Abstract
caudal (cad/Cdx) genes are essential for the formation of posterior structures in Drosophila, Caenorhabditis elegans, and vertebrates. In contrast to Drosophila, the majority of arthropods generate their segments sequentially from a posteriorly located growth zone, a process known as short-germ development. caudal homologues are expressed in the growth zone of diverse short-germ arthropods, but until now their functional role in these animals had not been studied. Here, we use RNA interference to examine the function of caudal genes in two short-germ arthropods, the crustacean Artemia franciscana and the beetle Tribolium castaneum. We show that, in both species, caudal is required for the formation of most body segments. In animals with reduced levels of caudal expression, axis elongation stops, resulting in severe truncations that remove most trunk segments. We also show that caudal function is required for the early phases of segmentation and Hox gene expression. The observed phenotypes suggest that in arthropods caudal had an ancestral role in axis elongation and segmentation, and was required for the formation of most body segments. Similarities to the function of vertebrate Cdx genes in the presomitic mesoderm, from which somites are generated, indicate that this role may also predate the origin of the Bilateria.
Keywords: caudal/Cdx genes, short-germ development, Artemia, Tribolium, evolution
The caudal (cad/Cdx) genes are homeobox genes involved in posterior patterning in diverse species (1–11). In early Drosophila embryos Caudal protein is distributed in a posterior to anterior concentration gradient that is needed for the activation of segmentation genes and segment formation in posterior parts of the animal. caudal mutant embryos have severe segmentation problems affecting posterior segments; in the most severely affected mutants, most abdominal segments are missing (1). This function of caudal is characteristic of long-germ development, found in Drosophila, where all of the body segments are molecularly determined during the blastoderm stage. This mode is thought to be evolutionarily derived within the higher insects and does not represent the ancestral mode of development (12). On the contrary, short-germ development is found in diverse and phylogenetically basal groups of insects and other arthropods, suggesting that this represents the ancestral mode for generating segments within the arthropods. In short-germ arthropods (which we take to include intermediate-germ species), only the most anterior segments are laid down in the blastoderm (e.g., Fig. 1A), whereas more posterior segments are generated sequentially from a posteriorly located presegmental zone, usually referred to as the “growth zone” (12–14). caudal homologues have been cloned in some short-germ arthropods and found to be expressed consistently in this presegmental zone (15–22), but until now their function in these species had not been studied.
Fig. 1.
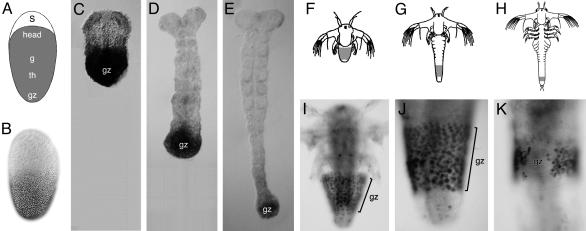
Caudal expression in Tribolium and Artemia.(A) Blastoderm fate map in Tribolium, indicating the position of the serosa (s), anterior head (head), gnathal (g), thoracic (th), and growth zone (gz) anlage. (B) Immunochemical staining showing the distribution of Caudal in the blastoderm of Tribolium. (C–E) Immunochemical staining showing Caudal expression in the growth zone (gz) of Tribolium, during successive stages of segmentation. The growth zone lies at the posterior-most end of the elongating germ band. (F–H) Illustrations of Artemia larvae, showing three successive stages of segmentation. The growth zone (indicated in gray) is located subterminally, lying anterior to the differentiated telson/anus. (I–K) Immunochemical stainings showing the expression of Caudal in the growth zone (gz) of Artemia, at similar stages to those shown in F–H. Anterior is up in all panels.
To explore the functional role of caudal genes in the growth zone of short-germ arthropods, we applied RNA interference (RNAi) to two short-germ arthropods, the branchiopod crustacean Artemia franciscana and the coleopteran insect Tribolium castaneum, and examined the phenotypes.
