Abstract
Background:
Colorectal cancer (CRC) is a heterogeneous disease in terms of molecular carcinogenic pathways. Based on recent findings regarding the multiple serrated neoplasia pathway, we revised an eight-marker panel for a new CIMP classification system.
Methods:
1370 patients who received surgical resection for CRCs were classified into three CIMP subtypes (CIMP-N: 0–4 methylated markers, CIMP-P1: 5–6 methylated markers and CIMP-P2: 7–8 methylated markers). Our findings were validated in a separate set of high-risk stage II or stage III CRCs receiving adjuvant fluoropyrimidine plus oxaliplatin (n=950).
Results:
A total of 1287/62/21 CRCs cases were classified as CIMP-N/CIMP-P1/CIMP-P2, respectively. CIMP-N showed male predominance, distal location, lower T, N category and devoid of BRAF mutation, microsatellite instability (MSI) and MLH1 methylation. CIMP-P1 showed female predominance, proximal location, advanced TNM stage, mild decrease of CK20 and CDX2 expression, mild increase of CK7 expression, BRAF mutation, MSI and MLH1 methylation. CIMP-P2 showed older age, female predominance, proximal location, advanced T category, markedly reduced CK20 and CDX2 expression, rare KRAS mutation, high frequency of CK7 expression, BRAF mutation, MSI and MLH1 methylation. CIMP-N showed better 5-year cancer-specific survival (CSS; HR=0.47; 95% CI: 0.28–0.78) in discovery set and better 5-year relapse-free survival (RFS; HR=0.50; 95% CI: 0.29–0.88) in validation set compared with CIMP-P1. CIMP-P2 showed marginally better 5-year CSS (HR=0.28, 95% CI: 0.07–1.22) in discovery set and marginally better 5-year RFS (HR=0.21, 95% CI: 0.05–0.92) in validation set compared with CIMP-P1.
Conclusions:
CIMP subtypes classified using our revised system showed different clinical outcomes, demonstrating the heterogeneity of multiple serrated precursors of CIMP-positive CRCs.
Keywords: colorectal cancer, CpG island methylator phenotype, microsatellite instability, prognosis, serrated neoplasia pathway
Comprehensive genomic and epigenomic studies have revealed the heterogeneity of colorectal cancers (CRCs; Hinoue et al, 2012; Network TCGA, 2012). Currently, three major subtypes are widely accepted in the classification of CRCs: phenotypes with chromosomal instability (CIN) or microsatellite instability (MSI) and the CpG island methylator phenotype (CIMP; Ogino et al, 2009; Boland and Goel, 2010). Virtually all CRCs arise from premalignant polyps. Fearon and Vogelstein (1990) previously revealed a multistep pathway of CIN tumorigenesis, termed the ‘adenoma-carcinoma sequence'. However, recent advances in molecular biology techniques for analysing serrated polyps have helped to elucidate an alternative multistep carcinogenesis model for MSI and CIMP CRCs, termed the ‘serrated neoplasia pathway' (Leggett and Whitehall, 2010; Rex et al, 2012).
Sessile serrated adenomas (SSAs) and traditional serrated adenomas (TSAs) are considered precursors of the serrated neoplasia pathway (Fujita et al, 2011; Kim et al, 2016). Sessile serrated adenomas occur more commonly in women and on the right side of the colon, possess BRAF mutations, have a positive MLH1 methylation status and often have a deficient DNA mismatch repair (dMMR) system. Traditional serrated adenomas occur more commonly in men and on the left side of the colon, possess KRAS or BRAF mutations, and have a proficient DNA mismatch repair (pMMR) system (Kim et al, 2010). Several recent studies investigating tubulovillous adenomas with serrated features (sTVA) suggest that sTVAs are involved as precursors in the serrated neoplasia pathway (Tsai et al, 2014; Bettington et al, 2016). This heterogeneity in serrated polyps may have different prognostic implication.
Studies into the prognostic value of individual genetic or epigenetic alteration, such as KRAS or BRAF mutations, MSI or CIMP, have shown inconsistent results due to the complex interactions of these molecular alterations that arise during colorectal carcinogenesis (Kim et al, 2009; Ogino et al, 2009; Andre et al, 2015; Taieb et al, 2016). In 2007, Jass classified CRCs into five molecular subtypes based on MSI and CIMP (Jass, 2007). In 2015, two studies showed an association of CRC patient outcomes with molecular subtypes based on this classification (Phipps et al, 2015). However, these classifications may need to be redefined based on recent findings regarding the multiple serrated neoplasia pathway.
In this study, we propose a revised CIMP classification system for CRCs based on the methylation level, which we found to correlate with molecular alterations in the precursors of multiple carcinogenic pathways in CRCs. Moreover, we clearly demonstrate the heterogeneity of the molecular characteristics and the corresponding prognostic implications of CIMP-positive CRCs.
Patients and methods
Study population
The discovery set consisted of 1853 CRC patients who underwent curative or palliative surgery at Seoul National University Hospital, Seoul, South Korea, between January 2004 and June 2008. After the exclusion of 483 patients (excluded based on refusal to participate in the study (N=174), non-invasive cancer (N=50), familial adenomatous polyposis (N=13), multiple occurrences (N=88), neoadjuvant chemo- and/or radiotherapy (N=118) or tumour recurrence (N=40)), formalin-fixed paraffin-embedded (FFPE) tissue samples from 1370 patients were collected for CIMP classification. Among these patients, 951 (69.4%) received postoperative chemotherapy and/or radiotherapy. Specifically, 531 patients received 5-fluorouracil (5-FU) with leucovorin, 365 received 5-FU and leucovorin plus oxaliplatin (FOLFOX), 49 received 5-FU and leucovorin plus irinotecan (FOLFIRI), and six received radiotherapy alone. Furthermore, 105 patients received concurrent chemoradiation therapy after the surgery, and 12 received additional treatment with bevacizumab. This study was approved by the Institutional Review Board of Seoul National University Hospital and was carried out in accordance with the recommendations of the Declaration of Helsinki for biomedical research involving human subjects.
Clinicopathological analysis
Clnicopathological characteristics, including age, sex, tumour location and TNM stage, were obtained from electronic medical records. Based on microscopic examination of representative tumour sections, two pathologists (JMB and GHK) who were blind to the molecular study results evaluated the following parameters: tumour differentiation, luminal necrosis, tumour budding, Crohn-like lymphoid reactions, number of tumour-infiltrating lymphocytes, luminal serration and mucin production (Bae et al, 2015).
