Abstract
Background
Variants of unknown significance (VUSs) have been identified in BRCA1 and BRCA2 and account for the majority of all identified sequence alterations. Notably, VUSs occur disproportionately in people of African descent hampering breast cancer (BCa) management and prevention efforts in the population. Our study sought to identify and characterize mutations associated with increased risk of BCa at young age.
Methods
In our study, the spectrum of mutations in BRCA1 and BRCA2 was enumerated in a cohort of 31 African American women of early age at onset breast cancer, with a family history of breast or cancer in general and/or with triple negative breast cancer. To improve the characterization of the BRCA1 and BRCA2 variants, bioinformatics tools were utilized to predict the potential function of each of the variants.
Results
Using next generation sequencing methods and in silico analysis of variants, a total of 197 BRCA1 and 266 BRCA2 variants comprising 77 unique variants were identified in 31 patients. Of the 77 unique variants, one (1.3%) was a pathogenic frameshift mutation (rs80359304; BRCA2 Met591Ile), 13 (16.9%) were possibly pathogenic, 34 (44.2%) were benign, and 29 (37.7%) were VUSs. Genetic epidemiological approaches were used to determine the association with variant, haplotype, and phenotypes, such as age at diagnosis, family history of cancer and family history of breast cancer. There were 5 BRCA1 SNPs associated with age at diagnosis; rs1799966 (P=.045; Log Additive model), rs16942 (P=.033; Log Additive model), rs1799949 (P=.058; Log Additive model), rs373413425 (P=.040 and .023; Dominant and Log Additive models, respectively) and rs3765640 (P=.033 Log Additive model). Additionally, a haplotype composed of all 5 SNPs was found to be significantly associated with younger age at diagnosis using linear regression modeling (P=.023). Specifically, the haplotype containing all the variant alleles was associated with older age at diagnosis (OR= 5.03 95% CI=.91-9.14).
Conclusions
Knowing a patient’s BRCA mutation status is important for prevention and treatment decision-making. Improving the characterization of mutations will lead to better management, treatment, and BCa prevention efforts in African Americans who are disproportionately affected with aggressive BCa and may inform future precision medicine genomic-based clinical studies.
Keywords: BRCA1, BRCA2, Familial Breast Cancer, Hereditary Breast Cancer, Next Generation Sequencing, African American, Precision Medicine, Cancer Disparities
Introduction
Mutations in BRCA1 and BRCA2 confer the greatest risk of breast cancer (BCa), 45%-65% by age 701 and approximately 5%-10% of all BCas are associated with inherited mutations.2 Although mutations in BRCA1 and BRCA2 are rare, occurring in <1% of the general population, they are more frequently present in individuals with a family history of breast and/or ovarian cancer, early-onset BCa, triple negative breast cancer (TNBC; estrogen receptor negative, progesterone receptor negative, and human epidermal growth factor negative) and those of Ashkenazi Jewish ancestry.3 BRCA-related BCa in Ashkenazi Jewish women can be mainly attributable to three well-documented founder mutations making prevention, management and treatment of BCa in the population more practical. However, the lifetime risk for non-White women remains unknown and may even differ due to variable European and African admixture. To date, several studies have identified recurrent BRCA1 and BRCA2 mutations in populations of African descent, such as the 943ins10 frameshift mutation,4-8 Y101X nonsense mutation9-11 and others.6,7,10,12-15 While the frequency of BRCA1 and BRCA2 mutations vary depending on the characteristics of the BCa patients (ie, early age at onset, having a family history of BCa), Hall et al6 demonstrated that high risk patients of African descent have a slightly higher frequency of deleterious mutations (15.6%) compared with Western European (12.1%), Eastern European (13.5%) and Latin American (14.8%) high-risk patients. Furthermore, Rummel et al showed that there was a higher proportion of clinically relevant germline BRCA1 mutations in African Americans (AA) with TNBC, accounting for 11% of TNBCs in AAs while accounting for only 7% of TNBCs in Caucasians.16
The rapidly decreasing price of complete genome, exome, and target-enriched sequencing, as well as decreased turn-around time, have provided a realistic alternative to genotyping array-based GWASs. However, for ethnically diverse populations, genetic testing remains futile in large part due to the increase reporting of variants of unknown significance (VUSs). VUSs in BRCA1 and BRCA2 account for up to 50% of all identified sequence alterations. Notably, they occur disproportionately in people of African descent hampering BCa management and prevention efforts in the population. Kurian et al15 and Hall et al6 showed an increased prevalence of VUSs in high-risk BCa patients of African descent (78%) compared with those of European descent (61%). Some of these VUSs have been termed innocuous, but their significance alone or in combination is truly unknown. Hence, it is important to validate the spectrum and frequency of mutations in the BRCA genes of BCa patients of different ethnicities.
