Abstract
Cyanobacteria possess a highly effective CO2-concentrating mechanism that elevates CO2 concentrations around the primary carboxylase, Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase). This CO2-concentrating mechanism incorporates light-dependent, active uptake systems for CO2 and HCO–3. Through mutant studies in a coastal marine cyanobacterium, Synechococcus sp. strain PCC7002, we identified bicA as a gene that encodes a class of HCO–3 transporter with relatively low transport affinity, but high flux rate. BicA is widely represented in genomes of oceanic cyanobacteria and belongs to a large family of eukaryotic and prokaryotic transporters presently annotated as sulfate transporters or permeases in many bacteria (SulP family). Further gain-of-function experiments in the freshwater cyanobacterium Synechococcus PCC7942 revealed that bicA expression alone is sufficient to confer a Na+-dependent, 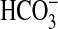 uptake activity. We identified and characterized three cyanobacterial BicA transporters in this manner, including one from the ecologically important oceanic strain, Synechococcus WH8102. This study presents functional data concerning prokaryotic members of the SulP transporter family and represents a previously uncharacterized transport function for the family. The discovery of BicA has significant implications for understanding the important contribution of oceanic strains of cyanobacteria to global CO2 sequestration processes.
uptake activity. We identified and characterized three cyanobacterial BicA transporters in this manner, including one from the ecologically important oceanic strain, Synechococcus WH8102. This study presents functional data concerning prokaryotic members of the SulP transporter family and represents a previously uncharacterized transport function for the family. The discovery of BicA has significant implications for understanding the important contribution of oceanic strains of cyanobacteria to global CO2 sequestration processes.
Keywords: SulP transporters, CO2 sequestration, photosynthesis
It is estimated that some 50% of global primary productivity occurs in the oceans, and marine cyanobacteria contribute significantly to this global CO2 sequestration process (1). For example, in open oceans located between 40°N and 40°S, photosynthetic CO2 fixation is dominated by marine cyanobacteria of the Synechococcus and Prochlorococcus genera, and together these species perform 30–80% of primary production (2, 3). The largest nutrient uptake flux encountered by marine cyanobacteria is for dissolved inorganic carbon (Ci), yet relatively little is known about the process of Ci accumulation for photosynthesis in the oceanic cyanobacteria, in contrast to our knowledge of freshwater strains.
In aquatic systems, Ci exists mainly as two slowly inter-convertible forms, CO2 and 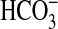 . In response to unique restrictions on the rate of CO2 supply in the aquatic environment, cyanobacteria have evolved a very efficient mechanism for capturing CO2 and HCO–3 for photosynthetic fixation into sugars. The Ci-capturing mechanism functions as a CO2-concentrating mechanism because it effectively concentrates CO2 around the main CO2-fixing enzyme, ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). In the best characterized species, mostly freshwater cyanobacterial strains, the CO2-concentrating mechanism consists of several active uptake systems for CO2 and
. In response to unique restrictions on the rate of CO2 supply in the aquatic environment, cyanobacteria have evolved a very efficient mechanism for capturing CO2 and HCO–3 for photosynthetic fixation into sugars. The Ci-capturing mechanism functions as a CO2-concentrating mechanism because it effectively concentrates CO2 around the main CO2-fixing enzyme, ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). In the best characterized species, mostly freshwater cyanobacterial strains, the CO2-concentrating mechanism consists of several active uptake systems for CO2 and  , plus a unique microcompartment called the carboxysome that contains the CO2-fixing enzyme, Rubisco (4–6).
, plus a unique microcompartment called the carboxysome that contains the CO2-fixing enzyme, Rubisco (4–6).
In recent years, the availability of a number of complete genomic databases for a range of cyanobacteria has made it possible to identify likely homologs of known Ci transporters in marine species of cyanobacteria (7, 8). An interesting observation is that marine cyanobacteria do not seem to possess the high affinity 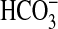 transporter (encoded by cmpABCD, a traffic ATPase) that is present in many freshwater species (7). Potential homologs of the Na+-dependent, SbtA-type
transporter (encoded by cmpABCD, a traffic ATPase) that is present in many freshwater species (7). Potential homologs of the Na+-dependent, SbtA-type 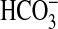 transporter have been noted as present in some oceanic strains (7, 8), but they share low sequence homology with SbtA proteins from Synechocystis PCC6803 (freshwater) and, as yet, have not been proven to transport
transporter have been noted as present in some oceanic strains (7, 8), but they share low sequence homology with SbtA proteins from Synechocystis PCC6803 (freshwater) and, as yet, have not been proven to transport 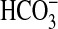 .
.
In this study, we have identified a previously undiscovered class of 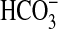 transporter, here named BicA, that is well represented in the genomes of oceanic cyanobacteria. BicA belongs to a large family of eukaryotic and prokaryotic anion transporters (9) where many of the eubacterial members are presently annotated as putative sulfate transporters (TC 2.A.53). Here, we report a previously uncharacterized function for a prokaryotic member of the SulP transporter family.
transporter, here named BicA, that is well represented in the genomes of oceanic cyanobacteria. BicA belongs to a large family of eukaryotic and prokaryotic anion transporters (9) where many of the eubacterial members are presently annotated as putative sulfate transporters (TC 2.A.53). Here, we report a previously uncharacterized function for a prokaryotic member of the SulP transporter family.
Materials and Methods
Cyanobacterial Strains and Culture Conditions. Synechococcus PCC7942 was cultured as described (10). Synechococcus PCC7002 was cultured in BG-11 medium supplemented with 16 μg·liter–1 biotin, 20 mM MgSO4, 8 mM KCl, and 300 mM NaCl, at 28°C with a light intensity of 120 μmol photons·m–2·s–1. Aeration of both strains was delivered through pipettes (1-mm annulus) at a flow rate of 0.15 liters·min–1.
Gene Inactivation in Synechococcus PCC7002. Genes for sbtA, sul2, and bicA (sul3), with flanking regions of 0.5–1.0 kb, were PCR amplified from Synechococcus PCC7002 genomic DNA and ligated into pGEM-T (Promega). Sequence data were obtained from the draft genome sequence for Synechococcus PCC7002 (GenBank genome NC_003488) compiled by D. A. Bryant and J. Zhou (Fig. 7, which is published as supporting information on the PNAS web site, shows the BicA protein sequence); primer sequences are included in Table 4, which is published as supporting information on the PNAS web site. The drug resistance markers kanamycin [Kan (11)], chloramphenicol [Cm (12)], and spectinomycin [Sp (13)] were cloned in the HincII site of sbtA, BalI-HpaI deletion in sul2, and EcoRI-BalI deletion of bicA (sul3), respectively, in parallel to the direction of transcription. Transformation of Synechococcus PCC7002 cells was as described (11). Levels of Kan, Cm, and Sp were 200, 8, and 10 μg·ml–1 in solid media and 150, 7, and 5 μg·ml–1 in liquid media.
