Abstract
Chromosomal rearrangements encoding oncogenic fusion proteins are found in a wide variety of malignancies. The use of programmable nucleases to generate specific double-strand breaks in endogenous loci, followed by non-homologous end joining DNA repair, has allowed several of these translocations to be generated as constitutively expressed fusion genes within a cell population. Here, we describe a novel approach that combines CRISPR-Cas9 technology with homology-directed repair to engineer, capture and modulate the expression of chromosomal translocation products in a human cell line. We have applied this approach to the genetic modelling of t(11;22)(q24;q12) and t(11;22)(p13;q12), translocation products of the EWSR1 gene and its 3’ fusion partners FLI1 and WT1, present in Ewing's sarcoma and desmoplastic small round cell tumour, respectively. Our innovative approach allows for temporal control of expression of engineered endogenous chromosomal rearrangements, and provides a means to generate models to study tumours driven by fusion genes.
Keywords: CRISPR-Cas9, genomic editing, chromosomal translocations, homology-directed repair, oncogene, Ewing sarcoma, Desmoplastic small round cell tumour (DSRCT)
Introduction
Recent advances in sequencing technologies have accelerated the identification of recurrent chromosomal rearrangements in several cancers [1]. In solid and haematological malignancies, the chimeric protein products of these fusion genes have been shown not only to be pathognomonic, but also to constitute the disease's primary drivers. The initiating event for these dramatic genomic alterations is the occurrence of double-strand breaks (DSBs) within chromosomal loci. Two concurrent DSBs at distinct loci, followed by illegitimate joining and repair lead to the generation of a variety of oncogenic fusion drivers in a wide array of cancers [2].
The development of designer programmable endonucleases has advanced our ability to study the aetiology and effects of chromosomal rearrangements by allowing the generation of faithful model systems of these genomic events. With programmable zinc finger nucleases, transcription activator-like effector nucleases, and the recently described clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) system, (CRISPR-Cas9) technology, it is possible to induce de novo translocations, deletions and inversions with synchronous introduction of two DSBs at two genomic loci. As a result, chromosomal rearrangements have been readily induced in human and mouse cell lines and in vivo [3-10]. The benefit of this approach is in the generation of an endogenous chromosomal rearrangement that recapitulates the properties of a genuine translocation present in a tumour, bypassing the necessity for the use of conventional cell line engineering approaches that are based upon introduction and subsequent over-expression of an exogenous fusion cDNA.
Irrespective of the choice of DSB-inducing enzyme system, isogenic clones with an induced translocation have not been readily identified. Moreover, generation of engineered genomic rearrangements through non-homologous end joining (NHEJ) leads to immediate, constitutive expression of the fusion products [11,12]. In this way, expression of the engineered fusion transcript is immediate and uncontrolled, and can therefore dramatically affect the biology of the targeted cell long before the correct clone could be identified and validated. Consequently, experimental outcomes using such models are harder to control. This obstacle is particularly troublesome for the engineering of oncogenic chimeric transcription factors that rewire global regulatory networks.
Here we present an approach for the temporal regulation of expression of the engineered endogenous genomic rearrangements. The strategy combines CRISPR-Cas9 technology, homology directed repair (HDR), and inclusion of a gene-trap to allow for the inducible expression of fusion transcripts. As proof of principle, we describe successful establishment of de novo engineered chromosomal translocations of the defining drivers for Ewing's sarcoma (ES), EWSR1-FLI1 t(11;22)(q24;q12), and desmoplastic small round cell tumour (DSRCT), EWSR1-WT1 t(11;22)(p13;q12), as conditional alleles in vitro.
Material and Methods
CRISPR-Cas9 Genomic Editing
The sequences for candidate guide RNAs (gRNAs) in this study were selected using http://crispr.mit.edu/. Oligonucleotides encoding the gRNA were cloned into the pSpCas9(BB)-2A-GFP (PX458) vector (Addgene #48138) (Supplementary Table S1). The EWSR1, FLI1 and WT1 gRNAs were transfected pairwise into HEK293T cells. After 72 h, cells were assayed for expression of type 1 EWSR1-FLI1 or EWSR1-WT1 mRNA by RT-PCR.
Generation of the Standard (NG1) and Conditional NG2 (TagRFP-Trap) HR Targeting Vector Constructs
EWSR1, FLI1 and WT1 homology arms (HA) (525 bp each) were PCR-amplified from HEK293T genomic DNA. The EWSR1 and FLI1 HA were cloned into the Footprint-Free™ Gene Editing Multivector™ (Transposagen), generating the pNG1-EWSR1-FLI1 (NG1) plasmid (Supplementary Table S2). For the generation of the TagRFP-Trap-Puro-T2A-TK (NG2) targeting plasmid, a TagRFP gene-trap cassette (TagRFP-Trap) was generated and inserted upstream of the Puro cassette of the NG1 construct. In brief, TagRFP (open reading frame, ORF)-PolyA (Evrogen) was PCR-amplified using a TagRFP-specific forward primer, modified to contain (from 5’ to 3’) a strong slice acceptor (SA) [13], and two additional nucleotides immediately upstream of the ATG codon of TagRFP to ensure in-frame splicing of EWSR1 into the TagRFP ORF. The reverse primer was designed to incorporate the PolyA addition signal. Correct orientation of the SA-TagRFP-PAS in pNG2-EF-TagRFP and pNG2-EW-TagRFP targeting vectors was confirmed by sequencing.
Mammalian Cell Culture and Cell Transfection
HEK293T cells were purchased from ATCC (ATCC® Number CRL-3216). Cells were maintained in high glucose Dulbecco's MEM with 10% fetal bovine serum and antibiotics. Cells were routinely subjected to Mycoplasma testing. To induce cuts in separate genomic loci, cells were transfected with 500 ng of each CRISPR-Cas9 construct for a total of 1 μg of vector. Transfections were carried out with FuGENE6 (Promega) according to the manufacturers’ instructions.
HDR Targeting and Selection of NG1 or NG2 Clones in HEK293T cells
For transfection of the HDR template, NG1 or NG2 targeting vectors were linearized with PciI enzyme (NEB), gel extracted and cleaned using a QIAquick Gel Extraction kit (Qiagen). Linearized NG1 and NG2 (500ng each) constructs were co-transfected with gRNA plasmids, for a total of 1.5 μg DNA per transfection. For NG1 targeting, cells were collected 72 h post transfection and reseeded at low density in puromycin containing medium (1.5 ug/ml). Following puromycin selection over 6 d, individual clones were picked using cloning discs (Sigma), expanded and analysed by genomic PCR, using primers listed in Supplementary Table S1.