KRAS and BRAF mutations
DNA was extracted from FFPE tumour specimens. The areas in which tumour cells were most dense were determined via light microscopy. The corresponding areas were marked on 10 serial unstained slides; the marked areas were then manually scraped from the glass slides and collected in microtubes containing a lysis buffer (50 mM Tris, 1 mM EDTA, pH 8.0 and 1% Tween-20) and proteinase K (3 mg ml−1). The samples were incubated at 55 °C for up to 48 h. After centrifugation, the supernatants were transferred into newly labelled microtubes. The samples were then placed into a 95 °C heating block for 10 min to inactivate the proteinase K. After the extraction of the genomic DNA, the KRAS exon 2 was amplified using a hemi-nested PCR method (with rTaq DNA Polymerase; Takara, Kyoto, Japan) with the following primer set: forward primer (5′-ACTGAATATAAACTTGTGGTAGTTGGCCCT-3′), reverse primer 1 (5′-TAATATGTCGACTAAAACAAGATTTACCTC-3′), and reverse primer 2 (5′-TCAAAGAATGGTCCTGGACC-3′). The first PCR reaction mixture contained the forward primer and reverse primer 1 at a concentration of 400 nM, 1 × rTaq PCR buffer, 3 mM MgCl2, 0.625 U of rTaq polymerase, 400 μM of each deoxynucleotide and 100 ng of genomic DNA in a total volume of 20 μl. The PCR reaction conditions were as follows: 95 °C initial denaturation, 25 cycles of amplification (denaturation at 94 °C for 30 s, annealing at 55 °C for 40 s and primer extension at 72 °C for 30 s), and a final extension step at 72 °C for 10 min. One microlitre of the first PCR product was used for the second PCR, which was conducted in a total volume of 25 μl containing 1 × PCR buffer (16.6 mM (NH4)2SO4; 67 mM Tris, pH 8.8; 6.7 mM MgCl2; and 10 mM β-mercaptoethanol), dNTP (each 1 mM) and primers (0.4 μM each of forward primer and reverse primer 2). The amplifications were performed in a thermal cycler for 35 cycles (30 s at 95 °C, 40 s at 57 °C and 30 s at 72 °C) and were allowed a final 10-min extension at 72 °C. Five microlitres of the PCR product were treated with 1.2 U of shrimp alkaline phosphatase and 6 U of exonuclease in a final volume of 10 μl. The PCR products were incubated at 37 °C for 15 min and then heat-inactivated at 80 °C for 15 min. The purified PCR products were sequenced with BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and analysed with a 3730 ABI capillary electrophoresis system (Applied Biosystems). All somatic mutations that were found were further validated via an independent amplification and sequencing experiment. BRAF mutations at codon 600 (V600E) were analysed by a real-time PCR-based allelic discrimination method, as previously described (Young et al, 2005).
Microsatellite analysis
The microsatellite status of each tumour was determined through an evaluation of five microsatellite markers (D2S123, D5S346, D17S250, BAT25 and BAT26), as standardised by the National Cancer Institute (NCI) (Boland et al, 1998). A fluorescent label was added to either the forward or the reverse primer for each marker, and the PCR products were electrophoresed and analysed. We classified MSI status as follows: MSI-high (instability at ⩾2 microsatellite markers), MSI-low (instability at 1 marker) or microsatellite stable (MSS, no instability; Ribic et al, 2003).
Methylation analysis
After sodium bisulfite conversion of DNA using an EZ DNA methylation kit (Zymo Research, Orange, CA, USA), a MethyLight assay was used to quantify the methylation status of the following eight CIMP markers: CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1. The primer sequences and PCR conditions have been previously described (Weisenberger et al, 2006). M.SssI-treated genomic DNA was used as a reference sample. The percentage of methylated reference (PMR) at a particular locus was calculated by 100 × (methylated reaction at the GENE/control reaction at the ALU ratio)sample/(methylated reaction at the GENE/control reaction at the ALU)M.SssI-treated placental genomic DNA. The MethyLight assay was performed in triplicate, and the median value was calculated as the representative value of the methylation level of each marker. A CpG island locus with a median PMR >4 was considered to be methylated.
Immunohistochemistry
Tissue microarray (TMA) construction using FFPE tissues from 1370 CRCs was performed. Two different tumour areas in the FFPE tissue samples of each individual CRC case were extracted as two tissue cores (2 mm in diameter) and were transferred to TMA blocks. Immunohistochemical analyses were performed with commercially available antibodies against cytokeratin 7 (CK7; clone OV-TL 12/30, DAKO), cytokeratin 20 (CK20; clone Ks20.8, DAKO) and nuclear protein CDX2 (clone EPR2764Y ready-to-use, CellMarque). For the interpretation of immunohistochemical stain results, cytoplasmic and/or membranous CK7, CK20 and nuclear CDX2 were scored as the percentage of positive tumour cells. The cutoff scores for increased CK7 expression, decreased CK20 expression and decreased CDX2 expression were 10%, 50% and 20%, respectively (Bae et al, 2015).
Validation cohort
To validate the prognostic utility of our CIMP classification, we examined an independent cohort of stage III or high-risk stage II CRC patients receiving adjuvant fluoropyrimidine plus oxaliplatin (N=988) in two hospitals (Seoul National University Hospital between April 2005 and December 2012 (N=655) and Seoul National University Bundang Hospital between January 2007 and December 2012 (N=333)). Patients received adjuvant chemotherapy consisting of either FOLFOX-4 (N=377), modified FOLFOX-6 (N=466) or capecitabine plus oxaliplatin (XELOX; N=145). Adjuvant FOLFOX and XELOX were planned for a total of 12 and 8 cycles, respectively. There were no significant differences in clinical outcomes related to the hospitals or protocols (data not shown). The main inclusion criteria for the retrospective selection of patients were as follows: over 18 years of age, adenocarcinoma histology, stage III or high-risk stage II CRC, complete resection of the tumour with negative margins and the completion of at least six cycles of adjuvant FOLFOX chemotherapy or four cycles of adjuvant XELOX therapy. High-risk stage II CRC was defined if the patient met any one of the following criteria: T4 lesion, obstruction or perforation, lymphovascular invasion, perineural invasion or poorly differentiated histology (Schmoll et al, 2012). None of the patients received anti-EGFR or anti-VEGFR treatment adjunct to FOLFOX. In total, 950 patients with complete data for their KRAS, BRAF and CIMP status were included in the validation set.
Statistical analysis
The clinical database was last updated in July 2016. The cancer-specific survival (CSS) time in the discovery set was calculated from the date of surgery to the date of death by CRC. The relapse-free survival (RFS) time in the validation set was calculated from the first date of chemotherapy to the date of documented relapse. The data from patients who did not experience cancer-specific death or relapse were censored at the date of the last follow-up visit to obtain the CSS and RFS. Categorical variables were compared using χ2-tests or Fisher's exact tests, as appropriate. For the comparison of clinicopathologic differences between CIMP subtypes, Kruskal–Wallis tests with post hoc Nemenyi tests were employed. Cancer-specific survival and RFS were calculated using a log-rank test with Kaplan–Meier curves. Hazard ratios were calculated using the Cox proportional hazard model. The assumption of proportional hazards was verified by plotting the log(-log(S(t)) against the time of the study. In the modelling process, stage, degree of differentiation and history of chemotherapy were adjusted. All statistical tests were two-sided, and statistical significance was defined as P<0.05. When multiple tests were used for a survival analysis, the level of significance was set more conservatively at P<0.017 (0.05/3). All statistical analyses were performed with SAS 9.4 software (Cary, NC, USA) and R software (www.r-project.org).