In our study, the spectrum of mutations in BRCA1 and BRCA2 was examined in a cohort of 31 African American women: 1) with early age at onset breast cancer; 2) with a family history of breast or cancer in general; and/or 3) with triple negative breast cancer in order to improve the characterization of the BRCA1 and BRCA2 variants and to identify mutations associated with increased risk of BCa in the group. Specifically, the mutational spectra of BRCA1 and BRCA2, the prevalence among high-risk African American BCa patients, and the pathogenicity or deleteriousness of identified mutations were assessed using next generation sequencing methods and in silico analysis of variants. Our study provides comprehensive insight valuable for this understudied population and with broad implications in familial and non-familial BCa across ethnic groups. Such information may facilitate mutation screening in a clinical setting and is needed to better manage resource allocation for genetic testing, genetic counseling, and planning of preventive interventions in all population subgroups.
Materials and Methods
Ascertainment of Probands, Cases and Controls
Our study was approved by the Howard University Institutional Review Board (09-MED-86). For this study, DNA samples from the African American Familial Breast Cancer Study (AAFBC)17 and the Breast Cancer Determinants in Women from Washington, DC (BCDC) study were utilized. Eligible study participants were African American women with a primary diagnosis of BCa and with a family history of cancer. Study participants allowed: 1) their medical and pathology records to be ascertained and the data abstracted and entered into a registry (database); 2) collection of personal data and family history by interview or by mail; 3) routine breast examinations; 4) routine screening mammography; 5) blood draws; 6) genetic screening of DNA; and 7) annual follow-up. For participants who were members of high-risk families, their family members were also enrolled into the study. Demographic information and family history were also collected from AAFBC and BCDC study participants. National Comprehensive Cancer Network (NCCN) Guidelines were utilized to determine eligibility for hereditary breast and ovarian cancer (HBOC).
Library Preparation, Template Preparation and Sequencing
DNA isolation was previously performed for all samples using the Gentra Puregene Blood Kit (www.qiagen.com; #158389) and the Qubit® 2.0 Fluorometer was utilized to determine the concentration of double stranded DNA. Library preparation of BRCA1 and BRCA2 was performed using Ion Ampliseq technology and the Ion Personal Genome Machine™ (PGM™) System (Life Technologies). The amplicon libraries for the PGM were constructed and generated with 20ng of genomic DNA using the Ion AmpliSeq™Library Kit 2.0. The three custom primer pools included primers for all BRCA1 and BRCA2 exons and intron/exon boundaries. Fragmentation was followed by end-repair, blunt-end ligation of the Ion Xpress Barcode and Ion P1 adaptors (Life Technologies) as well as nick translation. Library quantity for pooling 16 samples per chip was measured using the Ion Library Quantitation Kit for qPCR on the QuantStudio 7 Flex Real-Time PCR System (Life Technologies). Library templates were then prepared for sequencing using the Life Technologies Ion OneTouch 2™ protocols and reagents. For runs, template-positive ISPs per run, respectively, were deposited onto the Ion 318 chips by a series of centrifugation steps that were incorporated alternating the chip directionality. Sequencing was performed using the 500 flow (‘200 bp’) run format. There were 5,083,388 and 6,232,049 total usable reads per chip with a mean read length of 147 and 149 bases, respectively. The mean coverage per sample was 2,212.
Sequence Analysis Methods
Sequence data from the PGM was processed on the instrument server using the Torrent Suite v3.2 software package. These include signal processing, base calling, and mapping. Reads were mapped to the hg19 human reference genome assembly via the Torrent Mapping Alignment Program short read aligner. Single nucleotide variants (SNVs), multinucleotide polymorphisms (MNPs), insertions or deletions were identified using Torrent Variant Caller Plugin v.4.2.1.0.
In Silico Analysis of Variants
Utilizing ClinVar, SIFT (sorts intolerant from tolerant) and/or PolyPhen-2 (Polymorphism Phenotyping v2), the predicted deleterious effects of SNPs were determined. ClinVar reports the relationships among human variations and phenotypes, with supporting evidence and facilitates access to and communication about the relationships asserted between human variation and observed health status, and the history of that interpretation. ClinVar processes submissions reporting variants found in patient samples, assertions made regarding their clinical significance, information about the submitter, and other supporting data. Because the availability of supporting evidence may vary, particularly in regard to retrospective data aggregated from published literature, the archive accepts submissions from multiple groups, and aggregates related information, to reflect transparently both consensus and conflicting assertions of clinical significance.
