Abstract
Biological membranes are heterogeneous structures with complex electrostatic profiles arising from lipids, sterols, membrane proteins, and water molecules. We investigated the effect of cholesterol and its derivative 6-ketocholestanol (6-kc) on membrane electrostatics by directly measuring the dipole electric field (F⃗d) within lipid bilayers containing cholesterol or 6-kc at concentrations of 0–40 mol% through the vibrational Stark effect (VSE). We found that adding low concentrations of cholesterol, up to ~10 mol%, increases F⃗d, while adding more cholesterol up to 40 mol% lowers F⃗d. In contrast, we measured a monotonic increase in F⃗d as 6-kc concentration increased. We propose that this membrane electric field is affected by multiple factors: the polarity of the sterol molecules, the reorientation of the phospholipid dipole due to sterol, and the impact of the sterol on hydrogen bonding with surface water. We used molecular dynamics simulations to examine the distribution of phospholipids, sterol and helix in bilayers containing these sterols. At low concentrations, we observed clustering of sterols near the vibrational probe whereas at high concentration, we observed spatial correlation between the positions of the sterol molecules. This work demonstrates how a one-atom difference in a sterol changes the physicochemical and electric field properties of the bilayer.
Graphical Abstract
Introduction
Biological membranes are complex, heterogeneous structures composed of amphiphilic lipids that form a bilayer scaffold into which small molecules and proteins can intercalate and alter the physical and chemical properties of the membrane. Of special interest to many researchers is the effect of the intercalation of cholesterol, which provides mechanical strength and reduces fluidity of the cell membrane, a characteristic that distinguishes eukaryotic from prokaryotic cells. Cholesterol can constitute as much as 50% of total molecular composition of a plasma membrane in eukaryotic cells,1 and even more than 50% in human eye lens,2 thereby significantly influencing membrane organization, dynamics, and function. Experimental studies using nuclear magnetic resonance (NMR),3–5 electron paramagnetic resonance (EPR),6–9 and differential scanning calorimetry (DSC)9–12 have repeatedly shown that cholesterol induces temperature dependent “dual effects” on the phases of the lipid bilayer; cholesterol makes the gel phase more fluid below the transition temperature (Tm) and the liquid crystalline phase more rigid above Tm The later effect is significant for the role that this is believed to play in the formation of heterogeneous, dynamic, nanoscale domains enriched in glycolipids, sphingolipids, and cholesterol, often called “lipid rafts.” These domains are thought to be crucial for cellular functions including signal transduction, cell adhesion, and membrane trafficking.1,13 However, despite development of various in vivo visualization techniques of lipid rafts, there is still a lack of consensus about their size, definition and function.14–16
Cholesterol and its derivatives such as 6-ketocholestanol (6-kc), ergosterol, and 7-dehydrocholesterol have various physicochemical effects on lipid membranes stemming from excluded volume, steric, and electrostatic factors that collectively modify bilayer structure, fluidity, and function. Of these, the sterols’ affect on the membrane dipole electric potential (Vd) is the least understood.17, 18, 19 This electric potential is perpendicular to the plane of the membrane and is the result of the anisotropic orientation of molecular dipole moments of charged moieties in the head region of the lipid distributed between the lipid-water interface and the hydrocarbon interior of the membrane.20,21 The potential generated from these accumulated dipole moments, largely from the zwitterionic lipid head groups and from water molecules that are hydrogen bound to it, propagates a few nanometers through the low dielectric hydrophobic interior of a membrane, resulting in a large electrostatic dipole electric field (F⃗d) that a variety of experimental and computational techniques have estimated to be ~1–10 MV/cm.20,21 This is significantly larger than either of the other two electrostatic fields associated with the membrane structure, the transmembrane field (F⃗trans) which arises from the difference in ion concentration across the transmembrane region, and the surface field (F⃗s) due to the potential difference between the membrane-water interface and the bulk aqueous region, with magnitudes of 0.1–1 MV/cm and 0.01 MV/cm, respectively.20,21 Because of its magnitude, the dipole potential is believed to influence many aspects of membrane biological functions such as ion-transport rates across lipid membranes;22–26 membrane fusion;27 redox reaction kinetics;28 partitioning and translocation of small macromolecules such as Na+-K+-ATPase and phospholipase A2;29–32 and insertion and folding of membrane peptides like mitochondrial amphipathic signal peptide p2530 and simian immunodeficiency viral fusion peptide.25,33
In recent years, the effect of cholesterol on membrane dipole potential has been a subject of great interest. A substantial amount of experimental and computational research has focused on this subject, but the results of these efforts have occasionally been contradictory. Several experimental19,34 and computational35–38 results show that cholesterol increases the electric potential inside a membrane, but in contrast, additional studies have concluded that cholesterol also decreases the membrane electric potential.19,39 Researchers have proposed several hypotheses: 1) cholesterol increases the dipole potential by altering the strength and orientation of dipole moments associated with lipid head groups;35 2) cholesterol increases the dipole potential by increasing the volume of lipid head group, which in turn reorganizes the presence of water dipole moments at the membrane interface;34 or 3) the dipole moment of cholesterol itself adds incrementally to the magnitude of the electric potential dropped across the bilayer.19 These hypotheses have been proposed based on the results of a variety of indirect experimental techniques such as atomic force microscopy,40 ion-transport rates,41 ratiometric fluorescence measurements,42,43 and conductance method;44 or computationally through MD simulations 36,39,45 applied on lipid bilayers with different amounts of cholesterol. In this paper, we set out to test these hypotheses by measuring the magnitude of the electric field inside a bilayer composed of a saturated phospholipid, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 0–40 mol% of cholesterol using vibrational Stark effect (VSE) spectroscopy of a diatomic nitrile molecule and molecular dynamic simulations of model membranes. We also compared the effect of cholesterol with that of 6-kc, a cholesterol analog with an extra ketone group on the second ring (Figure 3), using the same methodology.
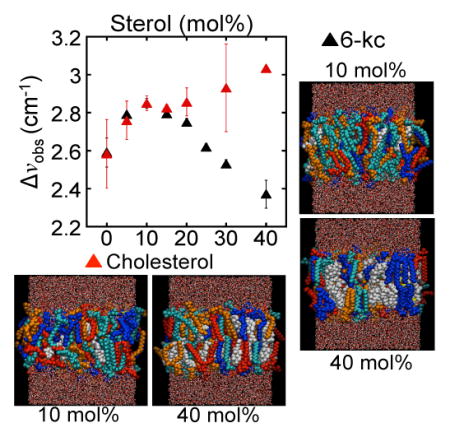
Figure 3.
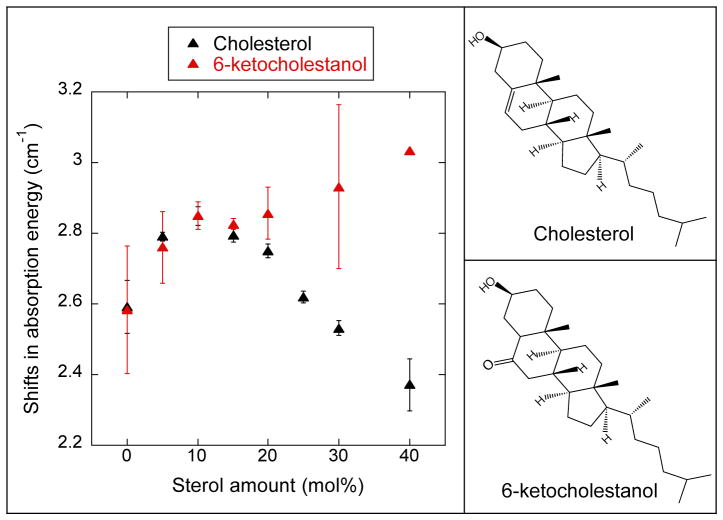
Left: The experimentally measured differences in the absorption energy (Δνobs ) of the nitrile when moved from αLAX(25) to αLAX(16) for DMPC vesicles containing varying concentrations of sterol and 1 mM peptide plotted as a function of sterol concentration. Error in Δνobs represents one standard deviation of at least three experimental measurements. Right: Chemical stuctures of the sterols considered in this manuscript, cholesterol and 6-ketocholestanol.
