Abstract
Werner syndrome (WS) is a progeroid-like syndrome caused by WRN gene mutations. WS cells exhibit shorter telomere length compared to normal cells, but it is not fully understood how WRN deficiency leads directly to telomere dysfunction. By generating localized telomere-specific DNA damage in a real-time fashion and a dose-dependent manner, we found that the damage response of WRN at telomeres relies on its RQC domain, which is different from the canonical damage response at genomic sites via its HRDC domain. We showed that in addition to steady state telomere erosion, WRN depleted cells are also sensitive to telomeric damage. WRN responds to site-specific telomeric damage via its RQC domain, interacting at Lysine 1016 and Phenylalanine1037 with the N-terminal acidic domain of the telomere shelterin protein TRF1 and demonstrating a novel mechanism for WRN's role in telomere protection. We also found that tankyrase1-mediated poly-ADP-ribosylation of TRF1 is important for both the interaction between WRN and TRF1 and the damage recruitment of WRN to telomeres. Mutations of potential tankyrase1 ADP-ribosylation sites within the RGCADG motif of TRF1 strongly diminish the interaction with WRN and the damage response of WRN only at telomeres. Taken together, our results reveal a novel mechanism as to how WRN protects telomere integrity from damage and telomere erosion.
INTRODUCTION
Cellular DNA is organized into 46 chromosomes that are capped by 92 telomeres in human cells. These telomeres form unique nucleoprotein structures to protect chromosome integrity. Reactive oxygen species (ROS) induce oxidative DNA damage leading to genome instability which contributes to cancer (1) aging and neurodegenerative disease (2) and which also accelerates telomere shortening (3–5). Werner syndrome (WS) is a segmental progeroid disorder characterized by the dramatic and rapid appearance of features and diseases associated with normal aging (6). In WS patients, cells with WRN protein defects caused by WRN mutations divide more slowly or stop dividing earlier than normal cells, and show accelerated telomere loss (7–9). Several lines of evidence suggest that telomere dysfunction in WS cells contributes to both the aging and cancer phenotypes of the syndrome (7,10). However, how the lack of WRN leads to telomere dysfunction and whether it is linked to oxidative damage are still unclear.
It has been shown that accelerated telomere shortening and telomere abnormalities can sensitize repair-deficient cell lines to radiation (11), suggesting that repair of oxidative DNA damage is necessary for maintaining telomeric integrity. Telomeric integrity is also fostered by large loop structures including T-loops, D-loop and G-quadruplex structures (12) that are unique among genome sites. Finally, telomeres are protected by a series of shelterin proteins, among them TRF1, which bind duplex TTAGGG repeats, localize specifically to telomeres, and form a complex with TRF2, TIN2, TPP1, POT1 and RAP1 that caps telomeres (13,14). Shelterin proteins prevent DNA double-strand break (DSB) repair by homologous recombination (HR) and non-homologous end joining (NHEJ), thereby avoiding chromosome fusions (15). This suggests that the repair mechanisms at telomeres would be different from those at other genome sites. In the repair of oxidative DNA damage via base excision repair (BER) at genome sites, poly (ADP-ribose) polymerase 1 (PARP1) is a sensor at such sites; DNA polymerase β (Polβ) is a major DNA polymerase during repair synthesis; and FEN1 is an endonuclease that removes the 5΄ flap after repair synthesis to complete the final ligation step. Telomeres contain large amounts of repetitive DNA sequences that are susceptible to ROS-induced DNA damage. Thus at telomeres, BER could be activated while NHEJ is suppressed. WRN interacts with PARP1, Polβ and FEN1 which are necessary for the repair of oxidative DNA damage (16–18), indicating that WRN plays a role in protecting cells from oxidative DNA damage. Furthermore, we previously showed that WRN is recruited to sites of oxidative DNA damage induced by laser micro-irradiation (19). Therefore, it is possible that WRN is an essential factor to regulate the repair of oxidative DNA damage at telomeric sites, thereby protecting cell senescence triggered by telomeric loss, possibly through a unique mechanism that involves cooperation with telomere shelterin proteins.
Despite the importance of maintaining telomere integrity in the face of oxidative DNA damage, the ability to associate oxidative base damage directly with telomere shortening and telomere dysfunction-induced senescence has been hampered by the lack of a methodology to confine oxidative damage to the telomeres. Previous approaches used chemical treatment or irradiation of cells to generate DNA damage throughout the genome, and the direct cellular effects of damage could not be ascribed to telomeric or genomic DNA. We established a novel methodology that involves DNA damage targeted at telomeres (20). In this method, ROS are specifically targeted to the telomeres by tagging the telomere binding protein, TRF1 or TRF2, with the photosensitizer protein KillerRed (KR-TRF1). Upon KR activation via exposure to visible light (520–590 nm) (21), ROS are produced at telomeres causing site-specific damage (20,22). In this study, we applied the oxidative telomere damage approach to elucidate the role of WRN at sites of telomeres upon oxidative DNA damage. We found that the WRN protein is localized and specifically responds to DNA damage at telomeric sites both in a telomerase positive HeLa cell line and a telomerase negative U2OS cell line. The recruitment of WRN to sites of telomeric damage is via its interaction with the N-terminal acidic domain of TRF1 through WRN's RQC domain. Importantly, tankyrase 1 (TNKS1)-mediated poly-ADP-ribosylation (PARylation) of the N-terminal acidic domain of TRF1 is necessary for both the interaction with WRN and its recruitment of WRN to oxidative damage at telomeres. Our results reveal a unique mechanism by which WRN is recruited to oxidative DNA damage specifically at telomeres via modification of a telomeric shelterin protein.
MATERIALS AND METHODS
Cell lines and transfections
U2OS, HeLa, Flp-in T-REx 293, MEF, Polymerase β-/- and PARP1-/- MEF cells were used. All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM, Lonza) with 10% fetal bovine sera (Atlanta Biologicals) at 37°C with 5% CO2 except that Polymerase β−/− MEF was cultured at 34°C with 10% CO2. Flp-in T-REx 293 cell lines (tet-on) were established by transfection of pcDNA5/FRT/TO RFP-TRF1, KR-TRF1 and NLS-KR, respectively, with 150 μg/ml Hygromycin B (Sigma) selection. KR-TRF1, DsR-TRF1 or NLS-KR stably expressing HeLa cell lines were established by infection with pLVX-IRES-Puro KR-TRF1 and DsR-TRF1 virus, respectively, and selection with 1 μg/ml Puromycin (Hyclone). The TRF1 TBM1 mutant fragment was first amplified by polymerase chain reaction (PCR) using a 5΄ primer containing mutation sites and a 3΄ primer with a NotI site. Then the PCR product served as a template to amplify the full length (FL) TBM1 mutant using a TRF1 5΄ primer with a SalI site and a TRF1 3΄ primer with a NotI site. The mutant was cloned into a pBS vector and selected with a Blue/white selection system. The pBS-TRF1 TBM1 mutant construct was then subcloned into the SalI–NotI sites of a pcDNA/FRT/TO/Flag-His vector (Invitrogen). TBM1 fragment forward primer: 5΄-GCGGCCCCGAGCCCGGCCGGCTGTGCGGATGCTAGGGAT-3΄; TBM1 fragment reverse primer: 5΄-TTTGCGGCCGCAGTCTTCGCTGTCTGAGGAAATCAG-3΄; TBM1 FL forward primer: 5΄-TTTTCTCGAGATGGCGGAGGATGTTTCCTCA-3΄; TBM1 FL reverse primer: 5΄-TTTGCGGCCGCAGTCTTCGCTGTCTGAGGAAATCAG-3΄. Plasmids were transfected with PolyJet (SignaGen) or electroporation (NEPAGENE, NEPA21, 2 mm gap cuvettes); 20 V, 50 ms at 50 ms intervals, 5 pulses, were applied for U2OS cells. siRNA oligonucleotides were transfected with DharmaFECT transfection regent 1 (Thermo Scientific, T-2001-02). siRNAs of siFEN1 (Thermo Scientific, A-010344-14), siPARP1 (Thermo Scientific, E-006656-00), siWRN (Santa Cruz, sc-36843) and siTRF2 (Santa Cruz Biotechnology, sc-38505) were used in this study.
Chemicals
The PARP inhibitors PJ34 (Sigma) and Olaparib (Selleckchem) with a final concentration of 4 and 10 μM were added into medium for 30 min, respectively. Tankyrase 1/2 inhibitor VI G007-LK (Millipore) was used with a 92 nM final concentration in medium for 24 h. Braco19 (Sigma) (10 μM for 3 days in medium), Trichostatin A (TSA) (Sigma) (10 μM for 6 h in medium) and CGK733 (Millipore) (10 μM for 24 h in media) were used for treatments. Induction of Flp-in T-REx 293 KR-TRF1 expression was done by adding 2 μg/ml tetracycline (Sigma). The PARG inhibitor ADP-HPD, dihydrate ammonium salt (CALBIOCHEM), was added to lysis buffer with a 960 μM final concentration. siTRF2 (Santa Cruz Biotechnology, sc-38505) and siTNKS1: 5΄-AACAAUUCACCGUCGUCCUCU-3΄ (23) were used in this study. Two TRF1 siRNA sequences targeting the 3΄UTR were used in imaging and the survival study of the TRF1 TBM1 mutant: 1.AGAGUAACCUAUAAGCAUG (J-010542-07-0005, Dharmacon), 2.UACCAGAGUUAAAGCAUAU (J-010542-08-0005, Dharmacon). These sequences were used for establishing the shTRF1 cell line. Three WRN siRNA sequences targeted to 5΄ UTR were designed by siDirect (http://sidirect2.rnai.jp) and synthesized by IDT (Integrated DNA Technologies; USA) (WRN siRNA 710–732; ACAAUAUCUGAAAGAAAACCC and GUUUUCUUUCAGAUAUUGUUU, WRN siRNA 719–741; AAAUACAAAACAAUAUCUGAA and CAGAUAUUGUUUUGUAUUUAC, WRN siRNA 740–762; AAAAACAAUGUCUUCAUGGGU and CCAUGAAGACAUUGUUUUUUG). The three duplex siRNAs were mixed and used in cell survival rescue studies. The multiple sequences were mixed in a 1:1 ratio and transfected into cells 48 h before analyzing.
