Abstract
Mutations in cis-regulatory sequences have been implicated as being the predominant source of variation in morphological evolution. We offer a hypothesis that gene-associated tandem repeat expansions and contractions are a major source of phenotypic variation in evolution. Here, we describe a comparative genomic study of repetitive elements in developmental genes of 92 breeds of dogs. We find evidence for selection for divergence at coding repeat loci in the form of both elevated purity and extensive length polymorphism among different breeds. Variations in the number of repeats in the coding regions of the Alx-4 (aristaless-like 4) and Runx-2 (runt-related transcription factor 2) genes were quantitatively associated with significant differences in limb and skull morphology. We identified similar repeat length variation in the coding repeats of Runx-2, Twist, and Dlx-2 in several other species. The high frequency and incremental effects of repeat length mutations provide molecular explanations for swift, yet topologically conservative morphological evolution.
Keywords: repeat, tandem
In an observation that has evolved into the modern theory of punctuated equilibrium, Darwin (1) inferred from the fossil record that evolution frequently occurs in rapid bursts. It is now well established that significant morphological evolution generally occurs on short time scales (2, 3). D'Arcy Thompson (4) described in his 1917 classic On Growth and Form how modern animal forms and fossils alike are related to one another by simple continuous mathematical transformations. The rapid pace of morphological evolution noted by Darwin along with Thompson's observations of topological conservation are not easily reconciled with the rates at which point mutations occur or with the sensitivity of complex organisms and their proteins to random point mutations. A broad consensus has emerged that contends with these difficulties by invoking mutations in cis-regulatory elements as the predominant source of the genetic diversity that underlies morphological variation and evolution (5, 6).
In this article we report on a series of comparative genomic and morphological studies revealing evidence supporting an alternative hypothesis that length variations in tandemly repeated sequences are a major source of morphological variation, both “Thompsonesque” and discontinuous, that permit rapid generation of useful alleles. Tandem repeats are abundant in the coding sequences of vertebrate genes, especially those involved in development (7, 8), and orthologous repeat tracts frequently are conserved across distantly related taxa (9). Repeat expansions or contractions vary in a locus-specific manner and occur at rates up to 100,000 times higher than point mutations, because of the distinct mutational mode of slipped-strand mispairing rather than an incorporation error (10). Transcription, mRNA processing, protein translation, folding, stability, and aggregation rates, as well as gross morphology, have been found to be incrementally affected by alterations in repeat tract length (9, 11–16). We hypothesize that gene-associated tandem repeats function as facilitators of evolution, providing abundant, robust variation and thus enabling extremely rapid evolution of new forms.
Materials and Methods
DNA Sequencing. Repeat-containing portions of developmental orthologs were PCR-amplified from total genomic DNA by using betaine and DMSO additives (Epicentre Technologies, Madison, WI). Sequences were determined by automated fluorescent sequencing of PCR products according to the manufacturers' specifications (Beckman Coulter and Applied Biosystems). Frozen tissue samples were obtained from the University of Alaska Museum in Fairbanks.
Morphometrics. High-resolution 3D models of skulls from purebred dogs were constructed by using a laser noncontact 3D digitizer (Vivid 700, Minolta, Wayne, NJ). The set of skulls chosen for modeling was selected such that a wide range of morphologies were sampled and to ensure that each overtly visible mode of variation was represented in variety. Each model was assembled from 25–50 individual scans comprising 400,000 to 2.2 million individual measurements, with a typical sampling rate of approximately five independent measurements per mm2. Facial length was measured as the distance from the intersection of the zygomatic, maxilla, and lacrimal bones to the point where the maxilla, nasal, and incisive bones intersect. Measurements from left and right sides were averaged and normalized for size by dividing by length of the Hirnstammbasis (the length from the oral edge of the foramen magnum to the boundary of the pterygoid and palatine where it joins the presphenoid), an established metric for size in canids. Clinorhynchy was defined as the angle between the Hirnstammbasis and the ventral surface of the maxilla. Models were created for the following breeds: Afghan hound SHSB330393, Airedale terrier SHSB357370, Borzoi SHSB295093, boxer SHSB350416, bull terrier SHSB259885, English bulldog SHSB290547, English setter SHSB224720, greyhound SHSB208660, lhasa apso PC, mastiff SHSB204638, New Guinea singing dog PC, Pembroke Welsh corgi SHSB177099, pharaoh hound SHSB111.183/1962, pit bull PC, Rhodesian ridgeback SHSB545437, saluki LACM22825, Skye terrier USMNH21988, standard poodle SHSB329337, vizsla SHSB338560, toy poodle SHSB343920, and seven mongrels (SU20792, SU20794, SU20795, SU20796, SU20797, SU20838, and SU20839), where USMNH is the Smithsonian Museum of Natural History (Washington, DC), SHSB is the Bern Museum of Natural History (Bern, Switzerland), LACM is the Los Angeles County Museum, and PC means private collection.
