Abstract
Testing thousands of chemicals to identify potential androgen receptor (AR) agonists or antagonists would cost millions of dollars and take decades to complete using current validated methods. High-throughput in vitro screening (HTS) and computational toxicology approaches can more rapidly and inexpensively identify potential androgen-active chemicals. We integrated 11 HTS ToxCast/Tox21 in vitro assays into a computational network model to distinguish true AR pathway activity from technology-specific assay interference. The in vitro HTS assays probed perturbations of the AR pathway at multiple points (receptor binding, coregulator recruitment, gene transcription, and protein production) and multiple cell types. Confirmatory in vitro antagonist assay data and cytotoxicity information were used as additional flags for potential nonspecific activity. Validating such alternative testing strategies requires high-quality reference data. We compiled 158 putative androgen-active and -inactive chemicals from a combination of international test method validation efforts and semiautomated systematic literature reviews. Detailed in vitro assay information and results were compiled into a single database using a standardized ontology. Reference chemical concentrations that activated or inhibited AR pathway activity were identified to establish a range of potencies with reproducible reference chemical results. Comparison with existing Tier 1 AR binding data from the U.S. EPA Endocrine Disruptor Screening Program revealed that the model identified binders at relevant test concentrations (<100 μM) and was more sensitive to antagonist activity. The AR pathway model based on the ToxCast/Tox21 assays had balanced accuracies of 95.2% for agonist (n = 29) and 97.5% for antagonist (n = 28) reference chemicals. Out of 1855 chemicals screened in the AR pathway model, 220 chemicals demonstrated AR agonist or antagonist activity and an additional 174 chemicals were predicted to have potential weak AR pathway activity.
Introduction
As many as 10,000 commercial substances in the environment lack data on their potential androgen receptor (AR) bioactivity with hundreds of new chemicals being added to this total each year.1,2 Testing to provide data on AR bioactivity using currently validated U.S. Environmental Protection Agency (EPA) and Organization for Economic Cooperation and Development (OECD) methods could cost millions of dollars and take decades to complete.3 Alternative approaches, such as those developed by the U.S. ToxCast and Tox21 programs,4−7 use high-throughput in vitro screening (HTS) assays and computational toxicology methods to rapidly and cost-effectively test chemicals for biological activity across a broad range of toxicologically relevant molecular targets and pathways. These approaches are currently accepted by the U.S. EPA for determining estrogen receptor (ER) bioactivity8,9 and could also be used to identify potential AR-active chemicals. However, application of alternative testing strategies for regulatory decision-making requires performance-based validation against a set of reference chemicals with reproducible responses over a range of potencies.
Here, we describe an integrated experimental and computational approach combining data from 11 ToxCast and Tox21 in vitro HTS assays measuring activity at multiple points along the androgen receptor (AR) pathway including receptor-binding, coregulator recruitment, chromatin-binding of the mature transcription factor, and gene transcription. A certain number of chemicals could be expected to act as true AR agonists or antagonists, but there are also chemicals that are known to interfere with these various assay technologies through false signals such as autofluorescence or cytostatic mechanisms.10−14 A well-accepted method of dealing with this issue is to leverage orthogonal assays that help distinguish nonspecific activity from interaction with the intended target.14,15 The approach is similar to that demonstrated for the ER pathway.16 Here, the data from 11 AR pathway assays were supplemented with an additional antagonist confirmation assay using a higher concentration of the activating ligand to characterize competitive binding. This battery of in vitro AR assays was used to screen a library of 1855 chemicals. Observed patterns of assay activity included no assays activated, all agonist assays activated, all antagonist assays activated, specific subsets of assays across technologies activated, and technology-specific assay activation. To navigate this complexity in the results, we developed a computational network model to infer whether chemicals that activate specific patterns of in vitro assays were more likely to be AR agonists, AR antagonists, false positives due to specific types of assay interference, or true negatives.
Evaluating and validating the AR pathway model requires high-quality reference data for AR agonist and antagonist activity. Unlike the ER pathway, which has a well-characterized set of in vitro and in vivo reference chemicals,8,16,17 the reference chemical set for the AR pathway is much less developed. Previous work focused on identifying chemicals that were positive or negative for (anti)androgenicity, without a specific emphasis on potency, and often included compounds that were “presumed” active or inactive.18 Using a comprehensive list of putative AR-active or -inactive chemicals from past and present international validation studies, we performed a literature search to compile high-quality published in vitro AR binding and transactivation (TA) assay data. To facilitate external validation of the AR pathway model results, no ToxCast or Tox21 assay data were included in the literature search. We identified a set of chemicals with reliable and reproducible in vitro results from the literature and binned the chemicals into defined potency categories. The list of proposed reference chemicals and the supporting data are provided and were used to evaluate the current computational model of AR pathway activity based on the Tox21 and ToxCast assays.
Methods
Workflow
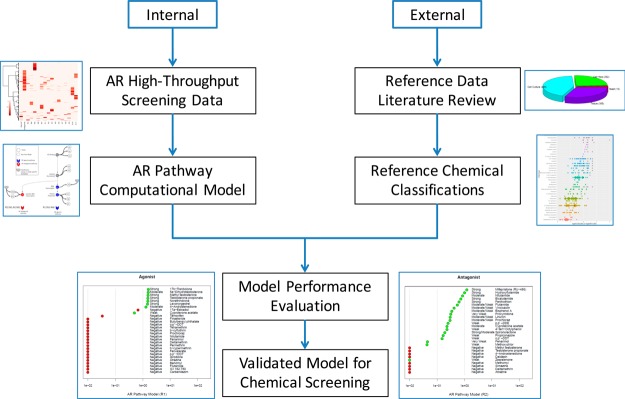
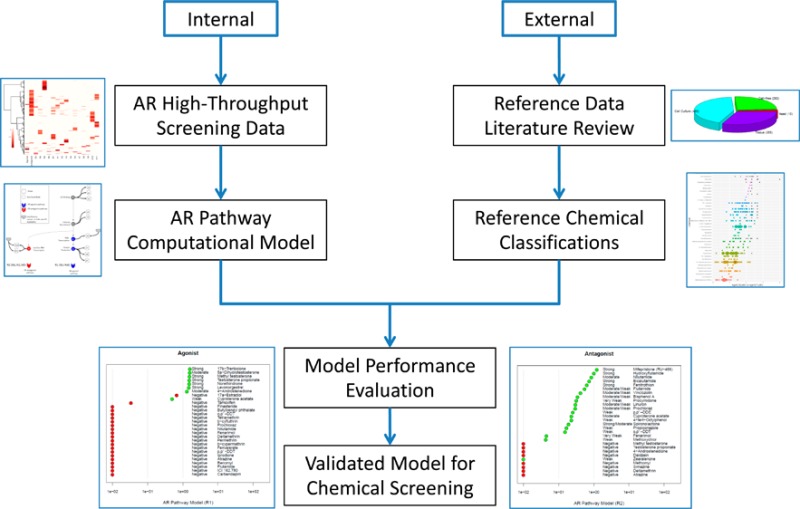
The workflow described in detail here is presented in Figure 1. Briefly, high-throughput screening data were generated on 1855 chemicals in 11 Tox21/ToxCast assays that map to key biological events along the AR pathway, and a computational model was used to integrate those data for each chemical to provide overall AR pathway activity predictions. In parallel, a systematic literature review was performed to characterize a list of reference chemicals using previously published work. These reference chemicals, classified based on the existing scientific literature, were used to evaluate the performance of the Tox21/ToxCast AR pathway model.
Figure 1.
Graphical representation of the AR pathway model workflow presented here. The internal process consisted of generating HTS data on 11 AR assays and building a computational model of AR pathway agonist/antagonist activity. The external process involved systematic literature review for reference data and curation of the reference chemical list based on published data. The model performance was evaluated against the reference chemicals, and screening results from the validated model on a large set of environmental chemicals are presented. AR = androgen receptor, HTS = high-throughput screening data.
High-Throughput Screening Data
Data on 1855 chemicals were generated during ToxCast Phases I and II and Tox21 screening using 11 AR-related in vitro assays (Table 1). These include three biochemical radioligand AR binding assays (Novascreen19−21), a coactivator recruitment assay measuring protein–protein interaction between AR and SRC1 at two different time points (Odyssey Thera), one transactivation assay measuring reporter RNA transcript levels (Attagene22), three transactivation assays measuring reporter protein level readouts (Odyssey Thera and Tox2123), and two transactivation antagonist assays (Tox2124−26). One of the transactivation antagonist assays, the Tox21 antagonist luciferase assay in the MDAKB2 cell line (A11), was run as a confirmation assay with a higher concentration of the synthetic ligand R1881 to verify chemical activity specific to the AR pathway. Higher concentration of the ligand should shift the potency of true competitive antagonists to higher concentrations. The chemicals were tested in concentration–response format in all assays except for the cell-free binding assays. The latter assays were initially tested at a single concentration (25 μM), and if significant activity was seen, the chemical was then tested in concentration–response mode. All concentration–response assay data27 were analyzed using the ToxCast data analysis pipeline, which automates the processes of baseline correction, normalization, curve-fitting, hit-calling, and AC50 (half-maximal activity) determination.28 All in vitro assays except the RNA transcript reporter assays (Attagene) were normalized to the range 0–100% using the positive control response. RNA transcript reporter data were normalized as a fold-change over the solvent control (0.5–1% DMSO, which has been determined to have no effect on assay performance) and then multiplied by a factor of 25 to yield a range of approximately 0–100. The data from each chemical assay pair was fit to three models: a constant model, a Hill model, and a gain-loss model, and the model with the lowest Akaike Information Criterion29 was selected. The pipeline also detects a variety of potential confounders, which are annotated as “caution flags”. For computational synthesis to be facilitated across different in vitro assays with different numbers of tested concentrations, a set of synthetic concentration–response activities was generated through interpolation for each chemical assay pair at standardized concentrations using a Hill equation based on the experimentally derived AC50, Hill slope, and Top parameters.16 All AC50 values were in μM, and the synthetic concentrations were a 1.5-fold dilutions series of 45 concentrations from 1 pM to 100 μM. The pipeline and all raw and processed data and annotations are publicly available (http://epa.gov/ncct/toxcast/data.html), and the data processing is described in detail elsewhere.16,28
Table 1. Tox21/ToxCast In Vitro Assays Used in AR Pathway Model.
| ID | node | assay name | source | genea | species | type | associated pathwaysb |
|---|---|---|---|---|---|---|---|
| A1 | N1 | NVS_NR_hAR | Novascreen | AR | Homo sapiens | receptor binding | R1; R2; R3 |
| A2 | N1 | NVS_NR_cAR | Novascreen | AR | P. troglodytes | receptor binding | R1; R2; R3 |
| A3 | N1 | NVS_NR_rAR | Novascreen | AR | Rattus norvegicus | receptor binding | R1; R2; R3 |
| A4 | N2 | OT_AR_ARSRC1_0480 | Odyssey Thera | AR; SRC | Homo sapiens | coregulator recruitment | R1; R2; R4 |
| A5 | N2 | OT_AR_ARSRC1_0960 | Odyssey Thera | AR; SRC | Homo sapiens | coregulator recruitment | R1; R2; R4 |
| A6 | N3 | ATG_AR_TRANS | Attagene | AR | Homo sapiens | RNA reporter gene | R1; R5 |
| A7 | N4 | OT_AR_ARELUC_AG_1440 | Odyssey Thera | AR; ARE | Homo sapiens | reporter gene | R1; R6 |
| A8 | N4 | Tox21_AR_BLA_Agonist_ratio | NCATS/NCGC | AR | Homo sapiens | reporter gene | R1; R6 |
| A9 | N4 | Tox21_AR_LUC_MDAKB2_Agonist | NCATS/NCGC | AR | Homo sapiens | reporter gene | R1; R6 |
| A10 | N5 | Tox21_AR_BLA_Antagonist_ratio | NCATS/NCGC | AR | Homo sapiens | reporter gene | R2; R7 |
| A11 | N5 | Tox21_AR_LUC_MDAKB2_Antagonist | NCATS/NCGC | AR | Homo sapiens | reporter gene | R2; R7 |
| A11c | N5 | Tox21_AR_LUC_MDAKB2_Antagonist-confirmation | NCATS/NCGC | AR | Homo sapiens | reporter gene | NA |
AR = androgen receptor; ARE = androgen response element; NCGC = NIH Chemical Genomics Center, now part of National Center for Advancing Translational Sciences (NCATS); SRC = c-Src tyrosine kinase.
