Abstract
Establishing ERBB2 [human epidermal growth factor receptor 2 (HER2)] amplification status in breast and gastric carcinomas is essential to treatment selection. Immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) constitute the current standard for assessment. With further advancements in genomic medicine, new clinically relevant biomarkers are rapidly emerging and options for targeted therapy are increasing in patients with advanced disease, driving the need for comprehensive molecular profiling. Next-generation sequencing (NGS) is an attractive approach for up-front comprehensive assessment, including ERBB2 status, but the concordance with traditional methods of HER2 assessment is not well established. The Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) assay, a hybrid capture-based NGS assay interrogating the coding regions of 410 cancer-related genes, was performed on manually macrodissected unstained sections from formalin-fixed, paraffin-embedded breast (n = 213) and gastroesophageal (n = 39) tumors submitted for clinical mutation profiling. ERBB2 status was assessed using a custom bioinformatics pipeline, and NGS results were compared to IHC and FISH. NGS ERBB2 amplification calls had an overall concordance of 98.4% (248/252) with the combined IHC/FISH results in this validation set. Discrepancies occurred in the context of low tumor content and HER2 heterogeneity. ERBB2 amplification status can be reliably determined by hybridization capture-based NGS methods, allowing efficient concurrent testing for other potentially actionable genomic alterations, particularly in limited material.
CME Accreditation Statement: This activity (“JMD 2017 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint providership of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“JMD 2017 CME Program in Molecular Diagnostics”) for a maximum of 36 AMA PRA Category 1 Credit(s)™. Physicians should only claim credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
Human epidermal growth factor receptor 2 (HER2) is a 185-kDa transmembrane tyrosine kinase receptor encoded by the ERBB2 gene (chromosome 17q12) that is a biomarker and therapeutic target in patients with breast and gastric cancers. Approximately 18% to 25%1, 2, 3 of all breast and 15% to 30%4, 5, 6, 7, 8 of all gastric or gastroesophageal (GE) cancers are classified as HER2-positive as a result of ERBB2 gene amplification and the subsequent overexpression of the HER2 protein on the surface of tumor cells. Therapies targeting HER2 have been approved for clinical use and include HER2-directed humanized monoclonal antibodies (ie, trastuzumab and pertuzumab) and small-molecule HER2 kinase inhibitors (ie, lapatinib).
In breast cancer, HER2 is both a predictive and prognostic biomarker. HER2-positive status predicts response to HER2-targeted therapy, and multiple prospective randomized trials have shown significant improvement in time to tumor progression.9, 10, 11 HER2-positive breast tumors have been associated with worse prognosis compared to HER2-negative tumors.1, 12, 13, 14, 15 Although the prognostic significance of the HER2 status in patients with GE adenocarcinoma has been the subject of multiple studies with varying results,4, 16, 17, 18 HER2-positive status is associated with response to targeted therapy in these tumors. Results of the randomized phase 3 Trastuzumab for Gastric Cancer trial demonstrated improved response and overall survival when trastuzumab was added to chemotherapy in patients with HER2-positive adenocarcinoma of the stomach and the esophagogastric junction.5
ERBB2 amplification is a potential therapeutic target in other solid tumors as well, including cancers of the lung, bladder, endometrium, ovary, colon, and rectum.19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30 The role of targeted therapy in these malignancies is an area of active study (National Clinical Trials Registry 02465060 and 02675829).
There are several methods recommended by the American Society of Clinical Oncology and the College of American Pathologists for evaluation of HER2 in breast and gastric cancers. These methods include immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), and bright field in situ hybridization (chromogenic in situ hybridization and silver in situ hybridization).31, 32, 33 IHC detects the expression of the HER2 protein in a tumor, whereas FISH and chromogenic in situ hybridization/silver in situ hybridization detect amplification of the ERBB2 gene.
With rapid advances in genomic medicine, new molecular alterations of clinical relevance are emerging, driving the need for broad comprehensive genotyping. With the advent of next-generation sequencing methods (NGS), the capability to concurrently derive copy number alteration (CNA) data from the same assays used to detect sequence alterations provides a cost- and tissue-efficient alternative to current single-gene assessment methods. To date, ERBB2 assessment by NGS methods has not been clinically validated, and the concordance with traditional methods is not well established.
The current study details our experience in the development and validation of an algorithm for assessing ERBB2 amplification status using a hybrid capture-based NGS assay [Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets assay (MSK-IMPACT)],34 and establishes the concordance with traditional methods.
Materials and Methods
Case Selection and Standard HER2 Testing
After obtaining institutional review board approval, the digital records of the Molecular Diagnostics service were searched for breast and GE specimens submitted for molecular analysis by the MSK-IMPACT assay,34 as part of an institutional clinical cancer genomics initiative.35 DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tissue after macrodissection, if necessary. Tumor percentages were recorded for each case as an estimate of tumor purity based on semiquantitative analysis of the hematoxylin and eosin–stained section by a pathologist. Specimens containing at least 10% tumor were suitable for analysis. Only cases with concurrent HER2 analysis previously performed by IHC [PATHWAY anti-HER-2/neu (4B5) (Ventana, Tucson, AZ); HercepTest (Dako, Carpinteria, CA)] and/or FISH [HER2 IQFISH pharmDx (Dako); PathVysion HER-2 DNA Probe Kit (Vysis, Downers Grove, IL)] using a Federal Drug Administration–approved method were included in this validation study. The IHC and FISH HER2 test results were reported in accordance with the guideline recommendations for breast cancer31 and gastric malignancies.32, 33 IHC and/or FISH results were both used to classify each tumor as HER2 amplified or nonamplified in accordance with the standard practice at our institution; IHC was performed first on all specimens, and only equivocal cases were reflexed to FISH. If there was a discordance between the IHC and the NGS result, FISH was performed when material was available.
The MSK-IMPACT Assay
Details of the MSK-IMPACT assay have been previously published.34 Briefly, MSK-IMPACT is a comprehensive molecular profiling assay that involves hybridization capture and deep sequencing of all exons and selected introns of up to 410 oncogenes and tumor-suppressor genes, allowing the detection of point mutations, small and large insertions or deletions, and rearrangements. In addition to capturing all coding regions of the genes, the assay also captures >1000 intergenic and intronic single-nucleotide polymorphisms (tiling probes), interspersed homogenously across the genome, aiding the accurate assessment of genome-wide copy number. In total, the probes target approximately 1.2 megabases of the human genome.
Genomic DNA was extracted from FFPE tumor tissue after manual macrodissection to ensure at least 10% tumor content. DNA input ranged from 100 to 200 ng, as measured by fluorometric methods. Matching peripheral blood (EDTA tube) from each patient was also extracted for use as a normal control. Sequence libraries were prepared (Kapa Biosystems, Wilmington, MA) through a series of enzymatic steps, including shearing of double-stranded DNA, end repair, A-base addition, ligation of bar-coded sequence adaptors, and low-cycle PCR amplification. Multiple bar-coded sequence libraries were pooled and captured using our custom-designed biotinylated probes (Roche NimbleGen, Madison, WI). Captured DNA fragments were sequenced on an Illumina HiSeq2500 (Illumina, San Diego, CA) as 100-bp paired-end reads that were then subjected to the bioinformatics analysis pipeline. Sequencing reads were aligned to the human genome (hg19) using Burrows-Wheeler Aligner software version 0.7.5a (http://arxiv.org/abs/1303.3997, last accessed October 1, 2013). ABRA36 was used for read realignment around indels, and Genome Analysis Toolkit37 was used for base quality score recalibration. Duplicate reads were marked and not used in the downstream analyses.
