Abstract
Next-generation sequencing has evolved technically and economically into the method of choice for interrogating the genome in cancer and inherited disorders. The introduction of procedural code sets for whole-exome and genome sequencing is a milestone toward financially sustainable clinical implementation; however, achieving reimbursement is currently a major challenge. As part of a prospective quality-improvement initiative to implement the new code sets, we adopted Agile, a development methodology originally devised in software development. We implemented eight functionally distinct modules (request review, cost estimation, preauthorization, accessioning, prebilling, testing, reporting, and reimbursement consultation) and obtained feedback via an anonymous survey. We managed 50 clinical requests (January to June 2015). The fraction of pursued-to-requested cases (n = 15/50; utilization management fraction, 0.3) aimed for a high rate of preauthorizations. In 13 of 15 patients the insurance plan required preauthorization, which we obtained in 70% and ultimately achieved reimbursement in 50%. Interoperability enabled assessment of 12 different combinations of modules that underline the importance of an adaptive workflow and policy tailoring to achieve higher yields of reimbursement. The survey confirmed a positive attitude toward self-organizing teams. We acknowledge the individuals and their interactions and termed the infrastructure: human pipeline. Nontechnical barriers currently are limiting the scope and availability of clinical genomic sequencing. The presented human pipeline is one approach toward long-term financial sustainability of clinical genomics.
Next-generation sequencing technologies are more powerful than traditional sequencing methods because they can provide an unbiased view of the whole genome.1, 2 Numerous studies have emphasized diagnostic advantages and there is an implicit understanding that exome and genome data will revolutionize clinical medicine.1, 2, 3, 4, 5 The American Medical Association recently introduced a Current Procedural Terminology (CPT) code set for exome and genome sequencing. The introduction of these codes acknowledges the clinical utility of these tests and generated the potential for insurance reimbursement. Moreover, applied within an appropriate infrastructure, these CPT codes should expand patient access to genomic sequencing.
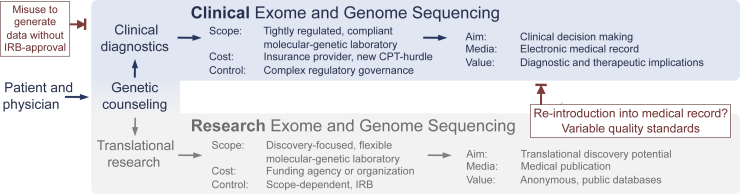
As US payment models rapidly evolve to incentivize value-based health care,6, 7 health systems need to emphasize optimal utilization of resources to remain financially viable.8, 9 Historic barriers to clinical application of genomics included sequencing cost and technical constraints. Rapidly improving sequencing technologies and analytic pipelines continue to overcome technical hurdles2 and significantly decrease costs (National Human Genome Research Institute, https://www.genome.gov/sequencingcosts, last accessed March 6, 2016). However, a key barrier to more wide-scale application remains: the lack of an operational infrastructure for logistics and revenue management in the clinical context.10 Current models lack efficient processes for utilization management, regulatory compliance, clinical quality assurance, and results reporting. In addition, current attempts to use genomic sequencing have failed to achieve long-term financial sustainability10 in that they do not include sufficient systems for adequate billing and reimbursement. In particular, clinical genomics requires navigating many nontechnical challenges beyond those required for research applications. Because of the insufficient availability of routine clinical genomic testing, clinicians often have substituted genomic testing performed in a research setting.4, 11 However, use of research test results performed outside a Clinical Laboratory Improvement Amendments–certified laboratory (CLIA1988)12 for use in patient diagnosis and clinical management is not only a violation of federal law [U.S. Code, Title 42, Chapter 6A, Subchapter II, Part F, Subpart 2, ×263a(a-q), Oct 31, 1988], but it also may endanger patients to the extent that nonclinical research testing is held to more variable quality standards (Figure 1). Even those research-focused laboratories that have obtained a Clinical Laboratory Improvement Amendments certification generally have not overcome the more significant barrier related to effective integration of genomic results into clinical diagnosis and continuous patient management.4, 11
Figure 1.
Comparison between clinical and research exome and genome sequencing. The scope, cost, and regulatory governance differ substantially between translational research (bottom) and clinical diagnostics (top). In contrast to the broad adoption of exome and genome sequencing by the scientific community, the administrative burden, regulatory complexity, and unresolved reimbursement situation in clinical diagnostics can appear as an unnecessary hurdle to an otherwise easily available test. The lack of a functioning clinical workflow has turned the institutionally review board–approved, patient-consented approach (bottom) into a pathway to re-introduce research-based findings into the medical record (bottom right, blocked red arrow). In reverse, utilization management is needed to oversee the clinical diagnostic route to prevent misuse (top left, blocked red arrow) of clinical data for research studies without appropriate institutional review. The comparison shows the need for a minimum viable workflow for clinical-grade, reimbursable exome and genome sequencing. CPT, Current Procedural Terminology (medical code set maintained by the American Medical Association through the CPT Editorial Panel); IRB, institutional review board/ethics committee.
To overcome the nontechnical barriers currently limiting the scope and availability of clinical genomic sequencing,10 we sought to define and implement systems and processes that could enable logistically scalable and financially sustainable clinical genomics. To facilitate the alignment of the numerous individuals and teams encompassing diverse skill sets and performing highly varied functions, we used an iterative development approach (Agile). Although Agile is derived from software development, it is not a software tool or a computer program. Agile is a set of development methodologies that have evolved into a standard used in many industries, with the notable exception of health care. By describing the systems and processes for a clinically valid workflow, and by reporting their performance in a prospective pilot, we exemplify one approach for financially sustainable clinical genomics.
Materials and Methods
Setting
The project sites were Massachusetts General Hospital, Brigham and Women's Hospital, and the Partners Laboratory for Molecular Medicine. The project, ongoing as a prospective quality-improvement initiative, does not require formal review or approval by the respective institutional review board for research activities per their policies (institutional checklist, Partners Human Research Committee, version May, 25th, 2012).
Design
After definition of project constraints (Supplemental Figure S1A), we established our management strategy13 (Supplemental Table S1) and chose and adopted Agile as our development methodology (Supplemental Figure S1B and Supplemental Tables S2 and S3). Agile is a development methodology that relies on a distinct combination of recursive and iterative delivery components. First, when the complexity requires recursion (a method in which the solution to a problem depends on solutions to smaller instances of the same problem), Agile is recursive and breaks down large projects into smaller components. Second, Agile is iterative and uses incremental delivery (eg, of the aforementioned smaller components) to add value immediately and allows for early discovery and correction of mistakes. Thereby, Agile is incorporating feedback-based improvements while delivering (additional) value earlier than with traditional delivery methodologies. An integral part of this development methodology is a specific terminology (Table 1) and two of the authors (J.K.L. and N.B.-L.) have formal Agile training (certified scrum master) and several authors have multiple years of experience working in Agile teams. The initial backlog (Table 1) was generated via one-on-one interviews with subject-matter experts including clinical geneticists, industry leaders, next-generation sequencing experts, big data analytic and genomics experts, payor-policy managers, preauthorization teams, and medical directors.
Table 1.
