Abstract
Background
Why schizophrenia has accompanied us throughout our history despite its negative effect on fitness remains an evolutionary enigma. It is proposed that schizophrenia is a by-product of the complex evolution of the human brain and a compromise for our language, creative thinking and cognitive abilities.
Method
We analyze recent large genome-wide association studies of schizophrenia and a range of other human phenotypes (anthropometric measures, cardiovascular disease risk factors, immune-mediated diseases) using a statistical framework that draws on polygenic architecture and ancillary information on genetic variants. We used information from the evolutionary proxy measure called Neanderthal selective sweep (NSS) score.
Results
We show that gene loci associated with schizophrenia are significantly (p = 7.30×10−9) more prevalent in genomic regions that are likely to have undergone recent positive selection in humans, i.e. with low NSS score. Variants in brain-related genes with low NSS score confer significantly higher susceptibility than variants in other brain-related genes. The enrichment is strongest for schizophrenia, but we cannot rule out enrichment for other phenotypes. The false discovery rate conditional on the evolutionary proxy, points to 27 candidate schizophrenia susceptibility loci, twelve of which are associated with schizophrenia and other psychiatric disorders, or linked to brain development.
Conclusion
The results suggest that there is a polygenic overlap between schizophrenia and NSS score, a marker of human evolution, which is in line with the hypothesis that the persistence of schizophrenia is related to the evolutionary process of becoming human.
Keywords: schizophrenia, GWAS, evolution, human, Neanderthal, polygenic
Introduction
Schizophrenia affects approximately 1% of the world’s population and has accompanied us through much of our recorded history (1–6). This seemingly human-specific disorder is characterized by hallucinations and delusions (often involving language), thought disorders and higher order cognitive dysfunctions. The mechanisms of schizophrenia are not well understood, but its heritability is high, between 60% and 80% (7), and the fecundity of affected people is reduced (8). Nevertheless, the prevalence of the disease seems to remain stable across generations giving rise to the yet unresolved “evolutionary enigma” of schizophrenia (3, 4, 9, 10). Large variations in incidence across populations argue for environmental causes. However, by using standard, precisely drawn diagnostic criteria the variation in incidence can be reduced (11). Classical explanations include a single, partially dominant gene with low penetrance giving slight physiological advantages (12), balanced selection, where the gene variants conferring risk of the disease provide an advantage in particular environments, and hitchhiking, where disease variants are passed along with advantageous neighboring gene variants. Newer studies have focused on the polygenic nature of schizophrenia and have attributed the disease’s prevalence to the sporadic nature of complex disorders (13).
Archaeological and paleontological evidence points to the appearance of various hominin forms like Homo habilis, Homo erectus, Homo neanderthalensis (Neanderthals) and modern Homo sapiens (Humans) over 2.5 million years from the Lower Paleolithic to the Neolithic. It is debated whether the emergence of the ‘modern human’ was a morphological or a behavioral process, a one-time event or a continuous process of adaptation and assimilation of different forms. Even as morphological changes stopped, behavioral changes continued, rapidly leading to the ultimate success of humans (14).
Over the Pleistocene period, we see the appearance of specialized tools, the introduction of decorative arts, burial practices (15) and possibly the development of language (16). Research suggests that language acquisition played an important role in shaping the brain, helping us to think abstractly and be more creative, but also made us vulnerable to psychiatric disorders like schizophrenia (17). Changes that contributed to our ability to think more creatively and to improve executive function (18) could have also harbored susceptibility to this pathology (19). However, while archeological evidence provides clues about other aspects of human evolution, it cannot offer insights into the origin of psychiatric disorders.
Recent developments in human genetics have provided unprecedented opportunities to investigate evolutionary aspects of schizophrenia. Genome-wide association studies (GWAS) have identified over 100 schizophrenia risk loci and highlighted the polygenic architecture of the disease (20). The genome sequence of Neanderthals (21, 22), close relatives of early modern humans, can help pinpoint the genomic regions affected by positive selection since the two species diverged. The genomic differences between the two homo species may help explain specific human features, and thus the relation between human evolution and schizophrenia.
Several lines of evidence indicate that schizophrenia is a polygenic disorder (23, 24) with a large number of risk loci, each with a small effect (20). We have recently developed statistical tools, building on an Empirical Bayesian framework (25), that are specifically designed for polygenic architectures. These tools have successfully been applied to investigate several complex human phenotypes (26–32) but have not yet been used to study the evolutionary features thereof. We hypothesize that schizophrenia is the result of human polygenic adaptation (24) and investigate if regions of the human genome, which may have undergone recent positive selection, are enriched of association with schizophrenia.
Methods and Materials
Samples
We obtained summary statistics for ~1.0–2.5 million single nucleotide polymorphisms (SNPs) from GWASs of schizophrenia (conducted by the Psychiatric Genomics Consortium (PGC)) and other phenotypes, including anthropometric measures (body mass index (BMI), height, waist-hip ratio (WHR)), cardiovascular disease risk factors (systolic blood pressure (SBP), total cholesterol (TC), triglycerides (TG)), immune-mediated diseases (celiac disease (CeD), Crohn’s disease (CD), rheumatoid arthritis (RA), ulcerative colitis (UC)) as well as other psychiatric and central nervous system disorders (attention deficit, hyperactivity disorder (ADHD), Alzheimer’s disease (AD), bipolar disorder (BD), and multiple sclerosis (MS)) (Table S1). In total, these studies included approximately 1.3 million phenotypical observations although overlap between samples makes the number of unique subjects lower.
Neanderthal selective sweep score
The Neanderthal selective sweep (NSS) score is obtained through alignment of human, Neanderthal and primate consensus sequences (21, 33) and is downloadable from the UCSC genome browser (http://genome.ucsc.edu, ntSssZScorePMVar track (S-scores)). This track consists of two entries per SNP, (z-score + sd) and (z-score − sd). The NSS score provides a likelihood index of positive selection in humans sometime after the divergence of humans and Neanderthals (21, 33) by measuring the relative abundance of ancestral/non-ancestral (i.e. aligned/non-aligned with primate consensus) alleles in these two lineages. A negative NSS score indicates scarcity of non-ancestral alleles in Neanderthal compared to modern humans and therefore possible positive selection in the latter. The (z-score + sd) entries in the genome track represent an upper bound on the statistic and are therefore conservative measures of positive selection likelihood. These were extracted for all SNPs in the GWASs of interest (Table S1 in Supplement 1) and follow the distribution illustrated in Fig. S1 in Supplement 1. The (z-score + sd) entries, termed NSS scores, were used as ancillary information or covariates in the enrichment analyses. Using the NSS scores, the authors of the two articles on Neanderthal genome identified regions of the human genome that are significantly likely to have undergone recent positive selection. The same analyses performed directly using the NSS scores were also performed using linkage disequilibrium (LD) weighted scores (see Analytical approach below) measuring affiliation to these regions.
Brain genes
In order to control the enrichment analyses for affiliation to brain genes, we identified genes with a known function in the brain using information from the NCBI (http://www.ncbi.nlm.nih.gov/gene). The query “human brain” in Homo sapiens revealed 2494 genes (March 2015). For comparison, we also used the list of brain genes from Kang et al.(34), which includes 1415 genes selected based on expression in various neural cells. The LD weighted procedure (see Analytical approach) applied to the NSS regions mentioned above was applied to these genes, yielding brain genes LD weighted affiliation scores.
Analytical approach
We employed a genetic enrichment method recently developed to dissect the genetic architecture of complex traits (26, 28, 29) (32, 35). Specifically, we investigated the enrichment of associations concurrent with the NSS score selection index in a covariate-modulated statistical approach (32). We investigated whether SNPs with low NSS score and therefore in regions possibly subjected to positive selection in humans, are more likely associated with schizophrenia or other phenotypes. All statistical analyses were carried out with a covariate-modulated enrichment analysis package developed on R (www.r-project.org) and MATLAB (www.mathworks.se/products/matlab/) programming platforms.
Quantile-Quantile (Q-Q) and Fold enrichment (36) plots
Q-Q plots are designed to compare two distributions; here we compared the nominal p-value distribution to the empirical distribution. In the presence of null relationships only, the nominal p-values form a straight line on a Q-Q plot when plotted against the empirical distribution. We plotted −log10 nominal p-values against −log10 empirical p-values for the two SNP strata determined by the NSS score (conditional Q-Q plots) as well as for all SNPs. Leftward deflections of the observed distribution from the null line reflect increased tail probabilities in the distribution of test statistics (z-scores) and consequently an over-abundance of low p-values compared to that expected under the null hypothesis.
To graphically assess genetic enrichment, we used conditional fold enrichment plots. Here, a direct measure of the enrichment is given by the degree of deflection from the expected null. The fold enrichment is derived as follows: first the empirical cumulative distribution of −log10(p)-values for SNP association is computed for a given phenotype for all SNPs, and for the two dichotomous SNPs strata determined by the NSS score. Each stratum’s fold enrichment is then calculated as the ratio CDFstratum/CDFall between the −log10(p) cumulative distribution for that stratum and the cumulative distribution for all SNPs. The nominal −log10(p) values are plotted on the x-axis, the fold enrichment in the y-axis. To assess polygenic effects below the standard GWAS significance threshold, we focused the fold enrichment plots on SNPs with nominal −log10(p) < 7.3 (corresponding to p > 5×10−8).
Binomial proportion test (BPT) (37)
Upon randomly subdividing a set of SNPs into two disjoint subsets, one would expect these to present similar p-values distributions. In particular, the proportion of SNPs with a p-value below a certain threshold should be the same in the two subsets. BPT measures deviations from this null hypothesis below a threshold of interest. We compared the proportions of SNPs in the top −log10(p) percentile within the two NSS strata. The BPT assumes independence of the data. Because of LD between SNPs, this independence requirement does not hold. We therefore subdivided the whole SNP set into blocks defined by 1Mb windows and an LD r2 threshold of 0.2 and randomly selected ten sets of SNP representatives from all blocks. Ten sets of BPTs were carried out on the approximately independent randomly chosen SNPs and the final p-value was calculated from the median of the BPT statistics.
LD weighted SNP annotation score
The use of GWAS SNPs in DNA regions of interest may underestimate the extent to which those regions are represented in the analysis. We used an LD weighted scoring algorithm developed in(26) order to identify SNPs that tag specific DNA regions even if they are not situated within them.