Despite its structural resemblance to cholesterol, experimental observations of 6-kc have consistently found it increases the membrane electric field.19,23,41,42,46 6-kc has been shown to sit further up in the bilayer compared to cholesterol because the additional oxygen forms hydrogen bonds with the interfacial waters and increases membrane fluidity (compared to membrane with similar composition of cholesterol).46 This is in contrast with cholesterol, which sits deeper in the bilayer under lipid head group “umbrellas,” making the bilayer less fluid. By measuring the electrostatic field in lipid bilayers containing these two highly similar sterols, we aim to elucidate and quantify how sterols affect membrane electrostatics by changes in local membrane structure and organization.
We do this using vibrational Stark effect (VSE) spectroscopy, in which a shift in the vibrational absorption frequency (Δνobs) of an oscillator probe is altered by changes in the local electrostatic environment in the vicinity of that probe.47–49 Spectral shifts of the oscillator, Δνobs, are due to the interaction of the field (F⃗d) with a known difference in dipole moment of the vibration (Δμ⃗), also known as Stark tuning rate. Equation 1 shows the relationship between the change in the absorption energy and the electrostatic field.
| (1) |
where h is Planck’s constant, θ is the angle between the field vector and the direction of the vibrational probe, and c is the speed of light. The brackets 〈…〉 denote an ensemble average. We choose our reference state to be at the center of the membrane. For the symmetric membranes used in these simulations, identical lipid compositions and ion concentrations in both leaflets results in an electric field of zero at the center of the membrane. Even in the presence of a transmembrane helix, which is not symmetric, the electric field at the center of the membrane is found to be low in the MD simulations, as we illustrate in the Results section. Hence in practice, the membrane electric field can be determined by the simplified expression in which the reference field at the membrane center is neglected: Δνobs = −|Δμ|〈|Fd|cos(θ)〉. The last expression is not a direct measurement of the electric field since it depends on cos (θ). However, it is possible to separate the averages of the field and the cosine of the angle if the two functions are uncorrelated, i.e. when 〈Fd · cos(θ)〉 ≅ 〈Fd〉 · 〈cos(θ)〉. As we show in the Results section, the simulations suggest that the two are indeed uncorrelated. It is thus possible to relate a measured change in absorption energy of the chromophore to the electric field at the probe, which is induced by the local environment.
In addition to experimental measurements, we computed the component of the electric field normal to the membrane plane from the simulation data. Sampled configurations from the Molecular Dynamics trajectories are used to calculate the charge densities that in turn are placed in Equation (2). We averaged over all the charges in the membrane plane to obtain the charge density, ρ(z):
| (2) |
where l denotes the boundary of the periodic box, and F(z) is the electric field at membrane depth z.
In this paper, we use VSE spectroscopy to measure the environment field inside a lipid bilayer containing different amounts of sterol molecules using a combined experimental-computational methodology that was developed and described previously.50,51 In our earlier work with 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) vesicles, we placed a nitrile oscillator into the bilayer interior by intercalating an α-helical transmembrane peptide containing an unnatural amino acid, p-cyanophenylalanine (p-CN-Phe).51 The nitrile molecule served as an excellent VSE probe, as is described extensively in earlier publications.48–52 We controlled the location of nitrile inside the bilayer by changing the position of p-CN-Phe along the amino acid sequence of the peptide. The four polypeptide sequences that we used in this study are given in Table 1.50,51 The peptide αLAX(25) places the nitrile near the lipid head group-water interface, where it was exposed to a very heterogeneous chemical environment containing charged groups and polar water molecules. At the other extreme, peptide αLAX(16) places the nitrile in a homogenous environment near the middle of the membrane that is mainly comprised of hydrocarbon chains. We confirmed the chemical differences in the local vicinity of these probes both experimentally (through differences in vibrational absorption line widths), and computationally (through MD simulations) and determined the magnitude of the dipole electric field in a DMPC bilayer to be 8 – 11 MV/cm, depending on a variety of factors such as vesicle size, probe concentration, and nitrile orientation.51
Table 1.
Amino acid sequences of the four polypeptides used for VSE studies in this work.
| Peptide | Sequence |
|---|---|
| αLAX(25) | HHGGPGLALALALALALALALALAXGPGGHH |
| αLAX(23) | HHGGPGLALALALALALALALAXALGPGGHH |
| αLAX(21) | HHGGPGLALALALALALALAXALALGPGGHH |
| αLAX(16) | HHGGPGLALALALALXLALALALALGPGGHH |
X = p-cyanophenylalanine
Here, we expand the use of VSE spectroscopy and MD simulation studies to understand the role of chemical complexity on the magnitude and function of the membrane electric field through direct measurements of the field upon addition of cholesterol and 6-kc at physiologically relevant concentrations up to 40 mol%. Experimental measurements show that cholesterol increases the electric field at lower concentrations (~10 mol%) and decreases it at higher concentrations (up to 40 mol%), while 6-kc increases the electric field monotonically through the range we investigated. We use molecular dynamics simulations to examine the lateral organization of these sterols within lipid-αLAX(16) bilayer at the sterol concentrations of 10, 20 and 40 mol%. We also evaluate the orientations of the helix and of the nitrile probe attached to the helix from the simulations. Using our experimental data and results from simulation studies, we demonstrate that both the chemical structure and the concentration of sterol affect the chemical environment of the bilayer, which in turn alters the membrane electric field in complex ways.
Materials and Methods
Materials
1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) powder was purchased from Avanti Polar Lipids, Inc. (Alabaster, AL) and was used without further purification. Cholesterol, 6-ketocholestanol, Hepes, NaN3, and D2O were purchased from Sigma Aldrich (St. Louis, MO). PrCl3.6H2O was purchased from Alfa Aesar (Ward Hill, MA). HPLC grade water and chloroform were purchased from Fisher Scientific (Pittsburg, PA). All peptides were synthesized using standard Fmoc solid-state peptide synthesis and obtained from Abgent Technologies (San Diego, CA) and InnoPep (San Diego, CA).
Sample preparation
Small unilamellar vesicles (SUV) were prepared using sonication as described in reference 53.53 For samples containing cholesterol, about 30 mg of DMPC powder and a correct amount of cholesterol powder and lyophilized peptide (determined as described below) were taken together in a vial and dissolved in approximately 1 mL of chloroform. For samples containing 6-kc, a stock solution of 6-kc in methanol was made, from which the correct volume was added to the lipid and peptide solution in chloroform. The sample was then vortexed for 5 min, dried under vacuum for 2 hr, then transferred into a N2(g)-purged glove box overnight. The dried sample was then hydrated with 1.5 mL of 10 mM Hepes buffer with 0.02% (w/v) NaN3, pH 7.2, and maintained at a temperature above 23°C, the gel-liquid crystal transition temperature (Tm ) of DMPC. This was vortex mixed for 5 min to get a homogeneous distribution of multilamellar vesicles. Small unilamellar vesicles were obtained by placing the multilamellar vesicle suspension in a sonication bath maintained at 35°C until the milky solution (indicative of multilamellar vesicles) cleared to slightly cloudy (indicative of light scattering by residual large particles remaining in the lipid suspension). These residuals were removed by centrifugation at 12000×g for approximately one hour to achieve a clear vesicle solution. The vesicle size distribution of each batch was determined using dynamic light scattering technique on a Malvern Zetasizer Nano ZS instrument equipped with He-Ne light source (633 nm) and photodiode detector. The average vesicle diameter determined from all batches of vesicle samples was determined to be ~73 nm with standard deviation of 35 nm. All samples were stored at a temperature above Tc and were stable for up to 5 days.
Sterols were added at concentrations of 0, 5, 10, 15, 20, 30 and 40 mol% and peptides were added to make a total concentration of 1 mM. On average, in a homogeneous distribution of 1 mM peptide results in a ratio of 33 peptides per 1000 lipids. The secondary structure of the peptides inside the vesicles was determined by circular dichroic (CD) spectroscopy using Jasco J-815 CD spectrometer. CD spectra were recorded using a 1 mm path length quartz cell over the range of 190 – 250 nm wavelength at 0.2 nm resolution, 50 nm/min scanning rate and 4 s response time and were background subtracted using 10 mM Hepes buffer as the background with Spectra Manager for Windows 95/NT Spectra Analysis software.
