Abstract
Femtosecond to nanosecond dynamics of O2 rebinding to human WT myoglobin and its mutants, V68F and I107F, have been studied by using transient absorption. The results are compared with NO rebinding. Even though the immediate environment around the heme binding site is changed by the mutations, the picosecond geminate rebinding of oxygen is at most minimally affected. On the other hand, the V68F (E11) mutation causes drastic differences in rebinding on the nanosecond time scale, whereas the effect of the I107F (G8) mutation remains relatively small within our 10-ns time window. Unlike traditional homogeneous kinetics and molecular dynamics collisional simulations, we propose a “bifurcation model” for populations of directed and undirected dynamics on the ultrafast time scale, reflecting the distribution of initial protein conformations. The major mutation effect occurs on the time scale on which global protein conformational change is possible, consistent with transitions between the conformations of directed and undirected population playing a role in the O2 binding. We discuss the relevance of these findings to the bimolecular function of the protein.
Keywords: femtobiology, molecular recognition
Myoglobin (Mb) is considered as the hydrogen atom of biology (1), and numerous studies have been carried out to elucidate its function, the storage of oxygen in vertebrate muscle cells. In addition to storing oxygen, it can interact with other important small molecules, for example NO and carbon monoxide (2, 3), making it a prototype system for the study of molecular recognition (4). One persistent question in the study of Mb concerns the ligand recognition and its correlation to protein dynamics. X-ray study has revealed that Mb is highly compact and has to undergo conformational fluctuation to form effective “channels” for the release of the ligand (ref. 1 and references therein).
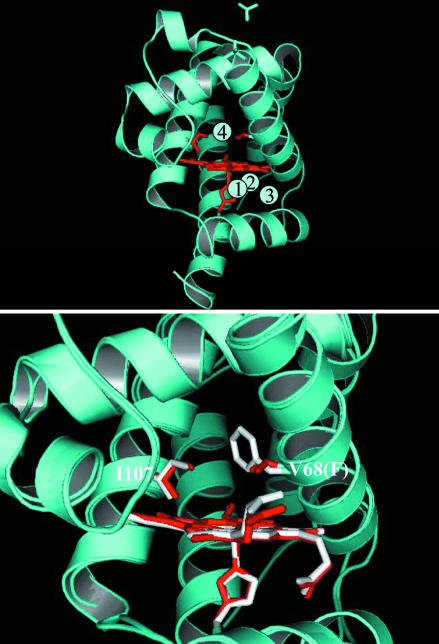
Tilton et al. (5) have shown that, in addition to the distal cavity next to the heme group, there are four folding defects inside the protein denoted as Xe1–Xe4 binding sites (Fig. 1 Upper), numbered in the decreasing order of affinity of Xe to these locations. These highly nonpolar cavities are believed to be involved in ligand migration (5). Cryogenic x-ray studies of photolysed WT and mutant carbonmonoxymyoglobin (MbCO) have indicated that CO ligand resides in the distal cavity (6, 7) as well as in the Xe4 (8, 9) and Xe1 (8, 10) sites. More recently, time-resolved x-ray studies on samples near room temperature revealed CO residence times of ligands in these sites, from 30 ns to 10 μs for WT and recombinant mutants (11–13).
Fig. 1.
X-ray protein structures. (Upper) Ribbon model of sperm whale WT Mb with Xe binding sites highlighted according to ref. 5. For clarity, two heme side chains are not drawn here, but can be seen in Lower. The two residues (107 and 68) indicated above the heme are the same in human and sperm whale Mb, but only the sperm whale Mb V68F mutant x-ray structure is available for the comparison in Lower.(Lower) Expanded view of the distal pocket of the superimposed structures of WT and V68F. The highlighted portions of the WT (Protein Data Bank ID code 1A6N) and V68F mutant (Protein Data Bank ID code 1MLK), including the side chains of residues 107 and 68, are shown in red and white, respectively. The remainders of both structures are shown in blue. I107F mutant is not shown because of a lack of crystal structure information.
Since the pioneering work of Frauenfelder and coworkers in 1975 (14) on ligand geminate rebinding at low temperature it is known that ligand–Mb interaction involves energy landscapes of multiple conformational substates and intermediates. Two perspectives usually are discussed in the literature to account for the rebinding of O2 and other small molecules. One of these, the multiple-site model, relates the rebinding rate to the physical location of the ligand inside the protein (15). The central theme is that discrete intermediates exist and can be mapped out on a static energy landscape. In contrast, rebinding may be controlled by time-dependent barrier changes involving transitions among conformational substates, as manifested in the nonexponential behavior of rates (16).
Protein engineering (mutagenesis), ultrafast spectroscopy, and computational simulations all have been invoked to study dynamics in Mb. Residues located near the heme iron center and bordering the Xe4 site are demonstrated to affect the ligand nanosecond geminate rebinding as well as the overall bimolecular rebinding kinetics (8, 13, 15, 17–37). It is generally believed that mutations at these positions alter the rebinding kinetics by enhancing or restricting the ligand movement according to the multiple-site model (15). Theoretical simulations have been used to investigate the effect of mutation on ligand trajectories upon photolysis (24). Those show the ligands being confined in a place that is within 4Åofthe iron center in a mutant where Val-68 has been mutated to phenylalanine (V68F mutant). This space restriction has been proposed to be the reason for the observed high geminate rebinding for the V68F mutant.