Results
Molecular alterations in relation to the number of methylated genes
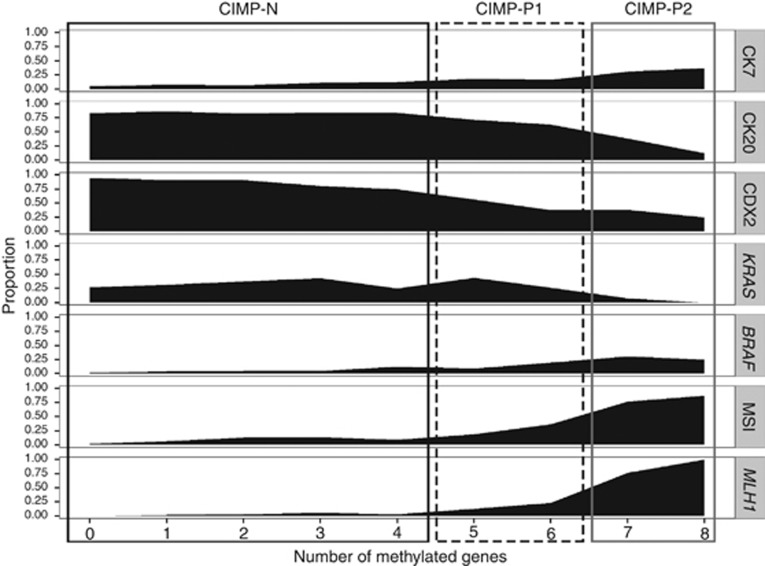
The proportions of molecular alterations in relation to the number of methylated genes in the discovery set are shown in Figure 1. The molecular characteristics of CRCs were not different between CRCs with no methylated genes and CRCs with four methylated genes. However, the frequency of alterations associated with the serrated neoplasia pathway, such as a gain of CK7 expression, decreased CK20 and CDX2 expression, an MSI-high status and MLH1 methylation, was moderately increased in CRCs with five or six methylated genes compared with CRCs with less than five methylated genes. Colorectal cancers with seven or eight methylated genes showed increased CK7 expression and decreased CK20 and CDX2 expression, were devoid of KRAS mutations, had a high rate of BRAF mutations, and had MSI-high and positive MLH1 methylation statuses. Moreover, synchronous BRAF mutation and MLH1 methylation were only observed in CRCs with seven or eight methylated genes (Supplementary Figure S1). Based on these trends, we classified CRCs into CIMP-negative (CIMP-N, 0–4 methylated genes), CIMP-positive 1 (CIMP-P1, 5–6 methylated genes) and CIMP-positive 2 (CIMP-P2, 7–8 methylated genes) categories. Although the five-marker panel introduced by Weisenberger et al. showed excellent concordance with the eight CIMP marker panel (the concordance rate between CIMP-P1 and CIMP-P2 in the eight-marker panel and CIMP in the five-marker panel: 99.3% Supplementary Table S1), we could not discriminate subtypes of CIMP-positive CRCs using Weisenberger's five-marker panel (Supplementary Figure S2).
Figure 1.
Molecular characteristics of colorectal cancers in the discovery set in relation to the number of methylated genes (N=1370).
Clinicopathologic characteristics of newly classified CIMP subtypes
With our refined CIMP classification system, CRCs from 1287 (93.9%), 62 (4.6%) and 21 (1.5%) patients were classified as CIMP-N, CIMP-P1 and CIMP-P2, respectively. Patients with CIMP-P1 CRCs showed more proximal location (P<0.001), higher T category (P=0.004), higher N category (P<0.001) and more advanced stage compared with CIMP-N CRCs, and were found to have poor differentiation, more lymphatic invasion, a higher number of tumour-infiltrating lymphocytes (TILs; P<0.001), greater luminal serration (P=0.024) and mucin production (P=0.013) compared with CIMP-N CRCs. Patients with CIMP-P2 CRCs showed older age at diagnosis compared with CIMP-N CRCs (P<0.001) and CIMP-P1 CRCs (P=0.002) and showed more proximal location (P<0.001), higher number of TILs (P=0.001) and more mucin production (P<0.001) compared with CIMP-N CRCs. Although the differences were not found to be statistically significant, CIMP-P2 CRCs showed a lower frequency of distant metastases (P=0.320) (Tables 1 and 2).
Table 1. Clinicopathologic and molecular characteristics of colorectal cancers from patients in the discovery set (N=1370).
| CIMP-N (N=1287, 93.9%) | CIMP-P1 (N=62, 4.6%) | CIMP-P2 (N=21, 1.5%) | P Overall | CIMP-N vs CIMP-P1 | CIMP-N vs CIMP-P2 | CIMP-P1 vs CIMP-P2 | |
|---|---|---|---|---|---|---|---|
| Age, median (min–max) | 62 (20–93) | 62 (39–80) | 71 (41–83) | 0.001 | > 0.999 | <0.001 | 0.002 |
| Sex | 0.046 | 0.110 | 0.800 | 0.870 | |||
| Female | 510 (39.6%) | 34 (54.8%) | 10 (52.4%) | ||||
| Male | 777 (60.4%) | 29 (45.2%) | 10 (47.6%) | ||||
| Location | <0.001 | <0.001 | <0.001 | 0.077 | |||
| Proximal colon | 290 (22.5%) | 35 (56.4%) | 17 (80.9%) | ||||
| Distal colon | 526 (40.9%) | 14 (22.6%) | 4 (19.0%) | ||||
| Rectum | 471 (36.6%) | 13 (21.0%) | 0 (0.0%) | ||||
| Gross pattern | 0.517 | 0.800 | 0.760 | 0.960 | |||
| Fungating | 851 (66.1%) | 38 (61.3%) | 12 (57.1%) | ||||
| Ulcerative | 436 (33.9%) | 24 (38.7%) | 9 (42.9%) | ||||
| T category | 0.001 | 0.004 | 0.343 | 0.895 | |||
| T1 | 56 (4.4%) | 0 (0.0%) | 0 (0.0%) | ||||
| T2 | 196 (15.2%) | 2 (3.2%) | 0 (0.0%) | ||||
| T3 | 913 (71.0%) | 47 (75.8%) | 19 (90.5%) | ||||
| T4 | 121 (9.4%) | 13 (21.0%) | 2 (9.5%) | ||||
| N category | <0.001 | <0.001 | 0.884 | 0.200 | |||
| N0 | 665 (51.7%) | 17 (27.4%) | 9 (42.9%) | ||||
| N1 | 354 (27.5%) | 17 (27.4%) | 8 (38.1%) | ||||
| N2 | 268 (20.8%) | 28 (45.2%) | 4 (19.0%) | ||||
| M category | 0.053 | 0.430 | 0.630 | 0.320 | |||
| M0 | 1075 (83.5%) | 46 (74.2%) | 20 (95.2%) | ||||
| M1 | 212 (16.5%) | 16 (25.8%) | 1 (4.8%) | ||||
| Stage | 0.001 | 0.001 | 0.904 | 0.300 | |||
| I | 205 (15.9%) | 0 (0.0%) | 0 (0.0%) | ||||
| II | 421 (32.7%) | 17 (27.4%) | 9 (42.9%) | ||||