Bioinformatics Pipeline to Detect CNAs
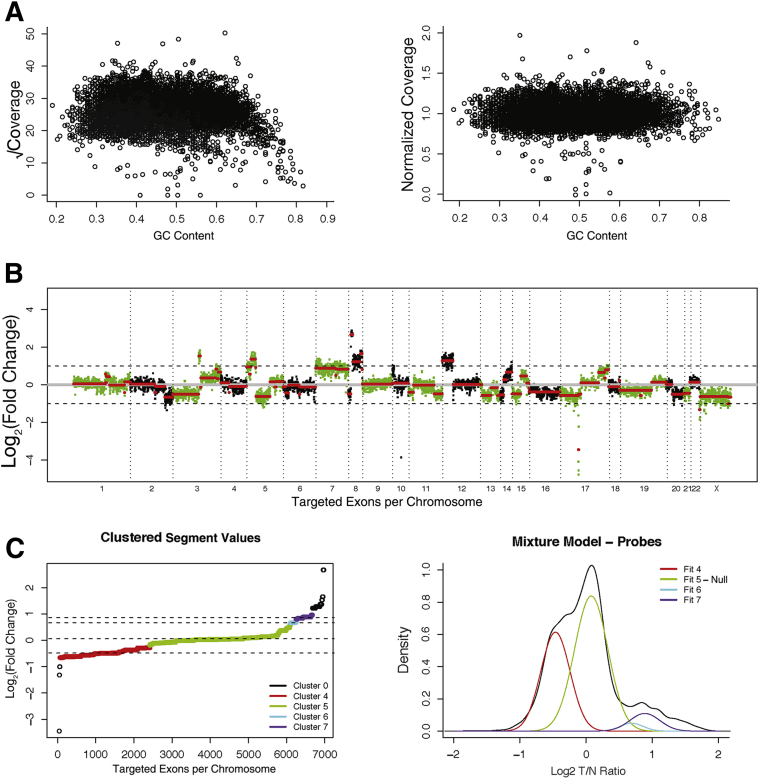
The copy number pipeline builds on the existing data management and analysis systems previously described for the MSK-IMPACT mutation profiling assay.34 The first step in the pipeline is to calculate the coverage of each captured target region (exons and tiling probes). After BAM files are generated, we calculate coverage using the Genome Analysis Toolkit Depth of Coverage tool, discarding reads with mapping quality <20.37 Coverage values are normalized for the overall coverage of the sample, square root transformed, and adjusted for the GC content of each target region using Loess normalization. This normalization removes the GC content dependency of the coverage values (Figure 1, A and B). Captured regions with coverage values in the lowest fifth percentile of coverage in >20% of the normal samples are excluded from the analysis. These regions usually correspond to genes with presence of pseudo copies and have a high rate of false-positive results.
Figure 1.
A: Square-root–transformed coverage value of each probe against the GC content of the probe shows the reduced coverage at high end of GC content (left panel). Loess normalization removes the dependency of coverage across different GC profiles (right panel). B: Example of a normalized log2 transformed fold change of coverage (log-ratio) values across all the captured regions in MSK-IMPACT assay. Circular binary segmentation algorithm is used to segment the data into discreet data points. Red lines indicate segmented regions determined by circular binary segmentation algorithm. C: Ordered log-ratio values are clustered on the basis of distance, and the cluster closest to 0 is selected as the null distribution (left panel). All other clusters are then tested against a model parameterized by the null distribution. Curves represent the distribution of each fit of different clusters (right panel).
Detection of CNAs requires a diploid normal genome as a comparator. For this purpose and to eliminate the sample type-dependent coverage differences, we use a series of FFPE normal samples captured and sequenced with the MSK-IMPACT assay. Normalized coverage values from each region in the tumor sample are divided by the normalized coverage values from each of the normal controls and are log-transformed. Using the log-ratios between a tumor and each normal control, we calculate the sum of square value as a proxy for the level of signal/noise ratio. The normal with the lowest sum-squared log-ratio value is chosen as the comparator for further downstream analysis.
Log-ratio values are then further segmented using the circular binary segmentation algorithm (Figure 1B).38 Segmented values are grouped into clusters with a minimum separation threshold of 0.08 and a minimum cluster membership of three segments (Figure 1C). Probe regions with segments belonging to the segment cluster where the mean log ratio is closest to 0 are used to parameterize a null distribution. This null distribution is used to generate P values for each segment, which are then adjusted for false-discovery rate using the Benjamini-Hochberg procedure. Genes in a given segment are assigned the log-ratio, and the adjusted P value of that segment and genes with significant deviance from this distribution are then identified as CNAs.
The following criteria were used to determine significance of whole-gene gain or loss events: fold change (FC) ≥ 2.0 (amplification), FC ≥ 1.5 but <2 (copy number gain/borderline amplification), or FC ≤ −2.0 (deletion) with P < 0.05. Copy number gains are interpreted along with overall copy number profile and tumor purity. See Results for further details.
Sensitivity Study
The sensitivity study was performed using three independent samples [one FFPE patient sample and two commercially available cell lines (MDA-MB-361 and MDA-MB-453, obtained from ATCC, Manassas, VA)] that showed varying degrees of ERBB2 amplification: markedly amplified, moderately amplified, and minimally amplified. Five serial dilutions were prepared (50%, 25%, 12.5%, 6.25%, and 3.13%). The cell lines were authenticated and cultured, as previously described,39 and cell blocks were prepared to perform HER2 FISH analysis.
Reproducibility Study
Three amplified samples from the data set were studied over three different runs for the inter-assay reproducibility study. The same samples were also studied in triplicate in the same run for the intra-assay reproducibility study.
Results
Establishment of Bioinformatics Thresholds for NGS-Based Assessment of ERBB2 Amplification
The percentage of breast cancers and GE adenocarcinomas submitted for MSK-IMPACT testing thus far at our institution is approximately 12% and 3%, respectively. In this validation study, tumor specimens were included from patients with breast cancer (n = 213) and GE adenocarcinoma (n = 39). The characteristics of the data set (procedure performed and sample type) are listed in Table 1. On the basis of the established reporting guidelines for HER2 IHC and/or FISH testing, 71 tumors (58 breast and 13 GE) were categorized as amplified, 178 tumors (152 breast and 26 GE) were categorized as nonamplified, and 3 breast tumors were categorized as equivocal. The three equivocal breast cases were treated as HER2 negative clinically and for the purpose of this study were included in the HER2 nonamplified category. All tumor samples showed adequate coverage, with an average depth of 727× and 689× for amplified and nonamplified samples, respectively.
Table 1.
