SUMMARY
Degradation of polysaccharides forms an essential arc in the carbon cycle, provides a percentage of our daily caloric intake, and is a major driver in the renewable chemical industry. Microorganisms proficient at degrading insoluble polysaccharides possess large numbers of carbohydrate active enzymes, many of which have been categorized as functionally redundant. Here we present data that suggests that carbohydrate active enzymes that have overlapping enzymatic activities can have unique, non-overlapping biological functions in the cell. Our comprehensive study to understand cellodextrin utilization in the soil saprophyte Cellvibrio japonicus found that only one of four predicted β-glucosidases is required in a physiological context. Gene deletion analysis indicated that only the cel3B gene product is essential for efficient cellodextrin utilization in C. japonicus and is constitutively expressed at high levels. Interestingly, expression of individual β-glucosidases in Escherichia coli K-12 enabled this non-cellulolytic bacterium to be fully capable of using cellobiose as a sole carbon source. Furthermore, enzyme kinetic studies indicated that the Cel3A enzyme is significantly more active than the Cel3B enzyme on the oligosaccharides but not disaccharides. Our approach for parsing related carbohydrate active enzymes to determine actual physiological roles in the cell can be applied to other polysaccharide-degradation systems.
Keywords: β-glucosidase, cellodextrin, cellulose, Cellvibrio japonicus, functional redundancy
Graphical Abstract
Bacteria that can degrade polysaccharides in the environment often encode hundreds of enzymes for this process. It is unclear, however, if all of these enzymes are essential for polysaccharide degradation or have overlapping functions. Our study of the four β-glucosidases from the saprophytic bacterium Cellvibrio japonicus indicate these enzymes share a few biochemical properties, but have little physiological functional overlap.
INTRODUCTION
Degradation of insoluble polysaccharides, particularly lignocellulose, is a critical biological process that substantially impacts life on a scale that ranges from individuals to ecosystems. A calculated 1011 tons of lignocellulose is annually turned over as part of global nutrient cycles (Lynd et al., 2002, Shin, 2010). Furthermore, an estimated 10% of the daily caloric intake of humans is derived from short chain fatty acids produced from lignocellulose degradation by gut microbiota (Lynd et al., 2002, Martens et al., 2014), the composition of which changes significantly depending on the type of plant biomass in the diet (Hehemann et al., 2010). The economics of polysaccharide degradation have also been of intense interest in the renewable fuel and chemical industry, as lignocellulose degradation is thought to be a sustainable alternative to petroleum (Bokinsky et al., 2011).
From both biotechnological and environmental perspectives, the key challenge in harnessing the energy in lignocellulose, or any other insoluble polysaccharide, is overcoming the so-called recalcitrance of the polymers (Marriott et al., 2014, Himmel et al., 2007). For example, cellulose comprises 30–50% of plant cell walls (Jordan et al., 2012), and while a single cellulose polymer is structurally simple (un-branched (1,4)-β-D-glucose), the extensive network of hydrogen bonding creates a crystalline matrix and renders this polysaccharide insoluble (Lei, 2014). To completely degrade cellulose, the activities of three Carbohydrate Active enZyme (CAZyme) families (Lombard et al., 2014) are required: endo-1,4-β-glucanases, exo-1,4-β-glucanases, and β-glucosidases (Horn et al., 2012, Jordan et al., 2012, Lombard et al., 2014). A fourth activity from lytic polysaccharide mono-oxygenases (LPMOs) has recently been shown to also be important for efficient cellulose depolymerization in aerobic systems (Vaaje-Kolsstad et al., 2012).
The microbes that have overcome the recalcitrance of insoluble polysaccharides often have genomes that encode a hundred or more CAZymes. A common characteristic of these polysaccharide-degrading microorganisms is the presence of multiple genes encoding enzymes that are predicted to have the same enzymatic activity (Mba Medie et al., 2012). In some model bacteria, several of these enzymes have been characterized and labeled as “redundant” (Couger et al., 2015, Gibson et al., 2011, Morgenstern et al., 2014, Schellenberger et al., 2011). However these assessments have been based exclusively on biochemical and structural data. This has proven problematic for industry efforts to find the most economical enzyme mixtures for biotechnological applications, or ecological studies to uncover the major factors driving lignocellulose degradation in a physiologically relevant manner. Several groups have expressed the necessity of in vivo studies because purely in vitro data have shown to be poor predictors of enzyme activities on physiologically relevant substrates (Zhang et al., 2002, Cartmell et al., 2011, Naas et al., 2014).
To overcome this challenge, a recent report employed a systems approach using physiologically relevant substrates and the genetically tractable model bacterium Cellvibrio japonicus to examine cellulose degradation (DeBoy et al., 2008, Gardner & Keating, 2012). The physiological relevance of biochemically important enzymes was shown, as well as the power of genetic analysis to uncover functions for “unknown” enzymes (Gardner et al., 2014). In this study, we examine the latter stages of cellulose degradation, focusing on cellodextrin utilization and the four β-glucosidases (GH3 family; Cel3A, Cel3B, Cel3D, and Cel3D) found in C. japonicus. Previous bioinformatics analysis of C. japonicus CAZymes predicted that Cel3D was secreted, Cel3A and Cel3C were located in the periplasm, and Cel3B was located in the cytoplasmic (DeBoy et al., 2008). The GH3 family of β-glucosidases was the focus of this study because C. japonicus does not have any GH1 β-glucosidases, which are common in other cellodextrin utilizing microorganisms (Ketudat Cairns & Esen, 2010). Our data indicate that these enzymes play unique roles within the cell based on differences in their predicted localizations, expression patterns, and specific activities despite the capacity of all these β-glucosidases to confer the ability to utilize cellobiose in a non-cellulolytic bacterium. In addition, we discuss the differences between functional redundancy and biochemical redundancy in the context of polysaccharide degradation. We argue that it is likely that most enzymes labeled as “functionally redundant” in polysaccharide degrading bacteria are merely biochemically redundant, and the true physiological function of these enzymes has yet to be uncovered.
