Abstract
Many bacterial type I toxin mRNAs possess a long 5΄ untranslated region (UTR) that serves as the target site of the corresponding antitoxin sRNA. This is the case for the zorO-orzO type I system where the OrzO antitoxin base pairs to the 174-nucleotide zorO 5΄ UTR. Here, we demonstrate that the full-length 5΄ UTR of the zorO type I toxin hinders its own translation independent of the sRNA whereas a processed 5΄ UTR (zorO Δ28) promotes translation. The full-length zorO 5΄ UTR folds into an extensive secondary structure sequestering the ribosome binding site (RBS). Processing of the 5΄ UTR does not alter the RBS structure, but opens a large region (EAP region) located upstream of the RBS. Truncation of this EAP region impairs zorO translation, but this defect can be rescued upon exposing the RBS. Additionally, the region spanning +35 to +50 of the zorO mRNA is needed for optimal translation of zorO. Importantly, the positive and negative effects on translation imparted by the 5΄ UTR can be transferred onto a reporter gene, indicative that the 5΄ UTR can solely drive regulation. Moreover, we show that the OrzO sRNA can inhibit zorO translation via base pairing to the of the EAP region.
INTRODUCTION
Across bacterial species, numerous toxin-antitoxin loci have been identified that are believed to function in stress responses (1). These two-gene loci are categorized by both the nature of the antitoxin and the mode of its action to repress the toxin. For a type I toxin-antitoxin locus, one gene encodes a small protein that is toxic when overproduced and the second gene encodes a small RNA (sRNA) that represses toxin production through base pairing [reviewed in (2)]. Most type I toxins share common biochemical features: they usually contain a single putative transmembrane domain and are hypothesized to function at the cytoplasmic membrane. Overproduction of type I toxins is detrimental to bacterial cells and results in growth stasis or cell death. For instance, overexpression of either the ibsC or shoB toxin gene in Escherichia coli inhibited cell growth and led to rapid membrane depolarization (3). Further, TisB toxin overproduction in E. coli caused similar membrane damage along with a dramatic decrease in cellular ATP levels (4). Although it is membrane-associated, overproduction of the type I toxin BsrG from Bacillus subtilis did not cause membrane depolarization. Instead, its association with the membrane led to mislocalization of cell wall synthesis machinery, triggering autolysin activity and subsequent cell death (5). Some other type I toxins can impair nucleoid segregation and cell division prior to damaging the cell membrane, as exemplified by the Fst toxin of Enterococcus faecalis (6).
Given the potential detrimental effects of producing such toxins, their production must be tightly controlled. For type I toxins, this can be achieved through regulation by its corresponding antitoxin sRNA. More specifically, the antitoxin sRNA base pairs with the toxin mRNA and inhibits translation of the mRNA and/or stimulates the degradation of the mRNA, thereby preventing toxin production. This is exemplified by the symE-symR locus of E. coli (7) in that the SymR sRNA base pairs to the translation initiation region of the symE mRNA and blocks its translation, therefore repressing SymE production (8). Further, in the txpA-ratA pair of B. subtilis, base pairing of the RatA sRNA to the txpA mRNA leads to RNase III-dependent degradation of txpA, rendering it untranslatable (9,10). Inhibition of bsrG by its antitoxin Sr4 in B. subtilis, however, is due to both the triggering of degradation of the toxin mRNA as well as a translational block (11).
Along with sRNA-mediated regulation, production of some type I toxins can be modulated by the 5΄ untranslated regions (UTRs) of the toxin mRNAs themselves, particularly for those toxin mRNAs that possess long and structured 5΄ UTRs [reviewed in (12)]. One well studied example is the type I toxin gene hok of plasmid R1 in E. coli (13). The hok mRNA has a 177-nt-long 5΄ UTR, which harbors a small leader gene termed mok (14). Translation of the hok mRNA is dependent upon translation of mok; however, the 5΄ UTR of hok forms into a secondary structure that occludes the mok RBS, preventing not only its translation but also that of Hok (15,16). These structural constraints are relieved upon processing of the 3΄ end of the hok mRNA, allowing translation to proceed (16). Although translation of the type I toxin TisB is also repressed by its long 5΄ UTR, the underlying mechanism is distinct from that of hok. In this case, the tisB RBS is structurally sequestered and translation of the tisB mRNA requires a single-stranded standby site upstream of the RBS that allows ribosomes to preload onto the mRNA (17). This standby site, though, is occluded in a stem structure and only becomes accessible upon processing of the 5΄ UTR; thus, the full-length tisB mRNA is translationally inactive.
We previously reported that the zorO–orzO gene pair in E. coli O157:H7 (EHEC) is a true type I toxin-antitoxin locus, with zorO encoding the toxic small protein and orzO encoding the antitoxin sRNA (18,19). The zorO mRNA has a long 5΄ UTR of 174 nts with the region of base pairing to OrzO located 60 nts upstream of the zorO RBS. Repression by OrzO is triggered by RNase III-dependent degradation of zorO (18). When examining the predicted structures of the zorO mRNA, we noted that the RBS is located in a stem structure (18), which could prevent ribosomal access. We hypothesized that translation of zorO may require ribosomal preloading onto a standby site to compensate for the closed RBS. Thus, disruption of this putative ribosome standby site of zorO would impair translation of the zorO mRNA and alleviate the toxic effects of zorO overexpression.
In this study, we confirm that the zorO RBS is occluded in stem structure and that processing of the 5΄ end is required for efficient translation. The 5΄ processing does not alter the structure at the RBS (+164 to +169 from the transcription start site), but leads to the opening of a structured region +73 to +102 from the transcription start site referred to as the EAP (Exposed After Processing) region that contains a putative standby site to promote translation. Optimal translation of zorO requires not only the EAP region, but also another region spanning from +35 to +50 of zorO 5΄ UTR. Moreover, we demonstrate that the OrzO sRNA can also affect translation of the zorO mRNA via base pairing at the EAP region of zorO, therefore inhibiting the production of ZorO toxin.
MATERIALS AND METHODS
Bacterial strains and plasmids
The strains and plasmids used in this study are listed in Supplementary Table S1, and the sequences of all oligonucleotides are given in Supplementary Table S2.
Growth conditions
Escherichia coli strains were grown in Luria–Burtani (LB) medium at 37°C with shaking. For strains carrying pAZ3-zorO Δ28 or pAZ3-zorO Δ34, 0.2% glucose was added to reduce leaky expression of the PBAD promoter (20). Antibiotics were used at the following concentrations when needed: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 25 μg/ml. Arabinose was added to a final concentration of 0.2%, 0.002%, 0.0001%, 0.00002% or 0.00001% as indicated. When used, IPTG was added to a final concentration of 1 mM.
Plasmid construction
For overexpression of the zorO gene under the PBAD promoter, the full 5΄ UTR of zorO along with the coding region was amplified from E. coli O157:H7 EDL933 genomic DNA, digested with EcoRI and HindIII, and cloned into the corresponding sites of pAZ3 (8), generating pAZ3-zorO. The pAZ3-zorO Δ28, pAZ3-zorO Δ34, pAZ3-zorO Δ50, and pAZ3-zorO Δ82 were constructed in the same fashion. The pEF21-zorO Δ28 was generated following the same scheme, but using a lower copy number vector pEF21 (pBAD33 derivative, pACYC origin) (3). The pAZ3-zorO Δ82 RBS was constructed through site-directed mutagenesis as previously described using pAZ3-zorO Δ82 as template (21).
For in vitro transcription assays, the full 5΄ UTR of zorO along with the coding region was amplified from E. coli O157:H7 EDL933 genomic DNA, digested with EcoRI and SmaI, and cloned into the corresponding sites of pGEM®-3Zf(+) Vector (Promega), generating pGEM-T7-zorO. Plasmids pGEM-T7-zorO Δ50, and pGEM-T7-zorO Δ82 were generated in the same manner. The pGEM-T7-zorO Δ82 RBS was generated through site-directed mutagenesis using pGEM-T7-zorO Δ82 as template.
The translational gfp fusion to zorO was generated using SOE PCR (22). PCR product A amplified the full length 5΄ UTR and the start codon of zorO from genomic DNA along with an additional 18 nts at the 3΄ end that overlapped with the PCR product B. PCR product B was the result of amplifying the coding sequence of gfp from vector pFA6a-GFP(S65T)-kanMX6 (23). The two products were spliced together using the external primers, digested with EcoRI and HindIII, and cloned into the corresponding sites of pAZ3, generating pAZ3-zorO UTR-gfp. The pAZ3-Δ28 UTR-gfp was constructed through amplifying the corresponding UTR-gfp region using pAZ3-zorO UTR-gfp as template via PCR, and cloned into pAZ3.
Overproduction of toxic proteins
ZorO toxicity assays were performed as described previously (18). Briefly, UTK007 was transformed with the indicated plasmid derivatives of pAZ3 via electroporation (20). The resulting transformants were grown overnight and then diluted to an OD600 of 0.01. When the OD600 reached 0.2–0.3, the culture was split and arabinose was added to half of the culture to a final concentration of 0.2%, 0.0001% or 0.00001% as indicated. Prior to arabinose induction, cells harboring either pAZ3-zorO Δ28 or pAZ3-zorO Δ34 were washed twice with 1×phosphate-buffered saline (PBS) and resuspended in LB medium to ensure no carryover of glucose (20). OD600 was measured every 30 min. Shown are averages ± standard deviations for a minimum of three independent experiments.