Results and Discussion
We examined the sequences of tandem repeats in coding regions of developmental genes in a large panel of dog breeds (breeds and numbers are listed in Supporting Text, which is published as supporting information on the PNAS web site) and compared polymorphisms in these sequences with interbreed variation in morphology. In the absence of purifying expansion and contraction mutations, a previously “pure” (i.e., devoid of interruptions of the canonical repeat sequence) tandem repeat will tend to degrade by the accumulation of point mutations, whereas repeat expansion and contraction mutations have the effect of removing imperfections from tandem repeats, caused by the “copy and paste” process by which they occur (10). Evidence of recent alterations in repeat copy number at a locus and selection for plasticity can be inferred by comparing the sequences of orthologous repetitive loci (7). We sequenced 37 repeat-containing regions (36 coding and one 3′ UTR) from the domestic dog orthologs of 17 human genes that are both likely to mutate frequently (7, 17) and are known or suspected to be involved in morphological, especially craniofacial, development. We then compared the sequence purity to the homologous repeats in humans (Table 1). Our hypothesis would predict that dogs would have higher repeat purity in these loci than humans if the intense selection for morphological variation in dogs has resulted in recent alterations in the lengths of the repeats in these genes. For 29 of the 36 loci compared, the sequences of the dog repeats had fewer interruptions than their human counterparts; dog and human had equal purity for the remaining seven and one repeat was found only in the dog; the interspecies differences in repeat purity was highly significant (P < 0.001, Wilcoxon test), indicative of significant recent fluctuation in dog allele lengths. The difference between the species was of such a magnitude that it begged the question of whether dogs had some general “defect” in their DNA replication or repair machinery that led to a genomewide increase in repeat purity. The timely publication of a partial dog genome sequence enabled us to address this “slippery replication” hypothesis by using a comparative genomic analysis. We compared 844 orthologous triplet microsatellite pairs identified within conserved portions of human and dog genomic sequence and identified no significant difference in repeat purity between the two species. Thus, the large increase in repeat purity observed in the developmental genes is caused by locus-specific forces (presumably selection) and not attributable to any genomewide mechanism.
Table 1. Elevated repeat purity and high polymorphism in dog developmental genes.
| Purity
|
||||
|---|---|---|---|---|
| Repeat locus | Repeat unit | Human | Dog | No. of alleles |
| Alx-4 | PQ | 0.75 | 1.00 | 2 |
| Alx-4 | PQ | 0.93 | 0.96 | 4* |
| Bmp-11 | A | 0.97 | 1.00 | 3 |
| Dlx-2 | G | 0.95 | 0.97 | 5 |
| Hlx-1 | Q | 0.85 | 0.85 | 1 |
| Hox-A2 | A | 0.93 | 1.00 | 2 |
| Hox-A7 | A | 0.83 | 1.00 | 2 |
| Hox-A7 | A | 0.81 | 0.96 | 5* |
| Hox-A7 | D/E | 0.79 | 0.81 | 1 |
| Hox-A7 | 3′utr | 0.67 | 0.82 | 2 |
| Hox-A10 | G | 0.67 | 0.67 | 1 |
| Hox-A11 | A | 0.94 | 1.00 | 5 |
| Hox-A11 | A | — | 1.00 | 2* |
| Hox-A11 | G | 1.00 | 1.00 | 1 |
| Hox-A11 | A | 0.80 | 1.00 | 4* |
| Hox-C13 | G | 0.88 | 1.00 | 4 |
| Hox-D8 | PH | 0.93 | 0.93 | 4 |
| Hox-D8 | P | 0.89 | 0.93 | 1 |
| Hox-D8 | G | 0.83 | 0.92 | 1 |
| Hox-D8 | GP | 0.89 | 0.93 | 3* |
| Hox-D8 | P | 0.88 | 0.92 | 2 |
| Hox-D11 | G | 0.83 | 0.88 | 2 |
| Hox-D11 | A | 0.92 | 0.97 | 2 |
| Hox-D13 | S | 0.89 | 1.00 | 3 |
| Hox-D13 | A | 0.94 | 1.00 | 5 |
| Hox-D13 | A | 0.91 | 0.95 | 5 |
| Runx-2 | Q | 0.93 | 0.96 | 6* |
| Runx-2 | A | 0.88 | 0.95 | 6 |
| Six-3 | G | 0.93 | 1.00 | 5 |
| Six-3 | G | 0.86 | 0.95 | 5 |
| Six-3 | G | 1.00 | 1.00 | 4 |
| Sox-9 | P | 0.87 | 0.96 | 2 |
| Sox-9 | PnQA | 0.86 | 0.94 | 4* |
| Twist | G | 0.90 | 0.95 | 7* |
| Twist | G | 1.00 | 1.00 | 5 |
| Zic-2 | A | 0.85 | 0.89 | 1 |
| Zic-2 | H | 1.00 | 1.00 | 2 |
Purity was computed by dividing the number of bases that deviated from the canonical repeat unit (borders defined by amino acid translation) by the total length of the repeat and subtracting from one.
