Abstract
Phosphorylation of the cAMP response element binding protein (CREB) at Ser-133 in response to hormonal stimuli triggers cellular gene expression via the recruitment of the histone acetylase coactivator paralogs CREB binding protein (CBP) and p300 to the promoter. The NMR structure of the CREB:CBP complex, using relevant interaction domains called KID and KIX, respectively, reveals a shallow hydrophobic groove on the surface of KIX that accommodates an amphipathic helix in phospho (Ser-133) KID. Using an NMR-based screening approach on a preselected small-molecule library, we identified several compounds that bind to different surfaces on KIX. One of these, KG-501 (2-naphthol-AS-E-phosphate), targeted a surface distal to the CREB binding groove that includes Arg-600, a residue that is required for the CREB:CBP interaction. When added to live cells, KG-501 disrupted the CREB: CBP complex and attenuated target gene induction in response to cAMP agonist. These results demonstrate the ability of small molecules to interfere with second-messenger signaling cascades by inhibiting specific protein–protein interactions in the nucleus.
Keywords: cAMP response element binding protein, transcription
Protein–protein interactions often serve as key regulatory points for signal propagation in response to extracellular stimuli. The formation of protein–protein complexes is of particular interest in the field of transcriptional regulation, where multiple low-affinity interactions appear to contribute to the recruitment of the transcriptional apparatus to the promoter (1). Initially thought to result from low energy interactions between unstructured regions, the recognition of an activator by its coactivator cognate often involves discrete surface contacts between well-folded protein domains. Moreover, the interaction surfaces between such protein pairs often are modulated by covalent modifications at residues near the protein–protein interface.
Phosphorylation of the cAMP response element binding protein (CREB) at Ser-133 stimulates its association with the coactivator paralogs CREB binding protein (CBP) and p300 via a direct mechanism (2); the Ser-133 phosphate forms a hydrogen bond with Tyr-658 and an ion pair with Lys-662 in the KIX domain (3). Binding of CREB to CBP/p300 is further stabilized by a random coil-to-helix transition in KID that provides the majority of hydrophobic surface contacts with residues lining a shallow groove in KIX (4, 5). The KIX domain folds into a three-helix structure with helices α1 and α3 aligned in nearly parallel fashion to form a groove that accommodates hydrophobic residues in KID. Helix α2 does not participate directly in surface contacts with KID but appears to stabilize KIX structure.
The CREB binding site in KIX also recognizes other activators, most notably the protooncogene c-Myb (6). Similar to phospho (Ser-133) KID, c-Myb binds to KIX via an amphipathic helix that forms numerous surface contacts with residues lining the shallow hydrophobic groove in KIX (7). Notably, surfaces distal to the CREB-binding site in KIX have been found to modulate KID:KIX complex formation. Binding of the mixed lineage leukemia (MLL) protein to KIX, for example, potentiates the interaction of phospho-CREB with KIX (8); MLL associates with a distinct surface on KIX and stabilizes a conformation that in turn favors binding of CREB (9). By contrast, methylation of KIX at Arg-600 by the arginine methylase CARM1 disrupts the CREB:CBP interaction (10). Arg-600 is located on helix α1 in KIX, distal to the CREB binding site; and mutations at this site alter KIX structure and block formation of the KID:KIX complex (2, 11).
Delineating the relative importance of the CREB:CBP and other activator–coactivator complexes for target gene activation in response to various stimuli requires the development of small-molecule antagonists that selectively block these interactions. Despite their key roles in signal transduction, however, most protein–protein complexes are thought to be refractory to such intervention, owing to the absence of deep enzymatic pockets along the interface that could readily accommodate a small molecule.
Using an NMR-based screening approach (12), we have identified several KIX binding compounds, including two that disrupt the KID:KIX interaction in vitro. Further, in vivo studies reveal that one of these (KG-501) inhibits the cAMP-dependent activation of cellular genes via CREB. Unexpectedly, this compound was found to recognize a surface in KIX adjacent to the shallow groove that binds P-CREB. These studies suggest that activator–coactivator complexes are amenable to small-molecule intervention, and the development of such antagonists provides a useful approach for dissecting regulatory networks that are activated in response to hormonal stimuli.
