Abstract
The ubiquitin‐proteasome pathway (UPP) plays a critical role in the degradation of proteins implicated in cell cycle control, signal transduction, DNA damage response, apoptosis and immune response. Proteasome inhibitors can inhibit the growth of a broad spectrum of human cancer cells by altering the balance of intracellular proteins. However, the targets of these compounds in acute myeloid leukemia (AML) cells have not been fully characterized. Herein, we combined large‐scale quantitative analysis by SILAC‐MS and targeted quantitative proteomic analysis in order to identify proteins regulated upon proteasome inhibition in two AML cell lines displaying different stages of maturation: immature KG1a cells and mature U937 cells. In‐depth data analysis enabled accurate quantification of more than 7000 proteins in these two cell lines. Several candidates were validated by selected reaction monitoring (SRM) measurements in a large number of samples. Despite the broad range of proteins known to be affected by proteasome inhibition, such as heat shock (HSP) and cell cycle proteins, our analysis identified new differentially regulated proteins, including IL‐32, MORF family mortality factors and apoptosis inducing factor SIVA, a target of p53. It could explain why proteasome inhibitors induce stronger apoptotic responses in immature AML cells.
Keywords: Apoptosis, Human acute myeloid leukemia (AML) cells, Proteasome inhibitor, SILAC, Targeted proteomics, Technology
Abbreviations
- AML
acute myeloid leukemia
- CE
collision energy
- E1
ubiquitin‐activating enzyme
- E2
ubiquitin carrier protein
- E3
ubiquitin‐protein isopeptide ligase
- MORF4
mortality factor 4
- MORF4L1
mortality factor 4‐like protein 1
- MRFAP1
MORF4 family‐associated protein 1
- MRGBP
MRG‐domain binding protein
- Q1
first quadrupole
- Q3
third quadruple
- RT
room temperature
- SILAC
stable isotope labeling with amino acids in cell culture
- UPP
Ubiquitin‐Proteasome Pathway
- UPS
Ubiquitin‐Proteasome System
1. Introduction
The 26S proteasome is a multi‐catalytic enzyme complex containing a 20S catalytic core and two 19S regulatory complexes 1. About 80% of intracellular proteins are tagged with ubiquitin for recognition and subsequent degradation by the Ubiquitin‐Proteasome Pathway (UPP). The degradation of cellular proteins is a complex and tightly controlled process that is central to regulating cellular function and maintaining protein homeostasis. Therefore, the UPP is a common regulatory modification system involved in the regulation of the cell cycle, apoptosis, and response to cellular stress, i.e., DNA damage, hypoxia, and intracellular signal transduction. Subsequently, abnormal regulation of cell cycle proteins can result in accelerated and uncontrolled cell division, leading to tumorigenesis and cancer formation 2.
Significance of the study
In this study we applied a quantitative proteomics approach to study the effect of proteasome inhibition in two human acute myeloid leukaemia (AML) cells at differential maturation stages. The global change in the proteome upon proteasomal inhibition in AML cells remains largely unexplored. Herein, we combined large‐scale quantitative analysis by SILAC‐MS and targeted quantitative proteomic analysis in order to identify proteins regulated upon proteasome inhibition in two AML cell lines displaying different stages of maturation: immature KG1a cells and mature U937 cells. In‐depth data analysis enabled accurate quantification of more than 7000 proteins in these two cell lines. Several candidates were validated by selected reaction monitoring (SRM) measurements in a large number of samples. Despite the broad range of proteins known to be affected by proteasome inhibition, such as heat shock (HSP) and cell cycle proteins, our analysis identified new differentially regulated proteins, including IL‐32, MORF family mortality factors and apoptosis inducing factor SIVA, a target of p53. Our results suggest that apoptosis induced by the proteasome inhibition in U937 cells is p53‐dependent pathway and might be different for KG1a cells. These results open up new avenues to investigate the mechanism of the proteasome inhibitors in AML.
Proteasome inhibitors have gained interest as promising chemotherapeutic agents based on their ability to inhibit the growth of cancer cells by altering the balance of intracellular proteins 3, 4. There are currently two small molecular inhibitors of this pathway approved for clinical use, bortezomib and carfilzomib, both of which function by inhibiting proteasomal activity 5, 6, 7. Bortezomib was the first proteasome inhibitor to enter the clinical arena, validating the UPP as a possible feasible therapeutic target and leading to a series of next generation proteasome inhibitors, many of which are in clinical development 8. Novel approaches for targeting the UPS are also being actively investigated, including the development of small molecule inhibitors of enzymes such as ATPases and deubiquitinase, which act at an earlier stage in the UPP 9, 10, 11. An alternative approach is to target key effectors that act earlier in signal transduction 12.
The proteasome plays a key role within the cell, so that inhibiting this mechanism would be expected to impact significantly on normal, as well as malignant cells. However, it is recognized that there is an elevated level of proteasome activity both in leukaemic cells and serum from patients with haematological malignancies, which is believed to increase their vulnerability to proteasome inhibition 7, 13. In haematological malignancies, the efficacy and safety of bortezomib alone or in combination with chemotherapy in multiple myeloma have been further investigated and bortezomib was shown to induce a higher level of apoptosis in malignant, compared to normal cells. Several reports, including our own previous study 14, demonstrated that proteasome inhibitors induce apoptosis in model leukemia cell lines 15, 16. Likewise, we have previously demonstrated that increased proteasome level and activity in leukemia cells lines, and primary human cells from patients, are correlated with increased sensitivity to proteasome inhibitors 14. However, the precise mode of action of such inhibitors still remains unclear.
Multiple techniques have been used to investigate the mechanism of action of proteasome inhibitors, each having its own strengths and limitations. Several studies used a large transcriptomic approach and measured the regulation of genes upon proteasome inhibition 17, 18. However genes regulation does not always correlated with change in protein abundance, which carry most biological activity in cells, and therefore does not elucidate the mechanism by which these molecules act on cancer cells. In contrast, proteomic studies present the advantage to take protein expression their post‐transcriptional events into account, particularly when altered protein degradation is expected to underlie studied mechanism, as upon treatments with proteasome inhibition. So far, very few proteomic studies have attempted to investigate the global cellular response to proteasome inhibition 19. Immunoblotting techniques have been widely used to measure protein abundance upon proteasome inhibition, but this strategy is restricted to well‐known pathway due to the availability of specific antibodies 18, 20, 21. One of the first large‐scale mass spectrometry (MS)‐based proteomic studies to decipher mechanism of proteasome inhibitors was performed in our lab by Uttenweiller and colleagues, and reported the effect of bortezomib on both differentiated and undifferentiated Acute Promyelocytic Leukemia (APL) cells and revealed that the differential apoptotic effects of bortezomib on these particular cells are mainly due to distinct protein toxicity levels 22. Quantitative proteomics analysis based on iTRAQTM peptide labeling allowed the quantification of 14 regulated proteins involved in protein stress response and apoptosis after proteasome inhibition.
With recent advances in instrumentation, new MS‐based proteomic approaches have been introduced, mainly differing with respect to their analytical performance in terms of reproducibility, dynamic range, limit of detection (LOD) and resolving power. To‐date, three main acquisition modes are used in bottom‐up proteomics field: Data Dependent Acquisition (DDA; frequently termed shotgun, or discovery), targeted data acquisition (Selected Reaction Monitoring and Parallel Reaction Monitoring) and Data Independent Acquisition (DIA, SWATH‐MS). Shotgun (or discovery) proteomics is the best method to identify large numbers of proteins found in complex mixtures, where no prior knowledge is available. Here, precursor ions are only selected for sequencing when they are detected at the MS level. This approach allows the identification of thousands of proteins in any cell type or organism and provides a comprehensive view of complex proteomes 23, 24, 25, 26, 27, 28. However, when multiple samples are compared, a major weakness of this technique is the selection of precursor ions that are biased towards abundant peptides, leading to irreproducible replicates of shotgun experiments, and thus, incomplete MS data sets. Lately, SWATH‐MS 27 was introduced to overcome this issue. Herein, the instrument is forced to fragment the whole precursor ion mass range continuously during the LC separation. Therefore, all peptides in the biological sample are fragmented, allowing for the complete identification and characterization of the (detectable) protein content. Peptides are subsequently identified using a reference spectral library. Consequently, the main limitation of this approach is a requirement for high‐quality assay libraries prior to quantification 29, 30. Targeted data acquisition method was developed for the systematic and precise detection and quantification of well‐defined sets of peptides and proteins in complex samples. This strategy allows the consistent monitoring of peptides with a high degree of specificity and sensitivity. In contrast to unsupervised approaches, targeted methods are hypothesis‐driven and require the careful selection of the peptides used to represent their parent proteins. The analyses are commonly performed in selected reaction monitoring (SRM) mode on triple quadrupole mass spectrometers (QQQ) 31. In SRM, the first (Q1) and the third (Q3) quadrupoles act as filters to specifically select predefined m/z values corresponding to a peptide ion and a specific fragment ion of the peptide, whereas the second quadrupole serves as a collision cell 32, 33, 34. Suitable sets of precursor and fragment ion masses for a given peptide, called SRM transitions, can be used in MS assays to identify each peptide and the corresponding proteins. The main limitations of this method are: the requirement for transition lists, which are time consuming to establish, the maximum number of targeted peptides that can be monitored in single analysis without jeopardising precision of the measurement 35. With its selectivity and sensitivity, SRM represents a powerful tool for the validation of selected candidates previously identified by shotgun across a large number of samples 35. This approach is frequently used in biomarker studies 33, 34. More recently, a new strategy has been introduced, named parallel reaction monitoring (PRM). In PRM mode, all ions resulting from the fragmentation of a single, or several, precursor ions are measured simultaneously in one MS/MS scan 7, 36.