| III | 450 (35.0%) | 29 (46.8%) | 11 (52.4%) | ||||
| IV | 211 (16.4%) | 16 (25.8%) | 1 (4.8%) | ||||
| KRAS | 0.103 | 0.799 | 0.103 | 0.088 | |||
| Wild type | 892 (69.3%) | 40 (64.5%) | 20 (95.2%) | ||||
| Mutated | 395 (30.7%) | 22 (35.5%) | 1 (4.8%) | ||||
| BRAF | <0.001 | 0.290 | 0.110 | 0.600 | |||
| Wild type | 1245 (96.7%) | 53 (85.5%) | 15 (71.4%) | ||||
| Mutated | 42 (3.3%) | 9 (14.5%) | 6 (28.6%) | ||||
| MSI | <0.001 | 0.008 | <0.001 | 0.001 | |||
| MSS | 1120 (87.0%) | 41 (66.1%) | 4 (19.1%) | ||||
| MSI-low | 96 (7.5%) | 4 (6.5%) | 0 (0.0%) | ||||
| MSI-high | 71 (5.5%) | 17 (27.4%) | 17 (80.9%) | ||||
| MLH1 | <0.001 | 0.073 | <0.001 | <0.001 | |||
| Unmethylated | 1270 (98.7%) | 51 (80.9%) | 3 (15.0%) | ||||
| Methylated | 17 (1.3%) | 11 (17.7%) | 18 (85.7%) | ||||
| CK7 expression | <0.001 | 0.062 | <0.001 | 0.020 | |||
| Not expressed | 1197 (93.0%) | 51 (82.3%) | 14 (66.7%) | ||||
| Expressed | 90 (7.0%) | 11 (17.7%) | 7 (33.3%) | ||||
| CK20 expression | <0.001 | 0.210 | 0.220 | 0.810 | |||
| Retained | 1091 (84.7%) | 42 (67.7%) | 6 (28.6%) | ||||
| Decreased | 196 (15.3%) | 20 (32.3%) | 15 (71.4%) | ||||
| CDX2 expression | <0.001 | <0.001 | <0.001 | 0.590 | |||
| Retained | 1172 (91.3%) | 30 (48.4%) | 7 (33.3%) | ||||
| Decreased | 111 (8.7%) | 32 (51.6%) | 14 (66.7%) |
Abbreviations: MSI=microsatellite instability; MSS=microsatellite stable.
Table 2. Histologic characteristics of colorectal cancers from patients in the discovery set.
| CIMP-N (N=1287, 93.9%) | CIMP-P1 (N=62, 4.6%) | CIMP-P2 (N=21, 1.5%) | P Overall | CIMP-N vs CIMP-P1 | CIMP-N vs CIMP-P2 | CIMP-P1 vs CIMP-P2 | |
|---|---|---|---|---|---|---|---|
| Differentiation | <0.001 | 0.022 | 0.217 | 0.996 | |||
| Well differentiated | 83 (6.5%) | 2 (3.2%) | 0 (0.0%) | ||||
| Moderately differentiated | 1165 (90.5%) | 47 (75.8%) | 17 (80.9%) | ||||
| Poorly differentiated | 39 (3.0%) | 13 (21.0%) | 4 (19.1%) | ||||
| Lymphatic invasion | <0.001 | <0.001 | 0.260 | 0.680 | |||
| Absent | 806 (62.6%) | 19 (30.7%) | 9 (42.9%) | ||||
| Present | 481 (37.4%) | 43 (69.3%) | 12 (57.1%) | ||||
| Venous invasion | 0.036 | 0.520 | 0.570 | 0.320 | |||
| Absent | 1122 (87.2%) | 49 (79.0%) | 21 (100.0%) | ||||
| Present | 165 (12.8%) | 13 (21.0%) | 0 (0.0%) | ||||
| Perineural invasion | 0.050 | 0.340 | 0.520 | 0.220 | |||
| Absent | 987 (76.7%) | 41 (66.1%) | 19 (90.5%) | ||||
| Present | 300 (23.3%) | 21 (33.9%) | 2 (9.5%) | ||||
| Luminal necrosis | <0.001 | 0.330 | 0.130 | 0.600 | |||
| Absent | 111 (8.6%) | 12 (19.4%) | 7 (33.3%) | ||||
| Present | 1176 (91.4%) | 50 (80.6%) | 14 (66.7%) | ||||
| Tumour budding | 0.038 | 0.120 | 0.900 | 0.800 | |||
| Absent | 377 (29.3%) | 9 (14.5%) | 5 (23.8%) | ||||
| Present | 910 (70.7%) | 53 (85.5%) | 16 (76.2%) | ||||
| Tumour-infiltrating lymphocytes | <0.001 | 0.013 | 0.001 | 0.252 | |||
| Low (<8/HPF) | 1001 (77.8%) | 35 (56.4%) | 7 (33.3%) | ||||
| High (⩾8/HPF) | 286 (22.2%) | 27 (43.6%) | 14 (66.7%) | ||||
| Crohn-like lymphoid reaction | 0.118 | 0.790 | 0.510 | 0.800 | |||
| Absent | 1101 (85.5%) | 50 (80.6%) | 15 (71.4%) | ||||
| Present | 186 (14.5%) | 12 (19.4%) | 6 (28.6%) | ||||
| Serration | <0.001 | 0.024 | 0.226 | 0.996 | |||
| Absent | 1250 (97.1%) | 48 (77.4%) | 16 (76.2%) | ||||
| Present | 37 (2.9%) | 14 (22.6%) | 5 (23.8%) | ||||
| Mucin production | <0.001 | 0.013 | <0.001 | 0.048 | |||
| Absent | 1144 (88.9%) | 42 (66.7%) | 7 (33.3%) | ||||
| Present | 143 (11.1%) | 21 (33.3%) | 14 (66.7%) |
Abbreviation: HPF=high-power field.
CIMP subtypes and patient survival
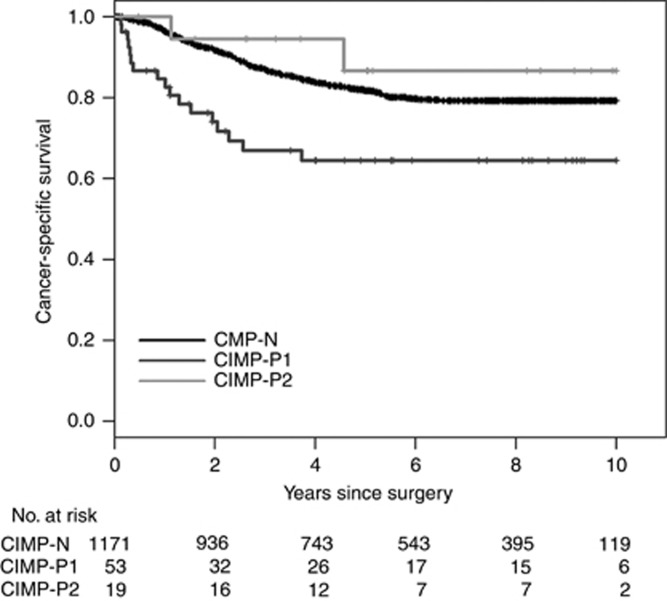
To investigate whether the revised CIMP classification system has prognostic implications, we analysed the CSS in the discovery set. In survival analysis, 127 patients who met the inclusion criteria of the validation set were eliminated. During the follow-up period, 223 (17.9%) of 1243 patients reported CSS events, and the median follow-up period was 85.9 months among the survivors. In a univariate analysis, patients with CIMP-P1 CRCs showed a worse 5-year CSS rate than those with CIMP-N (P<0.001) and CIMP-P2 CRCs (P<0.001; Figure 2). And this finding was observed in both adjuvant chemotherapy-treated and non-treated patients (Supplementary Figure S3). In a multivariate analysis, patients with CIMP-P1 CRCs showed a worse 5-year CSS rate than those with CIMP-N CRCs (P=0.004; Table 3). The statistical significance between the CSS rates of patients with CIMP-P1 CRCs and CIMP-P2 CRCs was marginal (P=0.090). Survival rates in relation to two previous classification systems showed similar trend in the discovery set (Supplementary Figure S4).