Materials and Methods
NMR Sample Preparation and Analysis. The gene sequence encoding the KIX domain of mouse CBP (residues 586–672) in the pET-21a(+) expression vector has been described (4). KIX protein was expressed in transformed Escherichia coli BL21(RIL) cells (Stratagene) growing in minimal M9 media containing 15N-labeled ammonium chloride. Methods for expression and purification of the KIX domain by RP-HPLC have been described (4). NMR experiments were recorded on a 500-MHz Unity Inova spectrometer (Varian) equipped with an automatic sample changer and a 50-position sample tray. Typical 15N/1H heteronuclear single quantum coherence experiments contained 250 μM of uniformly 15N-labeled KIX in 20 mM Tris-acetate, 50 mM NaCl, 10% D2O buffer at pH 5.5. Data were processed with nmrpipe (13) and analyzed with nmrview (14). The automated analysis of chemical-shift perturbations for hit identification was performed with an in-house written program adapted for use within nmrview. Resonances for the KIX protein were assigned based on previous NMR analyses (4).
In Vitro Binding and Transcription Assays. GST and GST-KIX proteins were purified from E. coli BL21(RIL) cells transformed with pGEX-4T1 and pGEX-KT KIX bacterial expression vectors (6) (Amersham Pharmacia). Proteins were purified with glutathione-sepharose 4B beads (Amersham Pharmacia) followed by elution with 20 mM glutathione by using standard protocols. Proteins were dialyzed (50 mM Tris, pH 7.5/150 mM NaCl) and stored in 40% glycerol. To produce 32P-labeled CREB, purified PKA catalytic subunit was incubated with His-tagged CREB and 32P-γATP in PKA buffer (50 mM Tris, pH 7.5/10 mM MgCl2/1 mM DTT) at 30°C for 30 min. The phosphorylation reaction was stopped with 100 mM EDTA, and the protein was purified on a Centrisep column according to the manufacturer's instructions (Princeton Separations, Adelphia, NJ). For the in vitro binding assays, purified GST fusion proteins were incubated with the compounds dissolved in DMSO (10 mM stock) or DMSO vehicle for 15 min in 200 μl of binding buffer (20 mM Tris, pH 7.5/100 mM NaCl/1 mM MgCl2/0.05% Nonidet P-40) at room temperature. Purified 32P-labeled CREB protein was added to the mixture and incubated for an additional 30 min. Samples were precipitated with glutathione-sepharose 4B beads for 30 min and then washed three times with binding buffer. Samples were analyzed by SDS-gel electrophoresis followed by autoradiography of either vacuum-dried gels or nitrocellulose-transferred gels. Quantitation of precipitated 32P-labeled CREB was done by using a Molecular Dynamics PhosphorImager and the imagequant program. In vitro transcription assays were performed as described (15).
Cell Culture and Transfection. HEK293T cells were maintained in DMEM, 10% FCS with antibiotics. Cells were plated in 12-well dishes for transfection of plasmid DNA by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. At 24 h after transfection, compounds dissolved in DMSO (10 mM stock) were added directly to the culture medium for 15 min. Cells were then stimulated with forskolin (10 μM) for a total incubation time of 4 h. Lysates were harvested and assayed for luciferase activity as described (16). The plasmids used for transfection included NF-κB luciferase (Clontech), pFR-Luc (Stratagene), evenskipped (EVX-1)-luciferase (16), GAL4-KID, KIX-VP16, and GAL4-CREB (17). Cotransfection of CMV β-galactosidase expression plasmid and measurement of β-galactosidase activity of the extracts was used to normalize for transfection efficiency.
Quantitative PCR. Total RNA was extracted from HEK293T cells or primary hepatocytes by using TRI Reagent (Molecular Research Center, Cincinnati) followed by an additional purification step using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. cDNA was prepared by reverse transcription of 100–500 ng of total RNA by using Superscript II enzyme and oligo(dT) primer. The resulting cDNAs were amplified by using the SYBR green PCR kit and measured with the ABIPRISM 7700 sequence detector (PerkinElmer). All mRNA expression data were normalized to GAPDH expression in the corresponding sample. Oligonucleotide sequences for the primers used are available on request.
Western Blot Analyses. To assay for CREB phosphorylation, cells were stimulated with forskolin (10 μM) after compound incubation for 30 min and then lysed in SDS lysis buffer (4 M urea/100 mM Tris, pH 8/500 mM NaCl/10 mM EDTA/1% SDS) containing protease and phosphatase inhibitors. Standard SDS-gel electrophoresis and Western blotting procedures were used to analyze the cell extracts. Nitrocellulose blots were probed with anti-CREB and anti-phospho-specific pSer-133 CREB antibodies (16).