Agile Glossary
| Term | Explanation |
|---|---|
| Agile | Similar to Six Sigma or Lean, Agile is a development methodology originally devised in the software industry. Agile is a group of development methodologies in which requirements and solutions evolve through collaboration between self-organizing teams. Agile emphasizes adaptive planning, evolutionary development, early delivery, and continuous improvement, and encourages rapid and flexible response to change. |
| Story | A statement that summarizes the work that must be performed to deliver a specific function. Stories are the basis for communication, planning, and describing requirements. Typically stories capture the who, what, and why in a simple and concise way. Example: instead of receiving unlabeled samples, as an extraction technician, I want appropriately labeled samples for extraction, shearing, and storage. |
| Task | Description of the actual work needed to complete a story; typically several tasks per story. Each team member owns at least one task (including test, inspection, and/or verification of the task). |
| Backlog | A collection of stories and tasks the team will work on at some point in the future (the to-do list).∗ |
| Epic (module)† | Very large story or sets of stories that can be broken down into smaller stories; typically applied for components in which full elaboration has been deferred until actually needed. We applied the term module instead of epic to emphasize interchangeability in the workflow. |
| Scrum | One of the most widely recognized Agile development frameworks. Scrum consists of a series of short iterations (sprints), each of which ends with the delivery of a valuable increment and review of the iteration (sprint review). |
| Sprint | Defined here as ultra-short, one-on-one meetings that determine workflow and tasks for each self-organizing team to deliver the next most valuable component (iterative delivery process). |
An excerpt of our current backlog is provided in Supplemental Table S4.
The term module is not part of the original Agile terminology; however, as stated above, we applied the term module to emphasize the interchangeability of the eight functionally distinct workflow components (Figure 2).
Workflow
To develop a series of semiautonomous, yet highly interoperable, modules (defined as workflow components organized in functional units that can be combined in different orders according to the specific needs per case), we applied Agile because it emphasizes self-organizing groups that optimize their respective processes according to functional specifications defined in stories (Table 1). Stories are short phrases and simplified descriptions that capture requirements in a succinct way14 and we followed a certain format: “instead of what not, as who I want what so that why.” An initial story following this format was as follows: “instead of not knowing how much a patient has to pay, as a physician, I want to know how much the test cost is so that I can tell the patient.” Throughout the project, stories develop and the wording is refined (eg, “Although it is not my responsibility to know details of the insurance plan of every patient, as the ordering physician, I want to provide the patient with a reliable maximum out-of-pocket estimate so that we will not be surprised about an unexpected bill”). These stories are high-level definitions of the required individual tasks and can be grouped together into functional modules. The modular approach allowed us to reduce a complex pipeline into subcomponents of manageable complexity. We considered tasks, stories, and modules as implemented when we received evidence of appropriate function from external sources (eg, payors or subsequent teams). We provided the implementation order and details about the timeline of delivering individual modules (Supplemental Figure S2).
Performance Assessment
We prospectively developed several key measures to assess performance of the workflow. One measure represented the proportion of cases initially submitted into the pipeline that were found to be inappropriate in prescreening, defining the utilization management fraction as the fraction of pursued-to-requested cases. We monitored cases passing institutional review by documenting the time necessary to complete each step, and by assessing the rates of successfully obtained insurance preauthorization and payment. We documented professional degrees and years of experience for each team member. When multiple individuals performed one function we noted the lowest educational and experience level. Recognizing variability between institutions, we provide these workforce measures (eg, cumulative years of experience per module) to allow some comparison of module complexity. For review of team satisfaction we used an anonymous survey,15 and for analysis of dichotomous answers we considered a preference toward one answer by more than 80% of the respondents as noteworthy.
Health Insurance Policy Review
For CPT codes corresponding to molecular testing (included within CPT81161 to 89291 and S3833 to S3870) we reviewed the specific coverage policies for the 12 most common private payors used by our patient population. We derived a seven-tiered classification scheme to describe coverage policies and used this to compare insurance policy content by CPT code across the 12 payors. We tracked policy changes over the course of the project and noted the changes on our classification scheme.
Results
Establishing the first operating workflow components and applying the new CPT code sets took 68 workdays (Supplemental Figure S2), and delivering and testing all modules in various combinations took another 85 days (Supplemental Figure S2). The established workflow consists of eight interoperable modules (Figure 2) that we deconstructed via sprints into 34 stories and 129 tasks provided in workflow diagrams (Supplemental Figures S3–S8). We detail the functions of each module along with the results from development testing performed using 50 clinical requests.
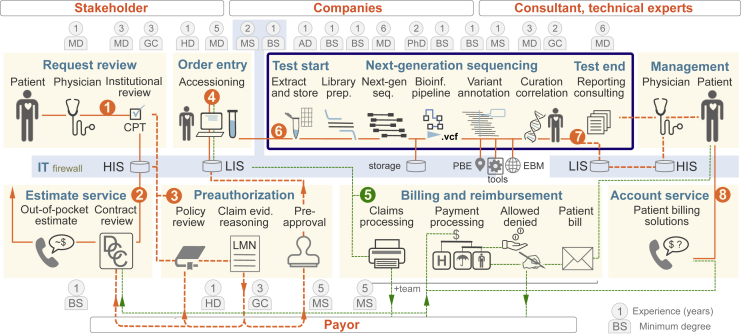
Figure 2.
The human pipeline of clinical exome and genome sequencing. The term pipeline is applied by bioinformaticians for a set of defined steps that lead from raw next-generation sequencing reads to variant call files (.vcf ± subsequent interpretation) as a central component of next-generation sequencing–based diagnostic tests (blue frame). To implement clinical exome and genome sequencing, we provide the current state of our pipeline from request to report and clinical management. The principle pathway is indicated by the following numbers in orange circles: (1) institutional review, (2) cost estimation, (3) preauthorization, (4) accessioning, (5) billing, (6) clinical exome and genome sequencing, (7) report and report consultation, and (8) patient financial counseling. Given that the personnel, their interactions, and professional qualifications play a central role in the clinical environment, we applied the term human pipeline and provide professional degrees and years of experience after training of the least experienced team member serving in each role. Detailed workflow diagrams can be found in Supplemental Figures S3–S8. AD, administrative personnel with various degrees; BS, Bachelor of Science; CPT, Current Procedural Terminology; DCC logo, deductible, copay, co-insurance; EBM, evidence-based medicine; GC, genetic counselor with board certification; HD, high-school diploma with on-the-job training; HIS, hospital information system; IT, information technology; LIS, laboratory information system; LMN, Letter of Medical Necessity; MD, medical school degree plus board examination; MS, Master of Science; PBE, practice-based evidence; PhD, doctor of philosophy in bioinformatics; Umbrella (logo), insurance covered part of the payment.
Module 1: Institutional Review Team
The function of this team is to ascertain the appropriateness of the test request and reject cases early to limit inefficient use of resources (Figure 2). Each request was screened by a series of questions (Supplemental Figure S3) and we rejected 35 of 50 clinical requests (utilization management fraction, 0.3) (Figure 3A). Our stringent review with 70% rejections goes beyond gatekeeping and recognizes that even experts rarely know all available test alternatives, or have the time to take payor rules into account. Thus, in all 35 rejected cases we offered a patient-tailored molecular-genetic consultation for appropriate work-up to the ordering physician (not shown) and listed consolidated rejection reasons (Figure 3A). The team, including the requesting physician, consists of three individuals and has a collective postgraduate experience of 7 years (Supplemental Figure S3).