Expression Constructs for Synechococcus PCC7942. Coding regions for bicA (sul3) and sul2 from Synechococcus PCC7002 (bicA-7002 and sul2-7002), Synechococcus WH8102 (bicA-WH and sul2-WH), and Synechocystis PCC6803 (bicA-6803 only) were PCR amplified from genomic DNA, ligated into pGEM-T, verified by DNA sequencing, and then cloned into the shuttle vector pSE4 (14). Sequence coordinates for Synechococcus PCC7002 clones are presented in Table 5, which is published as supporting information on the PNAS web site. Sequences for Synechococcus WH8102 and Synechocystis PCC6803 were obtained from Cyanobase (www.kazusa.or.jp); bicA-WH, sul2-WH, and bicA-6803 have gene identifiers of sll0834, slr0096, and SynW1524. Primer sequences and key cloning sites are shown in Table 6, which is published as supporting information on the PNAS web site. Synechococcus PCC7942 was transformed as described (15) with Sp selection at 10 and 7 μg·ml–1 on solid and liquid media, respectively. Cells usually were grown in the presence of 10 mM NH+4 to limit overexpression.
Physiological Measurements. Prepared cells were analyzed by mass spectrometry as described (10, 16). Assays for net O2 evolution were performed at 30°C at a chlorophyll density of 2 μg·ml–1 in the appropriate version of BG11 medium buffered with 50 mM BisTrisPropane-HCl (pH 8, 9, or 9.3), with 17 mM NaNO3 replaced with 20 mM NaCl. Changes for both O2 and CO2 were monitored at a light intensity of 600 μmol photons·m–2·s–1. Active species uptake of H14CO–3 was carried out at pH 9 (25°C) with 15-s uptake periods (17) terminated by silicone oil centrifugation-filtration (18). KH14CO3 aliquots were added from a 25 mM stock (pH 9.5). The rate of CO2 supply from 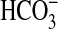 at pH 9 was calculated by applying an experimentally determined rate constant (19) of 0.603 min–1.
at pH 9 was calculated by applying an experimentally determined rate constant (19) of 0.603 min–1.
Real-Time RT-PCR Assays. All Synechococcus PCC7002 sequences were obtained from the draft genome sequence by comparisons with known genes from Synechococcus PCC7942 or Synechocystis PCC6803. Real-time RT-PCR assays using gene-specific primer pairs (Table 5), and with SYBR Green I to monitor product formation, were performed as described by Woodger et al. (10) except that a Rotorgene 3000 (Corbett Research, Sydney, Australia) was used and the accompanying software was used to analyze the second derivative of the raw fluorescence data collected for each sample during a run. Fold changes in transcript abundance relative to a basal condition were calculated after the method of Liu and Saint (20) by using two normalizing genes, rnpA (RNase P) and petB (Cyt b6).
Results
Identification of bicA as a Gene Encoding a Second  Transporter in Synechococcus PCC7002. Initial analysis of an sbtA mutant in Synechococcus PCC7002 grown under Ci-limitation showed a response to added Ci that was intermediate between the responses of WT cells grown under Ci-limitation and Ci-excess (Fig. 1), indicating the presence of another
Transporter in Synechococcus PCC7002. Initial analysis of an sbtA mutant in Synechococcus PCC7002 grown under Ci-limitation showed a response to added Ci that was intermediate between the responses of WT cells grown under Ci-limitation and Ci-excess (Fig. 1), indicating the presence of another 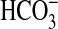 transporter moderate transport of affinity. Marine cyanobacteria so far sequenced (including Synechococcus PCC7002) do not possess a cmpABCD-type
transporter moderate transport of affinity. Marine cyanobacteria so far sequenced (including Synechococcus PCC7002) do not possess a cmpABCD-type  transporter (7). A draft genome sequence for Synechococcus PCC7002 has been constructed by D. A. Bryant and J. Zhou (GenBank genome NC_003488), and our survey of putative anion transporters indicated several potential candidates for another HCO– transporter. Three genes in particular have been annotated as sulfate transporters because of a distant homology to plant sulfate transporters; they were initially referred to as sul1, sul2, and sul3. It was thought significant that sul2 was located midway between the carboxysome-related operon (21) and genes encoding low-affinity CO2 uptake (7, 22, 23) because it is speculated that some CO2-concentrating mechanism-related gene clusters may have been transferred between cyanobacteria by lateral gene transfer (7). Accordingly, the sul2 gene and its closest homolog, sul3, were insertionally inactivated, and the resulting strains were analyzed for defects in
transporter (7). A draft genome sequence for Synechococcus PCC7002 has been constructed by D. A. Bryant and J. Zhou (GenBank genome NC_003488), and our survey of putative anion transporters indicated several potential candidates for another HCO– transporter. Three genes in particular have been annotated as sulfate transporters because of a distant homology to plant sulfate transporters; they were initially referred to as sul1, sul2, and sul3. It was thought significant that sul2 was located midway between the carboxysome-related operon (21) and genes encoding low-affinity CO2 uptake (7, 22, 23) because it is speculated that some CO2-concentrating mechanism-related gene clusters may have been transferred between cyanobacteria by lateral gene transfer (7). Accordingly, the sul2 gene and its closest homolog, sul3, were insertionally inactivated, and the resulting strains were analyzed for defects in 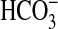 uptake.
uptake.
Fig. 1.
The rate of photosynthetic O2 evolution as a function of added Ci for bicA and sbtA knockout mutants of Synechococcus PCC7002 assayed at pH 9.3. Cells were grown at 2% CO2 and transferred to CO2-free air for 5 h. Typical responses are shown. The response of WT cells grown at 2% CO2 is shown as a dashed line (no symbol).
 uptake capacity was assessed at pH 9.3 as the response of photosynthetic net O2 evolution to added Ci (Fig. 1). Here, it is assumed that net uptake of Ci is 1:1 stoichiometric, with net O2 evolution during steady-state photosynthesis. At pH 9.3, the CO2 concentration at chemical equilibrium represents just 0.018% of total Ci, allowing the contribution of active CO2 uptake to photosynthesis to be minimized relative to
uptake capacity was assessed at pH 9.3 as the response of photosynthetic net O2 evolution to added Ci (Fig. 1). Here, it is assumed that net uptake of Ci is 1:1 stoichiometric, with net O2 evolution during steady-state photosynthesis. At pH 9.3, the CO2 concentration at chemical equilibrium represents just 0.018% of total Ci, allowing the contribution of active CO2 uptake to photosynthesis to be minimized relative to  uptake. Therefore, at this pH, the majority of photosynthetic O2 evolution is related to active, net uptake of
uptake. Therefore, at this pH, the majority of photosynthetic O2 evolution is related to active, net uptake of 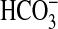 , especially at Ci levels of <1 mM. Carbonate (
, especially at Ci levels of <1 mM. Carbonate ( ), the other Ci species at this pH, is not considered to be a substrate for active uptake, and there is no case in the literature for active
), the other Ci species at this pH, is not considered to be a substrate for active uptake, and there is no case in the literature for active  uptake.
uptake.