For isolation of the NG2-TagRFP clones, cells were transfected as above. Puromycin resistant cells were FACs sorted using a BD FACSARIA cell sorter. TagRFP-positive cells were reseeded at low density in puromycin-containing medium as described above to obtain single colonies, and analysed for correct targeting using genomic PCR with primer pairs listed in Supplementary Table S2.
Genomic DNA Extraction and Standard PCR
The DNeasy Blood and Tissue Kit (Qiagen) was used for genomic DNA extractions following the manufacturer's instructions. Standard genomic PCR was carried out using either AmpliTaq Gold 360 Master Mix (Invitrogen) or Accuprime PFX (Invitrogen). Standard amplification was as following: 95 °C for 5 min, 35 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s and extension at 72 °C with duration depending upon expected product size, with a final extension of 10 min at 72 °C. No-template reactions were performed as negative controls. All PCR primer sequences are listed in Supplementary Table S2.
RNA Extraction, cDNA synthesis and Reverse Transcription PCR
Total RNA was extracted from cells using the RNAqueous-Micro Total RNA Isolation kit (Ambion). Samples of total RNA (500 ng) were reverse-transcribed using SuperScript VILO cDNA Synthesis Master Mix (Invitrogen). For RT-PCR reactions, PCR amplification was performed using either AmpliTaq Gold 360 Master Mix (Invitrogen) or Accuprime PFX (Invitrogen). Cycling conditions were the same as for standard genomic PCR (above), without reverse transcriptase (−RT) and no-template reactions as negative controls.
Quantitative Reverse Transcription PCR (RT-qPCR)
RT-qPCR was performed using the TaqMan® Gene Expression Master Mix and “inventoried” TaqMan® Gene Expression assays (all Applied Biosystems), using standard reaction-mix setups and cycling protocol for a StepOne Real-Time PCR System (Applied Biosystems). Quantification of gene expression was performed with the ΔΔCT method, using GAPDH as an internal control.
Transient Expression of Cre Recombinase
For excision of the Puro-T2A-TK and of the TagRFP-Trap-Puro-T2A-TK cassettes from the genomes of correctly targeted clones, Cre recombinase was transiently expressed from pCAG-Cre:GFP (Addgene#13776), Ganciclovir (Invivogen, San Diego, USA) but was added to the medium 72 h following transfection for negative selection of cells that retained the TK.
DNA Sequencing
PCR amplicons were sequenced either directly, using specific primers, or sub-cloned into TOPO vectors (Invitrogen) and then sequenced using universal vector primers.
Fluorescence In Situ Hybridization (FISH)
Cells from the NG1-EF-16.2 and NG1-EF-57.4 clones, as well as the parental HEK293 cell line, were harvested and fixed in Carnoy's fixative for FISH analysis. A dual-colour/dual-fusion FISH assay was performed to verify EWSR1-FLI1 fusion, the t(11;22)(q24;q12) translocation, using a customized EWSR1-FLI1 fusion probe set from Agilent Technologies (Santa Clara, CA). The green fluorochrome-labelled probe hybridizes to chromosome band 11q24 (Hg19 chr11:128274050-128867614) overlapping the entire FLI1 locus; the red fluorochrome-labelled probe hybridizes to chromosome band 22q12 overlapping the entire EWSR1 locus (Hg19 chr22:29380432-29980251). DAPI was used as DNA counterstain. FISH signals in both metaphase and interphase cells were analysed, and a minimum of 200 nuclei were scored.
Western Blot Analysis
Cells were lysed in RIPA buffer (Cell Signaling) containing protease inhibitors (Complete, Roche), and 20-40 μg of protein was separated on 4-12 % Bis-Tris NuPage gels (Invitrogen). Membranes were probed with the following antibodies: GAPDH (1:2000, Cell Signaling,14C10), Fli-1 (1:500, Santa Cruz Biotechnology, C-19, sc-356), WT1 (1:1000, Santa Cruz Biotechnology, C-19, sc-192).
Results
Generation of the type I EWSR1-FLI1 translocation by CRISPR-Cas9 and NHEJ
It has been reported previously that CRISPR-Cas9 genomic editing in combination with NHEJ produced a constitutively expressed type II EWSR1-FLI1 translocation, characteristic of Ewing's sarcoma [6]. Before engineering a conditional allele of the translocation, we proceeded to confirm whether this method can be applied for generation of a type I EWSR1-FLI1 fusion, the most frequently identified translocation event in patients with Ewing's sarcoma [14].
We designed and evaluated 5 pairwise combinations of gRNAs that directed the Cas9 endonuclease to each of the two introns involved in the translocation - intron 7 of EWSR1 on chromosome 22 and intron 5 of FLI1 on chromosome 11. The gRNAs were designed to induce DSBs within the first 500 bp downstream of the exon/intron boundary (Figure 1A). One gRNA combination in particular, E4-gRNA:F3-gRNA readily generated the fusion mRNA (Figure 1B). The resulting rearrangement was confirmed by sequencing of the corresponding amplicons at both mRNA and genomic DNA levels (Figure 1B and 1C). Furthermore, sequencing of the genomic DNA PCR amplicon confirmed that the fusion contained precise intronic end - joining, as well as microdeletions, a common by-product of the error-prone NHEJ pathway (Supplementary Figure S1).