Includes distinct alleles of equal length, identified by variable imperfections within the repeat consistent with independent expansion/contraction events.
If some of the capacity for rapid morphological diversification in the domestic dog is mediated through repeat variations in selected genes, then interbreed variations in repeat length at these loci should be found. We sequenced these 37 repeat loci of 142 dogs representing 92 different breeds for a total of 4,312 repeat genotypes (a complete list of the breeds and numbers examined is included in Supporting Text). For 15 of 17 genes, and 29 of 37 repeats tested, at least two different coding repeat alleles were identified among domestic dogs, with five genes having 12 or more alleles (Table 1).
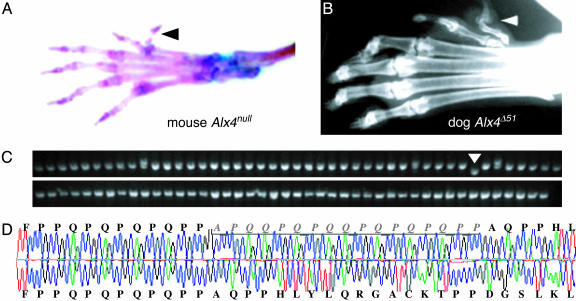
Most of the allelic variations observed were incremental differences in repeat length, within two or three repeat units of the length of the most common allele. However, five genes were found to have a rare repeat expansion or contraction alleles of large magnitude in coding sequences, including: Six-3 (Δ54 bp), Hox-a7 (Δ33 bp), Runx-2 (ins45 bp), Hox-d8 (Δ30 bp), and Alx-4 (Δ51 bp). Although these extreme alleles were atypical, in some cases they presented the opportunity to observe a large effect on the phenotype of the dog. The Alx-4Δ51 allele occurred in homozygosity in the Great Pyrenees breed (Fig. 1). An official characteristic of this breed is bilateral rear first digit polydactyly. All four Great Pyrenees with bilateral polydactyly examined were homozygous for this repeat contraction. None of the other 88 breeds examined for this locus had either polydactyly or a deletion in Alx-4. The Great Pyrenees breed exhibits some heterogeneity for this trait, and a single individual lacking extra dewclaws was identified and its Alx-4 repeat was sequenced. This individual was homozygous for the normal, full-length allele shared by all other nonpolydactylous breeds, making chance correlations caused by population subdivision (e.g., neutral founder effect) extremely unlikely. The form of polydactyly found in the hind limbs of this breed is similar to that observed in Alx-4 knockout mice (18), and it has been shown that deletion of this PQn repeat (but not other portions of the protein) specifically destroys the ability of Alx-4 to bind with lymphoid enhancer binding factor-1 and drive target gene expression in limb bud mesenchyme (19). Pedigree analysis in dogs indicates that the Great Pyrenees hind limb polydactyly is not a simple Mendelian trait; this finding is consistent with the genetic background dependence of the Alx-4 null mouse phenotype (20), and we conclude that it is likely caused in part by this 17-aa deletion (Fig. 1). The Alx-4-polydactyly relationship represents an extreme case of a coding repeat mutation resulting in gross morphological novelty, reminiscent of the saltatory genetic events Goldschmidt envisioned for his “hopeful monsters” (21). Although the fossil record indicates that this type of event occurred infrequently in evolution, it demonstrates the potential of repeat mutations to affect morphology in a manner distinct from that observed for polyglutamine expansion diseases (22).
Fig. 1.