RESULTS
Cel3B is the β-glucosidase that makes the most significant contribution to the utilization of cellobiose by C. japonicus
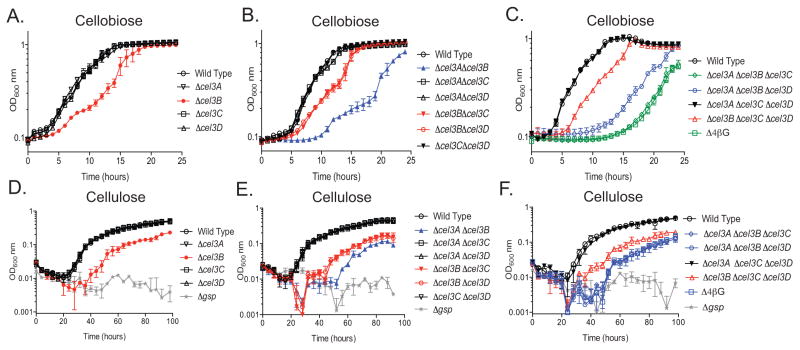
The four β-glucosidase genes in C. japonicus (cel3A, cel3B, cel3C, and cel3D) were deleted individually and in combination and the mutants were grown on glucose, cellobiose, or insoluble cellulose to assess growth fitness (Fig. 1). When grown on glucose, the growth rates for the single, double, triple, and quadruple β-glucosidase mutants were all similar to that of wild type as was the maximum growth attained (Fig. S1). When the single deletion mutants were grown using cellobiose as the sole carbon source, the Δcel3B mutant had a decreased growth rate compared to wild type, but was able to attain the same maximum optical density (OD) as the wild type bacterium. When grown with insoluble cellulose as the sole carbon source, the Δcel3B mutant had a decreased growth rate compared to wild type and also a decreased maximum OD (Table S2). In addition, the lag time of the cel3B mutant increased from three to four hours (Table S1). Single deletions of cel3A, cel3C, and cel3D resulted in no significant growth defect when grown with cellobiose or insoluble cellulose (Fig. 1A and D). A Δgsp mutant (defect in protein secretion) was used as a negative control for the insoluble cellulose experiments as it has previously been shown to be unable to utilize cellulose (Gardner & Keating, 2010).
Fig. 1. Deletion of multiple β-glucosidases enzymes shows varying contribution to cellobiose and filter paper utilization.
Growth analysis of all C. japonicus mutants grown using, 0.5% cellobiose or 1% cellulose as the sole carbon source. (A-C) Growth analysis of single (A), double (B), triple and quadruple (C) deletion mutants grown using cellobiose as the sole carbon source. (D-F) Growth analysis of single (D), double (E), triple and quadruple (F) deletion mutants grown using cellulose as the sole carbon source. All experiments were performed in biological triplicate where error bars represent standard deviation and are present for all data points, although sometimes too small to be observed. All strains grew similarly to wild type when glucose was the carbon source used, which is shown in Supplemental Figure S1.
Double deletion mutants were constructed combinatorially and tested as above (Fig. 1B and E). The Δcel3AΔcel3C, Δcel3AΔcel3D, and Δcel3CΔcel3D double mutants displayed no growth defect on cellobiose or insoluble cellulose. The Δcel3BΔcel3C and Δcel3BΔcel3D double deletion strains had growth rates similar to the Δcel3B single mutant on cellobiose and insoluble cellulose (Table S1 and Table S2). The Δcel3AΔcel3B double deletion had a more exaggerated growth defect than the Δcel3B single mutant, primarily due to a considerably longer lag phase compared to the Δcel3B single mutant (Fig. 1B). On insoluble cellulose the Δcel3AΔcel3B double mutant grew more slowly and reached a lower maximum OD than the Δcel3B single mutant (Table S2).
The combinatorial generation of triple deletion mutant strains were a powerful analytical tool as only one predicted β-glucosidase remained in these strains (Fig. 1C and F). The Δcel3AΔcel3CΔcel3D mutant displayed no growth defect on either cellobiose or insoluble cellulose. The growth of Δcel3BΔcel3CΔcel3D on cellobiose and insoluble cellulose was similar to the Δcel3B single mutant (Table S1 and Table S2). The Δcel3AΔcel3BΔcel3D mutant grew similar to the Δcel3AΔcel3B double mutant on both cellobiose and insoluble cellulose (Table S1 and Table S2) On cellobiose the Δcel3AΔcel3BΔcel3C mutant grew more slowly and had a longer lag phase than the wild type or the other triple mutant strains (Table S1 and Table S2). On insoluble cellulose however, the Δcel3AΔcel3BΔcel3C mutant grew similarly to the Δcel3AΔcel3B and Δcel3AΔcel3BΔcel3D mutant strains.
A strain with all four of the β-glucosidases deleted was constructed and grown in either cellobiose or insoluble cellulose containing media (Fig. 1C and F). When cultured on cellobiose as the sole carbon source, the Δcel3AΔcel3BΔcel3CΔcel3D (Δ4βG) strain displayed a similar phenotype to Δcel3AΔcel3BΔcel3C strain, both in terms of growth rate and maximum OD attained (Table S1). In addition, the Δ4βG strain had a very slow growth rate on insoluble cellulose and long lag phase on both cellobiose and insoluble cellulose. When using insoluble cellulose, the Δ4βG strain did not attain wild type levels of growth (Table S2). Overall, the Δ4βG strain behaved very similarly to the Δcel3AΔcel3BΔcel3C triple mutant strain.
β-glucosidase mutants exhibit decreased cellobiose consumption
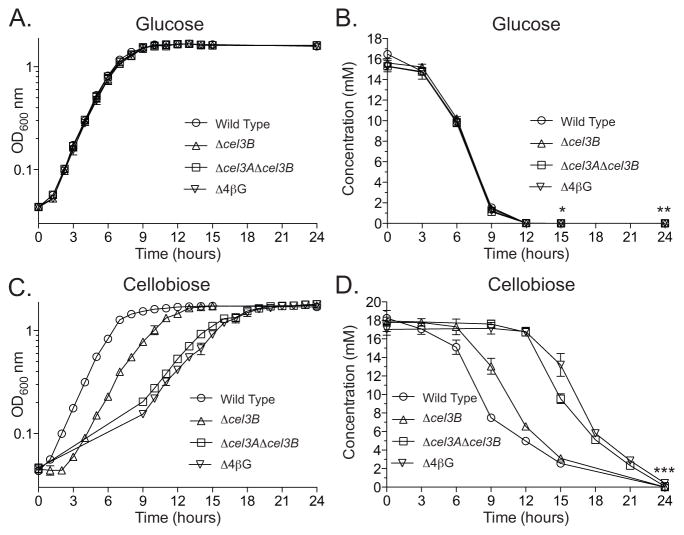
High performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) was performed to analyze glucose and cellobiose consumption rate changes of the Δcel3B, Δcel3AΔcel3B, and Δ4βG mutant strains compared to wild type. These strains were chosen for analysis based on the growth defect phenotypes observed (vide supra). As expected, the consumption of glucose was nearly identical between all mutant strains and wild type. All strains began to enter stationary phase nine hours after inoculation (Fig. 2A). By 15 hours after inoculation all of the strains had consumed nearly all of the glucose in the medium, and after 24 hours inoculation there was no detectable glucose in any of the samples (Fig. 2B).
Fig. 2. Decrease in rate of cellobiose consumption mirrors growth defects of β-glucosidase deletion mutants.
Quantification of 0.25% glucose and 0.5% cellobiose consumption during growth of wild type C. japonicus and Δcel3B, Δcel3AΔcel3B, and Δ4βG mutants. (A & B) All four strains were grown using glucose as the sole carbon source (A). During growth, samples were taken every three hours and the concentration of remaining glucose was quantified using HPAEC-PAD (B). (C & D) All four strains were grown using cellobiose as the sole carbon source (C). Samples were taken every three hours and quantified using HPAEC-PAD (D). The glucose and cellobiose experiments were both performed in biological triplicate. Error bars represent the standard deviation and are present for all data points. The limit of quantitation for glucose and cellobiose using the HPAEC-PAD parameters described in the Materials and Methods was 1 μM. The “*” symbol indicates Δcel3AΔcel3B was below the level of quantitation, “**” indicates all four strains were below the level of quantitation, and “***” indicates wild type and Δcel3B were below the level of quantitation.