Acetyltransferase Assay. Purified full-length p300 protein (30 ng) was incubated with KG-501 compound or the histone acetyl transferase (HAT) inhibitor compound, lysyl CoA (10 μM) in HAT assay buffer (10 μg of histones/50 mM Tris, pH 8.0/20 mM butyric acid/10% glycerol/1 mM PMSF/1 mM DTT) containing 14C-Acetyl CoA for 30 min at 30°C. Samples were analyzed on a 15% SDS-gel followed by autoradiography. Signal strength was amplified by incubation of the gel in Enlightening solution (NEN) before vacuum-drying the gel.
FRET Analysis. HEK293T or NIH 3T3 cells were transfected with nuclear-targeted YFP-KID (YKIDN) and KIX-CFP (KIXCN) fusion proteins and incubated with DMSO, Fsk, and/or KG-501 as indicated and transferred in a live cell observation chamber to a Leica TCS laser confocal microscope. Cyan fluorescent protein emission was measured before and after selective photobleaching of yellow fluorescent protein by using a 514-nm laser, and FRET efficiency was calculated as described (18).
Results
We screened for compounds that bind to KIX by using a preselected library of 762 compounds that was previously used for ligand discovery and binding site identification (19). Screens were conducted in a two-stage process, using 2D 15N-1H heteronuclear single quantum coherence spectra to monitor compound binding to uniformly 15N-labeled KIX. In the first stage, spectra were recorded on samples containing a pool of five compounds at 500 μM concentration. Samples were rankordered according to the extent of amide chemical shift changes, and the top 10 samples were further analyzed by recording spectra of KIX in the presence of each compound individually. Four library compounds that bound to distinct regions of the KIX domain were identified and selected for further study.
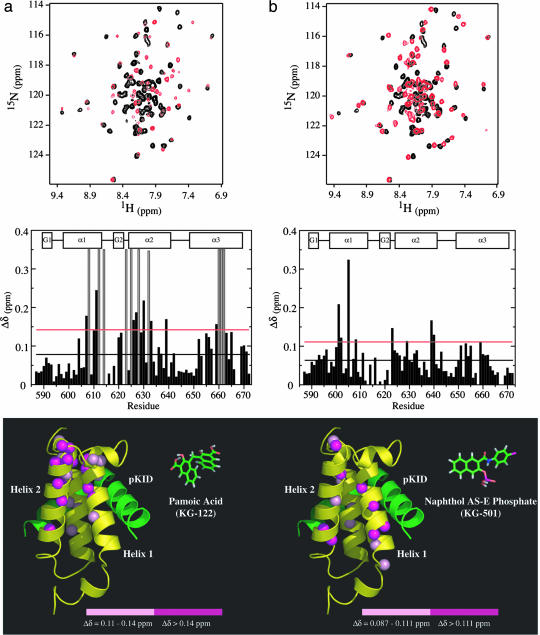
Fig. 1 shows data for two candidate compounds, pamoic acid (KG-122, Fig. 1a) and naphthol-AS-E-phosphate (KG-501, Fig. 1b). The chemical shift changes caused by each small molecule can be compared in the 15N-1H heteronuclear single quantum coherence spectra of 250 μM KIX in the absence (black) or presence of 500 μM (red) of each compound (Fig. 1 a and b Top). Mapping the most affected backbone amide sites onto the protein structure revealed two distinct modes for binding to KIX. KG-122 interacts with KIX through a face of the protein involving residues at the C-terminal fragments of helices α1 and α3, the N-terminal half of helix α2, and the small 3–10 helix G2. Chemical shift perturbations at residues further away from this area (residues 602, 639, 651, 654, and 655) may result from reorientation of the helices or long-range effects caused by the aromatic rings of the small molecule. This region overlaps with the interface used by KIX to bind the transcriptional factor c-Jun and coactivator protein MLL (9, 20). Notably, Lys-662, which forms an ion pair with the Ser-133 phosphate, is also shifted by KG-122, suggesting a potential inhibitory effect of this compound on the KID:KIX complex.
Fig. 1.