Materials and Methods
Double-Stranded RNA (dsRNA) Preparation. Single-stranded RNAs were produced from opposing strands of a full-length cDNA clone in pBluescript II, by in vitro transcription with the T3 and T7 polymerases (Ambion or Promega). The plasmid DNA was then removed by using DNaseI from the RNase-free kit (Ambion). The two single-stranded RNAs were allowed to anneal by mixing equal amounts of each strand, heating to 85°C, and cooling gradually to 40°C. The quality of the annealed dsRNA was checked by electrophoresis on an agarose gel.
Artemia Culture and Microinjections. Artemia franciscana diapause cysts from Great Salt Lake were hydrated, and larvae were raised in 3% artificial seawater, supplemented during later larval stages with brine shrimp food from NT Laboratories (Kent, U.K.). For Artemia larval microinjections, the larvae were placed on the surface of a Petri dish containing 2.5% agarose in seawater, immobilized by removing excess water with a paper towel, and injected into the body cavity, using a Narishige MN-151 micro-manipulator. The injection mix was prepared by adding an equal volume of Phenol Red (Sigma) to a solution containing 1 mg/ml dsRNA dissolved in water. The mix was centrifuged briefly to remove traces of solid materials. Approximately 5 ng dsRNA was injected per larva. The injected larvae were cultured for 1–2 weeks before collection and fixing.
Tribolium Culture and Microinjections. Tribolium were injected at pupal stages and reared as described in refs. 23–25.
Western Blots on Whole-Protein Extracts from Artemia. Three wild-type or phenotypically affected caudal RNAi Artemia of comparable developmental stage were used to prepare the sample loaded on each lane. The Artemia were fixed in 4% formaldehyde, washed in methanol, and homogenized by grinding in boiling SDS denaturing loading buffer. The extract was centrifuged briefly, and the supernatant was loaded on a 12.5% denaturing acrylamide gel. After electrophoresis the samples were transferred onto a Protran membrane (Schleicher & Schuell) and probed with an affinity-purified anti-AfCad antibody (20) at 1:3,000 dilution. Subsequently, the membrane was washed a few times in PTX (PBS with 0.1% Triton X-100) and reprobed with the E7 anti-β-tubulin monoclonal antibody (Developmental Studies Hybridoma Bank, Iowa City, IA) at 1:20,000 dilution.
Antibodies and Immunochemical Stainings. Immunochemical stainings in Artemia were carried out using specific polyclonal antibodies for Caudal and Eve (20), the monoclonal antibody 4F11 for En (26), and the monoclonal antibody FP6.87 for Ubx/AbdA (27). Whole-mount immunochemical stainings were carried out according to standard protocols (28), with sonication of varying strengths, depending on the developmental stage.
Immunochemical stainings in Tribolium were carried out using a specific polyclonal antibody for Caudal (16), the monoclonal antibody 4D9 for En (26), and the monoclonal antibody 2D8 for Eve (29) (Developmental Studies Hybridoma Bank).
Detection of Cell Proliferation and Apoptosis. Cell proliferation was detected by BrdUrd incorporation. Artemia larvae were fed with 0.2 mg/ml BrdUrd diluted in seawater for 2.5–3 h. After feeding, the larvae were washed extensively in seawater, fixed in 4% formaldehyde in seawater, washed extensively in methanol, washed in HCl/Triton solution (2.2 M HCl/0.1% Triton X-100), and processed according to standard immunochemical staining procedures, using an anti-BrdUrd monoclonal antibody (Becton Dickinson) at 1:50 dilution. Apoptosis was detected by using the In situ Cell Death Detection Kit, TMR Red (Roche). After fixation and methanol washes, the larvae were washed well in PTX (PBS with 0.1% Triton X-100) and incubated in the blocking solution (0.1 M Tris, pH 7.5/3% BSA/5% normal goat serum) for 1 h at room temperature. The larvae were then washed in PTX and incubated in the TUNEL enzyme reaction for 1–3 h at 37°C.