Champion and coworkers (38) have carefully studied, with femtosecond time resolution, the transient absorption dynamics of O2-bound native horse heart Mb and reported a geminate recombination lifetime of ≈5 ps, similar to the 6.4 ± 2-ps geminate recombination lifetime reported from an earlier picosecond study (39). For the mutants, no femtosecond O2 binding studies have been reported. Experiments on the sperm whale V68F/O2 system have been conducted with 35-ps laser pulses (30), but these could not resolve the ultrafast recombination process. Because the picosecond geminate rebinding is the ultimate step to form the protein–ligand bond, information in this time regime is critical for the understanding of the elementary steps of molecular recognition.
From this group, an earlier effort focused on the dynamics, kinetics, and thermodynamics of molecular recognition of O2 by a Mb and hemoglobin mimic, cobalt picket-fence porphyrin (40, 41). In this article, we report a study of oxygen recognition in real proteins, the recombinant human Mb and its mutants, from the femtosecond to nanosecond regime. We have chosen the mutants V68F and I107F (in which isoleucine at residue 107 is replaced by phenylalanine), which alter two of the residues that border the Xe4 site (Fig. 1 Lower). With femtosecond resolution, we were able to probe the mutant behavior with an extended window up to 10 ns. From the results, we propose a bifurcation dynamics model for which direct O2 rebinding occurs for some conformational populations on the femtosecond to picosecond time scales before protein conformational fluctuation can occur. This directed ultrafast recombination contrasts with the undirected diffusive ligand migration on the nanosecond to microsecond time scale.
Materials and Methods
Biochemistry: Protein Expression and Mutants. The synthetic gene for human WT Mb, which carries G80A and C110A mutations, was a generous gift from Steven Boxer (Stanford University, Stanford, CA). The gene was subcloned into pET-19b plasmid (Novagen), and V68F and I107F mutations were carried out with Quikchange (Stratagene). All protein sequences were verified by DNA sequencing. Proteins were expressed in BL21 (DE3) cells and purified by Ni-NTA (Qiagen, Valencia, CA) affinity column, and protein identities were confirmed by MS and N-terminal partial sequencing. All of the proteins have a His tag at the N terminus. MS spectra show that the His tag in V68F mutant remains mostly intact, but in WT and I107F 13 aa of the tag are cleaved. CD spectra of WT, V68F, and I107F were similar to each other. The ferric protein was then reduced with sodium dithionite and passed through a Sephadex G-25 column (Nap10, Amersham Pharmacia Biosciences) under oxygen-free condition. The quality of deoxymyoglobin (deoxyMb) was checked by UV-visible. Oxymyoglobin (MbO2) or nitrosylmyoglobin (MbNO) were obtained by bubbling pure oxygen or NO through the pure deoxyMb sample. All transient absorption measurements were carried out on samples in 0.1 M phosphate buffer, pH 7.0 at 21°C, at concentrations of ≈25 μM to give an OD ≈1.2 for the Soret band in a 5-mm optical cell. The cell was continuously stirred, and sample degradation was monitored during experiments by UV-visible of the Soret band as well as the Q band for traces of the ferric state generated by laser irradiation. New MbO2 solution was used when ≈5% deviation from the initial UV-visible spectrum was observed. Much larger spectral variations in the UV (250–400 nm) were seen among MbNO samples without noticeable change in the characteristic heme absorption bands or in transient absorption.
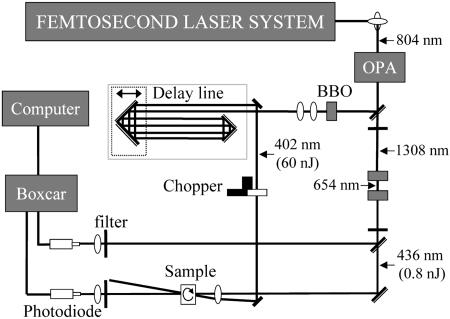
Femtosecond Spectroscopy. For these experiments, a Hurricane laser system (Spectra-Physics) produced ≈100-fs pulses at 804 nm (fundamental) with 900-Hz repetition rate and 1 mJ energy (Fig. 2). These pulses pumped an optical parametric amplifier (OPA), and probe pulses at 436 nm were generated by frequency tripling the OPA signal at 1,308 nm. The pump pulses at 402 nm were obtained by doubling the residual of the fundamental beam after the OPA in a β-barium borate crystal. The probe pulses were divided into two beams, one passing through the sample and the other serving as a reference. The time delay between pump and probe was controlled by a translation stage. By setting up a triple-pass delay line, we could scan up to 10 ns of delay. For the long time range data, transient shapes were very sensitive to imperfect laser collimation and alignment through the variable path of the delay line, so measurements were made of all of the different samples in this study under controlled conditions to allow unambiguous comparison of the transients recorded. For example, six samples, with O2 and NO bound to each of the three proteins, were run consecutively under the same conditions. Transients of samples with known long lifetimes (specifically, WT-O2 and I107F-O2 as seen in Fig. 3 Lower) also were used to calibrate for maximum influence of the delay line over the 9-ns delay range and the behavior is robust; that is, the conclusions reported here are totally independent of the effect of calibration. The data are shown in the figures only as recorded.
Fig. 2.