Analysis of KIX residues affected by addition of KG-122, pamoic acid (a), and KG-501, naphthol-AS-E-phosphate (b). (Top) The 15N-1H heteronuclear single quantum coherence spectra show KIX alone (black) and with compound added (red) at [protein] = 250 μM and [compound] = 500 μM. (Middle) The bar charts represent the combined 1H and 15N chemical shift changes (Δδ = [Δδ(1H)2 + 0.1*Δδ(15N)2]1/2) per residue. Helical regions of the KIX domain are represented as rectangles. Gray bars in a indicate amide residues whose peaks have broadened beyond detection because of addition of KG-122 and for which an arbitrary value of 0.35 ppm was assigned. The black line represents that average shift, and the red line indicates +1 SD. (Bottom) Ribbon representations of KIX, yellow (Protein Data Bank ID code 1KDX), highlighting backbone amides exhibiting significant chemical shift changes upon the addition of KG-122 or KG-501. For reference, the pKID polypeptide has been included and is shown in green. For KG-122, the average shift was 0.078 ppm with a SD of 0.064 ppm. Magenta spheres, Δδ > 0.142 ppm (1 SD more than average); pink spheres, 0.11 <Δδ < 0.142 ppm (0.5 SD). For KG-501 the average shift was 0.063 ppm with a SD of 0.048 ppm. Magenta spheres, Δδ > 0.111 ppm (1 SD); pink spheres, 0.087 <Δδ < 0.111 ppm (0.5 SD). KG-122 and KG-501 are shown on the same scale as the KIX structure (stick model) to compare their relative sizes.
In contrast to KG-122, KG-501 mainly perturbs backbone amides at sites along helices α1, α2, and, to a lesser extent, a few residues located in the central region of helix α3 (residues 651, 653, 655, and 659). Both helices α1 and α3 contribute a number of key residues to the interface used by KIX to bind the KID domain of CREB as well as the transactivation domain of c-Myb (3, 7). The perturbed sites include several residues between R600 and V608 (Fig. 1b), a region that also experiences significant chemical-shift changes upon complexation with Myb (21). As well, this area is adjacent to Arg-600, shown by previous mutagenesis studies to be critical for the CREB:CBP interaction (2, 11). Moreover, methylation of KIX at Arg-600 by the methyl-transferase CARM1/PRMT4 has been found to block CREB target gene activation in response to cAMP agonist by disrupting the CREB:CBP complex (10).
Based on their ability to perturb residues that are important for the formation of the KID:KIX complex, we hypothesized that KG-122 and KG-501 would disrupt the binding of phospho (Ser-133) CREB to KIX. To test this notion, we performed in vitro binding assays by using purified recombinant phospho (Ser-133) CREB and GST-KIX proteins. Both compounds inhibited the CREB:CBP interaction in pull-down assays, but KG-122 was eliminated from further study because of solubility problems (Fig. 2a). KG-501 disrupted phospho (Ser-133) CREB binding to KIX with a Ki of ≈90 μM, using concentrations of CREB that were within the linear range of the binding assay (Fig. 2b and Fig. 4, which is published as supporting information on the PNAS web site). In contrast, 2-naphthol, a parent molecule of KG-501, displayed little inhibitory effect. A similar Ki was observed for binding of phospho (Ser-133) CREB protein to full-length flag-tagged p300 (Fig. 2c). KG-501 does not appear to inhibit KIX domain function through general unfolding; the KIX NMR spectrum remained clearly defined in the presence of this small molecule at concentrations as high as 500 μM (Fig. 1b Top).
Fig. 2.
KG-501 inhibits the in vitro interaction between the CBP KIX domain and phospho (Ser-133) CREB. (a) Comparison of KG-501 (2-naphthol-AS-E-phosphate) and KG-122 (pamoic acid) effects on CREB:CBP interaction relative to 2-naphthol. Purified GST-KIX protein was preincubated with 100 μM of each compound. 32P-labeled Ser-133 phosphorylated CREB protein (0.2 pmol) was added to the binding reaction, and the bound proteins were precipitated with glutathione-agarose. Samples were analyzed by gel electrophoresis followed by autoradiography. In, 20% input 32P-CREB. (b) Purified GST-KIX protein was preincubated with increasing micromolar concentrations of KG-501 or 2-naphthol. In, 10% input 32P-CREB. (c) KG-501 inhibits binding of CREB to full-length p300. Purified flag-tagged full-length p300 protein was preincubated with the indicated micromolar concentrations of KG-501. 32P-labeled Ser-133 phosphorylated CREB protein was added to the binding reaction, and the bound proteins were precipitated with anti-Flag antibody-conjugated agarose. Samples were analyzed as in a. In, 20% input 32P-CREB. (d) KG-501 does not inhibit CREB dimer formation or complex formation between CREB and the coactivator TORC1. GST, GST-KIX, GST-CREB bZIP (amino acids 271–341) (Upper) or GST-TORC1 (Lower) fusion proteins were preincubated with DMSO vehicle (–) or increasing amounts of KG-501 compound [100 and 250 μM(Upper) and 100, 250, and 500 μM(Lower)]. Complexes of CREB bound to KIX were precipitated and analyzed as in a. In, 10% input 32P-CREB. (e) Effect of KG-501 on p300 HAT activity. Purified p300 was incubated with DMSO (–), KG-501, or the p300 HAT inhibitor lysyl CoA (28). Acetylase activity was measured by incubation with histones and 14C-acetyl CoA followed by gel electrophoresis (15% gel) and autoradiography.
