLA1 is necessary for development, reproduction, and energy metabolism in Drosophila. Sci. Rep. 7, 46516; doi: 10.1038/srep46516 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We are very thankful to Herbert Jäckle and colleagues from the Department of Molecular Developmental Biology for their help and support. We thank Fillip Port, Véronique Monnier, Hervé Tricoire, Jae Suh, Ralf Pflanz, Hugo Stocker, and the Vienna Drosophila Resource Center fly stocks. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. The Berkeley Drosophila Genome Project is acknowledged for the PAPLA1 cDNA. We also thank Jana Heidemann for technical assistance. This study was supported by the Max Planck Society.
Footnotes
The authors declare no competing financial interests.
Author Contributions M.G. and R.P.K. jointly conceived the study and designed the experiments; M.G., P.K. and J.M. performed the experiment; M.G. analyzed the data; M.G. and R.P.K. wrote the manuscript.
References
- Tappia P. S. & Dhalla N. S. Phospholipases in Health and Disease. (Springer Science & Business Media), doi: 10.1007/978-1-4939-0464-8(2014). [DOI] [Google Scholar]
- Arrese E. L., Patel R. T. & Soulages J. L. The main triglyceride-lipase from the insect fat body is an active phospholipase A(1): identification and characterization. J. Lipid Res. 47, 2656–2667, doi: 10.1194/jlr.M600161-JLR200 (2006). [DOI] [PubMed] [Google Scholar]
- Richmond G. S. & Smith T. K. Phospholipases A1. Int. J. Mol. Sci. 12, 588–612, doi: 10.3390/ijms12010588 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani K., Kogure T. & Inoue H. The intracellular phospholipase A1 protein family. Biomol. Concepts 3, 471–478, doi: 10.1515/bmc-2012-0014 (2012). [DOI] [PubMed] [Google Scholar]
- Mignarri A. et al. Mitochondrial dysfunction in hereditary spastic paraparesis with mutations in DDHD1/SPG28. J. Neurol. Sci. 362, 287–291, doi: 10.1016/j.jns.2016.02.007 (2016). [DOI] [PubMed] [Google Scholar]
- Schuurs-Hoeijmakers J. H. M. et al. Mutations in DDHD2, encoding an intracellular phospholipase A(1), cause a recessive form of complex hereditary spastic paraplegia. Am. J. Hum. Genet. 91, 1073–1081, doi: 10.1016/j.ajhg.2012.10.017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J. K. Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol. 126, 307–328, doi: 10.1007/s00401-013-1115-8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson C., Koht J. & Stevanin G. Delving into the complexity of hereditary spastic paraplegias: how unexpected phenotypes and inheritance modes are revolutionizing their nosology. Hum. Genet. 134, 511–538, doi: 10.1007/s00439-015-1536-7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas S., Proukakis C., Crosby A. & Warner T. T. Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol. 7, 1127–1138, doi: 10.1016/S1474-4422(08)70258-8 (2008). [DOI] [PubMed] [Google Scholar]
- Harding A. E. Hereditary ‘pure’ spastic paraplegia: a clinical and genetic study of 22 families. J. Neurol. Neurosurg. Psychiatr. 44, 871–883, doi: 10.1136/jnnp.44.10.871 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott C., White K., Bushby K. & Shaw P. Hereditary spastic paraparesis: a review of new developments. J. Neurol. Neurosurg. Psychiatr. 69, 150–160, doi: 10.1136/jnnp.69.2.150 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori R. et al. Impairment of brain and muscle energy metabolism detected by magnetic resonance spectroscopy in hereditary spastic paraparesis type 28 patients with DDHD1 mutations. J. Neurol. 261, 1789–1793, doi: 10.1007/s00415-014-7418-4 (2014). [DOI] [PubMed] [Google Scholar]
- Citterio A. et al. Mutations in CYP2U1, DDHD2 and GBA2 genes are rare causes of complicated forms of hereditary spastic paraparesis. J. Neurol. 261, 373–381, doi: 10.1007/s00415-013-7206-6 (2014). [DOI] [PubMed] [Google Scholar]
- de Bot S. T. et al. Rapidly deteriorating course in Dutch hereditary spastic paraplegia type 11 patients. Eur. J. Hum. Genet. 21, 1312–1315, doi: 10.1038/ejhg.2013.27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Yahyaee S. et al. A novel locus for hereditary spastic paraplegia with thin corpus callosum and epilepsy. Neurology 66, 1230–1234, doi: 10.1212/01.wnl.0000208501.52849.dd (2006). [DOI] [PubMed] [Google Scholar]