Large magnitude repeat length mutations can result in gross morphological change. (A) Alx-4–/– mice exhibit a duplication of the first digit (arrowhead). [Adapted with permission from ref. 18 (Copyright 1998, The Company of Biologists).] (B) A radiograph of the rear paw of a Great Pyrenees shows the typical double dewclaw phenotype specified in the breed standard (arrowhead). (C) Polydactylous Great Pyrenees are homozygous for a 51-nucleotide repeat contraction in the Alx-4 gene. PCR amplification of the repeat-containing regions of Alx-4 from 89 dog breeds reveals that this deletion is unique to the Great Pyrenees breed (arrow). Phenotypically normal basset hounds, flat-coated retrievers, and harriers were heterozygous for distinct two amino acid insertions (doublets). (D) DNA sequencing reveals that the deletion is caused by a contraction of the PQn repeat that results in the removal of 17 aa within the repeat.
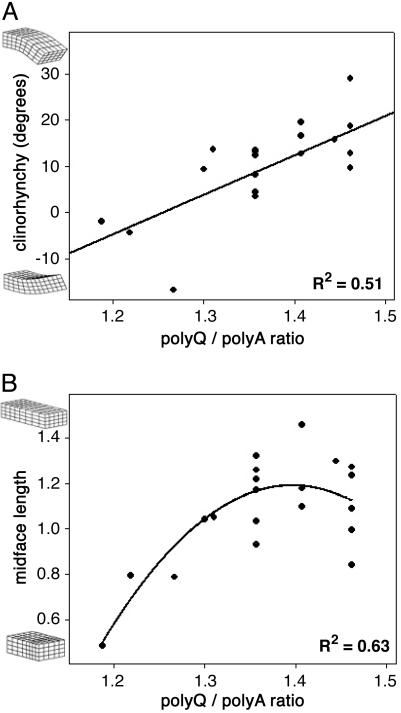
Incremental, topologically conservative transformations typify the succession of species in the fossil record and also typify more subtle intraspecies morphological variation. A quantitative effect of incremental repeat length variation on morphology was investigated for Runx-2, a master regulator of osteoblast differentiation. The human Runx-2 gene has a repeat encoding 23 glutamines followed by 17 alanines N-terminal to the DNA-binding domain, and inactivating mutations in Runx-2 cause cleidocranial dysplasia (CCD) in humans, a syndrome characterized by a variety of craniofacial and other skeletal malformations (23). The dog Runx-2 ortholog was identified, and a 330-bp segment spanning the repeat region was sequenced for 124 purebred dogs from 90 breeds. The dog Runx-2 gene encodes 18–20 glutamines followed by 12–17 alanines and is highly variable among breeds. Pearson correlation analysis revealed a modest correlation between total allele length (Q + A) and the degree of clinorhynchy (dorsoventral nose bend) and midface length, measured from high-precision 3D computer models of skulls from 20 morphologically diverse dog breeds and seven mongrels (24). Opposing effects on transcriptional activity have been reported for both polyglutamine and polyalanine homopolymers: polyglutamines have been observed to drive transcription and polyalanines to repress transcription in a length-dependent fashion (9, 11). Selection may have paired these repeats with opposing activities to facilitate the generation of alleles conferring precise modulation of transcription by the Runx-2 protein, and the difference between polyglutamine and polyalanine length may provide a superior indicator of allele activity. Repeating the correlation analysis by using the ratio of repeat lengths (Q/A) for each allele improved the explanatory power of repeat length for both clinorhynchy (Pearson one-sided significance P = 0.0001; Fig. 2A) and midface length (P = 0.0002; Fig. 2B). Clinical features of human CCD caused by haploinsufficiency of Runx-2 recapitulate the phenotypes of dog breeds with lower Q/A ratios, including brachycephaly, midface hypoplasia, and a low nasal bridge. A family with a mild form of CCD caused by a 10-alanine expansion in the Runx-2 polyalanine tract has been described (25). This finding and our data are consistent with longer polyalanine relative repeats (lower Q/A) being associated with hypomorphic alleles.
Fig. 2.