When grown on cellobiose, the wild type strain had a short one-hour lag and then grew rapidly before entry into stationary phase after nine hours. Upon entry of stationary phase there was still 7.49±0.17 mM left of the original 18.24±0.20 mM of cellobiose in the wild type samples. However, the cellobiose concentration continued to decrease through stationary phase in the wild type samples, dropping below 1 μM by 24 hours after inoculation. The Δcel3B mutant had a three-hour lag before reaching exponential growth and entered stationary phase 12 hours after inoculation, at which point ~40% of the cellobiose remained (Fig. 2C). After 24 hours the disaccharide was fully consumed by the Δcel3B mutant. The amount of cellobiose remaining in the medium of the Δcel3AΔcel3B and Δ4βG flasks changed little nine-hours post inoculation, with only a 0.33mM reduction for Δcel3AΔcel3B flasks and no measurable change in the Δ4βG flasks. Upon entering stationary phase, 18 hours after inoculation, there was ~28% cellobiose remaining in the Δcel3AΔcel3B flasks and ~30% in the media of the Δ4βG flasks (Fig. 2D). Unlike wild type and the Δcel3B strain, there was detectable cellobiose remaining in the media of Δcel3AΔcel3B and the Δ4βG flasks (0.098±0.11 mM and 0.43±0.1 mM, respectively), 24 hours post inoculation, indicating these mutants were unable to consume all of the cellobiose in the same timeframe as a wild type strain.
Individual β-glucosidases heterologously expressed in E. coli are sufficient for cellobiose utilization
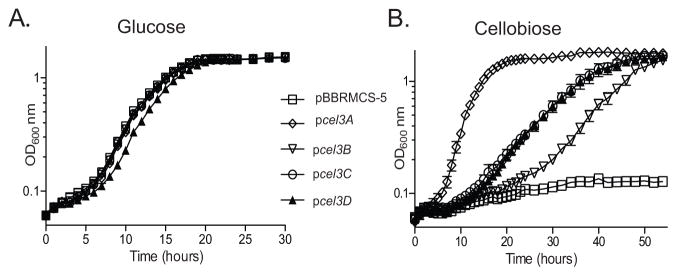
Our genetic analysis suggested that only cel3B is required for efficient cellobiose utilization as indicated by the Δcel3AΔcel3CΔcel3D mutant (Fig. 1C), however a previous enzymatic study showed that Cel3A is a highly active β-glucosidase (Rixon et al., 1992). This led us to hypothesize that perhaps all four of the predicted β-glucosidases in C. japonicus do indeed have sufficient β-glucosidase activity to support growth on cellobiose. To examine this hypothesis, the four β-glucosidases were cloned individually into the pBBRMCS-5 vector (Elzer et al., 1994) and expressed in E. coli K-12. As expected, the expression of these genes did not affect growth on glucose (Fig. 3A) and a strain containing an empty pBBRMCS-5 vector was unable to grow on cellobiose (Fig. 3B). When tested on cellobiose, three distinct phenotypes were displayed by the E. coli strains expressing the C. japonicus β-glucosidases. Expression of cel3A resulted in rapid growth, with a short lag of four hours and a growth rate of 0.34±0.01 generations per hour (gen hr−1). Strains expressing cel3C or cel3D grew similarly, with growth rates of 0.10±0.002 and 0.11±0.01 gen hr−1, respectively, and a lag of eight to ten hours. The strain expressing cel3B had a growth rate of 0.11±0.004 gen hr−1 and a lag of 24 hours before starting to grow. Despite the differences in growth rate, expression of the four β-glucosidase genes resulted in the same maximum OD attained for the E. coli strains. This suggests that while in C. japonicus the GH3 genes are not functionally equivalent, when expressed in E. coli, which removes much of the physiological and regulatory context, the GH3 genes all do have similar enzymatic action on the disaccharide cellobiose.
Fig. 3. All four β-glucosidases from C. japonicus are individually sufficient to confer cellobiose utilization to a non-cellulolytic organism.
The four β-glucosidases were cloned individually into the pBBRMCS-5 vector and transformed into E. coli K-12 and grown on 0.25% glucose (A) and 0.5% cellobiose (B). The empty pBBRMCS-5 vector was also transformed into K-12 as a control strain. Experiments were performed in biological triplicate and the error bars, in most cases too small to be seen, represent the standard deviation.
RNAseq analysis reveals cel3B is constitutively expressed at high levels
To examine the physiological extent for gene expression, we performed RNAseq analysis. Gene expression profiles for C. japonicus when using cellodextrins were collected with either glucose or cellobiose as the sole carbon source (Fig. S2). Samples were taken from either exponentially growing or stationary phase cells and subjected to RNAseq analysis. Using two carbon sources and two sampling points, there are four pair-wise comparative analyses possible: exponential (glucose) vs. exponential (cellobiose) (Fig. S2), exponential vs. stationary phase for glucose or cellobiose, or stationary (glucose) vs. stationary (cellobiose) (Fig. S3). The comparative analyses from the exponential vs. stationary phase for glucose or cellobiose are useful as control experiments to discover the genes that are up-regulated in response to changes in growth rate; none of the GH3 genes were regulated by growth rate.
Analysis of the exponential (glucose) vs. exponential (cellobiose) comparison (Fig. S2) revealed that only ten genes annotated as CAZymes were up-regulated and only one was a β-glucosidase (cel3A) (Table S3). The other up-regulated CAZymes were predicted to be active on a diverse range of substrates such as xylan, starch, and mannan. Bioinformatic analyses of these genes indicate they are transcribed alone (DeBoy et al., 2008), suggesting their up-regulation on cellobiose is not driven by co-expression in an operon. To test if these up-regulated CAZymes were playing a role in cellobiose utilization, we generated mutant strains for these genes and assessed the fitness of the strains using media containing cellobiose as the sole carbon source. All of these strains grew like wild type (Fig. S2). This result indicated that these genes were part of a regulatory program, rather than having a direct role in cellobiose utilization.
Comparative analysis is often useful to find differentially expressed genes; however, a drawback of this type of analysis is that it will not uncover constitutively expressed genes. An examination of the RNAseq data in the absence of comparison, as determined by the reads per kilobase per million mapped reads (RPKM), we found that cel3B is highly constitutively expressed (Table 1). The cel3A gene, while inducible by cellobiose, had an expression level that was 2.7-fold lower than cel3B expression. The cel3C and cel3D genes had low levels of expression under all growth conditions and were 27-fold and 15-fold lower than cel3B on cellobiose, respectively.
Table 1.