Our analysis of the sul3/sbtA double mutant revealed that the sul3 gene (hereafter as bicA for bicarbonate uptake) was found to be associated with expression of a low affinity HCO– uptake activity (Fig. 1 and Table 1). At pH 9.3, the sbtA/bicA 3double mutant, exposed to CO2-free air for 5 h (low-Ci cells), displayed a K0.5(Ci) (the Ci concentration required for half maximal response) of nearly 820 μM, indicating a very low capacity for  uptake. WT cells grown at 2% CO2 had a K0.5(Ci) that was similar to the double mutant (Table 1 and dotted line in Fig. 1). By contrast, low-Ci WT cells displayed a K0.5(Ci) of ≈18 μM, implying a high photosynthetic affinity for
uptake. WT cells grown at 2% CO2 had a K0.5(Ci) that was similar to the double mutant (Table 1 and dotted line in Fig. 1). By contrast, low-Ci WT cells displayed a K0.5(Ci) of ≈18 μM, implying a high photosynthetic affinity for 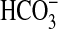 . In the sbtA mutant background, the BicA transporter displayed a K0.5(Ci) of 107 μM, indicative of a transporter with relatively low affinity for
. In the sbtA mutant background, the BicA transporter displayed a K0.5(Ci) of 107 μM, indicative of a transporter with relatively low affinity for  . Adjusted for the expected HCO– concentration salt-water medium, in the BicA transporter would have a K0.5(
. Adjusted for the expected HCO– concentration salt-water medium, in the BicA transporter would have a K0.5(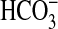 ) of 38 μM (Table 1). Interestingly, the bicA insertional mutant revealed the kinetics of the SbtA transporter to be biphasic, with the first component showing a K0.5(Ci) of 5 μM, saturating near 1 mM Ci (Fig. 1); the second component seems to be responsive to the action of the two CO2 uptake systems, particularly at Ci >1 mM. Importantly, BicA is apparently capable of supporting a relatively high uptake flux for HCO– because, at its intrinsic K0.5(Ci), it supports 50% of the maximum photosynthetic rate, whereas SbtA supports ≈25% of the maximum rate (Fig. 1). Growth of the bicA, sbtA, and sbtA/bicA mutants at high CO2 on plates and in liquid culture was indistinguishable from WT cells (data not shown). The mutants grew normally at pH 9.3 and 7.0 on plates in normal air (data not shown) due to a capacity for active CO2 uptake to support growth. The mutants grew normally in liquid medium at pH 8.0 bubbled with normal air; however, the double mutant grew more slowly than WT at pH 9.3 (data not shown).
) of 38 μM (Table 1). Interestingly, the bicA insertional mutant revealed the kinetics of the SbtA transporter to be biphasic, with the first component showing a K0.5(Ci) of 5 μM, saturating near 1 mM Ci (Fig. 1); the second component seems to be responsive to the action of the two CO2 uptake systems, particularly at Ci >1 mM. Importantly, BicA is apparently capable of supporting a relatively high uptake flux for HCO– because, at its intrinsic K0.5(Ci), it supports 50% of the maximum photosynthetic rate, whereas SbtA supports ≈25% of the maximum rate (Fig. 1). Growth of the bicA, sbtA, and sbtA/bicA mutants at high CO2 on plates and in liquid culture was indistinguishable from WT cells (data not shown). The mutants grew normally at pH 9.3 and 7.0 on plates in normal air (data not shown) due to a capacity for active CO2 uptake to support growth. The mutants grew normally in liquid medium at pH 8.0 bubbled with normal air; however, the double mutant grew more slowly than WT at pH 9.3 (data not shown).
Table 1. Photosynthetic affinities for Ci uptake in Synechococcus PCC7002 mutants determined as O2 evolution responses at pH 9.3 (salt water media).
| High-Ci cells
|
Low-Ci cells
|
||
|---|---|---|---|
| Cell type | K0.5(Ci), μM | K0.5(Ci), μM | K0.5(HCO3-), μM* |
| WT PCC7002 | 720 ± 43 | 17.9 ± 0.4 | 6.5 ± 0.2 |
| bicA (sul3) mutant | 1,263 ± 84 | 5.1 ± 0.2† | 1.8 ± 0.1 |
| sbtA mutant | 768 ± 29 | 107.0 ± 4.1 | 38.4 ± 1.5 |
| bicA/sbtA mutant | 1,960 ± 70 | 820.0 ± 51 | 295.0 ± 18 |
| sul2 mutant | 687 ± 25 | 18.5 ± 0.7 | 6.7 ± 0.3 |
| sul2/sbtA mutant | 700 ± 57 | 114.0 ± 11 | 41.2 ± 4.0 |
Data are shown as means ± SD (n = 3). Maximum photosynthetic rates were similar, with a mean of 846 μmol O2·mg Chl-1·h-1. Low-Ci cells were grown at 2% CO2 and transferred to CO2-free air for 5 h before assay.
For calculation of K0.5(HCO3-), the [HCO3-] at pH 9.3 was taken as 36% of total Ci species at chemical equilibrium.
K0.5(Ci) calculated for a maximum rate at 1 mM Ci (see Fig. 1).
In high-Ci cells, there is evidence that BicA is expressed at low levels because a bicA mutant exhibits a K0.5(Ci) that is greater than for WT cells (i.e., 1,263 vs. 720). Likewise, the SbtA transporter also may be active (Table 1). Consistent with this, sbtA and bicA mRNA were readily detected in high-Ci cells (data not shown). The closest homolog of bicA, namely sul2, does not seem to be associated with any appreciable 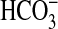 uptake activity because photosynthetic affinities for Ci in the sul2 and sul2/sbtA mutants (low-Ci) were similar to low-Ci WT (Table 1).
uptake activity because photosynthetic affinities for Ci in the sul2 and sul2/sbtA mutants (low-Ci) were similar to low-Ci WT (Table 1).
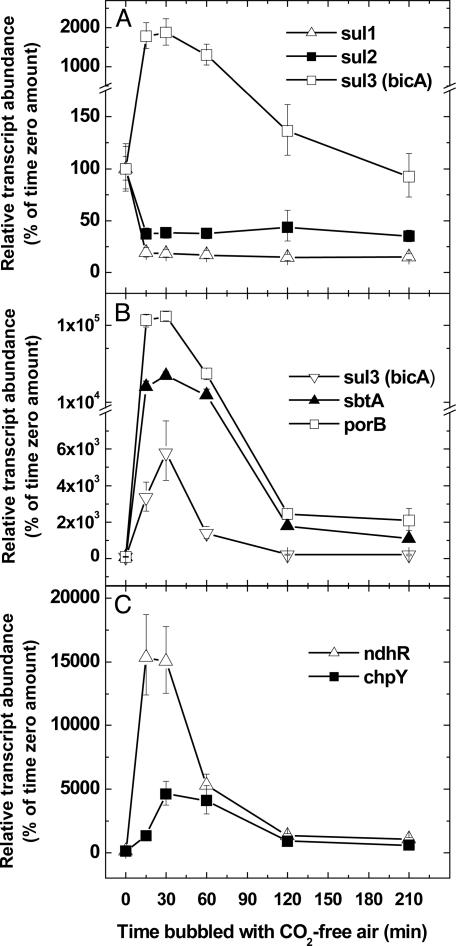
CO2-Responsive Expression of Genes Encoding Ci Transporters in Synechococcus PCC7002. The relative abundance of mRNA transcripts for sul1, sul2, bicA, and known Ci-responsive CO2-concentrating mechanism genes (Ci transporters sbtA and chpY; transcriptional regulator ndhR), as well as a possible  porin that we have termed porB, was determined after Ci-limitation. High-Ci cells of exponentially growing Synechococcus PCC7002 (bubbled with 1.7% CO2) were rapidly transferred to CO2-free air for 3.5 h. First-strand cDNA was generated from normalized total RNA extracts, and quantitative real-time RT-PCR assays were performed by using gene-specific primers and rnpA and petB as reference genes (Fig. 8, which is published as supporting information on the PNAS web site).
