Abstract
Phosphodiesterase (PDE) 8A1 is a cAMP-specific PDE isozyme characterized by the presence of a Per-Arnt-Sim (PAS) domain. However, the function(s) of the PAS domain has remained unknown. In this study, using a lysate of HEK293 cells overexpressing recombinant human PDE8A1, we detected a physical association between PDE8A1 and endogenous IκBβ by an antibody array technique. The association was specific for PDE8A1 and depended on the presence of the PAS domain. Subsequent coimmunoprecipitation experiments revealed that, in addition to IκBβ, other IκB family members examined (p105, p100, and IκBα) also associated with PDE8A1. Furthermore, it was found that PDE8A1 competed with the p65/p50 NF-κB for IκBβ binding. Taken together, these data indicate that PDE8A1, through its PAS domain, may bind with IκB proteins in a region containing their ankyrin repeats. Functionally, in vitro and in vivo experiments demonstrated that the association with IκB greatly enhanced the enzyme activity of PDE8A1. However, the PDE8A1–IκB association did not affect NF-κB activation. The biological role of the PDE8A1–IκB association remains to be elucidated.
Members of phosphodiesterase (PDE, EC 3.1.4.17) super-family, as the only cellular components for the degradation of the second messengers cAMP and cGMP, play a critical biological role (1–4). PDE8 is a recently discovered PDE family, characterized by its specificity for cAMP and the presence of a Per-Arnt-Sim (PAS) domain located in the N-terminal regulatory region (5–7). The PDE8 family comprises two genes: PDE8A and PDE8B. From each gene, multiple alternative splicing variants are derived, with PDE8A1 (6) and PDE8B1 (7) being the dominant variants from the respective genes in terms of expression level. Like other PDEs, PDE8 enzymes have their catalytic domains located toward the C-terminal regions. PDE8A is expressed ubiquitously (6), whereas PDE8B is expressed predominantly in the thyroid gland (7). The biological functions of PDE8 remain to be elucidated.
PAS domains were originally identified and characterized in several proteins in archaeal, eubacterial, and lower eukaryotic organisms and have been proposed to be involved in protein–protein interactions and small-molecule ligand binding (8–11). In many cases, the PAS domains are involved in sensory signal transduction. In eubacteria and plants many PAS domain proteins have been found to be protein kinases. In contrast, in mammals PAS domain proteins with known function frequently act as transcriptional regulators, although several others function as kinases (12) or ion channels (13). Some mammalian PAS domain proteins are involved in the circadian rhythm, cyclic patterns of hormone secretion, breeding, and locomotor activity (14, 15). However, the biological role of the PAS domains of PDE8 enzymes has remained completely unknown.
The transcription factor NF-κB is expressed in most cell types and plays a critical role in activation of a large, ever-increasing number of genes involved in inflammatory and immune responses, apoptosis, differentiation, and growth. NF-κB consists of homodimers or heterodimers of the Rel family proteins p65, p50, p52, c-Rel, and RelB (16–20). p65 and p50 are expressed ubiquitously, whereas the others are expressed in a cell- and tissue-specific pattern. The p65/p50 heterodimer is the most abundant NF-κB in various cell types and plays a more elaborate role than other NF-κB complexes in regulation of gene expression. The Rel proteins also differ in their transactivating abilities. Only p65 and c-Rel function as potent transcriptional activators in various cell types. In contrast, p50 and p52 lack a transactivating activity. The Rel family proteins share a 300-aa region called Rel homology domain, involved in dimerization, DNA binding, and association with the inhibitor proteins IκBs. In resting cells, NF-κB exists in the cytoplasm in an inactive state complexed with an inhibitory IκB protein.
IκB proteins include IκBα, IκBβ, p105, and p100 (16–20). p105 and p100 are precursor molecules of the NF-κB proteins p50 and p52, respectively. The N-terminal regions of p105 and p100 constitute the Rel homology domains of p50 and p52. Proteolytic cleavage of p105 and p100 releases p50 and p52. All IκB proteins contain multiple copies of ankyrin repeat sequences, which are crucial for IκB binding with NF-κB. Individual IκB proteins preferentially inhibit distinct NF-κBs.
In the activation of NF-κB pathways, IκBα is rapidly degraded (within minutes) upon cell stimulation, resulting in release and nuclear translocation of NF-κB to activate gene expression (16–20). IκBα is resynthesized within 1 h. The newly synthesized IκBα translocates to the nucleus and binds to nuclear NF-κB to terminate gene transcription. In contrast, IκBβ is slowly degraded (2 h after stimulation), resulting in persistent activation of NF-κB (>20 h).
In this study, we found that overexpressed recombinant human PDE8A1 physically associated with endogenous IκB proteins. The association may result from an interaction between the PAS domain of PDE8A1 and the ankyrin repeat domains of IκB proteins. Functionally, the IκB binding significantly increased the enzyme activity of PDE8A1. The association with IκB represents a function of the PAS domain of PDE8A1 and a function of a mammalian PAS domain.
Materials and Methods
Antibodies. Rabbit polyclonal antibodies against IκBβ (C-20), p65 (H-286), p100/p52 (K-27), p105/p50 (H-119), c-Rel (N-466), RelB (C-19), and IκBα (C-15) were obtained from Santa Cruz Biotechnology. These antibodies were used directly for immunoprecipitation. In addition, they were labeled with horseradish peroxidase (HRP) by using a Zenon Rabbit IgG Labeling kit (Molecular Probes) for Western blotting. Anti-V5–HRP and anti-Myc–HRP antibodies were from Invitrogen. Anti-V5-conjugated agarose was from Sigma, and anti-Myc-conjugated agarose was from Santa Cruz Biotechnology.