For each GWAS SNP a pairwise correlation coefficient approximation to LD (r2) was calculated for all 1KGP SNPs within a 1,000,000 base pairs (1Mb). All r2 values < 0.2 were set to 0 and each SNP was assigned an r2 value of 1.0 with itself. LD weighted region annotation scores for all DNA regions of interest were computed as the sum of LD r2 between the tag SNP and all 1KGP SNPs in those regions. Given SNPi, its LD weighted region annotation score was computed as LD scorei = Σj δj rij2, where rij2 is the LD r-squared between SNPi and SNPj and δj takes values of 1 or 0 depending on whether the 1KGP SNPj is within the region of interest or not.
Intergenic SNPs
Intergenic SNPs are defined as having LD weighted annotation scores for each of the genomic categories analyzed by Schork et al. (26) equal to zero and being in LD with no SNPs in the 1KGP reference panel located within 100,000 base pairs of a protein coding gene, within a non-coding RNA, within a transcription factor binding site or within a miRNA binding site. Those singled out in this way are expected to form a collection of non-genic SNPs not belonging to any (annotated) functional elements within the genome (including through LD) and therefore represent a collection of SNPs more likely to be null.
Intergenic correction
Intergenic SNPs were used to estimate the inflation of GWAS summary statistics due to cryptic relatedness. We used intergenic SNPs because their relative depletion of associations (26) suggests they provide a set of SNPs whose statistics are less inflated by polygenic associations. The inflation factor, λGC, was estimated as the median squared z-score of independent (LD r2 < 0.2) sets of intergenic SNPs across one hundred LD-pruning iterations, divided by the expected median of a chi-square distribution with one degree of freedom.
Squared z-score regression
The hypothesis here is that there is some proportionality between a continuous covariate of interest and the incidence of SNP association with a phenotype. A viable proxy for the latter is the extent of the association z-scores. We therefore regressed the squared z-scores against the NSS scores. Other covariates were included in the regression as well to account for possible confounding factors. These were exonic, intronic, 5′UTR, 3′UTR annotation scores (26, 38), brain gene affiliation scores, genotypic variance and total LD. As done for the BPT, regression analyses were performed on the ten sets of SNP representatives and the regression coefficient p-values were calculated from the median of the ten regression coefficient estimates.
Replication
The procedure used to compute the conditional rate of replication (for details see supplement 1) follows the one of Schork et al. (26). The 52 sub-studies were subdivided into two groups of 26 in 50 different ways, the first group, Dk, k = 1…50, serving as discovery group, the second, Rk, k = 1…50, as replication group. Cumulative replication rates were calculated over each of 1,000 equally-spaced bins spanning the range of negative log10(p-values) observed in the discovery group and for each of the 50 subdivisions. Every cumulative replication rate was calculated as the fraction of SNPs with a discovery negative log10(p-value) greater than the lower bound of the bin, that had a replication p-value smaller than 0.05. Average cumulative replication rates were subsequently computed across the 50 subdivisions.
Results
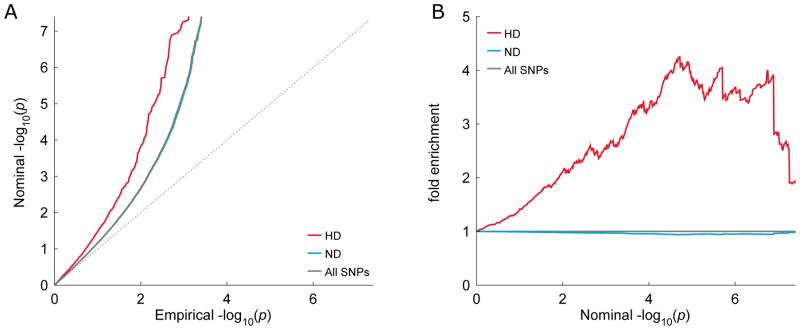
We first assessed the influence exerted on schizophrenia association propensity by the Neanderthal “character” of the SNP’s DNA region, as measured by the NSS score selection index (21) (Fig. S1). Using data from the recently published schizophrenia GWAS (20), we conditioned schizophrenia association p-values on the NSS score. The conditional Q-Q (Fig. 1A) and fold enrichment (Fig. 1B) plots show that SNPs with negative NSS scores are enriched for associations with schizophrenia compared to SNPs with positive NSS score. These results are nominally confirmed by the binomial proportion test (BPT), (p = 2.40×10−2) and more robustly by the squared z-score regression against the NSS score (β = −0.067, p = 7.30×10−9) (Table 1).
Fig. 1. Q-Q and fold enrichment plots of schizophrenia stratified according to Neanderthal selective sweep score.
Shown are A) quantile-quantile (Q-Q) and B) fold enrichment plots of GWAS summary statistics p-values for schizophrenia (SCZ), stratified based on Neanderthal selective sweep (NSS) score. The human divergent (HD) stratum comprises single nucleotide polymorphisms (SNPs) with negative NSS scores. The regions around these SNPs present fewer derived alleles in Neanderthal than expected given the frequency of derived alleles in humans, and may therefore have undergone recent positive selection in the latter. The non-divergent (ND) stratum comprises all SNPs with positive NSS scores. HD SNPs show a marked leftward (A) and upward (B) deflection from the lines corresponding to All SNPs. This signifies a comparatively higher proportion of low p-values among HD SNPs.
Table 1.
Neanderthal Selective Sweep Score; z squared logarithm regression and binomial proportion test (BPT).
| GWAS | β (Min, Max) | Std. error | p-value | C.I | BPT(p) 1%(Min, Max) |
|---|---|---|---|---|---|
| AD | −0.012(−0.029,0.012) | 0.016 | 5.00E-01 | −0.041,0.022 | 2.5E-01(8.0E-04,5.9E-01) |
| ADHD | 0.003(−0.013,0.017) | 0.013 | 8.40E-01 | −0.022,0.028 | 4.5E-01(3.6E-02,6.2E-01) |
| BD | −0.004(−0.020,0.013) | 0.01 | 7.10E-01 | −0.023,0.015 | 4.8E-01(1.4E-02,9.1E-01) |
| MDD | −0.018(−0.034,−0.006) | 0.012 | 2.00E-01 | −0.042,0.007 | 5.6E-01(1.1E-01,9.4E-01) |
| Migraine | −0.006(−0.029,−0.002) | 0.013 | 6.90E-01 | −0.032,0.020 | 7.4E-01(2.3E-01,9.2E-01) |
| MS | −0.003(−0.034,0.022) | 0.02 | 8.80E-01 | −0.042,0.035 | 5.1E-01(8.7E-03,8.5E-01) |
| SCZ 1 | −0.038(−0.052,−0.026) | 0.013 | 7.90E-03 | −0.063,−0.013 | 1.7E-01(1.8E-02,6.1E-01) |
| SCZ 2 | −0.067(−0.076,−0.056) | 0.01 | 7.30E-09 | −0.088,−0.047 | 2.4E-02(5.7E-06,3.7E-01) |
| BMI | −0.050(−0.061,−0.037) | 0.016 | 4.50E-03 | −0.079,−0.023 | 4.2E-01(6.2E-02,9.2E-01) |
| Height | −0.074(−0.098,−0.058) | 0.015 | 8.90E-06 | −0.104,−0.045 | 1.1E-01(3.9E-04,6.9E-01) |
| WHR | −0.026(−0.039,−0.019) | 0.011 | 2.50E-02 | −0.047,−0.006 | 2.3E-01(7.1E-03,5.1E-01) |
| SBP | −0.015(−0.024,−0.003) | 0.01 | 1.80E-01 | −0.036,0.005 | 3.7E-01(9.3E-02,7.3E-01) |
| TC | −0.001(−0.019,0.023) | 0.019 | 9.80E-01 | −0.037,0.039 | 5.3E-01(3.1E-01,8.6E-01) |
| TG | −0.017(−0.024,−0.003) | 0.015 | 3.30E-01 | −0.048,0.014 | 4.3E-01(8.2E-03,8.3E-01) |
| CD | −0.025(−0.049,0.003) | 0.019 | 2.50E-01 | −0.062,0.014 | 5.3E-01(2.6E-01,8.6E-01) |
| CeD | −0.000(−0.024,0.018) | 0.018 | 9.90E-01 | −0.037,0.035 | 3.7E-01(1.2E-01,8.4E-01) |
| RA | −0.004(−0.020,0.013) | 0.011 | 7.70E-01 | −0.025,0.017 | 5.7E-01(2.2E-02,8.8E-01) |
| UC | −0.017(−0.027,0.015) | 0.015 | 3.10E-01 | −0.047,0.014 | 5.3E-01(1.7E-01,9.2E-01) |
Phenotypes: psychiatric and other neurological diseases Alzheimer’s disease (AD), attention deficit hyperactivity disorder (ADHD), bipolar disorder (BD), major depressive disorder (MDD), migraine, multiple sclerosis (MS), first and second edition of the schizophrenia (SCZ) GWAS by the Psychiatric Genomic Consortium (SCZ1 and SCZ2), anthropometric measures (body mass index (BMI), height, waist-hip ratio (WHR)), cardiovascular risk factors (systolic blood pressure (SBP), total cholesterol (TC), triglycerides (TG)), immune-mediated diseases (Crohn’s disease (CD), celiac disease (CeD), rheumatoid arthritis (RA), ulcerative colitis (UC)). Squared z-scores logarithm versus Neanderthal selective sweep score (NSS) regression for various phenotypes controlling for other enrichment factors (genic annotation scores, genotypic variance, LD) and top 1% binomial proportion test (BPT) p-values.
SCZ is the only phenotype with a significant negative correlation between squared z-scores logarithms and NSS scores while controlling for other covariates. Also, in SCZ2 the top 1% SNPs include a nominally significant excess of SNPs with NSS score < 0 (Human Divergent) compared to any SNPs (BPT).
To control for the known effect of immune-related genes, all analyses were repeated after exclusion of SNPs in the MHC regions. These do not appear to affect the fold-enrichment to any measurable extent (Table S2, Fig. S2 in Supplement 1). Thus, it appears that the SNPs in human DNA regions that diverge from their Neanderthal counterparts have a higher propensity to be associated with schizophrenia. Similar analyses were repeated using affiliation to NSS regions that were deemed significantly likely (top 5%) to have undergone positive selection upon alignment with the Neanderthal genome. In this case, we investigated the original (21) as well as the more recently sequenced Neanderthal genome (22) and confirmed the initial results (Table S3, Fig. S3 in Supplement 1).