Figure 2.
Cancer-specific survival of patients in the discovery set (N=1243) in relation to the revised CIMP classification system.
Table 3. Multivariate survival analysis of the discovery and the validation sets.
| HR (95%) CI | P | |
|---|---|---|
|
5-year CSS in the discovery seta | ||
| CIMP-N | 0.47 (0.28–0.78) | 0.004 |
| CIMP-P1 | Reference | − |
| CIMP-P2 | 0.28 (0.07–1.22) | 0.090 |
|
5-year RFS in the validation setb | ||
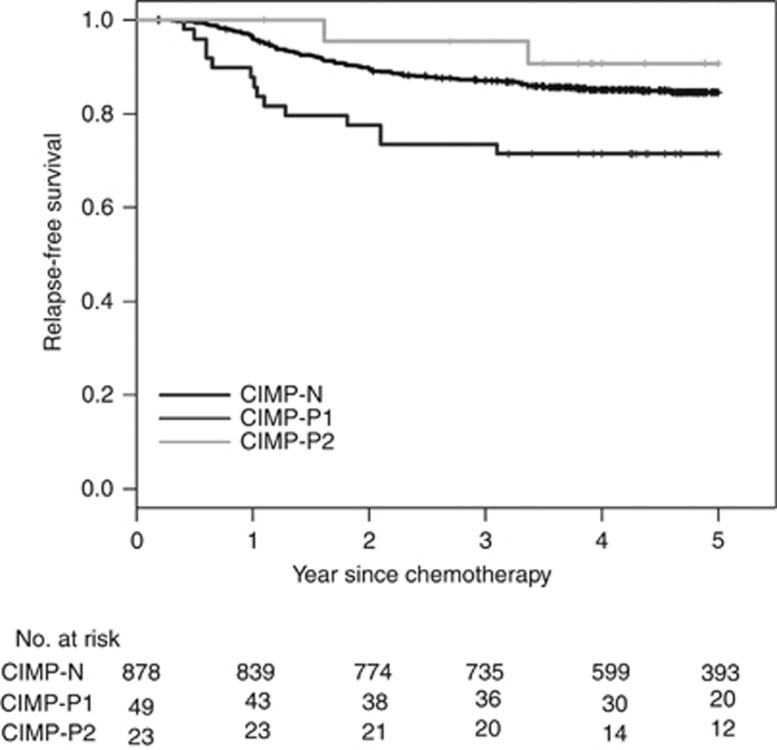
| CIMP-N | 0.50 (0.29–0.88) | 0.015 |
| CIMP-P1 | Reference | − |
| CIMP-P2 | 0.21 (0.05–0.92) | 0.038 |
Abbreviations: CI=confidence interval; CSS=cancer-specific survival; HR=hazard ratio; RFS=relapse-free survival.
Adjusted for stage, differentiation and chemotherapy status.
Adjusted for stage (III vs II, high risk) and differentiation.
Validation of the revised CIMP classification system
Survival analysis results from the discovery set could be biased by heterogeneity in cancer stages, adjuvant treatment modalities and general patient conditions. Therefore, we attempted to validate the prognostic utility of our revised CIMP classification system in an independent cohort of high-risk stage II or stage III CRC patients receiving adjuvant fluoropyrimidine plus oxaliplatin chemotherapy (FOLFOX or XELOX). Clinicopathologic and molecular characteristics of validation set are summarised in Supplementary Table S2. During the follow-up period, 155 (16.3%) out of 950 patients reported RFS events in the validation set. The median follow-up period was 60.9 months among the survivors. Survival rates in relation to two previous classification systems showed a similar trend in the validation set (Supplementary Figure S5). In our revised CIMP classification system, patients with CIMP-P1 CRCs showed a worse 5-year RFS rate than those with CIMP-N (P=0.018) and CIMP-P2 (P=0.006) in a univariate survival analysis (Figure 3). In a multivariate analysis, patients with CIMP-P1 showed a worse 5-year RFS rate than those with CIMP-N (P=0.015) and CIMP-P2 CRCs (P=0.038). Therefore, we conclude that our revised CIMP classification system can predict clinical outcomes of CRC patients. In addition, our data show that CIMP-P1 and CIMP-P2 CRCs exhibit different clinical behaviours. In a subgroup analysis, MSI/CIMP-P1 CRCs showed RFS trends that were more similar to those of MSS/CIMP-P1 CRCs than to those of CIMP-P2 CRCs, although the statistical significance was marginal (P=0.094 for MSS/CIMP-P1 vs CIMP-P2 and P=0.046 for MSI/CIMP-P1 vs CIMP-P2; Supplementary Figure S6). Among the KRAS-mutated/pMMR CRCs, patients with CIMP-P1 CRCs showed poorer RFS rates than those with CIMP-N CRCs (P=0.095; Supplementary Figure S7).
Figure 3.
Relapse-free survival of patients in the validation set (N=950) in relation to the revised CIMP classification system.
Discussion
The CIMP includes a subset of CRCs that exhibit a concordant hypermethylation of multiple genes or CpG island loci, as originally described by Toyota et al (1999). Although there are currently no consensus gene marker panels or marker threshold values for hypermethylation or standardised methodologies, CIMP-positive CRCs are strongly associated with female sex, proximal locations and MSI (Weisenberger et al, 2006; Kim et al, 2009; Ogino et al, 2009). Currently, the five-marker panel (CACNA1G, IGF2, NEUROG1, RUNX3 and SOCS1)-based CIMP classification system introduced by Weisenberger et al. and the 8-marker panel (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1)-based CIMP classification system that combines the two five-marker panel introduced by Weisenberger et al. and Ogino et al. are widely used (Weisenberger et al, 2006; Ogino et al, 2009). Because the five-marker panel introduced by Weisenberger et al. does not contain MLH1, it does not provide a detailed classification of CIMP-positive CRCs. In our present study, we successfully distinguished heterogeneous CIMP-positive CRCs using an eight-marker based CIMP classification system.