Activity in these assays/nodes could be associated with one or more of the following pathways: AR agonist (R1), AR antagonist (R2), or interference (R3–R7). Activity in individual assays could also be associated with assay-specific interference (A1–A11).
Confirmation assay data (overly high concentration of R1881) not used in AR pathway model scores.
AR Pathway Model
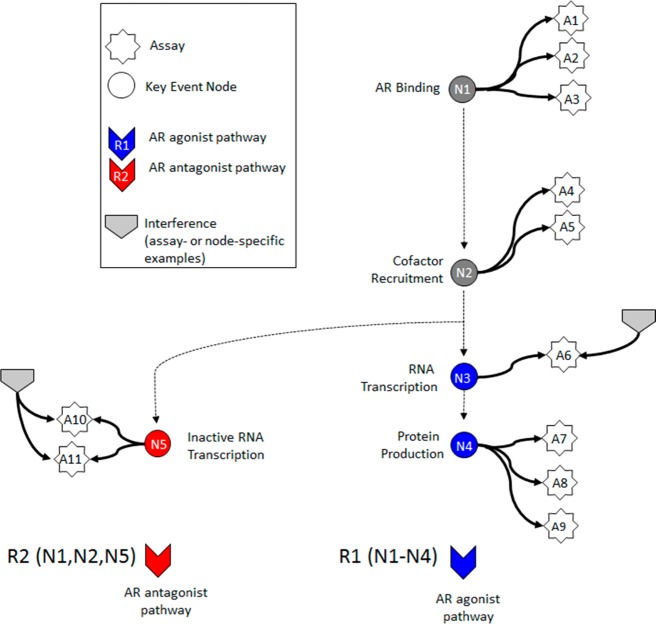
A computational network model for AR pathway activity (Figure 2) was built using 11 ToxCast and Tox21 in vitro assays (Table 1) that map to key events in the biological pathway. Figure 2 depicts the network model used to evaluate the integrated in vitro assay responses that mirrors previously published work on the ER pathway16 and is based on the series of molecular events that typically occur in a nuclear receptor-mediated response.30 An AR agonist will bind to the receptor monomer (node N1), cause the receptors to dimerize, and translocate to the nucleus and recruit coregulators (node N2) to form the complete, active transcription factor complex. The transcription factor complex binds to the chromatin DNA at specific response element sequences and initiates transcription of mRNA (node N3) and subsequent translation to protein (node N4). An AR antagonist acting through the receptor will bind to the receptor monomer (node N1), cause the receptors to dimerize, and translocate to the nucleus and recruit coregulators (node N2), forming a transcription factor complex that binds to the chromatin DNA at specific response element sequences, but is transcriptionally inactive and results in a lack of downstream protein production (node N5). Each of these key event nodes was assessed by one or more of the 11 in vitro assays listed in Table 1 (represented in Figure 2 as white stars). Figure 2 shows the two modes of the AR pathway: agonist (nodes associated with R1) and antagonist (nodes associated with R2). The model assumes that a chemical that interacts with the AR will bind and result in either or both of the agonist or antagonist conformations, triggering activity in the appropriate pathway. Each of the in vitro assays (A1–A11) is subject to processes that can lead to nonspecific activity independent of the AR pathway event that it is supposed to measure. These may be due to technological interference, artifacts, or other sources of experimental noise. Further, each group of assays that map to a key event node could be affected by non-AR-mediated activity specific to that key biological event (such as blocking transcription). Interference pathways R3–R7 correspond to nodes N1–N5 (detailed in Table 1). Two examples of interference pathways, one that is assay-specific (A6) and one that is node-specific (R7), are shown in Figure 2 as light gray arrow heads.
Figure 2.
Graphical representation of the AR pathway model based on Tox21/ToxCast assays: Circular nodes (N1–N5) represent key biological events along the pathway, where dark gray coloring indicates key events common to agonism and antagonism, and blue and red coloring indicates key events specific to agonism or antagonism, respectively. White stars (A1–A11) represent the in vitro assays that measure activity at the biological nodes. Colored arrow heads (R1/R2) represent true AR agonism/antagonism, respectively, and are comprised of the nodes listed in the diagram and their associated assays. Light gray arrow heads demonstrate examples of technology-specific interference or biological interference pathways, where individual assays or specific groups of assays are positive due to non-AR-mediated activity. Each in vitro assay and each key event node has an assay- or biology-specific interference pathway (defined in Table 1). Interference pathways R3–R7 correspond to nodes N1–N5, respectively, and interference pathways A1–A11 correspond to the respective assays. Two examples of interference pathways, one that is assay-specific (A6) and one that is node-specific (R7), are shown as light gray arrow heads. AR = androgen receptor.
Mathematical Representation of the Pathway Model
Following the ER pathway example presented in ref (16), a simple linear additive model is used to predict the relative AR agonist or antagonist activity of a test chemical based on data from the in vitro assays that map to the AR pathway in Figure 2. In the mathematical representation, the term “receptor” can refer to AR-mediated agonism, AR-mediated antagonism, or an interference pathway (mediated via biological activity or nontarget activity associated with a specific technology). The “receptors” R1–R7 associated with each assay or key event node are listed in Table 1. The model assumes that the value (the efficacy, A) returned by an assay at a given concentration is the sum of the contributions from the “receptors” that it measures, given as
| 1 |
where the index i ranges over the number of assays and index j over the number of “receptors” (where j = 1 for agonism, j = 2 for antagonism, and j > 2 for interference). The elements of the F matrix are 1 if there is a connection between a “receptor” j and an assay i and 0 otherwise. The model seeks a set of Rj values that minimizes the difference between the predicted assay values (Aipred) and the measured ones (Aimeas) for each chemical–concentration pair. A constrained least-squares minimization approach is used, where the function being minimized is
| 2 |
The term penalty(R) penalizes solutions that predict that many “receptors” are being simultaneously activated by the chemical, such that
| 3 |
| 4 |
The penalty term helps stabilize the solutions and is based on the assumption that it is unlikely that most chemicals will strongly and specifically interact with many dissimilar molecular targets.16 The model produces a response value (between 0 and 1) for each “receptor” at each concentration. These results are summarized as the integral across the concentration range expressed as area under the curve (AUC), such that
| 5 |
The biological response of greatest environmental concern is via antagonism of the AR pathway, which is also where most chemical activity is observed. Therefore, the AUC values were normalized to yield a value of 1 for the antagonist positive control. We used hydroxyflutamide, the antagonist positive control recommended by the OECD.31 The calibration curve plotting the relationship between AUC and activity concentration is given in Supplemental Figure S1. An AUC value of 0.1 corresponds to activity at ∼100 μM; because this was the top tested concentration of most assays (except the Attagene assays), we considered an AUC of ≥0.1 to be positive. AUC values between 0.001 and 0.1 indicate very weak potential activity and were considered inconclusive. AUC values were rounded to 3 significant digits, and values below 0.001 were truncated and set to zero.
Cytotoxicity Filter
Each antagonist assay that measured suppression of protein production (Tox21_AR_BLA_Antagonist_ratio and Tox21_AR_LUC_MDAKB2_Antagonist) also produced viability readouts measuring cell death. These cytotoxicity assays were analyzed using the ToxCast data analysis pipeline, as described above, and the cytotoxicity AC50 was used as a threshold filter for antagonist activity in a pairwise fashion. Any antagonist response with an AC50 greater than the cytotoxicity AC50 for that chemical assay combination was discarded. Additional filtering approaches that were both more permissive (no exclusion) and more restrictive (exclusion of AC50s within 20% of the cytotoxicity AC50) were investigated, and the corresponding results for the AR pathway model (as well as the paired cytotoxicity data) are included in Supplemental File 4. For ensuring removal of overtly cytotoxic compounds while still permitting analysis of chemicals that may show antagonist behavior at test concentrations immediately preceding cytotoxicity and for maintaining consistency with the criteria for the reference chemical data extracted from the literature, the threshold approach was chosen for this analysis.
Cell Stress Flags
In a global analysis of the ToxCast data set, it was observed that many different types of assays, both cell-based and cell-free, showed a rapid increase in the frequency of responses at concentrations corresponding to regions of cell stress/cytotoxicity.32 We flagged potential nonselective assay hits attributed to cell stress using the distance between the logAC50 (assay) and the median logAC50 (cytotox) with respect to the global cytotoxicity median of the median absolute deviation (MAD) of the logAC50 (cytotox) distributions across all chemicals. Details are given in ref (32). Briefly, for chemicals with two or more positive responses in assays measuring cytotoxicity or inhibition of proliferation, a “Z-score” was calculated for each AR pathway assay hit as
| 6 |
A large Z-score indicates an in vitro assay logAC50 at concentrations significantly below those causing cytotoxicity or inhibiting proliferation. Thus, a hit associated with this Z-score is unlikely to be caused by either cell stress or cytotoxicity-related processes and is more likely to be associated with a target-selective mechanism, e.g., interaction with the AR pathway.
Confirmation Flags
One of the transactivation antagonist in vitro assays, the Tox21 antagonist luciferase assay in the MDAKB2 cell line (Figure 2, A11), was run twice with two different concentrations of the stimulatory ligand R1881. These data were used to help confirm whether chemical activity was specific to the AR pathway. The first time the assay was run, the concentration of ligand R1881 was 10 nM (20× the EC50 of R1881), which resulted in saturation of the assay and a lack of activity for most chemicals, including known weak antagonists, based on the inability to displace the ligand, except for potent steroid antagonists (e.g., flutamide-like compounds). The second time the assay was run with 0.5 nM R1881 and was sensitive to a wider range of chemicals. This second run, with the appropriate R1881 concentration, was included in the AR pathway model, and the data from the first run, with the high R1881 concentration, were used in a paired fashion to examine compound specificity. A system of flags was applied to identify chemicals that may be activating the pathway through a nonreceptor-mediated mechanism. For true positives, it was expected that they would either be a hit in both runs, with a shift in the AC50 (from less to more potent), or they would be negative in the first run (when the assay was saturated with R1881) and a hit in the second run (weak antagonists). The data were flagged if a chemical was active in both runs at similar concentrations or if a potency shift was observed in the opposite direction than would be expected. Significance of the shift between AC50 values was determined using a bootstrapping approach across chemical replicates to define 95% confidence intervals as outlined below and in (Watt et al. 2016, manuscript in preparation), where overlapping confidence intervals were deemed a nonsignificant shift.
Uncertainty Quantification
All concentration–response curves used in the AR Pathway Model were analyzed using the R package toxboot v.0.1.0 (https://cran.r-project.org/web/packages/toxboot/index.html). One thousand bootstrap replicates were generated for each curve using smooth nonparametric bootstrap resampling to obtain a distribution of fit parameters, model selections, and activity calls. Each bootstrap sample was grouped by chemical and analyzed using the AR Pathway Model with the same workflow as used to generate the point estimates, resulting in a distribution of 1000 AUC values per chemical. The inner 95% confidence interval for each chemical AUC value was calculated on this distribution using the quantile function from the R stats package33 with probabilities 0.025 and 0.975 for the lower and upper thresholds of the confidence interval, respectively.