Schematic layout of the femtosecond transient absorption apparatus with pulse wavelengths indicated. OPA, optical parametric amplifier; BBO, β-barium borate.
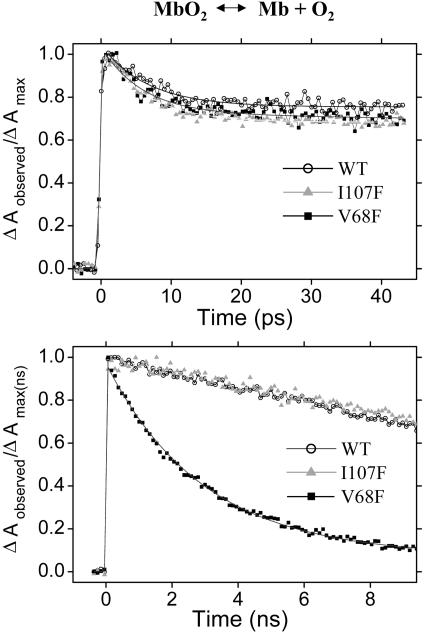
Fig. 3.
Femtosecond time-resolved geminate rebinding of O2 with Mb. The transient absorption difference is recorded at 436 nm after photolysis at 402 nm. Short (Upper, fs to ps) and long (Lower, ns) time scans are presented. Note that the nanosecond transients have been normalized to intensities measured at the plateau of the picosecond transients (t ≥ 60 ps). Fit curves are plotted with each of the short time transients and with the V68F data on the long time range.
Typical pump pulse energy was ≈60 nJ to minimize sample degradation, and probe energy was ≈0.8 nJ. These two beams were focused by a single 30-cm focal length lens and aligned to intersect within the sample cell, crossing at an angle of ≈10°. After the sample, the pump beam was blocked by two apertures and further reduced with a GG435 sharp-cutoff filter. The probe beam was focused at a photodiode for measurement. The output from the photodiode was gated and integrated by a boxcar integrator and normalized to the similarly processed reference. The log of the ratio between thus processed transmitted probe intensities with and without pump light generated the transient absorption, ΔA. The typical transient absorption maximum was ≈0.4%.
Experimental Results
Protein (Mb)–O2. The femtosecond-resolved transient absorption of WT, V68F, and I107F mutants of human MbO2 is shown in Fig. 3 for excitation at 402 nm and probing at 436 nm. The data sets compared for each time range were collected in the same experimental run, ensuring uniformity of conditions. On the picosecond time scale, we observed a decay component with a lifetime of 6 ± 1 ps and an amplitude of 30 ± 6% of the peak signal for all three Mb species (Fig. 3 Upper), similar to the picosecond behavior in horse heart Mb reported for 400-nm excitation and 438-nm probing (6.0 ps, 33%) (38). The differences in lifetimes and amplitudes among the transients shown in Fig. 3 are within the error range given and are typical of the experimental variability for any single mutant.
In contrast, the dynamics of the V68F mutant in the nanosecond regime is strikingly faster than that for WT and I107F mutants and the decay amplitude is much larger (Fig. 3 Lower). The decay rates for WT and I107F are too slow to be measured accurately in our 10-ns window, but literature values (33) and our measurements allow us to estimate the degree of distortion of the transients caused by delay line effects and calibrate the V68F data accordingly. With or without such calibration, all V68F mutant transients can be fit reasonably well to a single nanosecond lifetime (near 3 ns) with a baseline offset near 10% of the maximum signal (such a fit is plotted with the data in Fig. 3), although two lifetimes on the nanosecond time scale and a smaller offset clearly give a superior fit in all cases. However, given that even the 10-ns time range is too short to measure directly the long time plateau signal level, various combinations of parameters, with lifetimes ranging from 0.5 to 6 ns, produce equally satisfactory biexponential fits. Thus we can only say that the decay in our time window is apparently slightly nonexponential and note that such behavior is not unexpected in a dynamic protein environment. The decay on the nanosecond time scale measured here for human V68F/O2 has a corresponding component in the dynamics of sperm whale V68F/O2 previously reported as a single exponential of 7-ns lifetime after photolysis, but using pulses of 17 or 9 ns (24, 30).
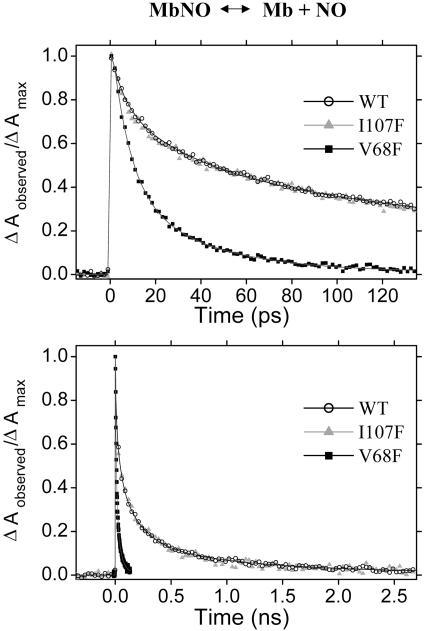
Protein (Mb)–NO. Experiments like those above on MbO2 also have been performed on MbNO. Transient absorption of WT and I107F MbNO is virtually indistinguishable on the picosecond and nanosecond time scales (Fig. 4). Four lifetimes were used in the global fit shown in Fig. 4 to the full time range of the WT measurements, with a 1% offset asymptote reached after several nanoseconds. Unlike for MbO2, the transient absorption of the V68F mutant of MbNO is significantly different from that of WT and I107F on the picosecond time scale and fits to two picosecond lifetimes (8 and 35 ps) with an offset of 1–2%. Further time dependence of the V68F signal is not measured because of the small amplitude remaining after 150 ps.