Although several studies report poor clinical outcomes for CIMP-positive CRCs, other studies have shown inconsistent results (Shen et al, 2007a; Ogino et al, 2007b, 2009; Barault et al, 2008; Bae et al, 2013; Juo et al, 2014). These inconsistencies may arise from the heterogeneity of CIMP-positive CRCs. Issa's group classified CIMP-positive CRCs into CIMP1 and CIMP2 CRCs by analysing 27 CpG sites in 97 cases of CRC (Shen et al, 2007b). These CIMP1 CRCs showed MLH1 methylation, frequent BRAF mutations, MSI, and rare KRAS mutation or TP53 mutation. The CIMP2 CRCs described by Issa's group show frequent KRAS mutations and rarely displayed MSI or BRAF and TP53 mutation. However, there were no significant differences observed in patient age and sex or stage between CIMP1 and CIMP2 CRCs. Our revised CIMP classification system showed similar spectra of molecular characteristics to that of the CIMP classification system described by Issa's group. However, in our present study, CIMP-P1 and CIMP-P2 CRCs exhibited several clinicopathologic differences. CIMP-P1 CRCs showed a strong association with aggressive clinicopathologic characteristics, such as nodal metastasis, distant metastasis and lymphovascular invasion. However, TNM stage of CIMP-P2 CRCs was not significantly different with CIMP-N CRCs. Moreover, CIMP-P2 CRCs showed more favourable clinical outcome than CIMP-P1 CRCs in the discovery and validation sets. Differential patient survival between those with CIMP-P1 and CIMP-P2 CRCs highlights the clinical relevance of this subclassification for CIMP-positive CRCs.
In 2007, Ogino et al (2007a) set a cutoff of ⩾6/8 methylated markers for CIMP-high based on the frequency of MSI-high, KRAS mutation and BRAF mutation. In that study, the frequency of MSI-high in CRCs with ⩾6/8 methylated markers was greater than that of CRCs with <6/8 methylated markers. Moreover, BRAF-mutated/MSI-high CRCs showed ⩾6/8 methylated markers. However, the frequency of KRAS mutation in CRCs with 0–6/8 methylated markers were greater than 30%, while the frequency of KRAS mutation in CRCs in CRCs with 7/8 or 8/8 methylated markers showed rare KRAS mutation. In our present study, cutoff of ⩾5/8 methylated markers for CIMP (including CIMP-P1 and CIMP-P2) showed excellent concordance with CIMP using the five-marker panel by Weisenberger et al (Supplementary Table S1). And CRCs with 7/8 or 8/8 methylated markers showed marked difference in frequency of KRAS mutation compared with CRCs with 0–6/8 methylated markers. Moreover, BRAF-mutated/MSI-high CRCs showed 7/8 or 8/8 methylated markers. BRAF-mutated CIMP-P1 CRCs (5/8 or 6/8 methylated markers) did not show MSI-high or MLH1 methylation.
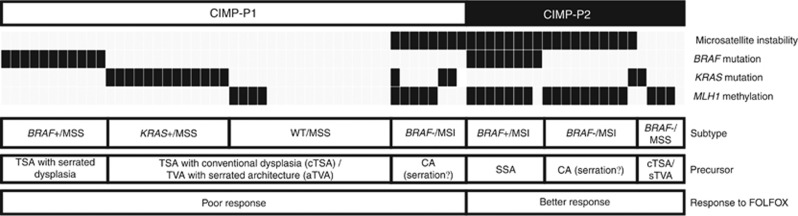
Recent advances in the characterisation of TSAs suggest that they can be classified into three distinct molecular subtypes: BRAF-mutated, KRAS-mutated and KRAS/BRAF-wild type (Tsai et al, 2014; Wiland et al, 2014; Bettington et al, 2015). BRAF-mutated TSAs are more common among women, predominantly found in the right colon, and have more extensive methylation and serrated dysplasia than KRAS-mutated or KRAS/BRAF-wild-type TSA (Kim et al, 2010; Bettington et al, 2015). Therefore, BRAF-mutated TSAs are regarded as a precursor of BRAF-mutated/pMMR/CIMP-positive CRCs, which show most aggressive clinical behaviour (Pai et al, 2012; Phipps et al, 2015; Sinicrope et al, 2015). However, the precursor of KRAS-mutated/CIMP-positive CRC has yet to be conclusively determined, as several subtypes of conventional adenomas, particularly TVAs, have been found to be precursors of CIMP-positive CRCs. Tubulovillous adenomas shows more methylation compared with tubular adenomas (Kakar et al, 2008). Several studies have demonstrated that sTVAs show more methylation than conventional TVAs (Tsai et al, 2014; Bettington et al, 2016). Because BRAF-wild-type TSAs and sTVAs show high frequencies of KRAS mutations, it is nearly impossible to distinguish between them as the potential precursors of BRAF-wild-type CIMP-positive CRCs (Bettington et al, 2016). Moreover, Farchoukh et al (2016) reported that BRAF-wild type/MLH1-methylated CRCs harbour KRAS mutations and arise from conventional adenomas. Based on recent findings regarding the serrated neoplasia pathway, we can make predictions about the putative precursors of the various CIMP-positive CRC subtypes (Figure 4).
Figure 4.
Molecular sub-classification of CIMP-positive colorectal cancers and their putative precursors.CA=conventional adenoma; MSS=microsatellite stable; MSI=microsatellite instability; SSA=sessile serrated adenoma; TSA=traditional serrated adenoma; TVA=tubulovillous adenoma.
Prospective clinical trials examining adjuvant FOLFOX and recent other two studies showed the adverse prognostic effect of KRAS mutations on CRC prognoses (Hutchins et al, 2011; Phipps et al, 2015; Sinicrope et al, 2015; Taieb et al, 2016). However, it is of interest whether the clinical outcomes of KRAS-mutated CRCs differ according to their methylation status. In a study by Phipps et al (2015), the authors excluded 37 KRAS-mutated CIMP-positive CRCs in their survival analysis. A study by Sinicrope et al (2015) only analysed the influence of MLH1 methylation status. Therefore, we investigated the effect of the methylation status on KRAS-mutated CRC prognoses. We found that KRAS-mutated/CIMP-positive CRCs showed worse clinical outcomes than KRAS-mutated/CIMP-negative CRCs in the validation set. This finding suggests that CRCs arising from KRAS-mutated TSAs or sTVAs are more aggressive than KRAS-mutated/CIMP-negative CRCs arising from conventional adenomas.
In our present study, CIMP-P1 CRCs were categorised as MSS/CIMP-P1 or MSI/CIMP-P1 CRCs. Because MSI/CIMP-P1 CRCs showed a high frequency of MLH1 methylation, it is of interest whether clinical outcomes of MSI/CIMP-P1 CRCs are more similar to those of MSS/CIMP-P1 or CIMP-P2 CRCs. Although the statistical significance was marginal, patients with MSI/CIMP-P1 CRC showed RFS that were similar to those of patients with MSS/CIMP-P1 CRC in the validation set. Recently, Farchoukh et al (2016) found no significant differences in DFS between patients with MLH1-hypermethylated/BRAF wild-type CRC and MLH1-hypermethylated/BRAF-mutated CRCs. Therefore, we can presume that the clinical outcome of patients with CIMP-positive CRC is more strongly influenced by the level of methylation than MMR or BRAF mutation status.