Reference Chemical Identification
We performed a targeted literature search for quantitative data to refine previously published reference chemical lists and provide potency characterization for AR agonism/antagonism. We identified 158 potential reference chemicals with AR agonist or antagonist activity (or lack of activity) from the following international assay validation efforts run by
• Interagency Coordinating Committee on the Validation of Alternative Methods18
• Organization for Economic Cooperation and Development31
• U.S. EPA Endocrine Disruptor Screening Program (EDSP)1
• European Union Reference Laboratory for Alternatives to Animal Testing (EURL ECVAM, ongoing)
• Korean Center for Validation of Alternative Methods (KoCVAM, ongoing)
We conducted semiautomated literature searches for in vitro androgen activity data on the superset of chemicals (n = 158) using PubMatrix (http://pubmatrix.grc.nia.nih.gov/) and Scopus (http://www.scopus.com/). Data from in vitro AR binding and TA assays were extracted from identified references and compiled into a single database (Supplemental File 1). Search keywords are listed in Supplemental File 1. Using a standardized ontology, the following information was recorded for each chemical–study combination:
• PubMed identifier, author, year
• Chemical tested, Chemical Abstracts Service Registry Number (CASRN)
• Table or figure where results were reported
• Hit, response, response notes
• Half-maximal activity concentration (AC50 or IC50), standard error measurement, units
• Assay type (tissue or cell culture), tissue of origin (for cell culture), species of origin
• Receptor information, species source
• Reference androgen or antiandrogen
• Number of concentrations tested, highest concentration tested, units, incubation time
• Binding assays only: binding affinity, dissociation constant, relative binding affinity (RBA)
• TA assays only: agonist or antagonist mode, whether cytotoxicity was evaluated, extent of cytotoxicity observed (i.e., at IC50)
• TA assays only: reporter type, reporter construct, whether construct was native, transient, or stable
Reference Chemical Criteria
To establish reference chemical lists, we examined high-quality AR binding and transactivation (TA) data from the literature search, filtered by conditions such as use of the full length receptor and concurrent measurement of cytotoxicity for antagonist-mode data (detailed in Results). To determine potency categories, we identified all quantitative AR TA assay data reported as AC50 or IC50 that could be converted to μM units and calculated mean, standard deviation, 95% confidence interval, and number of observations for each chemical. Binding data were used in a confirmatory fashion, where chemicals had to have positive binding results in the literature to be included as candidate positive agonist and antagonist reference chemicals. On the basis of the distribution of the results, we defined agonist and antagonist reference chemical lists and potency categories according to the following criteria:
Agonist
Positives: at least three TA experiments of which at least 70% yielded positive TA results and at least one positive binding result
• Strong: mean AC50 ≤ 0.1 μM
• Moderate: 0.1 μM < mean AC50 ≤ 1 μM
• Weak: 1 μM < mean AC50
Negatives: at least three TA experiments yielding negative results and no TA experiments yielding positive results
Antagonist
Positives: at least three TA experiments of which at least 70% yielded positive results that were not due to cytotoxicity and at least one positive binding result
• Strong: mean IC50 ≤ 0.5 μM
• Moderate: 0.5 μM < IC50 ≤ 5 μM
• Weak: 5 μM < mean IC50 ≤ 25 μM
• Very Weak: 25 μM < mean IC50
Negatives: at least two TA experiments yielding negative results and no TA experiments yielding positive results
Chemicals with upper 95% confidence intervals that spanned potency categories were given combined category designations such as “strong/moderate” or “moderate/weak”.
Results
Activity in the AR Pathway Model across the ToxCast Library
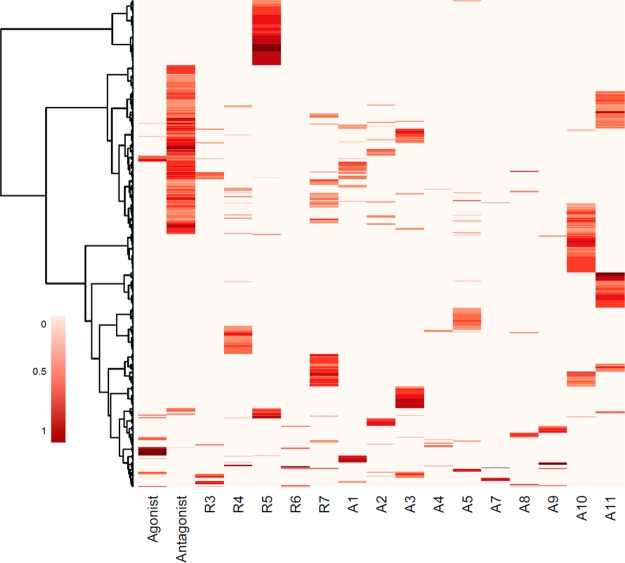
Of the 1855 chemicals tested in all 11 Tox21/ToxCast AR assays, 1461 (78.8%) were predicted to be inactive in the AR pathway model with both agonist (R1) and antagonist (R2) AUC values below 0.001, whereas 220 chemicals (11.9%) were predicted to be either androgen agonists (n = 33) or antagonists (n = 192) with R1 or R2 AUC values > 0.1. Five of the 220 chemicals had significant activity in both agonist (R1) and antagonist (R2) pathways. The remaining 174 chemicals (9.4%) had inconclusive low AR pathway model scores with R1 or R2 AUC values of 0.001 to 0.1. These chemicals were generally weakly active in a small number of assays and were usually also predicted by the model to be acting through interference pathways. Of the 1461 chemicals predicted to be inactive against the AR pathway, 1092 chemicals were inactive across all the assays, whereas 369 chemicals demonstrated activity associated with either assay interference or, less likely, weak activity only picked up in one technology type. Figure 3 shows the distribution of AR model pathway scores across the ToxCast chemical library for 763 chemicals that were active in at least one AR pathway assay. Chemicals were either predicted to act via AR agonism (R1), antagonism (R2), biology-specific interference (R3-R7), or assay-specific interference (A1–A11). Supplemental Figure S1 is a calibration curve to help interpret AUC values in terms of pathway activity concentration, and Supplemental File 2 contains the results for each assay and the AR pathway model (AUC values and associated confidence intervals for agonism, antagonism, and interference) for all 1855 chemicals. Results of the AR pathway model with uncertainty bounds corresponding to 95% confidence intervals are plotted in Supplemental Figure S2.
Figure 3.
Distribution of model AUC values across 763 chemicals. Heatmap shows the distribution of model area under the curve (AUC) values for 763 chemicals that were active in at least one AR pathway assay. The first two columns represent predictions for agonist (R1) and antagonist (R2) activity, and the remaining columns represent predicted assay (A1–11)- or biology (R3–7)-specific interference corresponding to the pathway diagram in Figure 2 and the interference pathways shown in Table 1. The darker red indicates higher AUC values corresponding to more potent activity (scale: 0.001–1). Clustering was done using Ward’s method.34
Literature Search Results
The targeted literature search for AR in vitro reference data yielded 4,795 chemical study pairs across 379 publications. Experimental protocol details and chemical effects were recorded in a standardized manner in a structured data table (Supplemental File 1). AR binding data were identified for 111 chemicals, and the data were compiled from 1261 experiments reported in 166 publications. Commonly used assay platforms included cell culture, tissue preparations, and cell-free systems (Supplemental Figure S3a). The majority of the binding assays used full-length receptors (Supplemental Figure S3b). A total of 26 species were represented among all binding assays with most using human (39%) or rat (33%) receptors. The four most commonly used reference androgens were methyltrienolone (R1881; 475 assays, 41%), 5α-dihydrotestosterone (DHT; 400 assays, 34%), testosterone (203 assays, 17%), and mibolerone (84 assays, 7%). Data from all assays returned in the literature search can be found in Supplemental File 1.
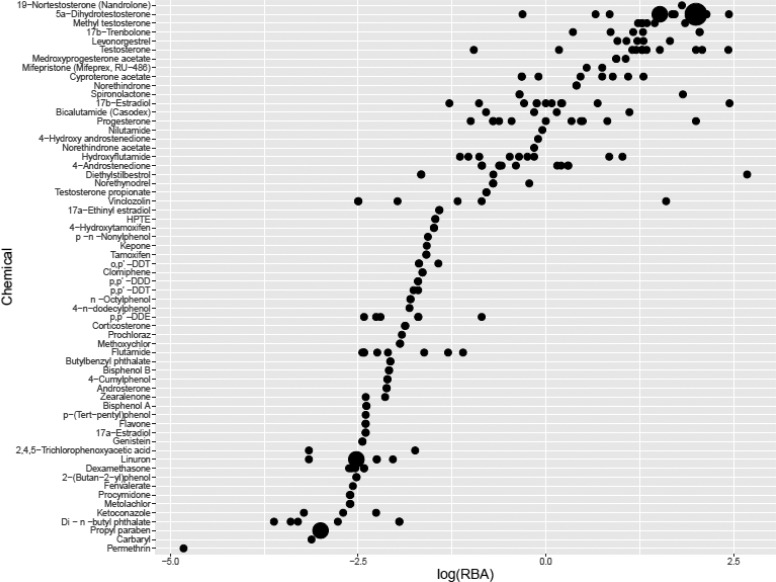
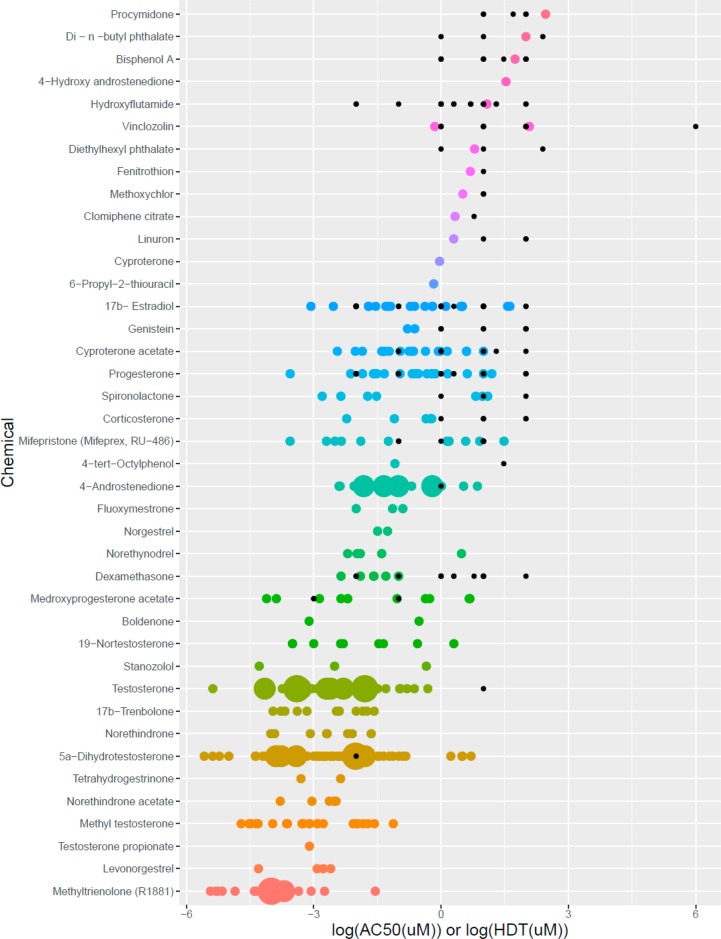
Results from experiments using mutant receptors were excluded. Further analyses were conducted on data from binding assays using the full-length receptor and the ligand-binding domain (957 experiments on 95 chemicals). Multiple positive binding results with no negative results were reported for 38 chemicals. Atrazine, cycloheximide, and 2,4-dinitrophenol had multiple negative binding results and no positive results. There were 14 chemicals with only one positive binding result (and no negatives) and six chemicals with only one negative binding result (and no positives). The remaining 34 chemicals had both positive and negative binding results reported, although there was usually a clear majority of positive or negative results for each chemical. Results for binding affinity were reported in many different formats, the most common being RBA or log RBA relative to a positive control. The relative binding data included R1881 (240 results), DHT (168 results), testosterone (97 results), and mibolerone (30 results). As an example, results for log RBA on 61 chemicals relative to the most common positive control compound, R1881, are shown in Figure 4.