Validation Data Set
| Type of cancer | Total | Procedure |
Sample type |
|||
|---|---|---|---|---|---|---|
| FNA | Biopsy | Excision | Primary | Metastasis | ||
| Breast | 213 | 7 | 125 | 81 | 106∗ | 107 |
| Gastroesophageal | 39 | 0 | 32 | 7 | 27 | 12 |
FNA, fine-needle aspiration.
Includes recurrences (n = 3).
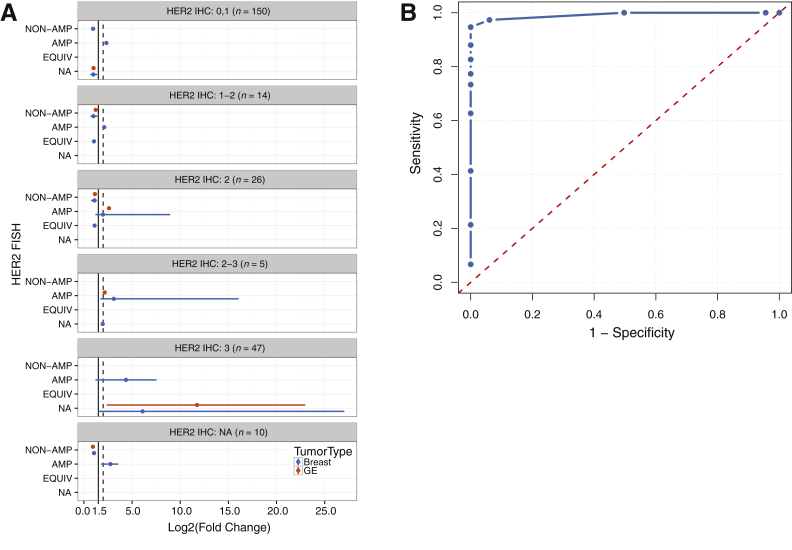
Assuming a tumor purity of 50% with a background of two copies per cell in normal tissue, six copies of a gene corresponds to a FC of 2. We used a FC of 2 with adjusted P < 0.05 as our initial cutoff criteria for calling amplification events. However, as shown in Figure 2A, this threshold failed to identify some true-positive cases. To increase our sensitivity for ERBB2 CN change when the tumor purity is <50% or the level of amplification is fairly low, we performed a series of different cutoff criteria and analyzed the results using a receiver operating characteristics curve (Figure 2B). By using log2 FC of 1.5 as the cutoff to detect ERBB2 amplification, we achieved 95% sensitivity and 100% specificity in our sample cohort.
Figure 2.
A: The range of fold change values for breast and gastroesophageal cases, separated according to the HER2 IHC result (score, 0 or 1, 1 to 2, 2, 2 to 3, and 3) and HER2 FISH result. The solid vertical line and dashed vertical line represent a fold change of 1.5 and 2.0, respectively. B: Receiver operating characteristics curve for different threshold values. For lower values of threshold, pipeline calls everything as amplified (AMP), which are all false positives (x axis); for higher values of cutoff threshold, our ability to call real events as amplified decreases. Threshold of 1.5 is the best in terms of sensitivity and specificity. EQUIV, equivalent; NA, not applicable.
Accuracy of NGS-Based Assessment of ERBB2 Amplification
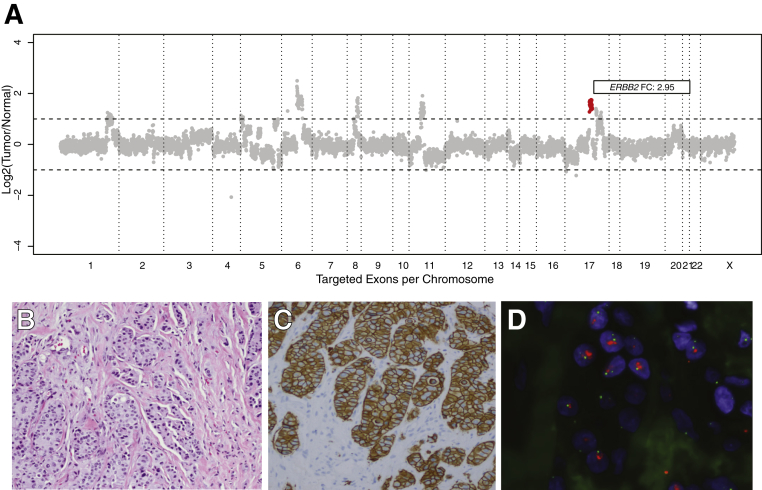
The concordance between IHC/FISH and the MSK-IMPACT assay for all samples is summarized in Table 2. The overall concordance was 98.4% (248/252). The concordance for the breast and GE samples was 98.1% (209/213) and 100% (39/39), respectively. Examples of the CN plot, HER2 IHC, and HER2 FISH for an amplified breast cancer are displayed in Figure 3.
Table 2.
Validation Data Set Concordance
| IHC/FISH |
MSK-IMPACT assay | |||
|---|---|---|---|---|
| Breast |
Gastroesophageal |
|||
| Nonamplified | Amplified | Nonamplified | Amplified | |
| 155 | 4 | 26 | 0 | Nonamplified |
| 0 | 54 | 0 | 13 | Amplified |
FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; MSK-IMPACT, Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets.
Figure 3.
Representative ERBB2 amplified case. A: Each dot represents a probe set, and the values on the y axis show the normalized log2 transformed fold change of coverage of tumor versus normal. ERBB2 fold change (FC) of 2.95. Additional amplification calls (cytoband, FC): PRDM1 (6q21, 3.0), ROS1 (6q22.1, 3.0), MYC (8q24.21, 2.8), CCND1 (11q13.3, 2.5), FGF19/4/3 cluster (11q13.3, 2.5), PAK1 (11q13.5, 2.5), and SPOP (17q21.33, 2.5). B: Hematoxylin and eosin–stained section of invasive carcinoma. C: HER2 immunohistochemical-stained slide showing strong and complete membranous staining in the invasive tumor cells (score, 3). D: HER2 FISH (red signal, HER2; green signal, CEP17). HER2/CEP17 ratio, 5.2; average HER2 CN, 9.9. Original magnification, ×200 (B and C).
Four discordant cases (all breast tumors) were amplified by the combined IHC/FISH algorithm but did not meet criteria for amplification by MSK-IMPACT (FC, 1.2 to 1.4). The characteristics of these samples are listed in Table 3. The estimated tumor percentages for these samples ranged from 10% to 60%. Background inflammation ranged from minimal to marked. One case showed heterogeneity on FISH, with only 10% of cells showing amplification. The remaining three samples were IHC equivocal, and FISH showed relatively low-level amplification.
Table 3.