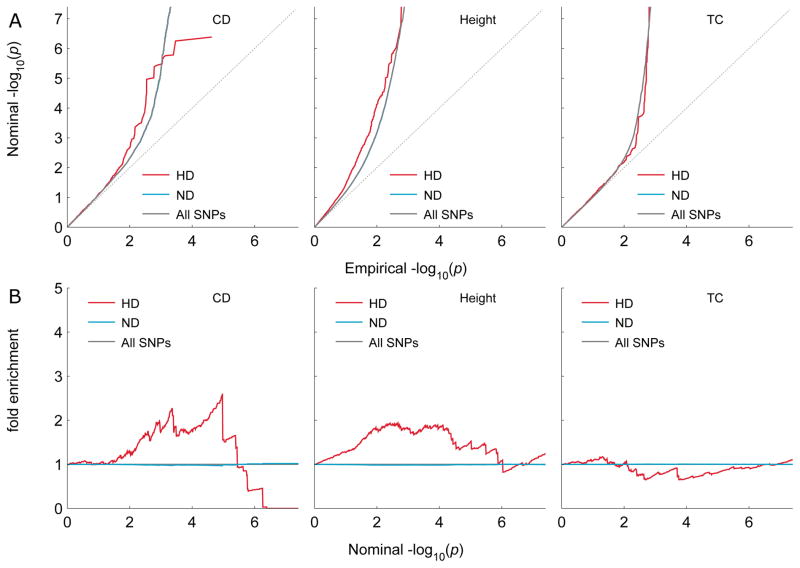
We carried out the same analyses on other phenotypes in order to assess the specificity of the evolutionary enrichment. As shown in Q-Q plots and fold enrichment plots (Fig. 2), other phenotypes show mostly modest or scarce enrichment as a function of NSS compared to schizophrenia. The only other significant excesses of low p-values were detected by the BPTs and the regression analyses for height and to some extent for BMI. (Table 1). Height in particular has effect size comparable to that of schizophrenia, and possibly larger still, but its standard error is somewhat larger. Targeted analyses of other psychiatric (ADHD, BD, MDD) and neurological (AD, Migraine, MS) disorders, revealed no measurable enrichment effect (Fig. S4–S5 in Supplement 1). Schizophrenia has by far the largest NSS effect size among the psychiatric and neurological GWASs, all of which have similar standard errors (Fig. S8). To test the extent to which the effect on schizophrenia depends on the power of the GWAS from 2014 (20), we performed the same analyses on the smaller schizophrenia GWAS from 2013(39), which is comparable in size to several of the other GWASs. The enrichment was somewhat diminished (Fig. S6 in Supplement 1) but remained nominally significant according to the regression analysis (β = −0.038, p = 7.93×10−3). We also tested the censored (methods in Supplement1) schizophrenia GWAS summary statistics and still found a significant (regression coefficient p=2.87×10−6) residual enrichment.
Fig. 2. Q-Q and fold enrichment plots of three non-schizophrenia phenotypes stratified according to Neanderthal selective sweep scores.
Phenotypes: Crohn’s disease (CD), Height and total cholesterol (TC). A) The quantile-quantile (Q-Q) plots show GWAS summary statistics p-values of SNPs tagging human divergent regions (HD), non-divergent (ND) regions as well as All SNPs. There is no indication of enrichment as seen in SCZ in (Fig. 1). B) The fold enrichment counterparts of the Q-Q plots in A) illustrate the lack of enrichment. The regression analysis however shows significant enrichment for Height. (Table 1).
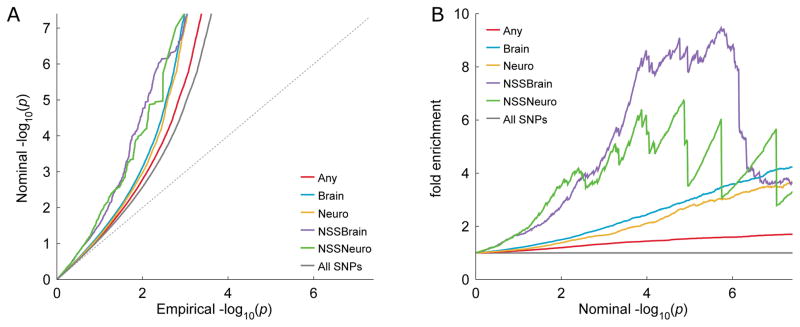
The effect of brain genes affiliation on enrichment was further investigated by testing whether brain genes with negative NSS scores are more enriched of associations with schizophrenia than any brain genes. The enrichment plots (Fig. 3) for brain genes with negative NSS show a wider deflection from baseline and the BPT shows a significant difference in the proportion of association p-values in the lowest percentile (p = 5.5×10−3).
Fig. 3. Q-Q and fold enrichment plots showing schizophrenia association enrichment of brain genes with negative Neanderthal selective sweep (NSS) score.
Shown are A) the quantile- quantile (Q-Q) and B) the fold enrichment plots for: SNPs annotated to generic genes (Any); SNPs annotated to genes associated with the brain, as established by an NCBI site search (Brain); SNPs annotated to genes associated with the brain, defined by Kang et al. (34) (Neuro); SNPs with negative NSS score and annotated to genes associated with the brain, as established by an NCBI site search (NSSBrain); or to genes defined by Kang et al. (34) (NSSNeuro); and all SNPs (All SNPs). The NSS Brain category is enriched (deflected left) compared to the other categories, i.e. presents a higher incidence of associations (lower p-values) with schizophrenia (SCZ). This is confirmed by the Binomial Proportion Test (BPT) comparing Brain and NSS Brain groups (p = 5.5×10−3).
We used the conditional FDR (condFDR) analysis (see methods in Supplement 1) to identify possible genomic loci associated with schizophrenia subject to the condition of having a negative NSS score. A total of 27 genomic loci were identified (condFDR<0.01). They are listed in Table S4 (Supplement 1) together with the annotated genes. A closer inspection of Table S4 (Supplement 1) reveals no preferential direction of effect (Fig. S7 in Supplement 1), i.e. positive and negative z-scores were equally represented. This lack of directionality is confirmed upon regressing the SNPs z-scores against their NSS score (regression data not shown), i.e. no significant association between the two could be detected. None of the loci are identified by the analyses involving NSS region affiliation scores. This is probably due to the dichotomous origin of this measure which is less well suited to the FDR lookup table smoothing procedure.
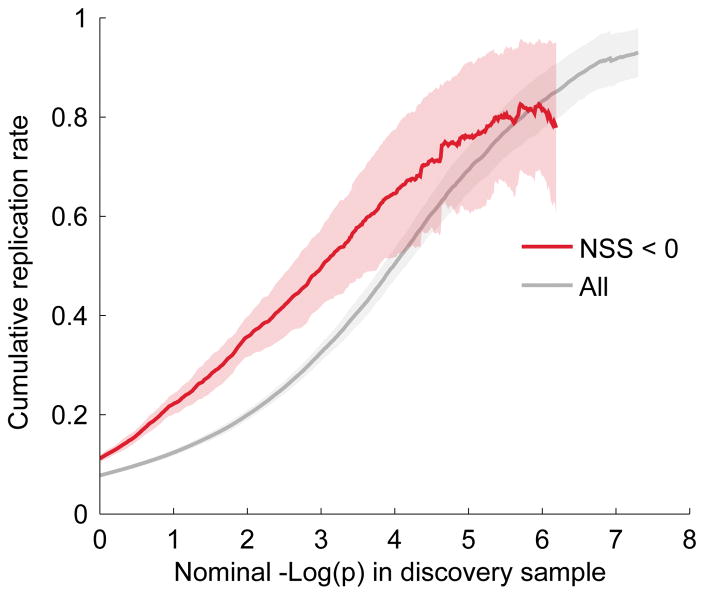
To assess the reliability of the genomic loci identified via condFDR, we investigated the association replication rates in independent schizophrenia sub-studies, defined as the proportion of SNPs declared significant in training samples with p-values below 0.05 in the replication sample and with z-scores with the same sign in both discovery and replication samples. We found that SNPs with NSS < 0 replicate at a higher rate than other SNPs (Fig. 4). This confirms that the observed enrichment is due to associations and not to population stratification or other potential sources of spurious effects. Replication rates were extrapolated for the 27 NSS candidate loci and reported in Table S4 as well.
Fig. 4. Replication plot for schizophrenia with and without conditioning on Neanderthal selective sweep score < 0.
SNPs with negative Neanderthal selective sweep score (NSS) score tend to replicate better than baseline SNPs across the 52 schizophrenia (SCZ) meta-analysis sub studies. For example, at a −log(p)-value level of 4 the cumulative replication rate improves from 60% to about 80% when restricting the choice to SNPs with negative NSS score. A negative NSS score seems therefore to be a viable aid to identify non-spurious schizophrenia associations.
Discussion
Applying a polygenic statistical approach, we leveraged recent large GWAS data and showed that schizophrenia associations have a higher propensity to be found in genomic regions that diverge from their Neanderthal counterparts (negative NSS score). Such polygenic overlap between schizophrenia and a marker of human evolution is in accordance with the hypothesis of Crow et al. (19), suggesting that a number of schizophrenia susceptibility factors might have arisen as a “side effect” of human achievements like language and creative thinking (17). The current findings support the view that this evolutionary process also made us vulnerable to schizophrenia.
Previous studies of evolutionary factors of schizophrenia have focused on small sets of genes (40, 41). Bigdeli et al.‘s analysis (42) was more systematic but applied human accelerated regions (HAR) as evolutionary proxy. Xu et al. (43), using special HARs, showed that genes next to HARs in primates were under greater selection pressure compared to other genes and are more likely to be associated with schizophrenia susceptibility loci. Green et al. (21), who reported the first Neanderthal draft sequence, introduced the selective sweep score and investigated its relationship with the disease association but only for the most significant genes singled out by their analysis. Here, we use the information from Green et al.’s original publication (21), as well as the more recent report by Prüfer et al. on the complete Neanderthal genome sequence (22), to identify evolutionary enrichment patterns with a polygenic approach. Another asset for our study was the availability of a large schizophrenia GWAS of more than 80,000 participants (20), which makes it feasible to investigate evolutionary factors in schizophrenia with adequate power.