Figure 4.
AR binding affinities relative to R1881 reference. Chemicals are listed along the x-axis; y-axis represents the log10 (RBA). The size of the dot increases with the number of observations (range: 1–15). Relative binding affinity decreases moving from top to bottom with a total of 61 chemicals described. AR = androgen receptor; R1881 = methyltrienolone; RBA = relative binding affinity.
AR transactivation data were compiled for 160 chemicals (3534 experiments from 287 papers). Although six different reporter types were used in the experiments, the majority of experiments used assays with a luciferase reporter (Supplemental Figure S4a). The use of a full-length receptor was also the most common (Supplemental Figure S4b). Many assays used a transiently transfected AR (46%) or stably integrated AR (39%) followed by native receptor expression (14%). Most TA assays used the human AR (93%), but receptors from a total of 14 species were represented among all assays in the database. The most common reference androgens were DHT (2262 assays, 64%), R1881 (703 assays, 20%), and testosterone (395 assays, 11%); the most common reference antiandrogens were flutamide (688 assays, 41%), hydroxyflutamide (487 assays, 30%), bicalutamide (220 assays, 13%), and cyproterone acetate (192 assays, 11%).
Further analyses were conducted on data from the TA assays using the full-length receptor and the ligand-binding domain. Positive and negative TA assay results were reported for 2393 experiments on 133 chemicals. Results were subdivided into modes measuring agonist activity (1447 experiments, 60%) and antagonist activity (946 experiments, 40%). There were 13 chemicals with multiple positive agonist results (i.e., increase in TA) and no antagonist results, all of which also had at least one negative result reported (i.e., no agonist or antagonist activity). However, for most of these chemicals, the number of positive agonist results far outnumbered the number of negative results, which tended to occur in specific cell or receptor types and/or at low concentrations. There were 32 chemicals with multiple positive antagonist results (i.e., decrease in TA) and no agonist results. All of these chemicals also had at least one negative TA result that tended to occur in specific cell types and/or at low concentrations. There were 17 chemicals with multiple negative (inactive for TA) results and no positive (agonist or antagonist) results. There were 15 chemicals with only one TA result in any category. The remaining 56 chemicals had a mix of positive (agonist and/or antagonist) and negative results. However, for most chemicals, there was a clear majority of either agonist or antagonist results.
Potency of Transactivation Agonists
Positive results for TA agonist activity were reported in many different formats and with many different units, the most common being lowest effect level (LEL; 415 results, 49%) and half-maximal activity concentration (AC50; 406 results, 48%). All TA agonist results were converted to log μM units where possible, and the respective agonist potencies based on AC50s for each chemical were compared to negative results in terms of highest dose tested (HDT). The distribution of activity for chemicals with both positive (AC50s, colored dots) and negative (HDTs, black dots) results is shown in Figure 5.
Figure 5.
Comparing AR Transactivation Agonist Results. Chemicals are listed along the x-axes, and the log transformed doses are listed along the y-axis. The colored dots represent positive results in log10 (AC50), and the black dots represent negative results in log10 (HDT). The size of the dot increases with the number of observations (range: 1–79). Agonist potency decreases moving from bottom to top, with a total of 40 chemicals described. AC50 = half-maximal activity concentration; AR = androgen receptor; HDT = highest dose tested.
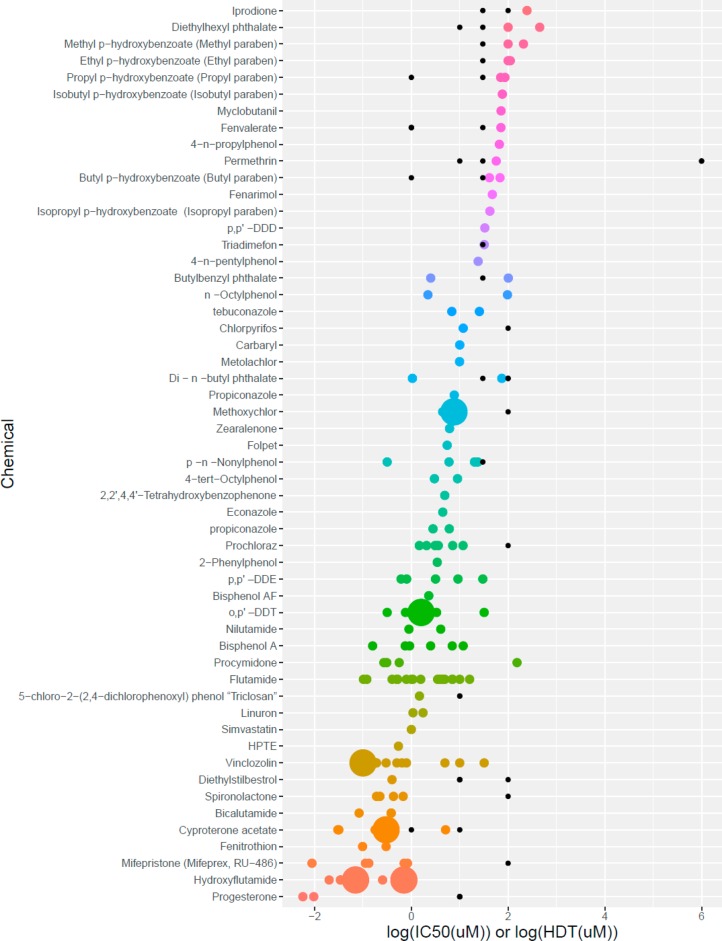
Potency of Transactivation Antagonists
We evaluated AR TA antagonist potency using only data from experiments that concurrently measured cytotoxicity (520 experiments [55%] representing 105 chemicals) with clearly stated acceptance criteria (e.g., <20% loss of viability). Positive results for antagonist activity were reported in many different formats and with many different units, the most common being half-maximal inhibition activity concentration (IC50; 224 results, 64%) and LEL (114 results, 33%). All TA antagonist results were converted to log μM units where possible, and the respective antagonist potencies based on IC50 were compared to the negative results in terms of HDT. The distribution of activity for chemicals with both positive (IC50s, colored dots) and negative (HDTs, black dots) results is shown in Figure 6.
Figure 6.
Comparing AR transactivation antagonist results. Chemicals are listed along the x-axes, and the log transformed doses are listed along the y-axis. The colored dots represent positive results in log10 (IC50), and the black dots represent negative results in log10 (HDT). The size of the dot increases with the number of observations (range: 1–21). Antagonist potency decreases moving from bottom to top with a total of 54 chemicals described. AR = androgen receptor; HDT = highest dose tested; IC50 = half-maximal inhibitory concentration.
AR Pathway In Vitro Reference Chemicals
Based on the criteria outlined in the Methods for reproducibility and consistency of response, we identified 37 reference chemicals for AR agonism and 28 reference chemicals for AR antagonism (Table 2). Initial reference chemical categorizations included strong, moderate, weak and very weak agonists and antagonists, and negative categorizations, all of which were based exclusively on the curated results from the published literature and did not include any information from the ToxCast or Tox21 assays. There were 11 chemicals that fulfilled reference criteria for both agonism and antagonism, usually as a positive reference in one and a negative reference in the other. Cyproterone acetate was classified as both a weak agonist and a moderate antagonist based on multiple literature results showing selective androgen receptor modulation with agonist and antagonist effects. Of the 54 reference chemicals classified based on data from the literature, 46 were among the 1855 chemicals tested in ToxCast/Tox21 and could be used for performance-based external validation of the AR pathway model results.
Table 2. AR Pathway In Vitro Reference Chemicals.
| CASRN | chemical name | agonist potency category | antagonist potency category | in ToxCast 10/2015 release |
|---|---|---|---|---|
| 52806-53-8 | hydroxyflutamide | NA | strong | yes |
| 90357-06-5 | bicalutamide | NA | strong | yes |
| 122-14-5 | fenitrothion | NA | strong | yes |
| 84371-65-3 | mifepristone | NA | strong/moderate | yes |
| 52-01-7 | spironolactone | NA | strong/moderate | yes |
| 63612-50-0 | nilutamide | negative | moderate | yes |
| 427-51-0 | cyproterone acetate | weak | moderate | yes |
| 80-05-7 | bisphenol A | NA | moderate/weak | yes |
| 330-55-2 | linuron | NA | moderate/weak | yes |
| 50471-44-8 | vinclozolin | NA | moderate/weak | yes |
| 13311-84-7 | flutamide | negative | moderate/weak | yes |
| 67747-09-5 | prochloraz | negative | moderate/weak | yes |
| 140-66-9 | 4-tert-octylphenol | NA | weak | yes |
| 72-43-5 | methoxychlor | NA | weak | yes |
| 72-55-9 | p,p′-DDE | NA | weak | yes |
| 60207-90-1 | propiconazole | NA | weak | yes |
| 17924-92-4 | zearalenone | NA | weak | yes |
| 789-02-6 | o,p′-DDT | negative | weak | yes |
| 32809-16-8 | procymidone | NA | very weak | yes |
| 60168-88-9 | fenarimol | negative | very weak | yes |
| 58-18-4 | methyl testosterone | strong | negative | yes |
| 58-22-0 | testosterone | strong | negative | propionate form |
| 63-05-8 | 4-androstenedione | moderate | negative | yes |
| 1912-24-9 | atrazine | negative | negative | yes |
| 52918-63-5 | deltamethrin | negative | negative | yes |
| 486-66-8 | daidzein | NA | negative | yes |
| 16752-77-5 | methomyl | NA | negative | yes |
| 122-34-9 | simazine | NA | negative | yes |
| 10161-33-8 | 17b-trenbolone | strong | NA | yes |
| 797-63-7 | levonorgestrel | strong | NA | yes |
| 965-93-5 | methyltrienolone (R1881) | strong | NA | no |
| 68-22-4 | norethindrone | strong | NA | yes |
| 51-98-9 | norethindrone acetate | strong | NA | no |
| 76-43-7 | fluoxymestrone | strong/moderate | NA | no |
| 434-22-0 | 19-nortestosterone | moderate | NA | no |
| 521-18-6 | 5a-dihydrotestosterone | moderate | NA | yes |
| 10418-03-8 | stanozolol | moderate | NA | no |
| 71-58-9 | medroxyprogesterone acetate | moderate/weak | NA | no |
| 68-23-5 | norethynodrel | moderate/weak | NA | no |
| 57-91-0 | 17a-estradiol | negative | NA | yes |
| 68359-37-5 | b-cyfluthrin | negative | NA | yes |
| 52315-07-8 | b-cypermethrin | negative | NA | yes |
| 17804-35-2 | benomyl | negative | NA | yes |
| 85-68-7 | butylbenzyl phthalate | negative | NA | yes |
| 10605-21-7 | carbendazim | negative | NA | yes |
| 51630-58-1 | fenvalerate | negative | NA | yes |
| 98319-26-7 | finasteride | negative | NA | yes |
| 129453-61-8 | ICI 182,780 | negative | NA | yes |
| 36734-19-7 | iprodione | negative | NA | yes |
| 50-29-3 | p,p′-DDT | negative | NA | yes |
| 52645-53-1 | permethrin | negative | NA | yes |
| 501-36-0 | resveratrol | negative | NA | no |
| 10540-29-1 | tamoxifen | negative | NA | yes |
| 7696-12-0 | tetramethrin | negative | NA | yes |
AR Pathway Model Performance
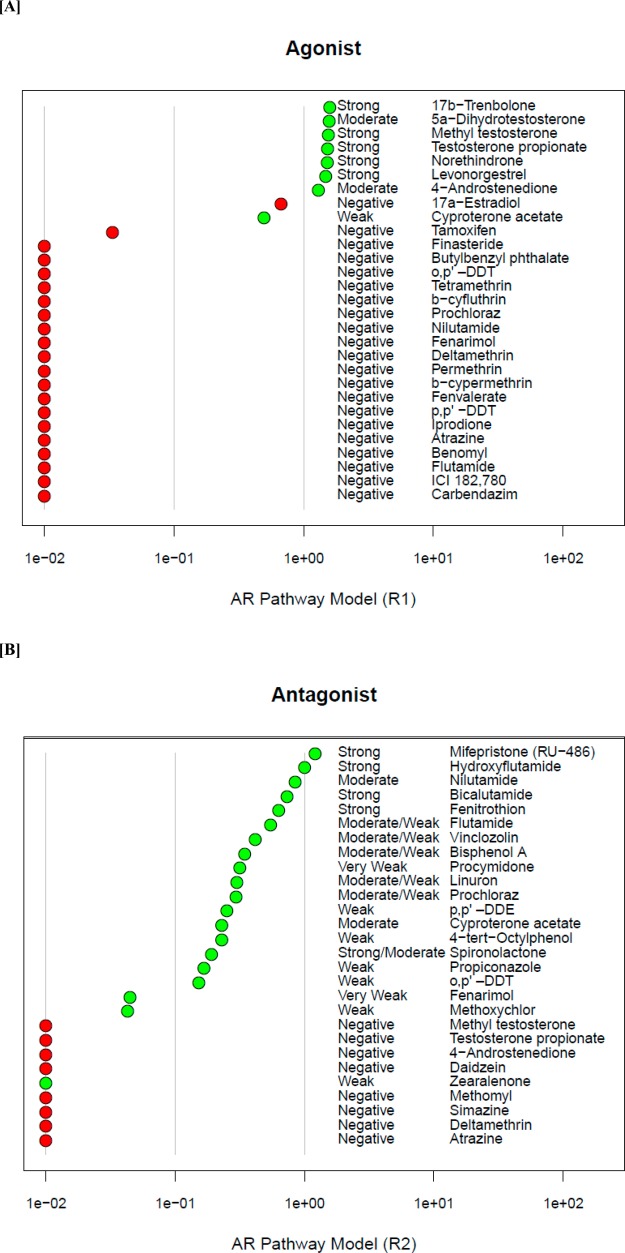