Like SIFT, Poly-Phen2 is a tool that predicts possible impact of an amino acid substitution on the structure and function of a human protein using straight forward physical and comparative considerations. Additional information for variant location with respect to coding length (amino acid number) and critical domains, such as protein binding or active sites, was determined. Potential stop codons possibly resulting in a truncated protein that lacks the binding site of a negative regulator will receive additional emphasis. Information on location was gathered through tools such as the Integrative Genomics Viewer (IGV) to confirm locations in genomic coordinates and the cBioPortal’s MutationMapper for visualization of critical protein domains.
Results
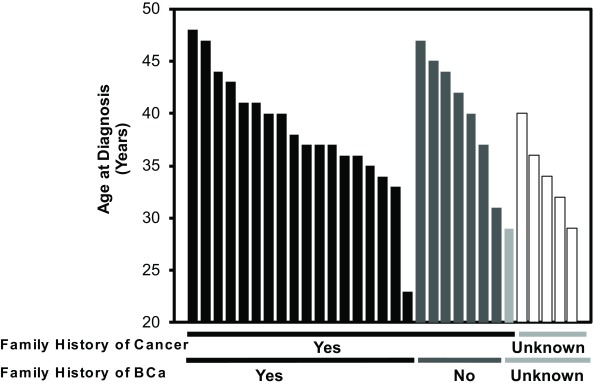
A total of 31 female African American patients were included in the study. All of the patients had an age at diagnosis <50 years and/or had a family history of cancer or BCa (Figure 1). The mean and median age at diagnosis was 38 and 37 years (range= 23-48), respectively. The mean age for those with a general history of cancer (ie, colon, prostate) was 38.6 (± 5.8 SD) years. For those with a family history of breast cancer, the mean age was 38.3 (± 5.7 SD) years. Those without a family history were slightly older at 40.7 (± 5.3 SD) years. Those with an unknown family history had a mean age of 33.1 (± 4.3 SD). After complete sequence analysis, a total of 197 BRCA1 (42.5%) and 266 BRCA2 (57.4%) variants comprising 77 unique variants were identified in the 31 patients. Notably, patients had an average of 15 BRCA1 and BRCA2 variants.
Figure 1. Age characteristics of 31 high-risk African American high-risk breast cancer patients that underwent exome sequencing of BRCA1 and BRCA2.
Black bars correspond to Family History of BCa-yes; dark grey bars correspond to Family History of BCa-no; light grey and white bars correspond to Family History of BCa-unknown.y
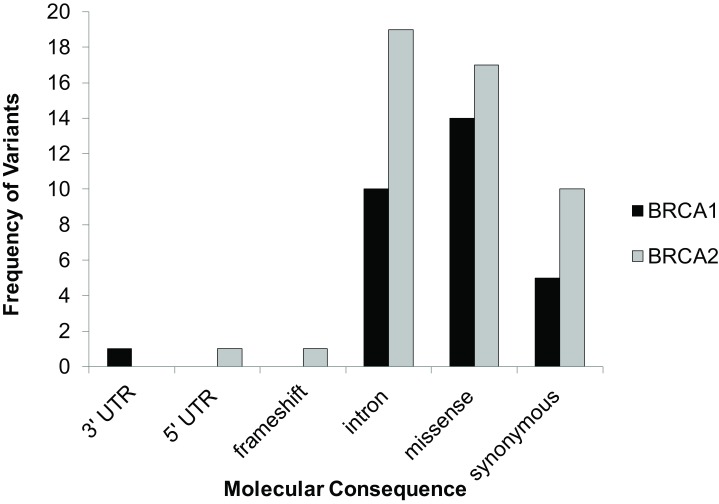
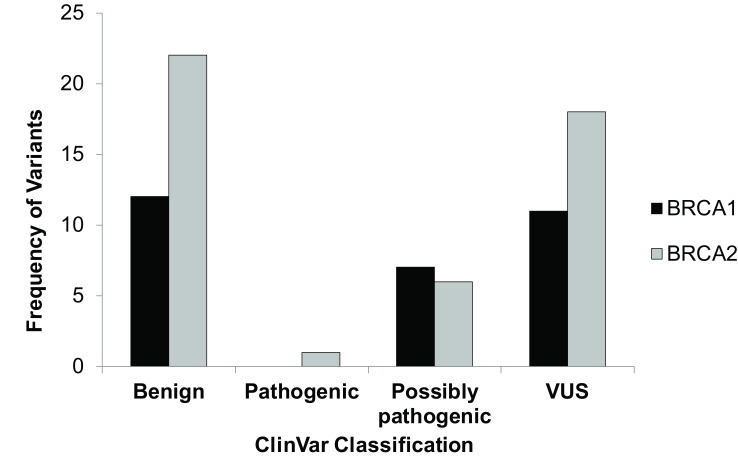
Of the 77 unique variants, 1 was a frameshift mutation in BRCA2 resulting in a pathogenic mutation. BRCA1 had 1 3’UTR, 10 intronic, 14 missense, and 5 synonymous SNPs (Table 1 and Figure 2). BRCA2 had 1 5’ UTR SNP, 1 frameshift, 19 intronic, 17 missense, and 9 synonymous SNPs (Table 1 and Figure 2). However, most variants were benign (Figure 3) and only 11 variants were either found to be pathogenic, variants of unknown significance or not found in ClinVar; SIFT and PolyPhen classified 4 ClinVar VUSs as possibly pathogenic and 3 SIFT/PolyPhen VUSs were not found in ClinVar (Table 1 and Figure 3).