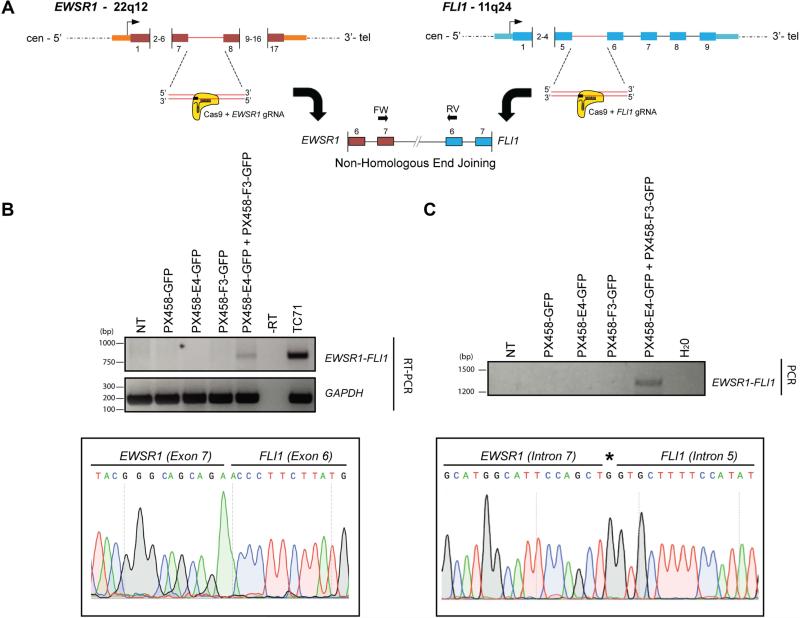
Figure 1. Generation of a de novo EWSR1-FLI1 t(22;11)(q12:q24) chromosomal translocation via CRISPR-Cas9 and NHEJ.
A. Schematic of the t(22;11)(q12:q24) rearrangement mediated by CRISPR-Cas9 genomic editing and non-homologous end-joining (NHEJ). gRNAs were designed to target intron 7 (E4) and intron 5 (F3) of EWSR1 and FLI1 loci, respectively.
B. RT-PCR confirming specific expression of the EWSR1-FLI1 fusion transcript between exon 7 of EWSR1 and exon 6 of FLI1 (top panel). TC71: fusion positive Ewing's sarcoma patient derived cell line. NT: non-transfected control. Sanger sequencing trace encompassing the fusion event is shown (lower panel).
C. Confirmation of the translocation at the genomic level. NT: non-transfected control. Sanger sequencing traces encompassing the genomic fusion event are shown (lower panel). Asterisk denotes the insertion of one additional nucleotide by NHEJ.
Importantly, the frequency of the translocation in this bulk cellular population was below the level of detection by dual colour/dual probe FISH, as we were unable to detect any EWSR1-FLI1 fusion positive nuclei from examination of 200 nuclei (data not shown). As further validation, and supporting the postulated low frequency of the event, we screened 100 single clones by RT-PCR and confirmed that none contained the EWSR1-FLI1 fusion product (data not shown).
In summary, as previously reported, our data confirmed that the CRISPR-Cas9 genome editing based on NHEJ can be used to generate constitutively active chromosomal translocations in vitro within a cellular population [5,6]. However, we found the frequency of the desired genomic rearrangement to be low.
Combination of CRISPR-Cas9 with Homology-directed repair (HDR) to generate hybrid fusions containing a drug selectable cassette
Induction of DSBs can stimulate DNA repair by at least two distinct mechanisms - NHEJ and HDR. As described above, the generation the EWSR1-FLI1 translocation following CRISPR-Cas9-induced DSBs was mediated by error-prone NHEJ. We thus hypothesized that providing a donor DNA template that bridges the fusion partners could lead to HDR-mediated generation of a chromosomal translocation following induction of CRISPR-Cas9 gRNA-specified DSBs (Figure 2A). We designed a HDR DNA donor template (NG1-EWSR1-FLI1, henceforth “NG1-EF”) containing homology arms (525 bp) corresponding to the respective 5’ and 3’ chromosomal arms flanking the DSBs induced by CRISPR-Cas9:EWSR1 (E4) and FLI1 (F3) gRNAs. Between the homology arms, the donor template includes a removable bi-functional cassette whose presence can be selected for or against - a puromycin (Puro) resistance gene and a truncated thymidine kinase (TK) gene (Figure 2A).
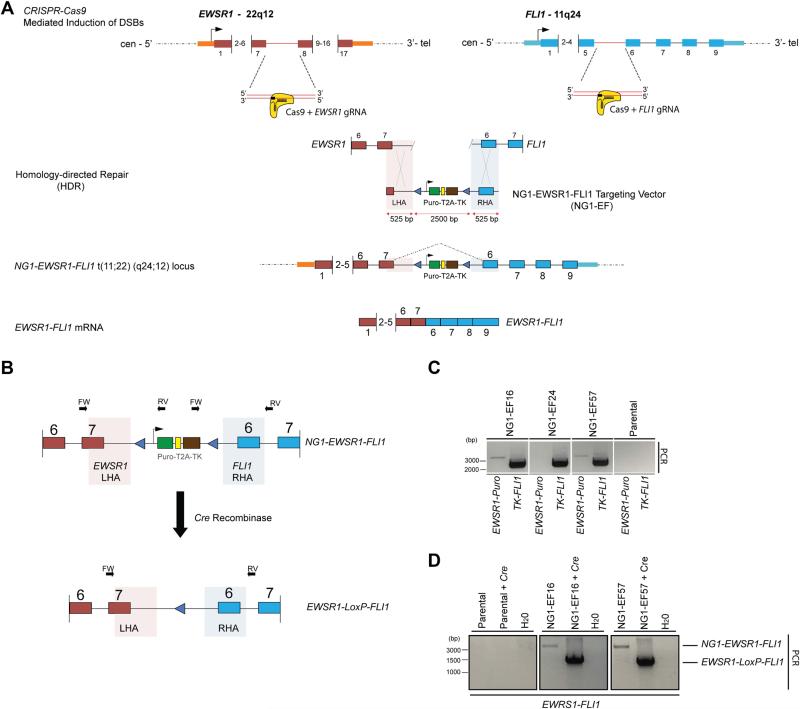
Figure 2. Generation of the EWSR1-FLI1 t(11;22)(q24;12) chromosomal translocation via CRISPR-Cas9 using homology-directed repair (HDR).
A. Schematic of the generation of the t(11;22)(q24:q12) translocation using CRISPR-Cas9 genomic editing and homology-directed repair (HDR). The introduction of a NG1-EWSR1-FLI1 (NG1-EF) targeting vector, which bridges the Cas9-gRNA directed double-strand breakpoints in EWSR1 and FLI1 loci, leads to the formation of a chimeric translocation, NG1-EWSR1-FLI1 t(11;22)(q24;12). The NG1-EF targeting vector contains arms of homology matching to EWSR1 and FLI1 (shaded, LHA and RHA).