Plasmids. A coding sequence of human PDE4B2 was generated from Marathon-ready brain cDNA (Clontech) by PCR with the primer sets AGTTAAGCTTACCATGAAGGAGCACGGGGGCACCT (sense) and AGTTCTAGACTCTGTATCCACGGGGGACTTGTCT (antisense). A resultant PCR product was digested with HindIII and XbaI, followed by ligation to HindIII/XbaI-digested pcDNA3.1/V5-HisA vector (Invitrogen). Human PDE4D3 was also cloned by PCR from Marathon-ready leukocyte cDNA (Clontech) by using the sense and antisense primers TAGT TA AGCT TGGACCATGATGCACGTGAATAATTTTC and GCAGAATTCCACCGTGTCAGGAGAACGATC, respectively. The vector used was Hin-dIII/EcoRI-digested pcDNA3.1/V5-HisA. An ORF clone (IOH11051, Invitrogen) containing an IκBβ insert was used as a PCR template for cloning IκBβ into HindIII/XbaI-digested pcDNA3.1/V5-HisA vector. The sense and antisense primers used were AGTTAAGCTTACCATGGCTGGGGTCGCGTGC and A AGT TCTAGACTCCACGGGGCGGGGGTCGTCAGGA, respectively. Human PDE9A1 construct was described previously (21). Human PDE8A1 and its PAS-deleted mutant (ΔPAS) were cloned by PCR by using a previously described PDE8A1 construct (6) as a template. For the cloning of PDE8A1, the sense and antisense primers used were CGATGGATCCGA ACCATGGGCTGTGCCCCGAGCAT and CGAGCGGCCGCCTTCAGGAGGTGGTCGGG, respectively. A resultant PCR product was digested with BamHI and NotI and then cloned into BamHI/NotI-digested pcDNA3.1/V5-HisB as well as pcDNA3.1/Myc-HisB. For the cloning of ΔPAS, two separate PCR fragments were first prepared. One fragment was generated by using the above sense primer for PDE8A1 and the antisense primer GTAAATTCCTTGCCACTCCTTGCCTATACAAGCCCTGAGTTTCAG. For the second fragment, a complementary sequence of the above antisense primer for the first fragment was used as sense primer, in combination with the above antisense primer for PDE8A1. The two fragments were combined and amplified by using the above primer set for PDE8A1. A resultant final PCR product was cloned into the two different vectors as with PDE8A1. All plasmid constructs were sequenced by double-stranded sequencing for sequence confirmation.
Cell Culture, Transient and Stable Transfection, and Stimulation. HEK293 cells were obtained from ATCC and maintained in Dulbecco's modified Eagle medium supplemented with 10% FCS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine (all from Invitrogen).
HEK293 cells were transfected by using Lipofectamine 2000 (Invitrogen). For transient transfection, cells were harvested 48 h after transfection. For stable transfection, 48 h after transfection cells were selected in a culture medium containing 0.5 mg/ml G418 (Invitrogen). Cells that survived from the first round of G418 selection were further subjected to limiting dilution for single clone selection.
For analyzing NF-κB activation, cells were stimulated with 20 ng/ml tumor necrosis factor (R & D Systems) for various periods of time.
Cellular Extract Preparation. Cell lysate was prepared either in a buffer containing 15 mM Tris·HCl (pH 7.5), 120 mM NaCl, 25 mM KCl, 2 mM EGTA, 2 mM EDTA, 0.5% Triton X-100, and a Protease Inhibitor mixture (Roche Applied Science, Indianapolis) for antibody array analysis or in a buffer containing 50 mM Tris·HCl (pH 8.0), 150 mM NaCl, 1% Nonidet P-40, and the Protease Inhibitor mixture for immunoprecipitation. Cell lysates were used after centrifugation at 16,000 × g for 10 min to remove particles.
Cytoplasmic and nuclear fractions were prepared by using a Nuclear Extract kit from Active Motif (Carlsbad, CA).
Antibody Array Analysis. An antibody array with 400 immobilized antibodies was obtained from Hypromatrix (Worcester, MA). The array was blocked with a TBST buffer (25 mM Tris·HCl, pH 7.5/150 mM NaCl/0.05% Tween 20) containing 4% BSA for 1 h at room temperature and then was incubated with cell lysate at 0.5 mg/ml (containing 0.5% BSA) for 2 h at room temperature. The membrane was washed three times with the TBST buffer and incubated with anti-V5–HRP for 1 h at room temperature. The membrane was then washed three times, and bound proteins were detected by a chemiluminescence method using a West Pico kit (Pierce, New York).
Quantification of NF-κB Family Members and Recombinant PDE8A1 and ΔPAS in Cellular Extracts. Quantification of various NF-κB family members was performed by ELISA using a TransAM NF-κB Family kit (Active Motif), and the expressed PDE8A1 and ΔPAS were quantified by ELISA as described previously (21).