Expression of the GH3 genes during exponential growth in defined minimal mediaa
| Gene | Glucose (RPKMb) | Cellobiose (RPKM) |
|---|---|---|
| cel3A | 183±17.9 | 522±34.1 |
| cel3B | 1420±70.7 | 1410±42.3 |
| cel3C | 42.1±21.7 | 52.2±5.64 |
| cel3D | 58.7±17.8 | 93.8±3.38 |
Experiment was performed in biological triplicate
Reads per kilobase per million mapped reads
The enzymatic activites of C. japonicus β-glucosidases indicates preference for different cellodextrins
The data described above indicated that of the four β-glucosidases Cel3B plays the most important role in cellobiose utilization by C. japonicus, although Cel3A also contributes to the degradation of the disaccharide. These results prompted detailed analysis of the kinetic properties of all four GH3 β-glucosidases. Cel3A, Cel3B, Cel3C, and Cel3D were produced in recombinant form from E. coli and purified to electrophoretic homogeneity. The kinetics of the four enzymes were evaluated against a series of β-1,4-glucooligosaccharides. The data are summarized in Table 2 with example Michaelis-Menten plots for Cel3A and Cel3B displayed in Fig. S4. Cel3B was ~50-fold more active than Cel3A and Cel3C against cellobiose, with Cel3D displaying very low activity against the disaccharide. Against substrates with a degree of polymerization ≥3, the activity of Cel3A and Cel3C increased while Cel3B decreased. Indeed, Cel3A and Cel3C appear to contain three glucose-binding subsites, extending from −1 to +2, as the activity of the enzyme against cellotriose, cellotetraose and cellopentaose were similar. Cel3D displayed only modest, catalytic efficiency against cellotetraose. It should be noted that the difference in activity against these substrates likely reflects their cellular location and contribution to cellulose degradation. The periplasmic location of Cel3A is consistent with exposure to cello-oligosaccharides derived from the extracellular degradation of β-1,4-glucans (Rixon et al., 1992, Gardner, 2016). The predicted cytoplasmic location of Cel3B (DeBoy et al., 2008) and its strong preference for cellobiose suggests that the inner membrane of C. japonicus contains a transporter that specifically imports the disaccharide into the cytoplasm.
Table 2.
Kinetics of C. japonicus GH3 β-glucosidases
| Enzyme | Substrate | kcat (min−1) | Km (mM) | kcat / Km (min−1 mM−1)a |
|---|---|---|---|---|
| Cel3A | Cellobiose | 117±2.5 | 1.8±0.094 | 65 |
| Cel3B | Cellobiose | 5385±175 | 1.5±0.13 | 3590 |
| Cel3C | Cellobiose | 168±6 | 1.9±0.14 | 89 |
| Cel3D | Cellobiose | 10±0.7 | 11.5±1.5 | 0.9 |
|
| ||||
| Cel3A | Cellotriose | 1240±37 | 0.23±0.02 | 5391 |
| Cel3B | Cellotriose | 731±42 | 1.5±0.16 | 487 |
| Cel3C | Cellotriose | 846±28 | 1.1±0.07 | 769 |
| Cel3D | Cellotriose | 227±18 | 6.2±0.94 | 37 |
|
| ||||
| Cel3A | Cellotetraose | ND | ND | 5306±105 |
| Cel3B | Cellotetraose | ND | ND | 522±12 |
| Cel3C | Cellotetraose | ND | ND | 856±22 |
| Cel3D | Cellotetraose | ND | ND | 91±2.5 |
|
| ||||
| Cel3A | Cellopentaose | ND | ND | 5126±133 |
| Cel3B | Cellopentaose | ND | ND | 464±16 |
| Cel3C | Cellopentaose | ND | ND | 865±14 |
| Cel3D | Cellopentaose | ND | ND | 44±1.7 |
In experiments where only kcat/Km was determined, substrate concentrations ≪Km were used as the initial rate provides a direct readout of kcat/Km and the rate is directly proportional to substrate concentration. To ensure the assays were in this linear region of Michaelis-Menten kinetics (enabling direct determination of kcat/Km), assays were carried out at three substrate concentrations to verify there was a linear relationship between rate and substrate concentration. For Cel3A, Cel3B, Cel3C and Cel3D the substrate ranges used to determine kcat/Km were 0.02 to 0.08 mM, 0.1 to 0.4 mM, 0.05 to 0.2 mM and 0.25 to 2 mM, respectively. The assays used to determine kcat and Km had three technical replicates. When only kcat/Km was determined nine technical replicates were used. Values are reported ± the standard error of the mean.
Not determined
DISCUSSION
Cellulose degradation is biochemically simple, requiring three to four enzymatic activities (Horn et al., 2012, Jordan et al., 2012, Vaaje-Kolsstad et al., 2012). However, the most effective cellulose degraders in the environment often possess dozens of CAZymes for cellulose degradation (Mba Medie et al., 2012). This has led to many statements in the literature about “functional redundancy” in regards to environmentally or biotechnologically relevant polysaccharide degradation (Couger et al., 2015, Gibson et al., 2011, Morgenstern et al., 2014, Schellenberger et al., 2011). However, this sentiment is formulated on almost exclusively biochemical and structural data and does not factor in physiologically relevant parameters. In this study, we use a systems approach to examine cellodextrin utilization, the final stages of cellulose degradation, in a saprophytic bacterium. Our data indicate that the four β-glucosidases present in C. japonicus are not “functionally redundant,” but share attributes that can be classified as “biochemically redundant”. This study may provide an example how to probe CAZymes in polysaccharide degrading microorganisms in a comprehensive manner to better classify, model, and predict CAZyme functions.
RNAseq analysis corroborated the mutational data and demonstrates the utility of both comparative and absolute expression analyses. When using a comparative approach with glucose as the reference condition, the cel3A gene was the only up-regulated β-glucosidase on cellobiose (~8-fold) during exponential growth. Several additional CAZyme genes were up-regulated on cellobiose, but this likely reflects a suite of genes under similar regulatory control, which is responsive to cellobiose in a manner similar to the cellulose regulon of Neurospora crassa (Znameroski et al., 2012). Deletion of these up-regulated CAZyme genes in C. japonicus did not influence cellobiose utilization (Table S2), again suggesting that these genes are part of a complex regulatory response, where cellodextrin utilization is only one part. The fact that more cellulases were not up-regulated was not surprising, as previous studies in N. crassa have shown that the lignocellulose degrading response was induced by cellobiose but was masked by the effects of carbon catabolite repression in the presence of β-glucosidases generating glucose (Znameroski et al., 2012). By examining the absolute level of expression (RPKM), it was revealed that the cel3B gene was highly expressed across all times and substrates, and nearly 2.7-fold higher than the induced expression of the cel3A gene in cellobiose. Expression of cel3C or cel3D genes was low and not induced to significant levels on cellobiose.