In this work, we employed quantitative proteomic approaches to achieve very extensive proteomic coverage of two human AML cells with differential maturation stages, before and after proteasome inhibition. We combined stable isotope labeling with amino acids in cell culture (SILAC‐based quantitative shotgun analysis, extensive subcellular and protein‐level fractionation, and high resolution MS, to study system‐wide effects of proteasome inhibition in KG1a cells and U937 cells that display a differential response to proteasome inhibition 14. Subsequently, a defined list of modulated proteins identified during the discovery phase, was validated by SRM across multiple samples and conditions.
2. Materials and methods
2.1. Drugs and reagents
Bortezomib (VELCADE®) was generously provided by Millennium Pharmaceuticals Inc. (Cambridge, MA, USA). MG‐132 (Z‐LLL‐CHO) and Lactacystin was purchased from Sigma Aldrich. Antibodies against human PARP, actin and α‐tubulin were purchased from Santa Cruz biotechnology 37, 38. Sequencing Grade Modified Trypsin V511A was obtained from Promega.
2.2. Cell culture
Human leukemic cell lines KG1a and U937 were purchased from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). U937 cell lines and KG1a cell lines were grown in RPMI 1640 media, depleted of arginine and lysine (Invitrogen), and supplemented with 10% or 20% of fetal bovine serum (FBS) dialyzed with a cutoff of 10 kDa (Invitrogen, 26400‐044), respectively. The media was supplemented with 100‐units/mL of penicillin / streptomycin, 2 mM L‐glutamine (Gibco). Arginine (Arg; R) and lysine (Lys; K) amino acid isotopes were added to a final concentration of 100 mg/L each in the culture medium: [12C]6, [14N]4‐L‐Arg (MW = 174.1117) plus [12C]6, [14N]2‐L‐Lys (MW = 146.1055) for R0K0 ‘light’ (L) medium; [13C]6, [15N]4‐L‐Arg (MW = 184.1241) plus [13C]6, [14N]2‐L‐Lys (MW = 152.1259) for R10K6 ‘heavy’ (H) medium. Cells were tested for incorporation of the labeled amino acids after six passages.
2.3. Whole proteome extraction and subcellular fractionation of SILAC labeled KG1a cells and U937 cells
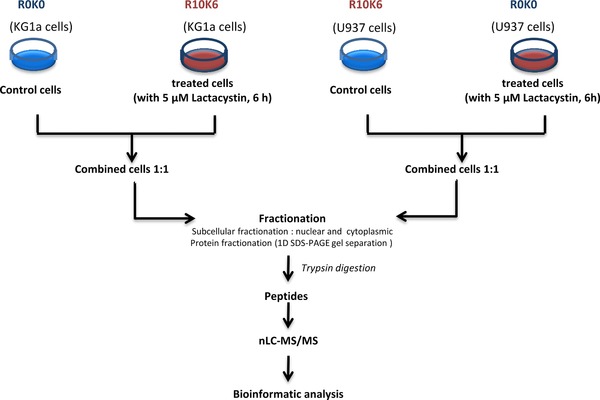
Cells were treated with either DMSO, or 5 μM Lactacytin or 10 nM bortezomib for 2, 6, 12 and 24 h. Whole proteome extract (For SRM analysis): all heavy isotope–labeled cells were mixed together to prepare the super SILAC mix for the SRM analysis 37, 38. Briefly, the light cells were treated separately (treated and untreated, see Fig. 3). Proteins were extracted from the heavy mix and light cells using 8 M urea in 0.1 M Tris‐HCl pH 8.0. After protein isolation, an equal protein amount from heavy mix cells were spiked into each light sample. The mixed samples were subjected to an in solution digestion following the protocol described by Matondo et al. 33, 39. Subcellular Fractionation (For Shotgun analysis): Treated and untreated cells were combined in a ratio 1:1 of cells. Mixed cells, for proteome variation study, were separated into cytoplasm and nuclear fractions using the following protocol. Briefly, 40×106 cells were washed three times with PBS, resuspended in 1 mL of buffer A (20 mM Tris‐HCl pH 7.5, 0.5 mM MgCl2, 0. % NP40, protease inhibitor (Roche)), and sonicated. The sonication efficiency was checked using phase‐contrast microscopy, ensuring that there were no intact cells and that the nucleoli were readily observed as dense, refractile bodies. The sonicated sample was centrifuged at 800 x g for 10 min at 4°C. The supernatant representing the cytoplasmic fraction was centrifuged at 15 000 x g for 45 min at 4°C. This was then washed three times with cooled acetone. The nuclear pellet was resuspended in 1 mL of buffer Bo (4.5 mM MgCl2; 5 mM CaCl2 and 1 μM DNase I) and incubated 10 min at 30°C, followed by centrifugation at 800 x g for 10 min at 4°C. The pellets were then resuspended in 1 mL buffer Bo with 1 M NaCl and centrifuged at 800 x g for 15 min at 4°C. Both nuclear extracts were pooled together. Proteins were quantified by the Bradford method. Equal amounts of total protein from each fraction were loaded onto a SDS‐PAGE gel and the purity of the isolated fractions was checked by Western blot, using antibodies against the nuclear PARP protein, cytoplasmic actin (see Supporting Information).
Figure 3.
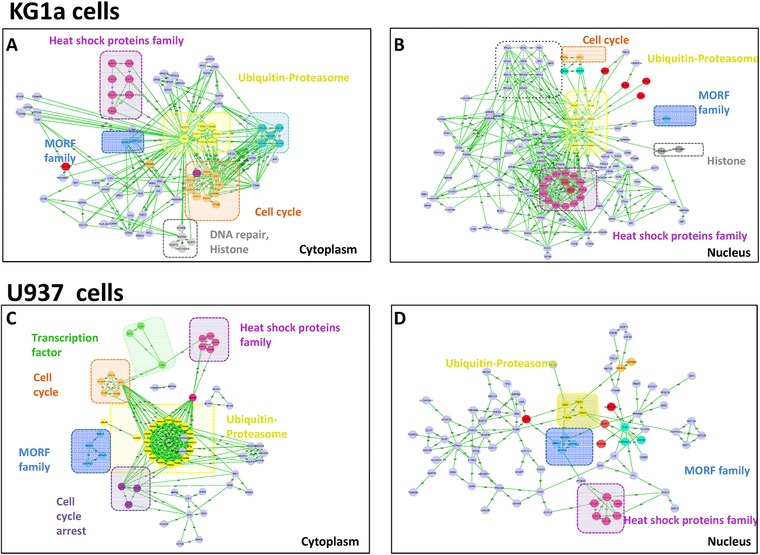
Functional analysis of regulated proteins after proteasome inhibition with Lactacystin in both KG1a and U937 cells. Figure displays significantly regulated proteins (H/L≥1.5) in cytoplasmic fraction (A,cC) and nuclear fraction (B, D) of KG1a and U937 cells respectively. Figures were made using String and Cytoscape. Colors indicate different process affected by the proteasome inhibition.
2.4. 1D SDS‐PAGE fractionation and Nano‐LC‐MS/MS analysis of proteins
After reduction and alkylation, 100 μg of proteins were separated on a 12% acrylamide SDS‐PAGE gel. Proteins were visualized by Coomassie Blue staining. Each lane was cut into 50 homogenous slices that were washed in 100 mM ammonium bicarbonate for 15 min, followed by a second wash in 100 mM ammonium bicarbonate, acetonitrile (1:1) for 15 min. Both washes were performed at 37°C. Second cycle of washes in ammonium bicarbonate and ammonium bicarbonate/acetonitrile was then performed. Proteins were digested by incubating each gel slice with 0.6 μg of modified sequencing grade trypsin in 50 mM ammonium bicarbonate overnight at 37°C. The resulting peptides were extracted from the gel in three steps: a first incubation in 50 mM ammonium bicarbonate for 15 min at 37°C and two incubations in 10% formic acid, acetonitrile (1:1) for 15 min at 37°C. The three collected extracts were pooled with the initial digestion supernatant, dried in a Speed‐Vac, and resuspended with 17 μL of 5% acetonitrile, 0.05% trifluoroacetic acid (TFA). The peptides mixtures were analyzed by nano‐LC‐MS/MS using an Ultimate 3000 system (Dionex) coupled to an LTQ‐Orbitrap Velos mass spectrometer or LTQ‐ Orbitrap XL (Thermo Fisher Scientific). Five microliters of each sample were loaded onto a C18 pre‐column (300 μm id, 5 mm; Dionex), at 20 μL/min, in 5% acetonitrile, 0.05W% TFA. After 5 min of desalting, the pre‐column was switched on line with the analytical C18 column (75 μm id x 15 cm C18 column; packed in‐house with Reprosil C18‐AQ Pur 3 μm resin, Dr. Maisch; Proxeon Biosystems), equilibrated in 95% solvent A (5 acetonitrile, 0.2 formic acid) and 5% solvent B (80 acetonitrile, 0.2 formic acid). Peptides were eluted using a 5–50% gradient of solvent B over 80 min and at a flow rate of 300 nL/min.
The LTQ‐Orbitrap Velos and XL were operated in DDA mode to automatically switch between full scan MS and MS/MS acquisition using Xcalibur software. Survey scan MS was acquired in the Orbitrap over the m/z 300–2000 range, with the resolution set to a value of 60 000 (m/z 400). The 20 most intense ions per survey scan were selected for CID fragmentation and the resulting fragments were analyzed in the linear trap (LTQ). Dynamic exclusion was employed within 60 seconds to prevent repetitive selection of the same peptide. Standard mass spectrometric conditions for all experiments were: spray voltage, 2.2 kV; no sheath and auxiliary gas flow; heated capillary temperature, 200°C; predictive automatic gain control (AGC) enabled, and an S‐lens RF level of 50–60%.