Validation Data Set Discordant Cases
| Breast sample type | Tumor (%) | Inflammation | HER2 IHC | HER2 FISH HER2/CEP17 (HER2 CN, CEP17 CN) |
ERBB2 fold change (range) |
|---|---|---|---|---|---|
| Bx | 50 | Minimal | 2 | 2.0 (5.0, 2.4) | 1.4 (1.38 to 1.97) |
| Ex | 10 | Marked | 3 | 2.3 (4.7, 2.05) | 1.2 (−1.04 to 1.35) |
| Ex | 60 | Moderate | 2 (Focal) | Heterogeneous | 1.4 (1.34 to 1.72) |
| 10%: 3.2 (6.8, 2.1) | |||||
| 90%: 1.5 (3.0, 2.0) | |||||
| Ex | 60 | Moderate | 2 | 2.9 (4.9, 1.7) | 1.2 (−1.02 to 1.33) |
Bx, biopsy; CEP17, chromosome 17 centromere marker; CN, copy number; Ex, excision; FISH, fluorescence in situ hybridization; HER2, human epidermal growth factor receptor 2; IHC, immunohistochemistry.
The amplification threshold criteria set forth by the validation data set were also applied to an independent set of consecutive breast (n = 599) and GE (n = 130) cancers submitted for MSK-IMPACT analysis over 1 year to monitor the performance of the assay across unselected samples. The overall concordance for the breast and GE cancers in this independent clinical data set was 99.3% (595/599) and 93.8% (122/130), respectively (Supplemental Table S1). For the breast cancers, the concordance was 97.1% (101/104) among cases amplified by IHC and/or FISH and 99.8% (494/495) among cases nonamplified by IHC and/or FISH. For the GE cancers, the concordance was 78.8% (26/33) among cases amplified by IHC and/or FISH and 99.0% (96/97) among cases nonamplified by IHC and/or FISH. Of the discordant GE cases, material was not available for confirmation of the HER2 status by FISH for three cases and the IHC status was not available for one case.
Reproducibility and Analytic Sensitivity of NGS-Based Assessment of ERBB2 Amplification
Data from the reproducibility studies showed excellent inter-assay and intra-assay reproducibility, with CVs ranging from 0.007 to 0.07. Results are summarized in Table 4.
Table 4.
Inter-Assay and Intra-Assay (Precision) Reproducibility Study for ERBB2 Amplification Detection
| Sample | Inter-Assay |
Intra-Assay |
|||
|---|---|---|---|---|---|
| Run 1 FC | Run 2 FC | Run 3a FC | Run 3b FC | Run 3c FC | |
| 1 | 7.02 | 7.10 | 6.95 | 6.93 | 6.69 |
| 2 | 2.80 | 2.93 | 2.84 | 2.84 | 2.84 |
| 3 | 2.33 | 2.33 | 2.36 | 2.36 | 2.08 |
FC, fold change.
Results of the sensitivity study are summarized in Table 5. One clinical FFPE sample and two cell lines (MDA-MB-361 and MDA-MB-453) with varying levels of ERBB2 amplification were used for this analysis. For the markedly amplified sample with a HER2/chromosome 17 centromere marker (CEP17) ratio of 13.7 by FISH, MSK-IMPACT showed a corresponding FC of 49.38. Subsequent dilutions with the matching nonamplified blood sample show a sequential linear decrease with detectable amplification even at the lowest dilution of approximately 3% tumor DNA. The moderately amplified sample with a HER2/CEP17 ratio of 4.2 by FISH showed unequivocal amplification down to the 12.5% dilution (FC, 1.61) but lower than the threshold for calling at the subsequent 6.25% dilution (FC, 1.33). For the minimally amplified sample, with a HER2/CEP17 ratio of 2.4 by FISH, the detection limit was seen at the 50% dilution (FC, 1.61). Although the FC values for the two cell lines lower than the above stated levels did not meet criteria for an amplification call, examination of the CN plots suggested CN gains at lower dilution levels (Supplemental Figure S1).
Table 5.
Sensitivity Study for ERBB2 Amplification Detection
| Sample | Dilution (%) | ERBB2 fold change | Result |
|---|---|---|---|
| FFPE (HER2/CEP17, 13.7) | 100 | 49.38 | AMP called |
| 50 | 14.44 | AMP called | |
| 25 | 6.90 | AMP called | |
| 12.50 | 3.73 | AMP called | |
| 6.25 | 2.34 | AMP called | |
| 3.13 | 1.56 | AMP called | |
| MDA-MB-361 (HER2/CEP17, 4.2) | 100 | 5.66 | AMP called |
| 50 | 3.32 | AMP called | |
| 25 | 2.16 | AMP called | |
| 12.50 | 1.61 | AMP called | |
| 6.25 | 1.33 | AMP not called | |
| MDA-MB-453 (HER2/CEP17, 2.4) | 100 | 2.31 | AMP called |
| 50 | 1.61 | AMP called | |
| 25 | 1.3 | AMP not called | |
| 12.50 | 1.15 | AMP not called | |
| 6.25 | 1.0 | AMP not called |
AMP, amplification; CEP17, chromosome 17 centromere marker; FFPE, formalin-fixed, paraffin-embedded; HER2, human epidermal growth factor receptor 2.
Concurrent Detection of Other Somatic Alterations in the Validation Data Set
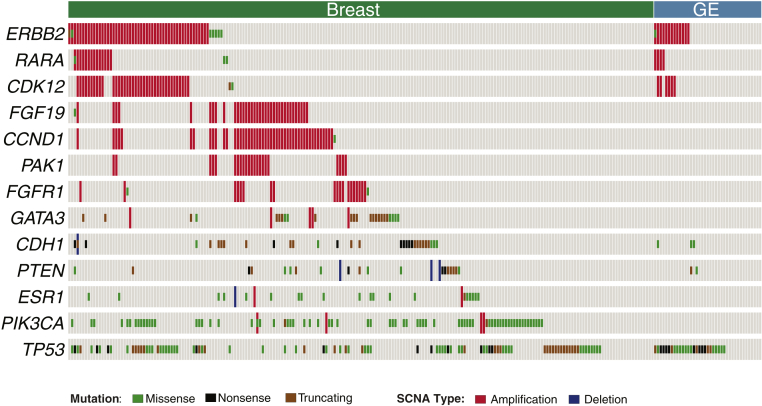
The concurrent somatic alterations for the breast and GE tumors in the validation data set are displayed in Figure 4. The most common alteration observed in both the breast and GE tumors was in the TP53 gene. Half of the TP53 mutations detected were loss-of-function alterations (frameshift indels, nonsense mutations, and splicing mutations). The other most common alterations in breast tumors were observed in PIK3CA, CDH1, GATA3, and ESR1. Most PIK3CA mutations encompassed the known activating mutations H1047R, E545K, and E542K. The CDH1 mutations consisted primarily of loss-of-function alterations.
Figure 4.
Concurrent somatic alterations in validation data set. The oncoprint displays the most common somatic alterations in the data set. Each column is a sample, and each row is a gene. Alterations are represented with various colors. GE, gastroesophageal; SCNA, somatic copy number alteration.
The most common CNAs detected in the validation cohort for breast tumors were amplifications of CCND1, CDK12, the FGF19/4/3 cluster, PAK1, FGFR1, and RARA. All RARA and CDK12 amplifications in both the breast and GE tumors were observed in cases with concurrent ERBB2 amplification. Most FGF19/3/4, CCND1, and PAK1 CNAs had a tendency of mutual exclusivity, with ERBB2 alterations in breast tumors; however, in this validation data set, there were no statistically significant patterns.