Immunoprecipitation. One milligram of cell lysate in 1 ml was preincubated with 100 μl of Protein A/G Agarose (50% slurry, Pierce) for3hat 4°C. Two micrograms of an antibody along with 50 μl of Protein A/G Agarose, or 50 μl of antibody-conjugated Protein A/G Agarose, was added to the precleared cell lysate. After incubation overnight at 4°C, the beads were washed three times with the buffer and then resuspended in 1× SDS gel sample buffer for SDS/PAGE. Western blotting was subsequently performed, and coimmunoprecipitated proteins were detected as described above by using appropriate HRP-labeled antibodies.
For PDE8A enzyme assay following immunoprecipitation procedures, the beads were washed three times with PBS and then resuspended in 10 mM Tris·HCl (pH 7.4) containing 1% BSA.
PDE8A Enzyme Assay. The enzyme activity of PDE8A and ΔPAS was assayed by using a [3H]cAMP-PDE Assay kit (Amersham Pharmacia, Arlington Heights, IL) in the presence of 50 nM unlabeled cAMP.
Results
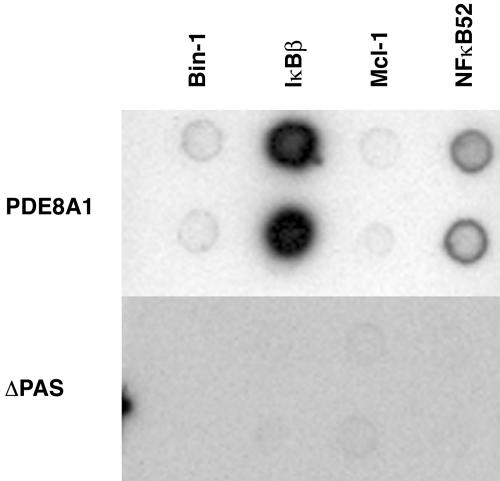
Identification of PAS Domain-Dependent PDE8A1 Association with IκBβ. To identify protein(s) that specifically interact with the PAS domain of human PDE8A1, expression plasmids for PDE8A1–V5 and PAS-deleted PDE8A1 (ΔPAS)–V5 were constructed and used to establish HEK293 stable cell lines. Cell lysates of the HEK293 cells expressing PDE8A1–V5 or ΔPAS–V5 were incubated with a commercially available antibody array containing 400 different antibodies. Proteins interacting with PDE8A1–V5 and/or ΔPAS–V5 were identified by using an anti-V5–HRP antibody. Similar expression levels of PDE8A1–V5 and ΔPAS–V5 were confirmed by Western blotting with an anti-V5–HRP antibody. Several antibodies were identified that detected proteins interacting with PDE8A1 but not ΔPAS. Such antibodies were used subsequently to make a custom array for a second antibody array analysis to confirm the PAS domain-dependent interactions. Fig. 1 shows a representative result using a custom array. Among the proteins identified in the initial screening (including Bin-1, IκBβ, Mcl-1, and p100/p52), IκBβ exhibited a much stronger signal than other proteins, suggesting that PDE8A1 may be associated with IκBβ in cells through the PAS domain. In addition, p100/p52, an IκB–NF-κB family protein (16–20), also showed a significant level of binding, albeit much weaker than that of IκBβ.
Fig. 1.
Detection of PAS-dependent PDE8A1 association with IκBβ by antibody array technique. Antibodies against Bin-1, IκBβ, Mcl-1, and NF-κB p100/p52, which showed specific association with PDE8A1–V5 but not with ΔPAS–V5 in preliminary screening using a commercially available antibody array, were used in this custom array. Cell lysates of HEK293 cells stably expressing PDE8A1–V5 or its PAS-deleted mutant ΔPAS–V5 were incubated with the antibody array, followed by detection of interacting proteins with anti-V5–HRP antibody. Equal expression levels between PDE8A1–V5 and ΔPAS–V5 were confirmed by ELISA (data not shown).
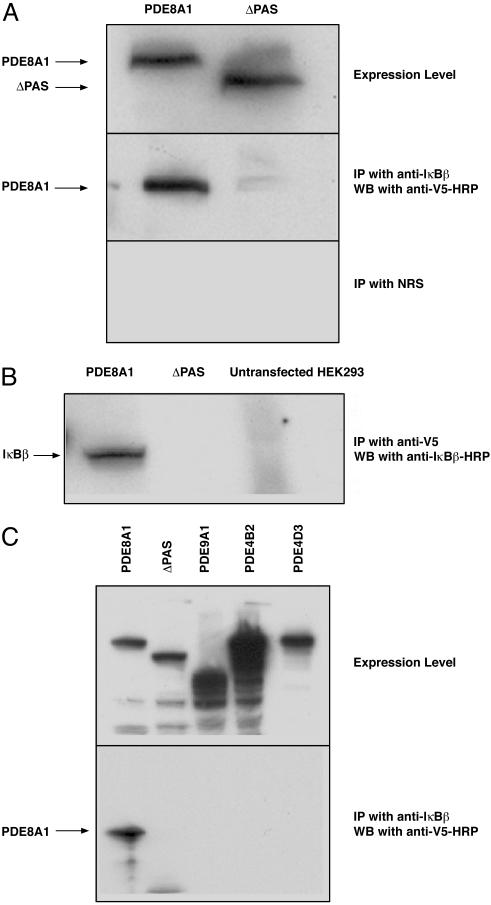
Confirmation of PAS Domain-Dependent, Specific PDE8A1 Association with IκBβ. To further confirm the PAS-dependent binding of PDE8A1 with IκBβ, the cell lysates of PDE8A1–V5- or ΔPAS–V5-expressing cells were subjected to coimmunoprecipitation. As shown in Fig. 2A, using an anti-IκBβ antibody in immunoprecipitation, PDE8A1 was detected with an anti-V5–HRP antibody in Western blot analysis. However, the coimmunoprecipitation signal was absent when cell lysates of ΔPAS–V5-expressing cells or a normal rabbit serum were used in the immunoprecipitation, indicating the requirement of PAS domain for the interaction of PDE8A1 and IκBβ. This interaction was confirmed by using an anti-V5 antibody in the immunoprecipitation and an anti-IκBβ–HRP antibody in Western blot analysis (Fig. 2B).