Rescue experiments
Rescue experiments were conducted as described previously (3,18,24). The pBR-plac plasmid carrying the orzO gene under an IPTG-induced PLlacO-1 promoter was transferred into either UTK007 (wild type) or UTK011 (Δrnc) through electroporation. Afterwards, the pEF21 plasmid containing the full-length 5΄ UTR of zorO and its coding sequence (zorO full-length) or the processed 5΄ UTR of zorO and its coding sequence (zorO Δ28) under the PBAD promoter was then transferred to the same cells. The resulting transformants harboring the two plasmids were grown overnight and then diluted to OD600 of 0.01. When the OD600 reached ≈0.1, the cultures were split and IPTG was added to half the culture to a final concentration of 1 mM. Thirty minutes after IPTG induction, arabinose was added to a final concentration of 0.002%, 0.0001% or 0.00002% as indicated for the overproduction of zorO. OD600 was measured every 30 min. Shown are averages ± standard deviations for a minimum of three independent experiments.
RNA extraction
To compare the RNA levels of zorO wild type versus zorO mutants, cells carrying the appropriate plasmid were grown as indicated in the toxicity assays to an OD600 ≈ 0.2–0.3, and arabinose (0.2% or 0.0001%) was added (time 0). Cells were harvested at time 0, 5, 15, 30 and 60 min post-arabinose induction and total RNA was isolated via hot acid-phenol or direct lysis as described previously (24,25). When needed, RNA was treated with TURBO™ DNase (Life Technologies) following the manufacturer's instructions.
Northern analysis
Total RNA (10 μg) was separated on a denatured 6% polyacrylamide-urea gel for detection of zorO wild type or mutant mRNA and transferred to a Zeta-Probe Genomic GT membrane (Bio-Rad). Specific oligonucleotide probes were 5΄ end-labeled with γ-32P by T4 polynucleotide kinase (New England Biolabs). Hybridization and washes of the membrane were performed as described previously (26). Northern analysis was conducted from a minimum of three independent experiments for every construct examined.
Flow cytometry analysis
Cells containing a translational gfp fusion to zorO were grown as previously indicated to an approximate OD600 ≈0.3 and split into two cultures: one was induced with 0.2% arabinose and the other served as a control. After the cultures were split, aliquots were taken for analysis at 0, 30 and 60 min. Due to cellular growth, 50 μl aliquots were taken at time 0 and 25 μl aliquots were taken at time 30 and 60. These aliquots were flooded with 4 ml of 1× PBS, centrifuged for 10 min at 4°C, and washed extensively twice. Cells were resuspended in 1ml 1× PBS and analyzed by flow cytometry in a LSR II flow cytometer (Becton Dickinson) with a 488-nm laser. Samples were run at an event rate of 3000 events per second. Green fluorescence was collected in the fluorescein isothiocyanate (FITC) channel. Data from the flow cytometer was analyzed using FlowJo software package (FlowJo LLC, Ashland, OR, USA). Fluorescence intensity was normalized by dividing the geometric mean fluorescence of the induced culture at each time point to its respective uninduced culture; shown are the averages ± standard deviation for three independent cultures.
In vitro transcription
T7 in vitro transcription was conducted as described previously (27) with modifications. Briefly, pGEM-T7-zorO or its derivatives was linearized with SmaI, purified using the Qiagen PCR Purification Kit, and 1 μg of digested plasmid was used as the DNA template for T7 transcription. A DNA template for zorO Δ28 mRNA was amplified from E. coli O157:H7 EDL933 genomic DNA via PCR with primers containing the T7 promoter. The DNA template, ribonucleotides (each at 2 mM, New England Biolabs), 8 mM DTT (Invitrogen) and 100 U T7 RNA polymerase (50 U/μl, New England Biolabs) were mixed in a 50 μl volume and incubated at 37°C for 16 h. Afterwards, 2 U of TURBO™ DNase (2 U/μl, Life Technologies) was added to the transcription reaction, followed by incubation at 37°C for an additional 15 min. Transcribed RNAs were then precipitated and gel purified (28).
RNA radiolabeling and in vitro structure probing
For 5΄ end labeling, the RNAs synthesized from T7 transcription were dephosphorylated and end-labeled with γ-32P by T4 polynucleotide kinase (New England Biolabs). For 3΄ end labeling, RNAs were labeled with [5- γ 32P] pCp and T4 RNA Ligase 1 (New England Biolabs). All radiolabeled RNAs were gel purified and folded prior to use.
In vitro structure probing was conducted in 10 μl reactions as described previously (29). Radiolabeled zorO RNA and its derivatives (0.2 pmol) were mixed with 1 × Structure Buffer (Ambion) and 1 μg yeast RNA. Unlabeled OrzO sRNA (250 nmol or 500 nmol) was added as indicated. For enzymatic probing, 2 μl of RNase T1 (0.01 U/μl; Ambion) were added to the reaction mixtures and incubated for 6 min at 37°C. For chemical probing, 2 μl of fresh lead(II) acetate (25 mM) were added to corresponding reactions and incubated for 1.5 min at 37°C. Reactions were stopped with the addition of 20 μl of Inactivation/Precipitation buffer (Ambion) mixed by vortexing, and precipitated. The RNA pellets were suspended in 7 μl Loading Buffer II (95% formamide, 18 mM EDTA, 0.025% SDS, 0.2% xylene cyanol, 0.2% bromophenol blue; Ambion).
To generate the RNase T1 ladder, radiolabeled RNA (0.4 pmol) was mixed with 1 × Sequencing Buffer (Ambion), denatured at 95°C for 1 min, and chilled on ice for 5 min. The mixture was incubated with RNase T1 (1 μl, 0.005 U/μl) at 55°C for 6 min (30). Reactions were stopped with the addition of 20 μl of inactivation/precipitation buffer (Ambion) mixed by vortexing, and precipitated. The RNA pellets were suspended in 7 μl Loading Buffer II. To generate the hydroxyl ladder, radiolabeled RNA (0.4 pmol) was mixed with 1× Alkaline Hydrolysis Buffer (Ambion) and incubated at 90°C for 5 min. The reaction was stopped with the addition of 12 μl of loading buffer II.
All samples were denatured at 95°C for 3 min and 3 μl of each sample was resolved on a denatured 6% polyacrylamide–urea gel. Gels were dried and then exposed to BioMax XAR film (Kodak).
In vitro translation
In vitro translation was performed using E. coli S30 Extract System (Promega) as described previously with modifications (17). Briefly, 0.1 μM or 0.25 μM of in vitro transcribed zorO RNA was mixed with 0.2 mM 35S-labeled methionine (>1000 Ci/mmol, PerkinElmer), 7.5 μl S30 extract, 10 μl S30 premix without amino acids, and 0.1 mM of each amino acid minus methionine. 5 μM of orzO RNA were added as indicated in the text. The reactions were incubated at 37°C for 30 min and chilled on ice for 5 min. For each reaction, 5 μl aliquots were mixed with 20 μl cold acetone and incubated on ice for an additional 15 min. Afterwards, the reaction mixtures were centrifuged for 10 min at 4°C at 12 000 g. The protein pellets were suspended in 15 μl of H2O, mixed with 4 × Bolt LDS sample buffer (Life Technologies), and resolved on NuPAGE Novex 4–12% Bis–Tris Plus Gels in MES Buffer (Life Technologies) alongside SeeBlue® Pre-Stained Protein Standard (Life Technologies). Gels were fixed in methanol/acetic acid solution (45% methanol, 10% acetic acid) for 30 min at room temperature and washed in 100 ml Amplify Fluorographic Reagent solution (GE Healthcare) for 60 min. Gels were subsequently dried and exposed to BioMax XAR film (Kodak).
RESULTS
5΄ processed form of zorO exhibits more efficient translation
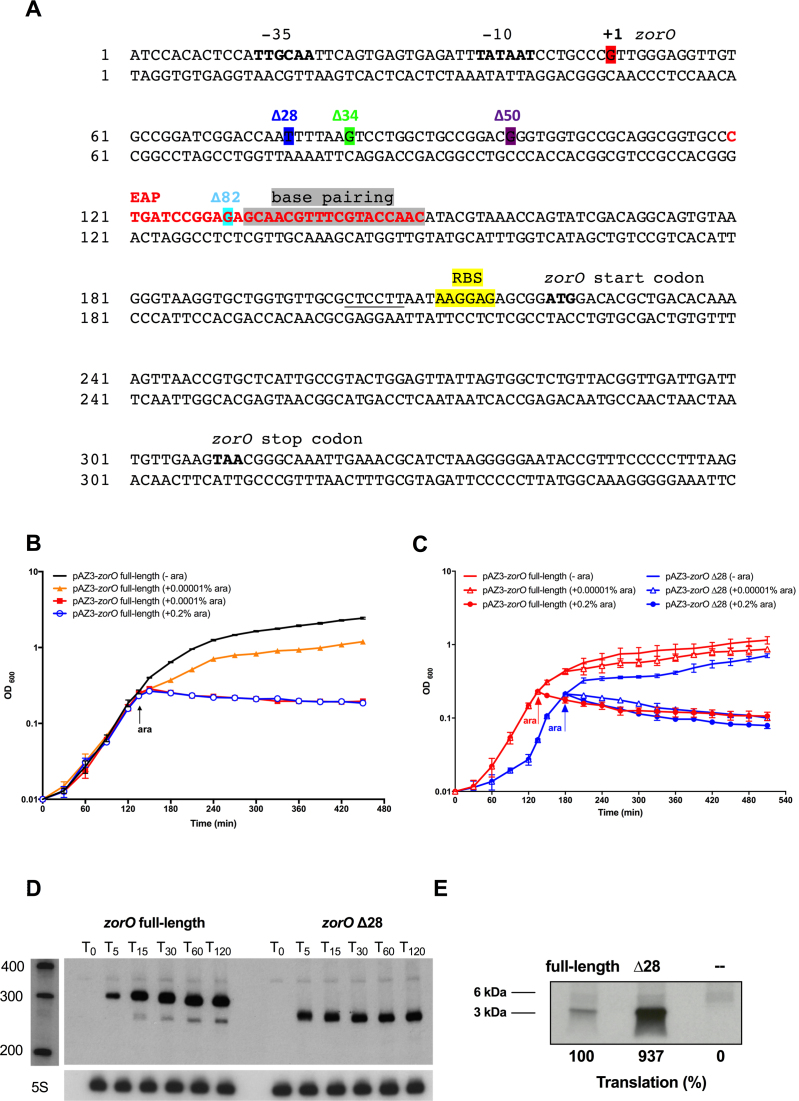
Previous primer extension analysis of the zorO mRNA revealed a processed form lacking the first 28 nucleotides of the full length mRNA (18). It has been shown that 5΄ processing of some type I toxin mRNAs increases translation of the mRNA [reviewed in (12)]. To test if processing of the zorO mRNA impacts its translation, we constructed a zorO Δ28 mutant under the control of the PBAD promoter in the plasmid pAZ3 (pAZ3-zorO Δ28) in which the first 28 nts of the zorO 5΄ UTR were removed (Figure 1A). This mutant reflects the native processed form of the zorO transcript. We predicted that if 5΄ processing of the zorO mRNA promotes zorO translation, the zorO Δ28 mutant would have a higher translation efficiency and exhibit a more pronounced toxic phenotype than the zorO full-length under the same induction condition.