The mutational analyses allowed us to determine a hierarchy of the β-glucosidases in C. japonicus in a physiological context, with cel3B being critical, cel3A having a supporting role, cel3C having a minor role, and cel3D having a negligible role in regards to cellobiose utilization. We observed a strong, reproducible growth defect with the single cel3B mutant strain when grown on cellobiose and insoluble cellulose (Fig. 1A and D). The HPAEC-PAD analysis of media during growth on cellobiose also indicated a decreased rate of cellobiose consumption directly correlated with the decreased growth rate observed for Δcel3B mutant strains (Fig. 2). The physiological roles of these genes were entirely consistent with their expression levels in response to cellobiose. Severe growth defects resulting from a single gene inactivation were also observed in our previous work on cellulose and xyloglucan degradation in C. japonicus (Gardner et al., 2014, Larsbrink et al., 2014). Single-gene mutants have been shown to elicit strong effects in other microbes; for example, the inactivation of a single GH9 endo-processive endo-glucanase gene in Clostridium phytofermentens was enough to abolish growth on cellulose in this anaerobe (Tolonen et al., 2009). However, the power of a combinatorial mutagenesis approach became evident with the creation of the double and triple β-glucosidase mutant strains. The analyses of the double mutant strains revealed a contribution to cellobiose utilization by cel3A but not cel3C or cel3D (Fig. 1B and E). Analysis of triple deletion strains, specifically the Δcel3AΔcel3CΔcel3D mutant that was able to grow like wild type on cellobiose and insoluble cellulose (Fig. 1C and F), gave strong evidence that cel3B is the dominant β-glucosidase in C. japonicus. The addition of the cel3C deletion to the Δcel3AΔcel3B background elicited another third growth phenotype that was absent from the Δcel3AΔcel3BΔcel3D triple mutant. Finally the quadruple mutant behaved nearly identical to the Δcel3AΔcel3BΔcel3C mutant, providing further evidence that the cel3D gene product was the least important for cellodextrin utilization in C. japonicus (Fig. 1C and F). Therefore, from a physiological perspective, the hierarchy for cellodextrin utilization in C. japonicus is cel3B > cel3A > cel3C ≫> cel3D.
Interestingly, the Δ4βG strain was still able to grow despite lacking all predicted β-glucosidases. This may be a result of other glucan 1,4-β-glucosidase activity from other cellulases. Glucan 1,4-β-glucosidase activity (EC 3.2.1.74) is found in GH3, GH5, and GH9 families, and C. japonicus has genes encoding 15 GH5 and 3 GH9 enzymes (Lombard et al., 2014). The GH5 family is large with enzymes that target a diverse range of substrates including cellulose, mannan, and xyloglucan. Of the GH5 genes encoded by C. japonicus there are nine enzymes that are predicted to play a role in cellulose degradation (Lombard et al., 2014). The GH5 family is further divided into subfamilies, many of which are poly-substrate specific, with more than one enzyme activity experimentally determined. The poly-substrate specificity could indicate that although many of the GH5 cellulases are not predicted to have the β-glucosidase activity, it is possible they have this activity in addition to, or instead of, their predicted activity. Aspeborg et. al. point out that annotated CAZyme activities could be absent in GH subgroups due to limitations in experimentally tested substrates (Aspeborg et al., 2012). This may hold true for the predicted cellulolytic GH5 and GH9 gene products in C. japonicus and will need to be further explored in the future.
Heterologous expression experiments in the non-cellulolytic E. coli indicated that all four of the GH3 genes of C. japonicus were sufficient to act as β-glucosidases. The expression of cel3A yielded rapid growth on cellobiose (Fig. 3B), which was not surprising as previous work indicated that Cel3A was a highly active enzyme (Rixon et al., 1992). The growth rate differences between the E. coli strains are likely consequences of a combination of 1) the GH3 genes being heterologously over-expressed, 2) catalytic parameters of the GH3 enzymes, and 3) poor transport of cellobiose into E. coli (Hall & Faunce, 1987). For example, Cel3A was determined to have low activity against cellobiose but substantially higher activity against cellotetrose and cellopentose (Table 2). When tested kinetically, Cel3B was determined to have high activity on cellobiose, although almost ~2-fold lower than the efficiency of Cel3A on cellotetrose and cellopentose. In addition, Cel3B has low activity on cellotriose, cellotetrose, and cellopentose. The differences in enzyme efficiencies may indicate specialization of the enzymes for the cellodextrins they encounter in the environment. Cel3A is membrane associated so it is positioned close to the cell and able to cleave glucose off of the larger cellodextrins, while Cel3B is cytoplasmic and most likely encounters cellobiose. Interestingly, E. coli cells expressing cel3C and cel3D also grew when provided cellobiose, suggesting that cel3C and cel3D can have β-glucosidase activity in vivo. However, it should be noted that growth observed in a heterologous system may not necessarily be biologically relevant because these experiments force expression higher than what is found in the native bacterium. For example, the enzymatic analysis of Cel3D found cellobiose to be a very poor substrate, which reinforces the C. japoincus mutant data that indicate that Cel3D does not play a role in cellobiose metabolism.
The synthesis of these data support the hypothesis that the β-glucosidases of C. japonicus are in fact playing different roles within the cell and are not functionally redundant (Fig. 4). This model is built from a previous bioinformatics study of C. japoncus CAZymes, which placed the GH3 enzymes in three distinct cellular spaces (DeBoy et al., 2008), however to date only one of these (Cel3A) has been experimentally confirmed (Rixon et al., 1992). This study adds to the model strong physiological data, including gene expression analysis, enzyme kinetics, and growth phenotypes of β-glucosidase mutant strains, and we argue that the four β-glucosidases of C. japonicus are playing unique roles within the cell despite their predicted redundancy. For this particular study, given that the activities of the GH3 enzymes are similar, the Cel3B β-glucosidase is dominant because of its high constitutive expression level and cellular location in the cytoplasm. We argue that to be considered “functionally redundant,” four physiologically relevant parameters must be considered: time (when the enzyme is expressed), space (cellular location), substrate specificity (including side activity), and kinetics (including enzyme synergy). Cel3B, the cytoplasmic constitutively expressed enzyme, appears to be responsible for degrading cellobiose that is transported into the cytoplasm. Cel3A while induced by cellobiose appears to be more specific for degrading cellotetrose and cellopentose. The lack of growth defects caused by the deletion of the cel3C and cel3D genes in C. japonicus, despite being able confer the ability to utilize cellobiose in E. coli, indicate these enzymes are biochemically able to act as β-glucosidases and we would term this “biochemical redundancy” not “functional redundancy.” The severe growth defects of the double deletions retaining only Cel3C and Cel3D indicate that these enzymes are unable to compensate for the lack of Cel3A and Cel3B when utilizing cellobiose. We argue that CelD is not functionally redundant because biochemical analysis indicates very low activity on cellobiose, and cel3D mutants are phenotypically wild type. Stated another way, Cel3D is not functionally redundant because it does not function physiologically (or enzymatically in a robust way) as a β-glucosidase. Therefore current investigations are looking for a biological role for Cel3D in the degradation of non-cellulose oligosaccharides.
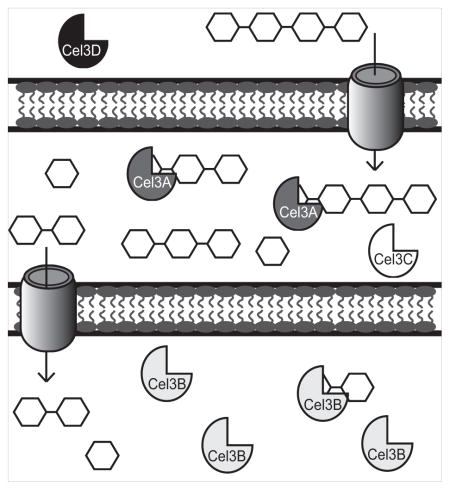
Fig. 4. Proposed model for soluble cellodextrin utilization in C. japonicus.