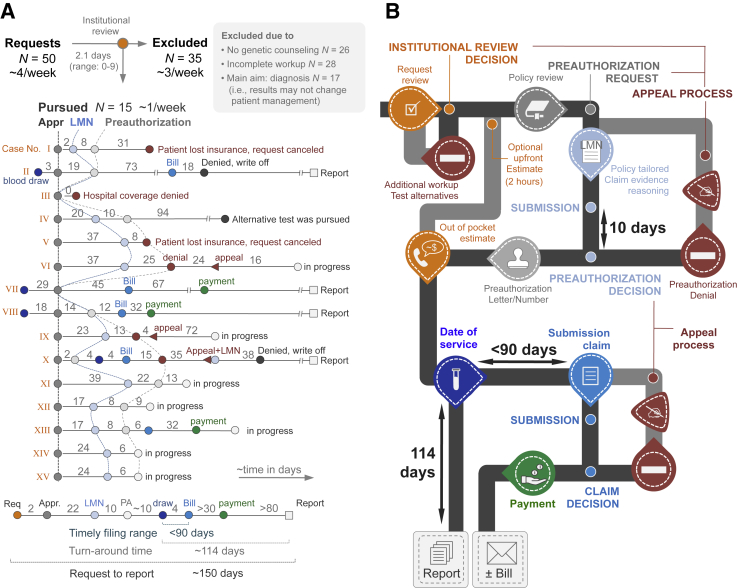
Figure 3.
Data from the pilot phase (A) and workflow infographics (B). Institutional review and cost estimation (orange) resulted in 35 rejections (selected exclusion criteria provided in gray box in panel A). The 15 pursued requests (labeled I to XV) are provided as line-dot plots providing cadence and timing of events. As a result of an unprepared delivery system and heterogeneity among payors, the process had to remain flexible and we tested 12 different combinations of modules. Connection lines between Letters of Medical Necessity and preauthorizations (hatched lines) are used to visualize the variability in timing between patients. The summary line-plot at the bottom of the panel illustrates the extension of the technical (laboratory) turnaround time by the addition of the preauthorization process. Based on this summary plot and the various possibilities (eg, preauthorization denial or reimbursement denials), we constructed the infographics (B). The colors correspond to those provided in panel A and fixed times (payor processing and test turnaround times) are provided. The typical cadence of modules is shown in black, optional and case-based pathways are shown in gray (eg, appeal process). Payment refers to reimbursement by insurance; Bill refers to a bill sent to the patient. Appr., approval; LMN, Letter of Medical Necessity; PA, preauthorization; Req, request.
Module 2: Cost Estimation Service
To account for the enormous financial burden imposed by unanticipated patient bills and the expressed need for cost transparency (based on Massachusetts Law for Health Care Cost Reduction. 2012, chapter 224, An Act Improving the Quality of Health Care and Reducing Costs through Increased Transparency, Efficiency and Innovation. 2012 Mass. Acts Chapter 224, Sec. 228, https://malegislature.gov/Laws/SessionLaws/Acts/2012/Chapter224, last accessed March 6, 2016), we installed a cost-estimation service (Figure 2) that integrates patient- and hospital-specific contracts into an out-of-pocket estimate (Supplemental Figure S3). Answering the question about test cost is not trivial and the patient's payment responsibility is contingent on hospital-payor contracts, payor rules, and specific insurance plans (to include deductibles, copays, and/or co-insurances, if applicable). It is inefficient to have the physicians bear responsibility to provide out-of-pocket estimates. The established counseling service entails one financial counselor (with 1 year of experience) who operates a cost estimator tool that integrates patient- and hospital-specific contracts and test-specific cost information (Supplemental Figure S3). Briefly, the tool is self-built (ie, currently not commercially available) and takes into account the patient's insurance plan and the experience from a CPT-code–based search in the revenue cycle management software to derive a hospital-based, out-of-pocket estimate. The value of the upfront estimation service is twofold: it clarifies that the role of the physician is to provide the risks and benefits of pursuing or not pursuing testing, and it empowers patients to make financially informed decisions.
Module 3: Preauthorization Team
The preauthorization team translates the screened and seemingly appropriate requests for clinical reimbursement into payor terms (Figure 2). To obtain preauthorization the clinical need has to be communicated to payors via complying with restrictions specified in payor policies (if applicable). The medium of translation is either a form or a Letter of Medical Necessity. Of the 13 patients who had an insurance plan that called for preauthorization, we obtained preapproval in 70% (N = 7/10) (Figure 3A). In our patient cohort, the time to draft a Letter of Medical Necessity took approximately 22 days (range, 1 to 37 days) and the payor decision added another approximately 10 days (range, 9 to 25 days) (Figure 3A). Even without the implemented appeal or exception process (for denied preauthorization requests) (Figure 3B), the addition of a month is unacceptable. When asking payors how to expedite the preauthorization process, their consistent response was to tell us to adhere strictly to their policies. These policies are carefully crafted CPT-matched documents that specify contractual rules and align the interests of patients, providers, and payors. The preauthorization team translates these policy rules into clinical practice, which required excellent communication skills, and a team of two individuals with 4 years of postgraduate experience (Supplemental Figure S4). For example, to many clinicians, the term Letter of Medical Necessity means a lengthy clinical note. To better capture the scope and core functionality of the letter, we prefer the term claim evidence reasoning and, in practical terms, we ask: “how does our clinical evidence (or lack thereof), support (in payor terms) our claim for exome testing?”
In conjunction, modules 1 to 3 embody pretest cost containment in our health care system.
Module 4: Accessioning Team
Accessioning splits physical samples (that go to the laboratory) from clinical information that once entered into the laboratory information system triggers billing before the test is completed. To accommodate evolving genomic knowledge, we incorporated the ability to re-analyze existing sequencing data (Supplemental Figure S5). Including the systems engineers for the laboratory information systems, the team of two individuals has >4 years of postgraduate experience and work in direct alignment with the sequencing team (module 6).
Module 5: Prebilling Team
The billing team (1 individual with 5 years of postgraduate experience) interfaces with the hospital finance division to submit claims to the health insurance provider. Specifically, prebilling (Figure 2) is necessary because the technical turnaround time of clinical exome and genome sequencing (in our series, approximately 114 days to final report) (Figure 3A) exceeds the payors' limit for timely filing of a claim (typically 90 days).
Module 6: Next-Generation Sequencing Team
The next-generation sequencing team handles all technical and analytic elements in the laboratory including bioinformatic work-up and report generation. Although technical specifications of sequencing, variant annotation, and interpretation (Figure 2) have been reported,1, 2, 4, 16, 17 here, we specify the tasks and qualifications from nucleic acid extraction to variant annotation (Supplemental Figures S5 and S6). Collectively, our team (11 individuals) has more than 20 years of postgraduate experience.
Module 7: Report Consultation Team
A geneticist (one individual with 6 years of experience) generates and signs the final report (posted to the electronic health record) and consults with the clinical team as needed (Figure 2).
Module 8: Reimbursement Consultation Team
The function of the reimbursement consultation team is to help patients understand the composition of their bill, if applicable (one individual with 1 year of experience) (Figure 2 and Supplemental Figure S7). Prebilling implies that the patient may have received a bill at the time of the visit to discuss test results and it is inefficient for the physicians to bear responsibility for the composition of the patient's bill. Moreover, reimbursement monitoring at the interface between departmental and hospital finance divisions is necessary because preauthorization does not imply reimbursement (Supplemental Figure S8). For example, we drew blood from one of our patients at the first visit and exceeded the timely filing limit (Figure 3A). Two other patients switched their insurance before a date-of-service was established (Figure 3A). There are numerous other hospital- and patient-plan–based reasons for denial of reimbursement, which implies a review and appeal process at the time of the reimbursement decision (Figure 3B and Supplemental Figure S8). When subtracting the cases in progress, we received reimbursement in three of six patients (50%; n = 2 partial; n = 1 full) (Figure 3A).