Plasmids
KR and DsRed with additional Age I and EcoRI sites were amplified by PCR and sub-cloned into a pYFP (Clontech) tagged TRF1 plasmid to generate pCMV KR-TRF1 and DsRed-TRF1 plasmids. The KR-TRF1 fragment was digested from a pCMV-KR-TRF1 construct by KpnI and SmaI and sub-cloned into the KpnI–EcoRV sites of pcDNA5/FRT/TO (Invitrogen). pLVX-IRES-Puro KR-TRF1 and DsRed-TRF1 were made by PCR of KR-TRF1, DsRed-TRF1 from pCMV-KR-TRF1 and pCMV-DsR-TRF1 constructs with additional SpeI and BamHI sites and sub-cloned into SpeI and BamHI sites of the pLVX-IRES-Puro (Clontech) vector. pEGFP-WRN, WRN N-terminal with the RQC domain (a.a. 1–1150), C-terminal with the HRDC domain (a.a. 1012–1432), C-terminal without the HRDC domain (a.a. 1229–1432), PARP1, FEN1 and polymerase β were described (19,24). The HRDC domain with an additional nuclear localization sequence (NLS) signal was used to insert the HRDC domain fragment into pEGFP-C1 2 × NLS using primers, WRN HRDC 5΄XhoI; TTT CTC GAG caagagcaggagactcagatt and WRN HRDC 3΄NotI; and TTT GCG GCC GCA ctgaacactatttgtttggca. GFP-WRN K1016A, F1037A and WRN HRDC deletion mutants were made with a Q5® Site-Directed Mutagenesis Kit using primers, WRN RQC K1016A F; GAGTTGGTGGgcgGCTTTTTCCC and WRN RQC K1016A R; TCTGTTTGATCCTTGCCAG, WRN RQC F1037A F; GTATAACAAAgctATGAAGATTTGCGCC and WRN RQC F1037A R; 5΄- CGAGAAACTTCTACCAAG, WRN HRDC del F; ACAGACCTCTTTTCAAGTAC and WRN HRDC del R; TGCCGAAATAACAGGCTG, respectively. TRF1 deletions were cloned into the pRK5 vector. The pEGFP E84A (Exo-Dead), K577M (Helicase-Dead) double mutant was made by excising the WRN E84A K577M fragment from pBK-WRN E84A K577M with SalI and EcoRV, ligating into the SalI and EcoRV sites of pEGFPC1-WRN, then digesting by SalI and PvuII, and ligating into the XhoI and SmaI sites of pEGFPC1. The vector harbors an extra nuclear localization signal. The pEGFP-NLS-WRN N-terminal without the RQC domain (a.a.1–966) fragment was excised from the pEGFP-WRN-N terminal with the RQC domain with XhoI and HindIII, and ligated into the XhoI and HindIII sites of pEGFPC1. The vector harbors an extra nuclear localization signal. The RQC domain was amplified by PCR using 5΄-CCGCTCGAGATGGATTGGATCATTGCTATTCCATG -3΄ and 5΄-GCGGCCGCACTCTGTACTTAATTCAACTGGTAC-3΄ as forward and reverse primers, respectively, and pEGFP-WRN as a template. The amplified product was sub-cloned into the XhoI and NotI sites of the pEGFP-C1 vector that harbors an extra nuclear localization signal to produce the pEGFP-WRN-RQC construct. All PCR products were confirmed with correct sequences.
Microscopy
The Olympus FV1000 confocal microscopy system (Cat. F10PRDMYR-1, Olympus) with FV1000 SIM Scanner and 405 nm laser diode (Cat. F10OSIM405, Olympus) was employed. FV1000 software was used for acquisition of images. For inducing DNA damage, a 405 nm laser was used with the indicated power; the output power of the 405 nm laser that passed through the lens is 5 mW/scan. Laser light was passed through a PLAPON 60× oil lens (super chromatic abe. corr. obj W/1.4NA FV, Cat. FM1-U2B990). Cells were incubated at 37°C on a thermo-plate (MATS-U52RA26 for IX81/71/51/70/50; metal insert, HQ control, Cat. OTH-I0126) in Opti-MEM during observation to avoid pH changes. For bleaching KR, a 559 nm laser was used. For assessing foci positive cells, cells containing >5 colocalized foci with KR-TRF1 were counted. For calculation of the percentage of colocalization with KR-TRF1, foci positive cells in 50 cells were counted in every experiment. Three independent experiments were performed, and representative data are shown. Fluoview Soft (Olympus) was used for data analysis. In cases of quantification of the intensity of the damage response of proteins, a ratio of enrichment of the same area in a single cell nucleus was used. Here, mean intensity of accumulated proteins at the sites of KR-TRF1/ mean intensity of proteins distant to the KR-TRF1 spot (background) in the same nucleus was calculated. Over a 1.5-fold increase of intensity was defined as colocalization. A total of 50 spots of 10 cells were calculated for the increase of intensity. The ±SD calculated in each case is shown in the Figure legend. The P-value is calculated by student's t-test using Stat Plus software; P < 0.005 is shown as **.
KR activation
KR activation was conducted in two ways. Activation of KR in a single cell was performed with a 559 nm laser for 20 scans (1 mW/scan) only for the selected cell nucleus. Local activation of one KR spot was performed with the same 559 nm laser in a selected area within a single cell nucleus. One scan takes <1 s. Activation of KR in bulk cells was done by exposing cells to a 15 watt SYLVANIA cool white fluorescent bulb for the indicated time (20 min to 4 h) in a stage UVP (Uvland, CA, USA). The dose of 559 nm laser light that was delivered to the KillerRed spot has been calculated previously (22). The KR-TRF1 (around 1 μm2 in diameter) spot is around 12 mJ/μm2. In the case of fluorescent light activation, the rate of light is 15 J/m2/s. With a 20 min–1 h light exposure, the final power delivered to each KR-TRF1 spot is around 20–60 mJ/μm2 upon light exposure. Cells were placed under a water bottle (height to light is 15 cm) to prevent an increase of temperature during light activation.
Immuno-assays
For immunofluorescence staining, cells were fixed with 3.7% (v/v) formaldehyde for 15 min at room temperature and followed by three washes with phosphate buffered saline (PBS). Cells were then permeabilized with 0.2% Triton X-100 for 5 min at RT, and two washes with PBS. Primary antibodies were diluted in DMEM+Azide and incubated overnight at 4°C. Afterward, cells were washed three times with PBS and incubated with secondary antibodies diluted in DMEM+Azide for 30 min at RT. Primary antibodies used in this research were: anti-WRN (1:100, Bethyl A300-239A), anti-γH2AX (1:400, Millipore 05636), anti-tankyrase 1 (1:100, sc-8337) and anti-Flag (1:100, M2, Kodak IB13026). Alexa Fluor 405/488/594 goat anti-mouse/rabbit immunoglobulin G or IgM (Invitrogen) were used. The samples were then mounted in drops of PermaFluor (Immunon). For immunoprecipitation, cells were prepared by lysis in lysis buffer (150 mM NaCl, 10 mM Hepes, 50 mM Tris–HCl, PH7.5, 0.1% NP-40, 1 mM ethylenediaminetetraacetic acid (EDTA)) containing complete protease inhibitor cocktail, EDTA-free (Roche). The PARG inhibitor ADP-HPD, dihydrate ammonium salt, was added to lysis buffer in the case of a detection of PAR signals. Immunoprecipitation of KR-TRF1, GFP-tagged and Myc-tagged proteins was performed using rProtein G beads (Invitrogen, 159-013), and mouse anti-TRF1 (sc-56807), mouse anti-GFP (Roche) and rabbit anti-Myc (sc-789), respectively. Sodium dodecyl sulphate-polyacrylamide gel electrophoresis and western blots were described in previous studies (25). The same antibodies were used for western blots to detect the respective fusion proteins. Three percent of lysate used in immunoprecipitation experiments was used as input control. The following primary antibodies were used: anti-FEN1 (sc-56675), anti-PARP1 (sc-56197), anti-TRF1 (sc-56807), anti-GFP (Roche), anti-Myc (sc-789), anti-poly ADP-ribose (Millipore MAB3192), anti-TRF2 (sc-9143) and anti-actin (Calbiochem, CP01).
Colony formation assays
KR-TRF1 and DsR-TRF1 stably expressing HeLa cell lines were used for this assay. Forty-eight hours after siWRN and plasmids transfection, 350 cells were seeded on a 60 mm petri dish 24 h before light irradiation. Cells were treated with or without 15 W white fluorescent light at indicated time periods in PBS. Cells were removed with PBS and DMEM was added after treatment. After 10 days culturing in the dark, cells were fixed and stained with 3.7% crystal violet in methanol. Colonies were counted and calculated.
Cell cycle synchronization and imaging
Normal DMEM + 2.5 mM thymidine was added to a 50% confluency of U2OS cells for 24 h; then thymidine was removed by washing with 1 × PBS, fresh DMEM was added and the cells were transfected with RT1/KT1 with lipofectamine 2000. Eight hours post-transfection, HU (2 uM) was added to cells. After 16 h incubation, cells were released by removing the drug, washing with 1 × PBS and adding fresh medium. Cells were fixed by 4% Paraformaldehyde to obtain G0/G1 phase cells. Synchronization of cells was confirmed and shown in our previous study (26). Cells were exposed to light for 20 min and recovered for 30 min in the dark before fixation to induce oxidative telomeric damage. After fixation, cells were permeabilized by 0.2% Triton X-100, blocked with 2% bovine serum albumin and incubated with WRN Ab for immunofluorescence and confocal imaging.