Fig. 4.
Femtosecond time-resolved geminate rebinding of NO with Mb. The transient absorption difference is recorded at 436 nm after photolysis at 402 nm. Short (Upper, fs to ps) and long (Lower, ns) time scans are presented for WT and I107F. A five-component global fit to the WT measurements and a three-component fit to the V68F transient are plotted with the data.
Discussion
Protein Dynamics and Recognition Landscape. In our measurements, the response to femtosecond photolysis of MbO2 is monitored at the peak of the deoxyMb Soret band and ΔA reflects, in general, the amount of unligated Mb. In the detailed transient absorption study of Champion and coworkers (38), it was found that vibrational cooling also contributed signals at a very early time (t < 2 ps) when probing at the wavelength of peak static absorption for horse heart deoxyMb, MbCO, MbO2, and MbNO. For MbO2, a 4–to 6-ps decay component was observed at all probe wavelengths and assigned as primarily caused by geminate recombination of oxygen to the ground state of the unbound protein for several reasons: (i) the 4–to 6-ps decay component did not appear in the transient absorption of deoxyMb; (ii) it did not appear in the transient absorption of MbCO, which has no picosecond geminate rebinding; (iii) it was observed with opposing signs when probing at 418 nm, the Soret band of MbO2, and 438 nm, the deoxyMb Soret band; and (iv) it was absent when probing at the isosbestic points of the Mb/O2 spectrum. The amplitude of this component relative to the residual ΔA changed with probe wavelength, which would not occur if it was totally caused by geminate recombination at all wavelengths, but its fraction was smallest for probing at the deoxyMb Soret band, as in our experiments. Accordingly, ΔA is considered to reflect the amount of unligated Mb over all time ranges, and as such, the experimental geminate rebinding rates and the fractions of ligand escaping on the picosecond and nanosecond time scales, from this work and the literature, are given in Table 1.
Table 1. Observed rates and fractions for O2 geminate rebinding to human WT, V68F, and I107F Mbs from transient absorption at 436 nm.
| This work
|
Literature*
|
|||||
|---|---|---|---|---|---|---|
| Mb | τ1, ps | Γesc(ps), % | τ2, ns | Γesc(ns), % | τ2, ns | Γesc(ns), % |
| WT | 6 ± 1 | 70 ± 6 | 55† | 55 | ||
| V68F | 6 ± 1 | 70 ± 6 | 2.7 ± 0.6‡ | 9 ± 5 | ||
| I107F | 6 ± 1 | 70 ± 6 | 45 | 27 | ||
Γesc(ps) and Γesc(ns) are the escape fractions in the picosecond and nanosecond regime. These are the ratios between the offset and the initial value of the transient absorption in the appropriate time range.
Data are from ref. 33.
The WT transient could be fit best with two nanosecond components with lifetimes of 27 and 135 ns.
The fit of all V68F nanosecond transients improved with the use of two exponential components and an offset, but fit parameters were not well determined in such fitting. See text.
The most significant observations made in this work can be summarized as follows: (i) As shown schematically in Fig. 5 Top, with the newly femtosecond-resolved transients of MbO2 reported here, the response to photolysis of the WT and mutants can be fully characterized, revealing that behaviors differ distinctly among mutants on long time scales, whereas on the ultrafast time scale they are essentially the same. The overall behavior is reflective of the dynamics of O2 binding and unbinding. (ii) Although the mutation at position 68 (Fig. 1 Bottom) introduces a structural blockage, through the phenyl group, in the immediate vicinity of the iron center, the picosecond behavior is not greatly altered. This response could not be observed in previous studies of the V68F mutant because of the 35-ps pulses used (30). (iii) The ultrafast response for the I107F mutant is likewise nearly the same as for the WT. (iv) On the nanosecond time scale, V68F mutation enhances the rates by more than an order of magnitude, qualitatively consistent with previous work on sperm whale Mb using 9- to 17-ns pulses (24, 30). For I107F, the nanosecond response is on a much longer time scale. (v) Finally, we repeated the same experiments with NO instead of O2 and obtained results in qualitative agreement with other work at different pump and probe wavelengths for WT and V68F sperm whale Mb (31), WT horse heart Mb (39), and WT human Mb (42). Our measured transient for WT over early time (Fig. 4 Upper) is very similar to that recorded for WT horse heart MbNO over the same time range and with the similar pump and probe wavelengths of 400 and 438 nm, respectively (38), as evidenced by a comparison of parameters obtained when fitting this restricted time range as a simple biexponential: 12.2 ps, 37% and 187 ps, 63% for our data vs. 13.0 ps, 35% and 190 ps, 65% in ref. 38.