To control for unanticipated effects of KG-501 on CREB, we tested whether KG-501 could modulate the association between CREB and the coactivator transducer of regulated CREB activity (TORC) (22, 23). In contrast to CBP/p300, TORC binds to the leucine zipper region of CREB via a CREB binding domain that displays no sequence similarity with KIX. Even at doses as high as 250 μM, KG-501 had no effect either on CREB homodimerization via its basic leucine zipper domain (Fig. 2d Upper) or CREB:TORC complex formation (Fig. 2d Lower) by pull-down assay of 32P-labeled CREB protein to GST-TORC (amino acids 1–46). Despite its ability to block the CREB:p300 interaction, KG-501 also had no effect on KIX-independent functions of p300, including its intrinsic HAT activity (Fig. 2e). Taken together these data indicate that the in vitro inhibitory effects of KG-501 on the CREB:CBP complex are mediated primarily through a specific action on the KIX domain.
Having seen that KG-501 blocks formation of the CREB:CBP complex in vitro, we examined whether this compound attenuates CREB target gene expression in vivo. Exposure of HEK293T cells to KG-501 blocked induction of the CREB-dependent evenskipped (EVX-1) promoter (16) by cAMP agonist in a dose-dependent manner; but KG-501 had no effect on the constitutive expression of a cotransfected Rous sarcoma virus-β-gal reporter (Fig. 3a). Removal of KG-501 from cells by replacement of the culture medium reversed the inhibitory effects of this compound on cAMP-inducible reporter activity, arguing against a general toxic effect of KG-501 on HEK293T cells (Fig. 5, which is published as supporting information on the PNAS web site). Treatment of HEK293T cells with KG-501 also blocked induction of endogenous CREB target genes (NR4A2, αCG, c-fos, and RGS2) by forskolin, indicating that KG-501 likely exerts a general effect on CREB activity (Fig. 3b and Fig. 6, which is published as supporting information on the PNAS web site). Addition of KG-501 did not interfere with phosphorylation of CREB at Ser-133 in response to forskolin, however, suggesting that this compound acts downstream of PKA to block CREB target gene expression (Fig. 3c).
Fig. 3.
KG-501 inhibits CREB signaling in vivo. (a) Effect of KG-501 on induction of the CREB-responsive promoter evenskipped (EVX-1) by cAMP agonist. EVX-1 luciferase reporter plasmid was transfected into HEK293T cells. At 24 h posttransfection, cells were pretreated with the indicated amount of KG-501 or DMSO vehicle for 20 min followed by coincubation with forskolin or DMSO. Cells were harvested at 4 h, and luciferase activity was measured. Luciferase activity was normalized to β-gal activity from cotransfected Rous sarcoma virus-β-gal expression plasmid. (b) Quantitative PCR analysis of mRNA levels for CREB target gene NR4A2. Relative mRNA levels were normalized to GAPDH expression. Effect of KG-501 (10 and 25 μM) on target gene expression in HEK293T cells under basal and forskolin (10 μM, 45 min) treated conditions are shown. (c) KG-501 does not interfere with CREB Ser-133 phosphorylation in response to forskolin. HEK293T cells were pretreated with 10 μM KG-501 or DMSO vehicle for 30 min and then coincubated with forskolin. (Upper) Cells were harvested at the indicated time points, and whole-cell lysates were analyzed by Western blot analysis using a specific anti-phospho-CREB antibody (5322). (Lower) Effect of pretreatment with DMSO, KG-501, or the PKA inhibitor H89 on CREB phosphorylation after addition of forskolin for 30 min. (d) Effect of KG-501 on GAL4 CREB (amino acids 1–283) activity in HEK293T cells cotransfected with GAL4-luciferase reporter plasmid and treated with forskolin as in a. (e) KG-501 inhibits the mammalian two-hybrid interaction between Gal4-KID with KIX-VP16. HEK293T cells were transfected with GAL4-KID, VP16-KIX, and GAL4-luciferase reporter plasmids and treated as in a with 10 μM KG-501. (f) Effect of KG-501 on the CREB:CBP interaction in living cells. FRET analysis of YFP-KID (YKIDN) with CFP-KIX (KIXCN) polypeptides. HEK293T cells were transfected with the YKIDN and KIXCN plasmids and preincubated with KG-501 (50 μM). The cells were then stimulated with forskolin, and FRET efficiency was measured as described (18).