Table 1. Germline mutations identified in 31 African American high-risk breast cancer patients.
| Gene ID | SNP Rs# | Nucleotide or | N | % | Mean age ± SD, years | Molecular consequences | ClinVar | Worst clinical significance (SIFT, PolyPhen) |
| Amino acid change | ||||||||
| BRCA1 | 55975699 | Asp232Asn | 1 | 3.23 | 36 ± na | missense | VUS | Possibly pathogenic |
| 397507258 | Asn319Ser | 1 | 3.23 | 44 ± na | missense | VUS | VUS | |
| 373202012 | c.441+52delC | 2 | 6.45 | 30.5 ± 2.1 | intron | Not found in ClinVar | VUS | |
| 578250989 | c.441+51T>C | 16 | 51.61 | 37.1 ± 6.3 | intron | Not found in ClinVar | VUS | |
| novel | c.441+54A>G | 1 | 3.23 | 37 ± na | intron | VUS | VUS | |
| BRCA2 | 80359304 | Met591Ile | 1 | 3.23 | 41 ± na | frameshift | Pathogenic | Pathogenic |
| 56328701 | Asp596His | 1 | 3.23 | 37 ± na | missense | VUS | Possibly pathogenic | |
| 45469092 | Arg2973Cys | 1 | 3.23 | 32 ± na | missense | VUS | Possibly pathogenic | |
| 206076 | Val2171Val | 31 | 100.0 | 37.9 ± 5.8 | synonymous | VUS | VUS | |
| 56014558 | c.7007+53G>A | 1 | 3.23 | 43 ± na | intron | Not found in ClinVar | VUS | |
| 76584943 | c.7008-62A>G | 1 | 3.23 | 32 ± na | intron | VUS | VUS |
VUS, variant of unknown significance.
Figure 2. Distribution of types of variants by molecular consequence.
Figure 3. Distribution of types of variants by ClinVar clinical significance.
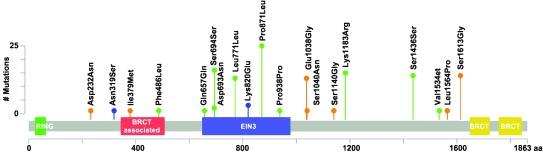
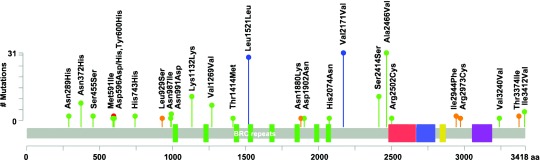
The variant location in regard to the gene domains can be found in Figure 4 and Figure 5. When variant location was considered, only the Ile379Met variant was found in a critical domain, the BRCT-associated serine-rich domain (Figure 4 and Figure 5). However, both Ser1613Gly and Leu1564Pro were located less than 100 amino acids from the BRCA1 C-terminus domain; both were labeled as benign by ClinVar and possibly pathogenic by SIFT/ PolyPhen. In BRCA2 both Leu929Ser and Asn1880Lys were located less than 100 amino acids from BRCA2 repeats 1 and 6; both were again labeled as benign by ClinVar and possibly pathogenic by SIFT/ PolyPhen. SNPs Ile2944Phe and Arg2973Cys were also located less than 100 amino acids from the BRCA2 OB3 pfama (oligonucleotide/oligosaccharide binding domain 3). The Arg2973Cys missense variant was considered of uncertain significance in ClinVar.