Discussion
Amplification/overexpression of the ERBB2/HER2 oncogene is an important biomarker for breast and gastric malignancies. Because of the potential targetability, it is also gaining importance in other cancers, such as lung, bladder, endometrium, ovary, colon, and rectum.19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30 As additional biomarkers of clinical significance continue to emerge for solid tumors, the applicability and sustainability of single-gene assessment is increasingly questionable, particularly for limited biopsy material. Although FISH and IHC are broadly used across clinical laboratories as robust methods for the assessment of ERBB2 status and are incorporated in current clinical guidelines, newer NGS methods provide a highly suitable and cost-effective solution for addressing the evolving needs of comprehensive genotyping. In the current study, we show that ERBB2 status can be reliably determined using our laboratory-developed hybridization capture-based assay. We developed and validated an algorithm for assessing CNAs in clinical samples routinely submitted for mutation profiling by MSK-IMPACT and critically compared the ERBB2 results with corresponding IHC and FISH studies.
Using 10% as the threshold for tumor content, our NGS assay had an overall concordance of 98.4% (248/252) in the validation data set and 98.4% (717/729) in the clinical data set compared to the combined IHC/FISH method. In the validation data set, the NGS method proved superior to IHC alone, allowing more accurate classification in equivocal categories (scores 1 to 2, 2, and 2 to 3) in 42 of 45 cases (93.3%). Only three cases characterized as equivocal by IHC and amplified by FISH were not resolved by the NGS assay: one with focal amplification in only 10% of the cells and two with low-level amplification by FISH. Although these cases did not meet the established threshold for amplification by the NGS method, review of the CN plots revealed low CN gains, with some probe sets reaching thresholds above a FC of 1.5 (Table 3), suggesting focal amplification that could prompt the further assessment of ERBB2 status by FISH.
We systematically examined the factors that influence CN results using NGS, including tumor purity, region coverage, heterogeneity, and level of amplification. Special attention was devoted to the assessment of tumor content. Not surprisingly, CN gain detection is heavily influenced by tumor content, with the highest impact in cases of low or borderline amplification. Cell line dilution studies allowed us to estimate the minimum tumor content necessary to detect ERBB2 amplification in the context of low or borderline level amplification. Using a cell line with moderate amplification (MDA-MB-361, HER2/CEP17 ratio, 4.2), the NGS method could confidently detect amplification down to the 12.5% dilution level (approximately 12.5% tumor DNA). With the low amplification cell line (MDA-MB-453, HER2/CEP17 ratio, 2.4), the established threshold for amplification is only met at the 50% dilution. Similar to our findings in the accuracy study, even at levels in which the FC threshold for amplification is not met, the CN plot still suggests CN gains at the 6.25% and 25% dilution levels for the moderate and low amplification cell lines, respectively (Supplemental Figure S1). Thus, for cases with CN gain in conjunction with a low tumor content, confirmation of the HER2 status by an alternate method would be recommended. An important caveat for low-level amplification calls is the requirement of a relatively clean/flat background on the CN plot. Samples with poor quality or low DNA content can yield noisy CN plots, limiting accurate assessment.
For GE tumors in particular, the assessment of the ERBB2 status by NGS can be limited by the high prevalence of intratumoral heterogeneity.40, 41 This degree of heterogeneity can be observed in IHC and FISH, with only focal or scattered areas of overexpression or amplification, respectively. In combination with the small size of the biopsy specimens often received for primary tumors and a background of inflammation, which can be marked, the overall concordance with standard methods of assessment, such as IHC and/or FISH, may be lower (Supplemental Table S1) and lead to false-negative results. This limitation is a reflection of the inherent nature of these tumors. In addition, sometimes there is the real-world limitation of only scant material remaining for molecular testing that may not be representative of the originally sampled tumor. In cases with discrepant results, reflex testing by FISH would be recommended on the same specimen if material remains or on an alternate tumor block.
Morphologic assessment before initiating testing is essential to achieve success in any molecular assay, not only for the assessment of tumor content but to ensure that the appropriate area of tumor is selected for testing. For breast carcinoma in particular, a potential pitfall of NGS assays can occur if areas of invasive carcinoma containing extensive in situ carcinoma are selected, leading to possible false-negative or false-positive results. HER2 overexpression/amplification is seen more frequently in ductal carcinoma in situ, particularly high-grade ductal carcinoma in situ (up to 60%),42, 43 compared to invasive carcinoma (18% to 25%). Avoiding areas with extensive ductal carcinoma in situ is therefore important to prevent false-positive results. An example of a case of invasive carcinoma with extensive ductal carcinoma in situ and the subsequent metastatic tumor submitted for NGS analysis is illustrated in Supplemental Figure S2.
Assessment of tumor content is critical for the accurate interpretation of molecular results. The estimation of tumor percentages based purely on review of hematoxylin and eosin–stained slides can be highly inaccurate because of many factors.44 An added advantage of broad NGS testing is the ability to more precisely estimate tumor purity on the basis of mutant allele fractions, allowing more confident recommendations for additional testing in cases with low tumor content.
In addition to tumor purity and tumor heterogeneity, there are technical factors that affect the CN calls. Adequate coverage of targeted exons is crucial and if the coverage of a region is much lower than the overall coverage of the sample, the variability of the FC values can increase substantially. During analysis, we remove regions where the coverage is in the lowest fifth percentile in 20% of the normals tested in a given pool. This reduces the number of regions with variable coverage and reduces false-positive FC values that could otherwise affect the normal distribution of FC values.
GC content of the captured targets is also an important factor to consider. Although GC content for a given region does not change across samples, the degree to which GC content affects capture efficiency changes on the basis of the starting DNA quality. This variability, unless corrected for, becomes an important factor when the null distribution of the FC values is used for model parametrization. Failure to correct for the GC content–based coverage variability can result in false-positive or false-negative results.
The sample type used for DNA extraction can be another important factor for detecting CNAs. FFPE-derived DNA-based sequencing data can produce false-positive results because of the highly degraded nature of the DNA molecules when the comparator is blood-derived DNA. We found that using an FFPE normal for comparison reduces the false-positive calls. Using a series of FFPE normals as candidates for best comparator increases the sensitivity of the calls by reducing the background noise dependent on the different qualities of FFPE-derived DNA.
Another important consideration of HER2 testing by NGS methods alone and up front is the turnaround time. Comprehensive NGS assays, which provide accurate assessment of CN, are not rapid assays and turnaround time is at least 10 working days. Although longer than the expected turnaround time for IHC or FISH alone, the overall time can be comparable to that expected for IHC and FISH if they are sequentially performed, taking into account the time allocated for recuts and transport if material is being sent out for testing. NGS testing is often requested on small samples, particularly aspirates of metastatic lesions, with material that is not suitable for extensive testing by multiple platforms. NGS in these cases would allow full comprehensive assessment up front and minimize the risk of tissue exhaustion because of multiple sections cut for IHC and FISH and refacing of the FFPE tissue block. In our experience, requests for HER2 IHC/FISH testing are increasingly being followed by requests for comprehensive genomic testing to assess for other potential targetable genomic alterations.