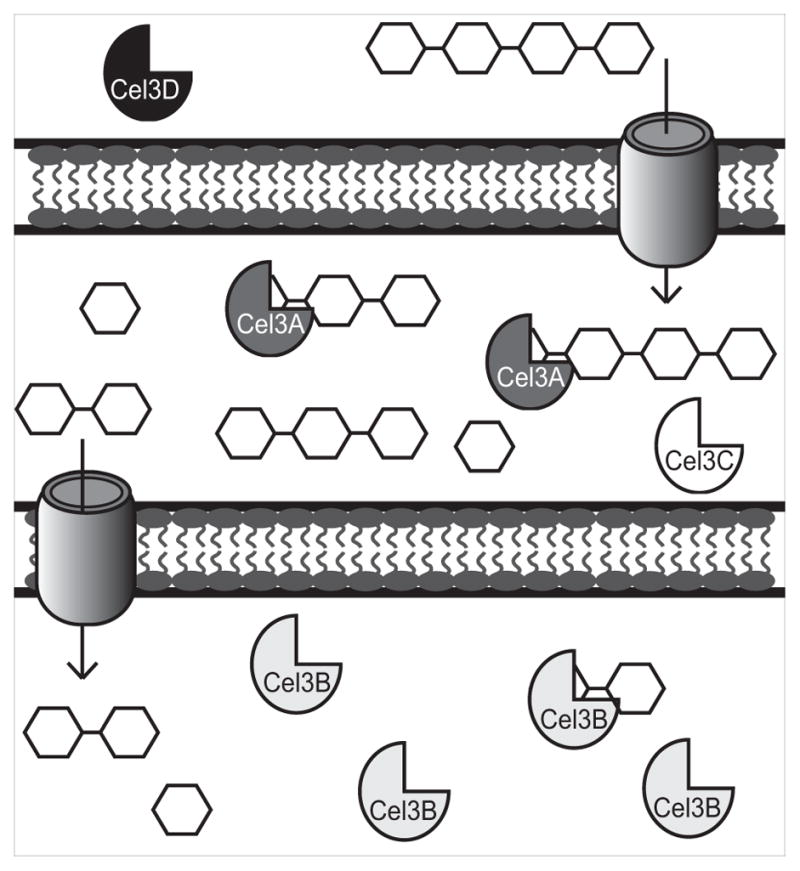
Cel3D, represented with black icons, is predicted to be located extracellularly. Cel3A, dark gray icons, and Cel3C, white icons, are predicted to be membrane associated which has been confirmed for Cel3A by Rixon et. al. (Rixon et al., 1992). Based off of enzyme activities, large cello-oligosaccharides are transported into the periplasm where Cel3A degrades them. Smaller cello-oligosaccharides like cellobiose are transported into the cytoplasm where Cel3B, which is represented with light gray icons, breaks them down into glucose. When cellobiose is transported into the cell, cel3A expression is induced while cel3B is constitutively expressed as indicated by RNAseq experiments.
Extrapolating beyond the utilization of cellodextrins, we believe that a “biochemical redundancy” and “functional redundancy” debate in regards to other CAZymes would have a similar outcome as suggested by this study. By calling into question if a set of CAZymes are truly “functional redundant” and striving to determine the physiological relevance of a gene product, we will better inform CAZyme capabilities and therefore better position them for environmental, biotechnological, and human health applications, as this approach will tease out which “redundant” enzymes are important in the degradation of a substrate they can all use.
EXPERIMENTAL PROCEDURES
Genetic techniques
Generation and verification of C. japonicus knockout mutants was done as previously described (Gardner & Keating, 2010). Deletion mutants were generated as previously described (Nelson & Gardner, 2015) with the following modifications. Briefly, the 500 bp upstream and downstream region of the targeted gene was amplified by PCR rather than being synthesized by GeneWIZ (South Plainfield, NJ) and assembled into the pK18mobsacB vector (Schafer et al., 1994) via the assembly method of Gibson et al. (Gibson, 2009). Primers used for the construction of vectors and PCR verification of C. japonicus mutants are listed in Table S4. For heterologous expression studies the cel3A, cel3B, cel3C, and cel3D genes were synthesized by GeneWIZ and cloned into the pBBRMCS-5 vector (Elzer et al., 1994). The pBBRMCS-5 vectors containing the individual β-glucosidases were transformed into chemically competent E. coli K-12 using standard protocols (Green & Rogers, 2013).
Growth analysis
C. japonicus strains were grown in MOPS defined medium (Neidhardt et al., 1974) containing glucose, cellobiose, or filter paper as the sole carbon source. E. coli strains were grown in lysogenic broth (LB) (Bertani, 1951) or M9 (Neidhardt et al., 1974) minimal media containing glucose or cellobiose. Cultures were grown as described by Nelson & Gardner (Nelson & Gardner, 2015). Briefly, strains were inoculated from a MOPS-glucose plate into 5 mL of MOPS-glucose broth and grown for 24 hours, then sub-cultured (1:100 dilution) into fresh medium used for growth analysis (e.g. MOPS-cellobiose). Growth was measured by optical density at 600 nm (OD600) using a Spec20D+ spectrophotometer (Thermo Scientific) or a Tecan M200Pro microtiter plate reader (TECAN Trading AG, Switzerland). Cultures were grown at 30°C with high aeration (220 rpm) in 18 mm culture tubes or in a 96 well flat bottom polystyrene assay plate (Corning Inc, Corning, NY). C. japonicus strains were grown in MOPS defined medium containing glucose (0.25% w:v), cellobiose (0.5% w:v), or filter paper (1% w:v) as the sole carbon source (Nelson & Gardner, 2015). All growth experiments were performed in biological triplicate with statistical significance calculated when appropriate using the GraphPad Prism 6 software package (La Jolla, CA). Exit from lag phase is a complex process and while many methods and models have been proposed (Zwietering et al., 1992), for this study it was defined simply as a >25% increase in OD from the starting OD. When required, kanamycin was used at a concentration of 50 μg mL−1, gentamycin at a concentration of 15 μg mL−1 and ampicillin at a concentration of 100 μg mL−1.