Tandem repeat length in a developmental gene is quantitatively correlated with continuous morphological features. (A and B) Reported (9, 11) effects on transcription of polyglutamine and polyalanine repeats suggested that these two domains may be involved in competitive activities and that the relative lengths of these domains may be more instructive than their aggregate length. A Pearson correlation test of this hypothesis revealed a significant correlation between Runx-2 polyglutamine to polyalanine ratio and clinorhynchy (D/V nose bend, P = 0.0001, Pearson one-sided significance, n = 27, A) and midface length (P = 0.0002, n = 27, B) (24). The nature and direction of these correlations is indicative of longer relative Runx-2 glutamine repeats resulting in increased midface growth, consistent with observations from human cleidocranial dysplasia patients (25). Published studies (9–16) indicate that amino acid repeat length-function relationships are typically nonlinear; however, fitting a quadratic or exponential to the clinorhynchy data (A) does not provide sufficient improvement in residuals to support the use of a nonlinear function over a simple line.
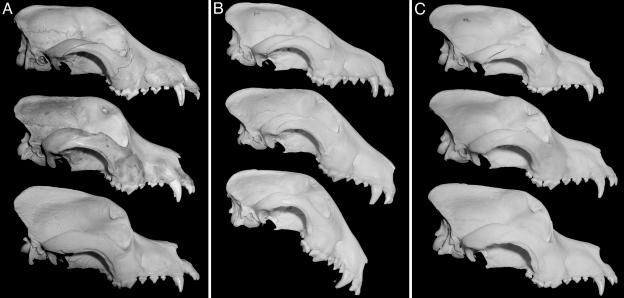
The dramatic changes that have occurred in domestic dog breeds in response to breeders' selection toward breed standards over the last 150 years demonstrate the potential of the mammalian genome to effect rapid morphological change in response to strong selection, even with small, closed gene pools (Fig. 3). Although mtDNA analyses suggest a common dog mitochondrial ancestor between 15,000 and 100,000 years ago, mitochondria are not as sensitive to the same purifying forces (particularly popular sire effects in which a single champion stud may sire or grandsire large portions of the breeding population) operating on nuclear alleles, and the last common ancestors for nuclear loci were in all likelihood much more recent (26, 27).
Fig. 3.
Rapid and sustained evolution of breeds. (A) Purebred St. Bernard skulls from ≈1850 (Top), 1921 (Middle), and 1967 (Bottom). (B) Purebred bull terrier skulls from 1931 (Top), 1950 (Middle), and 1976 (Bottom) (24). (C) Purebred Newfoundland skulls from 1926 (Top), 1964 (Middle), and 1971 (Bottom). Despite the lack of genetic diversity caused by population structure and history, these breeds are able to continually create new and more extreme morphological variations at a rapid and sustained pace. Analysis of the Runx-2 repeats in the 1931 bull terrier reveals a more intermediate allele (Q19A14) than is present in the modern bull terrier (Q19A13).
The rapid pace of change in morphology depicted in these breeds in Fig. 3 is even more remarkable when the limited capacity of these populations to harbor or maintain genetic diversity is considered (28). The tendency of the genetic diversity-reducing forces commonly operating in most breeds to quickly drive alleles to fixation in such small populations is overwhelming. To continually produce more extreme variation despite these purifying forces requires the dog genome to create new alleles at an extraordinary rate. With typical mutation rates in the 10–7 to 10–9 range, it is doubtful that point mutations could occur with sufficient frequency to provide breeders with adequate variation upon which selection can be applied. This is borne out in our sequencing results with only one common single nucleotide variant and two private alleles identified in the 7.2 kb of nonrepetitive DNA sequenced for our panel. The common allele, a silent C → A in Bmp-11, is likely of ancient origin given the diverse ancestries of the five breeds in which it occurred (treeing Walker coonhound, bluetick coonhound, Bedlington terrier, German shorthaired pointer, and pug); whereas the two rare alleles, a G → Tinthe 5′ UTR of Hox-d11 in the saluki and a silent C → G in Six-3 in the Maremma (both heterozygous), are more likely to have occurred after these breeds were founded. The paucity of single nucleotide diversity among breeds is consistent with other studies (29–32) and stands in stark contrast to the abundance of coding repeat variation we found in these same genes known to be involved in the developmental processes under selection in the radiation in dog breed morphologies.