Figure 4. BRCA1 and BRCA2 mutational spectra (x-axis) and frequency for study patients (y-axis) (Blue=variant of unknown significance; Green= Benign; Orange=possibly pathogenic; Red=pathogenic).
Figure 5. BRCA1 and BRCA2 mutational spectra (x-axis) and frequency for study patients (y-axis) (Blue=variant of unknown significance; Green= Benign; Orange=possibly pathogenic; Red=pathogenic).
Discussion
In this study, African American women with early onset breast cancer and a family history of cancer underwent next-generation sequencing to determine the spectrum of mutations and improve the clinical characterization of the BRCA1 and BRCA2 variants of unknown significance. Because certain mutations may be unique to specific populations and may have clinical consequences, it is imperative that the full spectrum of mutations be identified. In silico analysis of variants allowed us to make predictions on the SNPs’ pathogenicity and test for their association with BCa in young AA women with a family history of cancer. Our next generation sequencing results revealed a large range of variants in the small population. In particular, one unequivocal pathogenic BRCA2 mutation, Met591Ile, a frameshift mutation, was identified in a sole participant. ClinVar analysis also revealed that 11 (14.3%) of the 77 unique SNPs were variants of unknown significance or possibly pathogenic. These SNPs and mutations warrant functional analysis to definitively determine their pathogenicity. The results indicate that BRCA1 and BRCA2 mutations may contribute marginally to early-onset BCa in AA women and supports the assertions made by others that multi-gene testing may be more informative in the group.7
It is well-established that the prevalence of mutations varies by population. In recent studies, pathogenic BRCA1 and BRCA2 mutations were discovered in 10% and 8% of high-risk AA BCa participants,7 respectively; similarly, 9.6% and 6.5% of probands were discovered to be BRCA carriers in a study of US high-risk families.18 However, the North Carolina Breast Cancer Registry reports much lower rates of 1.3% in AAs.19 Comparatively, our study found a much lower incidence of pathogenic mutations in a high-risk AA BCa population. Given the percentages above, it was hypothesized that at least 3 (10%) participants would harbor unequivocally pathogenic BRCA1 or BRCA2 mutations. However, a pathogenic mutation (rs80359304; Ile591Met) was found only in one patient. This patient was found to have a frameshift mutation that results in a truncated protein at amino acid 622 of BRCA2. Our results show a rate of 3.2% (1 of 31 patients) that falls between reported frequencies. It is worth noting that that ClinVar classified 7 variants as VUSs and 3 variants were not reported in ClinVar. These underappreciated variants could be driving breast cancer predisposition in our young patients and need to be confirmed by linkage, segregation, or functional analysis. When the VUSs are considered, the rate of BRCA1/2 mutations increase to 22.58%, which is much higher than reported frequencies, but probably more reflective of the true frequency given their age and family history.
Notably, thousands of BRCA1 and BRCA2 genetic variants and polymorphisms have been reported and are included in the Breast Cancer Information Core (BIC; www.nchgr.gov/intramural_research/lab-transfer/Bic) database. The pathogenic frameshift variant found in one patient from this study was examined. The BIC reports only 1 patient with this 4 base pair TTTA deletion at amino acid site 591 (rs80359304; Ile591Met), and reports 2 TTAT deletions (rs80359305; Ile591Tyr) that result in truncated proteins; two additional mutations at the same site, rs80356859 (Ile591Thr) and 80357901 (Ile591Lys), result in amino acid changes suggesting that this site may be hypermutable although with unfavorable consequences. It is important to also note that this is the first report of this mutation in a self-reported African American woman; all others in the BIC were found in patients who self-reported being of European descent.
Nevertheless, questions about the function of many of the BRCA1 and BRCA2 variants remain. This is particularly important in AAs who have a greater number of variants in BRCA1 and BRCA2, which was also demonstrated in this study. The dearth of functional variant information in ethnically diverse populations may be due to the underrepresentation of ethnically diverse populations in genomic studies. Furthermore, the discovery of genetic variants of unknown significance in actionable genes hinders clinical decision making and will hamper prevention and cancer management efforts, especially in AA women who report a higher frequency of variants of unknown significance.15 Given the exponential growth of the unintended discovery of germline variants and the limited association of some of these genes with genetically defined cancer syndromes, annotation of large next generation sequencing panels will require different approaches.