Figure 1.
The 5΄ processed form of zorO (zorO Δ28) shows enhanced translation. (A) Sequence details of the zorO gene. Region of base pairing to OrzO is shaded in gray. The predicted −35 and −10 promoter elements, the start codon, and the stop codon of zorO are indicated in bold. Transcription start site of the full-length zorO full-length is highlighted in red. Transcription start site of each zorO truncation mutant is highlighted in color with the mutant name listed in the same color. The EAP region is labeled in red. The RBS of zorO is highlighted in yellow and the region base pairing with the RBS in the stem structure is underlined. (B) E. coli strain UTK007 harboring pAZ3-zorO was grown to mid-log and split in four cultures. Arabinose was added as indicated by the arrow. Shown are the averages ± standard deviations for three independent cultures. (C) UTK007 harboring pAZ3-zorO or pAZ3-zorO Δ28 was grown in LB supplemented with 0.2% glucose to exponential phase, washed, and split into three cultures. Arabinose was added as indicated by the arrow. Shown are the mean values ± standard deviations for three independent cultures. (D) Total RNA was isolated from E. coli MG1655 harboring pAZ3-zorO or pAZ3-zorO Δ28 at the indicated time points following the addition of 0.0001% arabinose to exponentially growing cultures (OD600 = 0.2–0.3). Shown is a representative of three independent northern blots detecting either zorO (upper panel) or the loading control 5S (lower panel). (E) In vitro translation assays with [35S]-Met were performed as described in the Material and Methods with 0.25 μM of zorO full-length or zorO Δ28 mRNAs. No RNA was added to the negative control. Quantification of band intensities were determined using ImageJ (54). The intensity of the band representing the zorO full-length was set to 100% translational efficiency.
To perform these in vivo analyses, we used a derivate of E. coli MG1655 (UTK007) as it does not naturally harbor the zorO–orzO locus; consequently, there is no endogenously produced toxin or antitoxin to confound our results. Additionally, given that the zorO and orzO genes are not directly antisense to each other (18,19), plasmids containing zorO do not contain any sequence corresponding to the orzO antitoxin. We noted that even in the absence of arabinose cells harboring the zorO Δ28 plasmid grew far slower than cells harboring the zorO full-length plasmid. After overnight growth, cellular lysis was evident in cultures containing the zorO Δ28 plasmid (data not shown). To further repress leaky expression from the PBAD promoter controlling zorO expression, glucose was supplemented to cultures harboring either vector; prior to induction with arabinose, cells were filtered and washed to remove excess glucose.
To better compare the toxicity of the zorO full-length and zorO Δ28 plasmids, we tested the effect of arabinose concentration (0.2, 0.0001 and 0.0001%) on stasis and found that the addition of 0.00001% arabinose to cells harboring the zorOΔ28 plasmid produced full stasis (Figure 1C), similar to what was seen with a higher concentration of arabinose (0.0001%) added to cells harboring the full length plasmid (Figure 1B and C). Given that the zorO coding sequence remains unchanged in zorO Δ28, this ‘hypertoxic’ phenotype observed is indicative of increased production of the ZorO toxin.
Since both the zorO full-length and the zorO Δ28 constructs examined above share the same promoter and coding sequence, the more pronounced toxic phenotype caused by zorO Δ28 is likely not due to increased transcription or protein stability. Rather, it could be due to greater RNA stability and/or higher translation efficiency. If the zorO Δ28 RNA is more stable, we would expect to see increased RNA levels compared to that of the zorO full-length under the same induction conditions. To test this, we examined the mRNA levels of the zorO full-length as well as the zorO Δ28 by northern blot analyses. As shown in Figure 1D, zorO Δ28 was expressed at levels equivalent to or even lower than the zorO full-length under the same induction condition. This suggested that the elevated toxic phenotype of the zorO Δ28 mutant is likely due to increased translation of the mRNA and not due to increased mRNA stability. To directly compare the translation efficiency of zorO full-length versus zorO Δ28, an in vitro translation assay was performed. Although both were translated, the zorO Δ28 mRNA was more robustly translated than the zorO full-length (Figure 1E). Therefore, the 5΄ processing of zorO mRNA facilitates the translation of zorO.
Enhanced translation by 5΄ processing can be transferred
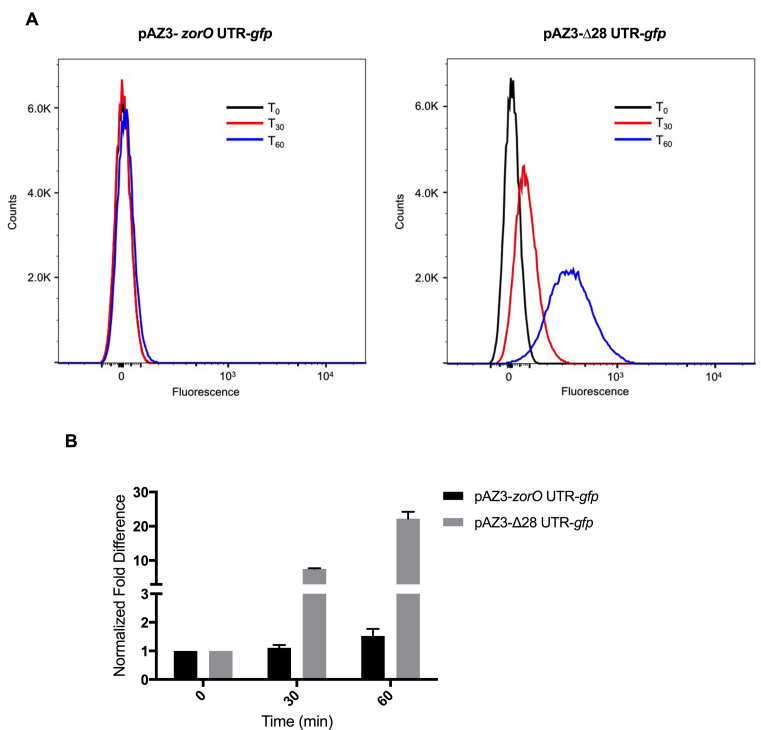
We next wanted to determine if the observed translational enhancement upon processing was due to changes in the 5΄ UTR. If changes in the UTR drove the effects on translation, then the transfer of the zorO 5΄ UTR onto another gene would mimic the above results. To test this hypothesis, we generated two translational gfp fusions with either the full-length UTR (zorO UTR) or the processed UTR (Δ28 UTR); both fusions were under control of the PBAD promoter. Using flow cytometry, we compared the production of GFP following arabinose induction in these constructs. We found that without induction, the two constructs showed similar background fluorescence throughout all analyzed time points (Supplemental Figure S1). When the full-length UTR-gfp was induced, we noted a very modest increase in fluorescence intensity over time (left panel, Figure 2A; see discussion). However, there was a far greater increase in fluorescence intensity in cells harboring Δ28 UTR-gfp (right panel, Figure 2A) as opposed to full-length UTR-gfp (left panel, Figure 2A). More specifically, 30 min post-induction, the Δ28 UTR-gfp showed a dramatic increase (∼8-fold) in geometric mean fluorescence intensity as compared to its uninduced control, whereas the geometric mean fluorescence intensity of the full-length UTR-gfp was similar to the control (Figure 2B). Further, after 60 min, fluorescence intensity in cells overexpressing the Δ28 UTR-gfp was 20-fold greater than its control, while fluorescence intensity in cells overexpressing the full-length UTR-gfp was only 1.5-fold greater than its control (Figure 2B). Overall, our data suggest that the processed UTR resulted in increased GFP production as compared to the full-length UTR.
Figure 2.
The 5΄ UTR-mediated translation effects can be transferred to gfp. (A) Shown are flow cytometry analyses of the fluorescence intensity of GFP from cells harboring either pAZ3-zorO UTR-gfp or pAZ3-Δ28 UTR-gfp after 0.2% arabinose induction. The lines indicate time in min post induction. Shown is a representative distribution plot of fluorescent intensity from three independent experiments. (B) Fold difference in gfp expression was determined using the geometric mean fluorescence normalized to to uninduced controls. Gray bars indicate pAZ3-zorO UTR-gfp and black bars indicate pAZ3-Δ28 UTR-gfp. Shown are the mean values ± standard deviations for three independent experiments.