Quantification of cellobiose consumption
C. japonicus wild type and deletion strains Δcel3B, Δcel3AΔcel3B, Δ4βG were grown in 250 mL baffled flasks as described above. Cultures were grown for 24 hours with OD600 read every hour for the first 15 hours and a final time point after 24 hours for wild type and Δcel3B. For Δcel3AΔcel3B and Δ4βG an initial OD600 reading taken after inoculation then OD600 was read every hour from nine hours post inoculation to 24 hours post inoculation. Samples were collected right after inoculation and 24 hours post inoculation for all samples. Additionally, samples for wild type and Δcel3B were taken every three hours from three hours post inoculation until 15 hours post inoculation and for Δcel3AΔcel3B and Δ4βG samples were taken every three hours from nine hours post inoculation until 21 hours post inoculation. Sampling was as follows: 5 mL was aseptically removed and transferred to 18 mm culture tubes for OD600 readings in a Spec20D+ spectrophotometer (Thermo Scientific) and returned to the original flask. A 1 mL sample was removed after OD600 readings and pelleted. The supernatant was filtered using 0.45 μM Spin-X column (Corning Inc., Corning, NY), aliquotted (200 μL), and stored at −20 °C. All experiments were performed in biological triplicate.
Samples were analyzed by HPAEC-PAD using a Dionex ICS-5000+ system (Thermo Scientific) equipped with a 4 × 250 mm SA-10 anion-exchange column (Thermo Scientific) and a 4 × 50 mm SA-10 guard column (Thermo Scientific). A flow rate of 1mL/min was used and the temperature of the column was kept at 30 °C. The 25 μL samples were injected and run isocratically with 20 mM NaOH. Data analysis was performed with the Dionex Chromeleon 7 (Thermo Scientific) software package.
RNAseq sampling and analysis
Gene expression analysis was performed similarly as that previously described (Gardner et al., 2014). Briefly, C. japonicus cultures were grown in 500 mL flasks and growth was monitored with OD600 readings every hour for the first 12 hours of growth and again at 24 hours. A 35 mL sample was taken during exponential growth phase and again during stationary phase as determined by OD600. Metabolism of the sample was stopped with 5ml of a phenol/ethanol solution (5%/95%) within 2 minutes of sampling. The samples were then immediately pelleted by centrifugation at 8000 × g for five minutes, the supernatant decanted, and the cell pellets flash frozen in a dry ice/ethanol bath for five minutes, then stored at −80°C. RNA extraction, library preparation, multiplexing, and sequencing was performed by GeneWIZ (South Plainfield, NJ). Illumina HiSeq2500 was performed in 50bp single-reads with at least 10 million reads generated per sample. The raw sequence data generated was converted into FASTQ files using Illumina CASSAVA 1.8.2, which were then imported into CLC Genomics Workbench 7.5.1. Low quality base pairs were trimmed from the ends and the sequence reads were mapped to the C. japonicus reference genome with RPKM values calculated for genes. Log2 transformation and quantile normalization was performed for the RPKM values and a Student’s T-test conducted to compare gene expression between various groups of samples. Additional analysis and visualization of the data was performed using the ArrayStar (DNASTAR, Madison, WI) software package. The raw data have been submitted to GEO (GSE90955).
Recombinant expression, purification, and kinetic analysis of Cel3A and Cel3B
C. japonicus genomic DNA was used for the PCR amplification of full length Cel3B and Cel3A lacking its signal peptide. The primers also contained sequence for the NdeI and XhoI restriction sites, and the PCR products were cloned into the E. coli expression vector pET28b. E. coli expression of Cel3A and Cel3B and the subsequent purification of the two enzymes to electrophoretic homogeneity by immobilized metal affinity chromatography using a Talon (immobilized cobalt) column was as described previously (Cuskin et al., 2015). The enzymes were assayed against β-1,4-cello-oligosaccharides obtained from Megazyme International using the Glucose Detection Kit supplied by the same company. The assays were carried out in 50 mM sodium phosphate buffer, pH 7.5 at 37°C. When measuring both Km and kcat the initial rate was determined at five or more concentrations of substrate. When determining only catalytic efficiency, kcat/Km, the initial rate at a substrate concentration ≪Km was determined.
Supplementary Material
Acknowledgments
This work was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC0014183 and the Wellcome Trust (grant No. WT097907AIA). Additional support to CEN came from a NIGMS Initiative for Maximizing Student Development under Award Number R25-GM55036. We wish to thank Dr. William LaCourse for helpful advice and technical assistance concerning HPAEC-PAD analysis.
Footnotes
AUTHOR CONTRIBUTIONS
CEN generated C. japonicus mutants and performed growth analysis experiments, generated heterologous expression plasmids for experiments in E. coli, performed HPAEC-PAD analysis, and contributed to writing the manuscript.
AR cloned and characterized the enzymes used in this study.
CM cloned and characterized the enzymes used in this study.
JAW assisted with HPAEC-PAD method development and data analysis.
HJG supervised the recombinant enzyme component of the study and contributed to writing the manuscript.
JGG designed the study, generated and analyzed the RNAseq data, supervised the remaining work, and contributed to writing the manuscript.
All authors read and approved the final submitted version of the manuscript.
DISCLAIMER
This report was prepared as an account of work sponsored by an agency of the United States Government. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof.
COMPLIANCE WITH ETHICAL STANDARDS
This article does not contain any studies with human participants or animals performed by any of the authors. In addition, the authors declare that they have no conflict of interest.
References
- Aspeborg H, Coutinho PM, Wang Y, Brumer H, 3rd, Henrissat B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5) BMC evolutionary biology. 2012;12:186. doi: 10.1186/1471-2148-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. Journal of bacteriology. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokinsky G, Peralta-Yahya PP, George A, Holmes BM, Steen EJ, Dietrich J, Lee TS, Tullman-Ercek D, Voigt CA, Simmons BA, Keasling JD. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. PNAS. 2011;108:19949–19954. doi: 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartmell A, McKee LS, Pena MJ, Larsbrink J, Brumer H, Kaneko S, Ichinose H, Lewis RJ, Vikso-Nielsen A, Gilbert HJ, Marles-Wright J. The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases. The Journal of biological chemistry. 2011;286:15483–15495. doi: 10.1074/jbc.M110.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couger MB, Youssef NH, Struchtemeyer CG, Liggenstoffer AS, Elshahed MS. Transcriptomic analysis of lignocellulosic biomass degradation by the anaerobic fungal isolate Orpinomyces sp. strain C1A. Biotechnol Biofuels. 2015:8. doi: 10.1186/s13068-015-0390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, Rogowski A, Hamilton BS, Chen R, Tolbert TJ, Piens K, Bracke D, Vervecken W, Hakki Z, Speciale G, Munoz-Munoz JL, Day A, Pena MJ, McLean R, Suits MD, Boraston AB, Atherly T, Ziemer CJ, Williams SJ, Davies GJ, Abbott DW, Martens EC, Gilbert HJ. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoy RT, Mongodin EF, Fouts DE, Tailford LE, Khouri H, Emerson JB, Mohamoud Y, Watkins K, Henrissat B, Gilbert HJ, Nelson KE. Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. Journal of bacteriology. 2008;190:5455–5463. doi: 10.1128/JB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzer PH, Kovach ME, Phillips RW, Robertson GT, Peterson KM, Roop RM. In vivo and in vitro stability of the broad-host-range cloning vector pBBR1MCS in six Brucella Species. Plasmid. 1994;33:51–57. doi: 10.1006/plas.1995.1006. [DOI] [PubMed] [Google Scholar]