RESULTS
WRN is recruited specifically to telomeres in response to DNA damage
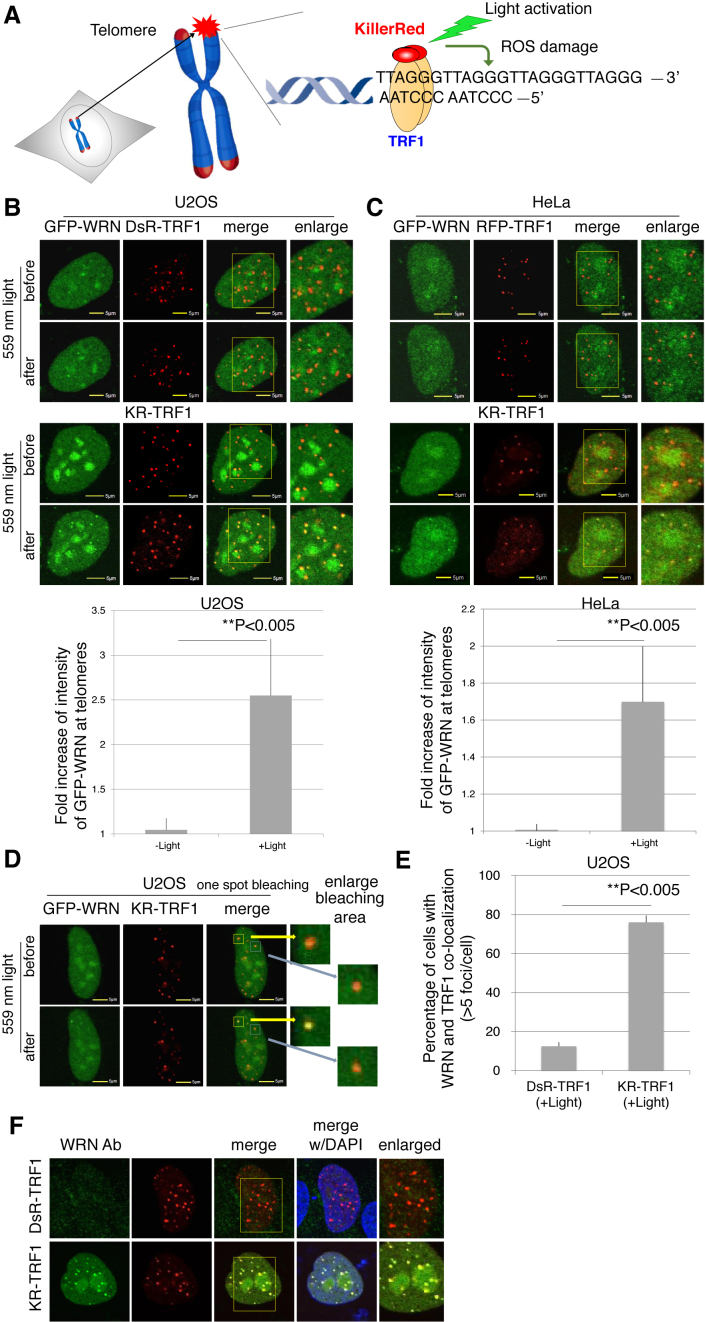
Based on the phenotype of shorter telomeres and telomere dysfunction shown in WS patients’ cells (10), we first explored whether WRN is recruited to oxidative DNA damage at sites of telomeres. By fusing KR with TRF1, a telomere shelterin protein that recognizes telomeric DNA repeats, ROS-induced oxidative DNA damage is specifically produced at sites of telomeres after light activation of KR (Figure 1A). DsRed-TRF1 (DsR-TRF1) and RFP-TRF1, two types of non-phototoxic red fluorescent proteins fused with TRF1, were used as a control (Figure 1B). To activate KR, we exposed a KR-TRF1 expressing cell to 559 nm laser light for 20 scans at a power rate of 1 mW/scan (equal to 20 mW) or we illuminated cells with a 15-W Sylvania cool white fluorescent bulb for 20 min (Figure 1B). Ten percent of WRN forms nuclear foci at telomeres during S phase in cells using the alternative lengthening of telomeres (ALT) pathway, but not in telomerase positive cells during replication (27). To examine whether the DNA damage response (DDR) of WRN is related to telomerase, we used both a telomerase negative U2OS cell line, which uses the ALT pathway for telomere elongation and a telomerase positive HeLa cell line. After laser light illumination, GFP-WRN was co-localized specifically at sites of telomeres in both cell lines. Quantification graphs (lower panel) indicate that the intensity of WRN at sites of KR-TRF1 in both cell lines was greatly increased after laser light activation of KR-TRF1 (Figure 1B and C). The recruitment of WRN to damaged telomeres was confirmed by activating one KR-TRF1 spot using the 559 nm laser to target a single telomere. WRN is specifically recruited to the activated KR spot (yellow square), but not to an inactivated spot (blue square) (Figure 1D). Moreover, we detected co-localization of endogenous WRN at either DsR-TRF1 or KR-TRF1 sites after light exposure in U2OS cells (Figure 1E and F). In KR-treated U2OS cells, the percentage of WRN co-localized with TRF1 at telomeres increased from 12 to 76% after damage in G0/G1 synchronized cells (Figure 1E). In conclusion, our data show that WRN is efficiently recruited to oxidative DNA damage at sites of telomeres in cells independently of cell cycle and telomerase.
Figure 1.
WRN is recruited to sites of oxidative DNA damage at telomeres. (A) Overview of the Damage Targeted at Telomeres method to induce ROS damage specifically at sites of telomeres. KillerRed (KR) is fused with TRF1 to bind to telomeric DNA repeats. Exposure of cells to visible light will activate KR to induce oxidative DNA damage at telomeres in living cells. (B and C) GFP-WRN and KR-TRF1, or DsR-TRF1 or RFP-TRF1, were co-transfected into U2OS cells (B) and HeLa cells (C). The single cell nucleus was scanned by a 559 nm laser; recruitment of WRN to KR-TRF1 damage sites in both U2OS (B) and HeLa cells (C) 3 min after activation is shown (upper panel). Quantification of the damage response of GFP-WRN at sites of KR-TRF1 (lower panel). The fold increase of GFP-WRN (mean intensity of WRN at KR-TRF1/mean intensity of GFP-WRN distant from KR-TRF1 in the nucleus) is shown in the U2OS cell line (B) and HeLa cell line (C). Data are represented as mean ± SEM, **P < 0.005. (D) GFP-WRN and KR-TRF1 were co-transfected into U2OS cells and one KR-TRF1 spot (indicated by a yellow square) was scanned by the 559 nm laser to activate KR; recruitment of WRN to the single KR-TRF1 spot is shown 3 min after activation. GFP-WRN at an inactivated KR-TRF1 spot is shown in the blue square. (E and F) Damage response of endogenous WRN at sites of telomeres is shown. U2OS cells synchronized at G0/G1 phase expressing either DsR-TRF1 or KR-TRF1 were exposed to a 15 W SYLVANIA cool white fluorescent bulb for 20 min and then immunostained with WRN antibody. Percentage of colocalization of WRN and DsR-TRF1 or KR-TRF1 (>5 colocalization spots were defined as positive) is shown. A total of 150 cells in total were counted in three independent experiments, data are represented as mean ± SEM.
The RQC domain of WRN is responsible for the selective damage response at telomeres but not at other genomic sites
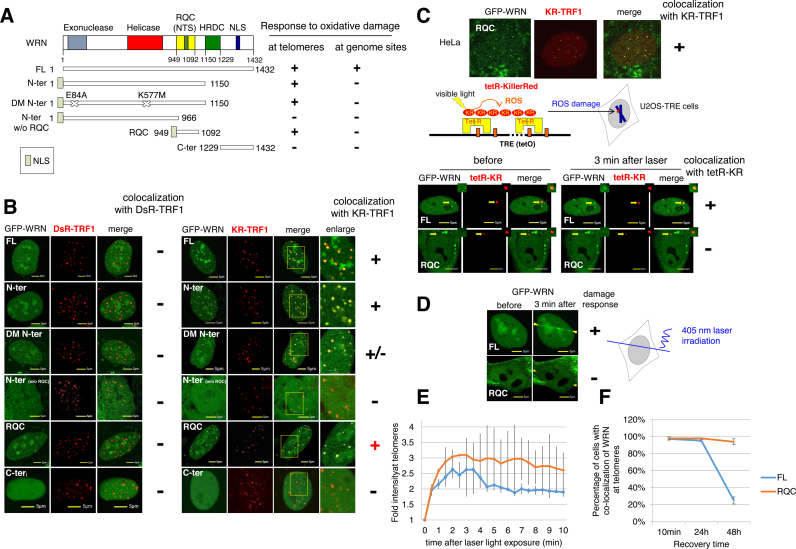
After showing the DDR of WRN at sites of telomeres, we next examined the functional domains of WRN that are responsible for the accumulation of WRN at telomeric damage. FL WRN consists of an exonuclease domain, a helicase domain, a C-terminal (RQC) domain that is conserved in the RecQ family together with a nucleolar targeting sequence (NTS), the helicase RNase D C-terminal conserved region (HRDC) and a nuclear localization sequence (NLS) (Figure 2A). We examined the recruitment of GFP-tagged WRN truncations or WRN mutants to DsR-TRF1 or KR-TRF1 sites in U2OS cells. All truncations without a C-terminus were tagged with an additional NLS signal to ensure their expression in nuclei. None of the WRN truncations shown in Figure 2A was recruited to DsRed-TRF1 before and after KR activation (Figure 2B, left panel). With activated KR-TRF1 damage induced by light illumination, the GFP-tagged WRN-N terminal (a.a. 1–1150, with the RQC domain) accumulated at KR-TRF1 sites, respectively (Figure 2B, right panel). N-terminal WRN with double mutants (E84A and K577M) that abolish exonuclease and helicase activity (28), respectively, was recruited to KR-TRF1 sites as well as FL WRN (Figure 2B), indicating that both enzymatic activities of WRN are not necessary for its recruitment to oxidative DNA damage at telomeres. Next we further divided the WRN-N terminal into a WRN-N terminal without the RQC domain (a.a.1–966) and with an RQC domain (a.a.949–1092). To our surprise, we found that the a.a.1–966 domain failed to respond to KR induced oxidative DNA damage at telomeres, while the RQC domain accumulated at sites of KR-TRF1 after light exposure both in U2OS and HeLa cells (Figure 2B and C). The WRN-C terminal (a.a.1229–1432) lost its ability to respond to telomeric damage (Figure 2B), supporting the conclusion that the RQC domain of WRN is responsible for recruitment to telomeric damage.
Figure 2.
The RQC domain of WRN responds to oxidative damage at telomeres but not at genome sites. (A) Schematic representation of the DDR of a full length (FL) GFP-tagged WRN and truncated or mutated WRN at telomeres and genome sites. (+) means a positive damage response and (−) means no damage response. (B) Three min after 559 nm laser light activation of KR, recruitment of GFP-tagged truncated or mutated WRN proteins to DsRed or KR-TRF1-induced damage at telomeres is shown in U2OS cells. The yellow rectangle indicates an enlarged area showing the colocalization of WRN at the sites of KR-TRF1. (C) Upper panel: recruitment of RQC domain of WRN at telomeres after KR-TRF1 activation in HeLa cells. Lower panel: Scheme of Damage Targeted at Genomic sites method to induce ROS damage specifically at one genome locus of a chromosome is shown. The tetracycline repressor (tetR) fused to KR (tetR-KR) binds a tetracycline response element (TRE) cassette in the defined genome site in U2OS cells. (D) Recruitment of GFP-tagged WRN FL and the RQC domain to 405 nm laser induced damage before (left) and 3 min after (right) 405 nm laser irradiation for 500 ms. The DDR of WRN FL but not the RQC domain to tetR-KR (D) or 405 nm laser (E) is shown with yellow arrowheads. (E) Quantification of accumulation kinetics of GFP-tagged WRN FL and the RQC domain at sites of telomeric damage by the fold increase of the relative intensity after 559 nm laser light irradiation. (F) Dissociation kinetics of GFP-tagged WRN FL and the RQC domain at sites of telomeric damage at 10 min, 24 and 48 h recovery time after 20 min cool white fluorescent bulb light activation of KR. The percentages of co-localization of WRN FL or the RQC domain with KR-TRF1 are shown in the graph. A total of 150 cells in total were counted in three independent experiments; data are represented as mean ± SEM.