B. Schematic representation of the 2-step PCR screening strategy. The external EWSR1 and FLI1 primers are specific for genomic DNA outside of the sequence retained in the left and right homology arms of the NG1-EF targeting vector (LHA and RHA, shaded areas). Treatment with Cre recombinase removes the selectable cassette leaving a single LoxP at the engineered locus.
C. A 2-Step PCR screening protocol on genomic DNA isolated from puromycin resistant clones identifies correctly targeted NG1-EF clones. Positioning of the primers is depicted in B. Combinational PCR screening using primers: (1) EWSR1-FW with Puro-RV (2) TK-FW with FLI1-RV identifies correctly targeted clones that contain both the left and the right homology arm and hence the NG1-EWSR1-FLI1 hybrid translocation.
D. Following Cre recombination and ganciclovir selection, genomic DNA was assayed to demonstrate the loss of the selectable cassette and the generation of the EWSR1-LoxP-FLI1 translocation by using a EWSR1-FW primer and a FLI1-RV primer.
HEK293T cells were transfected with the E4:F3-gRNA combination of CRISPR-Cas9 plasmids, along with the linearized NG1 donor template (Figure 2A). At 72 h post-transfection, cells were reseeded in puromycin-containing medium. Following selection, 100 puromycin-resistant clones were isolated for genomic analysis. Genomic PCR confirmed that two clones, NG1-EF-16 and NG1-EF-57, had undergone correct CRISPR-Cas9-mediated genomic editing via HDR, resulting in the generation of the EWSR1-FLI1 genomic fusion bridged by the NG1 template. This was confirmed by positive PCR amplicons from the insertion of the left and right arms of homology, and also a 3.4 kbp product generated from PCR primers residing outside of the homology arms in the targeted regions of EWSR1 and FLI1 (Figure 2B, Figure 2C and Figure 2D). Of note, inclusion of the NG1 donor template between the EWSR1 and FLI1 genomic fusion partners was not designed to inhibit the expression of the engineered fusion at the mRNA level. RT-PCR confirmed expression of the EWSR1-FLI1 fusion mRNA in the two positive clones, NG1-EF-16 and NG1-EF-57, but not in the negative clones, includi ng NG1-EF-24 (Supplementary Figure S2), which did not contain the full NG1-EF targeting vector.
We used Cre recombinase to remove the Puro/TK selection cassette. NG1-EF-16 and NG1-EF-57 cells were transiently transfected with a Cre recombinase expression plasmid and selected in ganciclovir to remove cells that retained the TK gene and thus had failed to undergo Cre-mediated recombination. Genomic PCR analysis of the ganciclovir-resistant clones confirmed selective removal of the bi-functional drug resistance cassette. Using the primer pairs that bind to sequences outside of homology arms, the original 3.4 kb hybrid fusion reverted to a 1.5 kb product post-Cre (Fig. 2D). Sanger sequencing confirmed the authenticity of this shorter PCR amplicon and demonstrated a CRISPR-Cas9-engineered EWSR1-FLI1 genomic fusion retaining a single LoxP site in the intron and no residual Puro/TK (Supplementary Figure S3).
From the ganciclovir-resistant populations of NG1-EF-16 and NG1-EF-57 cells, we selected two clones, NG1-EF-16.2 and NG1-EF-57.4, for dual-colour/dual-probe FISH analysis. This analysis revealed that, unlike the parental cells, which harboured separate red (EWSR1) and green (FLI1) signals, NG1-EF-16.2 and NG1-EF-57.4 cells displayed juxtaposed red and green signals, confirming the presence of the EWSR1-FLI1 fusion at the genomic level (Figure 3A). Moreover, RT-PCR (Figure 3B) and Western blot (Figure 3C) analyses of the NG1-EF-16.2 and NG1-EF-57.4 clones demonstrated that the chimeric EWSR1-FLI1 transcript and the EWSR1-FLI1 protein were expressed.
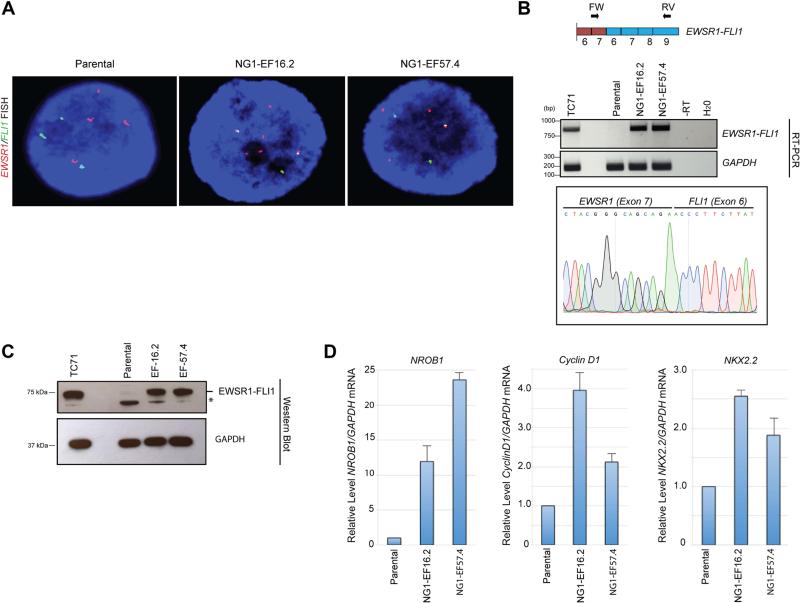
Figure 3. Faithful expression of a biologically active EWSR1-FLI1 translocation product engineered with CRISPR-Cas9 and HDR.
A. Verification of the EWSR1-FLI1 fusion by FISH. The EWSR1 locus-specific probe labelled in red, and the FLI1 locus-specific probe labelled in green. In the parental cells, distinct separate green and red signals were observed. In cells from the NG1-EF-16.2 and NG1-EF-57.4 clones, green/red fusion signals (appearing as partial or complete yellow dots) that represented EWSR1-FLI1 fusions were identified. DNA was counterstained with DAPI (Blue).
B. RT-PCR confirming specific expression of the EWSR1-FLI1 fusion transcript in the NG1-EF16.2 and NG1-EF57.4 engineered clones, but not in the parental cell line. Sanger sequencing trace encompassing the fusion event is shown (lower panel) confirming the EWSR1-FLI1 fusion mRNA. TC71: Fusion positive patient-derived Ewing's sarcoma cell line.