The predicted activity from the AR pathway model for the 46 reference chemicals was compared with the reference potency categories based on curated published results identified in the literature review. The results of the model predictions are shown in Figure 7a (29 agonist reference chemicals) and Figure 7b (28 antagonist reference chemicals). An AR pathway model score greater than 0.1 (approximate activity at concentrations less than 100 μM) was considered positive with higher model scores corresponding to stronger potency. With respect to the AR agonist reference chemicals, 17a-estradiol was the only false positive, and there were no false negatives. One negative agonist reference chemical, tamoxifen, had an inconclusive agonist AUC (R1) score of 0.0335. Following the example of Browne et al. 2015,8 we evaluated the model performance two ways. If inconclusive scores were considered positive, the AR pathway model had a balanced accuracy of 95.2% (100% sensitivity and 90.5% specificity) against the agonist reference chemicals, and if inconclusive results were excluded, the balanced accuracy was 97.5% (100% sensitivity and 95% specificity). Two of the antagonist reference chemicals, methoxychlor (weak potency) and fernarimol (very weak), had antagonist AUC (R2) scores in the inconclusive range of 0.0429 and 0.0446, respectively. Zearalenone, categorized in the literature review as a weak antagonist, was a false negative, and there were no false positives for antagonism. The model predicted that zearalenone was causing assay interference through R7 (corresponding to key event node N5 in Figure 2) because it hit both Tox21 antagonist assays but none of the upstream assays in the antagonist pathway (binding or coregulator recruitment). The AR pathway model had 97.5% balanced accuracy (95% sensitivity and 100% specificity) when predicting the antagonist reference chemicals and counting the inconclusive results as positive or 97.2% balanced accuracy (94.4% sensitivity and 100% specificity) if the inconclusive chemicals were excluded. Examples of the concentration–response curves for several reference chemicals are shown in Figure 8.
Figure 7.
AR pathway model results for reference chemicals. Reference chemicals and associated potency categories (from the literature search) are listed along the y-axes, and the AR pathway model AUC score for (a) agonism (R1) or (b) antagonism (R2) are listed along the x-axes. Green dots represent positive reference chemicals, and red dots represent negative reference chemicals. AR pathway model scores below 0.01 were truncated at 0.01 for plotting purposes. There was one false positive for agonism (17a-estradiol), and one negative agonist reference chemical with an inconclusive model score (tamoxifen). The initial false negative for antagonism (zearalenone) was confirmed as a potential true positive by the antagonist confirmation assay (Tox21_AR_LUC_MDAKB2_Antagonist-confirmation). Two antagonist reference chemicals had AUC scores in the inconclusive region.
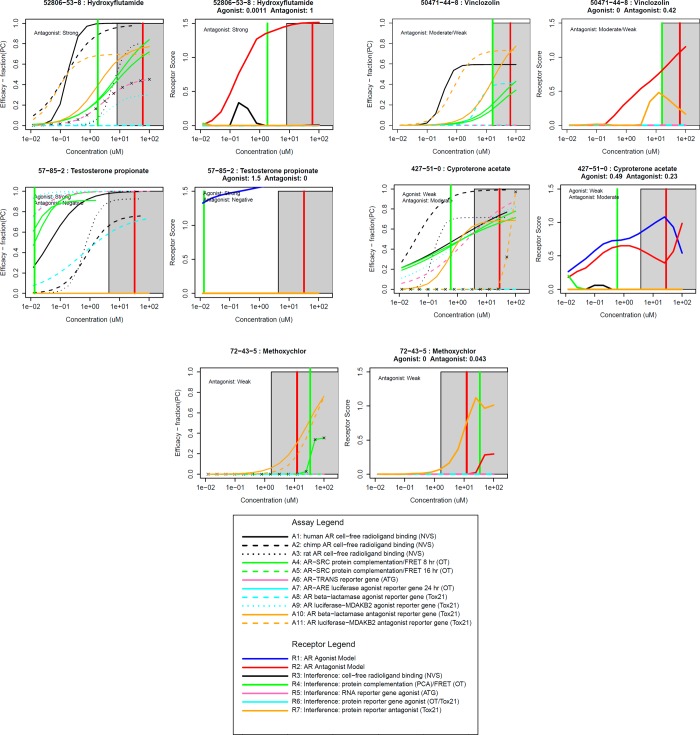
Figure 8.
Concentration response curves and AR pathway model results for selected reference chemicals. For each chemical, the left-hand panel shows the concentration response data for the 11 in vitro assays, colored by assay group as defined in the legend. The right-hand panel shows the magnitude of the modeled “receptor” responses, where the agonist pathway (R1) is in blue and the antagonist pathway (R2) is in red, and the other interference pathways (R3–R7) are colored as defined in the legend. Model AUC values are displayed below the chemical name, and literature-based reference classifications are displayed in the plot. The median cytotoxic concentration for each chemical is indicated by a vertical red line, and the cytotoxicity region (representing 3 median absolute deviations) is indicated by the gray shaded region. A green horizontal bar indicates the median AC50 of the active assays. Similar plots for all chemicals are given in Supplemental File 3.
Distinguishing Antagonism and Cell Stress
The Z-score provides a measure of proximity (how many median absolute deviations) for a chemical’s activity in a particular assay relative to the median concentration for that chemical across 33 viability and proliferation inhibition assays in the ToxCast library.32Z-scores for every chemical assay combination in the AR pathway model are reported in Supplemental File 2. A chemical-assay hit with a high Z-score (>3) indicates that AR-related activity occurred at concentrations far below the cytotoxicity threshold and suggests that there was no evidence of cell stress. These hits are more likely to be associated with specific biomolecular interactions with the intended biological target that the assays are designed to measure. Examples of chemicals with high AUC values for AR antagonism and high average Z-scores include hydroxyflutamide, nilutamide, vinclozolin, linuron, spironolactone, and apigenin. Hits with low Z-scores (activity concentrations in the cell stress/cytotoxicity region) are more likely to be associated with an interference process than hits with high Z-scores. However, because of the variable concentration spacing, quantitative uncertainties in AC50 values, and differential sensitivity among cell types, the Z-score cannot be used as a definitive filter and is instead valuable to provide context on the potential specificity of the results.
Antagonism Confirmation Assay Results
The confirmation assay data from the Tox21_MDAKB2_Luc_Antagonist assay with two different concentrations of stimulating ligand (R1881) provided additional insight into chemicals that were potentially acting via a nonreceptor-mediated mechanism (e.g., generalized cell stress or cytotoxicity) relative to chemicals that appeared to be acting via the AR ligand-binding domain. When considering these data, the one “false negative” reference chemical, zearalenone, displayed behavior indicative of true weak antagonist potential, where it was active in both screens and exhibited a potency shift in the expected direction, although the shift was flagged as not significant due to overlapping confidence intervals around the AC50 values. It is worth noting that zearalenone is predicted to be a fairly potent ER agonist (AUC model score of 0.7116). There were 128 chemicals that were only active when the assay was stimulated with the lower R1881 concentration, behavior that is consistent with the potential for weak antagonism. There were 57 chemicals that were active in both runs and exhibited the expected potency shift with nonoverlapping AC50 confidence intervals. Most of these were predicted as true antagonists by the model, including positive antagonist reference chemicals triclosan and bisphenols A/B/AF. Others (e.g., endosulfan sulfate, dinoseb, fenoxycarb) had inconclusive model scores or were predicted to act via interference pathways, such as suppression of protein production (R7, node N5) because they did not hit the binding or coregulator recruitment assays. There were 128 chemicals that were active in both runs and exhibited the expected potency shift but had overlapping AC50 confidence intervals. There were 65 chemicals that were active in both runs but exhibited a potency shift in the opposite direction (i.e., more potent in the assay with a higher R1881 concentration) and 22 chemicals that were only active in the assay with a higher R1881 concentration and inactive in the other run. These included potently cytotoxic compounds (e.g., gentian violet), cytotostatic compounds (e.g., cycloheximide), organometallics, and pesticides. There were 1455 chemicals that were inactive in both runs, most of which were also inactive against the AR pathway model. Each category of chemical activity is designated by the corresponding “Tox21 Antagonist Confirmation Assay Flag” in Supplemental File 2.
Antagonist Activity Confidence Scoring
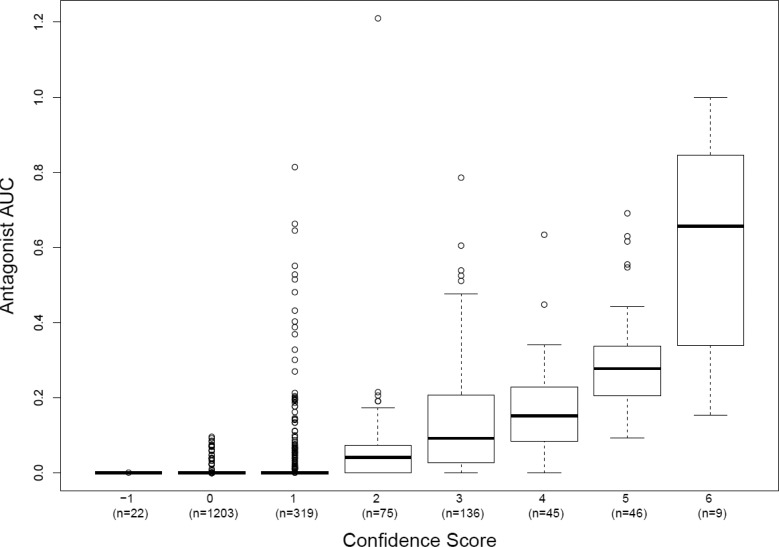
The AR pathway model AUC scores, cytotoxicity information, and confirmation flags were used to inform a simple summary score for each chemical that translates into confidence that the observed activity is via the AR pathway. The schema for assigning confidence scores is shown in Table 3. The default score for inactive chemicals was set to zero. Chemicals with high antagonist (R2) AR pathway model AUC scores were assigned higher confidence scores, as were those chemicals that were active in the concentration region prior to cell stress/cytotoxicity (high average Z-scores across the 11 assays). For potential antagonists, those exhibiting the expected potency shift in the confirmation assays were assigned higher confidence scores, whereas those with data indicating that the chemical was not acting via the receptor were assigned negative confidence scores. The confidence scores from each source were then summed to provide an overall confidence score to facilitate chemical prioritization in a manner that incorporates all the contributing data streams. The positive antagonist reference chemicals all had positive activity confidence scores. All 192 chemicals with R2 AUC values above 0.1 also had positive activity confidence scores, although there were 36 chemicals with low confidence scores (≤2) that were flagged based on the confirmation assay data and may be false positives. Out of the 170 chemicals with inconclusive model antagonist AUC scores (between 0.001 and 0.1), 144 chemicals had positive confidence scores and 61 of these had high confidence scores (≥3). There were 294 chemicals with positive confidence scores that were negative in the AR pathway model (R2 AUC values of 0), some of which were predicted agonists, and most of which were predicted to act via interference receptors. Of those 294 model negative chemicals, there were 26 chemicals with confidence scores ≥3, which may have been missed by the model and should be examined further for potential antagonist activity. There were 1225 chemicals with activity confidence scores ≤0, meaning that they were either inactive, caused technology-specific interference, or displayed activity indicative of a non-AR-mediated response (usually cytotoxicity driven). The distribution of AR pathway model antagonist AUC values across the different confidence scoring bins is shown in Figure 9.