porin that we have termed porB, was determined after Ci-limitation. High-Ci cells of exponentially growing Synechococcus PCC7002 (bubbled with 1.7% CO2) were rapidly transferred to CO2-free air for 3.5 h. First-strand cDNA was generated from normalized total RNA extracts, and quantitative real-time RT-PCR assays were performed by using gene-specific primers and rnpA and petB as reference genes (Fig. 8, which is published as supporting information on the PNAS web site).
Consistent with our data that the bicA gene product is a 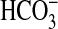 transporter, the expression of bicA mRNA was strongly CO2-responsive (Fig. 2A), increasing by ≈2,000% within 15 min of Ci-limitation, but returning to near basal levels between 90 and 120 min after the transition to Ci-limitation. By contrast, the expression of genes related to bicA, namely sul1 and sul2, was down-regulated under Ci-limitation, suggesting that these genes may encode transporters of nutrients other than Ci. As has been previously observed in Synechococcus PCC7942 or Synechocystis PCC6803, the abundance of ndhR mRNA was also strongly, but transiently, induced upon Ci-limitation whereas transcript levels for sbtA, chpY, and porB (slr0042) were strongly induced, but at more sustained levels (10, 24, 25).
transporter, the expression of bicA mRNA was strongly CO2-responsive (Fig. 2A), increasing by ≈2,000% within 15 min of Ci-limitation, but returning to near basal levels between 90 and 120 min after the transition to Ci-limitation. By contrast, the expression of genes related to bicA, namely sul1 and sul2, was down-regulated under Ci-limitation, suggesting that these genes may encode transporters of nutrients other than Ci. As has been previously observed in Synechococcus PCC7942 or Synechocystis PCC6803, the abundance of ndhR mRNA was also strongly, but transiently, induced upon Ci-limitation whereas transcript levels for sbtA, chpY, and porB (slr0042) were strongly induced, but at more sustained levels (10, 24, 25).
Fig. 2.
CO2-responsive expression of genes encoding putative Ci transporters and the transcriptional regulator ndhR in Synechococcus PCC7002 as determined by real-time RT-PCR. Symbols represent the extent of induction or repression after the shift to low Ci at each time point as a percentage of the high-Ci amount (set at 100%) + SE (n = 3). Representative results from two independent experiments are shown. Note y-axis break in A and B.
Gain-of-Function Expression of bicA and Close Homologs in Synechococcus PCC7942. A gain-of-function approach (17) was used to test whether expression of bicA from a plasmid was sufficient to confer  uptake activity in Synechococcus PCC7942. This host is ideal for this purpose because cells grown at high CO2 possess a very low capacity for
uptake activity in Synechococcus PCC7942. This host is ideal for this purpose because cells grown at high CO2 possess a very low capacity for 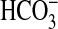 uptake at high pH (Fig. 3) and lack close homologs of bicA. In assays at pH 9.3, expression of bicA-7002 resulted in substantial capacity for photosynthetic Ci uptake, with a K0.5(Ci) of ≈102 μM; this result would equate to K0.5(
uptake at high pH (Fig. 3) and lack close homologs of bicA. In assays at pH 9.3, expression of bicA-7002 resulted in substantial capacity for photosynthetic Ci uptake, with a K0.5(Ci) of ≈102 μM; this result would equate to K0.5(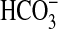 ) of ≈88 μM (Table 2 and Fig. 3). Clearly, expression of bicA-7002 gene product alone is sufficient to confer increased affinity and capacity for
) of ≈88 μM (Table 2 and Fig. 3). Clearly, expression of bicA-7002 gene product alone is sufficient to confer increased affinity and capacity for  uptake.
uptake.
Fig. 3.
The rate of photosynthetic O2 evolution as a function of added Ci for cells of Synechococcus PCC7942 expressing bicA-7002 or bicA-6803 genes. Cells were grown at 2% CO2 and assayed at pH 9.3. Typical responses are shown.
Table 2. Photosynthetic affinities for Ci uptake in cells of Synechococcus PCC7942 expressing bicA or sul2 genes, determined as O2 evolution responses at pH 9.3.
| Expression plasmid | K0.5(Ci), μM | K0.5(HCO3-)*, μM |
|---|---|---|
| No plasmid (WT cells) | 1,185 ± 77 | NA† |
| bicA-PCC7002 | 102 ± 10 | 88 ± 9 |
| bicA-PCC6803 | 198 ± 17 | 171 ± 15 |
| Sul2-PCC7002 | 1,100 ± 60 | NA |
Data are shown as means ± SD (n = 3-5). Maximum photosynthetic rates were similar, with a mean of 550 μmol O2·mg Chl-1·h-1. Cells were grown at 2% CO2 with 10 mM NH4+ in freshwater medium.
For calculation of K0.5(HCO3-), the [HCO3-] at pH 9.3 was taken as 86.3% of total Ci species at chemical equilibrium.
NA, not applicable.
Expression of bicA-6803 in Synechococcus PCC7942 also led to increased photosynthetic Ci uptake activity, but with a lower affinity than for BicA-7002: K0.5(Ci) of 198 μM and K0.5(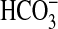 ) of 171 μM (Table 2). Expression of sul2-7002 did not confer Ci uptake activity that differed from the WT response. Expression of bicA-WH led to a low level of Ci uptake activity (not shown) that was difficult to quantify in pH 9.3 assays (see below), but could be demonstrated at pH 8 in the presence of a CO2 uptake inhibitor (Fig. 9, which is published as supporting information on the PNAS web site, and Table 5). BicA-7002 uptake activity was found to be Na+-dependent because the photosynthetic response to Ci was stimulated by NaCl (Fig. 4) but unaffected by comparable additions of KCl (data not shown). Na+ at 1.7 mM Na+ (SD ± 0.1; n = 3) was required for half maximal stimulation; activity was maximal at ≈20 mM NaCl. The BicA-6803 transporter was also found to have a similar Na+-dependence (data not shown). The response to Na+ is similar to the Na+-dependent SbtA transporter from Synechocystis PCC6803 (8). It is possible that BicA (or SbtA) are Na+-driven (e.g., symport), but this conclusion cannot yet be drawn with confidence from any data presented in this paper, or previous experiments, in the case of SbtA (8).