C. Expression of EWSR1-FLI1 fusion protein in the NG1-EF16.2 and NG1-EF57.4 clones as detected by an anti-FLI1 C-terminal antibody (top panel). The TC71 Ewing's sarcoma cell line was used a positive control. GAPDH was used as loading control (bottom panel). * non-specific band
D. Expression of EWSR1-FLI1 regulated transcripts in NG1-EF16.2 and NG1-EF57.4 clones as established by RT-qPCR. Significant upregulation of expression of NROB1, Cyclin D1 and NKX2.2 in NG1-EF16.2 and NG1-EF57.4 clones compared to the parental cell line. Data are presented as relative expression of a target transcript compared to the parental cell line. The data are derived from a total of 3 independent experiments (mean ± SD).
The EWSR1-FLI1 protein is a potent oncogenic transcription factor that drives Ewing's sarcoma tumourigenesis by reprogramming of the cellular gene expression profile [14]. To assess whether the engineered EWSR1-FLI1 in the NG1-EF-16.2 and NG1-EF-57.4 clones are biologically active, we performed RT-qPCR analysis of validated transcriptional targets of EWSR1-FLI1 [15-17]. Our analysis demonstrated robust up-regulation of several target transcripts, including NROB1, in NG1-EF-16.2 and NG1-EF-57.4 cells as compared to parental HEK293 cells (Figure 3D).
Thus, by combining CRISPR-Cas9 genomic editing and HDR, a DNA donor template can be used to bridge an interchromosomal translocation and simultaneously include a selectable cassette that allows for enhanced capture of the engineered genomic editing event.
Generation of a conditional inducible endogenous EWSR1-FLI1 chromosomal translocation product
We have shown that through combination of CRISPR-Cas9 genomic editing and HDR, a DNA donor template can be used to bridge and capture an interchromosomal translocation. In regards to EWSR1-FLI1, generation of the fusion by either CRISPR-Cas9/NHEJ or CRISPR-Cas9/HDR results in immediate expression of the genomic rearrangement (Figure 1B and Supplementary Figure S2). This phenomenon is particularly troublesome with engineered oncogenic transcription factors, such as EWSR1-FLI1, due to their hijacking of the gene expression networks in target cells, resulting in a dramatic change of the cellular phenotype [14,18,19]. To overcome this issue, temporal regulation of the engineered genomic rearrangements could provide the type of experimental control that would allow us to gain further insights into the gene networks controlled by these oncogenic drivers. To achieve this, we introduced a reversible gene-trap into the HDR donor template to enable conditional expression of the fusion mRNA (Figure 4A).
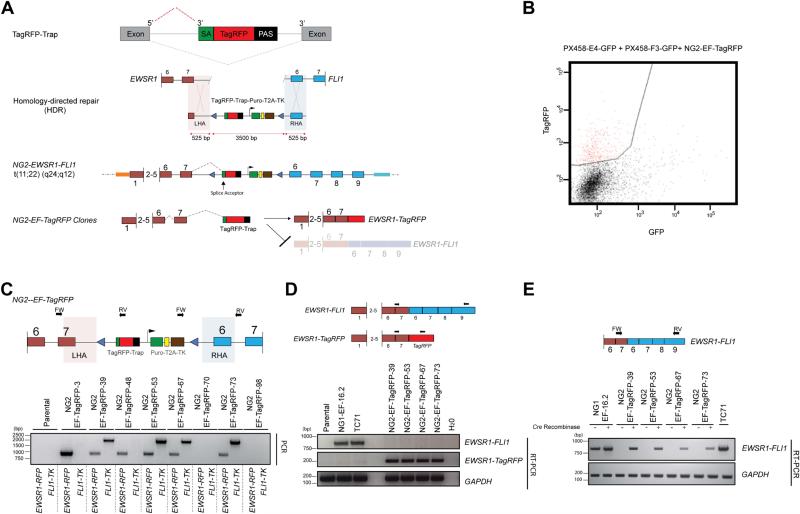
Figure 4. Generation of conditional endogenous EWSR1-FLI1 t(11;22)(q24;q12) chromosomal translocation via CRISPR-Cas9, HDR and inclusion of a gene-trap.
A. Schematic representation of a conditional chromosomal translocation with the incorporation of a gene-trap (TagRFP-Trap) in the engineered hybrid intron.
B. Isolation of TagRFP positive clones by FACS following HDR targeting with the NG2-EWSR1-FLI1 HDR template and co-transfection of EWSR1 and FLI1 gRNA and 6 days of puromycin selection.
C. Schematic representation of the primer positions used in the 2-step PCR screen (Top panel). The external EWSR1 and FLI1 primers are specific for genomic DNA outside of the sequence retained in the left and right homology arms of the NG2-EF-TagRFP targeting vector (shaded areas), whilst the internal primers were specific for TagRFP (RFP) and thymidine kinase (TK). Combinational PCR screening using primers in: (1) EWSR1-FW with a TagRFP-RV specific primer and (2) TK-FW primer with a FLI1-RV primer identifies correctly target clones that contain both the left and the right homology arm and hence the NG2-EWSR1-FLI1 hybrid translocation.
D. RT-PCR analysis confirms that the NG2-EF-TagRFP clones express the EWSR1-TagRFP transcript at the expense of the full-length EWSR1-FLI1 mRNA,
E. RT-PCR analysis confirms expression of the EWSR1-FLI1 fusion transcript in NG2-EF-TagRFP clones only in the presence of transiently expressed Cre recombinase, which excises the TagRFP-Trap, allowing for the transcription of the full-length fusion EWSR1-FLI1 mRNA.