Table 3. Schema for Antagonist Activity Confidence Scoring.
| source | criteria | confidence score contributiona |
|---|---|---|
| AR pathway model | AUC.R2 > 0.1 | 2 |
| 0.1 > AUC.R2 > 0.001 | 1 | |
| cell stress/cytotoxicity flag | average Z-score > 3 | 1 |
| confirmation assay data | true antagonist shift (hit/hit) | 3 |
| true antagonist shift (no hit/hit) | 2 | |
| FLAG: true antagonist shift but CI overlap | 1 | |
| FLAG: wrong direction shift (hit/hit) | –1 | |
| FLAG: wrong direction (hit/no hit) | –1 |
Contributions from the three source categories are summed to provide an overall antagonist activity confidence score ranging from −1 to 6.
Figure 9.
AR pathway model antagonist AUC distribution by confidence score.
Comparison with U.S. EPA EDSP Tier 1 AR Binding Assay
The current high-throughput AR pathway model, with 11 assays covering five key events, is intended as a potential alternative for the existing low-throughput EDSP Tier 1 AR binding assay covering one key event. Going beyond the binding assay, the model provides functional information, i.e., agonist versus antagonist activity. In addition, the complementarity of the assays helps overcome assay-specific interferences that can yield false positive and false negative results. There are a total of 101 chemicals with data from the EDSP Tier 1 AR binding assay and data from the current AR pathway model. Tier 1 AR binding data came from two sources: the ICCVAM assay validation document18 and results from the first set of test orders issued by the U.S. EPA EDSP, referred to as “List 1”.35 The Tier 1 assay measured binding rather than agonism or antagonism, so for comparison, we called a chemical model positive if the maximum of the agonist or antagonist AUC values was ≥0.1, negative if the maximum was <0.001, and inconclusive if the maximum AUC was between 0.001 and 0.1. For ICCVAM, RBA values were reported (IC50R1881 × 100/IC50test chemical), and for the List 1 chemicals, both RBA and IC50 values were reported. To facilitate comparison, we developed a calibration curve using the List 1 chemicals based on an observed linear relationship between log(IC50) and log(RBA), which allowed us to estimate IC50 values from RBAs for ICCVAM chemicals. A linear model (shown in Supplemental Figure S5) between the two yielded a root-mean-square error (RMSE) of 0.25 and coefficient of determination (R2) of 0.84 with both slope and intercept of approximately −1. All data on these comparisons is given in Supplemental File 5. Of the 101 chemicals, seven had equivocal calls in the Tier 1 data and six had inconclusive AR pathway model scores (1 chemical overlap), yielding 89 chemicals with comparable data.
Of the 39 List 1 chemicals with both List 1 AR binding assay data and AR model scores, two were positive in both, six were model positive and Tier 1 negative, seven were model negative and Tier 1 positive, and 24 were negative in both. The List 1 positive and AR model negative chemicals are 2-phenylphenol, carbaryl, diazinon, dichlobenil, metolachlor, myclobutanil, and phosmet. With the exception of phosmet, the IC50 values for these chemicals are well over 100 μM, and so would be expected to be negative in the model, as the top tested concentrations in ToxCast and Tox21 were ≤100 μM. The IC50 for phosmet for binding was 10 μM in Tier 1, in close agreement with the chimp AR binding assay (A2) AC50 of 18 μM in the AR model data; however, the human and rat binding assays did not yield positive hit calls when tested to 40 μM. Phosmet was negative in the AR model data transactivation assays in agreement with a previous published report.36 The model positive/List 1 negative chemicals are abamectin, captan, chlorothalonil, folpet, MGK-264, and propargite. All of these are classified as antagonists in the model with AUC antagonist values ranging from 0.09 to 0.48. However, all of these chemicals are flagged as potential false positives using the antagonist confirmation assay data based on either a potency shift in the wrong direction (abamectin, chlorothalonil, folpet, propargite) or no significant shift (captan, MGK-264). The two chemicals called positive in both approaches are propiconazole and tebuconazole. Both of these were classified in the model as antagonists, and both had significant shifts in the correct direction in the confirmation antagonist assay. In summary, the model positive/List 1 negative chemicals are likely all false positives in the model, but this was detected using the confirmation assay. The model negative, List 1 positive chemicals are all so weak that they would not be detected by the HTS assays used in the model because of the upper testing concentration of 100 μM with the possible exception of phosmet for which no clear call can be made. The model results, including uncertainty bounds, for all the List 1 chemicals are shown in Supplemental Figure S6.
There were 51 chemicals with data from the model and the ICCVAM validation set for the Tier 1 AR binding assay (atrazine was also on List 1). Of these, 22 were positive in both, 9 were model positive and Tier 1 negative, 1 was model negative and Tier 1 positive, and 19 were negative in both. This yields a sensitivity and specificity of 0.96 and 0.68, respectively. The single ICCVAM chemical that was model negative and Tier 1 positive was atrazine with an RBA of 0.0018, yielding a modeled IC50 of 53 μM, which is near the upper limit of HTS testing. Atrazine was also evaluated in the List 1 process using literature data, which yielded equivocal results but an ultimate List 1 call of inactive. The 10 model positive, Tier 1 negative chemicals are 17a-estradiol, 4-cumylphenol, apigenin, bisphenol B, clomiphene citrate, cycloheximide, fulvestrant, meso-hexestrol, oxazepam, and reserpine. All of these were classified as antagonists except for 17a-estradiol and oxazepam, although the former had an agonist AUC (R1) of 0.67 and antagonist AUC (R2) of 0.09. Of these chemicals, four had a significant shift in the correct direction in the antagonist confirmation assay (17a-estradiol, 4-cumylphenol, apigenin, bisphenol B), and three had a shift in the correct direction but with overlapping confidence intervals (clomiphene citrate, meso-hexestrol, reserpine). Cycloheximide had a shift in the wrong direction. Fulvestrant and oxazepam also had significant activity in interference channels and thus are likely active due to assay interference. In summary, among these model positive, Tier 1 negative chemicals, the model data support true activity for 17a-estradiol (mixed agonist/antagonist) and 4-cumylphenol, apigenin, and bisphenol B (antagonists). Note that these are all estrogen receptor agonists. Additionally, in the ICCVAM listing, these are noted as “presumed negative”. The remaining six chemicals show evidence for false-positive activity in the model. The model results, including uncertainty bounds, for all the ICCVAM chemicals are shown in Supplemental Figure S7.
Discussion
Implementation of HTS inToxCast and Tox21 has generated high-quality quantitative data on thousands of chemicals and potential environmental pollutants. The inclusion of orthogonal assays that query key events along a biological pathway in multiple ways has produced novel hazard screening capabilities. A similar mechanistic network model to the one presented here is already being used by the U.S. EPA EDSP to identify potential endocrine disruptors acting via estrogen agonism.9 The ER pathway model was validated against a well-defined set of reference chemicals,8,16 which heretofore was not possible for the AR pathway due to the lack of a well-characterized reference chemical set. In this study, we have reported the results from a comprehensive literature review on potential AR reference chemicals and used the resulting set to evaluate the performance of the AR pathway model based on 11 Tox21/ToxCast assays.
Every assay has inherent limitations driven by technological specifications and an applicability domain. A biological pathway-based approach that integrates multiple assays mapping to key upstream and downstream events provides a weight of evidence for the true potential of a chemical to activate or repress signaling, in this case via the AR. This type of additive model compensates for the individual shortcomings of any one assay. For example, there were 105 chemicals that were predicted to act through a receptor interference pathway (A7, Figure 2) because they were only active in the OT_AR_ARELUC_AG_1440 luciferase reporter gene assay measuring downstream transcriptional activation via protein production. None of these chemicals are known to be AR agonists, so it is likely that their activity was correctly flagged as interference and may have been a result of nonspecific transcriptional effects. Alternatively, these specific samples may have had cross-contamination from strong reference chemicals during the experimental protocol. There are also a large number of chemicals that produced hits in one or more of the cell-free receptor binding assays and were therefore predicted as A1–A3 or R3. Many of these chemicals are surfactants, indicating that these chemicals may have reacted with the proteins or otherwise caused denaturation, leading to displacement of the radioligand and a binding-like signal.
Cytotoxicity and response specificity were further considered and flagged based on chemical patterns across viability assays (i.e., Z-score) and confirmation assay data. An important point about the Z-score is that, in practice, it is more useful as a flag than an absolute cutoff. In the ToxCast data analysis pipeline, there are additional types of flags, e.g., to indicate noisy data or hits due to a single point crossing the statistical threshold for activity. These do not change the hit call but provide the user a set of cautions or warnings when evaluating data for a particular chemical assay pair.27 Similarly, the analysis of the confirmation assay data produces a set of flags that instills more or less confidence in true AR antagonist behavior. The initial Tox21_MDAKB2_Luc_Antagonist assay run with a stimulatory R1881 concentration of 10 nM (∼20× EC50) identified predominately only the strong antagonists, i.e., steroid pharmaceuticals, that could compete with the high agonist concentration, and many of the weak environmental antiandrogens were inactive. The assay run with 0.5 nM of R1881 (∼EC50) identified many more of the weak antagonists. The shift in potency between the two conditions was useful for identifying indirect inhibitors of the assay signal. Chemicals that had high model scores for antagonism (R2 AUC > 0.1) but were flagged for a lack of a potency shift in the confirmation results may not actually be acting through the AR but rather through generalized cell stress or technology interference. Examples of chemicals in this group include the dyes basic blue 7, rhodamine 6G, and FD&C green No. 3l, the organometallics tributyltin methacrylate and zinc pyrithione, and the pesticides abamectin and propargite. Conversely, chemicals that were missed by the binding (A1–A3) and coregulator recruitment (A4 and A5) assays, but exhibited a potency shift in the confirmation data, may have been incorrectly predicted by the model as acting through interference pathways (e.g., R7, corresponding to activity in only A10 and A11). It is also possible that some antagonists may bind outside the ligand binding domain, otherwise block dimerization, or act on some later step in the pathway. For example, a group of seven conazoles were classified as antagonists by the AR pathway model, had activity in both runs of the Tox21_MDAKB2_Luc_Antagonist assay, and a corresponding significant potency shift. Another six had a shift in the correct direction but the confidence intervals for the two AC50s overlapped. A clear shift in the confirmation assay data may be sufficient evidence of AR-mediated activity, regardless of model score. Chemicals with this type of response that may have been missed by the model were identified and prioritized by the activity confidence scoring system.