Infrared Spectroscopy
Infrared spectra of vesicle samples containing the VSE probe were recorded at room temperature (>Tm ) in a sample cell composed of two sapphire windows separated by 125 μm thick PETE spacers in a Bruker Vertex 70 FTIR instrument. The sample cell was illuminated with light in the range of 2000–2500 cm−1 selected by a broad band pass filter (Spectrogon, Parsippany, NJ) placed in front of the instrument’s IR source. Spectra were collected with a liquid nitrogen-cooled indium antimide (InSb) detector and were composed of 3000 scans at 2.0 cm−1 spectral resolution. Background-subtracted spectra were fit to a single Gaussian line shape with a custom least-squares fitting program to determine the peak center, obs, and full width at half maximum (fwhm). Uncertainty in absorption energy is reported as the standard deviation of at least three measurements.
Nuclear Magnetic Resonance Spectroscopy
The splitting and shifting of 31P NMR spectra were used to characterize vesicles as unilamellar, but also demonstrated the fluidity of bilayers containing 6-kc. Samples for NMR spectroscopy were prepared by adding 150 μL of D2O to 550 μL of vesicle sample in an NMR tube. 31P NMR spectra were obtained at 35 °C with 202.343 MHz on a Varian INOVA-500 NMR. For fluidity experiments, approximately 2.4 mg of PrCl3.6H2O was added to the NMR sample.
Molecular Dynamics Simulations
We computed molecular dynamics trajectories for the αLAX(16) at different compositions for the sterols in a bilayer of DMPC molecules. Three trajectories were computed for both cholesterol and 6-kc (Table 2). The total number of DMPC and sterol molecules was kept at 160 molecules in all the simulations.
Table 2.
Composition details of the simulations performed for cholesterol and 6-ketocholestanol.
| Sterol composition | # of DMPC | # of sterol | # of water | Total # atoms |
|---|---|---|---|---|
| 10 % | 144 | 16 | 7150 | 28785 |
| 20 % | 128 | 32 | 6909 | 27790 |
| 40 % | 96 | 64 | 6355 | 21362 |
We used the CHARMM-GUI54,55 membrane builder facility to build a starting configuration of the membrane system with the helical peptide initially embedded inside the membrane, along the membrane axis. Initially we prepared the three cholesterol/DMPC systems with the membrane builder tool and used these configurations to build the three 6-kc/DMPC systems by adding the carbonyl bond in the corresponding sterol ring. The simulations were performed with the molecular simulation code MOIL56,57 using a combination of the united-atom Berger force field for the acyl chain atoms of the lipid molecules and the OPLS force field for their head group region and the peptide atoms.58,59 For cholesterol we used parameters developed by Pasenkiewicz-Gierula et al.,60 for 6-kc we used the OPLS ketone charges, and for the nitrile bond of the probe we used parameters from Price et al.61 For water we used the SPC water model.62 By default the MOIL program adds improper torsions to the force field of the sterols. The improper torsions prevent chirality inversion at the C14 centers that could have been a concern in extended atom force field.74
After the initial setup, we heated the system to a target temperature of 298 K for a period of 2 ns and then performed 1 ns of equilibration at a constant pressure of 1 atm using a recently developed stochastic algorithm to sample the isobaric-isothermal ensemble.63 Production runs of 200 ns were performed in the NVT ensemble with the last 100 ns of simulation data used for analysis.
Periodic boundary conditions were imposed in the three spatial directions and the smooth particle mesh Ewald method64 was used for long-range electrostatic interactions with grid resolution of 1 mesh-point/Å. The cutoffs for van der Waals interactions and for the real space component of electrostatic forces were set to 9.5 Å. In all the simulations we constrained water bond length and angles with a matrix implementation of the SHAKE algorithm.65 The simulations were done with multiple time steps according to r-RESPA,66 with 1 fs time step to integrate the covalent, van der Waals, and real-space component of the Ewald sum, and a 4 fs time step to evaluate the reciprocal-space component of the Ewald sum. Configurations were saved every 1 ps for determination of the structural properties of the membrane systems.
The convergence of the simulations can be assessed by examining the ensemble averaged properties of functions of coarse variables. In the present manuscript the experiments focus on the probe, which is attached to the transmembrane helix. The convergence of the distribution of the helix tilt angle is relevant to the research presented below. Furthermore, the motions of the helix are slow variables and offer a strict convergence criterion. The orientation of the helix was examined using two different definitions: (i) The atomic positions of the Ca of histidine H1 and H31 were used to estimate the helix axis orientation and (ii) All the alpha carbons of the helix were overlapped with the initial helix configuration (that was oriented perpendicular to the membrane surface) with the Kabsch algorithm67 to determine tilt shift. Both approaches yield similar distributions.
Finally, we also performed a 150 ns simulation of a pure lipid bilayer containing 160 DMPC molecules for comparison to the results of the helix/sterol/membrane systems.
Results
Nuclear magnetic resonance spectroscopy
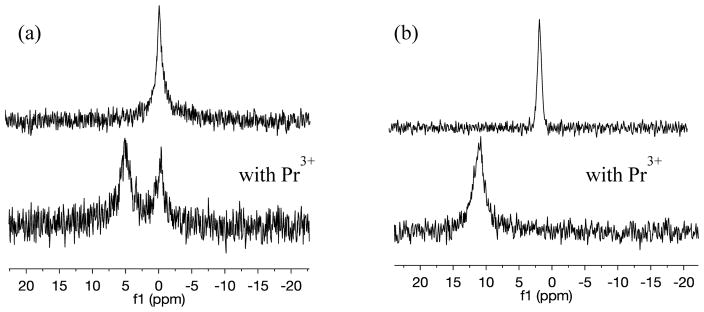
31P NMR has been used extensively in the literature to characterize the vesicle size and lamellarity. In 31P NMR spectroscopy, a “wide-line” or powder-line shape due to the restricted anisotropic motion is indicative of multilamellar vesicles whereas a single well-resolved peak at about 0.1 ppm is representative of unilamellar vesicles.68,69 In addition to providing qualitative information on the physical characteristics of the vesicles, 31P NMR experiments are also used to check the fluidity of the bilayer by adding external shift reagents. Paramagnetic lanthanide ions such as the praseodymium cation (Pr3+) have very short electron relaxation times and upon interaction with the phosphate ions in the lipid head group, perturb the nuclear spin relaxation times and move the chemical shift downfield.68,69 Typically, pure lipid bilayers are impermeable to lanthanide cations, hence Pr3+ only shifts the signal from the outer leaflet downfield, splitting the single narrow peak into two whereas the peak from the phospholipids in the inner leaflet remains unchanged. However, if the permeability of the bilayer increases upon intercalation of small sterols such as 6-kc, Pr3+ ions penetrate into the bilayer and interact with the lipid head groups in the inner leaflet as well. As a result the entire peak shifts downfield.
Figure 1(a) and Figure 1(b) show the representative 31P NMR spectra collected for 30 mM DMPC lipid vesicles only and vesicles with 20 mol% 6-kc respectively. In both figures, the top spectra represent vesicles in absence of external shift reagent and the bottom spectra represent vesicles with Pr3+ ions added. We obtained a single narrow peak characteristic of unilamellar vesicles before adding any shift reagent for both vesicles (top panels of Figure 1). When Pr3+ was added to vesicles composed of DMPC only, we observed the characteristic splitting of the phosphorus peak (Figure 1(a)). However, when Pr3+ was added to vesicles containing 20 mol% 6-kc, the entire peak was shifted downfield, shown in Figure 1(b). After confirming the vesicles were still intact through dynamic light scattering, we concluded that Pr3+ was able to permeate the bilayer when 6-kc was intercalated in the membrane, resulting in the entire peak shifting downfield to about 10 ppm. Pr3+ ions were interacting with phosphate groups in both outer and inner leaflet of the bilayer instead of splitting of the 31P NMR peak seen in empty DMPC vesicles. Based on this and previous experiments reported in the literature, we concluded that the presence of 6-kc in the lipid bilayer results in an increase in the bilayer permeability, which allowed for the passive translocation of Pr3+ through the bilayer to interact with the head group region of both leaflets of the membrane.
Figure 1.
Representative 31P NMR spectra of (a) 30 mM DMPC vesicles and (b) 30 mM DMPC and 20 mol% 6-kc (upper) and with addition of PrCl3.6H2O (lower). In pure DMPC vesicles, Pr3+splits the phosphorus signal into two, representative of inner and outer leaflets, while in vesicles containing 6-kc, it shifts the entire signal downfield indicating that it has permeated the bilayer.