) of 171 μM (Table 2). Expression of sul2-7002 did not confer Ci uptake activity that differed from the WT response. Expression of bicA-WH led to a low level of Ci uptake activity (not shown) that was difficult to quantify in pH 9.3 assays (see below), but could be demonstrated at pH 8 in the presence of a CO2 uptake inhibitor (Fig. 9, which is published as supporting information on the PNAS web site, and Table 5). BicA-7002 uptake activity was found to be Na+-dependent because the photosynthetic response to Ci was stimulated by NaCl (Fig. 4) but unaffected by comparable additions of KCl (data not shown). Na+ at 1.7 mM Na+ (SD ± 0.1; n = 3) was required for half maximal stimulation; activity was maximal at ≈20 mM NaCl. The BicA-6803 transporter was also found to have a similar Na+-dependence (data not shown). The response to Na+ is similar to the Na+-dependent SbtA transporter from Synechocystis PCC6803 (8). It is possible that BicA (or SbtA) are Na+-driven (e.g., symport), but this conclusion cannot yet be drawn with confidence from any data presented in this paper, or previous experiments, in the case of SbtA (8).
Fig. 4.
Na+-dependency of O2 evolution due to bicA-7002 expression in Synechococcus PCC7942 (pH 9.3). Cells were grown at 2% CO2 and washed in standard buffer with 0.1 mM Na+ (pH 9.3). O2 evolution was assayed at a Ci level of 270 μM KHCO3. Typical responses are shown. BicA-7002 responses were subtracted from WT responses to yield a C0.5(Na+) of 1.7 mM (SD = 0.1; n = 3).
Active Species Assays for  Uptake. Active species experiments for H14CO–3 uptake (pH 9 conditions) were undertaken to further test our view that BicA is specific for
Uptake. Active species experiments for H14CO–3 uptake (pH 9 conditions) were undertaken to further test our view that BicA is specific for 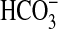 uptake. This view was supported. Experiments used short 15-s uptake periods after addition of H14CO–3 (pH 9.5) followed by rapid termination with silicone-oil centrifugation. Such an approach measures initial rates of gross Ci uptake before chemical equilibrium between
uptake. This view was supported. Experiments used short 15-s uptake periods after addition of H14CO–3 (pH 9.5) followed by rapid termination with silicone-oil centrifugation. Such an approach measures initial rates of gross Ci uptake before chemical equilibrium between 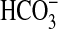 and CO2 is attained. Expression of bicA-7002 or bicA-6803 in Synechococcus PCC7942 led to enhanced rates of gross Ci uptake that approached substrate saturation around 1 mM Ci (Fig. 5). This response is indicative of active uptake of
and CO2 is attained. Expression of bicA-7002 or bicA-6803 in Synechococcus PCC7942 led to enhanced rates of gross Ci uptake that approached substrate saturation around 1 mM Ci (Fig. 5). This response is indicative of active uptake of  because the measured rates of uptake vastly exceeded the calculated maximum rate of CO2 supply from
because the measured rates of uptake vastly exceeded the calculated maximum rate of CO2 supply from  that could support CO2 uptake (Fig. 5A, dotted line). By contrast, the Ci uptake response in WT cells (Fig. 5A), and cells expressing sul2-7002 (data not shown), was low and responded linearly to added Ci; these control rates also indicate a background capacity for
that could support CO2 uptake (Fig. 5A, dotted line). By contrast, the Ci uptake response in WT cells (Fig. 5A), and cells expressing sul2-7002 (data not shown), was low and responded linearly to added Ci; these control rates also indicate a background capacity for 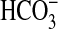 uptake because they were higher than the calculated rate of CO2 supply (Fig. 5A). Interestingly, Km (
uptake because they were higher than the calculated rate of CO2 supply (Fig. 5A). Interestingly, Km (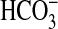 ) values (Fig. 5B and Table 3) were significantly higher than comparable K0.5(
) values (Fig. 5B and Table 3) were significantly higher than comparable K0.5(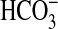 ) values inferred from steady-state photosynthetic measurements (Table 2). This finding might indicate that BicA is slow to respond to added
) values inferred from steady-state photosynthetic measurements (Table 2). This finding might indicate that BicA is slow to respond to added  when using the short uptake approach; it was observed that
when using the short uptake approach; it was observed that  -dependent O2 evolution took 2–3 min to reach steady rates after each Ci increment (data not shown). Largely as expected, gross rates of initial
-dependent O2 evolution took 2–3 min to reach steady rates after each Ci increment (data not shown). Largely as expected, gross rates of initial  uptake exceeded estimates of net
uptake exceeded estimates of net  uptake based on steady-state measurements (Fig. 3); reasons for this result include low 14Ci leakage and lack of feedback regulation during the initial uptake period. For cells washed with 0.1 mM Na+, it was found that
uptake based on steady-state measurements (Fig. 3); reasons for this result include low 14Ci leakage and lack of feedback regulation during the initial uptake period. For cells washed with 0.1 mM Na+, it was found that 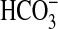 uptake in these assays due to BicA-7002 was stimulated by addition of 5 mM NaCl, but not by 5 mM KCl (data not shown). This finding indicates a potential role for Na+ coupling in
uptake in these assays due to BicA-7002 was stimulated by addition of 5 mM NaCl, but not by 5 mM KCl (data not shown). This finding indicates a potential role for Na+ coupling in 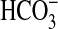 uptake but also could indicate a role in cellular pH regulation.
uptake but also could indicate a role in cellular pH regulation.
Fig. 5.
Assessment of 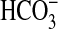 uptake capacity by using silicone oil filtration. (A) Rates of gross 14Ci uptake for bicA expression in Synechococcus PCC7942. Cells were grown at 2% CO2 and assayed at pH 9 by using silicone oil filtration; 15-s uptakes were extrapolated to an hourly basis. Data are shown as mean ± SD (n = 3). The calculated maximum rate of CO2 supply from HCO–3 is plotted as a dotted line. (B) Data for bicA responses replotted after subtraction of WT response and fitted to Michaelis–Menton functions.
uptake capacity by using silicone oil filtration. (A) Rates of gross 14Ci uptake for bicA expression in Synechococcus PCC7942. Cells were grown at 2% CO2 and assayed at pH 9 by using silicone oil filtration; 15-s uptakes were extrapolated to an hourly basis. Data are shown as mean ± SD (n = 3). The calculated maximum rate of CO2 supply from HCO–3 is plotted as a dotted line. (B) Data for bicA responses replotted after subtraction of WT response and fitted to Michaelis–Menton functions.