To rule out potential regulatory effects of KG-501 on other CBPs in vivo, we performed transient assays of HEK293T cells transfected with GAL4-CREB expression vector encoding the GAL4 DNA binding domain fused to the CREB transactivation domain (amino acids 1–283). Pretreatment with KG-501 disrupted cAMP-dependent induction of GAL4-CREB in a dose-dependent manner, demonstrating the ability of this compound to block CREB activity specifically (Fig. 3d). Despite its inhibitory effects on CREB, KG-501 did not appear to disrupt RNA polymerase II (pol II) activity; no changes in cellular levels of elongating pol II were observed by Western blot assay of HEK293T extracts with phospho (Ser-2) pol II-specific antisera (Fig. 7, which is published as supporting information on the PNAS web site). Indeed, at concentrations sufficient to eliminate CREB-dependent transcription in vivo (5–10 μM), KG-501 also had no effect on transcription from CREB-independent p21 and adenovirus major late promoters by in vitro transcription assay (Fig. 8, which is published as supporting information on the PNAS web site). These results indicate that KG-501 is unlikely to interfere with CREB-dependent transcription via direct inhibition of the transcriptional apparatus.
Having observed that KG-501 blocks CREB activity in HEK293T cells, we hypothesized that this compound inhibits target gene expression by disrupting the CREB:CBP interaction in vivo just as it did with the in vitro interaction between the isolated KID and KIX domains. To test this notion, we carried out mammalian two-hybrid assays with GAl4-KID and KIX-VP16 expression vectors. Exposure to forskolin (10 μM) strongly induced the KID:KIX interaction by this assay, and pretreatment with KG-501 (10 μM) reduced the interaction by 95% (Fig. 3e). We also used a FRET assay to monitor the CREB:CBP interaction in live cells by expressing KID and KIX polypeptides fused to enhanced yellow (EYFP:YKIDN) and enhanced cyan (ECFP: KIXCN) fluorescent proteins, respectively (18). No interaction between YKIDN and KIXCN polypeptides was detected in HEK293T cells under basal conditions (data not shown), but treatment with forskolin strongly promoted complex formation (Fig. 3f). Pretreatment with KG-501 (50 μM) inhibited the cAMP-induced KID:KIX interaction by ≈60% in HEK293T cells. Similar effects of KG-501 on KID:KIX complex were observed in NIH 3T3 cells, demonstrating that the inhibitory properties of this compound are not cell type-specific (Fig. 9, which is published as supporting information on the PNAS web site). Taken together, these data are consistent with the notion that KG-501 inhibits induction of cAMP-responsive genes, at least in part, by blocking the CREB:CBP interaction.
Discussion
The NMR-based screening approach enabled us to identify several classes of KIX binding compounds that recognize different surfaces on the KIX domain and to explore the functional importance of these surfaces for CREB:CBP complex formation and target gene activation. In particular, the ability of KG-501 to disrupt the CREB:CBP interaction illustrates the role of a distal surface on KIX for regulation of this complex.
Chemical-shift mapping suggests that KG-501 binds to a surface composed of residues in the α1 and α2 helices. This binding event could be responsible for a conformational change reported by chemical shift changes of amides in these two helices and to a lesser degree by several amides on residues in α3. Unlike mutation of Arg-600 at the N terminus of α1, which has been found to disrupt the CREB:CBP complex by unfolding the KIX domain (11), KG-501 does not cause any drastic, global conformational changes. Instead, KG-501 affects the environments of a group of residues in the α1 and α3 helices that are also perturbed by Myb binding (21). These results illustrate an underlying connection between KG-501 and CREB binding sites on KIX, such that binding to one surface causes reciprocal changes in the other by repositioning its helices with respect to each other.