Fig. 2.
Confirmation of PAS-dependent, specific PDE8A1 association with IκBβ. (A) Cell lysates of HEK293 cells stably expressing PDE8A1–V5 or its PAS-deleted mutant ΔPAS–V5 were subjected to immunoprecipitation (IP) with an anti-IκBβ antibody, followed by Western blotting (WB) with an anti-V5–HRP antibody (Middle). Similar expression levels of PDE8A1–V5 and ΔPAS–V5 are shown in Top. For negative control, normal rabbit serum (NRS) was used as shown in Bottom. (B) Cell lysates of HEK293 cells stably expressing PDE8A1–V5 or ΔPAS–V5 were subjected to immunoprecipitation with an anti-V5 antibody, followed by Western blotting with an anti-IκBβ–HRP antibody. (C) Cell lysates of HEK293 cells stably expressing human PDE9A1–V5, human PDE4B2–V5, or human PDE4D3–V5 were subjected to immunoprecipitation with the anti-IκBβ antibody, followed by Western blotting with the anti-V5 antibody. Expression levels of the various PDE–V5s were confirmed by Western blotting.
To further determine that this association with IκBβ is specific to PDE8A1, other PDEs were examined by using the same strategy. Cell lysates of HEK293 cells stably expressing human PDE9A1–V5, human PDE4B2–V5, or human PDE4D3–V5, all of which lack a PAS domain, were prepared as above. The anti-IκBβ antibody was used for immunoprecipitation, followed by Western blotting with the anti-V5–HRP antibody. The anti-IκBβ antibody was able to coprecipitate PDE8A1 but not any other PDE (Fig. 2C), demonstrating the specific, PAS-dependent association of overexpressed recombinant PDE8A1 with endogenous IκBβ.
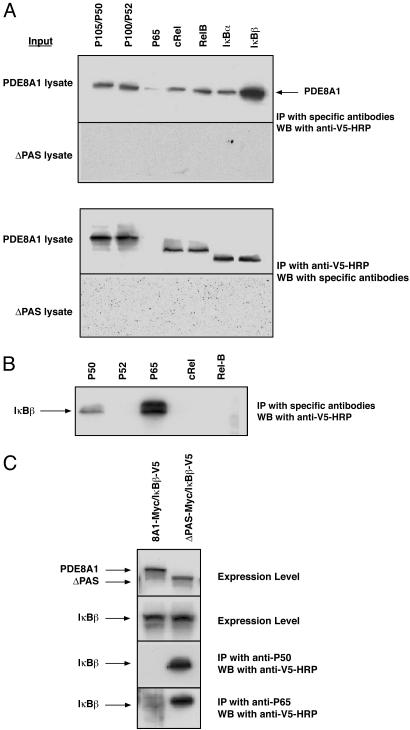
Mode of PDE8A1 Association with IκB. To understand how PDE8A1 associates with IκBβ, the cell lysate of PDE8A1–V5- or ΔPAS–V5-expressing cells was subjected to the immunoprecipitation procedures by using antibodies against various proteins in the IκB–NF-κB family including p105/p50, p100/p52, p65, c-Rel, RelB, IκBα, and IκBβ, followed by Western blotting with the anti-V5–HRP antibody. All antibodies, except for anti-p65 antibody, were able to coimmunoprecipitate PDE8A1 but not ΔPAS. Identical results were obtained by using an anti-V5 antibody for immunoprecipitation and the various anti-IκB–NF-κB antibodies (all labeled with HRP) for Western blot analysis (Fig. 3A). Note that the anti-V5 antibody coimmunoprecipitated p105 and p100 but not p50 and p52 (Fig. 3A Lower).
Fig. 3.
Mode of PDE8A1 association with IκB. (A) Cell lysates of HEK293 cells stably expressing PDE8A1–V5 or its PAS-deleted mutant ΔPAS–V5 were subjected to immunoprecipitation (IP) with antibodies against the various proteins in the IκB–NF-κB family, followed by Western blotting (WB) with an anti-V5–HRP antibody (Upper). Conversely, coimmunoprecipitation was performed with an anti-V5 antibody, followed by Western blotting with various HRP-labeled antibodies against IκB–NF-κB proteins (Lower). Note that the anti-V5 antibody coimmunoprecipitated p105 and p100 but not p50 and p52. (B) Cell lysates of HEK293 cells stably expressing IκBβ–V5 were subjected to immunoprecipitation with the antibodies against various NF-κB proteins, followed by Western blotting with the anti-V5–HRP antibody. (C) PDE8A1–Myc or ΔPAS–Myc was transiently expressed in the IκBβ–V5 stable cell line. Expression levels of PDE8A1–Myc, ΔPAS–Myc, and IκBβ–V5 were confirmed by Western blotting with appropriate HRP-labeled antibodies. Cell lysates of the cells were subjected to immunoprecipitation with the anti-p50 or anti-p65 antibody, followed by Western blotting with the anti-V5–HRP antibody.