The results presented here are in line with the idea of polygenic adaptation which is believed to play a role in the development of many complex human diseases, as it likely happened in our adaptation to pathogens and in the variation of morphological traits like height (44–46). Classic selective sweeps, originating from strong selective pressure, are relatively rare in modern humans (47) and natural selection is not the only factor shaping human variation. Instead, polygenic selection involving subtle shifts of allele frequencies at many loci simultaneously has been suggested to be common for complex traits in humans (47). Thus, selection acting simultaneously on many of standing variants could be an efficient mechanism for phenotypic adaptation (48, 49). Given these premises, it becomes desirable to employ analytical tools designed to capture polygenicity. The methods applied in our analyses have been useful in studying polygenic factors in schizophrenia before (28, 29, 50, 51). Our results indicate that many schizophrenia susceptibility factors in modern humans may have emerged after their divergence from Neanderthals.
Several of the genes found to likely have undergone positive selection in modern humans (21) are involved in cognitive functions. The enrichment of SNPs associations observed for schizophrenia may therefore be due to an overlap between swept genomic regions and brain and other CNS genes and the regulatory regions thereof. This question is addressed by the regression analysis in which protein coding annotations are accounted for (Table 1). Even the inclusion of brain genes annotation scores in the regression did not reduce the enrichment for schizophrenia. Interestingly however, among the brain genes themselves, the ones with a negative NSS score were more enriched of associations with schizophrenia compared to other brain genes, let alone just any genes (Fig. 3).
The loci identified by the conditional FDR analysis harbor many genes that could plausibly play a role in the etiology of schizophrenia. Genes like DPYD, ZNF804A, NRXN1, NRG3 and VRK2, which were previously known to be associated with the disease, confirm its potentially evolutionary nature (52–54). Other interesting patterns emerge from genes like AGBL4, CEP170, IFT81, and SDCCAG8, related to ciliogenesis and ciliary disorders (55–57), and DPP10 and FOXP1, related to autism (58, 59). The functional implications of the current associations based on tag SNPs need to be explored in experimental studies. It will be of interest at a later stage to investigate whether the current polygenic evolutionary signal in schizophrenia is associated with human specific brain structure variance. GWAS for relevant brain structures, however, are not yet adequately powered (n~15,000 – 21,000 participants) (60). Further, the interplay of the polygenic effects and de novo mutations, such as schizophrenia risk CNVs, should also be examined, even if the latter appear to explain a very small proportion of schizophrenia cases (61).
Notably, the enrichment found here seems to be related to schizophrenia, and some anthropomorphic human traits. However, we cannot rule out that there may be enrichment also in other disorders or diseases. The sample sizes available to some of the CNS GWAS might have limited the power to detect any enrichment. At any rate, the analysis of the smaller schizophrenia GWAS from 2013(39) also revealed a nominally significant enrichment effect, further supporting the notion of a specific association between schizophrenia and positive selection.
In conclusion, the present findings of a prevalence of schizophrenia risk loci overlapping with some genetic signatures of human evolution support the argument that both the emergence and the persistence of schizophrenia are connected to the human sapientia. This may help to explain the “evolutionary enigma” of schizophrenia.
Supplementary Material
Acknowledgments
We thank Dr. Tobias Kaufmann and Dr. Tuomo Mäki-Marttunen for technical advice and helpful support.
FB, SD and OAA designed the study. PGC, IHGC, BSW, JAZ, SD, IM, TW, DAC, OAA, MM provided data. MM, YW, AW, AJS, RD, WKT, VZ, AMD provided analytical tools or support. FB, SS, MM and AW performed the analyses. SS, FB, and OAA wrote the first draft of the paper. All authors commented on and approved the final manuscript.
We thank Cisca Wijmenga and David van Heel for access to Celiac Disease data.
We thank the International Genomics of Alzheimer’s Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i–Select chips was funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant n° 503480), Alzheimer’s Research UK (Grant n° 503176), the Wellcome Trust (Grant n° 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant n° 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG–12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC–10–196728.
Members of the Schizophrenia Working Group of the Psychiatric Genomics Consortium
Stephan Ripke1,2, Benjamin M. Neale1,2,3,4, Aiden Corvin5, James T. R. Walters6, Kai-How Farh1, Peter A. Holmans6,7, Phil Lee1,2,4, Brendan Bulik-Sullivan1,2, David A. Collier8,9, Hailiang Huang1,3, Tune H. Pers3,10,11, Ingrid Agartz12,13,14, Esben Agerbo15,16,17, Margot Albus18, Madeline Alexander19, Farooq Amin20,21, Silviu A. Bacanu22, Martin Begemann23, Richard A Belliveau Jr2, Judit Bene24,25, Sarah E. Bergen 2,26, Elizabeth Bevilacqua2, Tim B Bigdeli 22, Donald W. Black27, Richard Bruggeman28, Nancy G. Buccola29, Randy L. Buckner30,31,32, William Byerley33, Wiepke Cahn34, Guiqing Cai35,36, Murray J. Cairns39,120,170, Dominique Campion37, Rita M. Cantor38, Vaughan J. Carr39,40, Noa Carrera6, Stanley V. Catts39,41, Kimberly D. Chambert2, Raymond C. K. Chan42, Ronald Y. L. Chen43, Eric Y. H. Chen43,44, Wei Cheng45, Eric F. C. Cheung46, Siow Ann Chong47, C. Robert Cloninger48, David Cohen49, Nadine Cohen50, Paul Cormican5, Nick Craddock6,7, James J. Crowley51, David Curtis52,53, Michael Davidson54, Kenneth L. Davis36, Franziska Degenhardt55,56, Jurgen Del Favero57, Lynn E. DeLisi128,129, Ditte Demontis17,58,59, Dimitris Dikeos60, Timothy Dinan61, Srdjan Djurovic14,62, Gary Donohoe5,63, Elodie Drapeau36, Jubao Duan64,65, Frank Dudbridge66, Naser Durmishi67, Peter Eichhammer68, Johan Eriksson69,70,71, Valentina Escott-Price6, Laurent Essioux72, Ayman H. Fanous73,74,75,76, Martilias S. Farrell51, Josef Frank77, Lude Franke78, Robert Freedman79, Nelson B. Freimer80, Marion Friedl81, Joseph I. Friedman36, Menachem Fromer1,2,4,82, Giulio Genovese2, Lyudmila Georgieva6, Elliot S. Gershon209, Ina Giegling81,83, Paola Giusti-Rodríguez51, Stephanie Godard84, Jacqueline I. Goldstein1,3, Vera Golimbet85, Srihari Gopal86, Jacob Gratten87, Lieuwe de Haan88, Christian Hammer23, Marian L. Hamshere6, Mark Hansen89, Thomas Hansen17,90, Vahram Haroutunian36,91,92, Annette M. Hartmann81, Frans A. Henskens39,93,94, Stefan Herms55,56,95, Joel N. Hirschhorn3,11,96, Per Hoffmann55,56,95, Andrea Hofman55,56, Mads V. Hollegaard97, David M. Hougaard97, Masashi Ikeda98, Inge Joa99, Antonio Julià100, René S. Kahn34, Luba Kalaydjieva101,102, Sena Karachanak-Yankova103, Juha Karjalainen78, David Kavanagh6, Matthew C. Keller104, Brian J. Kelly120, James L. Kennedy105,106,107, Andrey Khrunin108, Yunjung Kim51, Janis Klovins109, James A. Knowles110, Bettina Konte81, Vaidutis Kucinskas111, Zita Ausrele Kucinskiene111, Hana Kuzelova-Ptackova112, Anna K. Kähler26, Claudine Laurent19,113, Jimmy Lee Chee Keong47,114, S. Hong Lee87, Sophie E. Legge6, Bernard Lerer115, Miaoxin Li43,44,116 Tao Li117, Kung-Yee Liang118, Jeffrey Lieberman119, Svetlana Limborska108, Carmel M. Loughland39,120, Jan Lubinski121, Jouko Lönnqvist122, Milan Macek Jr112, Patrik K. E. Magnusson26, Brion S. Maher123, Wolfgang Maier124, Jacques Mallet125, Sara Marsal100, Manuel Mattheisen17,58,59,126, Morten Mattingsdal14,127, Robert W. McCarley128,129, Colm McDonald130, Andrew M. McIntosh131,132, Sandra Meier77, Carin J. Meijer88, Bela Melegh24,25, Ingrid Melle14,133, Raquelle I. Mesholam-Gately128,134, Andres Metspalu135, Patricia T. Michie39,136, Lili Milani135, Vihra Milanova137, Younes Mokrab8, Derek W. Morris5,63, Ole Mors17,58,138, Kieran C. Murphy139, Robin M. Murray140, Inez Myin-Germeys141, Bertram Müller-Myhsok142,143,144, Mari Nelis135, Igor Nenadic145, Deborah A. Nertney146, Gerald Nestadt147, Kristin K. Nicodemus148, Liene Nikitina-Zake109, Laura Nisenbaum149, Annelie Nordin150, Eadbhard O’Callaghan151, Colm O’Dushlaine2, F. Anthony O’Neill152, Sang-Yun Oh153, Ann Olincy79, Line Olsen17,90, Jim Van Os141,154, Psychosis Endophenotypes International Consortium155, Christos Pantelis39,156, George N. Papadimitriou60, Sergi Papiol23, Elena Parkhomenko36, Michele T. Pato110, Tiina Paunio157,158, Milica Pejovic-Milovancevic159, Diana O. Perkins160, Olli Pietiläinen158,161, Jonathan Pimm53, Andrew J. Pocklington6, John Powell140, Alkes Price3,162, Ann E. Pulver147, Shaun M. Purcell82, Digby Quested163, Henrik B. Rasmussen17,90, Abraham Reichenberg36, Mark A. Reimers164, Alexander L. Richards6, Joshua L. Roffman30,32, Panos Roussos82,165, Douglas M. Ruderfer6,82, Veikko Salomaa71, Alan R. Sanders64,65, Ulrich Schall39,120, Christian R. Schubert166, Thomas G. Schulze77,167, Sibylle G. Schwab168, Edward M. Scolnick2, Rodney J. Scott39,169,170, Larry J. Seidman128,134, Jianxin Shi171, Engilbert Sigurdsson172, Teimuraz