NGS also overcomes some of the limitations of IHC and FISH, including the need for stringent fixation methods/times, variation among different antibody sensitivities and specificities, interobserver and intraobserver variabilities, and especially aneusomy of CEP17. On the basis of the American Society of Clinical Oncology/College of American Pathologists guidelines, ERBB2 amplification is defined relative to CEP17 when using dual-probe FISH assays.31, 32, 33 ERBB2 amplification status in GE tumors relies solely on the HER2/CEP17 ratio, whereas breast tumors take into account both the HER2/CEP17 ratio and HER2 CN. With NGS assays, one can derive reference CN information from across the genome rather than relying on a single locus, such as CEP17, to calculate a ratio. ERBB2 can be assessed in relation to other genes on chromosome 17 and, thus, the focality of the ERBB2 amplicon can be appreciated at the genomic level. In the absence of cytogenetic analysis, the number of CEP17 signals observed by ISH has been used as a surrogate for chromosome 17 CN. Using the definition of >3 CEP17 copies per cell for polysomy, the reported prevalence rates in breast tumors range between 3% and 46%.45 There are several reasons for variation in the CEP17 CN during FISH analysis, including section thickness, cell division, and chromosomal instability, and true polysomy is actually rare. In breast tumors, there is no clear relationship of polysomy 17 to HER2 protein status or benefit from HER2-targeted therapy at this time.45, 46 Loss or gain of the pericentromeric region of chromosome 17 is more commonly observed and can result in alterations in the HER2/CEP17 ratio and false-positive or false-negative results. With NGS assays, not only can information of simultaneous CNAs be assessed but also prognostic information and an overall better understanding of the molecular mechanisms.47, 48, 49
In conclusion, we find that the ERBB2 status can be reliably determined by NGS methods and allows the concurrent testing for other potentially actionable genomic alterations. Given our findings in this validation study, we propose that up-front assessment by an NGS method is a suitable solution in cases where both ERBB2 status and comprehensive molecular profiling is sought. This approach is particularly useful for cases with limited material up front, where sequential testing by IHC, reflex FISH, and further mutation analysis would lead to tissue exhaustion before completion of all necessary testing. Samples with low tumor content, heterogeneity, and low level of amplification may result in false-negative results, or low-level copy number gains, prompting reflex testing by FISH in a minority of cases to confirm the ERBB2 status. Although the use of NGS technology across laboratories continues to increase, intrinsic differences among platforms, assay design, and data analysis highlight the need for future updates of the HER2 guidelines to incorporate criteria for NGS-based assessment of ERBB2 status.
Footnotes
Supported in part through the NIH/National Cancer Institute Cancer Center Support grant P30 CA008748 and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2016.09.010.
Supplemental Data
Copy number plots from the sensitivity study for ERBB2-amplified cell lines MDA-MB-361 and MDA-MB-453. Each dot represents a probe set, and the values on the y axis show the normalized log2 transformed fold change (FC) of coverage of tumor versus normal. The red dots represent the ERBB2 gene and correspond to the FC listed in the title of each plot.
A: Copy number plot for a primary tumor with invasive carcinoma and extensive ductal carcinoma in situ (DCIS). ERBB2 fold change (FC) of 1.9. The genes CCND1 (FC, 1.5), FGF19 (FC, 1.5), FGF4 (FC, 1.5), and FGF3 (FC, 1.3) are highlighted with the segmented arrow, not reaching the cutoff for amplification in this material. B: Hematoxylin and eosin (H&E)–stained section of invasive carcinoma with DCIS. C: HER2 immunohistochemical-stained section showing strong and complete membranous staining in the DCIS (score, 3) and lack of membranous staining in the invasive carcinoma (score, 0). D: Copy number plot for metastatic carcinoma involving L3 soft tissue. ERBB2 FC of 1.1. The genes CCND1 (FC, 2.2), FGF19 (FC, 2.2), FGF4 (FC, 2.2), and FGF3 (FC, 2.2) are highlighted with the segmented arrow, reaching the cutoff for amplification in this material. E: H&E-stained section of metastatic carcinoma. F: HER2 immunohistochemical-stained section showing lack of membranous staining in the metastatic carcinoma (score, 0). Original magnification, ×100 (B, C, E, and F).
References
- 1.Slamon D.J., Clark G.M., Wong S.G., Levin W.J., Ullrich A., McGuire W.L. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Owens M.A., Horten B.C., Da Silva M.M. HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer. 2004;5:63–69. doi: 10.3816/cbc.2004.n.011. [DOI] [PubMed] [Google Scholar]
- 3.Yaziji H., Goldstein L.C., Barry T.S., Werling R., Hwang H., Ellis G.K., Gralow J.R., Livingston R.B., Gown A.M. HER-2 testing in breast cancer using parallel tissue-based methods. JAMA. 2004;291:1972–1977. doi: 10.1001/jama.291.16.1972. [DOI] [PubMed] [Google Scholar]
- 4.Jorgensen J.T. Targeted HER2 treatment in advanced gastric cancer. Oncology. 2010;78:26–33. doi: 10.1159/000288295. [DOI] [PubMed] [Google Scholar]
- 5.Bang Y.J., Van Cutsem E., Feyereislova A., Chung H.C., Shen L., Sawaki A., Lordick F., Ohtsu A., Omuro Y., Satoh T., Aprile G., Kulikov E., Hill J., Lehle M., Ruschoff J., Kang Y.K. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 6.Marx A.H., Tharun L., Muth J., Dancau A.M., Simon R., Yekebas E., Kaifi J.T., Mirlacher M., Brummendorf T.H., Bokemeyer C., Izbicki J.R., Sauter G. HER-2 amplification is highly homogenous in gastric cancer. Hum Pathol. 2009;40:769–777. doi: 10.1016/j.humpath.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Lee S., de Boer W.B., Fermoyle S., Platten M., Kumarasinghe M.P. Human epidermal growth factor receptor 2 testing in gastric carcinoma: issues related to heterogeneity in biopsies and resections. Histopathology. 2011;59:832–840. doi: 10.1111/j.1365-2559.2011.04017.x. [DOI] [PubMed] [Google Scholar]
- 8.Tafe L.J., Janjigian Y.Y., Zaidinski M., Hedvat C.V., Hameed M.R., Tang L.H., Hicks J.B., Shah M.A., Barbashina V. Human epidermal growth factor receptor 2 testing in gastroesophageal cancer: correlation between immunohistochemistry and fluorescence in situ hybridization. Arch Pathol Lab Med. 2011;135:1460–1465. doi: 10.5858/arpa.2010-0541-OA. [DOI] [PubMed] [Google Scholar]