Results and Discussion
caudal Is Expressed in the Growth Zone of Tribolium and Artemia. In Tribolium, caudal is uniformly expressed in the early blastoderm (likely to be of maternal origin), but as the blastoderm matures this expression changes into a posterior to anterior protein gradient that spans the anlagen of the gnathal and thoracic segments, and the growth zone (Fig. 1 A and B) (16). After the beginning of germ-band elongation, caudal expression becomes restricted to the growth zone and persists there until segmentation is completed (Fig. 1 C–E) (16, 17). Although the blastoderm phase of caudal expression in Tribolium is topologically similar to caudal expression in the Drosophila blastoderm, the expression domain covers different embryonic primordia. The later phase of expression in the growth zone cannot be easily compared to any aspect of caudal expression in Drosophila and seems to be associated specifically with short-germ development.
In Artemia, caudal is expressed in the growth zone starting from the earliest larval stages and persisting there until the last body segment is formed (20). The expression is restricted to the ectoderm and coincides with the elongation of the body axis and generation of all trunk segments from the growth zone (Fig. 1 F–K).
RNAi Can Be Applied to Obtain Specific Phenotypes in Artemia. The use of RNAi to inactivate genes appears to be applicable to a wide range of species (30, 31). In Tribolium RNAi has been successfully used to study the function of developmental genes. It has been shown to be effective when dsRNA is injected either into early embryos (embryonic RNAi) or into their mothers (parental RNAi). In the case of parental RNAi, it is likely that both maternal and zygotic mRNAs are effectively targeted (23, 24).
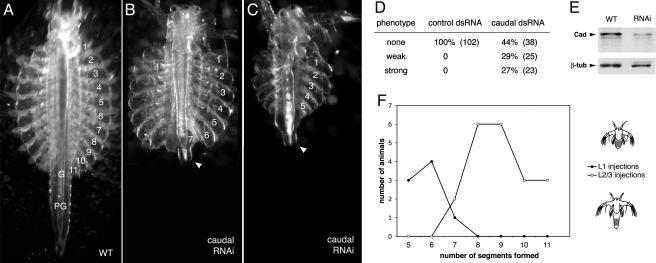
To study the function of caudal in Artemia, we developed a protocol for microinjecting dsRNA into the haemocoel of young larvae (stage L1–3) and observing the phenotype some days later. The technique is quite efficient and very specific for the injected RNA. Up to 50% of surviving larvae injected with caudal dsRNA show a strong phenotype, whereas injections with buffer or with an unrelated, control dsRNA give no specific effects (Fig. 3D).
Fig. 3.
caudal RNAi phenotype in Artemia.(A) Morphology of a normal fully segmented Artemia, with 11 thoracic segments (numbered), 2 genital segments (G), and 6 postgenital segments (PG). (B and C) Morphology of caudal RNAi-treated Artemia, showing severe truncations that remove many thoracic, genital, and postgenital segments. The telson/anus at the posterior end of the body is still present (arrowhead); the affected individuals are viable. (D) Frequency of phenotypes obtained in a typical RNAi experiment. Injection of a control dsRNA gives rise to 100% normal, fully segmented larvae. Injection of caudal dsRNAs gives rise to ≈30% larvae with severe truncations, in addition to larvae with minor phenotypes (few posterior segments missing, misshaped segments) and normal-looking larvae. Absolute numbers of surviving larvae are indicated in parentheses. (E) Western blots with extracts from wild-type and caudal RNAi-affected individuals, probed with an anti-Caudal and an anti-β-tubulin (control) antibody. Quantification of these bands shows that caudal RNAi has caused a 3- to 4-fold reduction in the levels of Caudal protein. (F) Graph depicting the association between the timing of caudal dsRNA injections and the extent of truncations. Individuals injected at the first larval stage (L1) usually develop five to six normal segments (filled circles). Individuals injected about a day later (stage L2–L3) develop a longer series of normal segments (open circles).
To examine the extent of gene inactivation caused by RNAi, we performed Western blots with whole-protein extracts from caudal dsRNA injected and uninjected larvae and probed them with an anti-Caudal antibody. We observe a 3- to 4-fold reduction in Caudal protein levels in animals that show a caudal RNAi phenotype, compared with uninjected controls (Fig. 3E). Thus, the phenotype we are observing results from a reduction in Caudal protein levels, but not from complete inactivation of gene expression. We have also performed whole-mount antibody stainings on individuals injected with caudal dsRNA and observed no strong Caudal staining among the treated animals (data not shown). These results show that RNAi in Artemia larvae is a powerful tool to reduce gene expression and to obtain specific phenotypes for the targeted genes.