Circular dichroic spectroscopy
In these experiments, we moved the nitrile infrared probe through the lipid bilayer by inserting a polypeptide containing the unnaturnal amino acid p-cyanophenylalanine at various locations along the sequence. The repeating leucine-alanine (LA) construct in the amino acid sequences shown in Table 1 is a strongly hydrophobic helical peptide that is insoluble in buffer but partitions into the self-assembling membrane bilayer during vesicle formation.50,51 We confirmed the helical secondary structure of these nitrile-containing peptides inside vesicles containing 0–40 mol% cholesterol and 6-kc with CD spectroscopy, shown in Figure S1 (Supporting Information). All peptides show two minima located near 208 and 222 nm, which are characteristics of helical secondary structure. On average, a homogeneous distribution of 1 mM peptide results in a ratio of 33 peptides per 1000 lipids. Although long range electrostatic interactions between peptides are possible at such concentrations; we did not observe any distortions in the CD spectra demonstrating that peptides did not aggregate within the bilayer at this low concentration.
FTIR spectroscopy
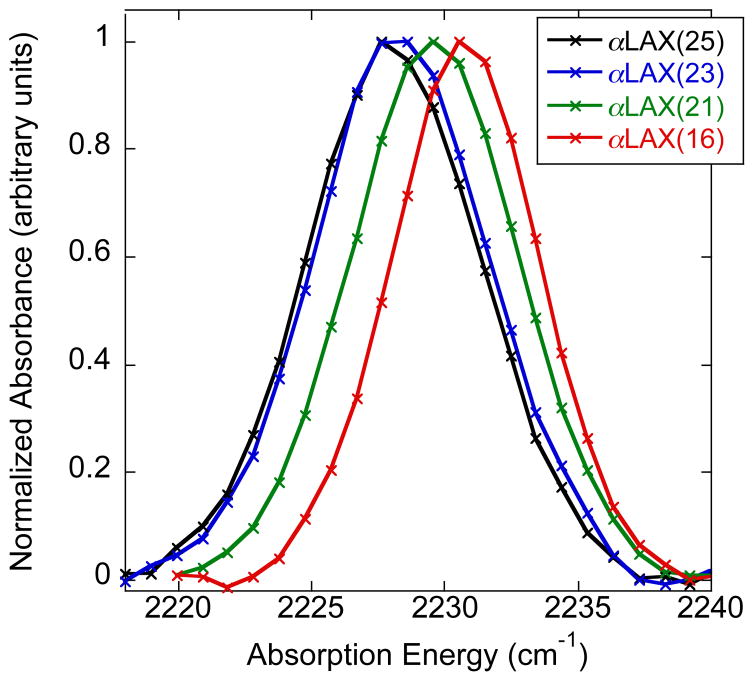
In our FTIR experiments, we used α-helical peptides containing a single p-CN-Phe unnatural amino acid to incorporate the nitrile oscillator at four different positions within the bilayer, beginning from the membrane-water interface with peptide αLAX(25) and progressing towards the middle of hydrophobic core with peptide αLAX(16). The potential gradient between the lipid’s charged head group and terminal alkyl chains creates an electric field that shifts the vibrational absorption energy of the nitrile oscillator between αLAX(25) and αLAX(16).20,21 In Figure 2, we show representative normalized infrared spectra of the nitrile stretching band in αLAX(25), αLAX(23), αLAX(21), and αLAX(16) placed into vesicles composed of 30 mM DMPC and 20 mol% cholesterol. In the figure, the absorption energy of the nitrile shifted by 2.7 cm−1 between αLAX(25) and αLAX(16). We carried out a series of FTIR measurements in vesicles containing 0 to 40 mol% cholesterol or 6-kc. We determined the vibrational energy shifts (Δνobs) at each composition and the results are provided in Table 3 and plotted as a function of sterol concentration in Figure 3 for both cholesterol (black) and 6-kc (red). We see two distinct trends in values as a function of sterol mole fraction in the bilayer. As we increased the concentration of cholesterol from 0 to 10 mol% in our vesicles, the Δνobs, increased from 2.59 cm−1 to 2.85 cm−1 but as we increased beyond 10 mol%, the Δνobs declined to as low as 2.37 cm−1 for 40 mol% cholesterol. In contrast, in vesicles containing 6-kc, the Δνobs increased monotonically with increasing concentration over the entire range examined.
Figure 2.
Normalized representative FTIR spectra of 1 mM peptides αLAX(25) (black), αLAX(23) (blue), αLAX(21) (green), and αLAX(16) (red) inserted in vesicles composed of 30 mM DMPC and 20 mol % cholesterol. Experimental data points are shown with “×”.
Table 3.
Experimentally measured differences in nitrile’s absorption energy (Δνobs ) when moved from αLAX(25) to αLAX(16) for DMPC vesicles containing varying concentrations of sterol and 1 mM peptide. Error in Δνobs represents one standard deviation of at least three experimental measurements.
| Sterol (mol %) | Cholesterol-DMPC | 6-ketocholestanol-DMPC |
|---|---|---|
| Δνobs (cm−1) | Δνobs (cm−1) | |
| 0 | 2.59 ± 0.08 | 2.6 ± 0.2 |
| 5 | 2.79 ± 0.01 | 2.8 ± 0.1 |
| 10 | 2.85 ± 0.03 | 2.85 ± 0.04 |
| 15 | 2.80 ± 0.02 | 2.83 ± 0.01 |
| 20 | 2.75 ± 0.02 | 2.86 ± 0.07 |
| 25 | 2.62 ± 0.02 | |
| 30 | 2.53 ± 0.02 | 2.9 ± 0.2 |
| 40 | 2.37 ± 0.07 | 3.03 |
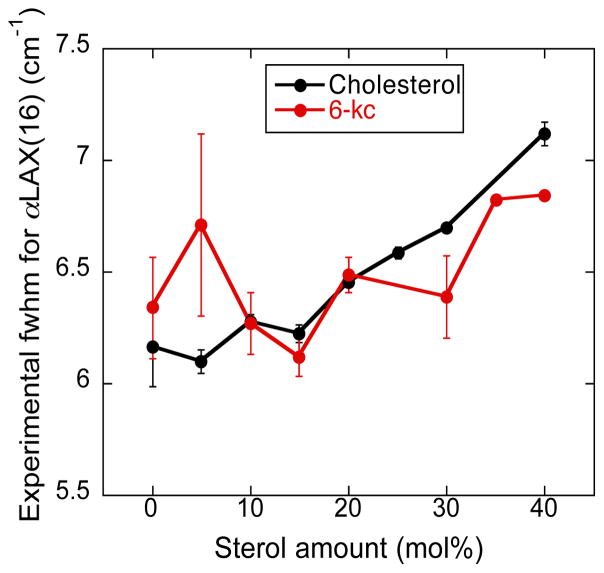
The addition of sterol molecules to a lipid bilayer increases heterogeneity of the system due to several factors, including by changing the order of the lipid alkyl chains. The nitrile probe on peptide αLAX(16) places the oscillator at the ends of these alkyl tails where the system is most perturbed. The oscillator is very sensitive to the local chemical environment, which is reflected in the full width half maximum (fwhm) values of its absorption peak. In Figure 4, we plot the experimental average fwhm values for the absorption peaks of nitrile attached to αLAX(16) that we placed in vesicles containing 0–40 mol% of each sterol. For cholesterol containing bilayers, the fwhm values of nitrile peaks, shown in black circles, rises monotonically with higher cholesterol concentration while the fwhm values of nitrile peaks in 6-kc containing bilayers, shown in red circles, do not exhibit any clear trend as a function of concentration, and appear to fluctuate around a value of ~6.5 cm−1. This result indicates that the hydrophobic core region of the bilayer, where the nitrile in αLAX(16) is placed, gets progressively more diverse in its chemical environment as the concentration of cholesterol is increased, but remains homogeneous as more 6-kc is added.
Figure 4.
Average full width half maximum values for vibrational absorption peaks of nitrile placed inside sterol-lipid-αLAX(16) bilayer for two different kinds of sterol, cholesterol (black) and 6-kc (red), at different concentrations. The values were average of at least three measurements.