Table 3. 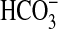 uptake data for cells of Synechococcus PCC7942 expressing bicA genes, determined by short-term uptake of
uptake data for cells of Synechococcus PCC7942 expressing bicA genes, determined by short-term uptake of 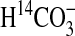 at pH 9 using silicone-oil-filtration.
at pH 9 using silicone-oil-filtration.
| Expression plasmid |
 , μM, for 2% CO2 cells , μM, for 2% CO2 cells |
Vmax (μmol·mg Chl-1·h-1) |
|---|---|---|
| No plasmid (WT) | NA* | NA |
| bicA-PCC7002 | 217 ± 23 | 1,333 ± 148 |
| bicA-PCC6803 | 353 ± 29 | 1,013 ± 73 |
| bicA-WH8102 | 75 ± 4 | 446 ± 36 |
| Sul2-PCC7002 | NA | NA |
Uptake periods of 15 s were employed. Michaelis—Menton parameters were obtained by fitting curves to the data in Fig. 5B. Data are shown as means ± SD (n = 3); cells were grown at 2% CO2.
NA, not applicable for background rates of 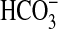 uptake.
uptake.
It is of particular interest that expression of BicA-WH, from the oceanic strain Synechococcus WH8102, displayed  uptake activity with a Vmax that was ≈33% of the BicA-7002 transporter (Fig. 5 and Table 3). Subtraction of the WT control response from the BicA-WH response yields uptake kinetics with a Km(
uptake activity with a Vmax that was ≈33% of the BicA-7002 transporter (Fig. 5 and Table 3). Subtraction of the WT control response from the BicA-WH response yields uptake kinetics with a Km( ) of ≈75 μM. Given that this assay yields higher affinity estimates than steady-state assays, it is possible that BicA-WH may have a K0.5(
) of ≈75 μM. Given that this assay yields higher affinity estimates than steady-state assays, it is possible that BicA-WH may have a K0.5( ) that is significantly greater than 75 μM. There is also the possibility that expression of bicA-WH is not fully activated in the heterologous host. Nevertheless, the data at hand clearly indicate that the BicA homolog from Synechococcus WH8102 is capable of supporting a significant rate of
) that is significantly greater than 75 μM. There is also the possibility that expression of bicA-WH is not fully activated in the heterologous host. Nevertheless, the data at hand clearly indicate that the BicA homolog from Synechococcus WH8102 is capable of supporting a significant rate of 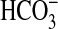 uptake.
uptake.
Discussion
Bicarbonate Transport in Synechococcus PCC7002. Cyanobacteria living in oligotrophic ocean environments play a critical role in global CO2 sequestration and primary photosynthetic productivity (1–3). However, relatively little is known about the genomics and physiology of active uptake processes for dissolved Ci, despite Ci uptake being the largest nutrient influx in these organisms. By contrast, linkages of specific gene products with physiological functions in active Ci uptake are well established in several freshwater cyanobacteria (4–6, 23).
In the present study we have identified BicA as a previously undiscovered class of Na+-dependent  transporter that is functionally active in the coastal marine cyanobacterium, Synechococcus PCC7002. BicA is distinguishable as an extant member of the SulP family of anion transporters in eukaryotes and prokaryotes (9). We also have confirmed that the other
transporter that is functionally active in the coastal marine cyanobacterium, Synechococcus PCC7002. BicA is distinguishable as an extant member of the SulP family of anion transporters in eukaryotes and prokaryotes (9). We also have confirmed that the other 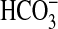 transporter present in Synechococcus PCC7002 is SbtA, an Na+-dependent transporter with high affinity for
transporter present in Synechococcus PCC7002 is SbtA, an Na+-dependent transporter with high affinity for 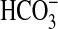 , first identified in Synechocystis (8). Together, BicA and SbtA account for most, if not all, of the
, first identified in Synechocystis (8). Together, BicA and SbtA account for most, if not all, of the 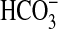 uptake capacity of low-CO2 adapted Synechococcus PCC7002 cells at pH 9.3 (Fig. 1). Gain-of-function analyses (Figs. 3, 4, 5) demonstrated that the single bicA-7002 gene product is sufficient to support Na+-dependent,
uptake capacity of low-CO2 adapted Synechococcus PCC7002 cells at pH 9.3 (Fig. 1). Gain-of-function analyses (Figs. 3, 4, 5) demonstrated that the single bicA-7002 gene product is sufficient to support Na+-dependent, 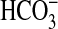 uptake in the freshwater cyanobacterium Synechococcus PCC7942, which lacks any close homologs of bicA. Analysis of gene transcripts in Synechococcus PCC7002 indicates that bicA, sbtA, and porB (coding for a putative porin) are highly up-regulated under Ci limitation (Fig. 2); this finding is consistent with all three genes playing a role in active Ci uptake. Although BicA-7002 has a moderate photosynthetic uptake affinity for
uptake in the freshwater cyanobacterium Synechococcus PCC7942, which lacks any close homologs of bicA. Analysis of gene transcripts in Synechococcus PCC7002 indicates that bicA, sbtA, and porB (coding for a putative porin) are highly up-regulated under Ci limitation (Fig. 2); this finding is consistent with all three genes playing a role in active Ci uptake. Although BicA-7002 has a moderate photosynthetic uptake affinity for 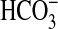 (K0.5 of ≈38 μM), it is able to support a high photosynthetic flux rate, unlike the SbtA transporter, which supports a low flux rate with high uptake affinity (K0.5 < 2 μM). This pairing of complementary transport physiologies may be functionally significant in the natural environment. Indeed, the two
(K0.5 of ≈38 μM), it is able to support a high photosynthetic flux rate, unlike the SbtA transporter, which supports a low flux rate with high uptake affinity (K0.5 < 2 μM). This pairing of complementary transport physiologies may be functionally significant in the natural environment. Indeed, the two  transporters acting together in fully induced WT cells are able to support a high flux rate with a net K0.5(
transporters acting together in fully induced WT cells are able to support a high flux rate with a net K0.5( ) of ≈6.5 μM (Table 1 and Fig. 1). This affinity for
) of ≈6.5 μM (Table 1 and Fig. 1). This affinity for  is similar to that previously measured in Synechococcus PCC7002 (16).
is similar to that previously measured in Synechococcus PCC7002 (16).
BicA in Other Cyanobacteria. BicA proteins are predicted to be plasma membrane-targeted, possessing 8–12 membrane-spanning domains, and bioinformatics searches show them to be members of an extensive anion transporter family that is present in eukaryotes and prokaryotes. BicA appears to be the only prokaryotic member with a proven transport function. In prokaryotes, many of the proteins are presently annotated as sulfate transporters, but without accompanying experimental verification. BicA homologs are highly represented among the 13 cyanobacterial genomes that have so far been completely or partially sequenced. BicA homologs are present in some freshwater strains, but it is of particular interest to note that all of the marine cyanobacteria so far sequenced have one or more BicA homologs that cluster in lobe 1 of the phylogenetic tree shown in Fig. 6. In the present study, we have shown that a functional BicA homolog is present in at least one oceanic strain of cyanobacteria, namely Synechococcus WH8102. The rates of 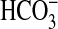 uptake supported by this transporter seem to be relatively low compared with BicA-7002 (33% of BicA-7002 rate; Table 3); however, oceanic strains typically grow at about one doubling per day (or less), compared with Synechococcus PCC7002, which can support doubling times of 4–5 h. Thus, the low rates so far demonstrated for the ectopically expressed BicA-WH transporter may be adequate to support typical rates of carbon gain in the euphotic zone where light intensities are typically 1–2% of full sunlight and carbon gain is light-limited. We speculate that BicA may act as a major route for Ci entry in oceanic cyanobacteria and thus have an important role in global CO2 sequestration. This view requires further testing, despite the difficulties of culturing oceanic cyanobacteria in the laboratory.