To compare the accumulation of WRN at sites of telomeric DNA damage and genomic DNA damage, we tested the DDR of WRN and its deletions to KR-induced damage at a defined genomic locus (22). We used tetR-KR (tetracycline repressor tagged KR), which binds to a tetracycline response element (TRE) array that was integrated into a genome site of 200 copies at a defined chromosome position in the U2OS cell line (Figure 2C and (22)). The length of the integrated TRE fragment is 10–12 kb, which is comparable to the telomere length in human cells, and the intensity of KR expression is similar in tetR-KR and TRF1-KR expressing cells (Figure 2A and C). In addition, we previously showed that the HRDC domain of WRN was necessary and sufficient for the response of WRN to 405 nm laser induced oxidative DNA damage (29). Here we used both tetR-KR and laser microirradiation to induce damage mainly at genome sites. As a result, the N-terminal WRN and RQC domain did not exhibit a damage response ability at either tetR-KR or 405 nm laser induced damage, suggesting that the DDR of the N-terminus of WRN at sites of telomeres is specifically mediated via its RQC domain both in U2OS and HeLa cells (Figure 2C and D; Supplementary Figure S1A and B). Meanwhile, the C-terminal with the HRDC domain was recruited to sites of tetR-KR and 405 nm laser damage as well as FL WRN (Figure 2C and Supplementary Figure S1). The WRN-N terminal without the RQC domain, and the C-terminal without the HRDC domain, failed to be recruited to damage sites in the genome (Supplementary Figure S1A and B). To further understand the role of the HRDC domain in the telomeric damage response, we constructed a truncated WRN with an internal deletion of HRDC1 (HRDC-Del) (Supplementary Figure S1A). HRDC-Del responds only to KR-TRF1 induced telomeric damage but not to 405 nm laser-induced non-telomeric damage (Supplementary Figure S1C). Importantly, the HRDC domain itself and fused to an NLS signal without any C-terminal extending sequences is not recruited to sites of telomeres (Supplementary Figure S1D). These results correspond to our previous observation that the HRDC domain is necessary for the DDR of WRN at genome loci (29) and further demonstrate a unique DDR of the RQC domain at telomeres.
Both FL WRN and the RQC domain were recruited to sites of damage as fast as within 1 min, reached a maximum at 3 min after damage induction (Figure 2E) and were retained at the sites of damage for at least 24 h (Figure 2F). Forty-eight hours after damage induction, FL WRN that had co-localized with KR-TRF1 had decreased to around 25%, while almost all of the RQC domain (94%) was still retained at sites of telomeres (Figure 2F). This observation indicates that although the RQC domain is responsible for the recruitment of WRN to telomeric damage, the dissociation of WRN needs a function of WRN other than that of RQC.
WRN and BER factors are recruited to oxidative damage at telomeres independently
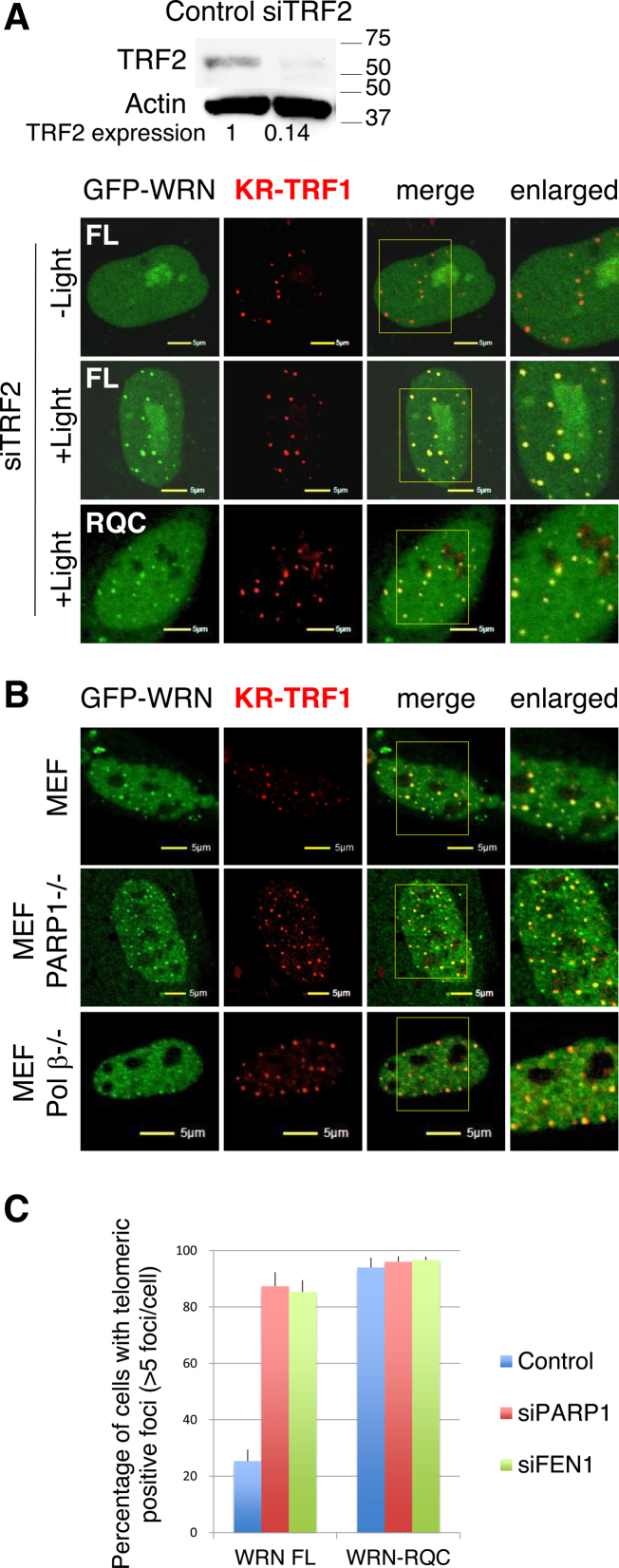
The recruitment of proteins to sites of DNA damage is based on protein-protein interactions, damage induced protein modifications, or signal transduction. To elucidate the mechanism of WRN recruitment to oxidative damage at telomeres, we examined the effects of inhibitors that affect WRN modifications or proteins reported to interact with WRN. It has been reported that WRN interacts with TRF2 in HeLa cells and seldom co-localizes with TRF2 during S phase (30). However, TRF2 knockdown did not affect the recruitment of FL WRN and RQC after light activation (Figure 3A). In addition, WRN did not show any localization with KR-TRF1 before light activation and with TRF2 suppression (Figure 3A). Since WRN functions as a helicase involved in resolving the G-quadruplex, we first tested the effect of the G-quadruplex stabilizing ligand Braco19, which has been shown to inhibit telomerase activity (31). Braco19 did not affect the DDR of WRN (Supplementary Figure S2A). WRN is acetylated by p300, deacetylated by SIRT1 (32,33) and phosphorylated by ataxia-telaniectasia mutated (ATM) and ataxia-telangiectasia and Rad3-related (ATR) proteins (32,34) under various cellular conditions. We therefore tested if the histone deacetylase inhibitor TSA and the ATM/ATR inhibitor CGK733 affect the DDR of WRN at telomeres. Neither TSA nor CGK733 affected the DDR of WRN at telomeres (Supplementary Figure S2A). As described previously, WRN interacts with a number of repair factors (e.g. PARP1, DNA Polβ and FEN1) (16–18), while the recruitment of both FL WRN and the RQC domain was not affected by siPARP or siFEN1 (Supplementary Figure S2B). In addition, the recruitment was not altered in PARP1 or Polβ knock-out mouse embryonic fibroblast (MEF) cells compared to the recruitment in WT MEF cells (Figure 3B). The recruitment of PARP1, FEN1 and Polβ was also not affected by siWRN in U2OS cells (Supplementary Figure S2C). These results indicate that WRN and repair factors are recruited to sites of oxidative damage at telomeres independently of each other. However, FL WRN was retained at telomeres 48 h after damage induction in either siPARP1 or siFEN1 treated cells as well as the RQC domain itself (Figure 3C), indicating that the dissociation of WRN needs the function of PARP1 and FEN1 mediated repair progression.
Figure 3.
WRN and BER factors are recruited to oxidative damage at telomeres independently. (A) Recruitment of WRN FL and RQC was observed in U2OS cells after siTRF2 treatment. Images before or 3 min after 559 nm laser light exposure or without laser light exposure of KR-TRF1 are shown. Yellow rectangles indicate the enlarged area. (B) Recruitment of WRN was observed 3 min after 559 nm laser light exposure of KR-TRF1 in WT and PARP1−/− and Polβ−/− MEF cells. (C) Dissociation of WRN from telomeres after damage induction was delayed by knocking down PARP1 or FEN1. Cells were exposed to light for 20 min and then placed in the dark for 48 h before fixation. The percentages of co-localization of WRN FL or RQC with KR-TRF1 are shown in the graph. A total of 150 cells in total were counted in three independent experiments; data are represented as mean ± SEM.
WRN interacts with TRF1 when oxidative DNA damage is induced at telomeres
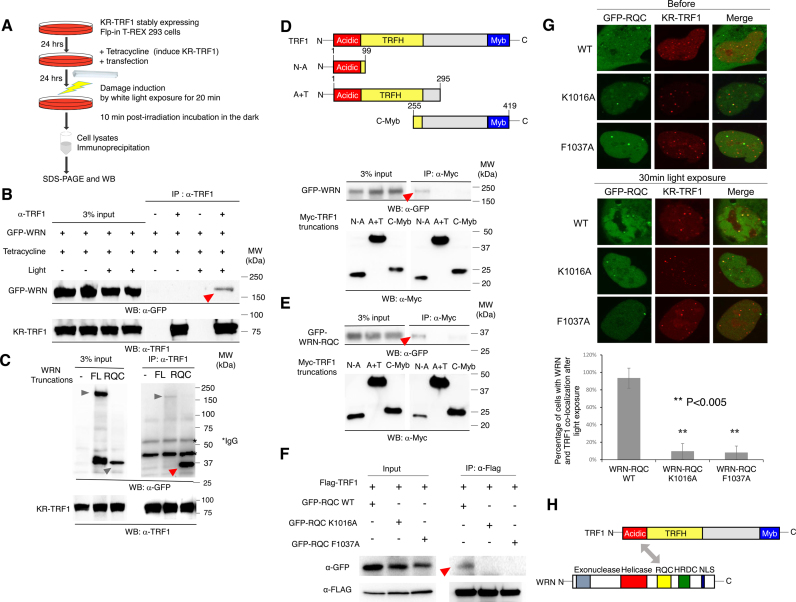
Having observed that WRN and BER factors are recruited to oxidative damage at telomeres independently, we next considered the possibility that WRN is specifically recruited to telomeres after damage by an interaction with telomere shelterin proteins. To examine this, we carried out immunoprecipitation (IP) in KR-TRF1 stably expressing Flp-in T-REx 293 cells after tetracycline induced expression (Figure 4A). WRN was pulled down by TRF1 only after light activation of KR-TRF1 in cells (Figure 4B), indicating that the interaction between WRN and TRF1 is induced at telomeres after damage.
Figure 4.