Silagadze173, Jeremy M. Silverman36,174, Kang Sim47, Petr Slominsky108, Jordan W. Smoller2,4, Hon-Cheong So43, Chris C. A. Spencer175, Eli A. Stahl3,82, Hreinn Stefansson176, Stacy Steinberg176, Elisabeth Stogmann177, Richard E. Straub178, Eric Strengman179,34, Jana Strohmaier77, T. Scott Stroup119, Mythily Subramaniam47, Jaana Suvisaari122, Dragan M. Svrakic48, Jin P. Szatkiewicz51, Erik Söderman12, Srinivas Thirumalai180, Draga Toncheva103, Paul A. Tooney39,120,170, Sarah Tosato181, Juha Veijola182,183, John Waddington184, Dermot Walsh185, Dai Wang86, Qiang Wang117, Bradley T. Webb22, Mark Weiser54, Dieter B. Wildenauer186, Nigel M. Williams6, Stephanie Williams51, Stephanie H. Witt77, Aaron R. Wolen164, Emily H. M. Wong43, Brandon K. Wormley22, Jing Qin Wu39,170, Hualin Simon Xi187, Clement C. Zai105,106, Xuebin Zheng188, Fritz Zimprich177, Naomi R. Wray87, Kari Stefansson176, Peter M. Visscher87, Wellcome Trust Case-Control Consortium 2189, Rolf Adolfsson150, Ole A. Andreassen14,133, Douglas H. R. Blackwood132, Elvira Bramon190, Joseph D. Buxbaum35,36,91,191, Anders D. Børglum17,58,59,138, Sven Cichon55,56,95,192, Ariel Darvasi193, Enrico Domenici194, Hannelore Ehrenreich23, Tõnu Esko3,11,96,135, Pablo V. Gejman64,65, Michael Gill5, Hugh Gurling53, Christina M. Hultman26, Nakao Iwata98, Assen V. Jablensky39,102,186,195, Erik G. Jönsson12,14, Kenneth S. Kendler196, George Kirov6, Jo Knight105,106,107, Todd Lencz197,198,199, Douglas F. Levinson19, Qingqin S. Li86, Jianjun Liu188,200, Anil K. Malhotra197,198,199, Steven A. McCarroll2,96, Andrew McQuillin53, Jennifer L. Moran2, Preben B. Mortensen15,16,17, Bryan J. Mowry87,201, Markus M. Nöthen55,56, Roel A. Ophoff38,80,34, Michael J. Owen6,7, Aarno Palotie2,4,161, Carlos N. Pato110, Tracey L. Petryshen2,128,202, Danielle Posthuma203,204,205, Marcella Rietschel77, Brien P. Riley196, Dan Rujescu81,83, Pak C. Sham43,44,116 Pamela Sklar82,91,165, David St Clair206, Daniel R. Weinberger178,207, Jens R. Wendland166, Thomas Werge17,90,208, Mark J. Daly1,2,3, Patrick F. Sullivan26,51,160 & Michael C. O’Donovan6,7
The International Headache Genetics Consortium
Consortium members listed by their main affiliated cohort:
AGES: Leonore Launer1
Australia ATM: Dale Nyholt3
Barcelona headache group: Alfons Macaya4, Patricia Pozo-Rosich5, Bru Cormand6, Jessica Fernandez5, Marta Vila-Pueyo4, Celia Sintas6
Danish Headache Center, Glostrup Hospital: Jes Olesen2, Anne Francke Christensen2, Ann-Louise Esserlind2
ERF: Najaf Amin7
Estonian Biobank: Tonu Esko8
Finnish MA: Aarno Palotie9, Mikko Kallela10, Maija Wessman11, Ville Artto10, Verneri Anttila12, Eija Hämäläinen13, Priit Palta13, Padhraig Gormley9, Ester Cuenca9
FinnTwin: Jaakko Kaprio13
German MO/MA: Martin Dichgans14, Hartmut Göbel15, Christian Kubisch16, Tobias Freilinger17, Rainer Malik14, Bertram Muller-Myhsok18
HUNT: John-Anker Zwart19, Bendik Winsvold19, Line Jacobsen19, Linda Pedersen19
Kaiser Permanente: Alice Pressman20
LUMINA MO/MA: Arn van den Maagdenberg21, Gisela Terwindt22, Boukje de Vries21, Rune R. Frants21, Michel Ferrari22
NTR/NESDA: Dorret I. Boomsma23, Lannie Ligthart23, Brenda Penninx24
Swedish Twin Registry: Andrea Carmine Belin27, Nancy Pedersen28
23&Me, Mountainview, California: Nick Eriksson35
Footnotes
The authors report no conflicts of interest.
Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, Massachusetts 02114, USA.
Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, Massachusetts 02142, USA.
Medical and Population Genetics Program, Broad Institute of MIT and Harvard, Cambridge, Massachusetts 02142, USA.
Psychiatric and Neurodevelopmental Genetics Unit, Massachusetts General Hospital, Boston, Massachusetts 02114, USA.
Neuropsychiatric Genetics Research Group, Department of Psychiatry, Trinity College Dublin, Dublin 8, Ireland.
MRC Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, School of Medicine, Cardiff University, Cardiff, CF24 4HQ, UK.
National Centre for Mental Health, Cardiff University, Cardiff, CF24 4HQ, UK.
Eli Lilly and Company Limited, Erl Wood Manor, Sunninghill Road, Windlesham, Surrey, GU20 6PH, UK.
Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, King’s College London, London, SE5 8AF, UK.
Center for Biological Sequence Analysis, Department of Systems Biology, Technical University of Denmark, DK-2800, Denmark.
Division of Endocrinology and Center for Basic and Translational Obesity Research, Boston Children’s Hospital, Boston, Massachusetts, 02115USA.
Department of Clinical Neuroscience, Psychiatry Section, Karolinska Institutet, SE-17176 Stockholm, Sweden.
Department of Psychiatry, Diakonhjemmet Hospital, 0319 Oslo, Norway.
NORMENT, KG Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo, 0424 Oslo, Norway.
Centre for Integrative Register-based Research, CIRRAU, Aarhus University, DK-8210 Aarhus, Denmark.
National Centre for Register-based Research, Aarhus University, DK-8210 Aarhus, Denmark.
The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH, Denmark.
State Mental Hospital, 85540 Haar, Germany.
Department of Psychiatry and Behavioral Sciences, Stanford University, Stanford, California 94305, USA.
Department of Psychiatry and Behavioral Sciences, Atlanta Veterans Affairs Medical Center, Atlanta, Georgia 30033, USA.
Department of Psychiatry and Behavioral Sciences, Emory University, Atlanta Georgia 30322, USA.
Virginia Institute for Psychiatric and Behavioral Genetics, Department of Psychiatry, Virginia Commonwealth University, Richmond, Virginia 23298, USA.
Clinical Neuroscience, Max Planck Institute of Experimental Medicine, Göttingen 37075, Germany.
Department of Medical Genetics, University of Pécs, Pécs H-7624, Hungary.
Szentagothai Research Center, University of Pécs, Pécs H-7624, Hungary.
Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm SE-17177, Sweden.
Department of Psychiatry, University of Iowa Carver College of Medicine, Iowa City, Iowa 52242, USA.
University Medical Center Groningen, Department of Psychiatry, University of Groningen NL-9700 RB, The Netherlands.
School of Nursing, Louisiana State University Health Sciences Center, New Orleans, Louisiana 70112, USA.
Athinoula A. Martinos Center, Massachusetts General Hospital, Boston, Massachusetts 02129, USA.
Center for Brain Science, Harvard University, Cambridge, Massachusetts, 02138 USA.
Department of Psychiatry, Massachusetts General Hospital, Boston, Massachusetts, 02114 USA.
Department of Psychiatry, University of California at San Francisco, San Francisco, California, 94143 USA.
University Medical Center Utrecht, Department of Psychiatry, Rudolf Magnus Institute of Neuroscience, 3584 Utrecht, The Netherlands.
Department of Human Genetics, Icahn School of Medicine at Mount Sinai, New York, New York 10029 USA.
Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, New York 10029 USA.
Centre Hospitalier du Rouvray and INSERM U1079 Faculty of Medicine, 76301 Rouen, France.
Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, California 90095, USA.
Schizophrenia Research Institute, Sydney NSW 2010, Australia.
School of Psychiatry, University of New South Wales, Sydney NSW 2031, Australia.
Royal Brisbane and Women’s Hospital, University of Queensland, Brisbane, St Lucia QLD 4072, Australia.
Institute of Psychology, Chinese Academy of Science, Beijing 100101, China.
Department of Psychiatry, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
State Key Laboratory for Brain and Cognitive Sciences, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
Department of Computer Science, University of North Carolina, Chapel Hill, North Carolina 27514, USA.
Castle Peak Hospital, Hong Kong, China.
Institute of Mental Health, Singapore 539747, Singapore.
Department of Psychiatry, Washington University, St. Louis, Missouri 63110, USA.
Department of Child and Adolescent Psychiatry, Assistance Publique Hopitaux de Paris, Pierre and Marie Curie Faculty of Medicine and Institute for Intelligent Systems and Robotics, Paris, 75013, France.
Blue Note Biosciences, Princeton, New Jersey 08540, USA
Department of Genetics, University of North Carolina, Chapel Hill, North Carolina 27599-7264, USA.
Department of Psychological Medicine, Queen Mary University of London, London E1 1BB, UK.
Molecular Psychiatry Laboratory, Division of Psychiatry, University College London, London WC1E 6JJ, UK.
Sheba Medical Center, Tel Hashomer 52621, Israel.
Department of Genomics, Life and Brain Center, D-53127 Bonn, Germany.
Institute of Human Genetics, University of Bonn, D-53127 Bonn, Germany.
Applied Molecular Genomics Unit, VIB Department of Molecular Genetics, University of Antwerp, B-2610 Antwerp, Belgium.
Centre for Integrative Sequencing, iSEQ, Aarhus University, DK-8000 Aarhus C, Denmark.
Department of Biomedicine, Aarhus University, DK-8000 Aarhus C, Denmark.
First Department of Psychiatry, University of Athens Medical School, Athens 11528, Greece.
Department of Psychiatry, University College Cork, Co. Cork, Ireland.
Department of Medical Genetics, Oslo University Hospital, 0424 Oslo, Norway.
Cognitive Genetics and Therapy Group, School of Psychology and Discipline of Biochemistry, National University of Ireland Galway, Co. Galway, Ireland.
Department of Psychiatry and Behavioral Neuroscience, University of Chicago, Chicago, Illinois 60637, USA.
Department of Psychiatry and Behavioral Sciences, NorthShore University HealthSystem, Evanston, Illinois 60201, USA.