uptake supported by this transporter seem to be relatively low compared with BicA-7002 (33% of BicA-7002 rate; Table 3); however, oceanic strains typically grow at about one doubling per day (or less), compared with Synechococcus PCC7002, which can support doubling times of 4–5 h. Thus, the low rates so far demonstrated for the ectopically expressed BicA-WH transporter may be adequate to support typical rates of carbon gain in the euphotic zone where light intensities are typically 1–2% of full sunlight and carbon gain is light-limited. We speculate that BicA may act as a major route for Ci entry in oceanic cyanobacteria and thus have an important role in global CO2 sequestration. This view requires further testing, despite the difficulties of culturing oceanic cyanobacteria in the laboratory.
Fig. 6.
Phylogenetic tree of sulfate transporter-like proteins in cyanobacteria. The clustalw program was used to align the protein sequences, and the tree was generated in the treeview program. Abbreviations for species names: slr/sll, Synechocystis PCC6803; alr/all, Anabaena PCC7120; tlr/tll, Thermosynechococcus BP1; Npun, Nostoc punctiforme; Tery, Trichodesmium erythraeum; PMT, Prochlorococcus marinus MIT9313; PMM, Prochlorococcus marinus MED4; Pro, Prochlorococcus marinus SS120; SynW, Synechococcus WH8102; 7002, Synechococcus PCC7002; Cwat, Crocosphaera watsonii WH8501; Avar, Anabaena variabilis ATCC29413; and Selo, Synechococcus elongatus PCC7942. Marine species are represented by 7002, Tery, Cwat, SynW, Pro, PMT, and PMM.
Synechococcus WH7803 (related to WH8102) exhibits a net efflux of CO2 under steady-state photosynthetic conditions (26), suggesting efficient 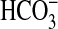 uptake but poor uptake of CO2. A related study (27) found that low-CO2 adapted cells of Synechococcus WH7803 have a good uptake affinity for Ci (K0.5 of 13 μM) and showed rates of photosynthesis up to 70 μmol O2·mg Chl–1·h–1. Synechococcus WH8102 possesses genes for low-affinity CO2 uptake (7) and BicA-type
uptake but poor uptake of CO2. A related study (27) found that low-CO2 adapted cells of Synechococcus WH7803 have a good uptake affinity for Ci (K0.5 of 13 μM) and showed rates of photosynthesis up to 70 μmol O2·mg Chl–1·h–1. Synechococcus WH8102 possesses genes for low-affinity CO2 uptake (7) and BicA-type 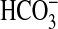 uptake (this study), but lacks genes for high-affinity CO2 uptake and SbtA-type
uptake (this study), but lacks genes for high-affinity CO2 uptake and SbtA-type 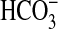 uptake (7). By contrast, the Prochlorococcus species lack all known genes specific to either CO2 uptake system, but possess a distant homolog of SbtA (7, 8) and close homologs of BicA (Fig. 6). It remains to be seen whether these distant homologs of SbtA are able to transport
uptake (7). By contrast, the Prochlorococcus species lack all known genes specific to either CO2 uptake system, but possess a distant homolog of SbtA (7, 8) and close homologs of BicA (Fig. 6). It remains to be seen whether these distant homologs of SbtA are able to transport  , but here it is expected that the Synechococcus PCC7942 expression system will be useful. In summary, there is potential for BicA transporters to feature significantly in Ci uptake in oceanic Synechococcus spp. and Prochlorococcus spp. as well as other marine species such as Crocosphaera watsonii WH8501 and Trichodesmium erythraeum (Fig. 6).
, but here it is expected that the Synechococcus PCC7942 expression system will be useful. In summary, there is potential for BicA transporters to feature significantly in Ci uptake in oceanic Synechococcus spp. and Prochlorococcus spp. as well as other marine species such as Crocosphaera watsonii WH8501 and Trichodesmium erythraeum (Fig. 6).
It is interesting that the BicA homolog from Synechocystis PCC6803 transports 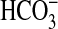 when expressed in Synechococcus PCC7942. This finding implies that BicA-6803 is an active transporter in Synechocystis. It is known that the gene is not up-regulated under Ci limitation (24), except for a modest up-regulation in an ndhR regulatory mutant, implying that the gene is normally constitutively expressed. This result would be consistent with the observation that high-CO2 grown cells possess constitutive
when expressed in Synechococcus PCC7942. This finding implies that BicA-6803 is an active transporter in Synechocystis. It is known that the gene is not up-regulated under Ci limitation (24), except for a modest up-regulation in an ndhR regulatory mutant, implying that the gene is normally constitutively expressed. This result would be consistent with the observation that high-CO2 grown cells possess constitutive  uptake (28). Other physiological evidence (see figure 4C of ref. 8) is consistent with the presence of significant
uptake (28). Other physiological evidence (see figure 4C of ref. 8) is consistent with the presence of significant 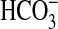 uptake capacity in a sbtA/cmpA deletion mutant (lacking two known
uptake capacity in a sbtA/cmpA deletion mutant (lacking two known  transporters) of Synechocystis PCC6803, although this result was interpreted initially as periplasmic conversion of
transporters) of Synechocystis PCC6803, although this result was interpreted initially as periplasmic conversion of 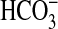 to CO2, followed by CO2 uptake.
to CO2, followed by CO2 uptake.
Wider Implications. Eukaryotic members of the SulP family have quite diverse functions, such as H+/sulfate symport activity in plants and anion exchange in mammals, with different members showing different specificities, e.g.,  /Cl– exchange in kidney cells; some members have been implicated in human genetic diseases, such as Pendred syndrome, diastrophic dysplasia, and congenital chloride diarrhea (29). We have identified a previously unknown transport function for this family by showing that some members of the bacterial sulP family can actively transport
/Cl– exchange in kidney cells; some members have been implicated in human genetic diseases, such as Pendred syndrome, diastrophic dysplasia, and congenital chloride diarrhea (29). We have identified a previously unknown transport function for this family by showing that some members of the bacterial sulP family can actively transport 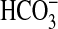 (possibly as Na+/
(possibly as Na+/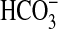 symport); however, given the diversity of functions in eukaryotic members, it is reasonable to expect that the sulP family in prokaryotes can transport or exchange a number of inorganic anions such as sulfate, nitrate, and chloride. There is also scope to identify other sulP members that transport or exchange
symport); however, given the diversity of functions in eukaryotic members, it is reasonable to expect that the sulP family in prokaryotes can transport or exchange a number of inorganic anions such as sulfate, nitrate, and chloride. There is also scope to identify other sulP members that transport or exchange 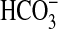 , and it is interesting to note that close homologs of BicA exist in several bacteria. For instance, Vibrio parahaemolyticus (and related Vibrio species) has a homolog that shares 59% identity (over a span of 538 aa) with BicA-7002. This level of homology is similar to the identity between BicA-7002 and BicA-WH (namely 66% over 551 aa), suggesting that Vibrio homologs could act as
, and it is interesting to note that close homologs of BicA exist in several bacteria. For instance, Vibrio parahaemolyticus (and related Vibrio species) has a homolog that shares 59% identity (over a span of 538 aa) with BicA-7002. This level of homology is similar to the identity between BicA-7002 and BicA-WH (namely 66% over 551 aa), suggesting that Vibrio homologs could act as 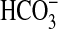 transporters with potential roles in
transporters with potential roles in 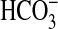 exchange/pH regulation or provision of
exchange/pH regulation or provision of  for anaplerotic metabolism. There is also a BicA homolog in the photosynthetic bacterium Rhodospirillum rubrum (47% identity over 545 aa).