Molecular dynamics simulations
Helix orientation
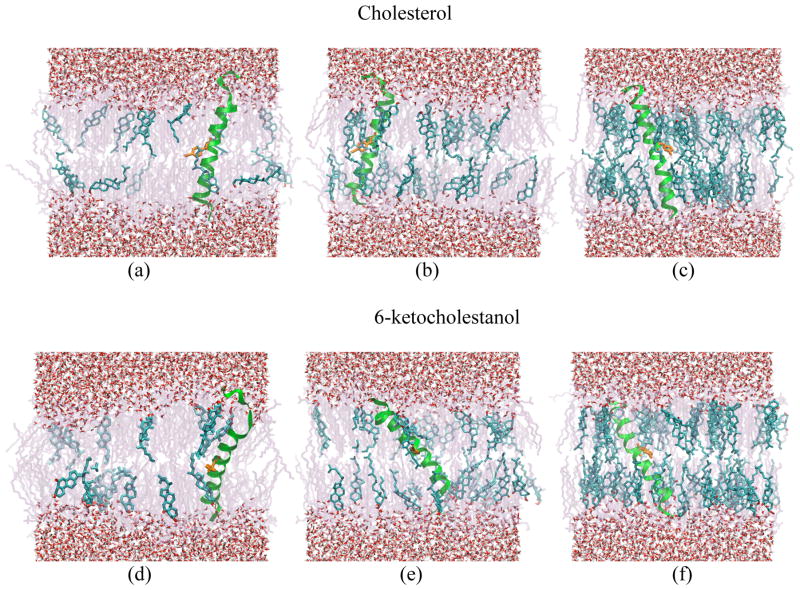
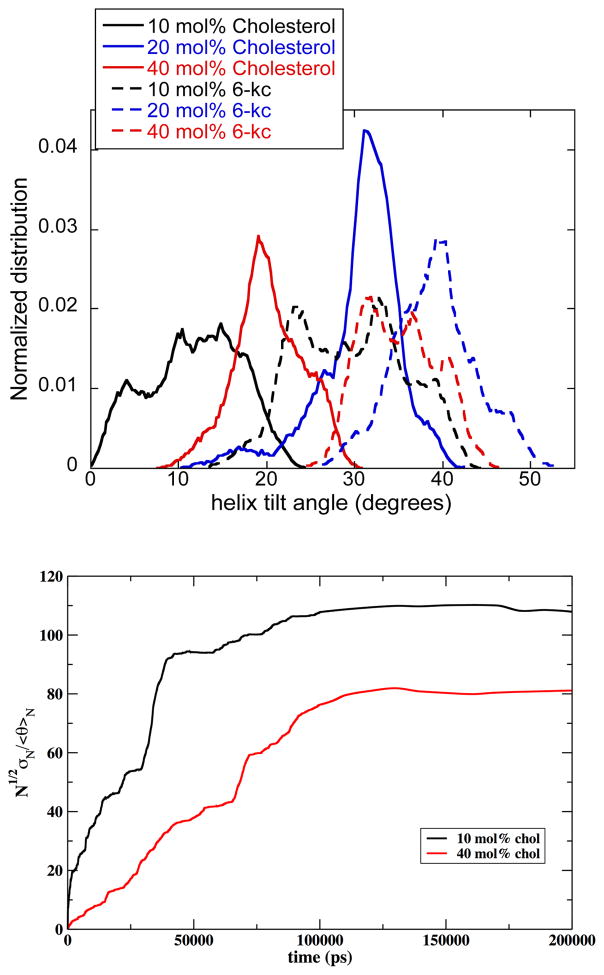
Although the peptide sequence should put the terminal glycine repeats in the head group region of the bilayer, and although the length of the peptide sequence (28.5 Å)50,51 was designed to be similar to the hydrophobic length of pure DMPC phospholipid bilayer (26 Å),70 our experimental methods provide no independent verification that the expectation of transmembrane insertion has been met. The set of CD spectra collected for αLAX(25), αLAX(23), αLAX(21) and αLAX(16) inserted into sterol-lipid bilayer shown in Figure S1 demonstrate the secondary structure of the peptide but they do not provide information on the orientation of the peptide (horizontal versus vertical) with respect to the bilayer normal. The helix orientation is affected by various physicochemical properties of the lipid bilayer for example the hydrophobic mismatch between the helical section of the peptide and the low dielectric hydrophobic alkyl chains, and the chemical interaction between the peptide and its neighboring membrane component. In Figure 5, we show representative snapshots taken from the equilibrated helix-membrane simulation of (a) 10 mol% cholesterol; (b) 20 mol% cholesterol; (c) 40 mol% cholesterol; (d) 10 mol% 6-kc; (e) 20 mol% 6-kc; and (f) 40 mol% 6-kc, run for a total of 200 ns each. For the results reported here, all analyses were made from the last 100 ns of simulation data to allow the system to equilibrate. These snapshots clearly show that the peptides are indeed helical and inserted parallel to the membrane normal with some degree of tilt. We calculated the distribution of helix tilt angles from the simulation trajectories for each bilayer composition, and results are shown in Figure 6. For cholesterol containing bilayers, the width of the helix tilt distribution became narrower with higher mole fraction of cholesterol whereas for 6-kc containing bilayers, the distribution widths were broader in general at all mole fractions of 6-kc. For each composition under investigation, the mean helix tilt angles were determined to be 12 ± 5°, 31 ± 5° and 20 ± 4° for 10, 20 and 40 mol% cholesterol, respectively, and 30 ± 6°, 39 ± 5° and 35 ± 4° for 10, 20 and 40 mol% 6-kc, respectively. To check the convergence of the calculation we also present computations of an ergodic measure for the tilt angle in Figure 6.
Figure 5.
Representative snapshots taken from molecular dynamic simulations of αLAX(16) aligned vertically inside a lipid bilayer composed of DMPC molecules and (a) 10 mol% cholesterol; (b) 20 mol% cholesterol; (c) 40 mol% cholesterol; (d) 10 mol% 6-kc; (e) 20 mol% 6-kc; and (f) 40 mol% 6-kc. SPC water molecules are shown in red and gray, sterol molecules are shown in cyan, the p-CN- probe is shown in orange and DMPC phospholipids are shown in light purple. Molecular snapshots were prepared with the program VMD.71
Figure 6.
Top: Normalized distribution of the helix tilt of αLAX(16) with respect to the membrane normal obtained from MD simulation of the peptide embedded into bilayer composed of DMPC and cholesterol (solid line) and DMPC and 6-ketocholestanol (dashed line) at concentrations of 10 mol% (black), 20 mol% (blue) and 40 mol% (red). Bottom: Ergodic measure calculations as a function of time. We illustrate that the standard deviation of the averaged tilt angle divided by the average of the tilt angle is proportional to N1/2 . This observation suggests that the average does not drift and is consistent with uniform sampling from the normal distribution (or Central Limit Theorem). Here the vector connecting histidine 1 and histidine 31 defines the orientation of the helix. Very similar results were obtained when all the alpha carbons of the helix were considered and the helix was overlapped with the initial configuration to determine the tilt angle. See text for more details. We have used this measure in the past for membrane simulations.72
Nitrile probe orientation
We also determined orientations of the nitrile inside the bilayer as the angle between the vector along the bond and the normal to the membrane. In our simulations, the nitrile on peptide αLAX(16) is located in the middle of hydrophobic core of the lipid bilayer, so any changes in the ordering of lipid alkyl chains induced by different amounts of sterol affect the orientation of the nitrile. Figure 7 shows the normalized distribution of this angle simulated for αLAX(16) in bilayers containing 10, 20 and 40 mol% sterol concentration. For cholesterol containing bilayers, we obtained unimodal distributions of nitrile angle with respect to the membrane normal for 10 and 40 mol% cholesterol with mean angles of 113 ± 13° and 125 ± 10° respectively. For 20 mol% cholesterol, we observed one prominent distribution of nitrile angle centered around 120° and another small yet distinguishable distribution centered around 85°. In the case of bilayers containing different mole fractions of 6-kc, the nitrile orientation remained unimodal at all sterol concentrations, with mean values of 90 ± 16°, 76 ± 16°, and 110 ± 10° for 10, 20 and 40% 6-kc respectively.
Figure 7.
Normalized distribution of all possible orientations that nitrile oscillator samples with respect to the membrane normal in MD simulations of αLAX(16) peptide embedded into bilayer composed of (a) DMPC and cholesterol (solid line), and (b) DMPC and 6-ketocholestanol (dashed line) at concentrations of 10 mol% (black), 20 mol% (blue) and 40 mol% (red). Note that the orientation of nitrile group does not necessarily correlate with the helix tilt.