Having 11 diverse orthogonal assays along the AR pathway protects against spurious results being driven by one particular technology type. This is evident when considering the excellent performance of the AR pathway model (>95% for both agonism and antagonism) against the reference chemicals. An interesting exception is the putative reference chemical 17a-estradiol, which was classified negative for AR agonism based on multiple literature results; however, the literature HDTs were ≤10 μM. All 11 Tox21/ToxCast AR assays were activated by 17a-estradiol (AC50/IC50 range 0.1–10 μM), resulting in a model prediction of both agonist and antagonist activity. These results could be indicative of true selective AR modulation by this chemical or heightened sensitivity of the HTS assays to strong steroid pharmaceuticals. With the publication of these analyses, and the availability of the ToxCast and Tox21 data (https://www.epa.gov/chemical-research/toxicity-forecasting), the reference chemical list can be updated to reflect the contribution of these assays to the body of published literature. We refrained from doing so here to provide an external validation for the current AR pathway model, but future work could incorporate the ToxCast, Tox21, and other assays into an expanded reference chemicals list. In that case, the contradictory results between the literature analysis and the ToxCast/Tox21 data would suggest removal of 17a-estradiol from future negative reference classifications if the source of crosstalk, whether it is biological or technological, can be determined. Another potential lesson learned from validating the AR pathway model against the reference chemicals concerns the threshold for positive activity. Two of the weak/very weak antagonist reference chemicals had AUC values in the inconclusive range, around 0.04, due to lack of activity in the binding assays. A limitation of the binding assays specifically is that chemicals were only tested in concentration response if they were active in a single high-concentration screen. Both of these chemicals (fenarimol and methoxychlor) had similar profiles against the remainder of the pathway with activity at 30–40 μM in one of the coregulator recruitment assays (A5) and both of the Tox21 antagonist assays (A10 and A11). Depending on the application and the desire to minimize false negatives in a regulatory setting, the threshold could be adjusted to consider all nonzero model values; chemicals with both inconclusive and positive AR pathway activity would then be prioritized for further testing.
Here, we presented a comparison of the AR pathway model integrating 11 HTS assays and the existing in vitro AR binding assay in the U.S. EPA EDSP Tier 1 battery. The overall summary of the comparison between the model and the Tier 1 AR binding assay is that the model correctly identifies binders with potency in the tested range (IC50 under 100 μM) but yields a significant number of false positives, especially as putative antagonists. However, most of these are identified as false positives using a combination of the antagonist confirmation assay and examination of assay interference channels. Finally, the model provides evidence in contradiction to the ICCVAM designations for at least four chemicals (17a-estradiol, 4-cumylphenol, apigenin, and bisphenol B), which should prompt further investigation. Further comparison with U.S. EPA EDSP Tier 1 results, including the in vivo Hershberger assay37 for AR agonists and antagonists, may help in understanding the relative performance of the AR pathway model based on ToxCast and Tox21 assays. Like the ER model,8 it appears the AR pathway model is more sensitive and also more quantitative than the EDSP Tier 1 assays based on the diversity of the 11 HTS assays and the computational network that integrates those data. The model and associated assays cover a broader range of biological processes than the Tier 1 binding assay and therefore yield a stronger weight of evidence for true AR agonist or antagonist activity.
Limitations of this model, and most HTS-based approaches, include the lack of or limited metabolic capacity of the systems and the restriction to chemicals that are DMSO soluble. There is a current challenge for the scientific community to tackle the issue of incorporating metabolism (http://www.transformtoxtesting.com/), and structure-based models are under development to identify chemicals predicted to undergo transformation to more bioactive metabolites. Future plans also include expanding chemical testing to a water-soluble library. Further, although the HTS results and computational model predictions have demonstrated the ability to effectively prioritize environmental compounds for endocrine disrupting potential, they should be integrated with exposure estimates for decision making in a risk assessment framework.9,38,39
For ultimately interpreting AR pathway activity and other mechanistic events in a biological framework that includes potentially adverse in vivo outcomes, efforts are underway to establish reference chemicals for additional end points and map these to adverse outcome pathways. It is important to note that the reference chemicals presented here are for agonist or antagonist behavior mediated through the AR, and some chemicals may have other endocrine relevant effects via pathways such as steroidogenesis. Following the example of the uterotrophic database,17 work is ongoing to compile in vivo androgen and antiandrogen data from the U.S. EPA EDSP Tier 1 Hershberger assay.37 Experimental reverse toxicokinetic measurements are being used to parametrize models for in vitro-to-in vivo extrapolation to facilitate a direct comparison to demonstrated effects in vivo and administered doses.40−44 These efforts can be used to validate additional high-throughput in vitro assays, and for some chemical classes, integration of HTS assays and computational models may be adequate to predict more apical developmental and reproductive effects.
Conclusions
We have compiled a database of literature results that includes a wide array of AR binding and transactivation data and used it to characterize a range of potential AR agonist and antagonist reference chemicals. The proposed reference chemical lists and associated potency categories can be used for current and future test method evaluations and will be submitted to OECD via the Validation Management Group–Non-Animal to facilitate international harmonization. The AR pathway model based on results from 11 Tox21/ToxCast HTS assays was validated against this independently curated set of reference chemicals and shown to be over 95% accurate for predicting both AR agonism and antagonism. The Tox21 confirmation assay data assisted in identifying chemicals that exhibited a shift in potency indicative of a true AR antagonist response and can be combined with cytotoxicity information to contextualize the AR pathway model results. A number of environmental chemicals were identified as potential AR antagonists, with varying degrees of confidence, and should be examined in the context of human and environmental exposures, metabolism, and persistence to characterize the risk of endocrine disruption and adverse outcomes in humans or wildlife.
Acknowledgments
The authors are grateful to Drs. Katie Paul-Friedman and Michael DeVito for their insightful comments, to Dr. Phillip Hartig for the material to run the chimpanzee AR assay, and to Drs. Robert Kavlock, Kevin Crofton, Stan Barone, and John Bucher for their feedback, leadership, and support. This manuscript has been cleared for publication by the U.S. EPA Office of Research and Development, the Office of Science Coordination and Policy, and the NIEHS Division of the National Toxicology Program. The opinions expressed and arguments employed herein are those of the authors and do not necessarily reflect the official views of the OECD, the governments of its member countries, or the official policy of any federal agency.
Glossary
Abbreviations
- AC50
half-maximal activity concentration
- AR
androgen receptor
- AUC
area under the curve
- EDSP
Endocrine Disruptor Screening Program
- EPA
U.S. Environmental Protection Agency
- ER
estrogen receptor
- HDT
highest dose tested
- HTS
high-throughput screening
- IC50
half-maximal inhibitory concentration
- ICCVAM
Interagency Coordinating Committee for the Validation of Alternative Methods
- OECD
Organization for Economic Cooperation and Development
- RBA
relative binding affinity
- TA
transactivation
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.chemrestox.6b00347.
Results for the AR pathway model on 1855 chemicals as well as the results from the literature search for AR reference data; calibration curves to facilitate translation of AR pathway model AUC values into AC50s and to estimate IC50 values from RBAs for the EDSP List 1 chemicals, as well as comparisons between alternate filtering approaches using paired cytotoxicity results; and uncertainty results given in the form of confidence intervals around AUC values for all 1855 chemicals, EDSP List 1, and ICCVAM chemicals (PDF)
Comparison of the results for the chemical groups (PDF)
AR reference literature database and associated literature search keywords used to identify references with in vitro AR binding and TA assays (XLSX)
AR pathway model (AUC values and associated confidence intervals for agonism, antagonism, and interference) for all 1855 chemicals (XLSX)
Results of the AR Pathway Model for all 1855 chemicals (PDF)
Cytotoxicity filtering information and additional filtering approaches (XLSX)
Data on comparisons between EDSP Tier 1 binding assays and AR Pathway model results (XLSX)
This work was funded by the U.S. EPA and the NIEHS. Contractual support was provided to NICEATM by Integrated Laboratory Systems under NIEHS Contract No. HHSN273201500010C.
The authors declare no competing financial interest.
Supplementary Material
References
- U.S. EPA, Endocrine Disruptor Screening Program (EDSP) Overview. Online: https://www.epa.gov/endocrine-disruption/endocrine-disruptor-screening-program-edsp-overview (accessed November 10, 2015), 2012.
- U.S. EPA, Endocrine Disruptor Screening Program Universe of Chemicals and General Validation Principles. Online: http://www.epa.gov/endo/pubs/edsp_chemical_universe_and_general_validations_white_paper_11_11.pdf (accessed November 10, 2015), 2012.
- U.S. EPA, The Incorporation of In Silico Models and In Vitro High Throughput Assays in the Endocrine Disruptor Screening Program (EDSP) for Prioritization and Screening. Summary Overview. A Part of the EDSP Comprehensive Management Plan Online: http://epa.gov/endo/pubs/edsp21_work_plan_summary_overview_final.pdf (accessed November 10, 2015), 2011.
- Kavlock R.; Chandler K.; Houck K.; Hunter S.; Judson R.; Kleinstreuer N.; Knudsen T.; Martin M.; Padilla S.; Reif D.; Richard A.; Rotroff D.; Sipes N.; Dix D. (2012) Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol. 25 (7), 1287–302. 10.1021/tx3000939. [DOI] [PubMed] [Google Scholar]
- Tice R. R.; Austin C. P.; Kavlock R. J.; Bucher J. R. (2013) Improving the human hazard characterization of chemicals: a Tox21 update. Environ. Health Perspect. 121 (7), 756–65. 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. S.; Philbert M. A.; Auerbach S. S.; Wetmore B. A.; Devito M. J.; Cote I.; Rowlands J. C.; Whelan M. P.; Hays S. M.; Andersen M. E.; Meek M. E.; Reiter L. W.; Lambert J. C.; Clewell H. J. 3rd; Stephens M. L.; Zhao Q. J.; Wesselkamper S. C.; Flowers L.; Carney E. W.; Pastoor T. P.; Petersen D. D.; Yauk C. L.; Nong A. (2013) Incorporating new technologies into toxicity testing and risk assessment: moving from 21st century vision to a data-driven framework. Toxicol. Sci. 136 (1), 4–18. 10.1093/toxsci/kft178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard A. M.; Judson R. S.; Houck K. A.; Grulke C. M.; Volarath P.; Thillainadarajah I.; Yang C.; Rathman J.; Martin M. T.; Wambaugh J. F.; Knudsen T. B.; Kancherla J.; Mansouri K.; Patlewicz G.; Williams A. J.; Little S. B.; Crofton K. M.; Thomas R. S. (2016) ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem. Res. Toxicol. 29 (8), 1225–1251. 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- Browne P.; Judson R. S.; Casey W. M.; Kleinstreuer N. C.; Thomas R. S. (2015) Screening Chemicals for Estrogen Receptor Bioactivity Using a Computational Model. Environ. Sci. Technol. 49 (14), 8804–14. 10.1021/acs.est.5b02641. [DOI] [PubMed] [Google Scholar]
- U.S. EPA, Use of High Throughput Assays and Computational Tools: Endocrine Disruptor Screening Program; Notice of Availability and Opportunity for Comment, 80 Fed. Reg. 118 (June 19, 2015). Prevention, O. o. C. S. a. P., Ed. Online: https://www.federalregister.gov/articles/2015/06/19/2015-15182/use-of-high-throughput-assays-and-computational-tools-endocrine-disruptor-screening-program-notice (accessed March 3, 2016), 2015.