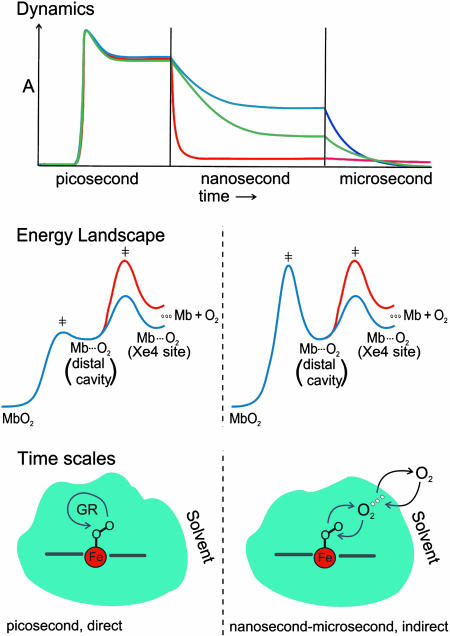
Fig. 5.
Dynamics and landscapes for Mb recognition of oxygen. (Top) Illustration of the time evolution of 436-nm transient absorbance of WT (blue), I107F (green), and V68F (red) MbO2 after femtosecond photolysis. The positive signal change, which is proportional to the population of unligated Mb, decays on multiple time scales as the protein rebinds O2. (Middle) Schematic free energy landscapes for O2 migration away from the heme binding site in WT (blue) and V68F (red) Mb. (Left) Picosecond, directed dynamics (unaffected by mutation). (Right) Nanosecond, undirected dynamics (affected by mutation). (Bottom) Schematic representation of a bifurcation of Mb conformations associated with directed (Left) and undirected (Right) ultrafast dynamics. GR, geminate recombination.
With these observations and comparisons, the following picture of the protein dynamics and energy landscape emerges. After the femtosecond initiation of O2 release, the ligand first dwells at a distal site close to the iron center, in a cavity with direct access to Xe4 (see Fig. 1). Then it could either geminately rebind or escape to sites of minimum energy, e.g., Xe4, identified by x-ray diffraction as one collection site for CO after photolysis of MbCO (11, 12). The observed geminate recombination is ultrafast (≈6 ps), and if it is driven by the kinetics of a homogeneous protein ensemble, then the V68F mutant (which changes the environment of the Xe4 site) should result in a significant change of the rate and the fraction rebound, contrary to our observation. For example, the blue energy landscape in Fig. 5 Middle Right is illustrative of one in which the forward migration to Xe4 would be faster than rebinding, consistent with the observed 70/30 escape vs. rebinding branching ratio. Because the V68F mutation reduces the Xe4 cavity to half of the space in WT (red potential curve in Fig. 5), the escape process is hindered, which should enhance the rebinding process. The molecular dynamics simulations using the locally enhanced sampling method predicted such a mutant effect (24). In those simulations, for V68F mutant, the phenylalanine at residue 68 occupies the same spot in the Xe4 cavity in which ligands gather in the WT. Consequently, the ligands are forced to reside in the primary distal site, causing more collisions between the ligands and the iron center.
The unchanging observed rate and fraction of the picosecond rebinding in our experiments would require, for a homogeneous system, a constant value for both the forward rate (to Xe4) and the rebinding rate, contradicting the above predictions of the V68F mutant effect. To account for the discrepancy, we propose that the ultrafast primary step follows the dynamics of the bifurcation model, which was found to be significant in ultrafast chemical and biological reactions (ref. 43 and references therein), such as DNA gating of electron transfer (44, 45). In this case, because of protein conformations with low free energy barriers to recombination, the directed population will rebind within the first few ps (Fig. 5 Middle Left and Bottom Left), whereas the undirected population will be in conformations capable of recombination after thermal and intramolecular motions on a longer time scale (Fig. 5 Middle Right and Bottom Right). Because the escape fraction is determined by the conformational bifurcation rather than the competition between forward and backward rates, as long as recombination is highly favored in the directed population of the WT, the obstruction of the Xe4 cavity in V68F will have little effect on the ultrafast time scale (Fig. 5 Middle Left).
The reasoning given above based on competition between forward and backward rates on a static energy landscape also should apply when photolysis-initiated local conformational dynamics (e.g., motion of the Fe atom relative to the heme plane) are important in the picosecond process (46, 47). That is, if mutation causes the pronounced change in the ligand/Fe collision frequency that is predicted by molecular dynamics simulations for V68F (24), the probability of reforming the ligand-Fe bond must change accordingly and at least the picosecond escape fraction would clearly reflect this change in a homogeneous sample.
This bifurcation picture raises an important point about the nature of the energy landscape and trajectory of motions. The instantaneous dissociation of O2 generates a nonequilibrium protein ensemble that then evolves toward equilibrium. On the femtosecond to picosecond time scale, the protein is essentially a frozen matrix and only small and local conformational changes (lower level of conformational tiers) may be involved. The directed geminate recombination takes place on one family of conformations that is adapted to ensure efficient reformation of the metal–ligand bond on a static or minimally fluctuating landscape, and we observe no change of rates or fractions with mutation.