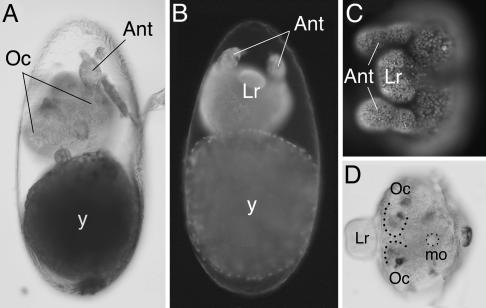
caudal RNAi Causes Severe Truncations in Tribolium and Artemia. Tribolium and Artemia injected with caudal dsRNA gave rise to individuals with severely truncated bodies. In Tribolium, caudal parental RNAi eliminated most body segments (including the gnathal, thoracic, and abdominal segments), giving rise to embryos that developed only the most anterior head segments (Fig. 2A). The morphology of these embryos was examined by DAPI staining to visualize nuclei or by immunohistochemical staining with the segmental marker Engrailed. The stainings confirm that Tribolium caudal RNAi individuals consist only of the pregnathal head, bearing the labrum, antennae, and eyes (Fig. 2 B–D). The same phenotype was consistently observed in all embryos derived from mothers injected with caudal dsRNA but was never seen when injections were carried out with buffer or other dsRNA molecules (24).
Fig. 2.
caudal RNAi phenotype in Tribolium. (A) Late embryo showing the caudal RNAi phenotype. Only the antenna (Ant) and pigmented eyes (Oc) can be identified. All of the gnathal, thoracic, and abdominal segments are missing. The yolk (y) lies posterior to the truncated embryo. (B) caudal RNAi embryo stained with DAPI. The labrum (Lr) and antennae (Ant) are visible anterior to the yolk (y). (C) Detail of DAPI-stained caudal RNAi embryo, focusing on the head. (D) caudal RNAi embryo immunochemically stained for the expression of Engrailed. The antennae (Ant) and the position of the mouth (mo) are outlined. The ocular segments (Oc) and the labrum (Lr) are also visible.
caudal RNAi in Artemia causes similar body truncations. The affected larvae have a normal head and anterior thoracic segments but fail to develop posterior thoracic, genital, and postgenital segments (Fig. 3 A–C). Because the injections of dsRNA in Artemia are carried out during early larval stages, RNAi is probably starting to have an effect after the first thoracic segments are already formed. Indeed, we observed that the extent of truncations varies depending on the larval stages at which injections were made (Fig. 3F). Injections at the earliest larval stage (L1) give rise to individuals with five to six unaffected thoracic segments, whereas injections carried out 1 day later (stage L2–L3) give rise to individuals with more unaffected segments, usually eight to nine. Thus, the severity of the phenotype reflects the onset of RNAi. We conclude that caudal is most likely required for the generation of all of the segments that arise from the growth zone, but the timing of injections and onset of RNAi in Artemia do not allow us to detect an effect in the head and most anterior thoracic segments.
Unlike Drosophila, where caudal is required exclusively in posterior structures (1), these phenotypes suggest that in Tribolium and Artemia caudal plays an essential role in the development of all trunk segments. The phenotypes are consistent with the caudal expression patterns. In Artemia, all of the affected segments arise from the growth zone that normally expresses caudal. In Tribolium, the lack of gnathal and thoracic segments corresponds to the early phase of expression in the blastoderm, whereas the lack of abdominal segments is linked to the disruption of caudal expression in the growth zone from which these segments arise.
caudal RNAi Truncations Are Caused by an Arrest of Axis Elongation. The truncations observed in Tribolium and Artemia are always associated with a shortening of the overall length of the body. The segments that are formed have a normal size, and the missing segments are not replaced by other structures, except for a mass of disorganized cells in the region that would normally correspond to the growth zone. Thus, caudal RNAi does not seem to cause the transformation of one type of tissue into another but a defect in generating or organizing the cells that would normally be required to form new segments.