Special consideration is required for the selection of chemotherapeutic agents for the treatment of CIMP-P1 CRCs. In our validation set, CIMP-P1 CRCs that received oxaliplatin-based adjuvant chemotherapy showed worst RFS among CIMP subtypes. Recently, we reported that metastatic CIMP-high CRCs showed poor overall survival when treated with FOLFOX (Cha et al, 2016). In that study, all of the CIMP-high CRC showed methylation of five or six markers, which corresponds to the CIMP-P1 in the present study. Additionally, Zhang et al (2016) reported that FOLFOX treatment followed by an irinotecan-based regimen shows unfavourable outcomes compared with the irinotecan-based regimen followed by FOLFOX treatment in metastatic CIMP-positive CRCs. Because stage III CIMP-positive CRCs can benefit from joint use of irinotecan with 5-FU (IFL) compared with 5-FU and leukovorin (FL), the CIMP could be used as a predictive marker for selecting oxaliplatin or irinotecan as an adjuvant or palliative chemotherapeutic agent for CRC treatment (Shiovitz et al, 2014).
Our discovery set comes with several limitations. First, our data were collected retrospectively at a single institution and from patients that received a variety of different post-operative treatments. Therefore, we aimed to validate the results from the discovery set in a validation set that controlled for tumour stage, performance status and adjuvant modality, and this set was collected from two institutions using the same patient care strategy. The results from the analysis of the discovery set were consistently reproduced by the analysis of the validation set, substantiating the results of our present study. Second, our study showed the lower frequency of KRAS mutation, BRAF mutation and MSI-high compared with those of Western countries. The frequencies of those molecular alterations in CRCs of Eastern Asia are lower than Western countries (Yuen et al, 2002; Nagasaka et al, 2004; Jeon et al, 2008; Shin et al, 2014). This discrepancy might originate from ethnic or environmental difference. However, we could not rule out the possibility of selection bias originated from hospital-based study.
Conclusion
CIMP-positive CRCs can be categorised as CIMP-P1 or CIMP-P2 based on the number of methylated markers. CIMP-P1 CRCs are more aggressive than CIMP-N and CIMP-P2 CRCs. Finally, our revised CIMP classification system reflects the biologic behaviours of multiple serrated precursors.
Acknowledgments
This work was supported by a grant from the National Research Foundation (NRF) funded by the Korean Ministry of Science, ICT and Future Planning (2011-0030049; 2016M3A9B6026921), a grant (2009-0093820) from the Priority Research Centers Program through the NRF, a grant from the SNUH Research Fund (0420150260 (2015-1174)) and a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute funded by the Korean Ministry of Health and Welfare (HI14C1277).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
The authors declare no conflict of interest.
Supplementary Material
References
- Andre T, de Gramont A, Vernerey D, Chibaudel B, Bonnetain F, Tijeras-Raballand A, Scriva A, Hickish T, Tabernero J, Van Laethem JL, Banzi M, Maartense E, Shmueli E, Carlsson GU, Scheithauer W, Papamichael D, Moehler M, Landolfi S, Demetter P, Colote S, Tournigand C, Louvet C, Duval A, Flejou JF, de Gramont A (2015) Adjuvant fluorouracil, leucovorin, and oxaliplatin in stage II to III colon cancer: updated 10-year survival and outcomes according to BRAF mutation and mismatch repair status of the MOSAIC Study. J Clin Oncol 33(35): 4176–4187. [DOI] [PubMed] [Google Scholar]
- Bae JM, Kim JH, Cho NY, Kim TY, Kang GH (2013) Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location. Br J Cancer 109(4): 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JM, Lee TH, Cho NY, Kim TY, Kang GH (2015) Loss of CDX2 expression is associated with poor prognosis in colorectal cancer patients. World J Gastroenterol 21(5): 1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barault L, Charon-Barra C, Jooste V, de la Vega MF, Martin L, Roignot P, Rat P, Bouvier AM, Laurent-Puig P, Faivre J, Chapusot C, Piard F (2008) Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res 68(20): 8541–8546. [DOI] [PubMed] [Google Scholar]
- Bettington M, Walker N, Rosty C, Brown I, Clouston A, McKeone D, Pearson SA, Klein K, Leggett B, Whitehall V (2016) Serrated tubulovillous adenoma of the large intestine. Histopathology 68(4): 578–587. [DOI] [PubMed] [Google Scholar]
- Bettington ML, Walker NI, Rosty C, Brown IS, Clouston AD, McKeone DM, Pearson SA, Klein K, Leggett BA, Whitehall VL (2015) A clinicopathological and molecular analysis of 200 traditional serrated adenomas. Mod Pathol 28(3): 414–427. [DOI] [PubMed] [Google Scholar]
- Boland CR, Goel A (2010) Microsatellite instability in colorectal cancer. Gastroenterology 138(6): 2073–2087 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S (1998) A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58(22): 5248–5257. [PubMed] [Google Scholar]
- Cha Y, Kim KJ, Han SW, Rhee YY, Bae JM, Wen X, Cho NY, Lee DW, Lee KH, Kim TY, Oh DY, Im SA, Bang YJ, Jeong SY, Park KJ, Kang GH, Kim TY (2016) Adverse prognostic impact of the CpG island methylator phenotype in metastatic colorectal cancer. Br J Cancer 115(2): 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farchoukh L, Kuan SF, Dudley B, Brand R, Nikiforova M, Pai RK (2016) MLH1-deficient colorectal carcinoma with wild-type BRAF and MLH1 promoter hypermethylation harbor KRAS mutations and arise from conventional adenomas. Am J Surg Pathol 40(10): 1390–1399. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B (1990) A genetic model for colorectal tumorigenesis. Cell 61(5): 759–767. [DOI] [PubMed] [Google Scholar]
- Fujita K, Yamamoto H, Matsumoto T, Hirahashi M, Gushima M, Kishimoto J, Nishiyama K, Taguchi T, Yao T, Oda Y (2011) Sessile serrated adenoma with early neoplastic progression: a clinicopathologic and molecular study. Am J Surg Pathol 35(2): 295–304. [DOI] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk CM, Tollenaar RA, Laird PW (2012) Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 22(2): 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins G, Southward K, Handley K, Magill L, Beaumont C, Stahlschmidt J, Richman S, Chambers P, Seymour M, Kerr D, Gray R, Quirke P (2011) Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clinl Oncol 29(10): 1261–1270. [DOI] [PubMed] [Google Scholar]
- Jass JR (2007) Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 50(1): 113–130. [DOI] [PubMed] [Google Scholar]
- Jeon CH, Lee HI, Shin IH, Park JW (2008) Genetic alterations of APC, K-ras, p53, MSI, and MAGE in Korean colorectal cancer patients. Int J Colorectal Dis 23(1): 29–35. [DOI] [PubMed] [Google Scholar]
- Juo YY, Johnston FM, Zhang DY, Juo HH, Wang H, Pappou EP, Yu T, Easwaran H, Baylin S, van Engeland M, Ahuja N (2014) Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta-analysis. Ann Oncol 25(12): 2314–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakar S, Deng G, Cun L, Sahai V, Kim YS (2008) CpG island methylation is frequently present in tubulovillous and villous adenomas and correlates with size, site, and villous component. Hum Pathol 39(1): 30–36. [DOI] [PubMed] [Google Scholar]