Additionally, we noted the full-length UTR-gfp was processed (data not shown) similarly as we have noted for zorO (18) indicating that processing can occur regardless of the downstream coding sequence. These results fully recapitulate the observed ‘hypertoxic’ phenotype of zorO Δ28 and confirm our findings that processing of the zorO 5΄ UTR promotes translation.
Opening of the +73 to +102 region in zorO Δ28
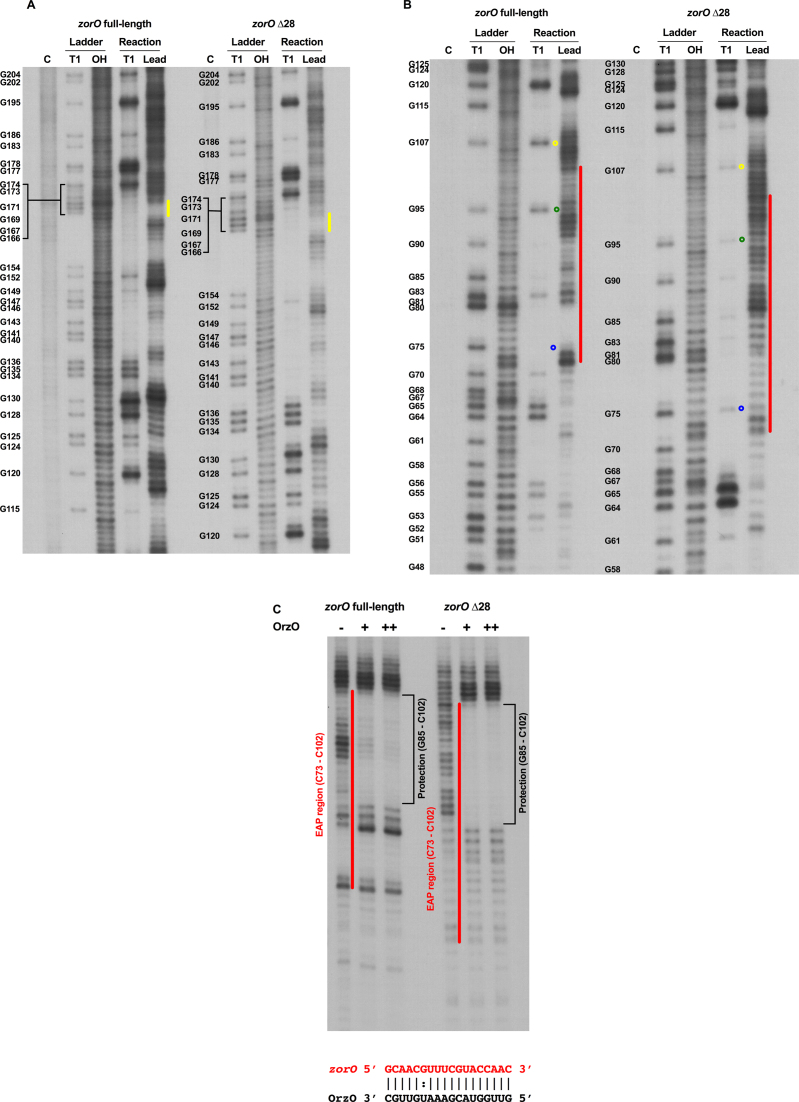
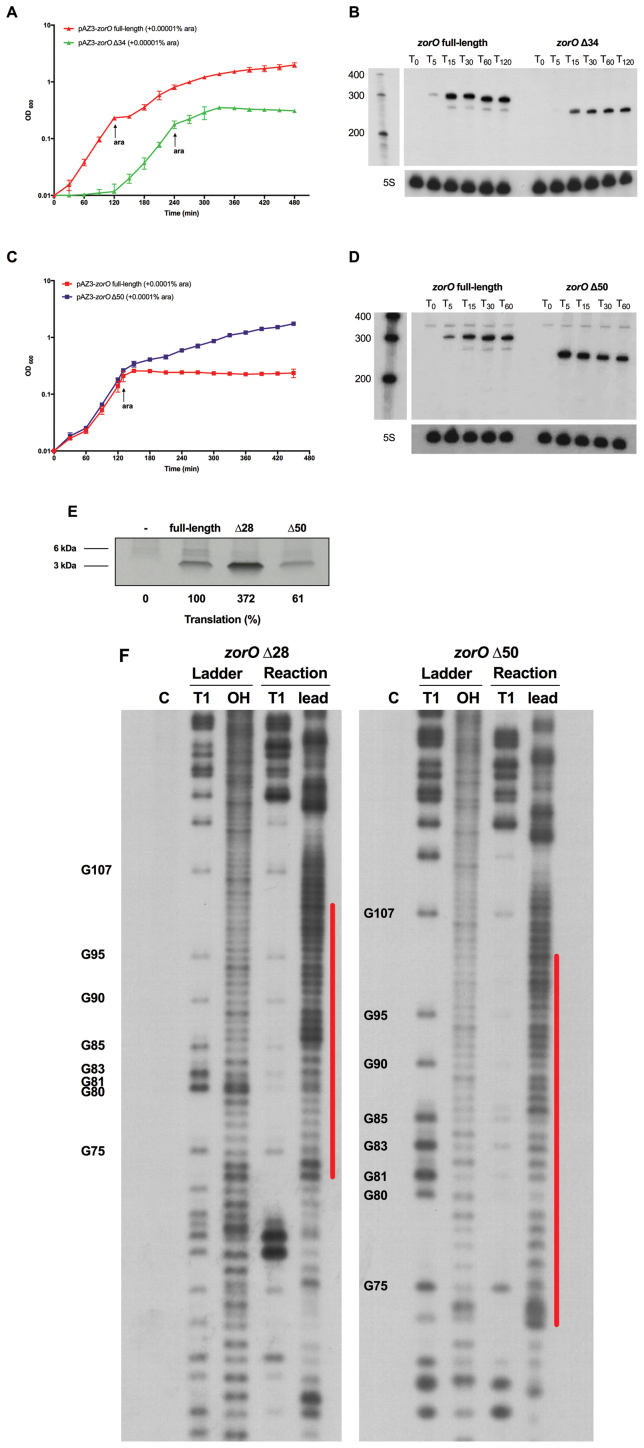
Our data suggested that removal of the first 28 nts of the 5΄ UTR promotes zorO translation, yet the mechanism underlying this increased translation remained unclear. The RBS of the zorO mRNA is predicted to be in a stem structure that may impede ribosome binding (18,31). This raised the possibility that the 5΄ processing could lead to an overall rearrangement of the structure of the mRNA, rendering the RBS more accessible. We thus employed in vitro structure probing to compare the zorO full-length and zorO Δ28 structures. We specifically used RNase T1, which cleaves single-stranded RNA at G residues and lead(II) acetate which predominantly cleaves at unpaired residues (32,33). Surprisingly, no structural differences were detected using either treatment at the RBS or the flanking regions of the zorO full-length and zorO Δ28 and both RBSs were in stem structures (Figure 3A and Supplemental Figure S2, indicated by yellow lines). This implies that the increased translation efficiency of zorO Δ28 was not due to structural changes that might alter RBS accessibility.
Figure 3.
The 5΄ processing of zorO does not alter RBS structure, but exposes the EAP region. (A) The RBS of both zorO full-length and zorO Δ28 are in a stem structure. 5΄-end-labeled zorO full-length or zorO Δ28 RNA was subjected to RNase T1 and lead(II) acetate cleavage as described in the Material and Methods. C, control RNA; Ladder/T1, RNase T1 ladder of indicated RNA under denaturing conditions; Ladder/OH, Alkaline ladder; Reaction/T1, RNase T1 digestion under native condition; and Reaction/Lead, Lead(II) acetate digestion under native condition. The position of selected cleaved G residues is listed at the left of each T1 ladder. For clarity, all positions are given as for zorO full-length mRNA from the 5΄ end of the transcript even though nucleotide numbers might be different in mutant RNAs. Yellow lines indicate the RBS region of each zorO transcript. (B) In vitro structure probing was conducted as in (A). The position of selected cleaved G residues is given at the left of each T1 ladder. Red lines indicate the corresponding EAP region (+73 to +102 from zorO full-length transcription start site) of zorO full-length and zorO Δ28. Sample positions that showed experimental variation between structure probing experiments are indicated as follows: yellow circles, G107: green circles, G95: the same corresponding regions are indicated in Supplemental Figure S3B, see text and supplemental figures for information. Position G75 (blue circles) is indicative of a residue whose cleavage was consistent across experiments. Note that the degree of sensitivity to cleavage may be variable between experiments, in all cases, residues indicated above were digested in zorO Δ28. (C) The OrzO sRNA can base pair with both the zorO full-length and zorO Δ28 at the 3΄ end of EAP region. Lead(II) acetate structure probing was conducted as in (A) with addition of 0 (−), 0.25 μM (+), and 0.5 μM (++) unlabeled OrzO sRNA. Red lines indicate the corresponding EAP region of the zorO full-length and zorO Δ28. Black brackets indicate the region protected by OrzO binding. Base pairing between zorO and OrzO is shown on the bottom.