WRN interacts with TRF1 after induction of oxidative damage at telomeres via the RQC domain of WRN and the N-terminal acidic domain of TRF1. (A) Schematic representation of the experimental procedure for IP. (B) WRN interacts with TRF1 after KR-TRF1 activation. KR-TRF1 stably expressing Flp-in T-REx 293 cells were transfected with GFP-WRN. Tetracycline was added to induce KR-TRF1 expression 48 h before light illumination. Cells were treated with or without 20 min cool white fluorescent bulb light exposure to activate KR and incubated in the dark for 10 min (all the same treatments in 4C-4F before IP). Cell lysates were immunoprecipitated with or without α-TRF1. The precipitates and 3% of the lysate (input) were immunoblotted with α-GFP and α-TRF1. (C) GFP-tagged WRN FL and RQC were transfected into KR-TRF1 stably expressing Flp-in T-REx 293 cells, respectively. Cell lysates were immunoprecipitated with α-TRF1 and immunoblotted with α-GFP and α-TRF1. Arrowheads indicate the bands of GFP-tagged WRN truncations. (D and E) Schematic representation of domains of TRF1 and truncations of Myc-tagged TRF1 are shown. Mapping the interaction domain of TRF1 with WRN and WRN RQC. KR-TRF1 stably expressing Flp-in T-REx 293 cells were cotransfected with Myc-tagged TRF1 deletions shown in 4C and GFP-WRN (D) or GFP-WRN-RQC (E). Cell lysates were immunoprecipitated with α-Myc and immunoblotted with α-myc and α-GFP. (F) GFP-tagged WRN RQC, RQC K1016A and Phe1037A were transfected into FLAG-KR-TRF1 stably expressing Flp-in T-REx 293 cells, respectively. Cell lysates were immunoprecipitated with α-FLAG and immunoblotted with α-GFP and α-FLAG. Arrowheads indicate the bands of GFP-tagged WRN mutants. (G) Upper: before and 3 min after 559 nm laser light activation of KR, recruitment of GFP-tagged WRN RQC (K1016A and Phe1037A) proteins to KR-TRF1-induced damage at telomeres is shown in U2OS cells. Middle: Quantification of accumulation of GFP-tagged WRN-RQC, RQC K1016A and Phe1037A at sites of telomeric damage by the fold increase of the relative intensity after 559 nm laser light irradiation (n = 10). (H) Schematic representation of the interaction between WRN and TRF1 mediated by the RQC domain of WRN and the N-terminal acidic domain of TRF1.
Since TRF1 binds to telomeric DNA repeats directly (35) and WRN shows affinity for certain DNA structures, e.g. Holliday junctions and G-quadruplex structures in vitro (36,37), we tested the possibility of a DNA-mediated association between TRF1 and WRN by performing IP with the addition of 100 μg/ml ethidium bromide (EtBr) to disrupt DNA–protein interactions (38). As shown in Supplementary Figure S3A, the damage-dependent interaction between TRF1 and WRN after KR-TRF1 activation was not decreased after EtBr treatment, therefore excluding a DNA-mediated interaction.
The RQC domain of WRN interacts with the N-terminal acidic domain of TRF1
To confirm and pursue the function of this damage-dependent interaction, we mapped the domains of WRN and TRF1 that are responsible for the interaction. Flp-in T-REx 293 cells with stably expressed KR-TRF1 were transfected with the deletions of WRN shown in Figure 4B. After 20 min of light exposure to activate KR and induce damage at telomeres, cell extracts were collected and TRF1 was pulled down. TRF1 co-precipitated with GFP-WRN FL (lane 2) and GFP-WRN-RQC (lane 3) (Figure 4C). We also compared the interaction between RQC-TRF1 and HRDC-TRF1 (Supplementary Figure S3B). Consistent with the result showing that the HRDC domain is not recruited to telomeric damage (Supplementary Figure S1D), the short HRDC domain fragment does not interact with TRF1 compared to the strong interaction between the RQC domain and TRF1 (Supplementary Figure S3B). The RQC-containing fragment exhibited a stronger interaction than a WRN FL fragment with TRF1 based on a similar expression level, shown in input. These data indicate that TRF1 binds to WRN primarily via its RQC domain. Considering that the RQC domain interacts with TRF2 (30) without induction of oxidative DNA damage, we examined whether the interaction of WRN and TRF1 was mediated via TRF2 by performing a pull-down assay of the GFP-WRN-RQC domain and TRF1 after knocking down TRF2. The RQC domain bound to TRF1 strongly in the siTRF2 treated cells, supporting the notion that the interaction between TRF1 and WRN is not mediated by TRF2 (Supplementary Figure S4).
We next mapped the TRF1 domain interacting with WRN by using a Myc tagged TRF1 N-terminal acidic domain (N-A: amino acid 1–99), TRF1 acidic and TRF homology (TRFH) (A+T: a.a 1–295) domains, and the TRF1 C-terminal Myb domain (C-Myb: a.a. 255–419) shown in Figure 4D. The TRF1 N-A domain interacts with both WRN FL (Figure 4D) and the WRN-RQC domain (Figure 4D). However, both the TRF1 A+T domain and the C-Myb domain, which is important for telomeric DNA binding, did not bind to either WRN (Figure 4D) or the WRN-RQC domain (Figure 4E). Since the TRFH domain is responsible for TRF1 dimerization (39), the fact that TRFH destroys the interaction indicates that dimerization of TRF1 might inhibit the interaction between WRN and TRF1. To further understand the function of the RQC domain, we have created site-directed mutant proteins of RQC (K1016A and Phe1037A) and examined their nuclear localization, damage response and interaction with TRF1 after damage. Lee et al. showed that Lys-1016 was important for the WRN–DNA interaction, and the K1016A mutation significantly decreased WRN binding to fork or bubble DNA substrates (40). Phe1037 is important for catalytic activity and protein–protein interaction in in vitro studies (41). Both WRN mutants lost TRF1 binding and do not bind to damaged telomeres efficiently, although they were well expressed in cells and localized in the nucleus (Figure 4F and G). Our results indicate that the Lys1016 and Phe1037 in the RQC domains are important for the telomere damage response of WRN. Thus, we conclude that the interaction between TRF1 and WRN is via the acidic domain of TRF1 and the RQC domain of WRN (Figure 4H).
TNKS1 mediated PARylation of TRF1 is necessary for WRN recruitment at telomeres after damage
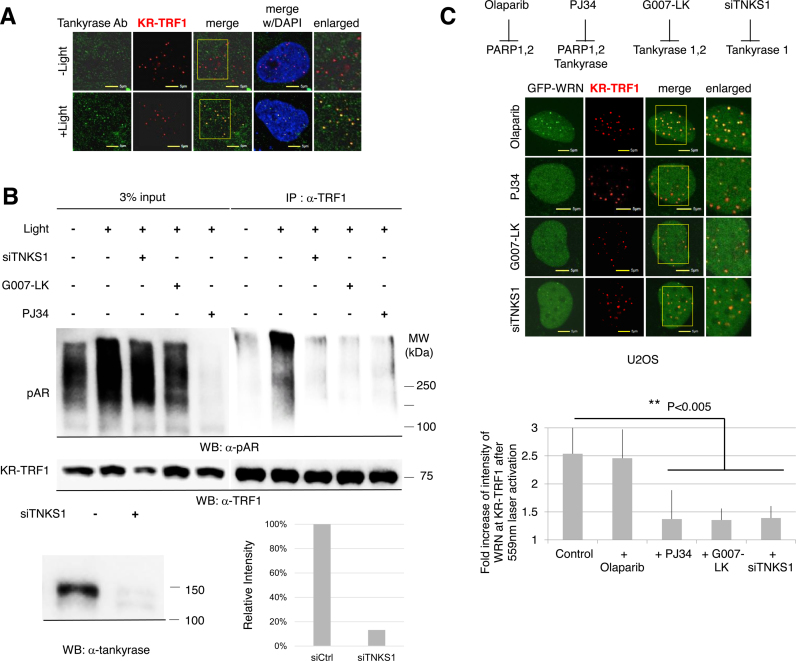
It has been reported that TNKS1 targets TRF1 for PARylation at its N-terminal acidic domain, which subsequently abolishes the DNA binding activity of TRF1 in vitro and removes the TRF1 complex from telomeres in vivo (42). Since the interaction of WRN with TRF1 is via the N-terminal acidic domain of TRF1 (Figure 4), next we examined whether TNKS1 is recruited to DNA damage at telomeres. Endogenous TNKS was recruited to damage at telomeres after KR-TRF1 activation, and rarely seen with KR-TRF1 before light activation (Figure 5A). This recruitment of TNKS1 to oxidative DNA damage at telomeres is independent of cell cycle progression and telomerase expression (submitted manuscript). To further identify whether TNKS1 mediated TRF1 PARylation is involved in the DDR at telomeres, we performed IP to compare the levels of PARylation of TRF1 with or without telomeric damage. Directly after light activation, the total level of PARylation in cell lysates dramatically increased compared to the level in cells without damage (Figure 5B), confirming the temporary induction of PARylation upon damage. We pre-treated cells with siTNKS1 or G007-LK (a specific TNKS1 inhibitor) or PJ34, which inhibits PARP1, 2 as well as TNKS1 (43). Signals of PARylation were slightly suppressed by siTNKS1 or G007-LK, the specific TNKS1 inhibitor, but were almost suppressed by PJ34, the universal PARP inhibitor (Figure 5B, left panel). This result indicates that both TNKS and PARP contribute to total PARylation in cells upon telomere damage. Next, TRF1 was pulled down to evaluate PARylation of TRF1 upon damage (Figure 5B, right panel). The level of TRF1 PARylation increased after KR activation to induce damage at telomeres (Figure 5B, right panel, lane 1 and 2). Importantly, PARylation of TRF1 upon KR-induced damage at telomeres was significantly decreased with TNKS1 inhibition either by siRNA or its specific inhibitor (Figure 5B, right panel, lane 3–5). Therefore, PARylation of TRF1 is stimulated by telomeric damage and this reaction is specifically mediated by TNKS1.
Figure 5.
TNKS1 mediated poly (ADP-ribosyl) ation of TRF1 after damage is necessary for the recruitment of WRN to sites of telomeric damage. (A) TNKS1 is recruited to sites of KR-TRF1 induced damage after light activation. KR-TRF1 transfected U2OS cells (left) were stained with α-TNKS1 after light exposure for 20 min. The yellow rectangle indicates the enlarged area. (B) TNKS1-mediated poly (ADP-ribosyl) ation of TRF1 is induced by telomeric oxidative damage. KR-TRF1 stably expressing HeLa cells were treated with or without siTNKS1 or 92 nM G007-LK for 24 h, or 4 μM PJ34 for 30 min. Cell lysates were collected with the lysis buffer containing 960 μM of the PARG inhibitor, ADP-HPD dihydrate ammonium salt, immediately after light exposure and immunoprecipitated with α-TRF1. The precipitates and 3% of the lysate (input) were immunoblotted with α-pAR and α-TRF1. (C) Recruitment of WRN to oxidative damage at telomeres is prevented by siTNKS1 or PARP inhibitors. Recruitment of GFP-WRN to KR-TRF1 induced damage at telomeres 3 min after 559 nm laser bleaching with treatment of Olaparib, PJ34, G007-LK or siTNKS1 in U2OS cells (left). Quantification of the damage response of GFP-WRN at sites of KR-TRF1 is shown (right). Data are represented as mean ± SEM of 10 cells. **P < 0.005.