The relative youth of the repeat alleles we discovered is underscored when the number of unique haplotypes is considered. In the Hox-A cluster, seven different repeat loci in the Hox-a2, Hox-a7, and Hox-a11 genes combine for at least 28 distinct haplotypes (Table 3, which is published as supporting information on the PNAS web site). In the Hox-D cluster, eight repeats in the Hox-d8, Hox-d11, and Hox-d13 genes combine for a minimum of 30 haplotypes, and if the tightly linked Dlx-2 locus is included, the lower bound rises to 36 (Table 4, which is published as supporting information on the PNAS web site). It is implausible that dozens of ancient repeat haplotypes, and few SNPs, predating domestication have been maintained through the ages for all of these loci; these represent repeat mutations that have occurred in modern dog breeds. In the case of the bull terriers pictured in Fig. 3, we were able to test this. The serendipitous discovery of preserved desiccated muscle tissue of a single purebred bull terrier maintained since 1931 by the Museum of Natural History in Bern, Switzerland enabled us to test this directly for Runx-2. We PCR-amplified this repeat from genomic DNA isolated from the individual whose skull is shown in Fig. 3B Top. Because of the uniqueness of this specimen we cannot ascertain that it is representative of the breed at the time genetically, we do, however, know from photographic evidence that it is an excellent representative of the bull terrier of that time period in its conformation. This individual was found to have a more intermediate Runx-2 repeat allele (Q19A14) than was found in the modern bull terrier (Q19A13, n = 6 individuals), as would be predicted from the genotype-phenotype relationship shown in Fig. 2 and the evolution of the breed illustrated in Fig. 3A. Although some modern breeds are ostensibly “recreations” of ancient breeds, the phenotypes of ancestral dog breeds as recorded in images and preserved skulls exhibit nothing approaching the spectrum of morphological extremes observed among their modern counterparts. Collectively, these data suggest that the conventional wisdom of attributing the morphological diversity among dogs to the partitioning of preexisting wolf allelic variation by selective breeding may require revision (33–35).
To determine whether this phenomenon is unique to the domestic dog, we genotyped the coding repeats of Runx-2, Twist, and Dlx-2 for individuals from several other species. Wolves, coyotes, Arctic foxes, swift foxes, red foxes, river otters, walruses, cottontail rabbits, silver-haired bats, and humans all were found to harbor variation in at least one of these amino acid repeats (Table 2); thus the rapid mutation rates of these loci renders the same types of variations perpetually available for selection in natural populations. With the exception of the silver-eared bat, allele lengths as well as the degree of morphological variation for all of these species fall within a more narrow range than observed among dog breeds. Selection for alterations in midface length or clinorhynchy in any of these species might be expected to act through altering allele frequencies of polyglutamine or polyalanine repeat variations at this locus.
Table 2. Alleles with modest alterations in developmental regulator gene coding repeat length are common in natural populations of placental mammals.
| Species | Panel size | No. of Runx-2 alleles | No. of Dlx-2 alleles | No. of Twist-1 alleles |
|---|---|---|---|---|
| Canis lupus (gray wolf) | 70 | 5 | 5 | 16 |
| Canis latrans (coyote) | 98 | 7 | 5 | 17 |
| Vulpes vulpes (red fox) | 10 | 4 | 1 | 1 |
| Vulpes velox (swift fox) | 1 | 1 | 2 | 2 |
| Alopex lagopus (Arctic fox) | 2 | 2 | 3 | 3 |
| Enhydra lutris (sea otter) | 3 | 2 | 2 | 3 |
| Lontra canadensis (river otter) | 1 | 2 | 1 | 2 |
| Ursus maritimus (polar bear) | 2 | 1 | 1 | 2 |
| Ursus arctos (brown bear) | 2 | 1 | 1 | 1 |
| Odobenus rosmarus (walrus) | 2 | 2 | 2 | 1 |
| Sylvilagus floridanus (Eastern cottontail rabbit) | 2 | 1 | 2 | 1 |
| Lasionycteris noctivagans (gray-eared bat) | 1 | 2 | 2 | 1 |
As evidenced by the fossil record, the pace of morphological evolution is often rapid, and new forms are generally described by topologically conservative geometric deformations of their ancestors. The changes that have occurred in many domestic dog breeds in recent history demonstrate just how swift mammalian evolution can be (Fig. 3). The data presented here show how our hypothesis may reconcile low single-nucleotide mutation rates, small population sizes, and the sensitivity of proteins to random point mutations with the pace and continuity of morphological change in vertebrates. We offer an alternative to the cis-regulatory element dogma: frequent length mutations in gene-associated tandem repeats generate copious robust morphological variation. Incremental changes in coding repeat lengths have been shown to cause similarly incremental changes in gene function and phenotype. The extraordinarily high length mutation rates of tandemly repeated sequences can thus result in abundant variation upon which selection may act. The evolution of a polyalanine stretch in a homeobox gene was recently shown to confer new activities instrumental in deep phyletic divergence in arthropods (9), and repeats frequently harbor most of the differences among developmental orthologs of closely related species. We find no evidence of functional interbreed single-nucleotide variation (only a few silent SNPs) in genes known to influence morphological traits, but find extraordinary levels of tandem repeat variation in the coding regions of these same genes, with almost every locus tested showing some coding length polymorphism, much of which appears to be of modern origin. The conservation of orthologous repeats across mammalian orders despite high mutation rates is indicative of strong stabilizing selection; thus, variation at these loci is not neutral. Recent studies have revealed roles for modest polyalanine expansions in the etiology of moderate-to-severe clinical phenotypes of nine human genetic diseases, and we and others have shown that many more such coding repeat loci exhibit considerable length polymorphism in humans (7, 36). We suggest that it is likely that hypermutation at these and other gene-associated repeat loci contributes some of the morphological variation necessary to yield the rates of morphological evolution depicted above. How broadly this mode of evolutionary change is exploited in nature remains to be seen, but if the prevalence of repetitive elements within genes is any indicator, then mammals, insects, plants, and other genomes throughout the natural world may use this mechanism to achieve evolutionary agility.