To study the actual cause of the truncations we looked for the changes in cell proliferation and programmed cell death (apoptosis) in the growth zone of caudal RNAi Artemia. No obvious changes in cell proliferation or apoptosis (visualized by BrdUrd incorporation and TUNEL staining) were detected in the region of the growth zone (data not shown), suggesting that the observed phenotype is not due to gross changes in cell proliferation and apoptosis. Nevertheless, the region corresponding to the growth zone has an abnormal appearance in RNAi-treated Artemia and Tribolium (see Figs. 3C and 4B). It is probable that these phenotypes are due to a deregulation of processes required for coordinated tissue growth, such as the orientation of growth and the ordered recruitment of cells into segments.
Fig. 4.
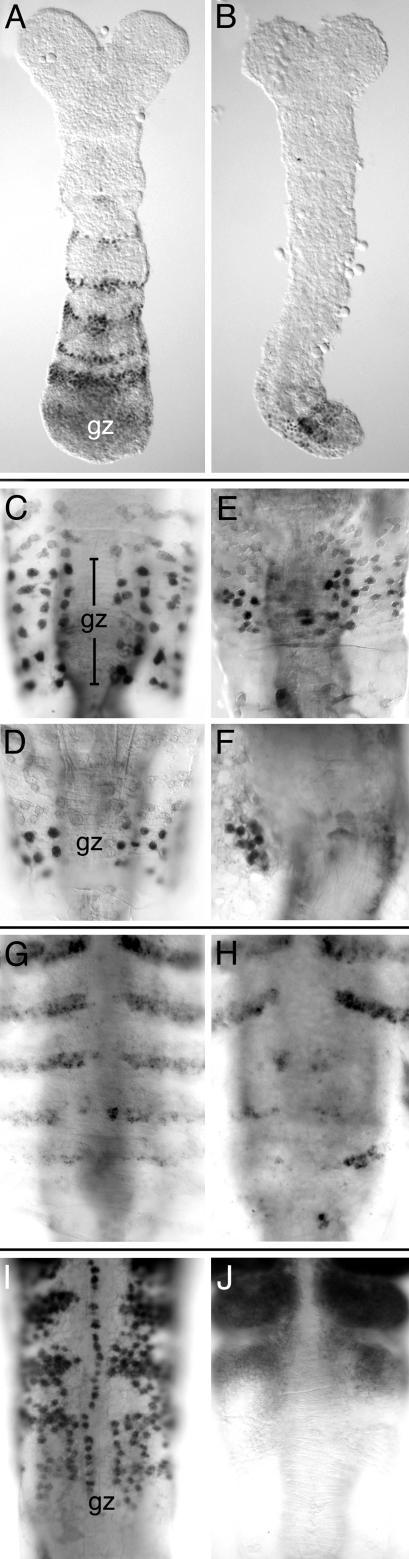
Effects of caudal RNAi on segmentation and Hox gene expression. (A) Wild-type Tribolium embryo immunochemically stained for the expression of Even-skipped (Eve). Expression is detected in the growth zone (gz) and in a series of stripes in the segments that arise from this zone. (B) Eve staining in a caudal RNAi-treated Tribolium embryo. Eve staining is irregular in the region of the growth zone, and the segmental expression is absent. This is a weakly affected embryo that has achieved some degree of axial elongation. (C and D) Eve expression in the growth zone (gz) of normal Artemia larvae. Eve is expressed in a solid band of cells in the growth zone; the band is broad in early stages (C) and becomes narrower with time (D). (E and F) Eve expression in the region of the growth zone of caudal RNAi-treated Artemia larvae. Expression is irregular; in some individuals the band has become discontinuous, with patches of cells not expressing Eve (E), and in others expression is only seen in isolated patches of cells (F). (G) Segmental pattern of Engrailed (En) expression in normal Artemia larva. The growth zone lies posterior to (below) the last stripes of En expression. (H) En expression in caudal RNAi-treated larva. Normal stripes of En expression are seen in mature segments, but the stripes in newly formed segments are replaced by isolated patches of cells expressing En. (I) Ubx/AbdA expression in normal Artemia larva, focusing on the segmentally repeated pattern in new segments arising from the growth zone (gz). As segments mature (toward to top of the panel) the expression becomes stronger and more uniform. (J) Ubx/AbdA expression in caudal RNAi-treated larva, showing strong uniform expression in mature segments but no early segmental expression in the region of the growth zone. Anterior is up in all panels.