When comparing the two RNA structures, the only major difference we observed was in the region spanning from the +73 to +102 from the transcription start site (Figure 3B, Supplemental Figure S3, indicated by red lines). This region in the zorO Δ28 mRNA was sensitive to lead digestion but the same region in the full-length zorO mRNA was protected from lead digestion, suggesting that this region is highly structured and perhaps helical in the full-length RNA (34). Thus, processing of the 5΄ end exposes this +73 to +102 region, which we refer to as the EAP (Exposure After Processing) region. We did note that there was altered sensitivity to RNase T1 for some residues in the full-length and processed form of the RNA for each experiment (G107 and G95 as examples, see Figure 3B and Supplemental Figure S3B; note the greater the band intensity, the more sensitive to digestion). Regardless, we observed digestion by RNase T1 in this region in the processed form for all of our experiments. There is also strong, consistent protection from lead digestion in the full-length zorO mRNA, specifically between the +80 to +74 region, that is not observed in the zorO Δ28 mRNA, supporting that there is a major structural change in the processed form. Interestingly, the EAP region encompasses the zorO base pairing region by the antitoxin sRNA OrzO (Figure 3C) (18). This result also indicates that the EAP region may be involved in sRNA dependent post-transcriptional regulation of zorO (see below).
Disruption of the EAP region impairs zorO translation
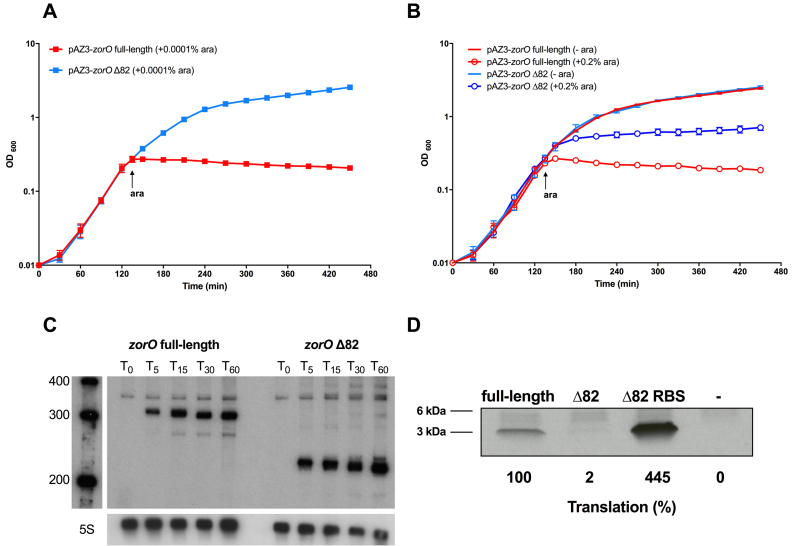
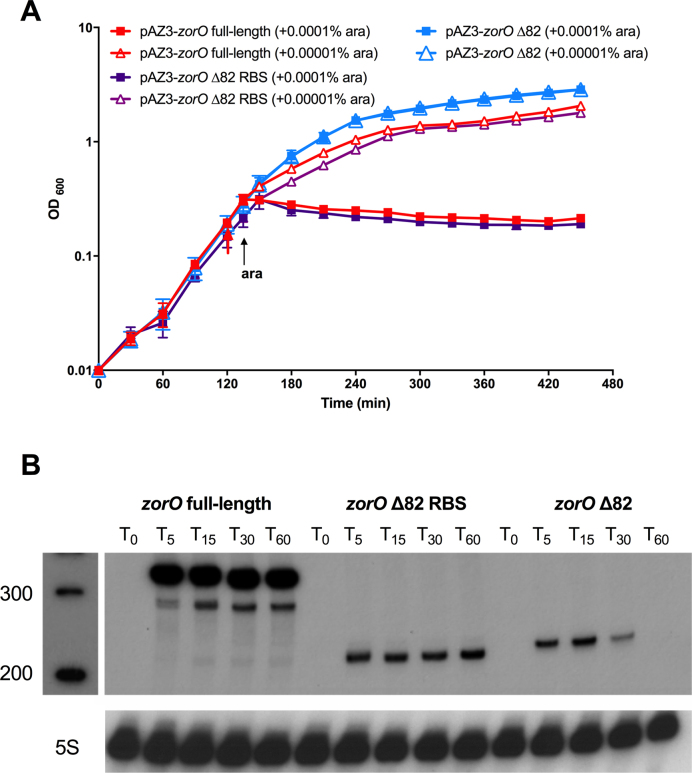
Since the EAP region is located upstream (∼100 nts) of the zorO RBS, we wanted to determine how exposure of this region impacted translation. Given that the RBS of zorO is structurally sequestered, direct ribosomal access to the RBS seems unlikely. Therefore, we hypothesized that optimal translation of zorO may require a ribosome standby site to facilitate ribosome preloading onto the mRNA. Owing to the more opened nature of the EAP region in zorO Δ28, we predicted that it may serve as a standby site. We thus generated a truncated EAP mutant by deleting the first 82 nts of the zorO 5΄ UTR (zorO Δ82) while maintaining the portion of the EAP region where the OrzO sRNA base pairs (Figure 1A). If the EAP region serves as a standby site, truncation of this region would interfere with ribosomal preloading and the subsequent translation, rendering the zorO Δ82 less toxic than the zorO full-length.
Indeed, the zorO Δ82 mutant was not toxic when induced by 0.0001% arabinose in contrast to full-length zorO (Figure 4A). Even when induced with saturating amounts of arabinose (0.2%), zorO Δ82 only showed impaired cellular growth (Figure 4B). To further clarify if we were observing growth stasis, versus a true decrease in cellular viability (i.e. cell death), cellular dilutions were plated. There were no viable colonies observed for cells harboring full-length zorO following 15 min of induction with 0.2% arabinose. However, colony forming units were detected even 2 h post-induction of zorO Δ82 using the same arabinose concentration (0.2%; data not shown). Taken together, these data suggested that cells overexpressing zorO Δ82 produced less ZorO toxin as compared to those overexpressing full-length zorO.
Figure 4.
Disruption of the EAP region of zorO leads to decreased translation. (A) Toxicity assay was performed as in Figure 1A with either zorO full-length or zorO Δ82 induced by 0.0001% arabinose. Shown are the mean values ± standard deviations for three independent cultures. (B) Toxicity assay was performed as in Figure 1A with either zorO full-length or zorO Δ82 induced by 0 (uninduced control) or 0.2% arabinose. Shown are the mean values ± standard deviations for three independent cultures. (C) Expression of zorO Δ82 induced by 0.2% arabinose is similar to or higher than zorO full-length induced by 0.0001% arabinose. Total RNA was isolated from E. coli MG1655 harboring pAZ3-zorO or pAZ3-zorO Δ82 at the indicated times following the addition of either 0.0001% (to induce zorO full-length) or 0.2% (to induce zorO Δ82) arabinose to exponentially growing cultures. Shown is a representative of three independent northern blots detecting zorO (upper panel) or the loading control 5S (lower panel). (D) In vitro translation assays with [35S]-Met were performed as described in the Material and Methods with 0.25 μM of zorO full-length, zorO Δ82, or zorO Δ82 RBS mRNAs. No mRNA was added to the negative control. Quantification of band intensities were determined using ImageJ (54). The intensity of the band representing the zorO full-length was set to 100% translational efficiency.
To separate the effect of RNA stability from that of translation efficiency, we determined the RNA levels of zorO Δ82 and zorO full-length by northern analysis. When both were induced with 0.2% arabinose, the levels of zorO Δ82 mRNA was lower than that of zorO full-length (data not shown), indicating that zorO Δ82 is less stable. We did note, though, that the levels of zorO Δ82 induced by 0.2% arabinose were equivalent to that of zorO full-length induced by 0.0001% arabinose (Figure 4C). However, despite the similar RNA levels, expression of only the zorO full-length mRNA and not zorO Δ82, conferred full growth stasis (Figure 4A, B). This suggested that in addition to decreased RNA stability, zorO Δ82 was not translated as efficiently.
To test whether the decreased toxicity was due in part to decreased translation of the zorO Δ82, we performed in vitro translation utilizing equal amounts of zorO full-length and zorO Δ82 mRNAs. As shown in Figure 4D, translation of the zorO Δ82 mRNA was barely detectable as compared to the zorO full-length. Together, these data suggest that disruption of the EAP region impairs translation efficiency.
Exposing the RBS rescues the translational defect of zorO Δ82
A standby site is thought to increase the concentration of ribosomes around the RBS (17,35). As a result, when the RBS is transiently exposed, the preloaded ribosomes can rapidly initiate translation. In line with this, we hypothesized that increased accessibility of the zorO RBS would obviate the need for a standby site. We thus disrupted the RBS-containing stem in the zorO Δ82 mRNA by mutating the region that base pairs with the zorO RBS (Supplemental Figure S4A, S4B). This resulted in a zorO mutant that harbors a constitutively exposed RBS (zorO Δ82 RBS). The toxicity of the zorO Δ82 RBS mutant mirrored that of the full-length zorO when induced by 0.0001% arabinose whereas zorO Δ82 was virtually nontoxic at this concentration (Figure 5A). Additionally, northern blot analysis revealed that the zorO Δ82 RBS was expressed at much lower levels than zorO full-length (Figure 5B), suggesting that the restored toxicity seen in the zorO Δ82 RBS mutant was due to more efficient translation. Further, translation of the zorO Δ82 RBS mRNA far exceeded that of both the zorO Δ82 and the full-length zorO mRNAs, indicating that the enhanced toxicity is due to increased translation (Figure 4D). Taken together, our data support the hypothesis that the EAP region of the zorO mRNA functions to compensate for a closed zorO RBS.
Figure 5.