Lateral organization of membrane components
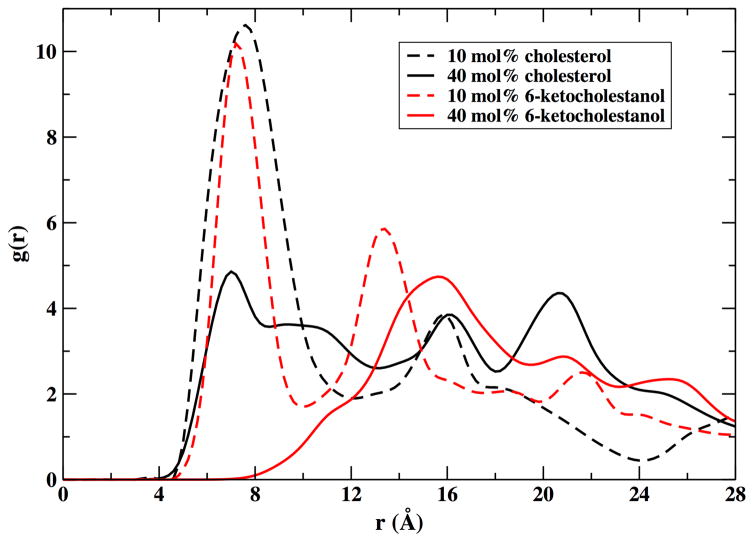
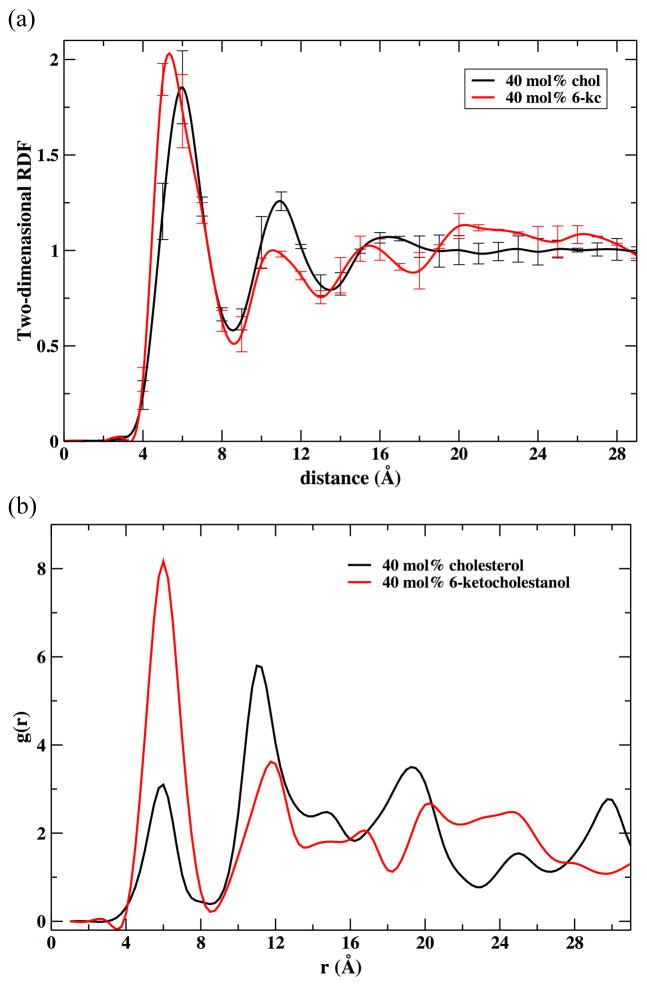
Molecular dynamics simulations allowed us to examine the lateral organization of membrane components including peptide, phospholipids and sterol molecules in our model sterol-lipid-αLAX(16) bilayer. In our computational model, we inserted one αLAX(16) helical peptide at each composition under investigation that are described in Table 2. We computed the three dimensional radial distribution function, g(r) , of the center of mass of the sterol molecules with respect to the nitrile probe attached to the helix for bilayers containing 10 mol% and 40 mol% sterol. These results are shown in Figure 8.
Figure 8.
The three-dimensional radial distribution function, g(r), between the position of the probe in the helix αLAX(16) and cholesterol (black), and αLAX(16) and 6-kc (red) calculated from the MD simulation of sterol-lipid- αLAX(16) containing 10 mol% sterol (dashed lines) and 40 mol% sterol (solid lines).
The pair correlation functions, shown in Figure 8, report the proximity of the probe and the sterol molecules. The following observations were made: (i) At 10% content of sterol we found a first density peak at about 7–8 Angstrom, indicating that the sterol molecules are near the probe. The first peak of 6-kc was slightly shifted to shorter distances. We also found a second significant peak for the 6-kc molecules at about 13 Angstrom. The second peak of the nitrile-cholesterol distribution was shifted to 16 Angstrom and was less pronounced. The reduction in the second peak of the 10% cholesterol was geometrical and is due to the use of three-dimensional distributions. (ii) At high concentrations of sterols, 6-kc was shifting away from the probe while cholesterol molecules remained closer to the helix. The cholesterol molecules at the higher concentrations are distributed more uniformly in the membrane but the absolute number of sterol molecules in the neighborhood of the probe (the first density peak) was larger than the number determined at the low concentrations.
We also computed sterol-sterol radial distribution function (RDF) for sterol-lipid-αLAX(16) bilayers containing 40 mol% cholesterol and 40 mol% 6-kc. In a lipid bilayer containing 40 mol% sterol, we distributed equally 64 total sterol molecules (shown in Table 2) among the outer and inner leaflets of the bilayer. For each layer we computed the distance between the center of mass of a reference sterol and the other 31 sterols in the same leaflet. We performed a similar calculation for the rest of the sterols in the bilayer. Figure 9 shows the results of the radial distribution analysis for both cholesterol and 6-ketocholestanol. We consider the distributions averaged over all sterol molecules in the leaflet (top) and the largest clusters only (bottom). The peak profile in cholesterol-cholesterol RDF plot (top, shown in black) was slightly different from the peak profile obtained for 6-kc-6-kc RDF plot (shown in red). The RDFs of the largest clusters (Figure 9(b)) had significant second and even third peaks. The most obvious observation of Figure 9(b) was the higher first peak of 6-kc compared to the cholesterol molecules, suggesting that 6-kc has stronger tendency to form short-range clusters. Taken together, the RDF results shown in Figures 8 and 9 indicate that that 6-kc is attracted to itself more than it is attracted to the probe. Cholesterol shows the opposite behavior; interactions with other cholesterol molecules are weaker compared to those with the helix. This may be due to the relative position of the two sterols. 6-ketocholestanol is placed higher in the membrane, a position with larger mass density.
Figure 9.
The sterol-sterol radial distribution function in the membrane plane. Top: The distribution is computed for all sterol molecules of sterol-lipid-aLAX(16) bilayers containing 40 mol% cholesterol (black) and 40 mol% 6-kc (red). Bottom: The distribution is computed for the largest clusters of sterol molecules. The largest cluster size for cholesterol molecules was 3, and for 6-kc it was 4. The number of molecules included in the first peak of the RDF determines the cluster size.
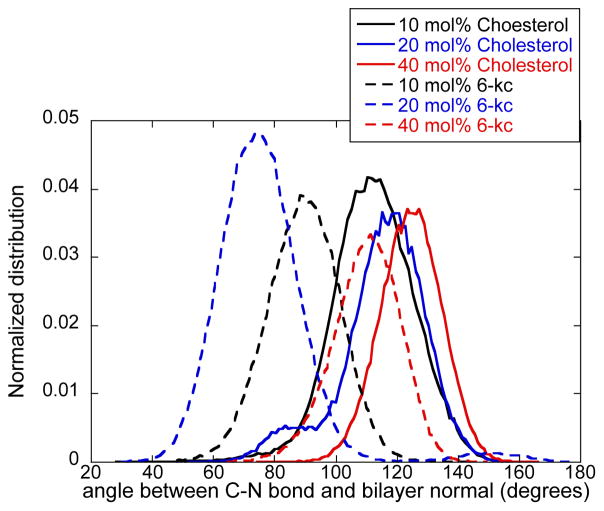
Pair correlation functions in the liquid phase generally approach rapidly asymptotic constant value as a function of distance. They are typically flat after the first or second peaks. Here, however, we observed more structure beyond the smallest clusters of molecules in direct contact to include sterols separated by a small number of lipid molecules. When the distribution was averaged over all cholesterol molecules a more homogeneous picture was obtained. Finally, we finally comment that the presence of the sterol molecules, (with the exception of 40% 6-ketocholestanol), does not change the orientation of the phospholipid dipoles significantly, measured by the P-N vector with respect to the membrane normal. This is illustrated in Figure S2 (Supporting Information).