2.5. Bioinformatics analysis
MS raw files were analyzed using the MaxQuant software version 1.5.3.8 40, 41. MS/MS spectra were searched in the Andromeda search engine against the forward and reverse Human Uniprot database (‘Human Reference Proteome’ retrieved on February 2014) combined with a commonly observed contaminants list. The digestion enzyme was set to trypsin/ with up to two missed cleavages. Methionine oxidation, ubiquitylation (GlyGly (K)) and N‐terminal acetylation were search as variable modifications and carbamidomethyl of cysteine as fixed modification. Parent peptide masses and fragment masses were searched with maximal initial mass deviation of 6–20 ppm, respectively. Mass recalibration was performed by a preceding Andromeda search with a mass window of 20 ppm Match between run was used. A first level of False Discovery Rate (FDR) filtration was done on the peptide‐spectrum match level, and this was followed by a second level of FDR control on the protein level. Both filtrations were performed at a 1% FDR. These filtrations were done using a standard target‐decoy database approach. When two proteins (isoforms and homologues with two Uniprot identifiers) could not be distinguished based on the identified peptides, they were merged by MaxQuant into one protein group (See Supporting Information table S2).
Statistical analysis of the data was performed using Perseus 1.5.2.6 42 (www.perseus‐framework.org), environment R, String (http://string‐db.org/), and cytoscape tools (http://www.cytoscape.org/) 43.
2.6. SRM‐LC‐MS/MS
Extracted proteins were solubilized in denaturation buffer (2‐amino‐2‐hydroxymethyl‐1,3‐propanediol (Tris) 100 mM pH 8.0, 8 M urea). Reduction of disulfide bridges was performed in 5 mM DTT for 30 min at room temperature; alkylation was performed in 20 mM iodocacetamide in the dark for 30 min at room temperature. Protein solutions was diluted in 50 mM ammonium bicarbonate (BA), until the urea concentration was below 1 M. Proteins were digested with trypsin (Promega, Madison, WI, USA) using a ratio of 1:100 enzyme / protein at 37°C overnight. Resulting peptides were purified using Sep‐Pak C18. Briefly, 50 mg of C18 phase was activated in methanol, rinsed once in 80% acetonitrile (ACN) 0.1% FA, and washed thrice in 0.1% FA. Tryptic peptides were analyzed on a TSQ Vantage Triple Quadrupole mass spectrometer (ThermoFisher scientific, San Jose, CA), equipped with a nanoelectrospray ion source. A spray voltage of 1.3 kV was used with a heated ion transfer tube set to a temperature of 280°C. Chromatographic separations of peptides were performed on a NanoLC‐2D plus HPLC system (Eksigent, Dublin, CA) coupled to a 10 cm fused silica emitter, 75 μm id, packed with a Magic C18 AQ 5 μm resin (Michrom BioResources, Auburn, CA, USA). Peptides were loaded onto the column from a cooled (4°C) Eksigent autosampler and separated with a linear gradient of acetonitrile/water, containing 0.1% formic acid, and at a flow rate of 300 nL/min. A gradient of 5 to 35% acetonitrile in 40 minutes was used. The mass spectrometer was operated with selected reaction monitoring (SRM) mode. For SRM acquisitions, Q1 and Q3 were measured at 0.7 unit mass resolution. Transitions were recorded for the endogenous (light) and the internal standard (heavy) peptides. Time‐based SRM was used to achieve a high dwell time, i.e > 50 ms, for each transition, where an acquisition time window of 3 min was set around their elution time. Argon was used as the collision gas at a nominal pressure of 1.5 mTorr. Collision energies (CE) for each transition were calculated according to the following equations: CE = 0.034 * m/z + 3.314 and CE = 0.044 * m/z + 3.314 (for doubly and triply charged precursor ions, respectively. The intensity of each SRM was extracted using Skyline (https://skyline.gs.washington.edu) and the relative protein abundances were summarized across all peptide intensities for each sample using MSstats 44, 45 (http://www.msstats.org).
For each target protein up to three signature peptides were selected for the SRM measurement. This selection was based on shotgun analysis performed previously, their proteotypicity (i.e., specific, to the protein of interest), their reported frequency in proteomic data repositories such as Peptide Atlas 46 and Human SRMAtlas 47. In total, three transitions per peptide were monitored resulting in a total of 1063 transitions (292 precursors, and 771 fragments) for the 66 targeted proteins (see Supporting Information Table S4).
3. Results and discussion
3.1. The level of apoptosis induced by Lactacystin and MG132 in immature KG1a AML (M0) cells is higher than in more mature U937 AML cells (M5)
KG1a and U937 cells were treated either with DMSO as a control or with proteasome inhibitors (Lactacystin or MG132) for 24 h and the percentage of apoptotic cells were measured as described in our previous study 14. Figure 1 displays results obtained using 5 μM MG132 (Fig. 1A) and 5 μM Lactacystin (Fig. 1B). These results show that both MG132 and Lactacystin induced high level apoptosis in immature KG1a AML cells (∼65% and ∼20% respectively) compared to U937 AML cells (∼25% and ∼10% respectively). The percentage of apoptotic cells induced by MG132 for both cell lines, less specific inhibitor of the proteasome, is higher than the percentage induced by Lactacystin, the specific inhibitor of the proteasome.
Figure 1.
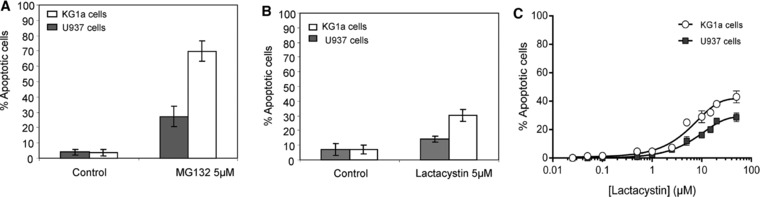
Comparison of the induction of apoptosis in KG1a and U937 AML cell lines. Induction of apoptosis in KG1a and U937 cells using two proteasome inhibitors, MG132 and Lactacystin. The percentage of apoptotic cells was measured using Annexin V assays performed 24 h after the treatment of KG1a and U937 AML cell lines with either DMSO or 5 μM MG132 (A) or 5 μM Lactacystin (B). Comparison of the induction of apoptosis in KG1a and U937 AML cell lines using increasing concentrations of Lactacystin (C). The percentage of apoptotic cells was measured using Annexin V assays performed 24 h after the treatment of KG1a and U937 cells with 0.1 μM to 50 μM Lactacystin as describded in our previous work Matondo et al., 2010 14. Results correspond to the mean ± SD of at least three independent experiments.
Dose‐response experiments were then performed with Lactacystin to better characterize the induction of apoptosis after proteasome inhibition using this specific inhibitor on KG1a and U937 AML cells. The corresponding curves were plotted (Fig. 1C). As described in our previous study for Bortezomib and MG132 proteasome inhibitor 14, apoptotic response induced by Lactacystin present the same trend that the two others inhibitors and was dose‐dependent. The results indicated that the proportion of apoptotic cells was higher for KG1a cells than for U937 cells at all inhibitor concentrations used (Fig. 1C).
3.2. Workflow to map protein expression changes associated with proteasome inhibition
To identify proteins regulated after inhibition of proteasome in AML cells at different stages of maturation, a treatment with a covalent and specific inhibitor of the proteasome, Lactacystin 48, was applied to KG1a and U937 cells. Duration of treatment and concentration of the drugs were chosen to study early events that could explain differential apoptotic level response between KG1a and U937 cells after proteasome inhibition (Supporting Information figure S1). Apoptotic response of both cells at 6 h and 5 μM of Lactacystin were compared (data not shown) and this condition was selected for the large‐scale shotgun analysis. As a control, cells were treated with DMSO for 6 h. The DMSO‐ control and Lactacystin‐treated cells were grown in either “light” (L) or “heavy” (H), respectively R0K0 or R10K6 SILAC media (see Materials and Methods). After 6 h of incubation, the two cell populations were mixed in equal proportions and processed for MS analysis as described below.
The two mixed‐cell populations were first fractionated into their subcellular compartments in order to reduce the complexity of sample protein content. Subsequently, nuclear and cytoplasmic subcellular fractions were analysed by SDS‐PAGE gel, then each gel lane was subdivided into 50 bands (Fig. 2). Proteins from each band were digested with trypsin and analysed by LC‐MS/MS. Samples were prepared twice and peptides analyzed by MS in a LTQ ‐Orbitrap mass spectrometer. The subcellular distribution of selected marker proteins known to be mainly present in vivo in either the cytoplasm (actin) or the nucleus (PARP) confirmed that each fraction was correctly prepared (Supporting Information figure S2). Extensive fractionation of our samples reduced their complexity and increased the chance to identify low abundant proteins by shotgun analysis.
Figure 2.
Overview of the experimental workflow used in this study. KG1a and U937 cells were grown in the presence of light (R0K0) or heavy (R10K6) stable isotopes of arginine and lysine (cf. Materials and Methods). Cells were incubated for 6 h with 5 μM Lactacystin, and the experiment was done in duplicate. Proteins were fractionated into cytoplasmic and nuclear fractions. Equal amount of each fraction was then separated by 1D SDS‐PAGE, proteins from each slice digested using trypsin. Resulting peptides were analyzed by nanoLC and measured on an LTQ‐Orbitrap instrument. Protein identification was accomplished with Maxquant and data analysis with Perseus.