The lack of association of p65 with PDE8A1 (Fig. 3A) was an unexpected result, because p65 is the most abundant NF-κB protein (16–20). When a similar experiment was performed by using cell lysates from HEK293 cells stably expressing IκBβ–V5 instead of lysates from PDE8A1–V5-expressing cells, there was a strong signal of p65 detected along with p50 (Fig. 3B). This finding is consistent with the fact that p65–p50 is the predominant NF-κB complex present in cells and bound with IκBβ (16–20). These results suggested that the failure of the anti-p65 antibody to coimmunoprecipitate PDE8A1 in PDE8A1-overexpressing cells might be caused by a competition of PDE8A1 with p65–p50 complexes in binding to IκB. To test this possibility, PDE8A1–Myc or ΔPAS–Myc was transiently expressed in the IκBβ–V5 stable cell line, and immunoprecipitation was performed with the anti-p50 and anti-p65 antibodies, followed by Western blotting with the anti-IκBβ–HRP antibody. Indeed, in the presence of overexpressed PDE8A1, but not of ΔPAS, there was no detectable binding between p50 or p65 and IκBβ, demonstrating that PDE8A1 competes with p65–p50 complexes for IκBβ binding (Fig. 3C). Because p65–p50 complex binds IκBs at their ankyrin repeat domains (16–20), the competition result suggests that PDE8A1, through its PAS domain, may also bind IκBs at the ankyrin repeat domains. This notion is further supported by the observation that all IκB proteins examined were coimmunoprecipitated with PDE8A1 (Fig. 3A).
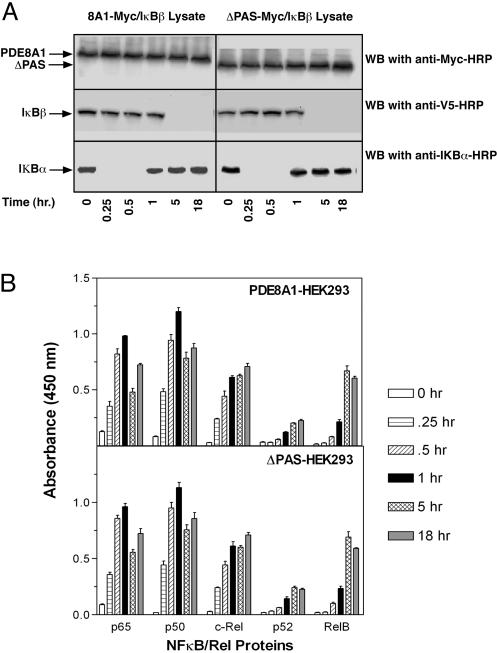
Effect of PDE8A1–IκB Association on NF-κB Activation. To elucidate the biological function(s) of the PDE8A1–IκB association, we first examined whether the association affects NF-κB activation. The IκBβ–V5 stable cell line transiently expressing PDE8A1–Myc or ΔPAS–Myc was stimulated with tumor necrosis factor for various periods of time, and then NF-κB activation was examined by analyzing the degradation of cytoplasmic IκBα and IκBβ–V5 and nuclear translocation of NF-κB proteins. In the cytoplasm of both PDE8A1–Myc- and ΔPAS–Myc-expressing cells, IκBα disappeared at 15 min and reappeared at 1 h, consistent with the literature in nontransfected cells (16–20). From1hto18hafter tumor necrosis factor stimulation, the IκBα level remained unchanged. In contrast, IκBβ–V5 was not degraded over at least the first hour after stimulation. Its degradation was observed only at 5 h after stimulation, and it remained undetectable from 5 h up to 18 h (Fig. 4A). Given the fact that consistent levels of PDE8A1–Myc or ΔPAS–Myc were present at the various time points, the similar signal patterns and intensities of IκBs detected in the two cell lines indicate that the PDE8A1–IκB association does not affect NF-κB activation. With regard to nuclear NF-κB proteins (as reported in refs. 16–20), p65, p50, and c-Rel exhibited a rapid nuclear translocation in relatively larger amounts, whereas p52 and RelB translocated with slower time courses in much smaller amounts (Fig. 4B). Note that for p65, p50, and c-Rel, the nuclear extract was diluted 20-fold for the quantification and hence the actual levels should be 20-fold higher than those shown. Nevertheless, the similar nuclear translocation profile of the NF-κB proteins in the PDE8A1–Myc- and ΔPAS–Myc-expressing cells indicates that the PDE8A1–IκB association does not affect NF-κB activation.
Fig. 4.
Effect of PDE8A1–IκB association on NF-κB activation. (A) HEK293 cells stably expressing IκBβ–V5 were transiently transfected with PDE8A1–Myc or its PAS-deleted mutant ΔPAS–Myc and then were stimulated with tumor necrosis factor for the times indicated. Cytoplasmic IκBβ and IκBα were analyzed by Western blotting (WB) with anti-V5–HRP and anti-IκBα–HRP antibodies, respectively. Similar amounts of PDE8A1–Myc or ΔPAS–Myc present at all time points examined are shown in Top. (B) Nuclear NF-κB proteins were analyzed by ELISA. Note that for p65, p50, and c-Rel, the nuclear extract was diluted 20-fold for the quantification and hence the actual levels should be 20-fold higher than those shown. Each data point represents the mean ± SE (n = 4).