- Kim JH, Bae JM, Cho NY, Kang GH (2016) Distinct features between MLH1-methylated and unmethylated colorectal carcinomas with the CpG island methylator phenotype: implications in the serrated neoplasia pathway. Oncotarget 7(12): 14095–14111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Shin SH, Kwon HJ, Cho NY, Kang GH (2009) Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch 455(6): 485–494. [DOI] [PubMed] [Google Scholar]
- Kim KM, Lee EJ, Kim YH, Chang DK, Odze RD (2010) KRAS mutations in traditional serrated adenomas from Korea herald an aggressive phenotype. Am J Surg Pathol 34(5): 667–675. [DOI] [PubMed] [Google Scholar]
- Leggett B, Whitehall V (2010) Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology 138(6): 2088–2100. [DOI] [PubMed] [Google Scholar]
- Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K, Kambara T, MacPhee DG, Young J, Leggett BA, Jass JR, Tanaka N, Matsubara N (2004) Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol 22(22): 4584–4594. [DOI] [PubMed] [Google Scholar]
- Network TCGA (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487(7407): 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS (2007. a) Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn 9(3): 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Meyerhardt JA, Kawasaki T, Clark JW, Ryan DP, Kulke MH, Enzinger PC, Wolpin BM, Loda M, Fuchs CS (2007. b) CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch 450(5): 529–537. [DOI] [PubMed] [Google Scholar]
- Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS (2009) CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut 58(1): 90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai RK, Jayachandran P, Koong AC, Chang DT, Kwok S, Ma L, Arber DA, Balise RR, Tubbs RR, Shadrach B, Pai RK (2012) BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol 36(5): 744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps AI, Limburg PJ, Baron JA, Burnett-Hartman AN, Weisenberger DJ, Laird PW, Sinicrope FA, Rosty C, Buchanan DD, Potter JD, Newcomb PA (2015) Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 148(1): 77–87.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex DK, Ahnen DJ, Baron JA, Batts KP, Burke CA, Burt RW, Goldblum JR, Guillem JG, Kahi CJ, Kalady MF, O'Brien MJ, Odze RD, Ogino S, Parry S, Snover DC, Torlakovic EE, Wise PE, Young J, Church J (2012) Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol 107(9): 1315–1329, quiz 1314, 1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, Tu D, Redston M, Gallinger S (2003) Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 349(3): 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoll HJ, Van Cutsem E, Stein A, Valentini V, Glimelius B, Haustermans K, Nordlinger B, van de Velde CJ, Balmana J, Regula J, Nagtegaal ID, Beets-Tan RG, Arnold D, Ciardiello F, Hoff P, Kerr D, Kohne CH, Labianca R, Price T, Scheithauer W, Sobrero A, Tabernero J, Aderka D, Barroso S, Bodoky G, Douillard JY, El Ghazaly H, Gallardo J, Garin A, Glynne-Jones R, Jordan K, Meshcheryakov A, Papamichail D, Pfeiffer P, Souglakos I, Turhal S, Cervantes A (2012) ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol 23(10): 2479–2516. [DOI] [PubMed] [Google Scholar]
- Shen L, Catalano PJ, Benson AB 3rd, O'Dwyer P, Hamilton SR, Issa JP (2007. a) Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res 13(20): 6093–6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, Hernandez NS, Chen X, Ahmed S, Konishi K, Hamilton SR, Issa JP (2007. b) Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci USA 104(47): 18654–18659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin US, Cho SS, Moon SM, Park SH, Jee SH, Jung EJ, Hwang DY (2014) Is microsatellite instability really a good prognostic factor of colorectal cancer? Ann Coloproctol 30(1): 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiovitz S, Bertagnolli MM, Renfro LA, Nam E, Foster NR, Dzieciatkowski S, Luo Y, Lao VV, Monnat RJ Jr, Emond MJ, Maizels N, Niedzwiecki D, Goldberg RM, Saltz LB, Venook A, Warren RS, Grady WM (2014) CpG island methylator phenotype is associated with response to adjuvant irinotecan-based therapy for stage III colon cancer. Gastroenterology 147(3): 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinicrope FA, Shi Q, Smyrk TC, Thibodeau SN, Dienstmann R, Guinney J, Bot BM, Tejpar S, Delorenzi M, Goldberg RM, Mahoney M, Sargent DJ, Alberts SR (2015) Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology 148(1): 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taieb J, Zaanan A, Le Malicot K, Julie C, Blons H, Mineur L, Bennouna J, Tabernero J, Mini E, Folprecht G, Van Laethem JL, Lepage C, Emile JF, Laurent-Puig P (2016) Prognostic effect of BRAF and KRAS mutations in patients with stage III colon cancer treated with leucovorin, fluorouracil, and oxaliplatin with or without cetuximab: a post hoc analysis of the PETACC-8 trial. JAMA Oncol 2(5): 643–652. [DOI] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP (1999) CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 96(15): 8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JH, Liau JY, Lin YL, Lin LI, Cheng YC, Cheng ML, Jeng YM (2014) Traditional serrated adenoma has two pathways of neoplastic progression that are distinct from the sessile serrated pathway of colorectal carcinogenesis. Mod Pathol 27(10): 1375–1385. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW (2006) CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38(7): 787–793. [DOI] [PubMed] [Google Scholar]
- Wiland HOt, Shadrach B, Allende D, Carver P, Goldblum JR, Liu X, Patil DT, Rybicki LA, Pai RK (2014) Morphologic and molecular characterization of traditional serrated adenomas of the distal colon and rectum. Am J Surg Pathol 38(9): 1290–1297. [DOI] [PubMed] [Google Scholar]
- Young J, Barker MA, Simms LA, Walsh MD, Biden KG, Buchanan D, Buttenshaw R, Whitehall VL, Arnold S, Jackson L, Kambara T, Spring KJ, Jenkins MA, Walker GJ, Hopper JL, Leggett BA, Jass JR (2005) Evidence for BRAF mutation and variable levels of microsatellite instability in a syndrome of familial colorectal cancer. Clin Gastroenterol Hepatol 3(3): 254–263. [DOI] [PubMed] [Google Scholar]
- Yuen ST, Davies H, Chan TL, Ho JW, Bignell GR, Cox C, Stephens P, Edkins S, Tsui WW, Chan AS, Futreal PA, Stratton MR, Wooster R, Leung SY (2002) Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res 62(22): 6451–6455. [PubMed] [Google Scholar]
- Zhang X, Shimodaira H, Soeda H, Komine K, Takahashi H, Ouchi K, Inoue M, Takahashi M, Takahashi S, Ishioka C (2016) CpG island methylator phenotype is associated with the efficacy of sequential oxaliplatin- and irinotecan-based chemotherapy and EGFR-related gene mutation in Japanese patients with metastatic colorectal cancer. Int J Clin Oncol 21(6): 1091–1101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.