We next tested if the recruitment of WRN to oxidative DNA damage at telomeres, mediated by the interaction with TRF1, is affected by inhibition of TNKS1. Based on a previous structure analysis, olaparib specifically suppresses the PARylation of PARP1 and PARP2 but not TNKS (44) while PJ34 inhibits all of the above polymerases (43). Interestingly, a different suppressing effect of olaparib and PJ34 on the recruitment of WRN to sites of oxidative damage at telomeres was seen: olaparib treatment of cells did not suppress the recruitment of WRN to oxidative damage but PJ34 did (Figure 5C), supporting the role of TNKS1 mediated PARylation in regulating the subsequent damage response at telomeres. As we expected, siTNKS1 affected the recruitment of WRN as well as the specific inhibitor G007-LK for TNKS (Figure 5C). Therefore, TNKS1 functions at telomeres much as PARylation of TRF1 functions upstream in the recruitment of WRN to oxidative DNA damage at telomeres. We also checked the effects of TNKS1 inhibition on the DDR of WRN at sites of 405 nm laser induced damage. G007-LK successfully suppressed the DDR of WRN at telomeres but not at the sites of 405 nm laser induced damage (Supplementary Figure S5A), again indicating that WRN is recruited to damage at genomic sites versus telomeric sites via a distinct mechanism. Taking the data together, we conclude that TNKS1 mediated PARylation of TRF1 is necessary for WRN recruitment at telomeres after damage.
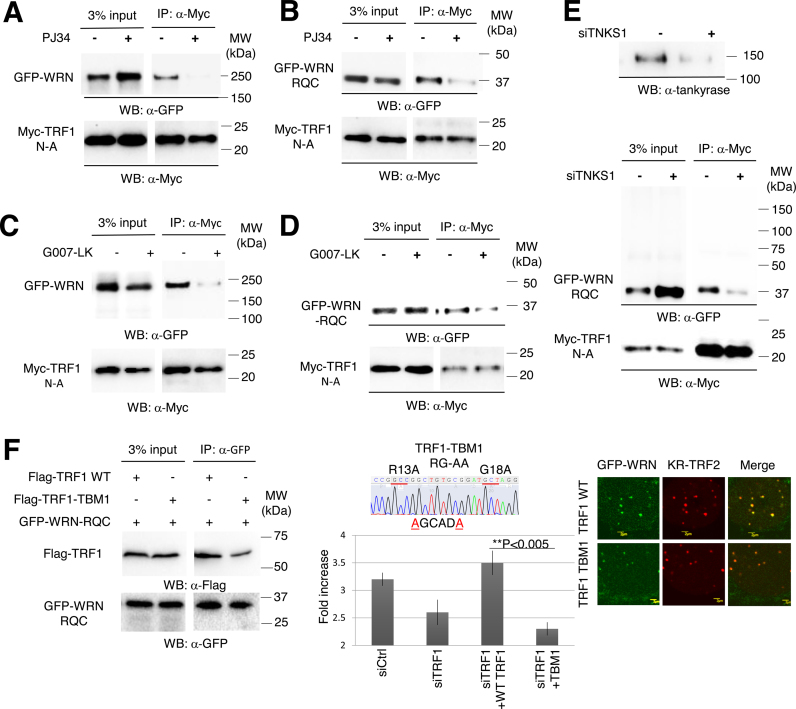
Since the recruitment of WRN to damage at telomeres is based on its interaction with TRF1, we next examined the interaction between FL WRN and the RQC domain with the TRF1 N-A domain after damage. Using PJ34, G007-LK or siTNKS1, treatments that block either function or expression of TNKS1, decreased the interaction between FL WRN and the RQC domain with the N-terminal acidic domain of TRF1 (Figure 6A–E, red arrows), indicating that TNKS1-mediated PARylation of TRF1 is necessary for the interaction between TRF1 and WRN, thereby serving as a signal to facilitate the recruitment of WRN to sites of damage at telomeres. To further strengthen this idea, we examined the effect of a mutant TRF1, which contains the RG to AA mutation at the 13-RGCADG-18 tankyrase binding motif (TBM), on the recruitment of WRN protein at damaged telomeres (Figure 6F). We recently found that the TBM mutant of TRF1 (TBM1) was barely PARylated upon telomere damage (manuscript submitted). In addition, TBM1 does not bind TNKS1 and suppresses the recruitment of TNKS1 to damaged telomeres in shTRF1 targeted cells at the 3΄-UTR region (manuscript submitted). Given that KR-TRF1 or KR-TRF2 induced telomeric damage following PAR formation to a similar extent in our previous study, to test the effect of the TBM mutant we utilized KR-TRF2 to induce telomere DNA damage. As shown in Figure 6F, we found a significant reduction of binding between TRF1-TBM with WRN. The remaining interaction might be mediated by the dimerization of TRF1-TBM with the endogenous TRF1. When we overexpressed TRF1 WT in the shTRF1 pretreated cells, the DNA damage response of WRN could be rescued; however, overexpression of the TBM mutant in these cells significantly suppresses the recruitment of WRN (Figure 6F, right). Together, these results suggest that TRF1 PARylation is important to recruit and interact with WRN after damage.
Figure 6.
The interaction between WRN or WRN-RQC and TRF1 is dependent on TNKS1-mediated PARylation. GFP-WRN (A and C) or GFP-WRN-RQC (B, D and E) and Myc-TRF1-N-A were co-transfected into KR-TRF1 stably expressing Flp-in T-REx 293 cells. After KR-TRF1 expression was induced by tetracycline, cells were treated with or without either PJ34 or G007-LK. Cells were treated with 20 min light exposure and recovered in the dark for 10 min. Cell lysates were immunoprecipitated with α-Myc. The precipitates and 3% of the lysate (input) were immunoblotted with α-GFP antibody and α-Myc antibody. Arrowheads indicate the bands of immunoprecipitated WRN FL or RQC. (F) TBM mutant overexpression diminished the interaction with and recruitment of WRN after telomeric damage. Left: 293 cells were transfected with FLAG-TRF1 or TBM and GFP-RQC. After damage induction, cell lysates were immunoprecipitated with α-GFP and immunoblotted with α-FLAG. Middle: schematic illustration of the TBM mutant is shown. Right: U2OS cells were co-transfected with GFP-WRN, KR-TRF2 and FLAG-TRF1 WT or TBM. Cells were treated with 20 min light exposure and recovered in the dark for 10 min. The percentages of co-localization of WRN FL or RQC with KR-TRF1 are shown in the graph. A total of 150 cells in total were counted in three independent experiments; data are represented as mean ± SEM.
Role of WRN in ensuring cell survival from ROS-induced DNA damage at telomeres
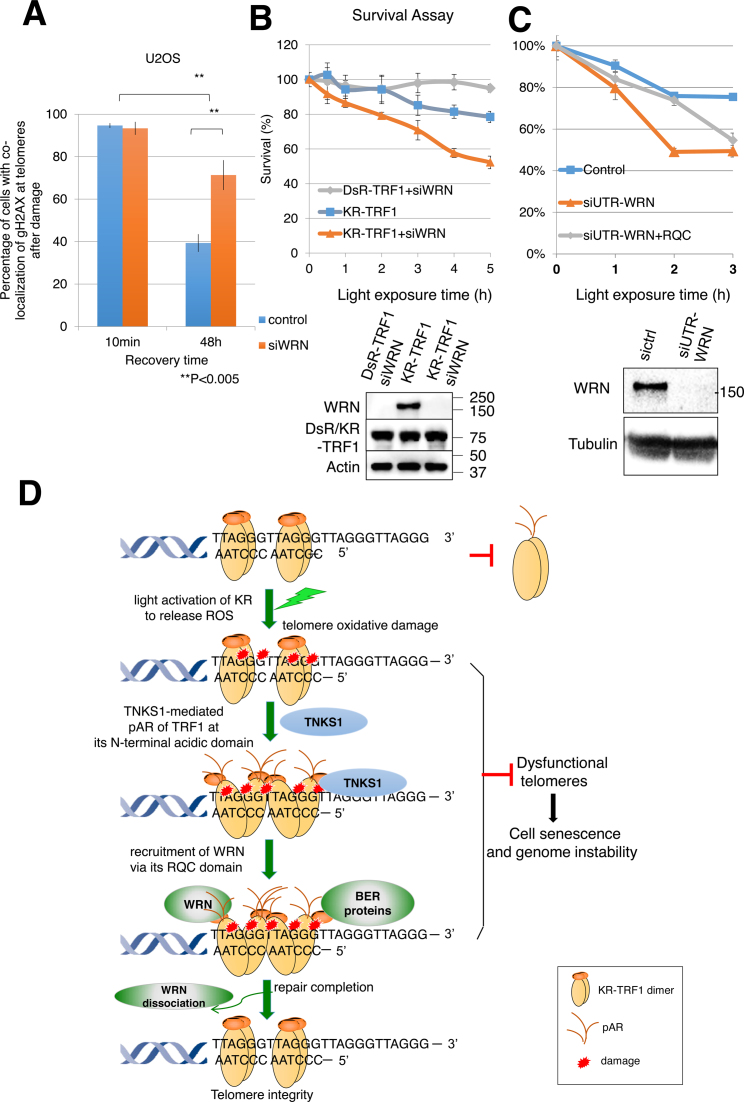
To test the role of WRN for protecting telomeres from damage, we monitored the γH2AX signal, which indicates unrepaired damage or dysfunctional telomeres at sites of KR-TRF1 after light activation, with the indicated recovery times with or without WRN knockdown (Figure 7A). Decreased percentages of cells with γH2AX foci colocalizing with KR-TRF1 were observed 48 h after damage induction, indicating decreased damage levels in cells with time. However, the percentage of γH2AX colocalizing with KR-TRF1 increased to 71% after WRN suppression, indicating delayed repair or an increased number of dysfuncional telomeres without WRN (Figure 7A). Importantly, siWRN treatment further sensitized cells to telomeric damage in a dose dependent manner (Figure 7B), demonstrating that WRN plays an important role in protecting cell survival upon oxidative damage at telomeres. We have suppressed WRN using 3΄ UTR siRNA and expressed GFP-RQC to test if RQC could rescue cell survival after damage. As shown in Figure 7C, expression of RQC partially rescued the siWRN treated cells under a low dose of damage, suggesting that the binding of RQC to PARylated TRF1 alone has functional significance. However, RQC expression does not rescue the cell death under high dose damage, indicating that other functions of WRN beyond RQC are required for cell survival. We also evaluated if the WRN helicase-dead and WRN-exonuclease dead mutants retain the telomere function or not. The helicase-dead WRN mutant does not rescue cell survival under a low dose of damage, and the WRN exonuclease mutant does not rescue cell survival under a high dose of damage compared to WT WRN (Supplementary Figure S5B). Thus, both helicase and exonuclease activities are necessary for the cell survival after telomeric damage, showing that the function of WRN is necessary for cell survival after telomeric damage.
Figure 7.