Supplementary Material
Acknowledgments
We thank M. Nussbaumer and the Albert Heim Foundation for Canine Research at the Berne Museum of Natural History for the generous assistance with canid morphometrics and access to collections and M. M. Horvath, E. Xu, and J. Laidlaw for technical assistance in DNA sequencing. H.R.G. is supported by the P. O'Bryan Montgomery Distinguished Chair in Human Growth and Development, the Evelyn Hudson Foundation, and National Institutes of Health/National Cancer Institute Grant CA096901.
Author contributions: J.W.F. designed research; J.W.F. performed research; H.R.G. contributed new reagents/analytic tools; J.W.F. analyzed data; and J.W.F. wrote the paper.
Abbreviation: Runx-2, runt-related transcription factor 2.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY308807–AY308823).
References
- 1.Darwin, C. (1859) On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life (Murray, London), 4th Ed., pp. 359–360. [PMC free article] [PubMed]
- 2.Mayr, E. (1954) in Evolution as a Process, ed. Huxley, J. (Allen and Unwin, London), pp. 157–180.
- 3.Eldredge, N. & Gould, S. J. (1972) in Models in Paleobiology, ed. Schopf, T. J. M. (Freeman & Cooper, San Francisco), pp. 305–332.
- 4.Thompson, D'A. W. (1917) On Growth and Form (Dover, New York).
- 5.Carroll, S. B. (2000) Cell 101, 577–580. [DOI] [PubMed] [Google Scholar]
- 6.Kirschner, M. & Gerhart, J. (1998) Proc. Natl. Acad. Sci. USA 95, 8420–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wren, J. D., Forgacs, E., Fondon, J. W., III, Pertsemlidis, A., Cheng, S. Y., Gallardo, T., Williams, R. S., Shohet, R. V., Minna, J. D. & Garner, H. R. (2000) Am. J. Hum. Genet. 67, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlin, S. & Burge, C. (1996) Proc. Natl. Acad. Sci. USA 93, 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galant, R. & Carroll, S. B. (2002) Nature 415, 910–913. [DOI] [PubMed] [Google Scholar]
- 10.Ellergen, H. (2000) Trends Genet. 16, 551–558. [DOI] [PubMed] [Google Scholar]
- 11.Gerber, H. P., Seipel, K., Georgiev, O., Hofferer, M., Hug, M., Rusconi, S. & Schaffner, W. (1994) Science 263, 808–811. [DOI] [PubMed] [Google Scholar]
- 12.Shimajiri, S., Arima, N., Tanimoto, A., Murata, Y., Hamada, T., Wang, K. Y. & Sasaguri, Y. (1999) FEBS Lett. 455, 70–74. [DOI] [PubMed] [Google Scholar]
- 13.Ladurner, A. G. & Fersht, A. R. (1997) J. Mol. Biol. 273, 330–337. [DOI] [PubMed] [Google Scholar]
- 14.Chen, S., Berthelier, V., Hamilton, J. B., O'Nuallain, B. & Wetzel, R. (2002) Biochemistry 41, 7391–7399. [DOI] [PubMed] [Google Scholar]
- 15.Goodman, F. R., Mundlos, S., Muragaki, Y., Donnai D, Giovannucci-Uzielli, M. L., Lapi, E., Majewski, F., McGaughran, J., McKeown, C., Reardon, W., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 7458–7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, K. R., Sweet, H. O., Donahue, L. R., Ward-Bailey, P., Bronson, R. T. & Davisson, M. T. (1998) Hum. Mol. Genet. 7, 1033–1038. [DOI] [PubMed] [Google Scholar]
- 17.Fondon, J. W., III, Mele, G. M., Brezinschek, R. I., Cummings, D., Pande, A., Wren, J., O'Brien, K. M., Kupfer, K. C., Wei, M. H., Lerman, M., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 7514–7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu, S., Tucker, S. C., Ehrlich, J. S., Levorse, J. M., Flaherty, L. A., Wisdom, R. & Vogt, T. F. (1988) Development (Cambridge, U.K.) 125, 2711–2721. [DOI] [PubMed] [Google Scholar]
- 19.Boras, K. & Hammel, P. A. (2002) J. Biol. Chem. 277, 1120–1127. [DOI] [PubMed] [Google Scholar]
- 20.Qu, S., Tucker, S. C., Zhao, Q., deCrombrugghe, B. & Wisdom, R. (1999) Development (Cambridge, U.K.) 126, 359–369. [DOI] [PubMed] [Google Scholar]
- 21.Goldschmidt, R. (1940) The Material Basis of Evolution (Yale Univ. Press, New Haven, CT).