There are strengths and weaknesses to our study. The cohort of African American women with early onset breast cancer was not very large. However, it is one of the few studies that have identified the spectrum of mutations in such a group. Even with this small cohort, there was family history data missing making it difficult to determine if the breast cancer is familial in nature. The limited information about family history reflects a larger issue in the African American community, which historically has had inequities in access to health care. This issue subsequently makes it more difficult to find and record family history data. Given these challenges, criteria for genomic testing should be reflective of the increased prevalence of early age at onset BCa in the population. In addition, the clinicopathological features associated with being a mutation carrier should be considered when available, especially for people of African descent who have the highest incidence of tumors associated with BRCA mutations. For example, it is well-established that BCa is a heterogeneous disease composed of several “intrinsic” molecular subtypes that have been classified using microarray analysis.20-26 Of the common molecular BCa subtypes, the triple negative BCa (TNBC) subtype, is considered highly aggressive25 and is characterized by Estrogen Receptor (ER), Progesterone Receptor (PR), and HER2/neu (Human Epidermal Growth Factor Receptor 2) negativity.20 TNBC has a significantly higher incidence in women of African descent27,28 and there is a link between TNBC and the hereditary breast and ovarian cancer (HBOC) syndrome gene BRCA1.29-36 Moreover, these receptors are excellent targets for BCa treatment and prevention. However, treatment options for tumors without the receptors are often limited. This is one of the reasons TNBC is associated with poor prognosis, accounts for its unresponsiveness to typical endocrine therapy, and may partially explain shortened survival.37-41 Having the clinicopathological information could have potentially aided in the prediction of the clinical significance of the identified variants.
Strengths of this study include the innovative use of next-generation technologies and novel bioinformatics approaches that assisted us with the in silico characterization of VUSs. This study is the second to utilize next generation sequencing technologies in the identification of BRCA1 and BRCA2 variants associated with BCa.7 It is now accepted that next generation sequencing is more sensitive, allowing for increased identification of rare variants given the ability to massively sequence samples for a given depth. This will facilitate multi-gene testing which will reduce the cost and time needed to identify patient mutations for cancer treatment and prevention. Given that Africa is the most genetically diverse region in the world42,43 and that people of African descent may present with greater variation in their genomes, clinical next generation sequencing approaches of multiple gene panels may be beneficial to the group compared with single gene tests such as the BRCA1 and BRCA2 test.
Acknowledgments
This project was supported in part or in whole by the Department of Defense grants (LSR pre-doctoral training grant BC030134, and BC02234, and DAMD179616292), the National Cancer Institute (CA092040 and AA09802), National Center for Research Resources (NCRR, National Institutes of Health, through the Clinical and Translational Science Awards Program (CTSA), a trademark of DHHS, part of the Roadmap Initiative, “Re-Engineering the Clinical Research Enterprise,” (UL1RR031975), the RCMI Program at Howard University, Division of Research Infrastructure, National Center for Research Resources (G12 RR003048), and the Howard University Cancer Center/ Johns Hopkins Cancer Center Partnership, National Cancer Institute, NIH (U54 CA091431). This work was also supported by Hampton University who funded the next generation sequencing analysis of the breast cancer samples and by the Georgetown-Howard Universities Center for Clinical and Translational Sciences (NCATS-UL1TR000101) who funded the pilot that allowed for the recruitment of patients and the improved characterization and annotation of the samples. We also thank the Howard University Research Centers for Minority Institutions (NIMHD-5G12MD007597) who provided the computational resources to record and analyze the data. Furthermore, we thank Dr. Simone Heyliger for editing and revising the manuscript critically for intellectual content.