Suppression of WRN causes delayed repair and increased cell death after oxidative damage at telomeres. (A) Quantification of the percentage of cells showing co-localization of KR-TRF1 and γH2AX with or without siWRN at the indicated recovery time point after 20 min of 15 W SYLVANIA cool white fluorescent bulb light activation of KR. Data are represented as mean ± SEM of three independent experiments with counting 150 cells/time. **P < 0.005 (B and C) Clonogenic survival assay of HeLa cells with stably expressed KR-TRF1 or DsR-TRF1 and treated with or without siWRN (B), siUTR-WRN (C) with or without expression of WRN-RQC. Cells were exposed to cool white fluorescent light for the indicated time. Data are represented as mean ± SEM of three independent experiments. Western blot analysis of WRN knockdown and DsR-TRF1 and KR-TRF1 expression in HeLa cells is shown. (D) A model of recruitment of WRN to oxidative DNA damage at telomeres via interaction with TRF1 upon TNKS1-mediated PARylation. Oxidative DNA damage at telomeres induced a TRF1 conformational change to expose its N-terminal acidic domain. The N-terminal acidic domain of TRF1 will be targeted for PARylation by TNKS1 and then WRN is recruited to sites of DNA damage mediated by interaction between the RQC domain of WRN and N-terminal PARylation of the acidic domain of TRF1. BER factors are recruited independently from WRN. WRN protein dissociates from the sites of damage after repair completion. The function of WRN at telomeres protects genome stability in the face of oxidative DNA damage.
DISCUSSION
The impact of WRN on genome stability is well established, and the maintenance of telomere integrity mediated by the WRN protein is likely to play a critical role as one considers the clinical and pathological manifestations of WS (9,45–48). However, it has not been understood if and how WRN protein protects telomere integrity in the face of oxidative DNA damage. Our unique telomere damage induction system provides a robust platform that enables visualization of the DNA damage response of WRN to oxidative damage at telomeres. In this study, we examined how WRN is recruited to oxidative DNA damage at telomeres to maintain telomere integrity. We have elucidated a unique mechanism by which WRN is recruited to oxidative DNA damage sites at telomeres by its interaction with the N-terminal acidic domain of TRF1 through its RQC domain, which is distinguished from its recruitment to genome damage via its HRDC domain. Thus, the damage at different genome loci will induce different repair responses.
The RQC domain binds to TRF1 after damage (Figures 4–6) and is responsible for the DDR of WRN to telomeric damage but not to genome damage (Figure 2). The RQC domain of WRN, containing α2–α3 loop and β-wing motifs, was also shown not only to be important for DNA binding (49) and many protein interactions (30,50), but also to be important for regulating the enzymatic activity of itself and the DNA repair proteins. For example, the Lys-1016 mutation of the RQC domain of WRN markedly reduced WRN helicase activity on fork, D-loop and Holliday junction substrates in addition to significantly reducing the ability of WRN to stimulate FEN1 incision activities (40). The RQC domain specifically interacted with a blunt end of the duplex and, surprisingly, unpaired a Watson-Crick base pair in the absence of an ATPase domain in addition to DNA helicase (51). It has been shown recently that WRN with RQC mutations was defective in DNA binding and helicase activity as well as exonuclease activity and strand annealing functions (41,51). Although the RQC domain of WRN has not previously been implicated in exonuclease or annealing activities, the RQC mutated WRN could not stimulate DNA glycosylase NEIL1 incision activity as did the wild-type WRN (41). More importantly, compared to the HRDC domain, which exhibits affinity for Holliday junctions, the RQC domain has an independent, high affinity for the conserved G4 DNA that potentially occurs at sites of telomeres (49). All of the above evidence, together with our findings, indicates that the RQC domain has the potential for recruitment to telomeric damage and then coordinates and regulates enzymatic activities of DNA repair proteins and WRN itself at sites of telomeres. The other important finding in the study is the TNKS1 mediated PARylation at telomeric damage, which is different from that at genome sites. ROS mainly induce oxidized bases and single-strand breaks (SSBs) in DNA that are repaired via the base excision/SSB repair (BER/SSBR) pathways (52,53). We previously showed that PARylation at sites of oxidative DNA damage amplifies the damage signal, especially in the context of a condensed chromatin structure (22), mainly dependent on the function of PARP1. Here, we showed that the TNKS1 mediated PARylation of TRF1 facilitates the interaction and recruitment of WRN to oxidative DNA damage at telomeres, suggesting that the mechanisms of PARylation at different genome loci are mediated by different ‘guards’ at distinct genome loci.
Although the DNA binding mode of TRF2 is likely identical to that of TRF1, TRF2 plays an important role in the t-loop formation that protects the ends of telomeres (54,55). In addition, the regulation of TRF1 and TRF2 is controlled by post-translational modifications affecting their functions such as DNA binding, dimerization, localization, degradation and interactions with other proteins (56). Within both proteins, dimerization involves the middle TRFH domain, which also mediates interactions with other telomeric proteins (39) and the C-terminal Myb domain can specifically and fully recognize the telomeric DNA sequence (57). The facilitation of TRF2 for the t-loop structure is mediated by the central TRFH domain. It condenses telomere structure and creates negative torsion on the adjacent DNA, which stimulates single-strand invasion and therefore favors telomeric DNA looping (54). On the other hand, although TRF1 contains the same TRFH domain, this activity was repressed in the full-length protein by the presence of an acidic domain at the N-terminus (58). Collectively, the N-terminal of TRF proteins originated from a gradual extension of the coding sequences of a duplicated ancestral gene, with a consequent and progressive alteration of the biochemical properties of these proteins, suggesting the importance of the N-terminal of TRF1 in finely regulating the ability of TRF proteins. In this study, we showed a functional interaction between WRN and the TRF1 N-terminal acidic domain independently of TRF2 (Figure 4). It is known that TRF1 is specifically PARylated by TNKS1 (59); here, we found that the 13-RGCADG-18 motif in the N-terminal of TRF1 is critical for the damage response of TNKS1 and WRN (Figure 6F). Remarkably, the interaction between WRN and TRF1 was suppressed in the presence of the TRFH domain (Figure 4), indicating that the dimerization of TRF1 might stabilize the TRF1 structure and prevent non-specific PARylation of TRF1 in the absence of damage. This also explains why we could not observe the predominant recruitment of TNKS1 and WRN at sites of telomeres without damage. Overall, the modification of TRF1 at the N-terminus is more likely to be involved in the DNA damage response (Figures 3–6), while the function of TRF2 is to maintain the t-loop structure (55). Our results also support the possibility of targeting TNKS1 for treatment of telomerase positive cancers (60) and demonstrate the molecular mechanism of the specific function of TNKS1 in protecting cells in the face of oxidative damage.
Accelerated loss of telomere length with age, mediated by WRN mutations, is likely to play a critical role in the clinical and pathological manifestations of WS. Deficiencies in repair factors such as Polβ or FEN1 result in lethal phenotypes (61,62), whereas deficiency of WRN results in an accelerated aging phenotype instead of lethality. This difference, on the one hand, might be due to the multiple functions of repair factors and ubiquitous DNA damage elsewhere in the genome, and on the other hand, indicates the role of WRN in regulating DNA repair pathways specifically at telomeric sites. We propose a model based on our results to identify the specific role of WRN protein at telomeres in the face of oxidative DNA damage (Figure 7C). First, TRF1 proteins form dimers at telomeres, while oxidative DNA damage at telomeres seems to induce a conformational change in the TRF1 dimer to expose its N-terminal acidic domain. The N-terminal acidic domain of TRF1 then will be targeted for PARylation by TNKS1, which subsequently recruits WRN to sites of damage at telomeres. The recruitment is via the binding of the RQC domain of WRN to the N-terminal PARylation acidic domain of TRF1. The PARylation reaction is a temporary phenomenon; the retention of WRN at sites of damage might be mediated by its interaction with telomere structure and repair factors, which are recruited to damage independently of WRN. The interactions with shelterin proteins and stimulation of BER enzymatic activities by shelterin proteins (16–18) support the notion that telomeres try to remove ROS damage by the cooperation of shelterin proteins and repair factors. The role of the RQC domain of WRN in facilitating the enzymatic activity of WRN and repair enzymes (41), and the decreased cell survival and delayed disappearance of γH2AX at sites of telomeres without WRN (Figure 7A, B), further support the role of WRN in protecting telomere function and preventing cell senescence. WRN proteins are dissociated from the sites of damage with a rate that corresponds to the kinetics of other repair factors, indicating repair completion.
In summary, by confining ROS to an individual genome locus or telomeres, we were able to determine for the first time a novel mechanism through which WRN is involved in the repair of oxidative DNA damage at telomeric sites, thereby protecting cells from senescence triggered by telomeric loss. Our study provides important insights into how oxidative DNA damage causes telomere defects and accelerates the aging process, and how the function of WRN prevents this process. This study may also lead to the development of new strategies for preserving telomere function so as to promote healthy aging.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: L.S. performed the major experiments. S.N., Y.T., H.C., L.Y., X.C. and B.G. provided help with the experiments. L.S. and L.L. designed the experiments and wrote the manuscript. A.S.L. discussed the experimental plans and interpretation of the results, and aided in the entire work. All authors helped in editing the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM118833, AG045545-01 to L.L., in part]; UPCI Imaging Facility and UPCI Cytometry Facility [P30CA047904]. Funding for the open access charge: NIH [AG045545-01].
Conflict of interest statement. None declared.