Two individual experiments were analysed on the same Maxquant search and statistical analysis performed with Perseus to allow the comparison between experiments 42. Our analysis gave a large coverage of AML cells proteomic, with approximatively 100 000 identified sequence‐unique peptides and more than 7 000 identified proteins with 1% FDR, both on the peptide‐spectrum match, and protein levels (Supporting Information Tables S1 and S2). Our results include the identification of low abundant proteins such as important transcription factors including JUN, c‐Myc, SP1 and ATF3 (Supporting Information Tables S1 and S2) or Interleukin family proteins. In‐depth comparison of the identified proteins from each fraction showed a high overlap (> 70%) between our two experiments. Due to its higher performance, the LTQ‐Orbitrap Velos leads to significantly more identifications at protein and peptides levels in the analysis of complex peptide mixtures (Supporting Information figure S3). About 3918 proteins were quantified in KG1a cytoplasmic fraction of the replicate 2 whereas 3070 proteins were quantified on replicate 1. From these quantified proteins, 2258 were common between the two replicates. The same trend was observed for U937 cells where 3708 proteins were quantified in the cytoplasmic fraction of replicate 2 and 2919 proteins were quantified of replicate 1. From these quantified proteins, 2267 were common between the two replicates. These results are particularly pronounced in the nuclear fraction. For both cell lines the LTQ‐Orbitrap Velos allows identification of approx. 50% more proteins in nuclear fraction (3958 versus 1477 proteins for KG1a, and 3495 versus 1809 proteins for U937) (Supporting Information figure S3). This trend might also be related to better extraction of the nuclear proteins for the second biological replicate. Conversely, a large majority of the quantified proteins were found in both replicates and allows for their comparison.
In addition, identified proteins from KG1a and U937 cells cellular models were similar and therefore allowed an unbiased comparison of their respective proteomes and their modulation after proteasome inhibition (Supporting Information Tables S1 and S2). The large number of proteins identified in this work offers a system‐wide quantitative view of the proteome variations in KG1a and U937 cells upon proteasome inhibition, which may serve as a basis for further downstream computational analysis and biological interpretation.
3.3. Functional discrimination between KG1a cells and U937 cells
Herein, we analysed the proteins that change in abundance and could explain the differential apoptotic responses to the proteasomal inhibition observed between immature KG1a and mature U937 AML cells. The total proteome dataset from each cell line was examined for responses to inhibition of proteasome by Lactacystin. Comparison of the quantified proteins from the two replicates of each cell line showed a high reproducibility (Supporting Information figure S4). These results validated our strategy.
To define a robust biological response, we set a conservative cutoff of a minimal 1.5‐fold change in the abundance ratio. We obtained three categories of proteins: up‐regulated (minimal ratio of heavy/light (H/L) of 1.5), down‐regulated (maximal ratio H/L of 0.7), and unchanged (0.7 < ratio H/L < 1.5). In both cell lines and subcellular fraction, less than 5% of the quantified proteins showed a significant change in abundance after proteasome inhibition, i.e., either increase, or decrease, greater than ∼1.5 fold after 6 h of Lactacystin treatment (Supporting Information Table S1 and S2). In total, 266 proteins were found significantly changed in cytoplasmic fraction and 134 proteins in the nuclear fraction of KG1a cells. Conversely, 103 and 148 proteins were found significantly changed in the cytoplasmic and nuclear fractions of U937 cells, respectively (Supporting Information Tables S3). Interestingly, some proteins that change in abundance were common to both cell lines, such as cyclin, heat shock and protein from UPP pathway, reflecting the global response of the cells to proteasome inhibition (Supporting Information Tables S3). These proteins are targets of the UPP. Others were specific to individual cell lines, for example IL32, and may potentially be involved in the differential apoptotic response to proteasome inhibition.
3.4. Proteasome inhibition induces the unfolded protein response and stress response
Protein abundance changes measured in the present study derive from two different processes following proteasome inhibition, i.e., decrease of protein degradation (accumulation of poly‐ubiquitinylated proteins) and regulation of specific genes. Indeed, it has been reported that proteasome inhibition induces the accumulation of k‐48‐linked ubiquitin protein targets and a loss of ubiquitin from a cohort of putatively monoubiquitylated proteins in the cells 49, 50. Herein, we quantified the up‐regulation (H/L>1.5) of Ubiquitin C (UBC), indicating the accumulation of polyubiquitylated proteins after 6 hours of proteasome inhibition in cytoplasmic fractions of both cell lines 49 (Supporting Information figure S5A). In addition, in order to cope with this inhibition 17, cells activate a specific gene expression response, leading to a change in abundance of some proteins.
As previously described in the literature, we quantified the de novo formation of the catalytic subunits of the 20S core complex to replace the inhibited forms, as well as the accumulation of the other subunits of the 20S core 51, 52, 53. This was particularly observed in the cytoplasm, confirming a recent result from our group showing that 20S proteasome is ∼1.5 higher in KG1a cells than in U937 cells 14, 54. Other compounds of the UPP were also found to be up‐regulated in both cell lines after proteasome inhibition, including E2 conjugating (UBE2C) and E3 ligase enzymes, proteasome regulators (PSMD3, PSME1…), and proteins from the cullin family (Fig. 3) (Supporting Information figure S5B).
Other well‐known pathways were found to be differentially regulated in this study and used as positive control to validate our results. A global increase of Heat‐Shock Proteins (HSP) (Supporting Information figure S5C), including HSP70 1A/1B, HSP105 HSPH1, HSPJ2 and HSP40 DNAJB1 was observed with a greater ratio in KG1a cells than U937 cells (Fig. 3, Supporting Information figure S5C). Our proteomics data showed an increase of proteins that are associated with the induction of the unfolded protein response, including ATF3, HSF2, PDRG1 and HSP 90AB1. Overall, these changes reflect a stress response in both cells lines, which would be consistent with the well‐documented role of these molecular chaperones in conferring protection against therapeutic agents 22, 55. However, these effects were found to be more pronounced in KG1a cells than in U937 cells, which shown to be more sensitive to proteasome inhibitors 14.
3.5. Cell cycle regulated proteins and pro‐apoptotic proteins
In agreement with several other studies, cyclins and the cell‐cycle progression pathway were found to be up‐regulated after proteasome inhibition in both nuclear and cytoplasmic fractions, for both cell lines (Fig. 3, Supporting Information figure S5D) 17, 18. Stathmin‐3 was found to be highly modulated in cytoplasm of KG1a cells (Supporting Information figure S5D). The pro‐apoptotic protein SIVA, previously described as a direct p53‐dependant gene selectively induced during apoptosis 55, was found specifically up‐regulated in the cytoplasmic fraction of KG1a cells (Supporting Information figure S6A). In contrast, cell cycle and apoptosis regulator proteins 1 and 2 (CARP‐1 and 2), an apoptosis inducer that regulates apoptosis signalling, were found to be down‐regulated only in U937 cells (Supporting Information Table S3). These regulated proteins tend to enhance the apoptotic effect of the proteasome inhibitor in KG1a cells.
3.6. MORF/MRG family of novel transcription factors
We quantified the accumulation of three well‐known interaction partners, i.e, two chromatin regulatory proteins, MORF 4L1 and L2, and MRFAP1, in both cell lines after proteasome inhibition (Supporting Information figure S6B). Recent studies have shown that MRFAP1 proteins have fast turnover rates, and are rapidly degraded via the ubiquitin‐proteasome system 56, 57. This protein and its interaction partner MORF4L1 were found amongst the most up‐regulated proteins after NEDD8 inhibition, in multiple human cell lines 57, 58, 59. The function of NEDD8, an ubiquitin‐like protein, is to promote the protein degradation of substrate through the UPP. NEDD8 is first activated by an E1 enzyme (NEDD8 activating enzyme (NAE; a heterodimer of NAE1 and UBA3 subunits)), transferred to an E2 enzyme (Ubc12, also known as UBE2M), and then conjugated to target substrates. To‐date, the most well characterized substrates of the NEDD8 pathway are the cullin family of proteins, including CUL4B 60.
Finally, Larance and colleagues have shown that MRFAP1 may regulate the ability for MORF4L1 to interact with chromatin modifying enzymes by binding to MORF4L1 in a mutually exclusive manner, with MRGBP part of the NuA4 complex 57. These proteins may play an important role in disease states, such as our cellular system, that could be influenced by alterations in histone acetylation and related modifications. This knowledge, combined with the fact that MORF proteins are rapidly accumulated after treatment with proteasome inhibitors used for the treatment of several types of myeloma and lymphoma, prompted development of specific inhibitors of the NEDD8 machinery and opens up new avenues of exploration for the mechanism of the proteasome inhibitor in haematological cells.
3.7. Cytokines responses: IL32, a potentially new pro‐apoptotic factor
Although a large number of interleukin family protein members were identified in this study, only IL32 was found to be specifically regulated with an average ratio of H/L greater than 6 in KG1a cells after inhibition.
IL‐32 is an inflammatory cytokine produced by T lymphocytes, natural killer (NK) cells, epithelial cells, and blood monocytes 61, 62. The expression of this protein is increased after the activation of T‐cells by mitogens or the activation of NK cells by IL‐2. IL‐32 induces tumor necrosis factor‐alpha (TNF‐α), IL‐1β, IL‐6, and IL‐10 and thereby may play an important role in tumor development. This protein encodes for six isoforms from alternate transcriptional splice variants 62, 63. One of the isoforms IL‐32 beta, has recently been described as a main player in the activation of NF‐kB and STAT3 pathways, which control tumor growth 64. In our study we covered about 60% of the sequence of IL32 protein but could not differentiated the isoform that changes in abundance after proteasome inhibition.
Even if we do not have functional validation of this candidate, we hypothesize that the up‐regulation of IL32 protein following proteasome inhibition, which appears from our result to be specific to KG1a cells, may be an important player in the differential apoptotic response 65 between both cell types. This may be related to the modulation of the NF‐kB pathway, although alternative mechanisms may also underly the role of IL32 in the proteasomal inhibition response.
3.8. Verification of new candidates by SRM
A targeted‐MS approach based on SRM 31, 35, 45 was used to follow the change of abundance of selected proteins over time after proteasome inhibition. Herein, we wanted to evaluate and compare the effects of bortezomib, used in the treatment of some AML patients, and Lactacystin (Fig. 4).