- Auld D. S.; Thorne N.; Nguyen D. T.; Inglese J. (2008) A specific mechanism for nonspecific activation in reporter-gene assays. ACS Chem. Biol. 3 (8), 463–70. 10.1021/cb8000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns R. F.; Watson I. A. (2012) Rules for identifying potentially reactive or promiscuous compounds. J. Med. Chem. 55 (22), 9763–72. 10.1021/jm301008n. [DOI] [PubMed] [Google Scholar]
- Hsieh J. H.; Sedykh A.; Huang R.; Xia M.; Tice R. R. (2015) A Data Analysis Pipeline Accounting for Artifacts in Tox21 Quantitative High-Throughput Screening Assays. J. Biomol. Screening 20 (7), 887–97. 10.1177/1087057115581317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese J.; Johnson R. L.; Simeonov A.; Xia M.; Zheng W.; Austin C. P.; Auld D. S. (2007) High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 3 (8), 466–79. 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- Thorne N.; Auld D. S.; Inglese J. (2010) Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr. Opin. Chem. Biol. 14 (3), 315–24. 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. R.; Thanabal V.; Melnick M. M.; Lall M.; Donovan C.; Sarver R. W.; Lee D. Y.; Ohren J.; Emerson D. (2010) The use of biochemical and biophysical tools for triage of high-throughput screening hits - A case study with Escherichia coli phosphopantetheine adenylyltransferase. Chem. Biol. Drug Des. 75 (5), 444–54. 10.1111/j.1747-0285.2010.00957.x. [DOI] [PubMed] [Google Scholar]
- Judson R. S.; Magpantay F. M.; Chickarmane V.; Haskell C.; Tania N.; Taylor J.; Xia M.; Huang R.; Rotroff D. M.; Filer D. L.; Houck K. A.; Martin M. T.; Sipes N.; Richard A. M.; Mansouri K.; Setzer R. W.; Knudsen T. B.; Crofton K. M.; Thomas R. S. (2015) Integrated Model of Chemical Perturbations of a Biological Pathway Using 18 In Vitro High-Throughput Screening Assays for the Estrogen Receptor. Toxicol. Sci. 148 (1), 137–54. 10.1093/toxsci/kfv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Masri H.; Kleinstreuer N.; Hines R. N.; Adams L.; Tal T.; Isaacs K.; Wetmore B. A.; Tan Y.-M. (2016) Integration of Life-Stage Physiologically Based Pharmacokinetic Models with Adverse Outcome Pathways and Environmental Exposure Models to Screen for Environmental Hazards. Toxicol. Sci. 152 (1), 230–243. 10.1093/toxsci/kfw082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICCVAM, ICCVAM Evaluation of In Vitro Test Methods for Detecting Potential Endocrine Disruptors: Estrogen Receptor and Androgen Receptor Binding and Transcriptional Activation Assays; NICEATM, Ed.: Research Triangle Park, NC, 2003. [Google Scholar]
- Knudsen T. B.; Houck K. A.; Sipes N. S.; Singh A. V.; Judson R. S.; Martin M. T.; Weissman A.; Kleinstreuer N. C.; Mortensen H. M.; Reif D. M.; Rabinowitz J. R.; Setzer R. W.; Richard A. M.; Dix D. J.; Kavlock R. J. (2011) Activity profiles of 309 ToxCast chemicals evaluated across 292 biochemical targets. Toxicology 282 (1–2), 1–15. 10.1016/j.tox.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Sipes N. S.; Martin M. T.; Kothiya P.; Reif D. M.; Judson R. S.; Richard A. M.; Houck K. A.; Dix D. J.; Kavlock R. J.; Knudsen T. B. (2013) Profiling 976 ToxCast Chemicals across 331 Enzymatic and Receptor Signaling Assays. Chem. Res. Toxicol. 26 (6), 878–895. 10.1021/tx400021f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig P. C.; Cardon M. C.; Blystone C. R.; Gray L. E. Jr.; Wilson V. S. (2008) High throughput adjustable 96-well plate assay for androgen receptor binding: a practical approach for EDC screening using the chimpanzee AR. Toxicol. Lett. 181 (2), 126–131. 10.1016/j.toxlet.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Martin M. T.; Dix D. J.; Judson R. S.; Kavlock R. J.; Reif D. M.; Richard A. M.; Rotroff D. M.; Romanov S.; Medvedev A.; Poltoratskaya N.; Gambarian M.; Moeser M.; Makarov S. S.; Houck K. A. (2010) Impact of environmental chemicals on key transcription regulators and correlation to toxicity end points within EPA’s ToxCast program. Chem. Res. Toxicol. 23 (3), 578–90. 10.1021/tx900325g. [DOI] [PubMed] [Google Scholar]
- Lynch C.; Sakamuru S.; Huang R.; Stavreva D.; Varticovski L.; Hager G.; Judson R.; Houck K.; Kleinstreuer N. C.; Casey W. M.; Paules R. S.; Simeonov A.; Xia M. (2016) Identifying Environmental Chemicals as Agonists of the Androgen Receptor by Applying a Quantitative High-throughput Screening Platform. Toxicology in submission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R.; Xia M.; Cho M. H.; Sakamuru S.; Shinn P.; Houck K. A.; Dix D. J.; Judson R. S.; Witt K. L.; Kavlock R. J.; Tice R. R.; Austin C. P. (2011) Chemical genomics profiling of environmental chemical modulation of human nuclear receptors. Environ. Health Perspect. 119 (8), 1142–8. 10.1289/ehp.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig P. C.; Bobseine K. L.; Britt B. H.; Cardon M. C.; Lambright C. R.; Wilson V. S.; Gray L. E. (2002) Development of Two Androgen Receptor Assays Using Adenoviral Transduction of MMTV-Luc Reporter and/or hAR for Endocrine Screening. Toxicol. Sci. 66 (1), 82–90. 10.1093/toxsci/66.1.82. [DOI] [PubMed] [Google Scholar]
- Wilson V. S.; Bobseine K.; Lambright C. R.; Gray L. E. (2002) A Novel Cell Line, MDA-kb2, That Stably Expresses an Androgen- and Glucocorticoid-Responsive Reporter for the Detection of Hormone Receptor Agonists and Antagonists. Toxicol. Sci. 66 (1), 69–81. 10.1093/toxsci/66.1.69. [DOI] [PubMed] [Google Scholar]
- EPA/NCCT iCSS ToxCast Dashboard: database dev_invitrodb. https://actor.epa.gov/dashboard/ (data extracted 8/4/2016).
- Filer D. The ToxCast Analysis Pipeline: An R Package for Processing and Modeling Chemical Screening Data. https://www3.epa.gov/research/COMPTOX/R_Programming_Language_Nov_2015/Pipeline_Overview.pdf (accessed August 10, 2016).
- Akaike H.Information theory and an extension of the maximum likelihood principle; Springer: New York, NY, 1998. [Google Scholar]
- Gronemeyer H.; Gustafsson J. A.; Laudet V. (2004) Principles for modulation of the nuclear receptor superfamily. Nat. Rev. Drug Discovery 3 (11), 950–64. 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- OECD, Draft Report of Pre-validation and Inter-laboratory Validation For Androgen Receptor (AR) Mediated Stably Transfected Transcriptional Activation (AR-STTA) Assay to Detect Androgenic and Anti-androgenic Activities Online: http://www.oecd.org/chemicalsafety/testing/46755800.pdf (accessed November 10, 2015), 2010.
- Judson R.; Houck K.; Martin M.; Richard A. M.; Knudsen T. B.; Shah I.; Little S.; Wambaugh J.; Woodrow Setzer R.; Kothya P.; Phuong J.; Filer D.; Smith D.; Reif D.; Rotroff D.; Kleinstreuer N.; Sipes N.; Xia M.; Huang R.; Crofton K.; Thomas R. S. (2016) Editor’s Highlight: Analysis of the Effects of Cell Stress and Cytotoxicity on In Vitro Assay Activity Across a Diverse Chemical and Assay Space. Toxicol. Sci. 152 (2), 323–39. 10.1093/toxsci/kfw092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R: CoreTeam R: A language and environment for statistical computing. https://www.r-project.org/.
- Ward J. H. (1963) Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 58 (301), 236–244. 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
- U.S. EPA, Endocrine Disruptor Screening Program Tier 1 Screening Determinations and Associated Data Evaluation Records. Online: https://www.epa.gov/endocrine-disruption/endocrine-disruptor-screening-program-tier-1-screening-determinations-and (accessed August 8, 2016), 2016.
- Kojima H.; Katsura E.; Takeuchi S.; Niiyama K.; Kobayashi K. (2004) Screening for estrogen and androgen receptor activities in 200 pesticides by in vitro reporter gene assays using Chinese hamster ovary cells. Environ. Health Perspect. 112 (5), 524–31. 10.1289/ehp.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA, Hershberger Assay, OCSPP Guideline 890.1400. Online: https://www.epa.gov/sites/production/files/2015-07/documents/final_890.1600_hershberger_assay_sep_10.6.11.pdf (accessed November 10, 2015), 2011.
- Paul Friedman K.; Papineni S.; Marty M. S.; Yi K. D.; Goetz A. K.; Rasoulpour R. J.; Kwiatkowski P.; Wolf D. C.; Blacker A. M.; Peffer R. C. (2016) A predictive data-driven framework for endocrine prioritization: a triazole fungicide case study. Crit. Rev. Toxicol. 46, 785–833. 10.1080/10408444.2016.1193722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeguarden J. G.; Tan Y.-M.; Edwards S. W.; Leonard J. A.; Anderson K. A.; Corley R. A.; Kile M. L.; Simonich S. M.; Stone D.; Tanguay R. L.; Waters K. M.; Harper S. L.; Williams D. E. (2016) Completing the Link between Exposure Science and Toxicology for Improved Environmental Health Decision Making: The Aggregate Exposure Pathway Framework. Environ. Sci. Technol. 50 (9), 4579–4586. 10.1021/acs.est.5b05311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X.; Kleinstreuer N.; Ceger P.; Hsieh J.-H.; Allen D.; Casey W. (2015) Application of Reverse Dosimetry to Compare In Vitro and In Vivo Estrogen Receptor Activity. Applied In Vitro Toxicology 1 (1), 33–44. 10.1089/aivt.2014.0005. [DOI] [Google Scholar]
- Rotroff D. M.; Wetmore B. A.; Dix D. J.; Ferguson S. S.; Clewell H. J.; Houck K. A.; Lecluyse E. L.; Andersen M. E.; Judson R. S.; Smith C. M.; Sochaski M. A.; Kavlock R. J.; Boellmann F.; Martin M. T.; Reif D. M.; Wambaugh J. F.; Thomas R. S. (2010) Incorporating human dosimetry and exposure into high-throughput in vitro toxicity screening. Toxicol. Sci. 117 (2), 348–58. 10.1093/toxsci/kfq220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wambaugh J. F.; Wang A.; Dionisio K. L.; Frame A.; Egeghy P.; Judson R.; Setzer R. W. (2014) High Throughput Heuristics for Prioritizing Human Exposure to Environmental Chemicals. Environ. Sci. Technol. 48 (21), 12760–12767. 10.1021/es503583j. [DOI] [PubMed] [Google Scholar]
- Wambaugh J. F.; Wetmore B. A.; Pearce R.; Strope C.; Goldsmith R.; Sluka J. P.; Sedykh A.; Tropsha A.; Bosgra S.; Shah I.; Judson R.; Thomas R. S.; Setzer R. W. (2015) Toxicokinetic Triage for Environmental Chemicals. Toxicol. Sci. 147 (1), 55–67. 10.1093/toxsci/kfv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore B. A.; Wambaugh J. F.; Allen B.; Ferguson S. S.; Sochaski M. A.; Setzer R. W.; Houck K. A.; Strope C. L.; Cantwell K.; Judson R. S.; LeCluyse E.; Clewell H. J.; Thomas R. S.; Andersen M. E. (2015) Incorporating High-Throughput Exposure Predictions With Dosimetry-Adjusted In Vitro Bioactivity to Inform Chemical Toxicity Testing. Toxicol. Sci. 148 (1), 121–136. 10.1093/toxsci/kfv171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.