caudal RNAi Disrupts the Early Phase of Segmentation and Hox Gene Expression. To further characterize the effects of caudal RNAi at the molecular level, we examined the expression of segmentation and Hox genes during the process of axis elongation and segmentation. In wild-type Tribolium and Artemia, Even-skipped is expressed in the growth zone and in stripes corresponding to newly formed segments (20, 29, 32). In Tribolium showing weak RNAi phenotypes (some elongation has taken place), Even-skipped expression in the region of the growth zone is weak and segmental expression is absent (Fig. 4B). In Artemia, the expression of Even-skipped becomes extremely patchy and disorganized in affected individuals (Fig. 4 E and F), reflecting an abnormal organization inside the growth zone.
Engrailed expression, which marks the posterior compartment of all segments soon after they have emerged from the growth zone (14, 33), is severely disrupted as a result of caudal RNAi. Sharp, newly formed Engrailed stripes are missing and are usually replaced by small and disordered patches of cells expressing the gene (Fig. 4H). Engrailed expression appears to be maintained normally in segments that were already established before the onset of RNAi, suggesting that caudal affects only the early steps of segment formation. These observations confirm at the molecular level what was already observed morphologically, that caudal RNAi severely disrupts the integrity of the growth zone and segment formation.
Hox genes mark the process by which segments acquire their distinct identities in different parts of the body. In normal Artemia development, the Hox genes Ubx and AbdA are expressed in the thoracic/trunk segments (34). This expression is activated very early, coinciding with the earliest signs of segmentation (Fig. 4I). We observe that this early phase of Ubx/AbdA expression is disrupted by caudal RNAi but that later expression in previously established segments is normal (Fig. 4J). This finding suggests that in short-germ arthropods caudal may have a role in regulating the Hox genes.
Ancestral Role of caudal/Cdx Genes in Axis Elongation and Segmentation. We have shown that disrupting the expression of caudal gives rise to very similar phenotypes in an insect and a crustacean, separated by ≈400–500 million years of evolution. The similarity of these phenotypes suggests that caudal genes have a common function in axis elongation and segmentation in diverse short-germ arthropods. Given that caudal is similarly expressed in short-germ insects, crustaceans, and myriapods (15–22), this function of caudal most probably represents an ancestral function, deriving from the common ancestor of all arthropods.
Some interesting parallels can also be found with the expression patterns and functions of caudal homologues in vertebrates, the Cdx genes. Similarly to short-germ arthropods, vertebrate somites are generated sequentially in anterior to posterior sequence from a posteriorly located presomitic zone (35). Cdx genes are expressed in the presomitic mesoderm and are known to play an important role in axis elongation, somitogenesis, and specification of somite identity, by maintaining the self-renewing potential of the presomitic cells and by regulating the expression of Hox genes (5–11). Cdx mutant mice show a reduction of presomitic mesoderm, posterior truncations, abnormal somitogenesis, and homeotic transformations of skeletal structures toward more anterior fates (10, 11). Combined with our results from short-germ arthropods, these observations suggest that caudal/Cdx genes have an evolutionarily conserved role in maintaining the integrity and function of the posterior generative zone, which is necessary for axis elongation and segmentation/somitogenesis. This ancestral role of caudal/Cdx genes is not apparent in the long-germ insect Drosophila and could only be revealed by functional data from short-germ arthropods.