- Baba T. et al. Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics. J. Biol. Chem. 289, 11497–11511, doi: 10.1074/jbc.M113.531921 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimitsu N. et al. p125/Sec23-interacting protein (Sec23ip) is required for spermiogenesis. FEBS Lett. 585, 2171–2176, doi: 10.1016/j.febslet.2011.05.050 (2011). [DOI] [PubMed] [Google Scholar]
- Orso G. et al. Disease-related phenotypes in a Drosophila model of hereditary spastic paraplegia are ameliorated by treatment with vinblastine. J. Clin. Invest. 115, 3026–3034, doi: 10.1172/JCI24694 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A. & Vale R. D. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr. Biol. 15, 650–655, doi: 10.1016/j.cub.2005.02.029 (2005). [DOI] [PubMed] [Google Scholar]
- Baxter S. L., Allard D. E., Crowl C. & Sherwood N. T. Cold temperature improves mobility and survival in Drosophila models of autosomal-dominant hereditary spastic paraplegia (AD-HSP). Dis. Model Mech. 7, 1005–1012, doi: 10.1242/dmm.013987 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füger P. et al. Spastic paraplegia mutation N256S in the neuronal microtubule motor KIF5A disrupts axonal transport in a Drosophila HSP model. PLoS Genet. 8, e1003066, doi: 10.1371/journal.pgen.1003066 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl F. L. W. et al. Genome-wide P-element screen for Drosophila synaptogenesis mutants. J. Neurobiol. 66, 332–347, doi: 10.1002/neu.20229 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunduri G. et al. Phosphatidic acid phospholipase A1 mediates ER-Golgi transit of a family of G protein-coupled receptors. J. Cell Biol. 206, 79–95, doi: 10.1083/jcb.201405020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon C. D. & Andrew D. J. Outside-in signaling–a brief review of GPCR signaling with a focus on the Drosophila GPCR family. J. Cell. Sci. 128, 3533–3542, doi: 10.1242/jcs.175158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel D. R. & Winther A. M. E. Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 92, 42–104, doi: 10.1016/j.pneurobio.2010.04.010 (2010). [DOI] [PubMed] [Google Scholar]
- Kondo S. & Ueda R. Highly Improved Gene Targeting by Germline-Specific Cas9 Expression in Drosophila. Genetics 195, 715–721, doi: 10.1534/genetics.113.156737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi Ram K. & Wolfner M. F. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr. Comp. Biol. 47, 427–445, doi: 10.1093/icb/icm046 (2007). [DOI] [PubMed] [Google Scholar]
- McCall K. Eggs over easy: cell death in the Drosophila ovary. Dev. Biol. 274, 3–14, doi: 10.1016/j.ydbio.2004.07.017 (2004). [DOI] [PubMed] [Google Scholar]
- Grönke S. et al. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 1, 323–330, doi: 10.1016/j.cmet.2005.04.003 (2005). [DOI] [PubMed] [Google Scholar]
- Gáliková M. et al. Energy Homeostasis Control in Drosophila Adipokinetic Hormone Mutants. Genetics 201, 665–683, doi: 10.1534/genetics.115.178897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricoire H. et al. The steroid hormone receptor EcR finely modulates Drosophila lifespan during adulthood in a sex-specific manner. Mech. Ageing Dev. 130, 547–552, doi: 10.1016/j.mad.2009.05.004 (2009). [DOI] [PubMed] [Google Scholar]
- Gáliková M., Klepsatel P., Xu Y. & Kühnlein R. P. The obesity‐related Adipokinetic hormone controls feeding and expression of neuropeptide regulators of Drosophila metabolism. Eur. J. Lipid. Sci. Tech., doi: 10.1002/ejlt.201600138 (2016). [DOI] [Google Scholar]
- Suh J. M. et al. Adipose is a conserved dosage-sensitive antiobesity gene. Cell Metab. 6, 195–207, doi: 10.1016/j.cmet.2007.08.001 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton S. J. et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. USA 102, 3105–3110, doi: 10.1073/pnas.0405775102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S., Clarke D.-F., Broughton S., Andrews T. D. & Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 6(2), e100085, doi: 10.1371/journal.pgen.1000857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch S., Melcher C., Bauer M., Katzenberger J. & Pankratz M. J. Opposing effects of dietary protein and sugar regulate a transcriptional target of Drosophila insulin-like peptide signaling. Cell Metab. 7, 321–332, doi: 10.1016/j.cmet.2008.02.012 (2008). [DOI] [PubMed] [Google Scholar]