Integration of Payor Rules into Clinical High-Complexity Testing
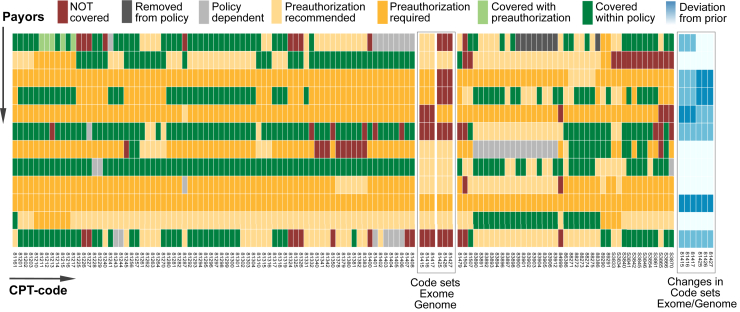
When we submitted our first claims in January 2015, we witnessed, as a response by payors to the introduced CPT code sets, the release of policies that classify clinical exome and genome sequencing as investigational and nonreimbursable. We approached this problem via cost-based claim evidence reasoning. For example, sequencing nine genes was covered by one payor; however, by showing that the associated cost exceeded exome testing with adequate coverage of these genes we received preauthorization (Figure 3A). As part of this cost-comparison approach, we reviewed policies for all molecular-genetic CPT code sets (Figure 4). During the pilot, policies changed and to visualize this additional level of complexity we included the number and relative degree of policy changes in our seven-tiered classification scheme for the new exome and genome CPT codes (Figure 4). The intricacy of these payor rules (and changes) indicates that cost comparison–based claim evidence reasoning has to be as personalized as molecular-genetic testing. Moreover, the adaptive design of our pipeline renders it useful for other genetic tests, which we regard as a utility breakpoint in terms of clinical value.
Figure 4.
Payor rules by CPT codes of selected molecular-genetic tests. The heatmap shows rules of 12 selected regional payors (rows) by selected molecular-genetic CPT code (columns). Each cell is colored assuming that the patients' main diagnosis (International Classification of Diseases code) matches the payors policy-specific rules; when multiple different policies exist for different plans (eg, bronze versus platinum-type plans) cells are colored in gray. The newly introduced exome and genome code sets show variability among payors (vertical comparison). Line-wise comparison allows comparison of payors. The matrix (ie, policy rules) changes on a regular basis (blue inset on right for exome/genome code sets) and the overall heterogeneity emphasizes the importance of patient- and policy-tailored claim evidence reasoning (Results) and the need for a flexible workflow that is adaptable to a wide array of other test types. CPT, Current Procedural Terminology.
Survey Results
Our survey captured the composition and satisfaction of our team (Figure 5). Twenty-five of 54 team members (46%) did not have personal experience with exome data. By perceived clinical value, the three most important teams were the clinicians, the preauthorization team, and the reporting team. Noteworthy responses included that team members were more satisfied with their tasks (in comparison with other work projects; 83%) and knew what was expected and completed it (94%). Project management (Agile) was perceived as different (86%), and 85% agreed that there was little waste of time. The quality of their respective supervisor was acknowledged by 95% of respondents and self-evaluation questions showed that 95% of respondents had a positive attitude toward self-organizing teams, and all respondents favored continuation of the approach.
Figure 5.
Survey results among team members. The figure offers an overview of the team size, composition, experience, as well as the overall attitude toward Agile and the project. Inst., institutional; IT, information technology; Preauth., preauthorization.
Discussion
Here, we describe a nontechnical pipeline to address some of the financial barriers previously limiting patient access to clinical genomics testing. Key features of the pipeline include the following: i) a seamless integration of financial and other nontechnical workflow components within an established analytic workflow,1, 2, 4, 16, 17 ii) a utilization management infrastructure to maintain the sustainability of our approach through the continued expansion of capitated and other value-based reimbursement models, and iii) an underlying Agile development methodology to enable adaptability to external clinical, technical, regulatory, or administrative advances.17, 18, 19
First, our approach focuses on nontechnical workflow components as an integral part of financially sustainable clinical genomics. Patients who previously were less exposed to their underlying health care expenses are increasingly facing greater out-of-pocket costs and physicians in turn are forced to provide additional consideration to the financial impacts of their care. Likewise, the financial loss associated with genomic testing has forced health care systems to restrict access. By coupling stringent clinically validated, pretest, cost-containment strategies with an infrastructure for maximizing the likelihood of insurance reimbursement, our approach helps to address these previous limitations. Although the number of cases in our pilot was relatively small, we nonetheless show the necessity of policy-tailored payor interaction to support financial viability. We consider a parallel to radiology in which certain high-cost imaging modalities (eg, high-resolution magnetic resonance imaging), once considered investigational, now have become the standard of care.20, 21 Key to this transformation was the meaningful adoption of these techniques by showing an added value. By facilitating the adoption of clinical genomics, our workflow may help to establish the role of clinical genomics as a standard of care, which may reduce the financial constraints further.
Second, our approach should remain viable as payment models evolve to emphasize value-based care. The current workflow establishes a preauthorization process with external payors. Given that in the near future capitated or bundled models may predominate,22, 23, 24 the hospitals themselves may be forced to take on roles in preauthorization and coverage determination traditionally performed by insurers.25 Therefore, the workflow for our clinical genomics project also outlines the need for utilization management and the resulting complexity is part of the care coordination process that necessitates a fundamental change in the delivery system.
Third, we see growth potential for Agile in health care. Managed care and utilization management are a first attempt to approach process improvements systematically as a scientific problem.26 Our basic intention is to reduce unnecessary costs and improve economic efficiency through subsequent approaches including quality assurance/quality control,27 operations management,28, 29, 30 re(verse) engineering,31 and today's Six Sigma.32, 33 Although Agile undoubtedly was used previously to develop many health care software applications, few, if any, examples of its application to genomic testing have been described previously. We thus hope that this report can provide a proof of concept to encourage further application of Agile to genomic and health care processes. By obtaining reimbursement in an unprepared delivery system we present evidence for successful implementation of Agile into health care. Our pilot project had several limitations including a small number of cases in one geographic region. Additional validation of this approach at other sites should help to further its generalizability. Nonetheless, we would consider these local results to be of significance, both as a proof of concept and as a potential model for utilization management and reimbursement with regard to high-cost testing. Indeed, in many cases national health care policies, protocols, and insurance coverage determinations originate as local policy initiatives and/or local coverage determinations.34 The goal to increase efficiency in an era of rapid technological change is challenging because formalizing details of a process carries the risk of fossilizing one state, causing rigidity, and preventing progress. We aligned a large number of systems and individuals with various functions and professional experience into an interoperable (modular) workflow that can serve its purpose only by remaining adaptable (agile). We regard the applied development methodology as an integral part of our project and hope to generate a positive attitude toward Agile or other development or management methods.35, 36, 37, 38, 39 Agile does not replace diligent planning but instead accounts for continuous changes in requirements. This may seem intuitive, however, when compared with the quantum leap in the computer coding industry (with computer games reaching complexity levels of >150 to 600 programmer years),40 the potential for Agile in health care is enormous. Our survey showed commitment to self-organization and we consider Agile to be particularly well suited to health care because it emphasizes the importance of human factors, their collaborative effectiveness, and communication.