Although substantial differences exist in the way arthropods and vertebrates generate their axial structures (arthropod segments are ectodermal, whereas vertebrate somites are mesodermal), there are important morphogenetic and molecular similarities that are difficult to explain by coincidence alone. In both phyla, axial structures are generated sequentially from a self-renewing posteriorly located population of cells and acquire their identities sequentially, as they emerge from this zone (12–14, 35) [a similar process also occurs in annelids (36)]. At the molecular level, we have shown that caudal/Cdx genes are likely to play a conserved role in this process. In addition, recent studies have argued for a conserved role of the Notch signaling pathway in somitogenesis/segmentation (35, 37), and it is clear that Hox genes play a conserved role in the specification of regional identities of segments and somites (38). These similarities suggest that a common mechanism for the sequential generation of axial structures could be an ancestral feature of all bilaterians.
Acknowledgments
We thank Tanja Mader for technical assistance, Mark Martindale and Tassos Pavlopoulos for help in setting up Artemia microinjections, Yiannis Leivadaras for providing microinjection needles, Nipam Patel and Rob White for providing antibodies, and Michael Akam for comments on the manuscript. This work was supported by a research fellowship from the Irakleitos program of the Greek Ministry of Education (to T.C.), a traveling fellowship from the Boehringer Ingelheim Foundation (to T.C.), a research grant from the Deutsche Forschungsgemeinschaft (to R.S.), and a European Molecular Biology Organization Young Investigators award (to M.A.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: dsRNA, double-stranded RNA; RNAi, RNA interference.
Note Added in Proof. An article by Shinmyo et al. (39) describes similar phenotypes obtained by caudal RNA interference in the cricket Gryllus bimaculatus.
References
- 1.Macdonald, P. M. & Struhl, G. (1986) Nature 324, 537–545. [DOI] [PubMed] [Google Scholar]
- 2.Moreno, E. & Morata, G. (1999) Nature 400, 873–877. [DOI] [PubMed] [Google Scholar]
- 3.Hunter, C. P. & Kenyon, C. (1996) Cell 87, 217–226. [DOI] [PubMed] [Google Scholar]
- 4.Edgar, L. G., Carr, S., Wang, H. & Wood, W. B. (2001) Dev. Biol. 229, 71–88. [DOI] [PubMed] [Google Scholar]
- 5.Meyer, B. I. & Gruss, P. (1993) Development (Cambridge, U.K.) 117, 191–203. [DOI] [PubMed] [Google Scholar]
- 6.Subramanian, V., Meyer, B. I. & Gruss, P. (1995) Cell 83, 641–653. [DOI] [PubMed] [Google Scholar]
- 7.Chawengsaksophak, K., James, R., Hammond, V. E., Köntgen, F. & Beck, F. (1997) Nature 386, 84–87. [DOI] [PubMed] [Google Scholar]
- 8.Marom, K., Shapira, E. & Fainsod, A. (1997) Mech. Dev. 64, 41–52. [DOI] [PubMed] [Google Scholar]
- 9.Epstein, M., Pillemer, G., Yelin, R., Yisraeli, J. K. & Fainsod, A. (1997) Development (Cambridge, U.K.) 124, 3805–3814. [DOI] [PubMed] [Google Scholar]
- 10.van den Akker, E., Forlani, S., Chawengsaksophak, K., de Graaff, W., Beck, F., Meyer, B. I. & Deschamps, J. (2002) Development (Cambridge, U.K.) 129, 2181–2193. [DOI] [PubMed] [Google Scholar]