Genome-wide gene-trap insertional mutagenesis is a powerful approach for loss-of-function studies [13,20]. Typically, a promoterless reporter and/or selectable marker flanked by an upstream 3' splice acceptor (SA) and a downstream transcriptional termination sequence (polyadenylation sequence signal; PAS) is randomly inserted into the genome to disrupt gene expression. In our work, we employed a fluorescent reporter, TagRFP. The cassette was adapted for in-frame expression of the TagRFP reporter under the control of the endogenous EWSR1 promoter, thereby producing EWSR1-TagRFP mRNA that replaces the expression of the full-length EWSR1-FLI1 mRNA (Figure 4A). We inserted the TagRFP cassette into the NG1 donor template, to generate the NG2-EWSR1-TagRFP-FLI1 (NG2-EF-TagRFP) targeting vector. Strategic positioning of LoxP sites allows for the Cre-mediated excision of the gene trap, eliminating expression of EWSR1-RFP and fully restoring EWRS1-FLI1 transcriptional activity (Figure 4A)
We co-transfected the NG2-EF-TagRFP donor template with the combination of E4 and F3 gRNA and selected in puromycin. Fluorescence-activated cell sorting (FACS) on the pool of puromycin-resistant cells was used to assay for expression and isolation of TagRFP+ cells (Figure 4B). TagRFP+ cells were diluted into plates, and 100 single cell clones were picked, expanded and analysed for correct targeting of the NG2-EF-TagRFP template. Genomic analysis by PCR (Figure 4C, Top panel) confirmed that 4 clones - NG2-EF-TagRFP-39, NG2-EF-TagRFP-53, NG2-EF-TagRFP-67 and NG2-EF-TagRFP-73 - contained the NG2-TagRFP vector correctly targeted to the CRISPR-Cas9-engineered EWSR1-FLI1 locus (Figure 4C, bottom panel).
To determine whether the SA site in the gene-trap cassette efficiently disrupts expression of EWSR1-FLI1, we employed an RT-PCR screen on the individual positive clones. Using a forward primer in the 5’ region of EWSR1 and a reverse primer in TagRFP, we detected efficient splicing of EWSR1 into TagRFP, with concomitant reduction of the EWSR1-FLI1 transcript (Figure 4D). Thus, our gene trap-based approach efficiently abrogates expression of EWSR1-FLI1 mRNA from the engineered locus in favour of an EWSR1-TagRFP reporter transcript.
To remove the TagRFP-Trap and release expression of the engineered EWSR1-FLI1 fusion mRNA, we transiently transfected Cre recombinase into the four positive clones. Following selection with ganciclovir, RT-PCR and RT-qPCR analysis confirmed transcriptional activation of EWSR1-FLI1 in all four clones (Figure 4E and Supplementary Figure S4), demonstrating the effectiveness of the conditional allele methodology.
Modelling of the EWSR1-WT1 t(11;22)(p13;q12) translocation as a conditional allele
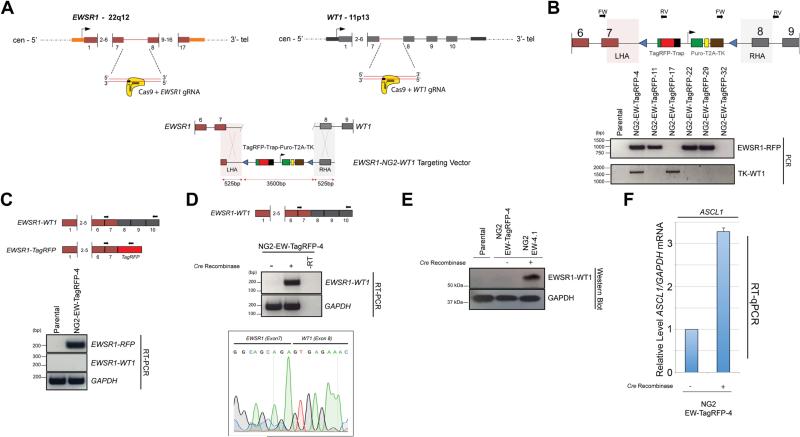
To demonstrate the generic adaptability of our approach, we generated an additional HDR donor template and gRNAs for establishment of a second translocation, EWSR1-WT1, which is the molecular hallmark of desmoplastic small round cell tumour (DSRCT). This sarcoma is defined by the presence of the chromosomal translocation t(11;22)(p13;q12), which results in the fusion of the N-terminal portion of EWSR1 and the C-terminal DNA-binding domain of WT1. The resulting hyperactive chimeric transcription factor, EWSR1-WT1, drives the oncogenesis of this very aggressive tumour in children and young adults [21-23].
For de novo generation of the DSRCT translocation, we designed and validated a series of gRNAs targeting intron 7 of WT1, and combined them with the EWSR1-E4 gRNA described previously. We identified a gRNA, W2, that in combination with E4, generated the desired fusion of exon 7 of EWSR1 to exon 8 of WT1 (Supplementary Figure S5). We adapted the NG2-EF-TagRFP HDR template described above to contain a right homology arm compatible with WT1 (NG2-EW-TagRFP) (Figure 5A). We then co-transfected it with the EWSR1/WT1 gRNAs and isolated TagRFP+ cells via FACS following puromycin selection. A genomic PCR screen of 25 clones identified one clone, NG2-EW-TagRFP-4, with the correct integration of the NG2-EW-TagRFP targeting cassette into the CRISPR-Cas9/HDR engineered EWSR1-WT1 translocation (Figure 5B). RT-PCR confirmed expression of the EWSR1-RFP reporter transcript, but not the EWSR1-WT1 fusion mRNA (Fig. 5C). Following transient expression of Cre recombinase and ganciclovir selection, RT-PCR and RT-qPCR analysis confirmed potent induction of EWSR1-WT1 mRNA (Figure 5D and Supplementary Figure S6). Sanger sequencing of the RT-PCR product confirmed authenticity of the EWSR1-WT1 fusion mRNA, with exon 7 of EWSR1 fused to exon 8 of WT1 (Figure 5D, bottom panel).
Figure 5. Generation of an conditional de novo EWSR1-WT1 t(11;22)(p13;q12) translocation, the hallmark of desmoplastic small round cell tumour (DSRCT).
A. Schematic representation of the generation of a conditional EWSR1-WT1 chromosomal translocation using the incorporation of a gene-trap (TagRFP-Trap) at the engineered intronic junction between the CRISPR-Cas9-mediated translocation of EWSR1 to WT1. gRNAs were directed against Intron 7 of EWSR1 and Intron 7 of WT1.