Figure 4.
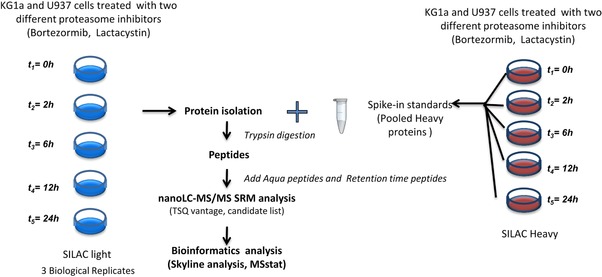
Workflow for the verification of potential candidates by SRM. Two cell populations of KG1a and U937 cells were SILAC‐encoded by growing them in the presence of (light and heavy) stable isotope‐labeled arginine and lysine. Cells were then treated either with 5 μM of lactacystin or 10 nM of bortezomib for 2, 6, 12 and 24 h. All heavy cells were mixed and used as standards for quantification. After protein isolation, the same amount of Heavy labeled proteins was added into each sample. Heavy and light‐labeled proteins were then digested using trypsin. Resulting peptides were analyzed by nanoLC–SRM on the TSQ vantage. A list of 66 proteins, identified previously by shotgun, was targeted by SRM (Table 1).
A total of 66 proteins that were identified in the shotgun experiment were selected for further verification by SRM (Table 1). This list of proteins included tubulin and HSP proteins, well‐known to be regulated after proteasome inhibition, as negative and positive controls, respectively. These proteins also served to evaluate the variation of sample preparation and MS measurement, and to normalize the data. The selected proteins for SRM quantification are involved in cell cycle control, apoptosis, transcription, DNA repair, and stress response and chromosome organization.
Table 1.
List of the 66 selected candidates from Shotgun analysis for the SRM measurement. Selected proteins are involved in the cell cycle apoptosis, transcription DNA repair, and stress response and chromosome organization
| GO Process | Protein names | Gene names | Uniprot Acc | Entry name |
|---|---|---|---|---|
| Apoptosis, Cell death | Apoptosis‐enhancing nuclease | AEN | Q8WTP8 | AEN_HUMAN |
| Apoptosis, Cell death | Phorbol‐12‐myristate‐13‐acetate‐induced protein 1 (PMA‐induced protein 1) | PMAIP1 | Q13794 | APR_HUMAN |
| Apoptosis, Cell death | Apoptosis regulator BAX (Bcl‐2‐like protein 4) (Bcl2‐L‐4) | BAX | Q07812 | BAX_HUMAN |
| Apoptosis, Cell death | Apoptosis regulator Bcl‐2 | BCL2 | P10415 | BCL2_HUMAN |
| Apoptosis, Cell death | BH3‐interacting domain death agonist (p22 BID) (BID) | BID | P55957 | BID_HUMAN |
| Apoptosis, Cell death | Bcl‐2‐interacting killer (Apoptosis inducer NBK) (BIP1) (BP4) | BIK | Q13323 | BIK_HUMAN |
| Apoptosis, Cell death | Baculoviral IAP repeat‐containing protein 3 (EC 6.3.2.‐) (Apoptosis inhibitor 2) | BIRC3 | Q13489 | BIRC3_HUMAN |
| Apoptosis, Cell death | Growth arrest and DNA damage‐inducible protein GADD45 beta | GADD45B | O75293 | GA45B_HUMAN |
| Apoptosis, Cell death | NF‐kappa‐B inhibitor alpha (I‐kappa‐B‐alpha) (IkB‐alpha) | NFKBIA | P25963 | IKBA_HUMAN |
| Apoptosis, Cell death | Induced myeloid leukemia cell differentiation protein Mcl‐1 | MCL1 | Q07820 | MCL1_HUMAN |
| Apoptosis, Cell death | Pleckstrin homology‐like domain family A member 3 | PHLDA3 | Q9Y5J5 | PHLA3_HUMAN |
| Apoptosis, Cell death | Apoptosis regulatory protein Siva | SIVA1 | O15304 | SIVA_HUMAN |
| Cell adhesion | Protocadherin alpha‐C2 (PCDH‐alpha‐C2) | PCDHAC2 | Q9Y5I4 | PCDC2_HUMAN |
| Cell cycle | G2/mitotic‐specific cyclin‐B1 | CCNB1 | P14635 | CCNB1_HUMAN |
| Cell cycle | Cell division cycle 7‐related protein kinase | CDC7 | O00311 | CDC7_HUMAN |
| Cell cycle | Cyclin‐dependent kinase 1 (CDK1) | CDK1 | P06493 | CDK1_HUMAN |
| Cell cycle | Cyclin‐dependent kinase inhibitor 1 (CDK‐interacting protein 1) | CDKN1A | P38936 | CDN1A_HUMAN |
| Cell cycle | Cyclin‐dependent kinase inhibitor 1B | CDKN1B | P46527 | CDN1B_HUMAN |
| Cell cycle | Cell cycle checkpoint control protein RAD9B | RAD9B | Q6WBX8 | RAD9B_HUMAN |
| Cell cycle | Kinesin‐like protein KIFC1 (Kinesin‐like protein 2) | KIFC1 | Q9BW19 | KIFC1_HUMAN |
| Cell cycle | Proline/serine‐rich coiled‐coil protein 1 | PSRC1 | Q6PGN9 | PSRC1_HUMAN |
| Cell cycle, apoptosis | NEDD8‐activating enzyme E1 regulatory subunit | NAE1 | Q13564 | ULA1_HUMAN |
| Cytoskeleton Organization, cell cycle | Tubulin alpha‐1A chain (Alpha‐tubulin 3) | TUBA1A | Q71U36 | TBA1A_HUMAN |
| DNA repair | Protein BTG2 (BTG family member 2) | BTG2 | P78543 | BTG2_HUMAN |
| DNA repair | Mortality factor 4‐like protein 1 | MORF4L1 | Q9UBU8 | MO4L1_HUMAN |
| DNA repair | Mortality factor 4‐like protein 2 | MORF4L2 | Q15014 | MO4L2_HUMAN |
| DNA repair | MORF4 family‐associated protein 1 | MRFAP1 | Q9Y605 | MOFA1_HUMAN |
| DNA repair | MRG/MORF4L‐binding protein | MRGBP | Q9NV56 | MRGBP_HUMAN |
| DNA repair | PCNA‐associated factor | KIAA0101 | Q15004 | PAF15_HUMAN |
| Histone deacetylase binding | Ankyrin repeat family A protein 2 (RFXANK‐like protein 2) | ANKRA2 | Q9H9E1 | ANRA2_HUMAN |
| Immune response, cell adhesion | Interleukin‐32 (IL‐32) | IL32 | P24001 | IL32_HUMAN |
| Others | Beta‐1,3‐N‐acetylglucosaminyltransferase lunatic fringe | LFNG | Q8NES3 | LFNG_HUMAN |
| Protein folding | p53 and DNA damage‐regulated protein 1 | PDRG1 | Q9NUG6 | PDRG1_HUMAN |
| Response to stress | Putative heat shock protein HSP 90‐beta 2 | HSP90A | Q58FF8 | H90B2_HUMAN |
| Response to stress | Putative heat shock protein HSP 90‐beta‐3 | HSP90BC | Q58FF7 | H90B3_HUMAN |
| Response to stress | Heat shock 70 kDa protein 1A | HSPA1A | P0DMV8 | HS71A_HUMAN |
| Response to stress | Heat shock 70 kDa protein 1B | HSPA1B | P0DMV9 | HS71B_HUMAN |
| Response to stress | Heat shock 70 kDa protein 1‐like (Heat shock 70 kDa protein 1L) (Heat shock 70 kDa protein 1‐Hom) (HSP70‐Hom) | HSPA1L | P34931 | HS71L_HUMAN |
| Response to stress | Heat shock protein HSP 90‐alpha A2 | HSP90AA2P | Q14568 | HS902_HUMAN |
| Response to stress | Putative heat shock protein HSP 90‐alpha A4 | HSP90AA4P | Q58FG1 | HS904_HUMAN |
| Response to stress | Heat shock protein HSP 90‐alpha | HSP90AA1 | P07900 | HS90A_HUMAN |
| Response to stress | Heat shock protein beta‐1 (HspB1) | HSPB1 | P04792 | HSPB1_HUMAN |
| Response to stress | DnaJ homolog subfamily B member 1 | DNAJB1 | P25685 | DNJB1_HUMAN |
| Signal transduction | NF‐kappa‐B inhibitor‐interacting Ras‐like protein 1 | KBRAS1 | Q9NYS0 | KBRS1_HUMAN |
| Signal transduction | NF‐kappa‐B inhibitor‐interacting Ras‐like protein 2 | KBRAS2 | Q9NYR9 | KBRS2_HUMAN |
| Transcription | Transcription factor E2F7 (E2F‐7) | E2F7 | Q96AV8 | E2F7_HUMAN |
| Transcription | Transcription factor MafF (U‐Maf) | MAFF | Q9ULX9 | MAFF_HUMAN |
| Transcription | Nuclear factor NF‐kappa‐B p105 subunit p50 subunit] | NFKB1 | P19838 | NFKB1_HUMAN |
| Transcription | Nuclear factor NF‐kappa‐B p100 subunit | NFKB2 | Q00653 | NFKB2_HUMAN |
| Transcription, DNA damage response | Cellular tumor antigen p53 | P53 | P04637 | P53_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit alpha type‐1 | PSMA1 | P25786 | PSA1_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit alpha type‐2 | PSMA2 | P25787 | PSA2_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit alpha type‐3 | PSMA3 | P25788 | PSA3_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit alpha type‐4 | PSMA4 | P25789 | PSA4_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit alpha type‐5 | PSMA5 | P28066 | PSA5_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit alpha type‐6 | PSMA6 | P60900 | PSA6_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit alpha type‐7 | PSMA7 | O14818 | PSA7_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit beta type‐10 | PSMB10 | P40306 | PSB10_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit beta type‐5 | PSMB5 | P28074 | PSB5_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit beta type‐6 | PSMB6 | P28072 | PSB6_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit beta type‐7 | PSMB7 | Q99436 | PSB7_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit beta type‐8 | PSMB8 | P28062 | PSB8_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome subunit beta type‐9 | PSMB9 | P28065 | PSB9_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome activator complex subunit 1 | PSME1 | Q06323 | PSME1_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome activator complex subunit 2 | PSME2 | Q9UL46 | PSME2_HUMAN |
| Ubiquitin Proteasome pathway | Proteasome activator complex subunit 3 | PSME3 | P61289 | PSME3_HUMAN |
Treated cells with either Lactacystin or bortezomib were harvested at 2, 6, 12 and 24 hours after inhibition. As a reference, we used the spike‐in method as described by T. Geider 66 and AQUA peptides. Mixtures of heavy‐ and light stable isotope‐labeled proteins were processed for MS analysis as described in Fig. 4. For the missing proteins in the spike‐in mixture, such as IL32, we used AQUA peptides to quantify the protein of interest (Supporting Information Table S4). Three individual biological replicates were prepared.