References
- 1. Easton DF, Hopper JL, Thomas DC, et al. Breast cancer risks for BRCA1/2 carriers. Science. 2004;306(5705):2187-2191. 10.1126/science.306.5705.2187c [DOI] [PubMed] [Google Scholar]
- 2. Claus EB, Schildkraut JM, Thompson WD, Risch NJ. The genetic attributable risk of breast and ovarian cancer. Cancer. 1996;77(11):2318-2324. 10.1002/(SICI)1097-0142(19960601)77:113.0.CO;2-Z [DOI] [PubMed] [Google Scholar]
- 3. Struewing JP, Hartge P, Wacholder S, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336(20):1401-1408. 10.1056/NEJM199705153362001 [DOI] [PubMed] [Google Scholar]
- 4. Stoppa-Lyonnet D, Laurent-Puig P, Essioux L, et al. ; Institut Curie Breast Cancer Group . BRCA1 sequence variations in 160 individuals referred to a breast/ovarian family cancer clinic. Am J Hum Genet. 1997;60(5):1021-1030. [PMC free article] [PubMed] [Google Scholar]
- 5. Nanda R, Schumm LP, Cummings S, et al. Genetic testing in an ethnically diverse cohort of high-risk women: a comparative analysis of BRCA1 and BRCA2 mutations in American families of European and African ancestry. JAMA. 2005;294(15):1925-1933. 10.1001/jama.294.15.1925 [DOI] [PubMed] [Google Scholar]
- 6. Hall MJ, Reid JE, Burbidge LA, et al. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer. 2009;115(10):2222-2233. 10.1002/cncr.24200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Churpek JE, Walsh T, Zheng Y, et al. Inherited predisposition to breast cancer among African American women. Breast Cancer Res Treat. 2015;149(1):31-39. 10.1007/s10549-014-3195-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olopade OI, Fackenthal JD, Dunston G, Tainsky MA, Collins F, Whitfield-Broome C. Breast cancer genetics in African Americans. Cancer. 2003;97(1)(suppl):236-245. 10.1002/cncr.11019 [DOI] [PubMed] [Google Scholar]
- 9. Zhang B, Fackenthal JD, Niu Q, et al. Evidence for an ancient BRCA1 mutation in breast cancer patients of Yoruban ancestry. Fam Cancer. 2009;8(1):15-22. 10.1007/s10689-008-9205-9 [DOI] [PubMed] [Google Scholar]
- 10. Zhang J, Fackenthal JD, Zheng Y, et al. Recurrent BRCA1 and BRCA2 mutations in breast cancer patients of African ancestry. Breast Cancer Res Treat. 2012;134(2):889-894. 10.1007/s10549-012-2136-z [DOI] [PubMed] [Google Scholar]
- 11. Fackenthal JD, Zhang J, Zhang B, et al. High prevalence of BRCA1 and BRCA2 mutations in unselected Nigerian breast cancer patients. Int J Cancer. 2012;131(5):1114-1123. 10.1002/ijc.27326 [DOI] [PubMed] [Google Scholar]
- 12. Oluwagbemiga LA, Oluwole A, Kayode AA. Seventeen years after BRCA1: what is the BRCA mutation status of the breast cancer patients in Africa? - a systematic review. Springerplus. 2012;1(1):83. 10.1186/2193-1801-1-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karami F, Mehdipour P.. A comprehensive focus on global spectrum of BRCA1 and BRCA2 mutations in breast cancer. Biomed Res Int. 2013;2013:928562. [DOI] [PMC free article] [PubMed]
- 14. Diez O, Pelegrí A, Gadea N, et al. Novel BRCA1 deleterious mutation (c.1949_1950delTA) in a woman of Senegalese descent with triple-negative early-onset breast cancer. Oncol Lett. 2011;2(6):1287-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kurian AW. BRCA1 and BRCA2 mutations across race and ethnicity: distribution and clinical implications. Curr Opin Obstet Gynecol. 2010;22(1):72-78. 10.1097/GCO.0b013e328332dca3 [DOI] [PubMed] [Google Scholar]
- 16. Rummel S, Varner E, Shriver CD, Ellsworth RE. Evaluation of BRCA1 mutations in an unselected patient population with triple-negative breast cancer. Breast Cancer Res Treat. 2013;137(1):119-125. 10.1007/s10549-012-2348-2 [DOI] [PubMed] [Google Scholar]
- 17. Panguluri RC, Brody LC, Modali R, et al. BRCA1 mutations in African Americans. Hum Genet. 1999;105(1-2):28-31. 10.1007/s004399900085 [DOI] [PubMed] [Google Scholar]
- 18. Walsh T, Casadei S, Coats KH, et al. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006;295(12):1379-1388. 10.1001/jama.295.12.1379 [DOI] [PubMed] [Google Scholar]
- 19. John EM, Miron A, Gong G, et al. Prevalence of pathogenic BRCA1 mutation carriers in 5 US racial/ethnic groups. JAMA. 2007;298(24):2869-2876. 10.1001/jama.298.24.2869 [DOI] [PubMed] [Google Scholar]
- 20. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938-1948. 10.1056/NEJMra1001389 [DOI] [PubMed] [Google Scholar]
- 21. Paik S. Molecular profiling of breast cancer. Curr Opin Obstet Gynecol. 2006;18(1):59-63. 10.1097/01.gco.0000192970.52320.29 [DOI] [PubMed] [Google Scholar]