B. Schematic representation of the primer positions used in the 2-step PCR screen (top panel). The external EWSR1-FW and WT1-RV primers are specific for genomic DNA outside of the sequence retained in the left and right homology arms of the NG2-EWSR1-WT1 targeting vector (shaded areas), whilst the internal primers were TagRFP and thymidine kinase (TK). Combinational PCR screening using primers in: (1) EWSR1-FW with a TagRFP-RV specific primer and (2) TK-FW with a WT1-RV primer combination identifies a correctly targeted clone that contains both the left and the right homology arm and hence the EWSR1-NG2-WT1 hybrid translocation (lower panel);
C. RT-PCR analysis confirms that the NG2-EW-TagRFP-4 clone expresses the EWSR1-TagRFP transcript at the expense of the full-length EWSR1-WT1 mRNA.
D. RT-PCR analysis confirms expression of the EWSR1-WT1 fusion transcript in NG2-EW-TagRFP-4 clone only in the presence of transiently expressed Cre recombinase, which excises the TagRFP-Trap, releasing transcription of the full-length EWSR1-WT1 fusion mRNA. Sanger sequencing (lower panel) confirms authenticity of the EWSR1-WT1 fusion mRNA.
E. Western Blot analysis confirms expression of EWSR1-WT1 protein following transient expression of Cre recombinase.
F. RT-qPCR confirms up-regulation in expression of ASCL1 following Cre-mediated activation of EWSR1-WT1. Data are presented as relative expression of a target transcript compared to the parental cell line. The data are derived from a total of 3 independent experiments (mean ± SD).
We then isolated a single clone following ganciclovir selection, NG2-EW-4.1. Genomic PCR and Sanger sequencing confirmed the presence of a CRISPR-Cas9/HDR engineered EWSR1-WT1 locus with retention of a single LoxP site in the chimeric intron, corroborating that the translocation was generated by CRISPR-Cas9/HDR and not by NHEJ (Supplementary Figure S7). Western blot analysis confirmed translation of the EWSR1-WT1 mRNA, with detectable induction of EWSR1-WT1 fusion protein following Cre-mediated activation (Figure 5E). Finally, we assessed the biological function of the engineered EWSR1-WT1 oncogene by assaying for induction of ASCL1, a recently validated downstream transcriptional target of EWSR1-WT1 [24]. RT-qPCR confirmed that ASCL1 was up-regulated following Cre-mediated activation of EWSR1-WT1 in NG2-EW-4 cells (Figure 5F).
Discussion
Faithful models of genomic rearrangements are a prerequisite for the study of their pathogenic roles in tumourigenesis. The ability to combine various programmable nucleases with NHEJ to generate endogenous genomic rearrangements in human cell lines has previously been documented [3-6,11,12]. Whilst this approach successfully generates de novo genomic rearrangements, expression of the resultant fusion gene is immediate and sustained, and isolation of fusion-positive clones remains challenging.
Here we have described a strategy for de novo genomic engineering of chromosomal translocations as conditionally expressed fusions in a single targeting step. We have successfully applied this methodology to generate conditionally inducible EWSR1-FLI1 and EWSR1-WT1 translocations in a human cell line. The novelty of our method resides in the generation of de novo engineered chromosomal translocations with the combination of CRISPR-Cas9 and HDR, allowing temporal control of their expression.
We first demonstrated that it is possible to introduce a DNA donor template containing a drug selection cassette (puromycin) to bridge the EWSR1 and FLI1 fusion partners to form an endogenous engineered EWSR1-FLI1 locus. This process allowed the addition of a drug selection step in the isolation of translocation-positive clones, selectively enriching for the desired event. To further enhance this methodology and generate conditional activation of our genomic editing, a gene-trap cassette was incorporated into the HDR donor template. This cassette allows for selective trapping of the 5’ fusion partner mRNA (EWSR1), thereby preventing the expression of the full-length engineered transcript (EWSR1-FLI1 or EWSR1-WT1). By designing the gene-trap to allow for in-frame expression of TagRFP under the control of the 5’ fusion partner (EWSR1), the expression of the EWSR1-TagRFP allowed us to enrich for targeted cells via FACS. Finally, temporal activation is provided by Cre-mediated excision of the gene-trap from the engineered locus, releasing the expression of the full-length EWSR1-FLI1 or EWSR1-WT1 mRNA.
Establishment of these model systems is of principal importance for rare translocation-driven malignancies such as Ewing's sarcoma and DSCRT - diseases in which identification of effective therapeutic options has been hampered by the paucity of available models, especially for DSCRT. EWSR1 represents one of the most commonly translocated gene partners in bone and soft tissue sarcomas [25,26]. The fusion of EWSR1 and its 3’ partner often joins the N-terminal transcription-activating domain of EWSR1 and the C-terminal, DNA-binding domain of the 3’ partner, yielding an oncogenic chimeric transcription factor.
Our system provides temporally controlled expression of these drivers in an isogenic, chemotherapy-naïve, cellular context. These features set our engineered models apart from conventional patient-derived cell lines, which are often complicated by additional alterations acquired during therapeutic intervention and/or prolonged culture. In fact, the low secondary mutation burden of these tumours highlights the need to avoid such alterations for faithful modelling of these malignancies [27]. Therefore using our approach in a physiologically relevant context, such as human mesenchymal stem cells, will permit the development of an isogenic in vitro platform for future studies.
However, the paucity of models is not restricted to rare tumour types. In fact, several subsets of common cancers are driven by fusion genes. For instance, non-small cell lung cancers harbouring fusion genes involving ALK, ROS1 or RET have no or few cell lines for functional studies, and the models currently available are not entirely representative of the spectrum of lesions harbouring these genomic events [28]. Therefore, our system has broad applicability beyond orphan malignancies and can be customized to a wider variety of chromosomal rearrangements, including intra-chromosomal rearrangements.
Our present study has its limitations. In generating a conditional chromosomal translocation, the insertion of the gene-trap into the 5’ fusion partner to interrupt expression may not be functionally neutral. A potential consequence would be expression of a non-physiological, truncated isoform of the 5’ fusion partner and/or reduced dosage of the full-length protein in the engineered cell. For EWSR1 in particular, heterozygous EWSR1 mice are viable and grossly normal, with no reported incidence of tumourigenesis [29]. However, in a Tp53-null zebrafish model, it has been demonstrated that reduction in EWSR1 expression can lead to tumourigenesis [30]. Secondly, although effectively silencing the fusion, engineered translocations result in the alteration in the chromosomal architecture within the nucleus via juxtaposition of chromosomal regions, which may perturb gene expression through topological changes in 3D genome structure by displacing non-proximal regulatory elements.