Out of the 66 human proteins, only 53% could be successfully detected and quantified in the majority of samples. Proteins which were not constituently detected in similar replicate were excluded for the further analysis. The success rate could be explained by the fact that we measured non‐fractionated samples by SRM and therefore lost the deeper analytical profile provided by extensive fractionation, as performed in the global shotgun approach. Indeed our selected proteins for the SRM measurement, such as transcription factors, are low abundant proteins. Therefore these proteins are not easy to detect in total extract proteome by SRM with sufficient number of transitions, even if the SRM is very sensitive. In addition, the spike‐in strategy used for the quantification also increases the complexity of the mixture and therefore reduced the ability to detect very low abundant proteins. After the extraction and integration of the peak intensity with Skyline, the statistical analysis of the data was performed with MSstats 34, 44, 45. The results of the statistical tests were summarized by Volcano plots (adjusted p‐value versus log fold change) for the treatment of both KG1a and U937 cells with bortezomib (Fig. 5A) and Lactacystin (Fig. 5B).
Figure 5.
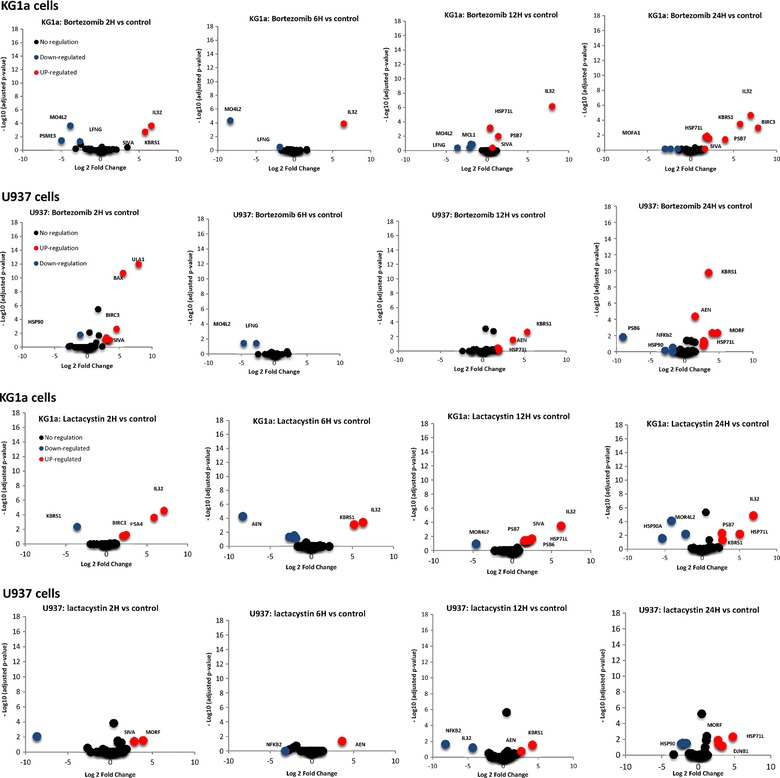
Quantitative analysis of proteins that change in abundance after proteasome inhibition with bortezomib (A) or Lactacystin (B) in both KG1a and U937 cells. The volcano plot represent the statistical significance expression level change (p‐value, SRM stat), as a function of protein expression ratio between control and treated cells. The red and blue dots indicate up‐ and down regulated proteins, respectively.
As anticipated, the abundance of HSP71 protein, our positive control, increases over time after the proteasome inhibition with bortezomib (Fig. 5A), while the abundance of tubulin proteins does not change, for both KG1a cells and U937 cells (Supporting Information figure S7). The same trend was observed after inhibition with Lactacystin. The abundance of HSP71 increase faster in KG1a cells which is more sensitive to proteasome inhibitor than in U937 (Supplemental figure S8). Other proteins selected from the shotgun experiment were also found up‐ or down‐regulated by SRM with the same behavior (Bax, PSB7, BIRC3.). Herein, in contrast to the shotgun analysis, cells were not fractionated into subcellular fractionation, therefore, modulation of the abundance this proteins reflects the change at the cellular level. Proteins found to change in abundance tend to be in favour of the induction of apoptosis in our cells. KBRS1 protein a Ras‐like protein that acts as a potent regulator of NF‐kappa‐B activity by preventing the degradation of NF‐kappa‐B inhibitor beta (NFKBIB) was found to be up‐regulated in both KG1a and U937 cells after proteasome inhibition with Lactacystin and bortezomib. Furthermore, Apoptosis‐enhancing nuclease protein (AEN), an exonuclease with an activity against single‐ and double‐stranded DNA and RNA, was found to be rapidly up‐regulated in U937 cells. This protein mediates p53‐induced apoptosis. This results suggest that apoptosis induced by proteasome inhibitor in U937 is mediated by p53‐dependent pathways of apoptosis.
In agreement with our global analysis, SIVA, and MORF proteins were found to be rapidly regulated after proteasome inhibition with either bortezomib or Lactacystin (Fig. 5A and B). Our data showed that the abundance of SIVA plateaus (Fig. 6) and is in agreement with the % rate of the apoptosis measured by FACS 14. IL32 was confidently quantified in KG1a cells. IL32 protein was found to be rapidly up‐regulated in KG1a, with a ratio higher than 10 after only 2 hours of inhibition with either bortezomib or Lactacystin (Fig. 5). Interestingly, IL32 up‐regulation occurs in KG1a cells, where an important apoptotic response is also observed (Fig. 6 and Supporting Information Figure 1). Inhibitor‐induced apoptosis is greatly delayed in U937 cells, where the IL32 expression level is also less affected. It has been established that IL‐32 may antagonize cancer growth and represent a useful target for human breast cancer 67. Therefore, we believe that this protein could be a potential key protein in the mechanism of action of proteasome inhibitors in KG1a cells and U937 cells. Complementary approaches, such as gene silencing, will help to better characterize the roles SIVA, MORF and IL32 in the mechanism of proteasome inhibitors in KG1a cells and U937 cells.
Figure 6.
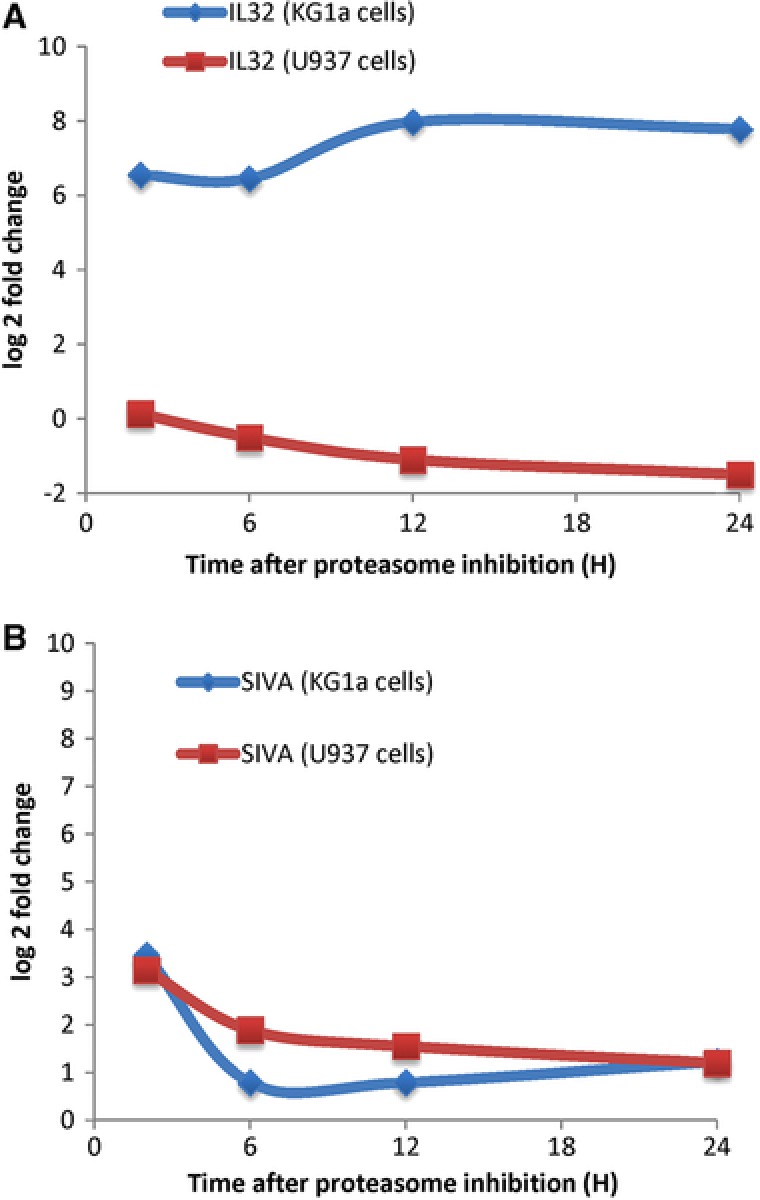
Differential regulation of IL32 and SIVA upon the proteasome inhibition measured by SRM. Cells were treated with 10 nM bortezomib and total protein were extracted and digested with trypsin and measured by SRM (see Materials and Methods). Graph reported the protein abundances (log 2 Fold change) over time (0 to 24 h) of IL32 (A) and SIVA (B) proteins after proteasome inhibition with bortezomib of both KG1a (blue) and U937 (red) cells. Results correspond to the mean ± SD of at least three independent experiments.