In summary, our approach, although viable, highlights the complexity of the challenge and the need for highly specialized and experienced personnel. We present a first set of operational steps (Figure 3B), personnel qualifications (Figures 2 and 5 and Supplemental Figures S3–S8), and a basic organizational infrastructure (Figure 2 and Supplemental Tables S1–S4) necessary to reproduce our human pipeline. With additional experience we hope to further simplify and improve this process; however, we do not anticipate any solution that will fundamentally circumvent the operational complexity or demand for motivated and highly skilled personnel. Nonetheless, the modular architecture should be highly amenable to optimization and readily adaptable to a wide array of other types of esoteric laboratory tests. Thus, we present our approach as a roadmap for overcoming the nontechnical barriers that currently limit the scope and availability of clinical genomics and other emerging diagnostic technologies.
Acknowledgments
J.K.L., J.A.G., D.N.L., S.T.W., and A.J.I. conceived the study; J.K.L., H.M.L., J.M.B., D.R., M.S.S., N.B.L., J.M.B., K.J.S., M.K.G., H.S.W., J.D.S., D.A.S., A.S., L.P.L., W.H., H.R., A.K., S.W.B.-S., J.A.G., D.N.L., S.W., and A.J.I. designed the experiments; J.K.L., J.M.B., H.M.L., D.M.R., M.S., J.M.B., H.S.W., L.P.L., and W.H. acquired data; J.K.L., H.M.L., D.M.R., M.S., and J.M.B. analyzed data; J.K.L., H.M.L., J.M.B., D.R., M.S.S., N.B.L., J.M.B., K.J.S., M.K.G., H.S.W., J.D.S., D.A.S., M.B., M.P., A.S., L.P.L., W.H., H.R., S.W.B.-S., and A.J.I. interpreted data; J.K.L., N.B.L., M.B., M.P., L.P.L., and A.J.I. helped with Agile implementation; J.K.L. wrote the initial manuscript and all authors approved the final version. All authors are accountable for the work and ensured the accuracy and integrity of the article.
Footnotes
Supported by a fellowship from the National Center of Tumor Diseases–Heidelberg School of Oncology, Heidelberg, Germany (A.S.); NIH grant U01 HG006500 from MedSeq (H.M.M., H.L.R.); and a Junior Faculty Award from the American College of Gastroenterology (M.K.G.). Partly supported by Harvard Catalyst/The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, NIH award UL1 TR001102), and financial contributions from Harvard University and its affiliated academic health care centers.
Disclosures: M.K.G. owns equity in New Amsterdam Genomics; H.R. is directing a fee-for-service clinical laboratory for exome sequencing (Laboratory for Molecular Medicine and Broad Clinical Laboratory Improvement Amendments–certified Clinical Research Platform); and L.P.L. and A.J.I. share equity in and act as consultants for ArcherDx. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University, and its affiliated academic health care centers, or the NIH.
Current address of H.M.L., GeneDX, Gaithersburg, MD.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2016.04.003.
Supplemental Data
Starting points and approach. A: Starting points and constraints. Overview attempts to summarize the most important constraints that were known upfront. B: Difference between project management and development methodology. From a team-leader perspective, the process of implementation comes with two responsibilities: organizing the people in terms of aligning their skills (project management) and the specific way to organize the work (development methodology). Details of the applied features are provided in Supplemental Table S1. Note, by specifying the ways to organize the work, the development methodology may influence project management (eg, communication style). CLIA, Clinical Laboratory Improvement Amendments.
Project schedule and examples of work in progress. A: The project was conducted as a clinical quality and improvement initiative and consisted of five main phases, planned around the CPT code implementation date (January 1, 2015). Milestones are indicated in orange, release dates of key workflow components are shown by rounded squares around numbers 1 to 8 (corresponding to the human pipeline diagram shown in Figure 2). Symbols represent 1-hour meetings of team lead with various team members to illustrate the time commitment (20% full-time equivalent effort) and distribution of development cycles (sprints) (Supplemental Table S3). B: Workflow board (status March 22, 2015). C: Kanban board of the initial project write-up (status May 29, 2015). CPT, Current Procedural Terminology (here for exome and genome sequencing).
Workflow diagram of the institutional review and cost estimation. The workflow contains two epics/modules: institutional review (1) and cost estimation (2). The numbers correspond to Figure 2. Note that in our pilot the subsequent steps of a rejection decision at the institutional review stage were not included in this report. The definitions of clinically unnecessary or not appropriately timed (from a utilization management standpoint) were as follows: i) lack of appropriate prior genetic work-up (eg, genetic counseling and at least one prior genetic test); ii) requests did not suggest that the exome or genome results could change medical management (end of diagnostic odyssey); and iii) when the results would be considered mostly intellectual curiosity or research-type interest without management or prognostic implications. Importantly, we view these stringent criteria as a means to put pressure on the workflow rather than preliminary reasons for or against exome or genome testing. ABMG, American Board of Medical Genetics; B.S., Bachelor of Science; CPT, Current Procedural Terminology; Exp., experience; FS, fellowship; GC, genetic counselor (ideally board certified); HIS, hospital information system; ID, identification number; MD, Medical Doctor; M.Sc., master of science; PG, postgraduate experience in years; R, residency; WT, wild-type. ∗Supplemental Figure S4; †Supplemental Figure S5; ‡Supplemental Figure S6; §Supplemental Figure S7; ‖Supplemental Figure S8.
Workflow diagram of the preauthorization process (3). The preauthorization process consists of seven separate steps: request (A), policy review (B), tailored draft of the preauthorization process and LMN (C), review of claim evidence reasoning (D), submission to payor (E), decision and preapproval or denial (F), and draft appeal or arguments for medical exemption (G). CPT, Current Procedural Terminology; D, degree; DOB, date of birth; Exp., experience; FS, fellowship; ICD, International Classification of Diseases; LMN, Letter of Medical Necessity or equivalent (eg, form, preauthorization document) that contains claim evidence reasoning; M.D., medical doctor; MRN, medical record number; M.S., master of science; N/A, not applicable; NGS, next-generation sequencing; PA/preauth., preauthorization; PG, postgraduate experience; R, residency. ∗Supplemental Figure S5; †Supplemental Figure S6.
Workflow diagram of accessioning and the wet-laboratory component of next-generation sequencing. The circled numbers correspond to epics/modules in Figure 2. Accessioning (4). There are two core functions of accessioning: first, the appropriate identification of patient and sample information; the second function of accessioning is an organized split of physical (blood or data file) and patient data into the LIS. Wet-laboratory component of next-generation sequencing (6). Each step is organized as a contained unit with defined performance characteristics. If these are not met the sample is re-run (kickback) to avoid waste of downstream efforts. Thus, certain sequencing metrics are integral components of a seamless wet-laboratory workflow, thus, sequencing quality assessment as well as basic assessment of the variant call files is shown here. Details of the sequencing analysis are provided in Supplemental Figure S8. Note that when the workflow is used for other assays with complex analytics, the performance/quality metrics should be an integral component of the wet-laboratory process. API, application programming interface (here used in terms of enabling communication between software applications); ASCP, American Society for Clinical Pathology; Assoc., Associates degree; B.A., bachelor of arts; BAM, binary alignment/map (without alignment data, compact version of Sequence Alignment/Map, plain-text version); B.Sc., Bachelor of Science; CPT, Current Procedural Terminology; D, degree; DOB, date of birth; Exp., experience; FASTQ, nucleotide sequence in text format; HIPAA, Health Insurance Portability and Accountability Act; ICD, International Classification of Diseases; ID, sample identifier (eg, barcode); IT, information technology team; lab, laboratory; LIS, laboratory information system; LMN, Letter of Medical Necessity or equivalent (eg, form, preauthorization document) that contains claim evidence reasoning; M.D., medical doctor; MRN, medical record number; M.S., master of science; NGS, next-generation sequencing; preauth., preauthorization; PG, postgraduate experience; QA/QC, quality assurance/quality control; qPCR, real-time quantitative PCR (part of quality assessment); R, residency; Raw, raw reads; SAM, Sequence Alignment/Map, plain-text version; SOP, standard operating procedure; vcf, variant call file. ∗Supplemental Figure S3; †Supplemental Figure S4; ‡Supplemental Figure S6; §Supplemental Figure S8.