Larger and global motions in the protein occur at longer time. Those motions involve transitions of higher tiers of conformational structures. On such time scales, the trajectories visit a large phase space and the landscape fluctuations play a role. Trajectories are the result of many collisions in the cavity and barriers and wells can fluctuate. For example, the landscape may evolve from one of undirected dynamics, as in Fig. 5 Right, to that in Fig. 5 Left, which will direct the ligand to immediate bond formation. In such a situation, a mutant effect on the protein global fluctuation will alter the rebinding dynamics. Indeed, the observed delayed mutant effect on recombination could be an indication of the time scale of protein global dynamics and thermal fluctuations and how they are affected by the mutations. For MbCO, time-resolved x-ray studies (11, 12) and spectroscopic studies (46, 48, 49) have indicated a wide range of time scales of photolysis-driven global conformational changes, from hundreds of picoseconds to microseconds. On the other hand, modification of the local energy landscape by steric hindrance undoubtedly also will have its effect on nanosecond and longer time dynamics. It is noteworthy that, even though residue 107 is located close to residue 68, mutant I107F does not cause as significant a deviation from the WT as V68F. This may be because these two spatially close residues are involved in the global dynamics differently, perhaps because they are on different helices.
When O2 rebinds from the solvent bimolecularly, it can be expected to be subject to the same processes as when photolytically released. O2 must reenter the distal cavity, possibly after passage through other intermediate states. On the time scale of this migration, protein conformational dynamics will clearly be important, as the energy landscape and barrier heights fluctuate. The V68F mutation slows down the initial process by a factor of 13 relative to WT (50), whereas the I107F mutation again has only a moderate effect (≈35% rate reduction) (33). The magnitudes of these changes are consistent with the influence of these specific mutations on protein dynamics inferred from the picture of dynamic control on the nanosecond rates.
In the Mb mimic, cobalt picket-fence porphyrin with methylimidazole base (B-CoP), analysis of the global rebinding dynamics showed that an intermediate B-CoP···O2 state was formed that had only a ≈2% probability of going on to form the ligand–metal bond (40). In addition, the ultrafast dynamics of this system showed no geminate recombination of O2 (41). Replacement of the cobalt center with iron results in a much higher affinity for O2 (51, 52) and 43% geminate recombination with a 25-ps lifetime (53). For the three Mb mutants studied here, with 30% recombination on the picosecond time scale, recombination from intermediate states on the nanosecond time scale ranges from 45% to 91% [1 -Γesc(ns) from Table 1]. These results all are consistent with the concept that the existence of conformations leading to directed ultrafast dynamics enhance the efficiency of the binding function, even in a relatively simple system like the picket fence.
Recombination to WT Mb on the picosecond time scale is much more efficient for NO than for O2. Because the intrinsic chemical reactivity, orientation when the ligand is bound to heme, and electrostatic and hydrogen-bonding interaction with the protein moiety all are so different for these two ligands, a difference in the binding process is not surprising. In contrast to O2, the V68F mutant effect for NO is dramatic within the first 10 ps of photolysis (Fig. 4). This behavior is consistent with the expectations from homogeneous kinetics and local structural changes, as modeled in the above-mentioned molecular dynamics simulations (24), and the role of conformations such as those associated with directed dynamics for O2 may be very different for NO because of the different binding. More recent theoretical work on NO rebinding using the reactive molecular dynamics method shows that on this short time scale fluctuations in the immediate vicinity of the binding site control the NO nonexponential rebinding (47).
Conclusion
With femtosecond time resolution, the picosecond to nanosecond geminate rebinding dynamics of oxygen to Mb WT, V68F, and I107F mutants has been studied. The lack of a pronounced mutation effect on the initial rebinding for either mutant shows that this process is kinetically isolated from the distal pocket environment through which the ligand moves on the nanosecond time scale. The bifurcation that allows this isolation is understood in terms of a distribution of conformational structures, with one set of conformations leading to directed O2-Fe bond formation without sampling a large phase space. These structures may be functionally important, with protein global conformational fluctuations into such structures guiding the O2 to efficient binding once it has reached the distal cavity from the solvent. The fact that V68F mutation causes a drastic effect on rebinding dynamics on the time scale of protein global fluctuation is consistent with this concept of directed populations. The existence of conformations optimized for directed bonding in MbO2 should be important for an efficient biological function (43, 54, 55).
Acknowledgments
We acknowledge the contribution of Dr. Qing-Bin Lu (California Institute of Technology) in developing the experimental apparatus used for the measurements reported here. This work was supported by the National Science Foundation.
Abbreviations: Mb, myoglobin; MbCO, carbonmonoxymyoglobin; MbO2, oxymyoglobin; MbNO, nitrosylmyoglobin; deoxyMb, deoxymyoglobin.