- Gardner JG. Polysaccharide degradation systems of the saprophytic bacterium Cellvibrio japonicus. World journal of microbiology & biotechnology. 2016;32:121. doi: 10.1007/s11274-016-2068-6. [DOI] [PubMed] [Google Scholar]
- Gardner JG, Crouch L, Labourel A, Forsberg Z, Bukhman YV, Vaaje-Kolstad G, Gilbert HJ, Keating DH. Systems biology defines the biological significance of redox-active proteins during cellulose degradation in an aerobic bacterium. Molecular microbiology. 2014 doi: 10.1111/mmi.12821. [DOI] [PubMed] [Google Scholar]
- Gardner JG, Keating DH. Requirement of the type II secretion system for utilization of cellulosic substrates by Cellvibrio japonicus. Applied and environmental microbiology. 2010;76:5079–5087. doi: 10.1128/AEM.00454-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JG, Keating DH. Genetic and functional genomic approaches for the study of plant cell wall degradation in Cellvibrio japonicus. Methods in enzymology. 2012;510:331–347. doi: 10.1016/B978-0-12-415931-0.00018-5. [DOI] [PubMed] [Google Scholar]
- Gibson DM, King BC, Hayes ML, Bergstrom GC. Plant pathogens as a source of diverse enzymes for lignocellulose digestion. Curr Opin Microbiol. 2011;14:264–270. doi: 10.1016/j.mib.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Gibson GGY, Chuang LR, Venter JC, Hutchison CA, III , Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Green R, Rogers EJ. Transformation of chemically competent E. coli. Methods in enzymology. 2013;529:329–336. doi: 10.1016/B978-0-12-418687-3.00028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG, Faunce W. Functional Genes for Cellobiose Untilization in Natural Isolates of Escherichia coli. Journal of Bateriology. 1987;169:2713–2717. doi: 10.1128/jb.169.6.2713-2717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- Horn SJ, Vaaje-Kolstad G, Westerng B, Eijsink V, GH Novel enzymes for the degradation of cellulose. Biotechnology for Biofuels. 2012:5. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DB, Bowman MJ, Braker JD, Dien BD, Hector RE, Lee CC. Plant cell walls to ethanol. Biochem, J. 2012;424:241–252. doi: 10.1042/BJ20111922. [DOI] [PubMed] [Google Scholar]
- Ketudat Cairns J, Esen A. B-Glucosidases. Cellular and molecular life sciences : CMLS. 2010;67:3389–3405. doi: 10.1007/s00018-010-0399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsbrink J, Thompson AJ, Lundqvist M, Gardner JG, Davies GJ, Brumer H. A complex gene locus enables xyloglucan utilization in the model saprophyte Cellvibrio japonicus. Molecular microbiology. 2014;94:418–433. doi: 10.1111/mmi.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei LLS, Bashline L, Gu Y. Dissecting the molecular mechanism underlying the intimate relationship between cellulose microfibrils and cortical microtubules. Fronteirs in Plant Science. 2014:5. doi: 10.3389/fpls.2014.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic acids research. 2014;42:D490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd LR, Weimer PJ, Pretorius IS. Microbial Cellulose Utilization: Fundamentals and Biotechnology. Microbiol Mol Biol Rev. 2002 doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott PE, Sibout R, Lapierre C, Fangel JU, Willats WG, Hofte H, Gomez LD, McQueen-Mason SJ. Range of cell-wall alterations enhance saccharification in Brachypodium distachyon mutants. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:14601–14606. doi: 10.1073/pnas.1414020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Kelly AG, Tauzin AS, Brumer H. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol. 2014;426:3851–3865. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mba Medie F, Davies GJ, Drancourt M, Henrissat B. Genome analyses highlight the different biological roles of cellulases. Nature reviews Microbiology. 2012;10:227–234. doi: 10.1038/nrmicro2729. [DOI] [PubMed] [Google Scholar]
- Morgenstern I, Powlowski J, Tsang A. Fungal cellulose degradation by oxidative enzymes: from dysfunctional GH61 family to powerful lytic polysaccharide monooxygenase family. Briefings in Functional Genomics. 2014;13:471–481. doi: 10.1093/bfgp/elu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas AE, Mackenzie AK, Mravec J, Schuckel J, Willats WG, Eijsink VG, Pope PB. Do rumen Bacteroidetes utilize an alternative mechanism for cellulose degradation? mBio. 2014;5:e01401–01414. doi: 10.1128/mBio.01401-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Block PL, Smith DF. Culture Medium for Enterobacteria. Journal of Bacteriology. 1974:119. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Gardner JG. In-frame deletions allow functional characterization of complex cellulose degradation phenotypes in Cellvibrio japonicus. Applied and environmental microbiology. 2015 doi: 10.1128/AEM.00847-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixon JE, Ferreira LM, Durrant AJ, Laurie JI, Hazlewood GP, Gilbert HJ. Characterization of the gene celD and its encoded product 1,4-beta-D-glucan glucohydrolase D from Pseudomonas fluorescens subsp. cellulosa. The Biochemical journal. 1992;285(Pt 3):947–955. doi: 10.1042/bj2850947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A, Tauch A, Jager W, Kalinowski J, Theirbach G, Puhler A. Small mobilization multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Schellenberger S, Drake HL, Kolb S. Functionally Redundant Cellobiose-Degrading Soil Bacteria Respond Differentially to Oxygen. Applied and environmental microbiology. 2011;77:6043–6048. doi: 10.1128/AEM.00564-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin MHL, DY, Skogerson K, Wohlgemuth G, Choi I, Fiehn O, Kim KH. Global metabolic Profiling of Plant Cell Wall Polysaccharide Degradation by Saccharophagus degradans. Biotechology and Bioengineering. 2010;105:477–488. doi: 10.1002/bit.22557. [DOI] [PubMed] [Google Scholar]
- Tolonen AC, Chilaka AC, Church GM. Targeted gene inactivation in Clostridium phytofermentans shows that cellulose degradation requires the family 9 hydrolase Cphy3367. Molecular microbiology. 2009;74:1300–1313. doi: 10.1111/j.1365-2958.2009.06890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaaje-Kolsstad G, Wetereng B, Horn SJ, Liu Z, Zhaai H, Sorlie M, Eijsink VGH. An Oxidative Enzyme Boosting the Enzymatic Conversion of Recalcitrnt Polysaccharides. Science. 2012;330:219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogowski A, Zhao L, MGH, Avci U, Knox JP, Gilber HJ. Understanding How Complex Molecular Architecture of Mannan-degrading Hydrolysis Contributes to Plant Cell Wall Degradation. Journal of Biological Chemistry. 2002;289:2002–2012. doi: 10.1074/jbc.M113.527770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Znameroski EA, Coradetti ST, Roche CM, Tsai JC, Iavarone AT, Cate JH, Glass NL. Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6012–6017. doi: 10.1073/pnas.1118440109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwietering MH, Rombouts FM, van’t Riet K. Comparison of definitions of the lag phase and the exponential phase in bacterial growth. Journal of Applied Microbiology. 1992;72:139–145. doi: 10.1111/j.1365-2672.1992.tb01815.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.