Workflow diagram next-generation sequencing analysis and report generation. The next-generation sequencing (after wet-laboratory) consists of three separate analytic components handled by three separate teams: the data analysis pipeline (left third of the diagram), via annotation tiers (middle third), to synthesis and report generation (right third with filled orange 7 and arrows to Supplemental Figure S7). The data analysis pipeline follows current workflows and produced a variant call and together with the phenotype (phenotype call file) these two files form the basis for annotation of variants by tiers. The variants are classified using (functional) prediction tools (open orange 1). Moving to higher annotation tiers (2 to 4) requires at least one other call file for comparisons (eg, tumor versus normal, trio sequencing, and so forth; red orange 2). When mining internal or external servers using phenotype or genotype data to generate worklists or candidate genes, in conjunction with the original call files, the resulting three-way lists/candidates are merged. We distinguish between local/institutional (practice-based evidence) databanks (open orange 3) and larger-scale interinstitutional or public databanks (open orange 4). The synthesis is made by comparing the results from different tiers for each variant, which ultimately culminates in assignments following the suggestions of the Mendelian disease variant terminology. Depending on the testing volume, some team members can be replaced using cross-functional assignments (bottom orange bracket). B.S., Bachelor of Science; BWA, Burrows Wheeler algorithm; CNV, copy number variant; CS, computer science; comp het., compound heterozygous; D, degree; Exp, experience; FastQC, quality control application for high-throughput sequence data; Funct., functional; GATKA, genome analysis toolkit; G.C., genetic counselor; HIS, hospital information system; homozyg., homozygous; InDel, insertion deletion (mutation type); Lab. Op., laboratory operations; LIS, laboratory information system; M.D., Medical Doctor; M.S., Master of Science; NGS, next-generation sequencing; .pcf, phenotype call files capturing clinical information; Ph.D., Doctor of Philosophy; Picard, set of Java command line tools; PG, postgraduate experience; QA/QC, quality assurance/quality control; SNP, single-nucleotide polymorphism; SOP, standard operating procedure; .vcf, variant call file. ∗Supplemental Figure S3; †Supplemental Figure S4; ‡Supplemental Figure S5; §Supplemental Figure S7.
Diagram of report consultation (7) and payment consultation (8). The primary responsibility to convey and explain the implications of the test results remains with the treating physician. However, we have implemented the opportunity to consult the reporting physician to acknowledge the content-related expertise necessary to interpret exome- or genome-type data. To make sure that the patient has a point person for billing-related questions (when applicable/necessary), we implemented payment consultation via a patient financial counselor in a separate process. B.S., Bachelor of Science; Exp, experience; FS, fellowship; LMN, Letter of Medical Necessity; M.D., Medical Doctor; Ph.D., Doctor of Philosophy; R, residency. ∗Supplemental Figure S8.
Diagram of billing, reimbursement, and appeal management. The entire billing process was not part of the workflow because the process of submission to the payor relies heavily on automated data-submission interfaces and the individual case review only holds value when dealing with denials. Briefly, when denied and hospital-based reasons are excluded, the uncovered test will be submitted as a full charge to the patient. The established appeal process takes the entire picture into account and also provides feedback to the cost-estimator group as well as the patient financial counselor. B.Sc., Bachelor of science; CPT, current procedural terminology; Dept, Department; DOB, date of birth; DOS, date of service; ExGS, whole exome and genome sequencing; GC, genetic counselor; ICD, international classification of diseases; Ins. Plan., patient-specific patient plan/policy; Inst., institutional; MGH, Massachusetts General Hospital (provided as an example); MRN, medical record number; path., Pathologic; Preauth., preauthorization. ∗Supplemental Figure S3; †Supplemental Figure S4; ‡Supplemental Figure S5; §Supplemental Figure S7.
References
- 1.Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L., Plon S.E., Ramos E.M., Sherry S.T., Watson M.S., ClinGen ClinGen–the clinical genome resource. N Engl J Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biesecker L.G., Green R.C. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370:2418–2425. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 3.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M., Fox M., Fogel B.L., Martinez-Agosto J.A., Wong D.A., Chang V.Y., Shieh P.B., Palmer C.G., Dipple K.M., Grody W.W., Vilain E., Nelson S.F. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green R.C., Berg J.S., Grody W.W., Kalia S.S., Korf B.R., Martin C.L., McGuire A.L., Nussbaum R.L., O'Daniel J.M., Ormond K.E., Rehm H.L., Watson M.S., Williams M.S., Biesecker L.G., American College of Medical Genetics and Genomics ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronson S.J., Clark E.H., Varugheese M., Baxter S., Babb L.J., Rehm H.L. Communicating new knowledge on previously reported genetic variants. Genet Med. 2012 doi: 10.1038/gim.2012.19. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumenthal D., Abrams M., Nuzum R. The Affordable Care Act at 5 years. N Engl J Med. 2015;372:2451–2458. doi: 10.1056/NEJMhpr1503614. [DOI] [PubMed] [Google Scholar]
- 7.Blumenthal D., Collins S.R. Health care coverage under the Affordable Care Act–a progress report. N Engl J Med. 2014;371:275–281. doi: 10.1056/NEJMhpr1405667. [DOI] [PubMed] [Google Scholar]
- 8.Abul-Husn N.S., Owusu Obeng A., Sanderson S.C., Gottesman O., Scott S.A. Implementation and utilization of genetic testing in personalized medicine. Pharmgenomics Pers Med. 2014;7:227–240. doi: 10.2147/PGPM.S48887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein D.A., Shaib W.L., Flowers C.R. Costs and effectiveness of genomic testing in the management of colorectal cancer. Oncology (Williston Park) 2015;29:175–183. [PubMed] [Google Scholar]
- 10.Deverka P.A., Dreyfus J.C. Clinical integration of next generation sequencing: coverage and reimbursement challenges. J Law Med Ethics. 2014;42(Suppl 1):22–41. doi: 10.1111/jlme.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennette C.S., Trinidad S.B., Fullerton S.M., Patrick D., Amendola L., Burke W., Hisama F.M., Jarvik G.P., Regier D.A., Veenstra D.L. Return of incidental findings in genomic medicine: measuring what patients value–development of an instrument to measure preferences for information from next-generation testing (IMPRINT) Genet Med. 2013;15:873–881. doi: 10.1038/gim.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B., Gagnon M., Shahangian S., Anderson N.L., Howerton D.A., Boone J.D., Centers for Disease Control and Prevention Good laboratory practices for molecular genetic testing for heritable diseases and conditions. MMWR Recomm Rep. 2009;58:1–37. quiz CE-1–4. [PubMed] [Google Scholar]