REFERENCES
- 1. Cooke M.S., Evans M.D., Dizdaroglu M., Lunec J.. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003; 17:1195–1214. [DOI] [PubMed] [Google Scholar]
- 2. Hegde M.L., Mantha A.K., Hazra T.K., Bhakat K.K., Mitra S., Szczesny B.. Oxidative genome damage and its repair: implications in aging and neurodegenerative diseases. Mech. Ageing Dev. 2012; 133:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Z., Rhee D.B., Lu J., Bohr C.T., Zhou F., Vallabhaneni H., de Souza-Pinto N.C., Liu Y.. Characterization of oxidative guanine damage and repair in mammalian telomeres. PLoS Genet. 2010; 6:e1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sahin E., Depinho R.A.. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010; 464:520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Richter T., von Zglinicki T.. A continuous correlation between oxidative stress and telomere shortening in fibroblasts. Exp. Gerontol. 2007; 42:1039–1042. [DOI] [PubMed] [Google Scholar]
- 6. Opresko P.L., Cheng W.H., von Kobbe C., Harrigan J.A., Bohr V.A.. Werner syndrome and the function of the Werner protein; what they can teach us about the molecular aging process. Carcinogenesis. 2003; 24:791–802. [DOI] [PubMed] [Google Scholar]
- 7. Crabbe L., Verdun R.E., Haggblom C.I., Karlseder J.. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004; 306:1951–1953. [DOI] [PubMed] [Google Scholar]
- 8. Wyllie F.S., Jones C.J., Skinner J.W., Haughton M.F., Wallis C., Wynford-Thomas D., Faragher R.G., Kipling D.. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nat. Genet. 2000; 24:16–17. [DOI] [PubMed] [Google Scholar]
- 9. Chang S., Multani A.S., Cabrera N.G., Naylor M.L., Laud P., Lombard D., Pathak S., Guarente L., DePinho R.A.. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat. Genet. 2004; 36:877–882. [DOI] [PubMed] [Google Scholar]
- 10. Crabbe L., Jauch A., Naeger C.M., Holtgreve-Grez H., Karlseder J.. Telomere dysfunction as a cause of genomic instability in Werner syndrome. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:2205–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cabuy E., Newton C., Joksic G., Woodbine L., Koller B., Jeggo P.A., Slijepcevic P.. Accelerated telomere shortening and telomere abnormalities in radiosensitive cell lines. Radiat. Res. 2005; 164:53–62. [DOI] [PubMed] [Google Scholar]
- 12. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palm W., de Lange T.. How shelterin protects mammalian telomeres. Annu Rev. Genet. 2008; 42:301–334. [DOI] [PubMed] [Google Scholar]
- 14. Martinez P., Blasco M.A.. Role of shelterin in cancer and aging. Aging Cell. 2010; 9:653–666. [DOI] [PubMed] [Google Scholar]
- 15. Sfeir A., de Lange T.. Removal of shelterin reveals the telomere end-protection problem. Science. 2012; 336:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brosh R.M. Jr, von Kobbe C., Sommers J.A., Karmakar P., Opresko P.L., Piotrowski J., Dianova I., Dianov G.L., Bohr V.A.. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001; 20:5791–5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harrigan J.A., Opresko P.L., von Kobbe C., Kedar P.S., Prasad R., Wilson S.H., Bohr V.A.. The Werner syndrome protein stimulates DNA polymerase beta strand displacement synthesis via its helicase activity. J. Biol. Chem. 2003; 278:22686–22695. [DOI] [PubMed] [Google Scholar]
- 18. Adelfalk C., Kontou M., Hirsch-Kauffmann M., Schweiger M.. Physical and functional interaction of the Werner syndrome protein with poly-ADP ribosyl transferase. FEBS Lett. 2003; 554:55–58. [DOI] [PubMed] [Google Scholar]
- 19. Lan L., Nakajima S., Komatsu K., Nussenzweig A., Shimamoto A., Oshima J., Yasui A.. Accumulation of Werner protein at DNA double-strand breaks in human cells. J. Cell Sci. 2005; 118:4153–4162. [DOI] [PubMed] [Google Scholar]
- 20. Sun L., Tan R., Xu J., LaFace J., Gao Y., Xiao Y., Attar M., Neumann C., Li G.M., Su B. et al. Targeted DNA damage at individual telomeres disrupts their integrity and triggers cell death. Nucleic Acids Res. 2015; 43:6334–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bulina M.E., Lukyanov K.A., Britanova O.V., Onichtchouk D., Lukyanov S., Chudakov D.M.. Chromophore-assisted light inactivation (CALI) using the phototoxic fluorescent protein KillerRed. Nat. Protoc. 2006; 1:947–953. [DOI] [PubMed] [Google Scholar]
- 22. Lan L., Nakajima S., Wei L., Sun L., Hsieh C.L., Sobol R.W., Bruchez M., Van Houten B., Yasui A., Levine A.S.. Novel method for site-specific induction of oxidative DNA damage reveals differences in recruitment of repair proteins to heterochromatin and euchromatin. Nucleic Acids Res. 2014; 42:2330–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dynek J.N., Smith S.. Resolution of sister telomere association is required for progression through mitosis. Science. 2004; 304:97–100. [DOI] [PubMed] [Google Scholar]
- 24. Lan L., Nakajima S., Oohata Y., Takao M., Okano S., Masutani M., Wilson S.H., Yasui A.. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:13738–13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lan L., Ui A., Nakajima S., Hatakeyama K., Hoshi M., Watanabe R., Janicki S.M., Ogiwara H., Kohno T., Kanno S. et al. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol. Cell. 2010; 40:976–987. [DOI] [PubMed] [Google Scholar]
- 26. Wei L., Nakajima S., Böhm S., Bernstein K.a., Shen Z., Tsang M., Levine A.S., Lan L.. DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3495–E3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Opresko P.L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kolvraa S., May A., Seidman M.M., Bohr V.A.. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004; 14:763–774. [DOI] [PubMed] [Google Scholar]
- 28. Wang L., Ogburn C.E., Ware C.B., Ladiges W.C., Youssoufian H., Martin G.M., Oshima J.. Cellular Werner phenotypes in mice expressing a putative dominant-negative human WRN gene. Genetics. 2000; 154:357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lan L., Nakajima S., Komatsu K., Nussenzweig A., Shimamoto A., Oshima J., Yasui A.. Accumulation of Werner protein at DNA double-strand breaks in human cells. J. Cell Sci. 2005; 118:4153–4162. [DOI] [PubMed] [Google Scholar]
- 30. Opresko P.L., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A.. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002; 277:41110–41119. [DOI] [PubMed] [Google Scholar]
- 31. Burger A.M., Dai F., Schultes C.M., Reszka A.P., Moore M.J., Double J.A., Neidle S.. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005; 65:1489–1496. [DOI] [PubMed] [Google Scholar]
- 32. Ammazzalorso F., Pirzio L.M., Bignami M., Franchitto A., Pichierri P.. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J. 2010; 29:3156–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li K., Casta A., Wang R., Lozada E., Fan W., Kane S., Ge Q., Gu W., Orren D., Luo J.. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J. Biol. Chem. 2008; 283:7590–7598. [DOI] [PubMed] [Google Scholar]
- 34. Cheng W.H., Muftic D., Muftuoglu M., Dawut L., Morris C., Helleday T., Shiloh Y., Bohr V.A.. WRN is required for ATM activation and the S-phase checkpoint in response to interstrand cross-link-induced DNA double-strand breaks. Mol. Biol. Cell. 2008; 19:3923–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Steensel B., de Lange T.. Control of telomere length by the human telomeric protein TRF1. Nature. 1997; 385:740–743. [DOI] [PubMed] [Google Scholar]
- 36. Mohaghegh P., Karow J.K., Brosh R.M. Jr, Bohr V.A., Hickson I.D.. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001; 29:2843–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Compton S.A., Tolun G., Kamath-Loeb A.S., Loeb L.A., Griffith J.D.. The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J. Biol. Chem. 2008; 283:24478–24483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lai J.S., Herr W.. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:6958–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fairall L., Chapman L., Moss H., de Lange T., Rhodes D.. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell. 2001; 8:351–361. [DOI] [PubMed] [Google Scholar]
- 40. Lee J.W., Kusumoto R., Doherty K.M., Lin G.X., Zeng W., Cheng W.H., von Kobbe C., Brosh R.M. Jr, Hu J.S., Bohr V.A.. Modulation of Werner syndrome protein function by a single mutation in the conserved RecQ domain. J. Biol. Chem. 2005; 280:39627–39636. [DOI] [PubMed] [Google Scholar]
- 41. Tadokoro T., Kulikowicz T., Dawut L., Croteau D.L., Bohr V.A.. DNA binding residues in the RQC domain of Werner protein are critical for its catalytic activities. Aging. 2012; 4:417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ye J.Z., de Lange T.. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat. Genet. 2004; 36:618–623. [DOI] [PubMed] [Google Scholar]
- 43. Simonelli V., Narciso L., Dogliotti E., Fortini P.. Base excision repair intermediates are mutagenic in mammalian cells. Nucleic Acids Res. 2005; 33:4404–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Narwal M., Venkannagari H., Lehtio L.. Structural basis of selective inhibition of human tankyrases. J. Med. Chem. 2012; 55:1360–1367. [DOI] [PubMed] [Google Scholar]
- 45. Ishikawa N., Nakamura K., Izumiyama-Shimomura N., Aida J., Ishii A., Goto M., Ishikawa Y., Asaka R., Matsuura M., Hatamochi A. et al. Accelerated in vivo epidermal telomere loss in Werner syndrome. Aging. 2011; 3:417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tahara H., Tokutake Y., Maeda S., Kataoka H., Watanabe T., Satoh M., Matsumoto T., Sugawara M., Ide T., Goto M. et al. Abnormal telomere dynamics of B-lymphoblastoid cell strains from Werner's syndrome patients transformed by Epstein-Barr virus. Oncogene. 1997; 15:1911–1920. [DOI] [PubMed] [Google Scholar]
- 47. Salk D., Au K., Hoehn H., Martin G.M.. Cytogenetics of Werner's syndrome cultured skin fibroblasts: variegated translocation mosaicism. Cytogenet. Cell Genet. 1981; 30:92–107. [DOI] [PubMed] [Google Scholar]
- 48. Moser M.J., Oshima J., Monnat R.J. Jr. WRN mutations in Werner syndrome. Hum. Mutat. 1999; 13:271–279. [DOI] [PubMed] [Google Scholar]
- 49. Huber M.D., Duquette M.L., Shiels J.C., Maizels N.. A conserved G4 DNA binding domain in RecQ family helicases. J. Mol. Biol. 2006; 358:1071–1080. [DOI] [PubMed] [Google Scholar]
- 50. von Kobbe C., Harrigan J.A., Schreiber V., Stiegler P., Piotrowski J., Dawut L., Bohr V.A.. Poly(ADP-ribose) polymerase 1 regulates both the exonuclease and helicase activities of the Werner syndrome protein. Nucleic Acids Res. 2004; 32:4003–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kitano K., Kim S.Y., Hakoshima T.. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 2010; 18:177–187. [DOI] [PubMed] [Google Scholar]
- 52. Berquist B.R., Wilson D.M. 3rd. Pathways for repairing and tolerating the spectrum of oxidative DNA lesions. Cancer Lett. 2012; 327:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Svilar D., Goellner E.M., Almeida K.H., Sobol R.W.. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid. Redox. Signal. 2011; 14:2491–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hanaoka S., Nagadoi A., Nishimura Y.. Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities. Protein Sci. 2005; 14:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Doksani Y., Wu J.Y., de Lange T., Zhuang X.. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013; 155:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Walker J.R., Zhu X.D.. Post-translational modifications of TRF1 and TRF2 and their roles in telomere maintenance. Mech. Ageing Dev. 2012; 133:421–434. [DOI] [PubMed] [Google Scholar]
- 57. Nishikawa T., Okamura H., Nagadoi A., Konig P., Rhodes D., Nishimura Y.. Solution structure of a telomeric DNA complex of human TRF1. Structure. 2001; 9:1237–1251. [DOI] [PubMed] [Google Scholar]
- 58. Poulet A., Pisano S., Faivre-Moskalenko C., Pei B., Tauran Y., Haftek-Terreau Z., Brunet F., Le Bihan Y.V., Ledu M.H., Montel F. et al. The N-terminal domains of TRF1 and TRF2 regulate their ability to condense telomeric DNA. Nucleic Acids Res. 2012; 40:2566–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Seimiya H., Muramatsu Y., Smith S., Tsuruo T.. Functional subdomain in the ankyrin domain of tankyrase 1 required for poly(ADP-ribosyl)ation of TRF1 and telomere elongation. Mol. Cell Biol. 2004; 24:1944–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lau T., Chan E., Callow M., Waaler J., Boggs J., Blake R.A., Magnuson S., Sambrone A., Schutten M., Firestein R. et al. A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res. 2013; 73:3132–3144. [DOI] [PubMed] [Google Scholar]
- 61. Gu H., Marth J.D., Orban P.C., Mossmann H., Rajewsky K.. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994; 265:103–106. [DOI] [PubMed] [Google Scholar]
- 62. Larsen E., Gran C., Saether B.E., Seeberg E., Klungland A.. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol. Cell. Biol. 2003; 23:5346–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.