Exposing the RBS rescues the translational defect of zorO Δ82. (A) Toxicity assay was performed as in Figure 1A with zorO full-length, zorO Δ82 or zorO Δ82 RBS by either 0.0001% or 0.00001% arabinose. Shown are the mean values ± standard deviations for three independent cultures. Note that there was no difference in toxicity observed when zorO Δ82 was induced by either 0.0001% or 0.00001% arabinose. (B) Total RNA was isolated from E. coli MG1655 harboring pAZ3-zorO, pAZ3-zorO Δ82 or pAZ3-zorO Δ82 RBS at the indicated times following the addition of 0.0001% arabinose to exponentially growing cultures. Shown is a representative of three independent northern blots detecting either zorO (upper panel) or the loading control 5S (lower panel).
An additional element in zorO that affects zorO translation efficiency
Our data indicate that the EAP region facilitates the translation of zorO. Since the zorO mRNA harbors a long (174 nts) structured 5΄ UTR, we next wanted to examine if other regions of the 5΄ UTR contribute to zorO translation. We thus constructed two additional zorO mutants in which either the first 34 nts (Δ34) or 50 nts (Δ50) from the transcription start site were deleted (Figure 1A). They were subjected to the same toxicity assays and northern blot analyses as described above. The zorO Δ34 mutant was similar to the aforementioned zorO Δ28 mutant in that it exhibited a hypertoxic phenotype and the RNA levels were lower than the full-length zorO under the same induction conditions (Figure 6A and B). This suggested that the translation efficiency of zorO Δ34 is also higher than that of zorO full-length.
Figure 6.
The 5΄ UTR contains an additional region that impacts zorO translation. (A) Toxicity assay was performed as in Figure 1B with either zorO full-length or zorO Δ34 induced by 0.00001% arabinose. Shown are the mean values ± standard deviations for three independent cultures. (B) Total RNA was isolated from E. coli MG1655 harboring pAZ3-zorO full-length or pAZ3-zorO Δ34 at the indicated times following the addition of 0.0001% arabinose to exponentially growing cultures. Shown is a representative of three independent northern blots detecting either zorO (upper panel) or the loading control 5S (lower panel). (C) Toxicity assay was performed as in Figure 1A with either zorO full-length or zorO Δ50 induced by 0.0001% arabinose. Shown are the mean values ± standard deviations for three independent cultures. (D) Total RNA was isolated from E. coli MG1655 harboring pAZ3-zorO or pAZ3-zorO Δ50 at the indicated times following the addition of 0.0001% arabinose to exponentially growing cultures. Shown is a representative of three independent northern blots detecting either zorO (upper panel) or the loading control 5S (lower panel). (E) In vitro translation assays with [35S]-Met were performed as described in the Material and Methods with 0.25 μM of zorO full-length, zorO Δ28, or zorO Δ50 mRNAs. No mRNA was added to the negative control. Quantification of band intensities were determined using ImageJ (54). The intensity of the band representing the zorO full-length was set to 100% translational efficiency. (F) In vitro structure probing was conducted as in Figure 3A. Red lines indicate the corresponding EAP region of zorO Δ28 and zorO Δ50. For simplicity, only the position of cleaved G residues within or flanking the EAP region is listed at the left of each T1 ladder and all positions are given as for zorO full-length mRNA from the 5΄ end of the transcript. Note that the samples were run on the same gel, but shown is two different exposures to better compare the cleavage patterns.
Interestingly, the zorO Δ50 mutant showed a decreased toxic phenotype as compared to the full-length when induced by 0.0001% arabinose (Figure 6C). Northern blot analysis indicated that zorO Δ50 was expressed at levels similar or even higher to that of the full-length, suggesting that the decreased toxicity was not due to differences in RNA levels (Figure 6D). When compared to the full-length zorO mRNA, there was a moderate reduction in ZorO protein production from the zorO Δ50 mRNA in vitro (Figure 6E). These data suggested that the zorO Δ50 mutant is translated less efficiently, leading to its overall decreased toxicity. Surprisingly, our structural probing results revealed that the zorO Δ50 mRNA is just as sensitive to lead digestion as the zorO Δ28 and thus possesses the same EAP region (Figure 6F) even though its translation efficiency is much lower than the zorO Δ28 (Figure 6E). Given that the zorO Δ34 is similar to that of the zorO Δ28, the reduced toxicity seen in zorO Δ50 is likely due to truncation of the sequence from +35 to +50. Therefore, these data imply that the region between +35 to +50 harbors an additional element that contributes to zorO translation (see discussion).
The OrzO sRNA can inhibit translation of zorO
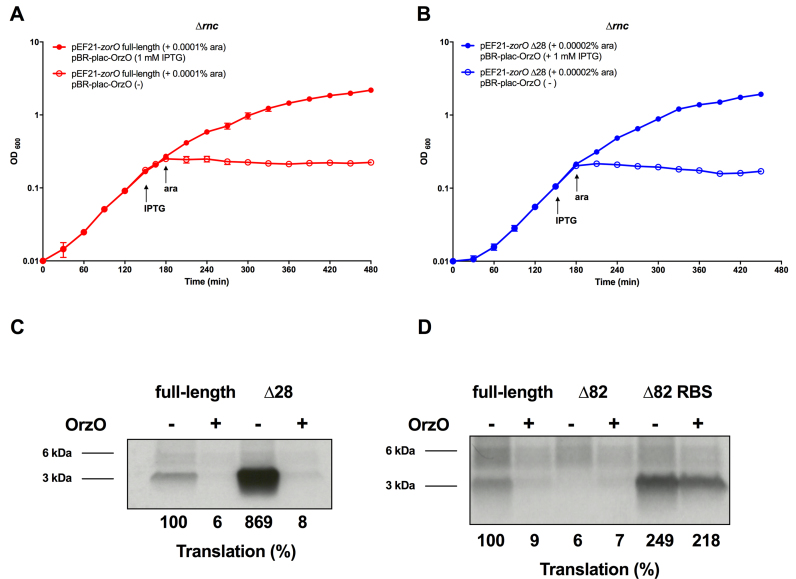
Our previous study demonstrated that base pairing of the OrzO sRNA to the zorO mRNA triggers RNase III-dependent degradation of the zorO mRNA (18). Whether this is the only mechanism by which the OrzO sRNA represses zorO expression is unclear. As shown, the structural probing data revealed that the OrzO sRNA can base pair with both the zorO full-length and the zorO Δ28 mRNAs at the 3΄ end of the EAP region (Figure 3C). If the EAP region acts as a standby site, base pairing by OrzO would prevent the ribosome from preloading onto the mRNA thus inhibiting translation of the zorO mRNA. Under this scenario, the OrzO sRNA would still be capable of repressing zorO even if RNase III is not present. To test this, we co-expressed the orzO sRNA gene along with either the zorO full-length or the zorO Δ28 gene in either a wild type strain (Supplemental Figure S5) or RNase III deletion (Δrnc) background to determine if OrzO could still restore bacterial growth from ZorO-induced toxicity. As we had previously noted, OrzO repression of ZorO-induced toxicity is partially dependent upon rnc since OrzO could not rescue cells from stasis induced by high levels of ZorO production (compare Supplemental Figure S5a and S5b). However, as shown in Figure 7A and B, bacterial growth stasis caused by overexpression of either the zorO full-length (0.0001% arabinose) or zorO Δ28 (0.00002% arabinose) was rescued by co-expression of orzO in a Δrnc strain. This indicates that the OrzO sRNA can repress ZorO toxin production in the absence of RNase III, likely through inhibiting zorO mRNA translation.
Figure 7.
OrzO sRNA inhibits translation of zorO transcripts. (A) E. coli strain UTK011 (Δrnc) carrying pBR-plac-orzO and pEF21-zorO was induced as indicated with arabinose (0.0001%) and/or IPTG (1 mM). Shown are the mean values ± standard deviations for three independent cultures. (B) Rescue experiment was performed as in (A) with zorO Δ28 expressed induced by 0.00002% arabinose. Shown are the mean values ± standard deviations for three independent cultures. (C) In vitro translation assays with [35S]-Met were performed as described in the Material and Methods with 0.1 μM of zorO full-length or zorO Δ28 mRNAs. 5 μM of OrzO RNA were added as indicated. Quantification of band intensities were determined using ImageJ (54). The intensity of the band representing the zorO full-length without the addition of OrzO was set to 100% translational efficiency. (D) In vitro translation assays with [35S]-Met were performed as described in the Material and Methods with 0.1 μM of zorO full-length, zorO Δ82, or zorO Δ82 RBS mRNAs. 5 μM of OrzO RNA were added as indicated. Quantification of band intensities were determined using ImageJ (54). The intensity of the band representing the zorO full-length without the addition of OrzO was set to 100% translational efficiency.
To directly assess the influence of the OrzO sRNA on zorO mRNA translation, we examined the in vitro translation efficiency of either zorO full-length or zorO Δ28 mRNA in the presence or absence of OrzO. The ZorO protein levels for either mRNA were reduced greatly in the presence of OrzO (Figure 7C). These results are concordant with the observed rescue from cellular toxicity in vivo (Figure 7A and B) and indicate that OrzO can regulate zorO expression at the translational level. Interestingly, the OrzO sRNA exhibited a more pronounced inhibitory effect on translation of zorO Δ28 mRNA as compared to zorO full-length mRNA. This corroborates our structural probing results that the zorO Δ28 mRNA has a more accessible EAP region for OrzO sRNA to bind. We also tested if the OrzO sRNA could impact the translation of zorO Δ82 RBS mRNA. Only a very mild reduction in ZorO protein levels were observed upon addition of OrzO sRNA (Figure 7D), which further supports the idea that an opened RBS would render the EAP region dispensable. Collectively, these data suggested that the OrzO sRNA represses ZorO production by affecting both translation and stability of the zorO mRNA.