- 13.Brown J.T. ed 2. McGraw-Hill; New York: 2014. The Handbook of Program Management: How to Facilitate Project Success with Optimal Program Management. [Google Scholar]
- 14.Adzic G., Evans D. ed 1. Neuri Coinsulting LLP; London: 2014. Fifty Quick Ideas to Improve Your User Stories. [Google Scholar]
- 15.Fanning E. Formatting a paper-based survey questionnaire: best practices. Pract Assess Res Eval. 2005;10:1–14. [Google Scholar]
- 16.McLaughlin H.M., Ceyhan-Birsoy O., Christensen K.D., Kohane I.S., Krier J., Lane W.J., Lautenbach D., Lebo M.S., Machini K., MacRae C.A., Azzariti D.R., Murray M.F., Seidman C.E., Vassy J.L., Green R.C., Rehm H.L., MedSeq Project A systematic approach to the reporting of medically relevant findings from whole genome sequencing. BMC Med Genet. 2014;15:134. doi: 10.1186/s12881-014-0134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gargis A.S., Kalman L., Bick D.P., da Silva C., Dimmock D.P., Funke B.H. Good laboratory practice for clinical next-generation sequencing informatics pipelines. Nat Biotechnol. 2015;33:689–693. doi: 10.1038/nbt.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curnutte M.A., Frumovitz K.L., Bollinger J.M., McGuire A.L., Kaufman D.J. Development of the clinical next-generation sequencing industry in a shifting policy climate. Nat Biotechnol. 2014;32:980–982. doi: 10.1038/nbt.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans J.P., Watson M.S. Genetic testing and FDA regulation: overregulation threatens the emergence of genomic medicine. JAMA. 2015;313:669–670. doi: 10.1001/jama.2014.18145. [DOI] [PubMed] [Google Scholar]
- 20.Hendee W.R., Becker G.J., Borgstede J.P., Bosma J., Casarella W.J., Erickson B.A., Maynard C.D., Thrall J.H., Wallner P.E. Addressing overutilization in medical imaging. Radiology. 2010;257:240–245. doi: 10.1148/radiol.10100063. [DOI] [PubMed] [Google Scholar]
- 21.Levin D.C., Rao V.M., Parker L. Physician orders contribute to high-tech imaging slowdown. Health Aff (Millwood) 2010;29:189–195. doi: 10.1377/hlthaff.2009.0528. [DOI] [PubMed] [Google Scholar]
- 22.Taheri P.A., Butz D.A., Greenfield L.J. Academic health systems management: the rationale behind capitated contracts. Ann Surg. 2000;231:849–859. doi: 10.1097/00000658-200006000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J. Bundled payment and enhanced recovery after surgery. J Med Pract Manage. 2015;30:349–353. [PubMed] [Google Scholar]
- 24.Mehrotra A., Hussey P. Including physicians in bundled hospital care payments: time to revisit an old idea? JAMA. 2015;313:1907–1908. doi: 10.1001/jama.2015.3359. [DOI] [PubMed] [Google Scholar]
- 25.Barnes A.J., Unruh L., Chukmaitov A., van Ginneken E. Accountable care organizations in the USA: types, developments and challenges. Health Policy. 2014;118:1–7. doi: 10.1016/j.healthpol.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 26.Derksen M. Turning men into machines? Scientific management, industrial psychology, and the “human factor”. J Hist Behav Sci. 2014;50:148–165. doi: 10.1002/jhbs.21650. [DOI] [PubMed] [Google Scholar]
- 27.Evans J.R., Lindsay W.M. ed 6. Thomson/South-Western; Mason, OH: 2005. The Management and Control of Quality. [Google Scholar]
- 28.Chase R.B., Jacobs F.R. ed 11. McGraw-Hill/Irwin; Boston: 2006. Operations Management for Competitive Advantage; p. xix. McGraw-Hill/Irwin Series Operations and Decision Sciences. [Google Scholar]
- 29.Jacobs F.R., Chase R.B., Aquilano N.J., Chase R.B. ed 12. McGraw-Hill; Boston: 2009. Operations and Supply Management. [Google Scholar]
- 30.Malakooti B. John Wiley & Sons Inc.; Hoboken, NJ: 2014. Operations and Production Systems with Multiple Objectives. [Google Scholar]
- 31.Messler R.W. McGraw-Hill Education; New York: 2014. Reverse Engineering: Mechanisms, Structures, Systems, and Materials. [Google Scholar]
- 32.Suneja A., Suneja C. ASQ Quality Press; Milwaukee, WI: 2010. Lean Doctors: A Bold and Practical Guide to Using Lean Principles to Transform Healthcare Systems, One Doctor at a Time. [Google Scholar]
- 33.Zidel T. ASQ Quality Press; Milwaukee, WI: 2006. A Lean Guide to Transforming Healthcare: How to Implement Lean Principles in Hospitals, Medical Offices, Clinics, and Other Healthcare Organizations. [Google Scholar]
- 34.Steingisser L., Acker B., Berman S., Brenner M.J., Bornstein B.A., Busse P., FitzGerald T.J., Jacobson J.O., Jekowsky E., Kachnic L.A., Mamon H., McKee A., Shulman L.N., Stevenson M.A., Wazer D., Fallon J.A. Bending the cost curve: a unique collaboration between radiation oncologists and Blue Cross Blue Shield of Massachusetts to optimize the use of advanced technology. J Oncol Pract. 2014;10:e321–e327. doi: 10.1200/JOP.2014.001473. [DOI] [PubMed] [Google Scholar]
- 35.Brechner E. Microsoft Press; Redmond, WA: 2015. Agile Project Management with Kanban. [Google Scholar]
- 36.Hammarberg M., Sundén J. Manning; Shelter Island, NY: 2014. Kanban in Action. [Google Scholar]
- 37.Kniberg H., Beck K., Keppler K. Pragmatic Bookshelf; Dallas, TX: 2011. Lean from the Trenches: Managing Large-Scale Projects with Kanban. [Google Scholar]
- 38.Reddy A. Addison-Wesley; New York: 2016. The Scrumban [r]evolution: Getting the Most Out of Agile, Scrum, and Lean Kanban. [Google Scholar]
- 39.Vatalaro J.C., Taylor R.E. Productivity Press; New York: 2003. Implementing a Mixed Model Kanban System: The Lean Replenishment Technique for Pull Production. [Google Scholar]
- 40.Hall R., Novak J. Delmar/Cengage Learning; Clifton Park, NY: 2008. Chapter 1 History: Where Did Online Games Come from. Game Development Essentials Online Game Development; p. xxii. 20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Starting points and approach. A: Starting points and constraints. Overview attempts to summarize the most important constraints that were known upfront. B: Difference between project management and development methodology. From a team-leader perspective, the process of implementation comes with two responsibilities: organizing the people in terms of aligning their skills (project management) and the specific way to organize the work (development methodology). Details of the applied features are provided in Supplemental Table S1. Note, by specifying the ways to organize the work, the development methodology may influence project management (eg, communication style). CLIA, Clinical Laboratory Improvement Amendments.
Project schedule and examples of work in progress. A: The project was conducted as a clinical quality and improvement initiative and consisted of five main phases, planned around the CPT code implementation date (January 1, 2015). Milestones are indicated in orange, release dates of key workflow components are shown by rounded squares around numbers 1 to 8 (corresponding to the human pipeline diagram shown in Figure 2). Symbols represent 1-hour meetings of team lead with various team members to illustrate the time commitment (20% full-time equivalent effort) and distribution of development cycles (sprints) (Supplemental Table S3). B: Workflow board (status March 22, 2015). C: Kanban board of the initial project write-up (status May 29, 2015). CPT, Current Procedural Terminology (here for exome and genome sequencing).