- 9.Seidman A.D., Berry D., Cirrincione C., Harris L., Muss H., Marcom P.K., Gipson G., Burstein H., Lake D., Shapiro C.L., Ungaro P., Norton L., Winer E., Hudis C. Randomized phase III trial of weekly compared with every-3-weeks paclitaxel for metastatic breast cancer, with trastuzumab for all HER-2 overexpressors and random assignment to trastuzumab or not in HER-2 nonoverexpressors: final results of Cancer and Leukemia Group B protocol 9840. J Clin Oncol. 2008;26:1642–1649. doi: 10.1200/JCO.2007.11.6699. [DOI] [PubMed] [Google Scholar]
- 10.Konecny G.E., Pegram M.D., Venkatesan N., Finn R., Yang G., Rahmeh M., Untch M., Rusnak D.W., Spehar G., Mullin R.J., Keith B.R., Gilmer T.M., Berger M., Podratz K.C., Slamon D.J. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 11.Di Leo A., Gomez H.L., Aziz Z., Zvirbule Z., Bines J., Arbushites M.C., Guerrera S.F., Koehler M., Oliva C., Stein S.H., Williams L.S., Dering J., Finn R.S., Press M.F. Phase III, double-blind, randomized study comparing lapatinib plus paclitaxel with placebo plus paclitaxel as first-line treatment for metastatic breast cancer. J Clin Oncol. 2008;26:5544–5552. doi: 10.1200/JCO.2008.16.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteva F.J., Hortobagyi G.N. Prognostic molecular markers in early breast cancer. Breast Cancer Res. 2004;6:109–118. doi: 10.1186/bcr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowinsky E.K. The erbB family: targets for therapeutic development against cancer and therapeutic strategies using monoclonal antibodies and tyrosine kinase inhibitors. Annu Rev Med. 2004;55:433–457. doi: 10.1146/annurev.med.55.091902.104433. [DOI] [PubMed] [Google Scholar]
- 14.Hynes N.E., Lane H.A. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 15.Borg A., Tandon A.K., Sigurdsson H., Clark G.M., Ferno M., Fuqua S.A., Killander D., McGuire W.L. HER-2/neu amplification predicts poor survival in node-positive breast cancer. Cancer Res. 1990;50:4332–4337. [PubMed] [Google Scholar]
- 16.Boku N. HER2-positive gastric cancer. Gastric Cancer. 2014;17:1–12. doi: 10.1007/s10120-013-0252-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann M., Stoss O., Shi D., Buttner R., van de Vijver M., Kim W., Ochiai A., Ruschoff J., Henkel T. Assessment of a HER2 scoring system for gastric cancer: results from a validation study. Histopathology. 2008;52:797–805. doi: 10.1111/j.1365-2559.2008.03028.x. [DOI] [PubMed] [Google Scholar]
- 18.Janjigian Y.Y., Werner D., Pauligk C., Steinmetz K., Kelsen D.P., Jager E., Altmannsberger H.M., Robinson E., Tafe L.J., Tang L.H., Shah M.A., Al-Batran S.E. Prognosis of metastatic gastric and gastroesophageal junction cancer by HER2 status: a European and USA International collaborative analysis. Ann Oncol. 2012;23:2656–2662. doi: 10.1093/annonc/mds104. [DOI] [PubMed] [Google Scholar]
- 19.Santin A.D., Bellone S., Roman J.J., McKenney J.K., Pecorelli S. Trastuzumab treatment in patients with advanced or recurrent endometrial carcinoma overexpressing HER2/neu. Int J Gynaecol Obstet. 2008;102:128–131. doi: 10.1016/j.ijgo.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Heinmoller P., Gross C., Beyser K., Schmidtgen C., Maass G., Pedrocchi M., Ruschoff J. HER2 status in non-small cell lung cancer: results from patient screening for enrollment to a phase II study of herceptin. Clin Cancer Res. 2003;9:5238–5243. [PubMed] [Google Scholar]
- 21.Gatzemeier U., Groth G., Butts C., Van Zandwijk N., Shepherd F., Ardizzoni A., Barton C., Ghahramani P., Hirsh V. Randomized phase II trial of gemcitabine-cisplatin with or without trastuzumab in HER2-positive non-small-cell lung cancer. Ann Oncol. 2004;15:19–27. doi: 10.1093/annonc/mdh031. [DOI] [PubMed] [Google Scholar]
- 22.Serrano-Olvera A., Duenas-Gonzalez A., Gallardo-Rincon D., Candelaria M., De la Garza-Salazar J. Prognostic, predictive and therapeutic implications of HER2 in invasive epithelial ovarian cancer. Cancer Treat Rev. 2006;32:180–190. doi: 10.1016/j.ctrv.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kris M.G., Johnson B.E., Berry L.D., Kwiatkowski D.J., Iafrate A.J., Wistuba I.I., Varella-Garcia M., Franklin W.A., Aronson S.L., Su P.F., Shyr Y., Camidge D.R., Sequist L.V., Glisson B.S., Khuri F.R., Garon E.B., Pao W., Rudin C., Schiller J., Haura E.B., Socinski M., Shirai K., Chen H., Giaccone G., Ladanyi M., Kugler K., Minna J.D., Bunn P.A. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pellegrini C., Falleni M., Marchetti A., Cassani B., Miozzo M., Buttitta F., Roncalli M., Coggi G., Bosari S. HER-2/Neu alterations in non-small cell lung cancer: a comprehensive evaluation by real time reverse transcription-PCR, fluorescence in situ hybridization, and immunohistochemistry. Clin Cancer Res. 2003;9:3645–3652. [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network. Kandoth C., Schultz N., Cherniack A.D., Akbani R., Liu Y., Shen H., Robertson A.G., Pashtan I., Shen R., Benz C.C., Yau C., Laird P.W., Ding L., Zhang W., Mills G.B., Kucherlapati R., Mardis E.R., Levine D.A. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grushko T.A., Filiaci V.L., Mundt A.J., Ridderstrale K., Olopade O.I., Fleming G.F., Gynecologic Oncology Group An exploratory analysis of HER-2 amplification and overexpression in advanced endometrial carcinoma: a Gynecologic Oncology Group study. Gynecol Oncol. 2008;108:3–9. doi: 10.1016/j.ygyno.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McAlpine J.N., Wiegand K.C., Vang R., Ronnett B.M., Adamiak A., Kobel M., Kalloger S.E., Swenerton K.D., Huntsman D.G., Gilks C.B., Miller D.M. HER2 overexpression and amplification is present in a subset of ovarian mucinous carcinomas and can be targeted with trastuzumab therapy. BMC Cancer. 2009;9:433. doi: 10.1186/1471-2407-9-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolff A.C., Hammond M.E., Hicks D.G., Dowsett M., McShane L.M., Allison K.H., Allred D.C., Bartlett J.M., Bilous M., Fitzgibbons P., Hanna W., Jenkins R.B., Mangu P.B., Paik S., Perez E.A., Press M.F., Spears P.A., Vance G.H., Viale G., Hayes D.F. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 32.Ajani J.A., Bentrem D.J., Besh S., D'Amico T.A., Das P., Denlinger C., Fakih M.G., Fuchs C.S., Gerdes H., Glasgow R.E., Hayman J.A., Hofstetter W.L., Ilson D.H., Keswani R.N., Kleinberg L.R., Korn W.M., Lockhart A.C., Meredith K., Mulcahy M.F., Orringer M.B., Posey J.A., Sasson A.R., Scott W.J., Strong V.E., Varghese T.K., Jr., Warren G., Washington M.K., Willett C., Wright C.D., McMillian N.R., Sundar H., National Comprehensive Cancer Network Gastric cancer, version 2.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw. 2013;11:531–546. doi: 10.6004/jnccn.2013.0070. [DOI] [PubMed] [Google Scholar]