DISCUSSION
In this study, we show that the 5΄ UTR of the zorO mRNA regulates the production of ZorO toxin through more than one mechanism. The 5΄ UTR affects the stability of the zorO mRNA as almost all zorO truncation mutants tested exhibited decreased RNA levels as compared to the zorO full-length. Additionally, the 5΄ UTR modulates the translation of the zorO mRNA through two distinct regions. One region, EAP, is less structured only upon processing and may serve as a putative ribosome standby site that likely facilitates ribosome preloading onto the mRNA, thereby promoting zorO translation. The other region spans from +35 to +50 of zorO and is required for optimal translation of zorO, although the underlying mechanism remains unclear (see below). Furthermore, the OrzO antitoxin can block translation of zorO, likely by competing with ribosomes for the EAP region. This base pairing was also previously shown to stimulate zorO degradation in an RNase III-dependent fashion (18), thereby preventing accumulation of the zorO mRNA.
Optimal translation of zorO requires a standby site
The 5΄ UTR mediated translational regulation of ZorO toxin synthesis is reminiscent to that of the TisB toxin of the type I pair tisAB-istR of E. coli. The tisAB mRNA possesses two forms: a full-length form (+1) and a processed form (+42) (17). The RBS of both forms of tisAB are folded in stems. Our work revealed that the zorO mRNA also exists in a full-length form and a processed form (Δ28) (18). Structural probing data demonstrated that the RBS is shielded in stem structures in both the zorO full-length and the Δ28 mRNAs. The processed tisAB undergoes structural changes at the 5΄ UTR as compared to full-length tisAB and opens a standby site that was initially sequestered in the full-length transcript (17). Similar structural rearrangement also occurs in zorO upon processing of the zorO 5΄ UTR, which leads to the opening of region in zorO Δ28 that spans from +73 to +102 (the EAP region, Supplemental Figure S3). Additionally, the processed tisAB exhibited robust translation whereas the full-length tisAB is translationally inert (17). Our results from the toxicity and in vitro translation assays also showed that zorO Δ28 has increased translation relative to full-length zorO. Given these extensive similarities between zorO and tisAB, it is likely that the translation of zorO follows the standby model and the EAP region serves as a standby site.
Unlike tisAB, both the zorO full-length and the zorO Δ28 forms are translatable (Figure 1E). Hence, the putative standby region of zorO can promote translation, but is not required for translation. As proposed by de Smit et al., a standby site operates by increasing the local concentration of ribosomes given that the time window during which a folded RBS is transiently opened is too short to efficiently recruit ribosomes (35). Thus, the effect of a standby site is dependent on the opening of the closed RBS. Our data comparing the zorO Δ82 versus zorO Δ82 RBS mutant showed that despite the perturbation of the standby site, translation of the toxin mRNAs were recovered after permanently exposing the RBS. This suggests the standby site is dispensable when the RBS is exposed. Therefore, although the zorO full-length mRNA may not harbor an accessible standby site, transitory opening of the RBS in zorO may provide an opportunity for translation to occur in vivo.
As noted above, the full-length zorO UTR is translated in vitro, yet we observed very little GFP fluorescence when the reporter gene was fused to the full-length zorO UTR (Figure 2A, left panel). Overproduction of ZorO, though, leads to membrane depolarization within 5 min of induction (Wen, Bogati, Harp, Fozo, unpublished data). We hypothesize that ZorO may form a small pore within the membrane, leading to ion leakage, as has been suggested for TisB (4,36–38). Consequently, if only a few individual proteins are needed to form a pore to induce ion leakage and eventual cell stasis, this could be reflective in the low levels of GFP detected. Furthermore, our previous results (18) suggested that processing is a rare event, which could contribute to these observations. Experiments are ongoing to test this hypothesis.
Potential requirement for ribosome to preload onto a standby site
As mentioned above, the translational regulation of zorO by its 5΄ UTR likely follows the standby model as proposed in tisAB, though the actual lengths of the standby sites may vary for each. The features of a standby site that allow ribosome preloading remain unclear. For normal translation initiation, the 30S subunit of the ribosome anchors onto the mRNA through base pairing interactions between the 3΄ terminus of 16S rRNA and the RBS of the mRNA. We do not recognize any obvious complementary sequence in zorO or tisB that could base pair with the 3΄ of 16S rRNA. However, it has been shown that alternative sequence features of the mRNA, other than complementarity to the 16S rRNA, can permit 30S subunit preloading onto mRNA. Studies by Barendt et al. revealed that translation initiation of non-Shine Dalgarno-led genes exhibit unique preferences on certain purines or pyrimidines at the 5΄ UTR (39,40). We note that both mRNAs contain a stretch of 5-nt-long sequence (5΄ CAACA 3΄) at the 3΄ end where the corresponding antitoxin sRNA targets. Whether this stretch of sequence plays a role in anchoring the ribosome is currently under investigation.
A more widely accepted feature of a standby site suggests that preloading of the ribosome onto a standby site is predominantly dependent on the single stranded nature of the site. Using fluorescence energy transfer experiments on designed mRNAs containing different secondary structures, Studer et al revealed that a single-stranded region in a structured mRNA allows the binding to the 30S subunit of the ribosome in a sequence independent manner (41). Another recent study also suggests that hairpin structures harboring single-stranded surface can function as standby sites regardless of their sequences (42).
Given that the association affinity between a standby site and the 30S subunit may not be as strong as that in the translation initiation complex, direct biochemical detection of a bound form of 30S with the standby site can be challenging. It is noteworthy that the sequence and the structure requirements of a standby site may not be mutually exclusive. Hence, it is possible that one standby site can harbor both specific sequence features as well as structure features.
Optimal translation of zorO requires an additional site
Along with the EAP standby region, we identified another region spanning from +35 to +50 that contributes to zorO translation. Our lead(II) digestion indicated that the +35 to +50 region comprises a highly structured region (Supplemental Figure S6). A study by Borujeni et al suggested that any single hairpin structure surrounded by distal and proximal single-stranded binding sites could serve as a standby site and contribute to translation (42). Additionally, specific sequence motifs have also been shown to enhance translation. For example, the ACA motif, found in the UTR of the conserved, putative amino acid transporter yfiK in Salmonella, was critical for efficient translation and served as a target of the sRNA repressor GcvB (43); this motif is conserved across other RNAs (44–46), but is not present in the +35 to +50 region of zorO. The ribosomal protein S1, which can interact with AU-rich regions, is also known to enhance translation upon interacting with such regions and may stimulate ribosomal loading (47,48), but this particular motif is lacking in ZorO. Regardless of the lack of these specific sequences, it is still possible that the may be a sequence motif in the +35 to +50 region of the zorO mRNA that is critical for ribosomal loading and/or translation enhancement. The more opened nature of the +35 to +50 region could alternatively promote ribosome loading, independent of the specific sequence. Importantly, though, a synthetic sRNA that can base pair with this region failed to rescue cells from zorO induced toxicity in a Δrnc strain (data not shown), implying that sRNA pairing to this region does not interfere with zorO translation. Further investigations on the significance/role of this region are warranted.
Another possibility is that this region may impact the kinetics of RNA unfolding. RNA can adopt different structures before reaching its native fold (49). The folding process can occur in a sequential fashion in which the first stable structure formed directs subsequent folding (50). In line with this, one may imagine that there could be many alternative ways for a structured RNA to reverse to a ‘functional’ unfolded state. Perhaps the presence of the +35 to +50 region confines the choices of possible unfolding pathways and streamlines this process, therefore allowing a more rapid translation of zorO. Additionally, it has been suggested that the folding of some RNAs occurs in a cooperative fashion that requires interactions of different portions of the RNA, as exemplified by the RNase P of B. subtilis (51). This raises the possibility that the +35 to +50 region may accelerate the folding/unfolding of the zorO RNA but may not impact the final structure. This could explain why lack of this region results in reduced translation efficiency (Figure 6E) although the overall structure was not affected. Additional studies of zorO RNA unfolding could provide mechanistic insights on how this region affects zorO translation.
Alternative model for zorO translation
Another mode of regulation of type I toxins is the translation coupling model represented by mok-hok (13,52). In this model, translation of the hok toxin mRNA is dependent on the translation of a leader peptide, Mok, encoded in the long 5΄ UTR of hok. The RBS of mok is sequestered in secondary structure and only exposed upon 3΄ processing of hok (15,16). In zorO, structural alteration is mediated by 5΄ processing instead of the 3΄ processing seen in hok. Further, no AUG start codon is found within the zorO 5΄ UTR. Although there are several GUG or UUG sequences that may serve as an alternative start codon, no canonical RBS is detected upstream of these sequences nor are they located within the EAP. Given that the translation coupling model requires the presence of a leader peptide, it is unlikely that the zorO mRNA uses this mechanism to initiate its translation.
Besides ZorO and TisB, 5΄ processing has been observed in two other type I toxins in E. coli, ShoB and DinQ, and appears linked to the translational efficiency of those toxin mRNAs. Specifically, two forms of shoB transcripts were detected: a full-length form (∼320 nts) and a 5΄ processed form (∼280 nts) (7), but only the processed form showed measurable β-galactosidase activities when fused translationally to lacZ (3). Further, dinQ possesses four processed forms yet only one transcript (+44) was translationally active (53). These observations suggest that optimal translation of shoB or dinQ requires 5΄ processing, likely through de-repressing the inhibitory effects caused by their respective 5΄ UTRs. This raises the possibility that similar to zorO and tisB, shoB and dinQ may use standby sites to facilitate translation; furthermore, their respective antitoxins, OhsC and AgrB, may act by targeting the standby sites. Surprisingly, the zorO-orzO, tisB-istR, shoB-ohsC, and dinQ-agrB type I pairs share distinct features: the toxin is encoded divergently from the corresponding antitoxin sRNA gene and base pairing occurs distal from the RBS at the toxin 5΄ UTR (3,18,53,55). Indeed, these four type I loci are the only ones identified to date that follow such genetic arrangement and base pairing pattern. It would be interesting to see whether the shoB-ohsC and dinQ-agrB pairs employ the standby mechanism to regulate toxin production, and if they do, could we predict the regulatory mechanisms of a type I pair based on its intrinsic genetic arrangement and the base pairing pattern?