Department of Non-Communicable Disease Epidemiology, London School of Hygiene and Tropical Medicine, London WC1E 7HT, UK.
Department of Child and Adolescent Psychiatry, University Clinic of Psychiatry, Skopje 1000, Republic of Macedonia.
Department of Psychiatry, University of Regensburg, 93053 Regensburg, Germany.
Department of General Practice, Helsinki University Central Hospital, University of Helsinki P.O. Box 20, Tukholmankatu 8 B, FI-00014, Helsinki, Finland
Folkhälsan Research Center, Helsinki, Finland, Biomedicum Helsinki 1, Haartmaninkatu 8, FI-00290, Helsinki, Finland.
National Institute for Health and Welfare, P.O. BOX 30, FI-00271 Helsinki, Finland.
Translational Technologies and Bioinformatics, Pharma Research and Early Development, F. Hoffman-La Roche, CH-4070 Basel, Switzerland.
Department of Psychiatry, Georgetown University School of Medicine, Washington DC 20057, USA.
Department of Psychiatry, Keck School of Medicine of the University of Southern California, Los Angeles, California 90033, USA.
Department of Psychiatry, Virginia Commonwealth University School of Medicine, Richmond, Virginia 23298, USA.
Mental Health Service Line, Washington VA Medical Center, Washington DC 20422, USA.
Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, University of Heidelberg, Heidelberg, D-68159 Mannheim, Germany.
Department of Genetics, University of Groningen, University Medical Centre Groningen, 9700 RB Groningen, The Netherlands.
Department of Psychiatry, University of Colorado Denver, Aurora, Colorado 80045, USA.
Center for Neurobehavioral Genetics, Semel Institute for Neuroscience and Human Behavior, University of California, Los Angeles, California 90095, USA.
Department of Psychiatry, University of Halle, 06112 Halle, Germany.
Division of Psychiatric Genomics, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, New York 10029, USA.
Department of Psychiatry, University of Munich, 80336, Munich, Germany.
Departments of Psychiatry and Human and Molecular Genetics, INSERM, Institut de Myologie, Hôpital de la Pitiè-Salpêtrière, Paris, 75013, France.
Mental Health Research Centre, Russian Academy of Medical Sciences, 115522 Moscow, Russia.
Neuroscience Therapeutic Area, Janssen Research and Development, Raritan, New Jersey 08869, USA.
Queensland Brain Institute, The University of Queensland, Brisbane, Queensland, QLD 4072, Australia.
Academic Medical Centre University of Amsterdam, Department of Psychiatry, 1105 AZ Amsterdam, The Netherlands.
Illumina, La Jolla, California, California 92122, USA.
Institute of Biological Psychiatry, Mental Health Centre Sct. Hans, Mental Health Services Copenhagen, DK-4000, Denmark.
Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, New York 10029, USA.
J. J. Peters VA Medical Center, Bronx, New York, New York 10468, USA.
Priority Research Centre for Health Behaviour, University of Newcastle, Newcastle NSW 2308, Australia.
School of Electrical Engineering and Computer Science, University of Newcastle, Newcastle NSW 2308, Australia.
Division of Medical Genetics, Department of Biomedicine, University of Basel, Basel, CH-4058, Switzerland.
Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA.
Section of Neonatal Screening and Hormones, Department of Clinical Biochemistry, Immunology and Genetics, Statens Serum Institut, Copenhagen, DK-2300, Denmark.
Department of Psychiatry, Fujita Health University School of Medicine, Toyoake, Aichi, 470-1192, Japan.
Regional Centre for Clinical Research in Psychosis, Department of Psychiatry, Stavanger University Hospital, 4011 Stavanger, Norway.
Rheumatology Research Group, Vall d’Hebron Research Institute, Barcelona, 08035, Spain.
Centre for Medical Research, The University of Western Australia, Perth, WA 6009, Australia.
The Perkins Institute for Medical Research, The University of Western Australia, Perth, WA 6009, Australia.
Department of Medical Genetics, Medical University, Sofia1431, Bulgaria.
Department of Psychology, University of Colorado Boulder, Boulder, Colorado 80309, USA.
Campbell Family Mental Health Research Institute, Centre for Addiction and Mental Health, Toronto, Ontario, M5T 1R8, Canada.
Department of Psychiatry, University of Toronto, Toronto, Ontario, M5T 1R8, Canada.
Institute of Medical Science, University of Toronto, Toronto, Ontario, M5S 1A8, Canada.
Institute of Molecular Genetics, Russian Academy of Sciences, Moscow123182, Russia.
Latvian Biomedical Research and Study Centre, Riga, LV-1067, Latvia.
Department of Psychiatry and Zilkha Neurogenetics Institute, Keck School of Medicine at University of Southern California, Los Angeles, California 90089, USA.
Faculty of Medicine, Vilnius University, LT-01513 Vilnius, Lithuania.
Department of Biology and Medical Genetics, 2nd Faculty of Medicine and University Hospital Motol, 150 06 Prague, Czech Republic.
Department of Child and Adolescent Psychiatry, Pierre and Marie Curie Faculty of Medicine, Paris 75013, France.
Duke-NUS Graduate Medical School, Singapore 169857, Singapore.
Department of Psychiatry, Hadassah-Hebrew University Medical Center, Jerusalem 91120, Israel.
Centre for Genomic Sciences, The University of Hong Kong, Hong Kong, China.
Mental Health Centre and Psychiatric Laboratory, West China Hospital, Sichuan University, Chengdu, 610041, Sichuan, China.
Department of Biostatistics, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland 21205, USA.
Department of Psychiatry, Columbia University, New York, New York 10032, USA.
Priority Centre for Translational Neuroscience and Mental Health, University of Newcastle, Newcastle NSW 2300, Australia.
Department of Genetics and Pathology, International Hereditary Cancer Center, Pomeranian Medical University in Szczecin, 70-453 Szczecin, Poland.
Department of Mental Health and Substance Abuse Services; National Institute for Health and Welfare, P.O. BOX 30, FI-00271 Helsinki, Finland
Department of Mental Health, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland 21205, USA.
Department of Psychiatry, University of Bonn, D-53127 Bonn, Germany.
Centre National de la Recherche Scientifique, Laboratoire de Génétique Moléculaire de la Neurotransmission et des Processus Neurodégénératifs, Hôpital de la Pitié Salpêtrière, 75013, Paris, France.
Department of Genomics Mathematics, University of Bonn, D-53127 Bonn, Germany.
Research Unit, Sørlandet Hospital, 4604 Kristiansand, Norway.
Department of Psychiatry, Harvard Medical School, Boston, Massachusetts 02115, USA.
VA Boston Health Care System, Brockton, Massachusetts 02301, USA.
Department of Psychiatry, National University of Ireland Galway, Co. Galway, Ireland.
Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, Edinburgh EH16 4SB, UK.
Division of Psychiatry, University of Edinburgh, Edinburgh EH16 4SB, UK.
Division of Mental Health and Addiction, Oslo University Hospital, 0424 Oslo, Norway.
Massachusetts Mental Health Center Public Psychiatry Division of the Beth Israel Deaconess Medical Center, Boston, Massachusetts 02114, USA.
Estonian Genome Center, University of Tartu, Tartu 50090, Estonia.
School of Psychology, University of Newcastle, Newcastle NSW 2308, Australia.
First Psychiatric Clinic, Medical University, Sofia 1431, Bulgaria.
Department P, Aarhus University Hospital, DK-8240 Risskov, Denmark.
Department of Psychiatry, Royal College of Surgeons in Ireland, Dublin 2, Ireland.
King’s College London, London SE5 8AF, UK.
Maastricht University Medical Centre, South Limburg Mental Health Research and Teaching Network, EURON, 6229 HX Maastricht, The Netherlands.
Institute of Translational Medicine, University of Liverpool, Liverpool L69 3BX, UK.
Max Planck Institute of Psychiatry, 80336 Munich, Germany.
Munich Cluster for Systems Neurology (SyNergy), 80336 Munich, Germany.
Department of Psychiatry and Psychotherapy, Jena University Hospital, 07743 Jena, Germany.
Department of Psychiatry, Queensland Brain Institute and Queensland Centre for Mental Health Research, University of Queensland, Brisbane, Queensland, St Lucia QLD 4072, Australia.
Department of Psychiatry and Behavioral Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205, USA.
Department of Psychiatry, Trinity College Dublin, Dublin 2, Ireland.
Eli Lilly and Company, Lilly Corporate Center, Indianapolis, 46285 Indiana, USA.
Department of Clinical Sciences, Psychiatry, Umeå University, SE-901 87 Umeå, Sweden.
DETECT Early Intervention Service for Psychosis, Blackrock, Co. Dublin, Ireland.
Centre for Public Health, Institute of Clinical Sciences, Queen’s University Belfast, Belfast BT12 6AB, UK.
Lawrence Berkeley National Laboratory, University of California at Berkeley, Berkeley, California 94720, USA.
Institute of Psychiatry, King’s College London, London SE5 8AF, UK.
A list of authors and affiliations appear in the Supplementary Information.
Melbourne Neuropsychiatry Centre, University of Melbourne & Melbourne Health, Melbourne, Vic 3053, Australia.
Department of Psychiatry, University of Helsinki, P.O. Box 590, FI-00029 HUS, Helsinki, Finland.
Public Health Genomics Unit, National Institute for Health and Welfare, P.O. BOX 30, FI-00271 Helsinki, Finland
Medical Faculty, University of Belgrade, 11000 Belgrade, Serbia.
Department of Psychiatry, University of North Carolina, Chapel Hill, North Carolina 27599-7160, USA.
Institute for Molecular Medicine Finland, FIMM, University of Helsinki, P.O. Box 20 FI-00014, Helsinki, Finland
Department of Epidemiology, Harvard School of Public Health, Boston, Massachusetts 02115, USA.
Department of Psychiatry, University of Oxford, Oxford, OX3 7JX, UK.
Virginia Institute for Psychiatric and Behavioral Genetics, Virginia Commonwealth University, Richmond, Virginia 23298, USA.
Institute for Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York 10029, USA.
PharmaTherapeutics Clinical Research, Pfizer Worldwide Research and Development, Cambridge, Massachusetts 02139, USA.
Department of Psychiatry and Psychotherapy, University of Gottingen, 37073 Göttingen, Germany.
Psychiatry and Psychotherapy Clinic, University of Erlangen, 91054 Erlangen, Germany.
Hunter New England Health Service, Newcastle NSW 2308, Australia.