Although a number of compounds with related chemical structures to KG-501 (naphthol-AS, naphthol-AS-TR, naphthol-AS-phenylacetate, naphthol-AS-OL-acetate, naphthol-AS-MX-butyrate, naphthol-AS-MX-acetate, naphthol-AS-E-acetate, naphthol-AS-d-chloroacetate, and naphthol-AS-D-acetate) were tested for their ability to inhibit the KID:KIX interaction, all of these show low solubility in water, leading to inconsistent results in binding and transactivation studies (J.L.B. and M.M., unpublished work). Future efforts in this direction will reveal, however, the extent to which various moieties in KG-501 promote binding to the KIX domain.
Commensurate with its effects on the CREB:CBP complex, KG-501 interfered with the induction of CREB target genes by cAMP. Notably, KG-501 appeared more potent at disrupting CREB-dependent transcription (Ki = 10 μM) compared to the CREB:CBP interaction (Ki = 50 μM) by FRET assay. These differences may reflect reduced affinity of KG-501 for tagged versions of KIX used in our experiments compared with the full-length CBP. Alternatively, a partial reduction in CREB:CBP complex formation may be sufficient to completely disrupt target gene activation in response to cAMP.
CBP is recruited to a wide variety of transcriptional complexes, and the presence of the KIX domain has often been cited as critically important for in vivo transcriptional activation (24). Demonstrating the inherent limitations of targeting a domain with numerous binding partners, we also have observed inhibitory effects of KG-501 on other factors that associate with the KIX domain, such as NF-κB, although the extent of these effects relative to CREB is unclear (Fig. 10, which is published as supporting information on the PNAS web site). Additional chemical refinements, perhaps involving covalent links between KG-501 with small molecules like KG-122 that target other surfaces of KIX, may allow for the development of compounds with greater affinity and selectivity for CREB.
The inhibition of protein–protein interactions by small-molecule antagonists offers possibilities in approaches to disease intervention. In a recent study, for example, antagonists that block binding of MDM2 to the tumor suppressor p53 were found to stabilize p53 and enhance its growth inhibitory properties, offering potential treatment for certain cancers (25). Given its ability to regulate hepatic gluconeogenesis during fasting and in diabetes, CREB has also been identified as a potential therapeutic target (26, 27). By disrupting the expression of target genes involved in hepatic glucose production, small-molecule CREB:CBP antagonists may offer potential therapeutic benefit to type II diabetic patients.
Supplementary Material
Acknowledgments
We thank Steve McKnight for helpful suggestions. This work was supported by National Institutes of Health Grants CA-90601 (to K.H.G.) and GM-37828 (to M.M.), the Wellcome Trust, and the Hillblom and Keikhefer foundations.
Author contributions: J.L.B., L.F., C.M.M., B.E., T.Z., K.H.G., and M.M. designed research; J.L.B., C.A.A., B.M., L.F., C.M.M., T.Z., and K.H.G. performed research; J.L.B., C.A.A., B.M., L.F., C.M.M., B.E., K.H.G., and M.M. analyzed data; and J.L.B., C.A.A., K.H.G., and M.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CREB, cAMP response element binding protein; CBP, CREB binding protein; HAT, histone acetyl transferase; TORC, transducer of regulated CREB activity.
References
- 1.Ptashne, M. & Gann, A. (1997) Nature 386, 569–577. [DOI] [PubMed] [Google Scholar]
- 2.Parker, D., Ferreri, K., Nakajima, T., LaMorte, V., Evans, R., Koerber, S., Hoeger, C. & Montminy, M. (1996) Mol. Cell. Biol. 16, 694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radhakrishnan, I., Perez-Alvarado, G. C., Parker, D., Dyson, H. J., Montminy, M. & Wright, P. E. (1997) Cell 91, 741–752. [DOI] [PubMed] [Google Scholar]