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Z. Li for technical assistance. We also are grateful to L. Waters, M. Thomason, M. Chua, G. Storz and T. Reynolds for comments on the manuscript.
Footnotes
Present address: Jia Wen, Department of Molecular Genetics and Microbiology, Duke University, Durham, NC, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH/NIGMS [R15GM106291 to E.M.F.]. Funding for open access charge: NIH/NIGMS [R15GM106291 to E.M.F.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Goeders N., Van Melderen L.. Toxin-antitoxin systems as multilevel interaction systems. Toxins (Basel). 2014; 6:304–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fozo E.M., Hemm M.R., Storz G.. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol Rev. 2008; 72:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fozo E.M., Kawano M., Fontaine F., Kaya Y., Mendieta K.S., Jones K.L., Ocampo A., Rudd K.E., Storz G.. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008; 70:1076–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Unoson C., Wagner E.G.. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 2008; 70:258–270. [DOI] [PubMed] [Google Scholar]
- 5. Jahn N., Brantl S., Strahl H.. Against the mainstream: the membrane-associated type I toxin BsrG from Bacillus subtilis interferes with cell envelope biosynthesis without increasing membrane permeability. Mol. Microbiol. 2015; 98:651–666. [DOI] [PubMed] [Google Scholar]
- 6. Patel S., Weaver K.E.. Addiction toxin Fst has unique effects on chromosome segregation and cell division in Enterococcus faecalis and Bacillus subtilis. J. Bacteriol. 2006; 188:5374–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kawano M., Reynolds A.A., Miranda-Rios J., Storz G.. Detection of 5΄- and 3΄-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005; 33:1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kawano M., Aravind L., Storz G.. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007; 64:738–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Silvaggi J.M., Perkins J.B., Losick R.. Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 2005; 187:6641–6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Durand S., Gilet L., Condon C.. The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet. 2012; 8:e1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jahn N., Brantl S.. One antitoxin–two functions: SR4 controls toxin mRNA decay and translation. Nucleic Acids Res. 2013; 41:9870–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brantl S., Jahn N.. sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 2015; 39:413–427. [DOI] [PubMed] [Google Scholar]
- 13. Gerdes K., Larsen J.E., Molin S.. Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol. 1985; 161:292–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gerdes K., Thisted T., Martinussen J.. Mechanism of post-segregational killing by the hok/sok system of plasmid R1: Sok antisense RNA regulates formation of a hok mRNA species correlated with killing of plasmid-free cells. Mol. Microbiol. 1990; 4:1807–1818. [DOI] [PubMed] [Google Scholar]
- 15. Franch T., Gerdes K.. Programmed cell death in bacteria: translational repression by mRNA end-pairing. Mol. Microbiol. 1996; 21:1049–1060. [DOI] [PubMed] [Google Scholar]
- 16. Franch T., Gultyaev A.P., Gerdes K.. Programmed cell death by hok/sok of plasmid R1: processing at the hok mRNA 3΄-end triggers structural rearrangements that allow translation and antisense RNA binding. J. Mol. Biol. 1997; 273:38–51. [DOI] [PubMed] [Google Scholar]
- 17. Darfeuille F., Unoson C., Vogel J., Wagner E.G.. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007; 26:381–392. [DOI] [PubMed] [Google Scholar]
- 18. Wen J., Won D., Fozo E.M.. The ZorO-OrzO type I toxin-antitoxin locus: repression by the OrzO antitoxin. Nucleic Acids Res. 2014; 42:1930–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fozo E.M., Makarova K.S., Shabalina S.A., Yutin N., Koonin E.V., Storz G.. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010; 38:3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guzman L.M., Belin D., Carson M.J., Beckwith J.. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995; 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kunkel T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horton R.M., Hunt H.D., Ho S.N., Pullen J.K., Pease L.R.. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989; 77:61–68. [DOI] [PubMed] [Google Scholar]
- 23. Bahler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A. 3rd, Steever A.B., Wach A., Philippsen P., Pringle J.R.. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998; 14:943–951. [DOI] [PubMed] [Google Scholar]
- 24. Kawano M., Oshima T., Kasai H., Mori H.. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002; 45:333–349. [DOI] [PubMed] [Google Scholar]
- 25. Masse E., Escorcia F.E., Gottesman S.. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003; 17:2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Opdyke J.A., Kang J.G., Storz G.. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004; 186:6698–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Opdyke J.A., Fozo E.M., Hemm M.R., Storz G.. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J. Mol. Biol. 2011; 406:29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nilsen T.W. Gel purification of RNA. Cold Spring Harb. Protoc. 2013; 2013:180–183. [DOI] [PubMed] [Google Scholar]
- 29. Beisel C.L., Updegrove T.B., Janson B.J., Storz G.. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012; 31:1961–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trotochaud A.E., Wassarman K.M.. A highly conserved 6S RNA structure is required for regulation of transcription. Nat. Struct. Mol. Biol. 2005; 12:313–319. [DOI] [PubMed] [Google Scholar]
- 31. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lindell M., Romby P., Wagner E.G.. Lead(II) as a probe for investigating RNA structure in vivo. RNA. 2002; 8:534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoshida H. The ribonuclease T1 family. Methods Enzymol. 2001; 341:28–41. [DOI] [PubMed] [Google Scholar]
- 34. Malmgren C., Wagner E.G., Ehresmann C., Ehresmann B., Romby P.. Antisense RNA control of plasmid R1 replication. The dominant product of the antisense RNA-mRNA binding is not a full RNA duplex. J. Biol. Chem. 1997; 272:12508–12512. [DOI] [PubMed] [Google Scholar]
- 35. de Smit M.H., van Duin J.. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J. Mol. Biol. 2003; 331:737–743. [DOI] [PubMed] [Google Scholar]
- 36. Dorr T., Vulic M., Lewis K.. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010; 8:e1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gurnev P.A., Ortenberg R., Dorr T., Lewis K., Bezrukov S.M.. Persister-promoting bacterial toxin TisB produces anion-selective pores in planar lipid bilayers. FEBS Lett. 2012; 586:2529–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walther T.H., Gottselig C., Grage S.L., Wolf M., Vargiu A.V., Klein M.J., Vollmer S., Prock S., Hartmann M., Afonin S. et al. Folding and self-assembly of the TatA translocation pore based on a charge zipper mechanism. Cell. 2013; 152:316–326. [DOI] [PubMed] [Google Scholar]
- 39. Barendt P.A., Shah N.A., Barendt G.A., Sarkar C.A.. Broad-specificity mRNA-rRNA complementarity in efficient protein translation. PLoS Genet. 2012; 8:e1002598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barendt P.A., Shah N.A., Barendt G.A., Kothari P.A., Sarkar C.A.. Evidence for context-dependent complementarity of non-Shine-Dalgarno ribosome binding sites to Escherichia coli rRNA. ACS Chem. Biol. 2013; 8:958–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Studer S.M., Joseph S.. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell. 2006; 22:105–115. [DOI] [PubMed] [Google Scholar]
- 42. Espah Borujeni A., Channarasappa A.S., Salis H.M.. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2014; 42:2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang Q., Figueroa-Bossi N., Bossi L.. Translation enhancing ACA motifs and their silencing by a bacterial small regulatory RNA. PLoS Genet. 2014; 10:e1004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Komarova A.V., Tchufistova L.S., Supina E.V., Boni I.V.. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA. 2002; 8:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McCarthy J.E., Schairer H.U., Sebald W.. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 1985; 4:519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tzareva N.V., Makhno V.I., Boni I.V.. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 1994; 337:189–194. [DOI] [PubMed] [Google Scholar]
- 47. Gualerzi C.O., Pon C.L.. Initiation of mRNA translation in bacteria: structural and dynamic aspects. Cell Mol. Life Sci. 2015; 72:4341–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hajnsdorf E., Boni I.V.. Multiple activities of RNA-binding proteins S1 and Hfq. Biochimie. 2012; 94:1544–1553. [DOI] [PubMed] [Google Scholar]
- 49. Li P.T., Vieregg J., Tinoco I. Jr. How RNA unfolds and refolds. Annu. Rev. Biochem. 2008; 77:77–100. [DOI] [PubMed] [Google Scholar]
- 50. Mahen E.M., Watson P.Y., Cottrell J.W., Fedor M.J.. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 2010; 8:e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baird N.J., Westhof E., Qin H., Pan T., Sosnick T.R.. Structure of a folding intermediate reveals the interplay between core and peripheral elements in RNA folding. J. Mol. Biol. 2005; 352:712–722. [DOI] [PubMed] [Google Scholar]
- 52. Gerdes K., Poulsen L.K., Thisted T., Nielsen A.K., Martinussen J., Andreasen P.H.. The hok killer gene family in gram-negative bacteria. New Biol. 1990; 2:946–956. [PubMed] [Google Scholar]
- 53. Weel-Sneve R., Kristiansen K.I., Odsbu I., Dalhus B., Booth J., Rognes T., Skarstad K., Bjoras M.. Single transmembrane peptide DinQ modulates membrane-dependent activities. PLoS Genet. 2013; 9:e1003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vogel J., Argaman L., Wagner E.G., Altuvia S.. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004; 14:2271–2276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.