School of Biomedical Sciences and Pharmacy, University of Newcastle, Callaghan NSW 2308, Australia.
Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, Maryland 20892, USA.
University of Iceland, Landspitali, National University Hospital, 101 Reykjavik, Iceland.
Department of Psychiatry and Drug Addiction, Tbilisi State Medical University (TSMU), N33, 0177 Tbilisi, Georgia.
Research and Development, Bronx Veterans Affairs Medical Center, New York, New York 10468, USA.
Wellcome Trust Centre for Human Genetics, Oxford, OX3 7BN, UK.
deCODE Genetics, 101 Reykjavik, Iceland.
Department of Clinical Neurology, Medical University of Vienna, 1090 Wien, Austria.
Lieber Institute for Brain Development, Baltimore, Maryland 21205, USA.
Department of Medical Genetics, University Medical Centre Utrecht, Universiteitsweg 100, 3584 CG, Utrecht, The Netherlands.
Berkshire Healthcare NHS Foundation Trust, Bracknell RG12 1BQ, UK.
Section of Psychiatry, University of Verona, 37134 Verona, Italy.
Department of Psychiatry, University of Oulu, P.O. BOX 5000, 90014, Finland
University Hospital of Oulu, P.O. BOX 20, 90029 OYS, Finland.
Molecular and Cellular Therapeutics, Royal College of Surgeons in Ireland, Dublin 2, Ireland.
Health Research Board, Dublin 2, Ireland.
School of Psychiatry and Clinical Neurosciences, The University of Western Australia, Perth WA6009, Australia.
Computational Sciences CoE, Pfizer Worldwide Research and Development, Cambridge, Massachusetts 02139, USA.
Human Genetics, Genome Institute of Singapore, A*STAR, Singapore 138672, Singapore.
A list of authors and affiliations appear in the Supplementary Information.
University College London, London WC1E 6BT, UK.
Department of Neuroscience, Icahn School of Medicine at Mount Sinai, New York, New York 10029, USA.
Institute of Neuroscience and Medicine (INM-1), Research Center Juelich, 52428 Juelich, Germany.
Department of Genetics, The Hebrew University of Jerusalem, 91905 Jerusalem, Israel.
Neuroscience Discovery and Translational Area, Pharma Research and Early Development, F. Hoffman-La Roche, CH-4070 Basel, Switzerland.
Centre for Clinical Research in Neuropsychiatry, School of Psychiatry and Clinical Neurosciences, The University of Western Australia, Medical Research Foundation Building, Perth WA 6000, Australia.
Virginia Institute for Psychiatric and Behavioral Genetics, Departments of Psychiatry and Human and Molecular Genetics, Virginia Commonwealth University, Richmond, Virginia 23298, USA.
The Feinstein Institute for Medical Research, Manhasset, New York, 11030 USA.
The Hofstra NS-LIJ School of Medicine, Hempstead, New York, 11549 USA.
The Zucker Hillside Hospital, Glen Oaks, New York,11004 USA.
Saw Swee Hock School of Public Health, National University of Singapore, Singapore 117597, Singapore.
Queensland Centre for Mental Health Research, University of Queensland, Brisbane 4076, Queensland, Australia.
Center for Human Genetic Research and Department of Psychiatry, Massachusetts General Hospital, Boston, Massachusetts 02114, USA.
Department of Child and Adolescent Psychiatry, Erasmus University Medical Centre, Rotterdam 3000, The Netherlands.
Department of Complex Trait Genetics, Neuroscience Campus Amsterdam, VU University Medical Center Amsterdam, Amsterdam 1081, The Netherlands.
Department of Functional Genomics, Center for Neurogenomics and Cognitive Research, Neuroscience Campus Amsterdam, VU University, Amsterdam 1081, The Netherlands.
University of Aberdeen, Institute of Medical Sciences, Aberdeen, AB25 2ZD, UK.
Departments of Psychiatry, Neurology, Neuroscience and Institute of Genetic Medicine, Johns Hopkins School of Medicine, Baltimore, Maryland 21205, USA.
Department of Clinical Medicine, University of Copenhagen, Copenhagen 2200, Denmark.
Departments of Psychiatry and Human Genetics, University of Chicago, Chicago, Illinois 60637, USA.
Laboratory of Epidemiology, Demography and Biometry, National Institute on Aging, Bethesda, Maryland, USA.
Medical Research Council (MRC) Integrative Epidemiology Unit at the University of Bristol, Bristol, UK.
Queensland Institute of Medical Research, Brisbane, Queensland, Australia.
Pediatric Neurology Research Group, Institut de Recerca (VHIR), Universitat Autònoma de Barcelona, Barcelona, Spain.
Headache and Neurological Pain Research Group, Institut de Recerca (VHIR), Universitat Autònoma de Barcelona, Barcelona.
Departament de Genètica, Facultat de Biologia, Universitat de Barcelona, Barcelona, Spain.
Department of Epidemiology, Erasmus University Medical Centre, Rotterdam, The Netherlands.
Estonian Genome Center, University of Tartu, Tartu, Estonia.
Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA, USA.
Department of Neurology, Helsinki University Central Hospital, Helsinki, Finland.
Institute of Genetics, Folkhälsan Research Center, Helsinki, Finland.
Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA.
Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland.
Institute for Stroke and Dementia Research, Klinikum der Universität München, Ludwig-Maximilians-Universität, Munich, Germany.
Kiel Pain and Headache Center, Kiel, Germany.
Institute of Human Genetics, University of Ulm, Ulm, Germany.
Department of Neurology and Epileptology and Hertie-Institute for Clinical Brain Research, University of Tübingen.
Max Planck Institute of Psychiatry, Munich, Germany.
FORMI, Oslo University Hospital, Oslo, Norway.
Division of Research, Kaiser Permanente, Oakland, CA, USA.
Department of Human Genetics, Leiden University Medical Centre, Leiden, The Netherlands.
Department of Neurology, Leiden University Medical Centre, Leiden, The Netherlands.
Department of Biological Psychology, VU University, Amsterdam, The Netherlands.
Department of Psychiatry, VU University Medical Center, Amsterdam, The Netherlands.
Biocenter Oulu, University of Oulu, Oulu, Finland.
Institute of Health Sciences, University of Oulu, Oulu, Finland.
Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden.
Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden.
Department of Twin Research and Genetic Epidemiology, King’s College London, London, UK.
Division of Preventive Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA.
Institut National de la Santé et de la Recherche Médicale (INSERM) Research Center for Epidemiology and Biostatistics (U897) Team–Neuroepidemiology, Bordeaux, France.
Department of Neurology, University Hospital Essen, Essen, Germany.
Department of Clinical Chemistry, Fimlab Laboratories, Tampere, Finland.
Department of Clinical Physiology and Nuclear Medicine, Turku University Hospital, Turku, Finland.
23andMe, Mountain View, California, USA.
Financial Disclosure
The study was supported by the Research Council of Norway (#213837, #223273, #225989) and South-East Norway Health Authority (# 2013-123). The funding agencies had no role in the conception, design of the study or collection of data.
References
- 1.Allen JS, Sarich VM. Schizophrenia in an Evolutionary Perspective. Perspect Biol Med. 1988;32:132–153. doi: 10.1353/pbm.1988.0039. [DOI] [PubMed] [Google Scholar]
- 2.Crow TJ. A Darwinian Approach to the Origins of Psychosis [Published Erratum Appears in Br J Psychiatry 1995 Sep;167:414] The British Journal of Psychiatry. 1995;167:12–25. doi: 10.1192/bjp.167.1.12. [DOI] [PubMed] [Google Scholar]
- 3.Burns JK. Psychosis: A Costly by-Product of Social Brain Evolution in Homo Sapiens. Prog Neuropsychopharmacol Biol Psychiatry. 2006;30:797–814. doi: 10.1016/j.pnpbp.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Polimeni J, Reiss JP. Evolutionary Perspectives on Schizophrenia. Can J Psychiatry. 2003;48:34–39. doi: 10.1177/070674370304800107. [DOI] [PubMed] [Google Scholar]
- 5.Lewis DA, Hashimoto T, Volk DW. Cortical Inhibitory Neurons and Schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 6.Brune M. Schizophrenia-an Evolutionary Enigma? Neurosci Biobehav Rev. 2004;28:41–53. doi: 10.1016/j.neubiorev.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, et al. Common Genetic Determinants of Schizophrenia and Bipolar Disorder in Swedish Families: A Population-Based Study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Power RA, Kyaga S, Uher R, MacCabe JH, Langstrom N, Landen M, et al. Fecundity of Patients with Schizophrenia, Autism, Bipolar Disorder, Depression, Anorexia Nervosa, or Substance Abuse Vs Their Unaffected Siblings. JAMA Psychiatry. 2013;70:22–30. doi: 10.1001/jamapsychiatry.2013.268. [DOI] [PubMed] [Google Scholar]
- 9.Uher R. The Role of Genetic Variation in the Causation of Mental Illness: An Evolution-Informed Framework. Mol Psychiatry. 2009;14:1072–1082. doi: 10.1038/mp.2009.85. [DOI] [PubMed] [Google Scholar]
- 10.Pearlson GD, Folley BS. Schizophrenia, Psychiatric Genetics, and Darwinian Psychiatry: An Evolutionary Framework. Schizophr Bull. 2008;34:722–733. doi: 10.1093/schbul/sbm130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jablensky A, Sartorius N, Ernberg G, Anker M, Korten A, Cooper J, et al. Schizophrenia: Manifestations, Incidence and Course in Different Cultures a World Health Organization Ten-Country Study. Psychol Med Monogr Suppl. 1992;20:1–97. doi: 10.1017/s0264180100000904. [DOI] [PubMed] [Google Scholar]
- 12.Huxley J, Mayr E, Osmond H, Hoffer A. Schizophrenia as a Genetic Morphism. Nature. 1964;206:220–221. doi: 10.1038/204220a0. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Visscher PM, Wray NR. Sporadic Cases Are the Norm for Complex Disease. Eur J Hum Genet. 2010;18:1039–1043. doi: 10.1038/ejhg.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stringer C. The Morphological and Behavioral Origins of Modern Humans. In: Crow TJ, editor. The Speciation of Modern Homo Sapiens. Oxford: Oxford University Press; 2002. [Google Scholar]
- 15.Hublin JJ, Spoor F, Braun M, Zonneveld F, Condemi S. A Late Neanderthal Associated with Upper Palaeolithic Artefacts. Nature. 1996;381:224–226. doi: 10.1038/381224a0. [DOI] [PubMed] [Google Scholar]