- 11.Chawengsaksophak, K., de Graaff, W., Rossant, J., Deschamps, J. & Beck, F. (2004) Proc. Natl. Acad. Sci. USA 101, 7641–7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, G. K. & Patel, N. H. (2002) Annu. Rev. Entomol. 47, 669–699. [DOI] [PubMed] [Google Scholar]
- 13.Anderson, D. T. (1967) Aust. J. Zool. 15, 47–91. [Google Scholar]
- 14.Patel, N. H., Kornberg, T. B. & Goodman, C. S. (1989) Development (Cambridge, U.K.) 107, 201–212. [DOI] [PubMed] [Google Scholar]
- 15.Xu, X., Xu, P.-X. & Suzuki, Y. (1994) Development (Cambridge, U.K.) 120, 277–285. [DOI] [PubMed] [Google Scholar]
- 16.Schulz, C., Schröder, R., Hausdorf, B., Wolff, C. & Tautz, D. (1998) Dev. Genes Evol. 208, 283–289. [DOI] [PubMed] [Google Scholar]
- 17.Schroder, R., Eckert, C., Wolff, C. & Tautz, D. (2000) Proc. Natl. Acad. Sci. USA 97, 6591–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dearden, P. K. & Akam, M. (2001) Development (Cambridge, U.K.) 128, 3435–3444. [DOI] [PubMed] [Google Scholar]
- 19.Rabet, N., Gibert, J.-M., Quéinnec, É., Deutsch, J. S. & Mouchel-Vielh, E. (2001) Dev. Genes Evol. 211, 172–178. [DOI] [PubMed] [Google Scholar]
- 20.Copf, T., Rabet, N., Celniker, S. E. & Averof, M. (2003) Development (Cambridge, U.K.) 130, 5915–5927. [DOI] [PubMed] [Google Scholar]
- 21.Akiyama-Oda, Y. & Oda, H. (2003) Development (Cambridge, U.K.) 130, 1735–1747. [DOI] [PubMed] [Google Scholar]
- 22.Chipman, A. D., Arthur, W. & Akam, M. (2004) Curr. Biol. 14, 1250–1255. [DOI] [PubMed] [Google Scholar]
- 23.Bucher, G., Scholten, J. & Klingler, M. (2002) Curr. Biol. 12, R85–R86. [DOI] [PubMed] [Google Scholar]
- 24.Schröder, R. (2003) Nature 422, 621–625. [DOI] [PubMed] [Google Scholar]
- 25.Berghammer, A., Bucher, G., Maderspacher, F. & Klingler, M. (1999) Dev. Genes Evol. 209, 382–389. [DOI] [PubMed] [Google Scholar]
- 26.Patel, N. H., Martin-Blanco, E., Coleman, K. G., Poole, S. J., Ellis, M. C., Kornberg, T. B. & Goodman, C. S. (1989) Cell 58, 955–968. [DOI] [PubMed] [Google Scholar]
- 27.Kelsh, R., Weinzieri, R. O., White, R. A. & Akam, M. (1994) Dev. Genet. 15, 19–31. [DOI] [PubMed] [Google Scholar]
- 28.Patel, N. H. (1994) Methods Cell Biol. 44, 445–487. [DOI] [PubMed] [Google Scholar]
- 29.Patel, N. H., Condron, B. G. & Zinn, K. (1994) Nature 367, 429–434. [DOI] [PubMed] [Google Scholar]
- 30.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 31.Zamore, P. D. (2001) Nat. Struct. Biol. 8, 746–750. [DOI] [PubMed] [Google Scholar]
- 32.Brown, S. J., Parrish, J. K., Beeman, R. W. & Denell, R. E. (1997) Mech. Dev. 61, 165–173. [DOI] [PubMed] [Google Scholar]
- 33.Manzanares, M., Marco, R. & Garesse, R. (1993) Development (Cambridge, U.K.) 118, 1209–1219. [DOI] [PubMed] [Google Scholar]
- 34.Averof, M. & Akam, M. (1995) Nature 376, 420–423. [DOI] [PubMed] [Google Scholar]
- 35.Pourquie, O. (2003) Int. J. Dev. Biol. 47, 597–603. [PubMed] [Google Scholar]
- 36.Irvine, S. M. & Martindale, M. Q. (1996) Semin. Cell Dev. Biol. 7, 593–604. [Google Scholar]
- 37.Stollewerk, A., Schoppmeier, M. & Damen, W. G. (2003) Nature 423, 863–865. [DOI] [PubMed] [Google Scholar]
- 38.McGinnis, W. & Krumlauf, R. (1992) Cell 68, 283–302. [DOI] [PubMed] [Google Scholar]
- 39.Shinmyo, Y., Mito, T., Matsushita, T., Sarashina, I., Miyawaki, K., Ohuchi, H. & Noji, S. (2004) Mech. Dev., in press. [DOI] [PubMed]