Hydrogen bond network
The membrane-water interface is extremely heterogeneous, due in large part to a network of hydrogen bonds between phospholipid head groups, water molecules and sterols. As sterol concentration changes, the distribution of these hydrogen bonds will change as well, altering the arrangement of dipoles at the membrane-water interface. We calculated the average number of hydrogen bonds between different hydrogen bond donor and acceptor chemical pairs in the sterol-lipid-αLAX(16) bilayer model. In Figure 10, we present the results for phospholipid:water (black); phospholipid:phospholipid (blue); phospholipid:sterol (green); sterol:sterol (red); and sterol:water (magenta) calculated from MD simulations of lipid bilayer containing cholesterol (solid circles) or 6-kc (open circles) plotted as a function of sterol concentration. Figure 10 highlights two important observations: (1) the number of hydrogen bonds between phospholipids and water were largest compared to other molecules because of the greater number of hydrogen bond donors and acceptors located at the phospholipid head group; and (2) the only significant difference between cholesterol and 6-kc in this analysis was the extent of hydrogen bonding between the sterol and the water (data shown in magenta). Because of the extra ketone functional group on 6-kc, this sterol is susceptible to significantly more hydrogen bonds to water than cholesterol (1 versus 0.6 hydrogen bonds per sterol, respectively) at all concentrations. This observation is significant because water dipoles are thought to be the major contributor to the magnitude of the electric field. We will return to this observation below.
Figure 10.
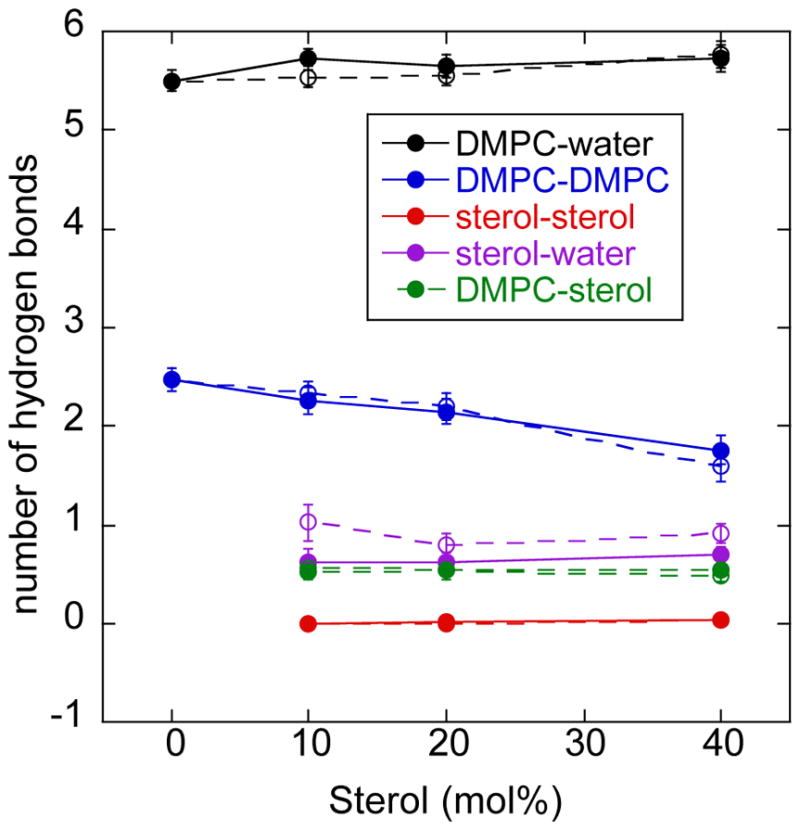
Molecular interactions in lipid bilayer containing cholesterol (solid lines) and 6-kc (dashed lines) at different concentrations. The average number of hydrogen bonds (per lipid) calculated between phospholipid:water (black); phospholipid:sterol (green); sterol:sterol (red); and sterol:water (purple). We used a distance cutoff of 2.4 Å between the donor oxygen and acceptor hydrogen atoms to define a hydrogen bond. We also consider the formation of salt bridges between phospholipid molecules. In the blue line we counted the number of salt bridges formed between choline and phosphate of two different phospholipid molecules. A salt bridge is assumed to form when the distance between a non-ester oxygen of the phosphate and the carbon of the choline was less than 4 Angstroms.
Electric field
Computer simulations
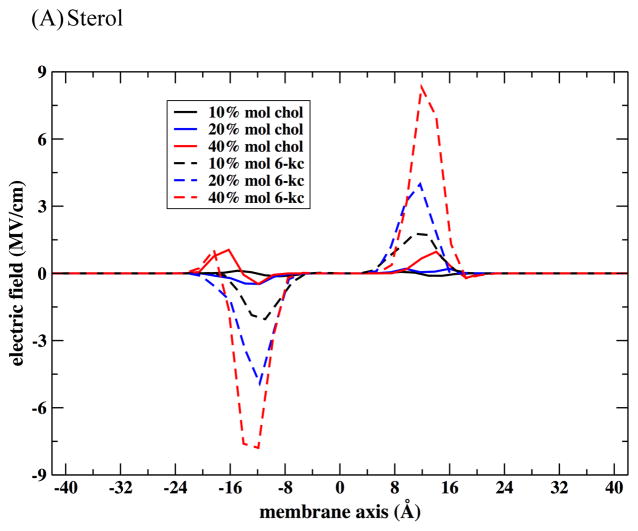
We used the configurations sampled in MD simulations to compute the electric field in the direction normal to the membrane plane following Equation (2). In Figure 11 we present the electric field computed at several conditions. We also evaluated the contributions to the electric field of different membrane components.
Figure 11.
The contributions of the different membrane components to the electric field across the membrane. Note the different scales of the electric fields at different plots. The dipole moment of the phospholipid shows the largest deviation between the highest concentration of sterol (40%) and the lowest concentration (10%). The water contribution is smaller and has little contributions of the variations between cholesterol and 6-kc. The sterol themselves induce significant variations between their corresponding fields. Note also the exceptionally low electric field at the membrane center.
Experiments
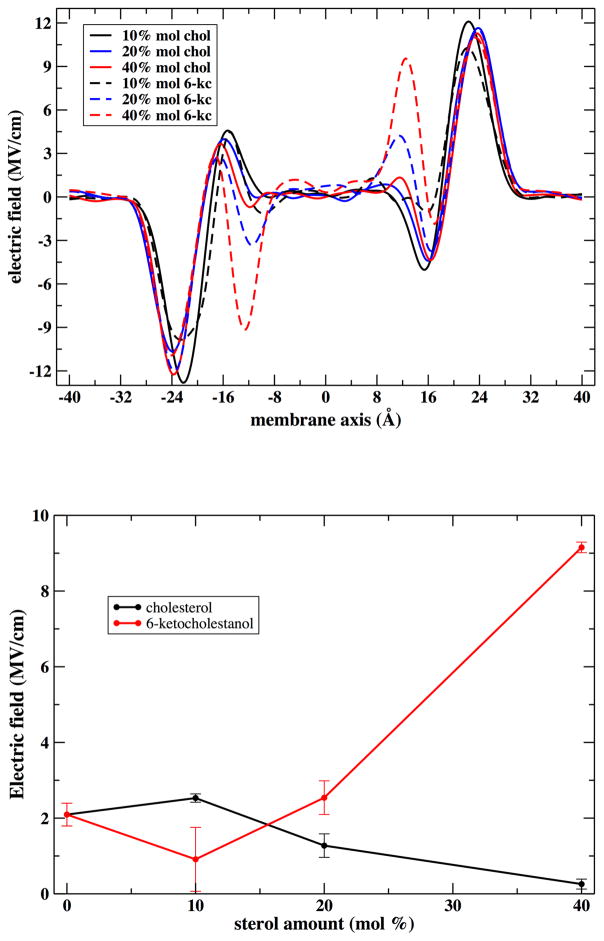
Using Equation 1, we calculated the membrane electric field for bilayers containing 0, 10, 20 and 40 mol% sterol from the experimentally measured Δνobs values, nitrile’s Stark tuning rate of 0.67 cm−1/ (MV/cm),47,48 and the average cosine of the nitrile angle obtained from MD simulations of lipid-αLAX(16) bilayers. In the introduction we noted that the above analysis requires the ensemble average 〈Fd · cos(θ)〉 of the electric field multiplied by the cosine angle of the vibrational probe vector. Here we approximate the above expression by 〈Fd〉 · 〈cos(θ)〉. The approximation is sound only if the field and the probe directions are uncorrelated. In Figure S3 (Supporting Information) we show a scatter plot of the z components of the two vectors, extracted from the MD simulations, and supporting the hypothesis of no correlation.