- 22.Bates, G. & Lehrach, H. (1994) BioEssays 16, 277–284. [DOI] [PubMed] [Google Scholar]
- 23.Lee, B., Thirunavukkarasu, K., Zhou, L., Pastore, L., Baldini, A., Hecht, J., Geoffroy, V., Ducy, P. & Karsenty, G. (1997) Nat. Genet. 16, 307–310. [DOI] [PubMed] [Google Scholar]
- 24.Nussbaumer, M. (1982) Zool. Anz. 209, 1–32. [Google Scholar]
- 25.Mundlos, S., Otto, F., Mundlos, C., Mulliken, J. B., Aylsworth, A. S., Albright, S., Lindhout, D., Cole, W. G., Henn, W., Knoll, J. H., et al. (1997) Cell 89, 773–779. [DOI] [PubMed] [Google Scholar]
- 26.Goto, S. G. & Kimura, M. T. (2001) Mol. Phylogenet. Evol. 18, 404–422. [DOI] [PubMed] [Google Scholar]
- 27.Vila, C., Maldonado, J. E. & Wayne, R. K. (1999) J. Hered. 90, 71–77. [DOI] [PubMed] [Google Scholar]
- 28.Schleger, A. (1983) Graduate dissertation (Univ. of Vienna, Vienna).
- 29.Haworth, K., Breen, M., Binns, M., Hopkinson, D. A. & Edwards, Y. H. (2001) Anim. Genet. 32, 32–36. [DOI] [PubMed] [Google Scholar]
- 30.Aguirre, G. D., Baldwin V., Pearce-Kelling S., Narfstrom K., Ray K. & Acland, G. M. (1998) Mol. Vis. 4, 23. [PubMed] [Google Scholar]
- 31.Kirkness, E. F., Bafna, V., Halpern, A. L., Levy, S., Remington, K., Rusch, D. B., Delcher, A.L., Pop, M., Wang, W., Fraser, C.M., et al. (2003) Science 301, 1898–1903. [DOI] [PubMed] [Google Scholar]
- 32.Parker, H. G., Kim, L. V., Sutter, N. B., Carlson, S., Lorentzen, T. D., Malek, T. B., Johnson, G. S., DeFrance, H. B., Ostransder, E. A. & Kruglyak, L. (2004) Science 305, 1160–1164. [DOI] [PubMed] [Google Scholar]
- 33.Savolainen, P., Zhang, Y. P., Luo, J., Lundeberg, J. & Leitner, T. (2002) Science 298, 1610–1613. [DOI] [PubMed] [Google Scholar]
- 34.Vila, C., Savolainen, P., Maldonado, J. E., Amorim, I. R., Rice, J. E., Honeycutt, R. L., Crandall, K. A., Lundeberg, J. & Wayne, R. K. (1997) Science 276, 1687–1689. [DOI] [PubMed] [Google Scholar]
- 35.Wayne, R. K. & Ostrander, E. A. (1999) BioEssays 21, 247–257. [DOI] [PubMed] [Google Scholar]
- 36.Lavoie, H., Debeane, F., Trinh, Q. D., Turcotte, J. F., Corbeil-Girard, L. P., Dicaire, M. J., Saint-Denis, A., Page, M., Rouleau, G. A. & Brais, B. (2003) Hum. Mol. Genet. 12, 2967–2979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.