- Synofzik M. et al. PNPLA6 mutations cause Boucher-Neuhauser and Gordon Holmes syndromes as part of a broad neurodegenerative spectrum. Brain 137, 69–77, doi: 10.1093/brain/awt326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P. K. & Rajasekharan R. Misregulation of a DDHD Domain-containing Lipase Causes Mitochondrial Dysfunction in Yeast. J. Biol. Chem. 291, 18562–18581, doi: 10.1074/jbc.M116.733378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson C. et al. Alteration of fatty-acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia. Am. J. Hum. Genet. 91, 1051–1064, doi: 10.1016/j.ajhg.2012.11.001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inloes J. M. et al. The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc. Natl. Acad. Sci. USA 111, 14924–14929, doi: 10.1073/pnas.1413706111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach J. et al. A Drosophila in vivo screen identifies store-operated calcium entry as a key regulator of adiposity. Cell Metab. 19, 331–343, doi: 10.1016/j.cmet.2013.12.004 (2014). [DOI] [PubMed] [Google Scholar]
- Pospisilik J. A. et al. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell 140, 148–160, doi: 10.1016/j.cell.2009.12.027 (2010). [DOI] [PubMed] [Google Scholar]
- Rossi A. et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233, doi: 10.1038/nature14580 (2015). [DOI] [PubMed] [Google Scholar]
- Di Cara F. & King-Jones K. The Circadian Clock Is a Key Driver of Steroid Hormone Production in Drosophila. Curr. Biol. 26, 2469–2477, doi: 10.1016/j.cub.2016.07.004 (2016). [DOI] [PubMed] [Google Scholar]
- Danielsen E. T. & Rewitz K. F. Developmental Biology: When Less Damage Causes More Harm. Curr. Biol. 26, R855–858, doi: 10.1016/j.cub.2016.07.068 (2016). [DOI] [PubMed] [Google Scholar]
- Gonzalez M. et al. Mutations in phospholipase DDHD2 cause autosomal recessive hereditary spastic paraplegia (SPG54). Eur. J. Hum. Genet. 21, 1214–1218, doi: 10.1038/ejhg.2013.29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K. R. et al. Defining the genetic basis of early onset hereditary spastic paraplegia using whole genome sequencing. Neurogenetics 17, 265–270, doi: 10.1007/s10048-016-0495-z (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M. et al. The Drosophila gene collection: identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 12, 1294–1300, doi: 10.1101/gr.269102 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. F. et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 17, 1823–1836, doi: 10.1101/gr.6679507 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoue R., Slaidina M. & Léopold P. The steroid hormone ecdysone controls systemic growth by repressing dMyc function in Drosophila fat cells. Dev. Cell 18, 1012–1021, doi: 10.1016/j.devcel.2010.05.007 (2010). [DOI] [PubMed] [Google Scholar]
- Klepsatel P., Gáliková M., Huber C. D. & Flatt T. Similarities and differences in altitudinal versus latitudinal variation for morphological traits in Drosophila melanogaster. Evolution 68, 1385–1398, doi: 10.1111/evo.12351 (2014). [DOI] [PubMed] [Google Scholar]
- Yatsenko A. S., Marrone A. K., Kucherenko M. M. & Shcherbata H. R. Measurement of metabolic rate in Drosophila using respirometry. J. Vis. Exp. e51681, doi: 10.3791/51681 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. M., Barry W. E., Cox J. & Thummel C. S. Methods for studying metabolism in Drosophila. Methods 68, 105–115, doi: 10.1016/j.ymeth.2014.02.034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ja W. W. et al. Prandiology of Drosophila and the CAFE assay. Proc. Natl. Acad. Sci. USA 104, 8253–8256, doi: 10.1073/pnas.0702726104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø., Harper D. A. T. & Ryan P. D. Paleontological Statistics Software: Package for Education and Data Analysis. Palaeontol. Electron. 4, 9 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.