References
- 1.Frauenfelder, H., McMahon, B. H. & Fenimore, P. W. (2003) Proc. Natl. Acad. Sci. USA 100, 8615–8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olson, J. S. & Phillips, G. N. (1996) J. Biol. Chem. 271, 17593–17596. [DOI] [PubMed] [Google Scholar]
- 3.Gibson, Q. H. (1989) J. Biol. Chem. 264, 20155–20158. [PubMed] [Google Scholar]
- 4.Springer, B. A., Sligar, S. G., Olson, J. S. & Phillips, G. N. (1994) Chem. Rev. 94, 699–714. [Google Scholar]
- 5.Tilton, R. F., Kuntz, I. D. & Petsko, G. A. (1984) Biochemistry 23, 2849–2857. [DOI] [PubMed] [Google Scholar]
- 6.Schlichting, I., Berendzen, J., Phillips, G. N. & Sweet, R. M. (1994) Nature 371, 808–812. [DOI] [PubMed] [Google Scholar]
- 7.Teng, T. Y., Schildkamp, W., Dolmer, P. & Moffat, K. (1994) J. Appl. Crystallogr. 27, 133–139. [Google Scholar]
- 8.Ostermann, A., Waschipky, R., Parak, F. G. & Nienhaus, G. U. (2000) Nature 404, 205–208. [DOI] [PubMed] [Google Scholar]
- 9.Brunori, M., Vallone, B., Cutruzzola, F., Travaglini-Allocatelli, C., Berendzen, J., Chu, K., Sweet, R. M. & Schlichting, I. (2000) Proc. Natl. Acad. Sci. USA 97, 2058–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu, K., Vojtchovsky, J., McMahon, B. H., Sweet, R. M., Berendzen, J. & Schlichting, I. (2000) Nature 403, 921–923. [DOI] [PubMed] [Google Scholar]
- 11.Srajer, V., Ren, Z., Teng, T. Y., Schmidt, M., Ursby, T., Bourgeois, D., Pradervand, C., Schildkamp, W., Wulff, M. & Moffat, K. (2001) Biochemistry 40, 13802–13815. [DOI] [PubMed] [Google Scholar]
- 12.Schotte, F., Lim, M. H., Jackson, T. A., Smirnov, A. V., Soman, J., Olson, J. S., Phillips, G. N., Wulff, M. & Anfinrud, P. A. (2003) Science 300, 1944–1947. [DOI] [PubMed] [Google Scholar]
- 13.Bourgeois, D., Vallone, B., Schotte, F., Arcovito, A., Miele, A. E., Sciara, G., Wulff, M., Anfinrud, P. & Brunori, M. (2003) Proc. Natl. Acad. Sci. USA 100, 8704–8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Austin, R. H., Beeson, K. W., Eisenstein, L., Frauenfelder, H. & Gunsalus, I. C. (1975) Biochemistry 14, 5355–5373. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, Q. H., Regan, R., Elber, R., Olson, J. S. & Carver, T. E. (1992) J. Biol. Chem. 267, 22022–22034. [PubMed] [Google Scholar]
- 16.Frauenfelder, H., Sligar, S. G. & Wolynes, P. G. (1991) Science 254, 1598–1603. [DOI] [PubMed] [Google Scholar]
- 17.Nienhaus, K., Deng, P. C., Kriegl, J. M. & Nienhaus, G. U. (2003) Biochemistry 42, 9633–9646. [DOI] [PubMed] [Google Scholar]
- 18.Kriegl, J. M., Nienhaus, K., Deng, P. C., Fuchs, J. & Nienhaus, G. U. (2003) Proc. Natl. Acad. Sci. USA 100, 7069–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nienhaus, G. U. & Nienhaus, K. (2002) J. Biol. Phys. 28, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb, D. C., Nienhaus, K., Arcovito, A., Draghi, F., Miele, A. E., Brunori, M. & Nienhaus, G. U. (2002) J. Biol. Chem. 277, 11636–11644. [DOI] [PubMed] [Google Scholar]
- 21.Uchida, T., Ishimori, K. & Morishima, I. (2000) J. Biol. Chem. 275, 30309–30316. [DOI] [PubMed] [Google Scholar]
- 22.Adachi, S., Sunohara, N., Ishimori, K. & Morishima, I. (1992) J. Biol. Chem. 267, 12614–12621. [PubMed] [Google Scholar]
- 23.Carver, T. E., Brantley, R. E., Singleton, E. W., Arduini, R. M., Quillin, M. L., Phillips, G. N. & Olson, J. S. (1992) J. Biol. Chem. 267, 14443–14450. [PubMed] [Google Scholar]
- 24.Quillin, M. L., Li, T. S., Olson, J. S., Phillips, G. N., Dou, Y., Ikedasaito, M., Regan, R., Carlson, M., Gibson, Q. H., Li, H. Y. & Elber, R. (1995) J. Mol. Biol. 245, 416–436. [DOI] [PubMed] [Google Scholar]
- 25.Smerdon, S. J., Dodson, G. G., Wilkinson, A. J., Gibson, Q. H., Blackmore, R. S., Carver, T. E. & Olson, J. S. (1991) Biochemistry 30, 6252–6260. [DOI] [PubMed] [Google Scholar]
- 26.Dou, Y., Admiraal, S. J., Ikedasaito, M., Krzywda, S., Wilkinson, A. J., Li, T. S., Olson, J. S., Prince, R. C., Pickering, I. J. & George, G. N. (1995) J. Biol. Chem. 270, 15993–16001. [DOI] [PubMed] [Google Scholar]