- 22. Perou CM, Jeffrey SS, van de Rijn M, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96(16):9212-9217. 10.1073/pnas.96.16.9212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747-752. 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 24. Pusztai L. Molecular heterogeneity of breast cancer: implications for treatment and clinical trial design. Breast Cancer Res. 2009;11(S1)(suppl 1):S4. 10.1186/bcr226520030879 [DOI] [Google Scholar]
- 25. Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869-10874. 10.1073/pnas.191367098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100(14):8418-8423. 10.1073/pnas.0932692100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lund MJ, Trivers KF, Porter PL, et al. Race and triple negative threats to breast cancer survival: a population-based study in Atlanta, GA. Breast Cancer Res Treat. 2009;113(2):357-370. 10.1007/s10549-008-9926-3 [DOI] [PubMed] [Google Scholar]
- 28. Lund MJ, Butler EN, Hair BY, et al. Age/race differences in HER2 testing and in incidence rates for breast cancer triple subtypes: a population-based study and first report. Cancer. 2010;116(11):2549-2559. [DOI] [PubMed] [Google Scholar]
- 29. Greenup R, Buchanan A, Lorizio W, et al. Prevalence of BRCA mutations among women with triple-negative breast cancer (TNBC) in a genetic counseling cohort. Ann Surg Oncol. 2013;20(10):3254-3258. 10.1245/s10434-013-3205-1 [DOI] [PubMed] [Google Scholar]
- 30. Hartman AR, Kaldate RR, Sailer LM, et al. Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer. 2012;118(11):2787-2795. 10.1002/cncr.26576 [DOI] [PubMed] [Google Scholar]
- 31. Evans DG, Howell A, Ward D, Lalloo F, Jones JL, Eccles DM. Prevalence of BRCA1 and BRCA2 mutations in triple negative breast cancer. J Med Genet. 2011;48(8):520-522. 10.1136/jmedgenet-2011-100006 [DOI] [PubMed] [Google Scholar]
- 32. Comen E, Davids M, Kirchhoff T, Hudis C, Offit K, Robson M. Relative contributions of BRCA1 and BRCA2 mutations to “triple-negative” breast cancer in Ashkenazi Women. Breast Cancer Res Treat. 2011;129(1):185-190. 10.1007/s10549-011-1433-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gonzalez-Angulo AM, Timms KM, Liu S, et al. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res. 2011;17(5):1082-1089. 10.1158/1078-0432.CCR-10-2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Linn SC, Van ’t Veer LJ. Clinical relevance of the triple-negative breast cancer concept: genetic basis and clinical utility of the concept. Eur J Cancer. 2009;45(suppl 1):11-26. 10.1016/S0959-8049(09)70012-7 [DOI] [PubMed] [Google Scholar]
- 35. Young SR, Pilarski RT, Donenberg T, et al. The prevalence of BRCA1 mutations among young women with triple-negative breast cancer. BMC Cancer. 2009;9(1):86. 10.1186/1471-2407-9-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Atchley DP, Albarracin CT, Lopez A, et al. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol. 2008;26(26):4282-4288. 10.1200/JCO.2008.16.6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dent R, Hanna WM, Trudeau M, Rawlinson E, Sun P, Narod SA. Time to disease recurrence in basal-type breast cancers: effects of tumor size and lymph node status. Cancer. 2009;115(21):4917-4923. 10.1002/cncr.24573 [DOI] [PubMed] [Google Scholar]
- 38. Lachapelle J, Foulkes WD. Triple-negative and basal-like breast cancer: implications for oncologists. Curr Oncol. 2011;18(4):161-164. 10.3747/co.v18i4.824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lønning PE, Sørlie T, Perou CM, Brown PO, Botstein D, Børresen-Dale AL. Microarrays in primary breast cancer--lessons from chemotherapy studies. Endocr Relat Cancer. 2001;8(3):259-263. 10.1677/erc.0.0080259 [DOI] [PubMed] [Google Scholar]
- 40. Lønning PE, Sørlie T, Børresen-Dale AL. Genomics in breast cancer-therapeutic implications. Nat Clin Pract Oncol. 2005;2(1):26-33. 10.1038/ncponc0072 [DOI] [PubMed] [Google Scholar]
- 41. Paik S. Clinical trial methods to discover and validate predictive markers for treatment response in cancer. Biotechnol Annu Rev. 2003;9:259-267. 10.1016/S1387-2656(03)09005-7 [DOI] [PubMed] [Google Scholar]
- 42. Tishkoff SA, Varkonyi R, Cahinhinan N, et al. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science. 2001;293(5529):455-462. 10.1126/science.1061573 [DOI] [PubMed] [Google Scholar]
- 43. Gurdasani D, Carstensen T, Tekola-Ayele F, et al. The African Genome Variation Project shapes medical genetics in Africa. Nature. 2015;517(7534):327-332. 10.1038/nature13997 [DOI] [PMC free article] [PubMed] [Google Scholar]