- 16.d’Errico F, Henshilwood C, Lawson G, Vanhaeren M, Tillier AM, Soressi M, et al. Archaeological Evidence for the Emergence of Language, Symbolism, and Music - an Alternative Multidisciplinary Perspective. Journal of World Prehistory. 2003;17:1–70. [Google Scholar]
- 17.Crow TJ. Schizophrenia as the Price That Homo Sapiens Pays for Language: A Resolution of the Central Paradox in the Origin of the Species. Brain Research Reviews. 2000;31:118–129. doi: 10.1016/s0165-0173(99)00029-6. [DOI] [PubMed] [Google Scholar]
- 18.Wynn T, Coolidge FL. The Implications of the Working Memory Model for the Evolution of Modern Cognition. Int J Evol Biol. 2011;2011:741357. doi: 10.4061/2011/741357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crow TJ. Is Schizophrenia the Price That Homo Sapiens Pays for Language? Schizophr Res. 1997;28:127–141. doi: 10.1016/s0920-9964(97)00110-2. [DOI] [PubMed] [Google Scholar]
- 20.Schizophrenia Working Group of the Psychiatric Genomics C. Biological Insights from 108 Schizophrenia-Associated Genetic Loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, et al. A Draft Sequence of the Neandertal Genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, et al. The Complete Genome Sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner GP, Zhang J. The Pleiotropic Structure of the Genotype-Phenotype Map: The Evolvability of Complex Organisms. Nat Rev Genet. 2011;12:204–213. doi: 10.1038/nrg2949. [DOI] [PubMed] [Google Scholar]
- 24.International Schizophrenia C. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. Common Polygenic Variation Contributes to Risk of Schizophrenia and Bipolar Disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Efron B. Large-Scale Inference : Empirical Bayes Methods for Estimation, Testing, and Prediction. Cambridge ; New York: Cambridge University Press; 2010. [Google Scholar]
- 26.Schork AJ, Thompson WK, Pham P, Torkamani A, Roddey JC, Sullivan PF, et al. All Snps Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated Snps. PLoS Genet. 2013;9:e1003449. doi: 10.1371/journal.pgen.1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu JZ, Hov JR, Folseraas T, Ellinghaus E, Rushbrook SM, Doncheva NT, et al. Dense Genotyping of Immune-Related Disease Regions Identifies Nine New Risk Loci for Primary Sclerosing Cholangitis. Nat Genet. 2013;45:670–675. doi: 10.1038/ng.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreassen OA, Thompson WK, Schork AJ, Ripke S, Mattingsdal M, Kelsoe JR, et al. Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate. PLoS Genet. 2013;9:e1003455. doi: 10.1371/journal.pgen.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreassen OA, Djurovic S, Thompson WK, Schork AJ, Kendler KS, O’Donovan MC, et al. Improved Detection of Common Variants Associated with Schizophrenia by Leveraging Pleiotropy with Cardiovascular-Disease Risk Factors. Am J Hum Genet. 2013;92:197–209. doi: 10.1016/j.ajhg.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreassen OA, Zuber V, Thompson WK, Schork AJ, Bettella F, et al. Consortium P. Shared Common Variants in Prostate Cancer and Blood Lipids. Int J Epidemiol. 2014;43:1205–1214. doi: 10.1093/ije/dyu090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andreassen OA, McEvoy LK, Thompson WK, Wang Y, Reppe S, Schork AJ, et al. Identifying Common Genetic Variants in Blood Pressure Due to Polygenic Pleiotropy with Associated Phenotypes. Hypertension. 2014 doi: 10.1161/HYPERTENSIONAHA.113.02077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andreassen OA, Harbo HF, Wang Y, Thompson WK, Schork AJ, Mattingsdal M, et al. Genetic Pleiotropy between Multiple Sclerosis and Schizophrenia but Not Bipolar Disorder: Differential Involvement of Immune-Related Gene Loci. Mol Psychiatry. 2015;20:207–214. doi: 10.1038/mp.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burbano HA, Hodges E, Green RE, Briggs AW, Krause J, Meyer M, et al. Targeted Investigation of the Neandertal Genome by Array-Based Sequence Capture. Science. 2010;328:723–725. doi: 10.1126/science.1188046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. Spatio-Temporal Transcriptome of the Human Brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andreassen OA, Djurovic S, Thompson WK, Schork AJ, Kendler KS, O’Donovan MC, et al. Improved Detection of Common Variants Associated with Schizophrenia by Leveraging Pleiotropy with Cardiovascular-Disease Risk Factors. Am J Hum Genet. 2013;92:197–209. doi: 10.1016/j.ajhg.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium TEP. The Encode (Encyclopedia of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 37.Ryan TP. Modern Engineering Statistics. Hoboken, N.J: Wiley-Interscience; 2007. [Google Scholar]
- 38.Zablocki RW, Schork AJ, Levine RA, Andreassen OA, Dale AM, Thompson WK. Covariate-Modulated Local False Discovery Rate for Genome-Wide Association Studies. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, et al. Genome-Wide Association Analysis Identifies 13 New Risk Loci for Schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamm GB, Pisciottano F, Kliger R, Franchini LF. The Developmental Brain Gene Npas3 Contains the Largest Number of Accelerated Regulatory Sequences in the Human Genome. Mol Biol Evol. 2013;30:1088–1102. doi: 10.1093/molbev/mst023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison PJ, Owen MJ. Genes for Schizophrenia? Recent Findings and Their Pathophysiological Implications. The Lancet. 2003;361:417–419. doi: 10.1016/S0140-6736(03)12379-3. [DOI] [PubMed] [Google Scholar]
- 42.Bigdeli TB, Fanous AH, Riley BP, Reimers M, Chen X, et al. Schizophrenia Psychiatric Genome-Wide Association Study C. On Schizophrenia as a “Disease of Humanity”. Schizophr Res. 2013;143:223–224. doi: 10.1016/j.schres.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu K, Schadt EE, Pollard KS, Roussos P, Dudley JT. Genomic and Network Patterns of Schizophrenia Genetic Variation in Human Evolutionary Accelerated Regions. Mol Biol Evol. 2015;32:1148–1160. doi: 10.1093/molbev/msv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daub JT, Hofer T, Cutivet E, Dupanloup I, Quintana-Murci L, Robinson-Rechavi M, et al. Evidence for Polygenic Adaptation to Pathogens in the Human Genome. Mol Biol Evol. 2013;30:1544–1558. doi: 10.1093/molbev/mst080. [DOI] [PubMed] [Google Scholar]
- 45.Perry GH, Foll M, Grenier JC, Patin E, Nedelec Y, Pacis A, et al. Adaptive, Convergent Origins of the Pygmy Phenotype in African Rainforest Hunter-Gatherers. Proc Natl Acad Sci U S A. 2014;111:E3596–3603. doi: 10.1073/pnas.1402875111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turchin MC, Chiang CW, Palmer CD, Sankararaman S, Reich D, et al. Genetic Investigation of ATC. Evidence of Widespread Selection on Standing Variation in Europe at Height-Associated Snps. Nat Genet. 2012;44:1015–1019. doi: 10.1038/ng.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu W, Akey JM. Selection and Adaptation in the Human Genome. Annu Rev Genomics Hum Genet. 2013;14:467–489. doi: 10.1146/annurev-genom-091212-153509. [DOI] [PubMed] [Google Scholar]
- 48.Burger R, Gimelfarb A. Genetic Variation Maintained in Multilocus Models of Additive Quantitative Traits under Stabilizing Selection. Genetics. 1999;152:807–820. doi: 10.1093/genetics/152.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pritchard JK, Di Rienzo A. Adaptation - Not by Sweeps Alone. Nat Rev Genet. 2010;11:665–667. doi: 10.1038/nrg2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andreassen OA, Thompson WK, Dale AM. Boosting the Power of Schizophrenia Genetics by Leveraging New Statistical Tools. Schizophr Bull. 2014;40:13–17. doi: 10.1093/schbul/sbt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andreassen OA, Harbo HF, Wang Y, Thompson WK, Schork AJ, Mattingsdal M, et al. Genetic Pleiotropy between Multiple Sclerosis and Schizophrenia but Not Bipolar Disorder: Differential Involvement of Immune-Related Gene Loci. Mol Psychiatry. 2014 doi: 10.1038/mp.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, et al. Disc1 and Pde4b Are Interacting Genetic Factors in Schizophrenia That Regulate Camp Signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 53.Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y, et al. De Novo Gene Mutations Highlight Patterns of Genetic and Neural Complexity in Schizophrenia. Nat Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cacabelos R, Martínez-Bouza R. Genomics and Pharmacogenomics of Schizophrenia. CNS Neuroscience & Therapeutics. 2011;17:541–565. doi: 10.1111/j.1755-5949.2010.00187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guarguaglini G, Duncan PI, Stierhof YD, Holmström T, Duensing S, Nigg EA. The Forkhead-Associated Domain Protein Cep170 Interacts with Polo-Like Kinase 1 and Serves as a Marker for Mature Centrioles. Molecular Biology of the Cell. 2005;16:1095–1107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lucker BF, Behal RH, Qin H, Siron LC, Taggart WD, Rosenbaum JL, et al. Characterization of the Intraflagellar Transport Complex B Core: Direct Interaction of the Ift81 and Ift74/72 Subunits. J Biol Chem. 2005;280:27688–27696. doi: 10.1074/jbc.M505062200. [DOI] [PubMed] [Google Scholar]
- 57.Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, Stoetzel C, et al. Candidate Exome Capture Identifies Mutation of Sdccag8 as the Cause of a Retinal-Renal Ciliopathy. Nat Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Girirajan S, Dennis Megan Y, Baker C, Malig M, Coe Bradley P, Campbell Catarina D, et al. Refinement and Discovery of New Hotspots of Copy-Number Variation Associated with Autism Spectrum Disorder. Am J Hum Genet. 2013;92:221–237. doi: 10.1016/j.ajhg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bacon C, Schneider M, Le Magueresse C, Froehlich H, Sticht C, Gluch C, et al. Brain-Specific Foxp1 Deletion Impairs Neuronal Development and Causes Autistic-Like Behaviour. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of Common Variants Associated with Human Hippocampal and Intracranial Volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large Recurrent Microdeletions Associated with Schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.