4. Concluding remarks
In conclusion, we have used SILAC‐based quantitative MS combined with targeted approaches to characterize proteins that change in abundance upon proteasomes inhibition in two different AML cells and at differential stages of maturation. Our data establish for the first time a global profiling of these cells after proteasome inhibition with high quantification accuracy, allowing a deeper exploration of the biological processes involved. Proteins found to change in abundance are in agreement with high apoptosis levels induced in KG1a cells by all the proteasome inhibitors tested. In addition to the well‐known regulated proteins upon proteasome inhibition, we identified novel players, including IL32, SIVA, and MORF family proteins, that could potentially explain the differential apoptotic response observed. Our results suggest that apoptosis induced by the proteasome inhibition in U937 cells is p53‐dependent pathway and might be different for KG1a cells. These results open up new avenues to investigate the mechanism of the proteasome inhibitors in AML, but also for the development of inhibitors with higher therapeutic potency and minimal toxicity to normal cells.
The studies were designed by MaMat, KC, MaMar, AG, MB, BM and OS. Experimental work and data analysis were performed by MaMat, KC and MaMar. MaMat wrote the paper. All authors contributed to discussion of the results and critical review of the manuscript. O.S and B.M supervised the project.
The authors declare that they have no conflict of interest.
Supporting information
figS1
figS2
figS3
figS4
figS5
figS6
figS7
figS8
Table‐S1‐S2
Table S3
Table S4
Supplementary Figures and Tables
Western Blot and antibodies
Acknowledgments
We thank members of the group for insightful comments related to this work. We would like to thank David Bouyssié for MFPAQ software and technical support. We would like also to thank our collaborators Stephane Manenti, for discussion. We Thanks Pr. Dr. Ruedi Aebersold from IMSB (ETH Zurich) for allowing the SRM measurement on the TSQ Vantage. The work was supported in part by grants from the Région Midi‐Pyrénées, the “Fondation pour la Recherche Médicale” (programme Grands Equipements), European Fonds (FEDER), Toulouse metropole, “Fond Social Européen” (FSE) and the “Ligue contre le cancer” Grants. We thank Dr. Darragh O'Brien from Institut Pasteur for the careful reading of this manuscript.
Matondo M., Marcellin M., Chaoui K., Bousquet‐Dubouch M., Gonzalez‐de‐Peredo A., Monsarrat B., Burlet‐Schiltz O., Proteomics 2017, 17, 1600089.
Colour Online: See the article online to view Figs. 2 and 4 in colour.
This article is part of the Special Issue on “Applications and Developments in Targeted Proteomics” which can be accessed at: http://onlinelibrary.wiley.com/doi/10.1002/pmic.v16.15‐16/issuetoc.
5 References
- 1. Voges, D. , Zwickl, P. , Baumeister, W. , The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999, 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- 2. Gillessen, S. , Groettup, M. , Cerny, T. , The proteasome, a new target for cancer therapy. Onkologie 2002, 25, 534–539. [DOI] [PubMed] [Google Scholar]
- 3. Mato, A. R. , Feldman, T. , Goy, A. , Proteasome inhibition and combination therapy for non‐Hodgkin's lymphoma: from bench to bedside. Oncologist 2012, 17, 694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leonard, J. P. , Furman, R. R. , Coleman, M. , Proteasome inhibition with bortezomib: a new therapeutic strategy for non‐Hodgkin's lymphoma. Int. J. Cancer 2006, 119, 971–979. [DOI] [PubMed] [Google Scholar]
- 5. Gu, J. J. , Hernandez–Ilizaliturri, F. J. , Kaufman, G. P. , Czuczman, N. M. et al., The novel proteasome inhibitor carfilzomib induces cell cycle arrest, apoptosis and potentiates the anti‐tumour activity of chemotherapy in rituximab‐resistant lymphoma. Br. J. Haematol. 2013, 162, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McBride, A. , Klaus, J. O. , Stockerl‐Goldstein, K. , Carfilzomib: a second‐generation proteasome inhibitor for the treatment of multiple myeloma. Am. J. Health. Syst. Pharm. 2015, 72, 353–360. [DOI] [PubMed] [Google Scholar]
- 7. Citrin, R. , Foster, J. , Teachey, D. , The role of proteasome inhibition in the treatment of malignant and non‐malignant hematologic disorders. Expert Rev. Hematol. 2016, 9, 873–889. [DOI] [PubMed] [Google Scholar]
- 8. Kisselev, A. F. , van der Linden, W. A. , Overkleeft, H. S. , Proteasome inhibitors: an expanding army attacking a unique target. Chem. Biol. 2012, 19, 99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D'Arcy, P. , Linder, S. , Molecular pathways: translational potential of deubiquitinases as drug targets. Clin. Cancer. Res. 2014, 20, 3908–3914. [DOI] [PubMed] [Google Scholar]
- 10. D'Arcy, P. , Linder, S. , Proteasome deubiquitinases as novel targets for cancer therapy. Int. J. Biochem. Cell Biol. 2012, 44, 1729–1738. [DOI] [PubMed] [Google Scholar]
- 11. Lim, K. H. , Baek, K. H. , Deubiquitinating enzymes as therapeutic targets in cancer. Curr. Pharm. Des. 2013, 19, 4039–4052. [DOI] [PubMed] [Google Scholar]
- 12. Lamsoul, I. , Uttenweiler‐Joseph, S. , Moog‐Lutz, C. , Lutz, P. G. , Cullin 5‐RING E3 ubiquitin ligases, new therapeutic targets? Biochimie 2016, 122, 339–347. [DOI] [PubMed] [Google Scholar]
- 13. Fabre, B. , Fabre, B. , Lambour, T. , Garrigues, L. et al., Label‐free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. J. Proteome Res. 2014, 13, 3027–3037. [DOI] [PubMed] [Google Scholar]
- 14. Matondo, M. , Bousquet‐Dubouch, M. P. , Gallay, N. , Uttenweiler‐Joseph, S. et al., Proteasome inhibitor‐induced apoptosis in acute myeloid leukemia: a correlation with the proteasome status. Leuk. Res. 2010, 34, 498–506. [DOI] [PubMed] [Google Scholar]
- 15. Cortes, J. , Thomas, D. , Koller, C. , Giles, F. et al., Guerciolini, R. , Wright, J. , Kantarjian, H. , Phase I study of bortezomib in refractory or relapsed acute leukemias. Clin. Cancer. Res. 2004, 10, 3371–3376. [DOI] [PubMed] [Google Scholar]
- 16. Attar, E. C. , Amrein, P. C. , Fraser, J. W. , Fathi, A. T. et al., Phase I dose escalation study of bortezomib in combination with lenalidomide in patients with myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML). Leuk. Res. 2013, 37, 1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mitsiades, N. , Mitsiades, C. S. , Poulaki, V. , Chauhan, D. et al., Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc. Natl. Acad. Sci. USA 2002, 99, 14374–14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bieler, S. , Meiners, S. , Stangl, V. , Pohl, T. , Stangl, K. , Comprehensive proteomic and transcriptomic analysis reveals early induction of a protective anti‐oxidative stress response by low‐dose proteasome inhibition. Proteomics 2009, 9, 3257–3267. [DOI] [PubMed] [Google Scholar]
- 19. Ge F., Xiao C. L., Bi L. J., Tao S. C. et al., Quantitative phosphoproteomics of proteasome inhibition in multiple myeloma cells. PLoS One 2010, 5, e13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilde I. B., Brack M., Winget J. M., Mayor T. et al., Proteomic characterization of aggregating proteins after the inhibition of the ubiquitin proteasome system. J. Proteome Res. 2011, 10, 1062–1072. [DOI] [PubMed] [Google Scholar]
- 21. Choi M. R., Najafi F., Safa A. R., Drexler H. C. et al., Analysis of changes in the proteome of HL‐60 promyeloid leukemia cells induced by the proteasome inhibitor PSI. Biochem. Pharmacol. 2008, 75, 2276–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uttenweiler‐Joseph S., Bouyssié D., Calligaris D., Lutz P. G., Monsarrat B., Burlet‐Schiltz O. et al., Quantitative proteomic analysis to decipher the differential apoptotic response of bortezomib‐treated APL cells before and after retinoic acid differentiation reveals involvement of protein toxicity mechanisms. Proteomics 2013, 13, 37–47. [DOI] [PubMed] [Google Scholar]
- 23. Kim, M. S. , Lee J. G., McKinney K. Q., Lee Y. Y. et al., A draft map of the human proteome. Nature 2014, 509, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilhelm M., Schlegl J., Hahne H., Gholami A. M. et al., Mass‐spectrometry‐based draft of the human proteome. Nature 2014, 509, 582–587. [DOI] [PubMed] [Google Scholar]
- 25. Rosenberger G., Koh C. C., Guo T., Röst H. L. et al., A repository of assays to quantify 10,000 human proteins by SWATH‐MS. Sci. Data 2014, 1, 140031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schubert O. T., Ludwig C., Kogadeeva M., Zimmermann M. et al., Absolute proteome composition and dynamics during dormancy and resuscitation of Mycobacterium tuberculosis. Cell Host Microbe 2015, 18, 96–108. [DOI] [PubMed] [Google Scholar]