- 27.Ikedasaito, M., Dou, Y., Yonetani, T., Olson, J. S., Li, T. S., Regan, R. & Gibson, Q. H. (1993) J. Biol. Chem. 268, 6855–6857. [PubMed] [Google Scholar]
- 28.Krzywda, S., Murshudov, G. N., Brzozowski, A. M., Jaskolski, M., Scott, E. E., Klizas, S. A., Gibson, Q. H., Olson, J. S. & Wilkinson, A. J. (1998) Biochemistry 37, 15896–15907. [DOI] [PubMed] [Google Scholar]
- 29.Nienhaus, K., Deng, P. C., Olson, J. S., Warren, J. J. & Nienhaus, G. U. (2003) J. Biol. Chem. 278, 42532–42544. [DOI] [PubMed] [Google Scholar]
- 30.Carver, T. E., Rohlfs, R. J., Olson, J. S., Gibson, Q. H., Blackmore, R. S., Springer, B. A. & Sligar, S. G. (1990) J. Biol. Chem. 265, 20007–20020. [PubMed] [Google Scholar]
- 31.Kholodenko, Y., Gooding, E. A., Dou, Y., Ikeda-Saito, M. & Hochstrasser, R. M. (1999) Biochemistry 38, 5918–5924. [DOI] [PubMed] [Google Scholar]
- 32.Nienhaus, K., Deng, P. C., Kriegl, J. M. & Nienhaus, G. U. (2003) Biochemistry 42, 9647–9658. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa, H., Uchida, T., Takahashi, S., Ishimori, K. & Morishima, I. (2001) Biophys. J. 80, 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rector, K. D., Rella, C. W., Hill, J. R., Kwok, A. S., Sligar, S. G., Chien, E. Y. T., Dlott, D. D. & Fayer, M. D. (1997) J. Phys. Chem. B 101, 1468–1475. [Google Scholar]
- 35.Rector, K. D., Engholm, J. R., Hill, J. R., Myers, D. J., Hu, R., Boxer, S. G., Dlott, D. D. & Fayer, M. D. (1998) J. Phys. Chem. B 102, 331–333. [Google Scholar]
- 36.Rector, K. D. & Fayer, M. D. (1999) Laser Chem. 19, 19–34. [Google Scholar]
- 37.Suzuki, T., Watanabe, Y., Nagasawa, M., Matsuoka, A. & Shikama, K. (2000) Eur. J. Biochem. 267, 6166–6174. [DOI] [PubMed] [Google Scholar]
- 38.Ye, X., Demidov, A. & Champion, P. M. (2002) J. Am. Chem. Soc. 124, 5914–5924. [DOI] [PubMed] [Google Scholar]
- 39.Walda, K. N., Liu, X. Y., Sharma, V. S. & Magde, D. (1994) Biochemistry 33, 2198–2209. [DOI] [PubMed] [Google Scholar]
- 40.Zou, S. Z., Baskin, J. S. & Zewail, A. H. (2002) Proc. Natl. Acad. Sci. USA 99, 9625–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steiger, B., Baskin, J. S., Anson, F. C. & Zewail, A. H. (2000) Angew. Chem. Int. Ed. 39, 257–260. [PubMed] [Google Scholar]
- 42.Petrich, J. W., Lambry, J. C., Balasubramanian, S., Lambright, D. G., Boxer, S. G. & Martin, J. L. (1994) J. Mol. Biol. 238, 437–444. [DOI] [PubMed] [Google Scholar]
- 43.Zewail, A. H. (2000) Angew. Chem. Int. Ed. 39, 2587–2631. [Google Scholar]
- 44.Wan, C. Z., Fiebig, T., Kelley, S. O., Treadway, C. R., Barton, J. K. & Zewail, A. H. (1999) Proc. Natl. Acad. Sci. USA 96, 6014–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruinsma, R., Grüner, G., D'Orsogna, M. R. & Rodnick, J. (2000) Phys. Rev. Lett. 85, 4393–4396. [DOI] [PubMed] [Google Scholar]
- 46.Lim, M. H., Jackson, T. A. & Anfinrud, P. A. (1993) Proc. Natl. Acad. Sci. USA 90, 5801–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meuwly, M., Becker, O. M., Stote, R. & Karplus, M. (2002) Biophys. Chem. 98, 183–207. [DOI] [PubMed] [Google Scholar]
- 48.Xie, X. L. & Simon, J. D. (1991) Biochemistry 30, 3682–3692. [DOI] [PubMed] [Google Scholar]
- 49.Goodno, G. D., Astinov, V. & Miller, R. J. D. (1999) J. Phys. Chem. A 103, 10630–10643. [Google Scholar]
- 50.Egeberg, K. D., Springer, B. A., Sligar, S. G., Carver, T. E., Rohlfs, R. J. & Olson, J. S. (1990) J. Biol. Chem. 265, 11788–11795. [PubMed] [Google Scholar]
- 51.Collman, J. P., Brauman, J. I., Doxsee, K. M., Halbert, T. R., Hayes, S. E. & Suslick, K. S. (1978) J. Am. Chem. Soc. 100, 2761–2766. [Google Scholar]
- 52.Collman, J. P. & Fu, L. (1999) Acc. Chem. Res. 32, 455–463. [Google Scholar]
- 53.Grogan, T. G., Bag, N., Traylor, T. G. & Magde, D. (1994) J. Phys. Chem. 98, 13791–13796. [Google Scholar]
- 54.McCammon, J. A. (2000) in Simplicity and Complexity in Proteins and Nucleic Acids, eds. Frauenfelder, H., Deisenhofer, J. & Wolynes, P. G. (Dahlem Univ. Press, Berlin), pp. 193–198.
- 55.Karplus, M. (2000) in Simplicity and Complexity in Proteins and Nucleic Acids, eds. Frauenfelder, H., Deisenhofer, J. & Wolynes, P. G. (Dahlem Univ. Press, Berlin), pp. 139–177.