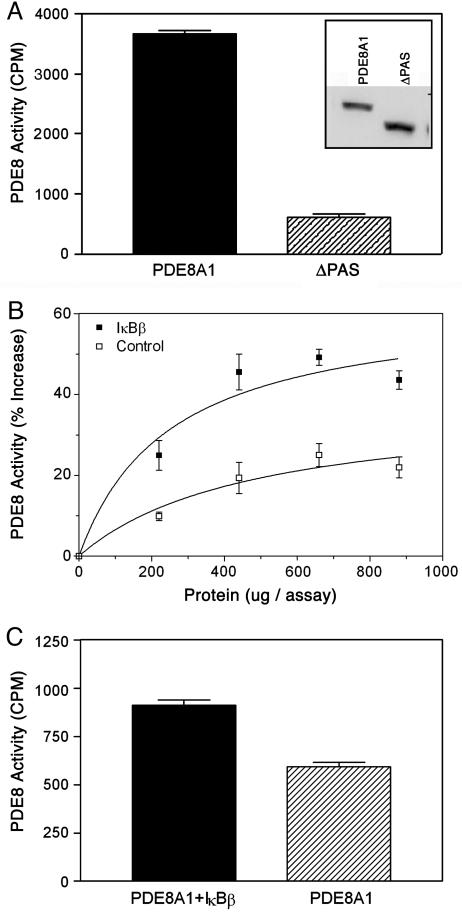
Effect of PDE8A1–IκB Association on PDE8A1 Enzyme Activity. Equal amounts of PDE8A1–V5 and ΔPAS–V5 from the cell lysates as determined by ELISA (data not shown) were subjected to immunoprecipitation with the anti-V5 antibody, followed by PDE8 enzyme assay. Surprisingly, the enzyme activity of PDE8A1–V5 was 6-fold higher than that of ΔPAS–V5. This higher enzyme activity was not caused by a proportionally larger amount of PDE8A1–V5 precipitated, because Western blot analysis confirmed a level of PDE8A1–V5 similar to (if not lower than) that of ΔPAS–V5 (Fig. 5A).
Fig. 5.
Effect of PDE8A1–IκB association on PDE8A1 enzyme activity. (A) The concentrations of PDE8A1–V5 and ΔPAS–V5 in the cell lysates of HEK293 cells stably expressing PDE8A1–V5 or its PAS-deleted mutant ΔPAS–V5 were measured by ELISA (data not shown). Lysates containing equal amounts of PDE8A1–V5 and ΔPAS–V5 were subjected to immunoprecipitation with an anti-V5 antibody. The immunoprecipitates were subjected to Western blotting with an anti-V5–HRP antibody (to confirm similar amounts of PDE8A1–V5 and ΔPAS–V5 immunoprecipitated; see Inset) and PDE8 enzyme assay. (B) Cell lysates of HEK293 cells stably expressing PDE8A1–Myc were incubated with an increasing amount of cell lysate derived from HEK293 cells stably expressing IκBβ–V5 or control (nontransfected) cell lysate. PDE8A1–Myc was immunoprecipitated with an anti-Myc antibody, followed by PDE8A1 enzyme assay. (C) Wild-type HEK293 cells or the cells stably expressing IκBβ–V5 were transiently transfected with PDE8A1–Myc, and then cell lysates of the cells were subjected to immunoprecipitation with the anti-Myc antibody. Resultant antibody beads were subjected to a PDE8A1 enzyme assay. Each data point represents the mean ± SE (n = 4).
To confirm that this higher enzyme activity was caused by the PDE8A1–IκB association, the effect of IκBβ on PDE8A1 enzyme activity was examined both in vitro and in vivo. In the in vitro experiments, cell lysates of PDE8A1–Myc-expressing cells were incubated with an increasing amount of the cell lysate of IκBβ–V5 stable cell line, followed by immunoprecipitation with an anti-Myc antibody. Enzyme assay of the precipitated PDE8A1–Myc showed that the enzyme activity was significantly increased by increasing the amount of IκBβ–V5 cell lysate, as compared with control (nontransfected) cell lysate (Fig. 5B). The small increases of PDE8A1 enzyme activity by control cell lysate were presumably caused by endogenous IκB proteins.
In the in vivo experiments, HEK293 or the IκBβ–V5 stable cell line was transiently transfected with the PDE8A1–Myc, and then the cell lysates were subjected to immunoprecipitation with the anti-Myc antibody. Similar to the in vitro experiments, enzyme assay showed a significantly higher activity in the presence of overexpressed IκBβ–V5 (Fig. 5C). These results demonstrate that the IκB-PAS domain association increases the enzyme activity of PDE8A1.
Discussion
PAS domain-dependent, specific physical association of over-expressed recombinant human PDE8A1 with endogenous IκBβ was first detected by using an antibody array technique. The association of PDE8A1 with all IκB proteins examined (IκBβ, IκBα, p105, and p100) was confirmed by coimmunoprecipitation technique. The commercially available antibody array used in the initial screening contained antibodies against IκBα, p105/p50, and p100/p52 in addition to IκBβ. The failure to detect IκBα or p105/p50 in the initial screening was probably caused by lower binding efficiencies of the corresponding immobilized antibodies.