- 4.Radhakrishnan, I., Perez-Alvarado, G. C., Parker, D., Dyson, H. J., Montminy, M. R. & Wright, P. E. (1999) J. Mol. Biol. 287, 859–865. [DOI] [PubMed] [Google Scholar]
- 5.Parker, D., Jhala, U., Radhakrishnan, I., Yaffe, M., Reyes, C., Shulman, A., Cantley, L., Wright, P. & Montminy, M. (1998) Mol. Cell 2, 353–359. [DOI] [PubMed] [Google Scholar]
- 6.Parker, D., Rivera, M., Zor, T., Henrion-Caude, A., Radhakrishnan, I., Kumar, A., Shapiro, L., Wright, P., Montminy, M. & Brindle, P. (1999) Mol. Cell. Biol. 19, 5601–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zor, T., De Guzman, R. N., Dyson, H. J. & Wright, P. E. (2004) J. Mol. Biol. 337, 521–534. [DOI] [PubMed] [Google Scholar]
- 8.Ernst, P., Wang, J., Huang, M., Goodman, R. H. & Korsmeyer, S. J. (2001) Mol. Cell. Biol. 21, 2249–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goto, N. K., Zor, T., Martinez-Yamout, M., Dyson, H. J. & Wright, P. E. (2002) J. Biol. Chem. 277, 43168–43174. [DOI] [PubMed] [Google Scholar]
- 10.Xu, W., Chen, H., Du, K., Asahara, H., Tini, M., Emerson, B. M., Montminy, M. & Evans, R. M. (2001) Science 294, 2507–2511. [DOI] [PubMed] [Google Scholar]
- 11.Wei, Y., Horng, J. C., Vendel, A. C., Raleigh, D. P. & Lumb, K. J. (2003) Biochemistry 42, 7044–7049. [DOI] [PubMed] [Google Scholar]
- 12.Shuker, S. B., Hajduk, P. J., Meadows, R. P. & Fesik, S. W. (1996) Science 274, 1531–1534. [DOI] [PubMed] [Google Scholar]
- 13.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, B. A. & Blevins, R. A. (1994) J. Biomol. NMR 4, 603–614. [DOI] [PubMed] [Google Scholar]
- 15.Espinosa, J. M. & Emerson, B. M. (2001) Mol. Cell 8, 57–69. [DOI] [PubMed] [Google Scholar]
- 16.Conkright, M. D., Guzmán, E., Flechner, L., Su, A. I., Hogenesch, J. & Montminy, M. (2003) Mol. Cell 11, 1101–1108. [DOI] [PubMed] [Google Scholar]
- 17.Du, K., Asahara, H., Jhala, U., Wagner, B. & Montminy, M. (2000) Mol. Cell. Biol. 20, 4320–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayr, B., Canetierri, L. & Montminy, M. (2001) Proc. Natl. Acad. Sci. USA 98, 10936–10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amezcua, C. A., Harper, S. M., Rutter, J. & Gardner, K. H. (2002) Structure (London) 10, 1349–1361. [DOI] [PubMed] [Google Scholar]
- 20.Campbell, K. M. & Lumb, K. J. (2002) Biochemistry 41, 13956–13964. [DOI] [PubMed] [Google Scholar]
- 21.Zor, T., Mayr, B. M., Dyson, H. J., Montminy, M. R. & Wright, P. E. (2002) J. Biol. Chem. 277, 42241–42248. [DOI] [PubMed] [Google Scholar]
- 22.Conkright, M. D., Canettieri, G., Screaton, R., Guzman, E., Miraglia, L., Hogenesch, J. B. & Montminy, M. (2003) Mol. Cell 12, 413–423. [DOI] [PubMed] [Google Scholar]
- 23.Iourgenko, V., Zhang, W., Mickanin, C., Daly, I., Jiang, C., Hexham, J. M., Orth, A. P., Miraglia, L., Meltzer, J., Garza, D., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 12147–12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong, H., Voll, R. E. & Ghosh, S. (1998) Mol. Cell 1, 661–671. [DOI] [PubMed] [Google Scholar]
- 25.Vassilev, L. T., Vu, B. T., Graves, B., Carvajal, D., Podlaski, F., Filipovic, Z., Kong, N., Kammlott, U., Lukacs, C., Klein, C., et al. (2004) Science 303, 844–848. [DOI] [PubMed] [Google Scholar]
- 26.Herzig, S., Long, F., Jhala, U., Hedrick, S., Quinn, R., Bauer, A., Schutz, G., Yoon, C., Puisgever, P., Spiegelman, B. & Montminy, M. (2001) Nature 413, 179–183. [DOI] [PubMed] [Google Scholar]
- 27.Yoon, J., Puigserver, P., Chen, G., Donovan, J., Wu, Z., Rhee, J., Adelmant, G., Stafford, J., Kahn, C., Granner, D., et al. (2001) Nature 413, 131–138. [DOI] [PubMed] [Google Scholar]
- 28.Lau, O. D., Kundu, T. K., Soccio, R. E., Ait-Si-Ali, S., Khalil, E. M., Vassilev, A., Wolffe, A. P., Nakatani, Y., Roeder, R. G. & Cole, P. A. (2000) Mol. Cell 5, 589–595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.