- 27. Gillet L.C., Navarro P., Tate S., Röst H. et al., Targeted data extraction of the MS/MS spectra generated by data‐independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 2012, 11, O111 016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tyanova S., Albrechtsen R., Kronqvist P., Cox J. et al., Proteomic maps of breast cancer subtypes. Nat. Commun. 2016, 7, 10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gillet, L. C. , Leitner, A. , Aebersold, R. , Mass spectrometry applied to bottom‐up proteomics: entering the high‐throughput era for hypothesis testing. Annu. Rev. Anal. Chem. (Palo Alto Calif) 2016, 9, 449–472. [DOI] [PubMed] [Google Scholar]
- 30. Schubert O. T., Gillet L. C., Collins B. C., Navarro P. et al., Building high‐quality assay libraries for targeted analysis of SWATH MS data. Nat. Protoc. 2015, 10, 426–441. [DOI] [PubMed] [Google Scholar]
- 31. Picotti P., Rinner O., Stallmach R., Dautel F. et al., High‐throughput generation of selected reaction‐monitoring assays for proteins and proteomes. Nat. Methods 2010, 7, 43–46. [DOI] [PubMed] [Google Scholar]
- 32. Lange V., Picotti P., Domon B., Aebersold R. et al., Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst Biol. 2008, 4, 222, doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Selevsek N., Matondo M., Sanchez Carbayo M., Aebersold R., Domon B., Systematic quantification of peptides/proteins in urine using selected reaction monitoring. Proteomics 2011, 11, 1135–1147. [DOI] [PubMed] [Google Scholar]
- 34. Surinova S., Choi M., Tao S., Schüffler P. J. et al., Prediction of colorectal cancer diagnosis based on circulating plasma proteins. EMBO Mol. Med. 2015, 7, 1166–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gallien, S. , Duriez, E. , Domon, B. , Selected reaction monitoring applied to proteomics. J. Mass Spectrom. 2011, 46, 298–312. [DOI] [PubMed] [Google Scholar]
- 36. Kim Y. J., Gallien S., El‐Khoury V., Goswami P. et al., Quantification of SAA1 and SAA2 in lung cancer plasma using the isotype‐specific PRM assays. Proteomics 2015, 15, 3116–3125. [DOI] [PubMed] [Google Scholar]
- 37. Tebbe A., Klammer M., Sighart S., Schaab C., Daub H., Systematic evaluation of label‐free and super‐SILAC quantification for proteome expression analysis. Rapid Commun. Mass Spectrom. 2015, 29, 795–801. [DOI] [PubMed] [Google Scholar]
- 38. Shenoy, A. , Geiger, T. , Super‐SILAC: current trends and future perspectives. Expert Rev. Proteomics 2015, 12, 13–19. [DOI] [PubMed] [Google Scholar]
- 39. Court M., Selevsek N., Matondo M., Allory Y. et al., Toward a standardized urine proteome analysis methodology. Proteomics 2011, 11, 1160–1171. [DOI] [PubMed] [Google Scholar]
- 40. Tyanova, S. , Mann, M. , Cox, J. , MaxQuant for in‐depth analysis of large SILAC datasets. Methods Mol. Biol. 2014, 1188, 351–364. [DOI] [PubMed] [Google Scholar]
- 41. Cox, J. et al., Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- 42. Tyanova S., Temu T., Sinitcyn P., Carlson A. et al., The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [DOI] [PubMed] [Google Scholar]
- 43. Shannon P., Markiel A., Ozier O., Baliga N. S. et al., Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Surinova S., Hüttenhain R., Chang C. Y., Espona L. et al., Automated selected reaction monitoring data analysis workflow for large‐scale targeted proteomic studies. Nat. Protoc. 2013, 8, 1602–1619. [DOI] [PubMed] [Google Scholar]
- 45. Chang C. Y., Picotti P., Hüttenhain R., Heinzelmann‐Schwarz V. et al., Protein significance analysis in selected reaction monitoring (SRM) measurements. Mol. Cell. Proteomics 2012, 11, M111 014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deutsch, E. W. , Lam, H. , Aebersold, R. , PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008, 9, 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kusebauch U., Campbell D. S., Deutsch E. W., Chu C. S. et al., Human SRMAtlas: a resource of targeted assays to quantify the complete human proteome. Cell 2016, 166, 766–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adams, J. , The proteasome: a suitable antineoplastic target. Nat. Rev. Cancer 2004, 4, 349–360. [DOI] [PubMed] [Google Scholar]
- 49. Kim H. J., Jamart C., Deldicque L., An G. L. et al., Endoplasmic reticulum stress markers and ubiquitin‐proteasome pathway activity in response to a 200‐km run. Med. Sci. Sports Exerc. 2011, 43, 18–25. [DOI] [PubMed] [Google Scholar]
- 50. Gatti L., Hoe K. L., Hayles J., Righetti S. C. et al., Ubiquitin‐proteasome genes as targets for modulation of cisplatin sensitivity in fission yeast, BMC Genomics, 2011, 12, 44. doi 10.1186/1471-2164-12-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Meiners S., Heyken D., Weller A., Ludwig A. et al., Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. J. Biol. Chem. 2003, 278, 21517–21525. [DOI] [PubMed] [Google Scholar]
- 52. Glickman M. H., Rubin D. M., Coux O., Wefes I. et al., A subcomplex of the proteasome regulatory particle required for ubiquitin‐conjugate degradation and related to the COP9‐signalosome and eIF3. Cell 1998, 94, 615–623. [DOI] [PubMed] [Google Scholar]
- 53. Baumeister W., Walz J., Zühl F., Seemüller E. et al., The proteasome: paradigm of a self‐compartmentalizing protease. Cell 1998, 92, 367–380. [DOI] [PubMed] [Google Scholar]
- 54. Fabre B., Lambour T., Delobel J., Amalric F. et al., Subcellular distribution and dynamics of active proteasome complexes unraveled by a workflow combining in vivo complex cross‐linking and quantitative proteomics. Mol. Cell. Proteomics 2013, 12, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mitsiades N., Mitsiades C. S., Richardson P. G., Poulaki V. et al., The proteasome inhibitor PS‐341 potentiates sensitivity of multiple myeloma cells to conventional chemotherapeutic agents: therapeutic applications. Blood 2003, 101, 2377–2380. [DOI] [PubMed] [Google Scholar]
- 56. Gupta P., Leahul L., Wang X., Wang C. et al., Proteasome regulation of the chromodomain protein MRG‐1 controls the balance between proliferative fate and differentiation in the C. elegans germ line. Development 2015, 142, 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Larance M., Kirkwood K. J., Xirodimas D. P., Lundberg E. et al., Characterization of MRFAP1 turnover and interactions downstream of the NEDD8 pathway. Mol. Cell Proteomics 2012, 11, M111 014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bailly A., Perrin A., Bou Malhab L. J., Pion E. et al., The NEDD8 inhibitor MLN4924 increases the size of the nucleolus and activates p53 through the ribosomal‐Mdm2 pathway. Oncogene 2016, 35, 415–426. [DOI] [PubMed] [Google Scholar]
- 59. Soucy, T. A. , Smith, P. G. , Rolfe, M. , Targeting NEDD8‐activated cullin‐RING ligases for the treatment of cancer. Clin. Cancer Res. 2009, 15, 3912–3916. [DOI] [PubMed] [Google Scholar]
- 60. Pan Z. Q., Kentsis A., Dias D. C., Yamoah K., Wu K. et al., Nedd8 on cullin: building an expressway to protein destruction. Oncogene 2004, 23, 1985–19897. [DOI] [PubMed] [Google Scholar]
- 61. Li W., Yang F., Liu Y., Gong R., Liu L., Feng Y., Hu P., Sun W., Hao Q., Kang L., Wu J., Zhu Y. et al., Negative feedback regulation of IL‐32 production by iNOS activation in response to dsRNA or influenza virus infection. Eur. J. Immunol. 2009, 39, 1019–1024. [DOI] [PubMed] [Google Scholar]
- 62. Hong J., Bae S., Kang Y., Yoon D. et al., Suppressing IL‐32 in monocytes impairs the induction of the proinflammatory cytokines TNFalpha and IL‐1beta. Cytokine 2010, 49, 171–176. [DOI] [PubMed] [Google Scholar]
- 63. Dinarello, C. A. , Kim, S. H. , IL‐32, a novel cytokine with a possible role in disease. Ann. Rheum. Dis. 2006, 65(Suppl 3), iii61–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yun H. M., Oh J. H., Shim J. H., Ban J. O. et al., Antitumor activity of IL‐32beta through the activation of lymphocytes, and the inactivation of NF‐kappaB and STAT3 signals. Cell Death Dis. 2013, 4, e640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hodge D. L., Yang J., Buschman M. D., Schaughency P. M. et al., Interleukin‐15 enhances proteasomal degradation of bid in normal lymphocytes: implications for large granular lymphocyte leukemias. Cancer Res. 2009, 69, 3986–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., Mann M., Super‐SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 2010, 7, 383–385. [DOI] [PubMed] [Google Scholar]
- 67. Wang, S. , Chen, F. , Tang, L. , IL‐32 promotes breast cancer cell growth and invasiveness. Oncol. Lett. 2015, 9, 305–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
figS1
figS2
figS3
figS4
figS5
figS6
figS7
figS8
Table‐S1‐S2
Table S3
Table S4
Supplementary Figures and Tables
Western Blot and antibodies