- 33.Bartley A.N., Christ J., Fitzgibbons P.L., Hamilton S.R., Kakar S., Shah M.A., Tang L.H., Troxell M.L., Members of the Cancer Biomarker Reporting Committee, College of American Pathologists Template for reporting results of HER2 (ERBB2) biomarker testing of specimens from patients with adenocarcinoma of the stomach or esophagogastric junction. Arch Pathol Lab Med. 2015;139:618–620. doi: 10.5858/arpa.2014-0395-CP. [DOI] [PubMed] [Google Scholar]
- 34.Cheng D.T., Mitchell T.N., Zehir A., Shah R.H., Benayed R., Syed A., Chandramohan R., Liu Z.Y., Won H.H., Scott S.N., Brannon A.R., O'Reilly C., Sadowska J., Casanova J., Yannes A., Hechtman J.F., Yao J., Song W., Ross D.S., Oultache A., Dogan S., Borsu L., Hameed M., Nafa K., Arcila M.E., Ladanyi M., Berger M.F. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyman D.M., Solit D.B., Arcila M.E., Cheng D.T., Sabbatini P., Baselga J., Berger M.F., Ladanyi M. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. Drug Discov Today. 2015;20:1422–1428. doi: 10.1016/j.drudis.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mose L.E., Wilkerson M.D., Hayes D.N., Perou C.M., Parker J.S. ABRA: improved coding indel detection via assembly-based realignment. Bioinformatics. 2014;30:2813–2815. doi: 10.1093/bioinformatics/btu376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 39.Weigelt B., Warne P.H., Downward J. PIK3CA mutation, but not PTEN loss of function, determines the sensitivity of breast cancer cells to mTOR inhibitory drugs. Oncogene. 2011;30:3222–3233. doi: 10.1038/onc.2011.42. [DOI] [PubMed] [Google Scholar]
- 40.Yang J., Luo H., Li Y., Li J., Cai Z., Su X., Dai D., Du W., Chen T., Chen M. Intratumoral heterogeneity determines discordant results of diagnostic tests for human epidermal growth factor receptor (HER) 2 in gastric cancer specimens. Cell Biochem Biophys. 2012;62:221–228. doi: 10.1007/s12013-011-9286-1. [DOI] [PubMed] [Google Scholar]
- 41.Kim M.A., Lee H.J., Yang H.K., Bang Y.J., Kim W.H. Heterogeneous amplification of ERBB2 in primary lesions is responsible for the discordant ERBB2 status of primary and metastatic lesions in gastric carcinoma. Histopathology. 2011;59:822–831. doi: 10.1111/j.1365-2559.2011.04012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnes D.M., Bartkova J., Camplejohn R.S., Gullick W.J., Smith P.J., Millis R.R. Overexpression of the c-erbB-2 oncoprotein: why does this occur more frequently in ductal carcinoma in situ than in invasive mammary carcinoma and is this of prognostic significance? Eur J Cancer. 1992;28:644–648. doi: 10.1016/s0959-8049(05)80117-0. [DOI] [PubMed] [Google Scholar]
- 43.van de Vijver M.J., Peterse J.L., Mooi W.J., Wisman P., Lomans J., Dalesio O., Nusse R. Neu-protein overexpression in breast cancer: association with comedo-type ductal carcinoma in situ and limited prognostic value in stage II breast cancer. N Engl J Med. 1988;319:1239–1245. doi: 10.1056/NEJM198811103191902. [DOI] [PubMed] [Google Scholar]
- 44.Smits A.J., Kummer J.A., de Bruin P.C., Bol M., van den Tweel J.G., Seldenrijk K.A., Willems S.M., Offerhaus G.J., de Weger R.A., van Diest P.J., Vink A. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod Pathol. 2014;27:168–174. doi: 10.1038/modpathol.2013.134. [DOI] [PubMed] [Google Scholar]
- 45.Hanna W.M., Ruschoff J., Bilous M., Coudry R.A., Dowsett M., Osamura R.Y., Penault-Llorca F., van de Vijver M., Viale G. HER2 in situ hybridization in breast cancer: clinical implications of polysomy 17 and genetic heterogeneity. Mod Pathol. 2014;27:4–18. doi: 10.1038/modpathol.2013.103. [DOI] [PubMed] [Google Scholar]
- 46.Lal P., Salazar P.A., Ladanyi M., Chen B. Impact of polysomy 17 on HER-2/neu immunohistochemistry in breast carcinomas without HER-2/neu gene amplification. J Mol Diagn. 2003;5:155–159. doi: 10.1016/S1525-1578(10)60467-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung S.H., Lee A., Yim S.H., Hu H.J., Choe C., Chung Y.J. Simultaneous copy number gains of NUPR1 and ERBB2 predicting poor prognosis in early-stage breast cancer. BMC Cancer. 2012;12:382. doi: 10.1186/1471-2407-12-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwei K.A., Kung Y., Salari K., Holcomb I.N., Pollack J.R. Genomic instability in breast cancer: pathogenesis and clinical implications. Mol Oncol. 2010;4:255–266. doi: 10.1016/j.molonc.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sircoulomb F., Bekhouche I., Finetti P., Adelaide J., Ben Hamida A., Bonansea J., Raynaud S., Innocenti C., Charafe-Jauffret E., Tarpin C., Ben Ayed F., Viens P., Jacquemier J., Bertucci F., Birnbaum D., Chaffanet M. Genome profiling of ERBB2-amplified breast cancers. BMC Cancer. 2010;10:539. doi: 10.1186/1471-2407-10-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Copy number plots from the sensitivity study for ERBB2-amplified cell lines MDA-MB-361 and MDA-MB-453. Each dot represents a probe set, and the values on the y axis show the normalized log2 transformed fold change (FC) of coverage of tumor versus normal. The red dots represent the ERBB2 gene and correspond to the FC listed in the title of each plot.
A: Copy number plot for a primary tumor with invasive carcinoma and extensive ductal carcinoma in situ (DCIS). ERBB2 fold change (FC) of 1.9. The genes CCND1 (FC, 1.5), FGF19 (FC, 1.5), FGF4 (FC, 1.5), and FGF3 (FC, 1.3) are highlighted with the segmented arrow, not reaching the cutoff for amplification in this material. B: Hematoxylin and eosin (H&E)–stained section of invasive carcinoma with DCIS. C: HER2 immunohistochemical-stained section showing strong and complete membranous staining in the DCIS (score, 3) and lack of membranous staining in the invasive carcinoma (score, 0). D: Copy number plot for metastatic carcinoma involving L3 soft tissue. ERBB2 FC of 1.1. The genes CCND1 (FC, 2.2), FGF19 (FC, 2.2), FGF4 (FC, 2.2), and FGF3 (FC, 2.2) are highlighted with the segmented arrow, reaching the cutoff for amplification in this material. E: H&E-stained section of metastatic carcinoma. F: HER2 immunohistochemical-stained section showing lack of membranous staining in the metastatic carcinoma (score, 0). Original magnification, ×100 (B, C, E, and F).