Despite these limitations, our approach represents a significant advance in modelling chromosomal translocations by allowing for precise and rapid generation of in vitro systems that afford temporal control of fusion genes resulting from chromosomal rearrangements of interest.
Supplementary Material
Acknowledgements
We thank Professor Nick Hastie, Dr Jason Huse and Dr Alex Kentsis for critical reading of this manuscript. We thank Dr Huichun Tai for technical assistance. We thank Mr Ethan Rai-Spraggon and Ms Anya Rai-Spraggon for helpful comments throughout the course of this work. We also thank Dr Maria Jasin and Dr Fabio Vanoli for helpful and constructive comments.
The authors wish to thank the following for reagents: the pSpCas9(BB)-2A-GFP (PX458) was a gift from Dr Feng Zhang (Addgene plasmid # 48138), pCAG-Cre:GFP was a gift from Dr Connie Cepko (Addgene plasmid # 13776).
This work was supported in part by an Alex's Lemonade Stand Innovation Award (to M.L). Research reported in this publication was supported in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (Grant No. P30CA008748). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interests statement: The authors declare no competing financial interests
Statement of Author Contributions
L.S. and L.G.M., conceived, initiated, designed and performed most of the experiments in the paper.
J.H., T.H., H.T., P.D.F, J.W., L.W., E.S. assisted in the remaining experiments and collected data. J.H and J.S.R.-F. commented on the manuscript.
L.S., L.G.M, and M.L. wrote the manuscript
Reference List
- 1.Mertens F, Johansson B, Fioretos T, et al. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15:371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 2.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 3.Brunet E, Simsek D, Tomishima M, et al. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci USA. 2009;106:10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piganeau M, Ghezraoui H, De Cian A, et al. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res. 2013;23:1182–1193. doi: 10.1101/gr.147314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torres R, Martin MC, Garcia A, et al. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat Commun. 2014;5:3964. doi: 10.1038/ncomms4964. [DOI] [PubMed] [Google Scholar]
- 7.Blasco RB, Karaca E, Ambrogio C, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014;9:1219–1227. doi: 10.1016/j.celrep.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 8.Maddalo D, Manchado E, Concepcion CP, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Gaj T, Yang Y, et al. Efficient delivery of nuclease proteins for genome editing in human stem cells and primary cells. Nat Protoc. 2015;10:1842–1859. doi: 10.1038/nprot.2015.117. [DOI] [PubMed] [Google Scholar]
- 10.Lagutina IV, Valentine V, Picchione F, et al. Modeling of the human alveolar rhabdomyosarcoma Pax3-Foxo1 chromosome translocation in mouse myoblasts using CRISPR-Cas9 nuclease. PLoS Genet. 2015;11:e1004951. doi: 10.1371/journal.pgen.1004951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghezraoui H, Piganeau M, Renouf B, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burckstummer T, Banning C, Hainzl P, et al. A reversible gene trap collection empowers haploid genetics in human cells. Nat Methods. 2013;10:965–971. doi: 10.1038/nmeth.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lessnick SL, Ladanyi M. Molecular pathogenesis of Ewing sarcoma: new therapeutic and transcriptional targets. Annu Rev Pathol. 2012;7:145–159. doi: 10.1146/annurev-pathol-011110-130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendiola M, Carrillo J, Garcia E, et al. The orphan nuclear receptor DAX1 is up-regulated by the EWS/FLI1 oncoprotein and is highly expressed in Ewing tumors. Int J Cancer. 2006;118:1381–1389. doi: 10.1002/ijc.21578. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez G, Bittencourt D, Laud K, et al. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc Natl Acad Sci USA. 2008;105:6004–6009. doi: 10.1073/pnas.0710748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith R, Owen LA, Trem DJ, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell. 2006;9:405–416. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Riggi N, Knoechel B, Gillespie SM, et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;26:668–681. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selvanathan SP, Graham GT, Erkizan HV, et al. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc Natl Acad Sci USA. 2015;112:E1307–1316. doi: 10.1073/pnas.1500536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanford WL, Cohn JB, Cordes SP. Gene-trap mutagenesis: past, present and beyond. Nat Rev Genet. 2001;2:756–768. doi: 10.1038/35093548. [DOI] [PubMed] [Google Scholar]
- 21.Ladanyi M, Gerald W. Fusion of the EWS and WT1 genes in the desmoplastic small round cell tumor. Cancer Res. 1994;54:2837–2840. [PubMed] [Google Scholar]
- 22.Gerald WL, Rosai J, Ladanyi M. Characterization of the genomic breakpoint and chimeric transcripts in the EWS-WT1 gene fusion of desmoplastic small round cell tumor. Proc Natl Acad sci USA. 1995;92:1028–1032. doi: 10.1073/pnas.92.4.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerald WL, Ladanyi M, de Alava E, et al. Clinical, pathologic, and molecular spectrum of tumors associated with t(11;22)(p13;q12): desmoplastic small round-cell tumor and its variants. J Clin Oncol. 1998;16:3028–3036. doi: 10.1200/JCO.1998.16.9.3028. [DOI] [PubMed] [Google Scholar]
- 24.Kang HJ, Park JH, Chen W, et al. EWS-WT1 oncoprotein activates neuronal reprogramming factor ASCL1 and promotes neural differentiation. Cancer Res. 2014;74:4526–4535. doi: 10.1158/0008-5472.CAN-13-3663. [DOI] [PubMed] [Google Scholar]
- 25.Sankar S, Lessnick SL. Promiscuous partnerships in Ewing's sarcoma. Cancer Genet. 2011;204:351–365. doi: 10.1016/j.cancergen.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor BS, Barretina J, Maki RG, et al. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer. 2011;11:541–557. doi: 10.1038/nrc3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shukla N, Ameur N, Yilmaz I, et al. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18:748–757. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Watford W, Li C, et al. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J Clin Invest. 2007;117:1314–1323. doi: 10.1172/JCI31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park H, Galbraith R, Turner T, et al. Loss of Ewing sarcoma EWS allele promotes tumorigenesis by inducing chromosomal instability in zebrafish. Sci Rep. 2016;6:32297. doi: 10.1038/srep32297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.