Workflow diagram of the institutional review and cost estimation. The workflow contains two epics/modules: institutional review (1) and cost estimation (2). The numbers correspond to Figure 2. Note that in our pilot the subsequent steps of a rejection decision at the institutional review stage were not included in this report. The definitions of clinically unnecessary or not appropriately timed (from a utilization management standpoint) were as follows: i) lack of appropriate prior genetic work-up (eg, genetic counseling and at least one prior genetic test); ii) requests did not suggest that the exome or genome results could change medical management (end of diagnostic odyssey); and iii) when the results would be considered mostly intellectual curiosity or research-type interest without management or prognostic implications. Importantly, we view these stringent criteria as a means to put pressure on the workflow rather than preliminary reasons for or against exome or genome testing. ABMG, American Board of Medical Genetics; B.S., Bachelor of Science; CPT, Current Procedural Terminology; Exp., experience; FS, fellowship; GC, genetic counselor (ideally board certified); HIS, hospital information system; ID, identification number; MD, Medical Doctor; M.Sc., master of science; PG, postgraduate experience in years; R, residency; WT, wild-type. ∗Supplemental Figure S4; †Supplemental Figure S5; ‡Supplemental Figure S6; §Supplemental Figure S7; ‖Supplemental Figure S8.
Workflow diagram of the preauthorization process (3). The preauthorization process consists of seven separate steps: request (A), policy review (B), tailored draft of the preauthorization process and LMN (C), review of claim evidence reasoning (D), submission to payor (E), decision and preapproval or denial (F), and draft appeal or arguments for medical exemption (G). CPT, Current Procedural Terminology; D, degree; DOB, date of birth; Exp., experience; FS, fellowship; ICD, International Classification of Diseases; LMN, Letter of Medical Necessity or equivalent (eg, form, preauthorization document) that contains claim evidence reasoning; M.D., medical doctor; MRN, medical record number; M.S., master of science; N/A, not applicable; NGS, next-generation sequencing; PA/preauth., preauthorization; PG, postgraduate experience; R, residency. ∗Supplemental Figure S5; †Supplemental Figure S6.
Workflow diagram of accessioning and the wet-laboratory component of next-generation sequencing. The circled numbers correspond to epics/modules in Figure 2. Accessioning (4). There are two core functions of accessioning: first, the appropriate identification of patient and sample information; the second function of accessioning is an organized split of physical (blood or data file) and patient data into the LIS. Wet-laboratory component of next-generation sequencing (6). Each step is organized as a contained unit with defined performance characteristics. If these are not met the sample is re-run (kickback) to avoid waste of downstream efforts. Thus, certain sequencing metrics are integral components of a seamless wet-laboratory workflow, thus, sequencing quality assessment as well as basic assessment of the variant call files is shown here. Details of the sequencing analysis are provided in Supplemental Figure S8. Note that when the workflow is used for other assays with complex analytics, the performance/quality metrics should be an integral component of the wet-laboratory process. API, application programming interface (here used in terms of enabling communication between software applications); ASCP, American Society for Clinical Pathology; Assoc., Associates degree; B.A., bachelor of arts; BAM, binary alignment/map (without alignment data, compact version of Sequence Alignment/Map, plain-text version); B.Sc., Bachelor of Science; CPT, Current Procedural Terminology; D, degree; DOB, date of birth; Exp., experience; FASTQ, nucleotide sequence in text format; HIPAA, Health Insurance Portability and Accountability Act; ICD, International Classification of Diseases; ID, sample identifier (eg, barcode); IT, information technology team; lab, laboratory; LIS, laboratory information system; LMN, Letter of Medical Necessity or equivalent (eg, form, preauthorization document) that contains claim evidence reasoning; M.D., medical doctor; MRN, medical record number; M.S., master of science; NGS, next-generation sequencing; preauth., preauthorization; PG, postgraduate experience; QA/QC, quality assurance/quality control; qPCR, real-time quantitative PCR (part of quality assessment); R, residency; Raw, raw reads; SAM, Sequence Alignment/Map, plain-text version; SOP, standard operating procedure; vcf, variant call file. ∗Supplemental Figure S3; †Supplemental Figure S4; ‡Supplemental Figure S6; §Supplemental Figure S8.
Workflow diagram next-generation sequencing analysis and report generation. The next-generation sequencing (after wet-laboratory) consists of three separate analytic components handled by three separate teams: the data analysis pipeline (left third of the diagram), via annotation tiers (middle third), to synthesis and report generation (right third with filled orange 7 and arrows to Supplemental Figure S7). The data analysis pipeline follows current workflows and produced a variant call and together with the phenotype (phenotype call file) these two files form the basis for annotation of variants by tiers. The variants are classified using (functional) prediction tools (open orange 1). Moving to higher annotation tiers (2 to 4) requires at least one other call file for comparisons (eg, tumor versus normal, trio sequencing, and so forth; red orange 2). When mining internal or external servers using phenotype or genotype data to generate worklists or candidate genes, in conjunction with the original call files, the resulting three-way lists/candidates are merged. We distinguish between local/institutional (practice-based evidence) databanks (open orange 3) and larger-scale interinstitutional or public databanks (open orange 4). The synthesis is made by comparing the results from different tiers for each variant, which ultimately culminates in assignments following the suggestions of the Mendelian disease variant terminology. Depending on the testing volume, some team members can be replaced using cross-functional assignments (bottom orange bracket). B.S., Bachelor of Science; BWA, Burrows Wheeler algorithm; CNV, copy number variant; CS, computer science; comp het., compound heterozygous; D, degree; Exp, experience; FastQC, quality control application for high-throughput sequence data; Funct., functional; GATKA, genome analysis toolkit; G.C., genetic counselor; HIS, hospital information system; homozyg., homozygous; InDel, insertion deletion (mutation type); Lab. Op., laboratory operations; LIS, laboratory information system; M.D., Medical Doctor; M.S., Master of Science; NGS, next-generation sequencing; .pcf, phenotype call files capturing clinical information; Ph.D., Doctor of Philosophy; Picard, set of Java command line tools; PG, postgraduate experience; QA/QC, quality assurance/quality control; SNP, single-nucleotide polymorphism; SOP, standard operating procedure; .vcf, variant call file. ∗Supplemental Figure S3; †Supplemental Figure S4; ‡Supplemental Figure S5; §Supplemental Figure S7.
Diagram of report consultation (7) and payment consultation (8). The primary responsibility to convey and explain the implications of the test results remains with the treating physician. However, we have implemented the opportunity to consult the reporting physician to acknowledge the content-related expertise necessary to interpret exome- or genome-type data. To make sure that the patient has a point person for billing-related questions (when applicable/necessary), we implemented payment consultation via a patient financial counselor in a separate process. B.S., Bachelor of Science; Exp, experience; FS, fellowship; LMN, Letter of Medical Necessity; M.D., Medical Doctor; Ph.D., Doctor of Philosophy; R, residency. ∗Supplemental Figure S8.
Diagram of billing, reimbursement, and appeal management. The entire billing process was not part of the workflow because the process of submission to the payor relies heavily on automated data-submission interfaces and the individual case review only holds value when dealing with denials. Briefly, when denied and hospital-based reasons are excluded, the uncovered test will be submitted as a full charge to the patient. The established appeal process takes the entire picture into account and also provides feedback to the cost-estimator group as well as the patient financial counselor. B.Sc., Bachelor of science; CPT, current procedural terminology; Dept, Department; DOB, date of birth; DOS, date of service; ExGS, whole exome and genome sequencing; GC, genetic counselor; ICD, international classification of diseases; Ins. Plan., patient-specific patient plan/policy; Inst., institutional; MGH, Massachusetts General Hospital (provided as an example); MRN, medical record number; path., Pathologic; Preauth., preauthorization. ∗Supplemental Figure S3; †Supplemental Figure S4; ‡Supplemental Figure S5; §Supplemental Figure S7.