PDE8A1 was coimmunoprecipitated with all IκB proteins examined and with the NF-κB proteins c-Rel and RelB but not p65, p50, or p52 (Fig. 3A). The failure of p65 or p50 to be coimmunoprecipitated with PDE8A1 was found to be caused by competition between PDE8A1 and p65/p50 for IκB binding (Fig. 3C), suggesting that PDE8A1 binds to the NF-κB-binding site of IκB. The IκB family proteins all have ankyrin repeat domains as their binding sites for NF-κB (16–20), which may explain the association of all IκB proteins with PDE8A1. The exact binding site can be confirmed/determined in future studies by mutational analysis. The NF-κB proteins c-Rel and RelB are known to associate with p105 and p100 through their Rel homology domains (20, 22, 23), which may explain why c-Rel and RelB were coimmunoprecipitated with PDE8A1. p65 and p50 also can form heterodimers with p105 and p100 (23, 24). However, the failure of p50 or p65 to be coimmunoprecipitated with PDE8A1 (Fig. 3A) may be explained by the fact that they preferentially form the p65–p50 complex in vivo because of a relatively higher affinity between these two proteins (25). With regard to the lack of detectable coimmunoprecipitation between p52 and PDE8A1 (Fig. 3A), it was presumably caused by the fact that p52 is in much lower abundance (26, 27).
The enzyme activity of the immunoprecipitated PDE8A1 was at least 6-fold higher than that of its PAS domain-deleted version (Fig. 5A). This result suggests that the higher enzyme activity may be caused by the PDE8A1–IκB association. Furthermore, in vitro and in vivo addition of IκBβ to PDE8A1 increased the enzyme activity (Fig. 5 B and C). The relatively smaller increases in enzyme activity observed in the IκBβ-adding experiments (Fig. 5 B and C) were probably caused by the fact that the majority of PDE8A1 molecules without the addition of IκBβ were already in complex with endogenous IκB proteins. Thus, despite the smaller effect, these results confirm the notion that the association with IκB enhances the enzyme activity of PDE8A1. These data are not very surprising, because it has been known that IκB binds with and modulates the enzyme activity of protein kinase A (28) and cyclin-dependent kinase 4 (29). The association with IκBβ may result in a conformational change of PDE8A1 so that its catalytic site is more accessible to cAMP and/or the cAMP hydrolysis is more efficient.
Extensive evaluation did not indicate a direct effect of the PDE8A1–IκB association on NF-κB activation (Fig. 4). These data were consistent with the observation that PDE8A1 competed with NF-κB for IκB binding (Fig. 3C). Because of the competition, PDE8A1 would not be present in NF-κB–IκB complexes involved in cell stimulation and gene expression. These data suggest that the biological role of the IκB association may be related to compartmentalization and activation of PDE8A1 but not to the NF-κB pathway. However, the possibility of an involvement of the PDE8A1–IκB association in the NF-κB pathway cannot be ruled out. Several proteins have been reported to physically associate with IκB, including protein kinase A (28), cyclin-dependent kinase 4 (29), Tax protein (30), κB-Ras (31), and retinoid X receptor (32). The IκB binding by each of these proteins has certain impact on NF-κB pathway. It has been shown that NF-κB activation can be regulated by protein kinase A by both cAMP-dependent and cAMP-independent mechanisms (33). Moreover, it has been established that cAMP signaling is highly compartmentalized (34, 35). It is conceivable that, by IκB association, PDE8A1 may be recruited to and sequestered in the “microcompartment” of cAMP-dependent NF-κB activation, thereby playing an indirect role in the NF-κB activation by regulating local cAMP levels. How endogenous PDE8A1 and IκB interact with each other and, particularly, whether the interaction plays a regulatory role in NF-κB activation in various cells would be interesting subjects for future studies.
In summary, in this study we demonstrate that overexpressed recombinant human PDE8A1 physically associates with endogenous IκB and that the IκB association increases the enzyme activity of PDE8A1. The IκB association requires the PAS domain of PDE8A1, and thus this association represents a function of the PAS domain of PDE8A1 and a function of a mammalian PAS domain. Among the PDE superfamily, the presence of a PAS domain is unique to PDE8, as was the association with IκB (Fig. 2C). Based on the IκB association, PDE8 may have a unique biological role among various PDEs.
Acknowledgments
We thank Dr. Daniel Lundell for critical reading of the manuscript.
Author contributions: P. Wu and P. Wang designed research; P. Wu performed research; P. Wu and P. Wang analyzed data; and P. Wang wrote the paper.
Abbreviations: PAS, Per-Arnt-Sim; PDE, phosphodiesterase; ΔPAS, PAS-deleted PDE8A1; HRP, horseradish peroxidase.