for anaplerotic metabolism. There is also a BicA homolog in the photosynthetic bacterium Rhodospirillum rubrum (47% identity over 545 aa).
Another interesting class of BicA homologs exists in several bacteria, namely Streptomyces coelicolor, Streptomyces avermitilis, Leptospira interrogans, Pirellula sp, Mycobacterium tuberculosis, and Cytophaga hutchinsonii. These bacteria possess homologs that are only 27–29% identical to BicA-7002, but are intriguing in that they possess a C-terminal fusion for a beta carbonic anhydrase protein involved in catalyzed interconversion of CO2 and 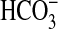 species. This carbonic anhydrase fusion would make logical sense if the transporter domain of the protein were able to transport
species. This carbonic anhydrase fusion would make logical sense if the transporter domain of the protein were able to transport 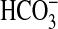 . The physiological function of any such
. The physiological function of any such 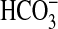 transporter remains unclear at this stage.
transporter remains unclear at this stage.
Supplementary Material
Acknowledgments
We thank Ms. B. Dixon for expert technical assistance. Transcript analyses were supported with funding from an Australian Research Council Discovery Grant (to G.D.P.).
Author contributions: G.D.P. designed research; G.D.P., F.J.W., and L.T. performed research; L.T. contributed new reagents or analytic tools; G.D.P., F.J.W., M.R.B., and S.M.H. analyzed data; and G.D.P., F.J.W., M.R.B., and S.M.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ci, inorganic carbon; K0.5, concentration required for half maximal response.
References
- 1.Field, C. B., Behrenfeld, M. J., Randerson, J. T. & Falkowski, P. (1998) Science 281, 237–240. [DOI] [PubMed] [Google Scholar]
- 2.Liu, H. B., Nolla, H. A. & Campbell, L. (1997) Aquat. Microb. Ecol. 12, 39–47. [Google Scholar]
- 3.Partensky, F., Hess, W. R. & Vaulot, D. (1999) Microbiol. Mol. Biol. Rev. 63, 106–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan, A. & Reinhold, L. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 539–570. [DOI] [PubMed] [Google Scholar]
- 5.Price, G. D., Maeda, S., Omata, T. & Badger, M. R. (2002) Funct. Plant Biol. 29, 131–149. [DOI] [PubMed] [Google Scholar]
- 6.Badger, M. R. & Price, G. D. (2003) J. Exp. Botany 54, 609–622. [DOI] [PubMed] [Google Scholar]
- 7.Badger, M. R., Hanson, D. & Price, G. D. (2002) Funct. Plant Biol. 29, 161–173. [DOI] [PubMed] [Google Scholar]
- 8.Shibata, M., Katoh, H., Sonoda, M., Ohkawa, H., Shimoyama, M., Fukuzawa, H., Kaplan, A. & Ogawa, T. (2002) J. Biol. Chem. 277, 18658–18664. [DOI] [PubMed] [Google Scholar]
- 9.Saier, M. H. (2000) Microbiol. Mol. Biol. Rev. 64, 354–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodger, F. J., Badger, M. R. & Price, G. D. (2003) Plant Physiol. 133, 2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klughammer, B., Sultemeyer, D., Badger, M. R. & Price, G. D. (1999) Mol. Microbiol. 32, 1305–1315. [DOI] [PubMed] [Google Scholar]
- 12.Dzelzkalns, V. A., Owens, G. C. & Bogorad, L. (1984) Nucleic Acids Res. 12, 8917–8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elhai, J. & Wolk, C. P. (1988) Gene 68, 119–138. [DOI] [PubMed] [Google Scholar]
- 14.Maeda, S., Kawaguchi, Y., Ohe, T. & Omata, T. (1998) J. Bacteriol. 180, 4080–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price, G. D. & Badger, M. R. (1989) Plant Physiol. 91, 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sültemeyer, D., Price, G. D., Yu, J. W. & Badger, M. R. (1995) Planta 197, 597–607. [Google Scholar]
- 17.Omata, T., Price, G. D., Badger, M. R., Okamura, M., Gohta, S. & Ogawa, T. (1999) Proc. Natl. Acad. Sci. USA 96, 13571–13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price, G. D. & Badger, M. R. (1989) Plant Physiol. 89, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Badger, M. R., Palmqvist, K. & Yu, J. W. (1994) Physiol. Plant 90, 529–536. [Google Scholar]
- 20.Liu, W. H. & Saint, D. A. (2002) Biochem. Biophys. Res. Commun. 294, 347–353. [DOI] [PubMed] [Google Scholar]
- 21.Ludwig, M., Sultemeyer, D. & Price, G. D. (2000) J. Phycol. 36, 1109–1118. [Google Scholar]
- 22.Maeda, S., Badger, M. R. & Price, G. D. (2002) Mol. Microbiol. 43, 425–435. [DOI] [PubMed] [Google Scholar]
- 23.Shibata, M., Ohkawa, H., Katoh, H., Shimoyama, M. & Ogawa, T. (2002) Funct. Plant Biol. 29, 123–129. [DOI] [PubMed] [Google Scholar]
- 24.Wang, H. L., Postier, B. L. & Burnap, R. L. (2004) J. Biol. Chem. 279, 5739–5751. [DOI] [PubMed] [Google Scholar]
- 25.McGinn, P. J., Price, G. D., Maleszka, R. & Badger, M. R. (2003) Plant Physiol. 132, 218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tchernov, D., Hassidim, M., Vardi, A., Luz, B., Sukenik, A., Reinhold, L. & Kaplan, A. (1998) Can. J. Bot. 76, 949–953. [Google Scholar]
- 27.Hassidim, M., Keren, N., Ohad, I., Reinhold, L. & Kaplan, A. (1997) J. Phycol. 33, 811–817. [Google Scholar]
- 28.Benschop, J. J., Badger, M. R. & Price, G. D. (2003) Photosynth. Res. 77, 117–126. [DOI] [PubMed] [Google Scholar]
- 29.Mount, D. B. & Romero, M. F. (2004) Eur. J. Physiol. 447, 710–721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.