References
- 1.Beavo, J. A. (1995) Physiol. Rev. 75, 725–748. [DOI] [PubMed] [Google Scholar]
- 2.Manganiello, V. C., Murata, T., Taira, M., Belfrage, P. & Degerman, E. (1995) Arch. Biochem. Biophys. 322, 1–13. [DOI] [PubMed] [Google Scholar]
- 3.Conti, M. & Jin, S.-L. C. (2000) Prog. Nucleic Acid Res. Mol. Biol. 63, 1–38. [DOI] [PubMed] [Google Scholar]
- 4.Francis, S. H., Turko, I. V. & Corbin, J. D. (2001) Prog. Nucleic Acid Res. Mol. Biol. 65, 1–52. [DOI] [PubMed] [Google Scholar]
- 5.Soderling, S. H., Bayuga, S. J. & Beavo, J. A. (1998) Proc. Natl. Acad. Sci. USA 95, 8991–8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang, P., Wu, P., Egan, R. W. & Billah, M. M. (2001) Gene 280, 183–194. [DOI] [PubMed] [Google Scholar]
- 7.Gamanuma, M., Yuasa, K., Sasaki, T., Sakurai, N., Kotera, J. & Omori, K. (2002) Cell. Signalling 15, 565–574. [DOI] [PubMed] [Google Scholar]
- 8.Taylor, B. L. & Zhulin, I. B. (1999) Microbiol. Mol. Biol. Rev. 63, 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semenza, G. L. (1999) Annu. Rev. Cell Dev. Biol. 15, 551–578. [DOI] [PubMed] [Google Scholar]
- 10.Taylor, B. L., Zhulin, I. B. & Johnson, M. S. (1999) Annu. Rev. Microbiol. 53, 103–128. [DOI] [PubMed] [Google Scholar]
- 11.Gu, Y. Z., Hogenesch, J. B. & Bradfield, C. A. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 519–561. [DOI] [PubMed] [Google Scholar]
- 12.Rutter, J., Michnoff, C., Harper, S. M., Gardner, K. H. & McKnight, S. L. (2001) Proc. Natl. Acad. Sci. USA 98, 8991–8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabral, J. H. M., Lee, A., Cohen, S. L., Chait, B. T., Li, M. & Mackinnon, R. (1998) Cell 95, 649–655. [DOI] [PubMed] [Google Scholar]
- 14.Reppert, S. M. (1998) Cell 21, 1–4. [DOI] [PubMed] [Google Scholar]
- 15.Sassone-Corsi, P. (1998) Nature 392, 871–874. [DOI] [PubMed] [Google Scholar]
- 16.Rothwarf, D. M. & Karin, M. (1999) Science's STKE 1999, RE1. [DOI] [PubMed] [Google Scholar]
- 17.May, M. J. & Ghosh, S. (1997) Semin. Cancer Biol. 8, 63–73. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh, S., May, M. J. & Kopp, E. B. (1998) Annu. Rev. Immunol. 16, 225–260. [DOI] [PubMed] [Google Scholar]
- 19.Li, Q. & Verma, I. M. (2002) Nature Rev. Immunol. 2, 725–734. [DOI] [PubMed] [Google Scholar]
- 20.Baldwin, A. S., Jr. (1996) Annu. Rev. Immunol. 14, 649–683. [DOI] [PubMed] [Google Scholar]
- 21.Wang, P., Wu, P., Egan, R. W. & Billah, M. M. (2003) Gene 314, 15–27. [DOI] [PubMed] [Google Scholar]
- 22.Solan, N. J., Miyoshi, H., Carmona, E. M., Bren, G. D. & Paya, C. V. (2002) J. Biol. Chem. 277, 1405–1418. [DOI] [PubMed] [Google Scholar]
- 23.Mercurio, F., DiDonato, J. A., Rosette, C. & Karin, M. (1993) Genes Dev. 7, 705–718. [DOI] [PubMed] [Google Scholar]
- 24.Lin, L., DeMartino, G. N. & Greene, W. C. (2000) EMBO J. 17, 4712–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, F. E., Huang, D.-B., Chen, Y.-Q. & Ghosh, G. (1998) Nature 391, 410–413. [DOI] [PubMed] [Google Scholar]
- 26.Betts, J. C. & Nabel, G. J. (1996) Mol. Cell. Biol. 16, 6363–6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heusch, M., Lin, L., Geleziunas, R. & Greene, W. C. (1999) Oncogene 18, 6201–6208. [DOI] [PubMed] [Google Scholar]
- 28.Zhong, H., SuYang, H., Erdjument-Bromage, H., Tempst, P. & Ghosh, S. (1997) Cell 89, 413–424. [DOI] [PubMed] [Google Scholar]
- 29.Li, J., Joo, S. H. & Tsai, M.-D. (2003) Biochemistry 42, 13476–13483. [DOI] [PubMed] [Google Scholar]
- 30.Hirai, H., Suzuki, T., Fujisawa, J., Inoue, J. & Yoshida, M. (1994) Proc. Natl. Acad. Sci. USA 91, 3584–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen, Y., Wu, J. & Ghosh, G. (2003) J. Biol. Chem. 278, 23101–23106. [DOI] [PubMed] [Google Scholar]
- 32.Na, S.-Y., Kim, H.-J., Lee, S.-K., Choi, H.-S., Na, D.-S., Lee, M.-O., Chung, M., Moore, D. D. & Lee, J. W. (1998) J. Biol. Chem. 273, 3212–3215. [DOI] [PubMed] [Google Scholar]
- 33.Torgersen, K. M., Vang, T., Abrahamsen, H., Yaqub, S. & Tasken, K. (2002) Cell. Signaling 14, 1–9. [DOI] [PubMed] [Google Scholar]
- 34.Houslay, M. D. (2001) Prog. Nucleic Acid Res. Mol. Biol. 69, 249–315. [DOI] [PubMed] [Google Scholar]
- 35.Conti, M., Tichter, W., Mehats, C., Livera, G., Park, J.-Y. & Jin, C. (2003) J. Biol. Chem. 278, 5493–5496. [DOI] [PubMed] [Google Scholar]