We determined the absolute electric field, F⃗d, of 10.3 ± 20 MV/cm, 10.9 ± 16 MV/cm, 9.7 ±14 MV/cm and 6.2 ± 8 MV/cm inside the lipid bilayers containing 0, 10, 20 and 40 mol%cholesterol respectively. Similarly, we calculated F⃗d magnitudes of 10.9 ± 20 MV/cm, 11.4 ± 21 MV/cm, 17.8 ± 3 MV/cm and 13.2 ± 20 MV/cm inside lipid bilayers with 0, 10, 20 and 40 mol% 6-kc respectively. The large errors reported for F⃗d do not reflect experimental errors, but rather represent the distribution of field values arising from the distribution of the nitrile angle computed from MD simulations and carried over through the dot product in VSE equation. For both cholesterol and 6-kc, the incorporation of low concentration of sterol (10 mol%) increased the F⃗d values by ~1 MV/cm compared to bilayer with no sterol. As the cholesterol concentration increased up to 40 mol%, the F⃗d magnitude declined to as low as ~6 MV/cm. For 6-kc, the absolute F⃗d value increased up to 18 MV/cm with incorporation of 20 mol% of 6-kc and then dropped down to 13 MV/cm at 40 mol% 6-kc. This decline in the field is due to the small cosine value of the nitrile angle of 110° calculated computationally for bilayers containing 40 mol% 6-kc. The shift in the nitrile absorption frequency (Δνobs ) that we measured experimentally is a more direct reporter of the electric field, and it increases monotonically with higher 6-kc mole fraction.
Discussion
The purpose of this work is to elucidate the effect of cholesterol and an analogue sterol, 6-ketocholestanol on the magnitude of the membrane’s electric field. In our experiments, we determined that cholesterol and 6-kc sterols increased the membrane electric field at lower concentrations, whereas at higher concentration, they had opposite effect; cholesterol lowered the magnitude of the field and 6-kc increased the field. Sterols can impact the membrane’s field in two ways: (1) directly through the inclusion of its own molecular dipole moment (~2.01 D for cholesterol and ~4 D for 6-kc)19 into the system of ordered dipoles; or (2) indirectly by inducing changes in membranes physical properties including fluidity, stiffness, and packing.35–39 Here we report different organization of cholesterol and 6-kc near each other and the helix. This organization alters the electric field that the probe, which is attached to the helix, experiences when the concentration and/or type of sterol are modified.
Evidence reported elsewhere suggests that the intercalation of cholesterol into a lipid bilayer membrane induces ordering in the alkyl chains of lipid membranes,70,73 increasing membrane density,74–77 decreasing fluidity,78,79 and increasing mechanical strength.80 One of the most widely observed impacts of cholesterol in lipid membranes is its condensing effect because of the non-ideal interaction of cholesterol with phospholipids.81 Cholesterol participates in hydrogen bonding with water molecules and other phospholipids through a polar hydroxyl group or via water bridges with adjacent phospholipids, thus anchoring itself to the membrane-aqueous interface.82 Molecular simulations have shown that upon incorporation of cholesterol into the pure lipid bilayer, there is a decrease in the formation of gauche rotamers in the lipid alkyl chains and a substantial reduction in the average tilt of the lipid chains with respect to the plane of the membrane bilayer.39 These two structural effects straighten the hydrocarbon tails of the lipid, which in turn increases the packing density of the lipid molecules and condenses the area per molecule.39,74,75,83 6-kc, on the other hand, is known to increase the membrane permeability and does not impact the ordering of the alkyl tails.33,84 The additional ketone group on the second ring structure allows 6-kc hydrogen bond to greater number of water molecules, thus influencing hydration levels at the membrane-water interface.
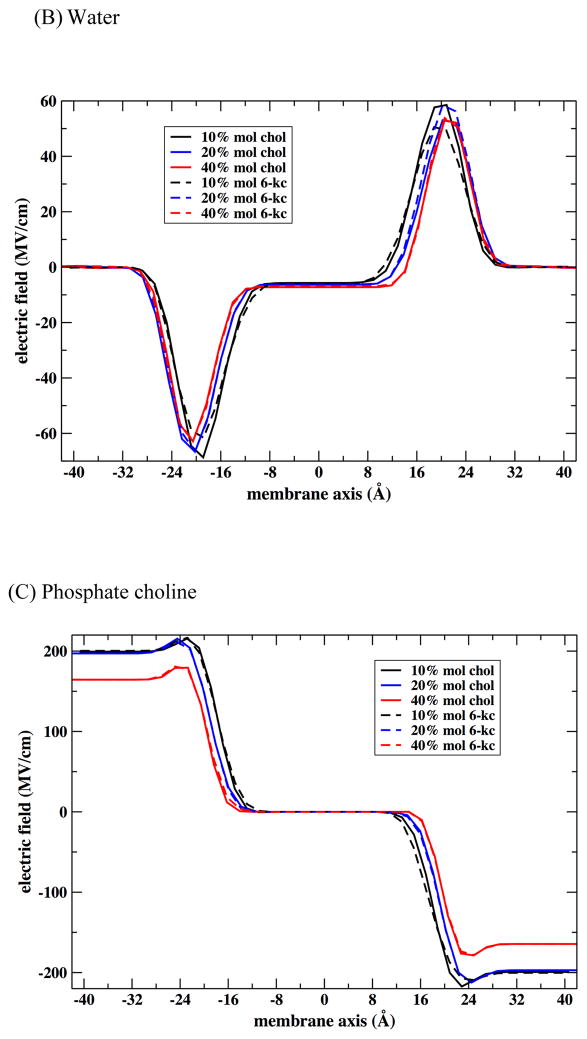
With no sterol, the lipid bilayer above the transition temperature is in a fluid-like disordered phase and has a membrane electric field of 10.3 MV/cm.51 Upon incorporation of a small amount of sterol up to ~10 mol% into the bilayer, the membrane field increases for both sterols. At this low concentration, according to our simulations, the sterols are more concentrated near the helix while at higher concentrations their distribution is more uniform (Figure 8). The sterol coverage of the first solvation shell around the helix is frequently incomplete. Slightly more order lipid molecules replace missing sterol molecules in the helix solvation shell resulting in a stronger local electric field at the probe. As the concentration of 6-kc increases from 10 to 40 mol%, phospholipid dipole ordering increases and the electric field near the boundary of the layer increases consistent with the experimental observation. The total electric filed is displayed below in Figure 12.
Figure 12.
Top: Calculations of the total electric field for different concentrations of cholesterol and 6-kc. Bottom: Electric field dependence of the sterol concentration at fixed membrane depth of 13 Å. Note the significant increase in the absolute magnitude of the electric field for 6-kc, demonstrating that the monotonic increase in the electric field observed experimentally for 6-kc is reproduced in the simulations. The differences for cholesterol are smaller; nevertheless, examination of the peak heights and locations for cholesterol suggests that the results are consistent with a decrease in the electric field as a function of the cholesterol, again consistent with experiment.
We considered three hypotheses for the origin of the monotonic increase in the measured and computed electric fields as a function of sterol concentration. The first hypothesis points to the enhanced ordering of the lipid head group dipole moments. The changes in the field are significant especially between the 40% 6-kc dipoles and the rest. The second hypothesis is the direct contribution of the additional carbonyl group of 6-kc to the electric field. As illustrated in Figure 11, this contribution is also significant. Finally the third hypothesis suggests that the water molecules that are hydrogen bonded to the extra carbonyl in 6-kc cause an increase in the electric field. Indeed the water contribution shows decrease in the electric field for the cholesterol. Smaller changes are observed for 6-kc.
This phenomena is exhibited by the broad distribution of helix tilt angle of αLAX(16) shown in Figure 7. The helix itself is also homogeneously distributed in both bilayers, thus probing the greater electric field coming from the increased dipole density at the membrane-water interface from the phospholipid as well as the water molecules hydrogen bound to each phospholipid head group in bilayers. The homogeneous distribution of helix in bilayers containing both type of sterols is indicated by the similar RDF plots of helix-sterol separation shown in Figure 8 for 10 mol% cholesterol and 10 mol% 6-kc. Hence, at 10 mol% sterol, the elevated packing of phospholipids, sterol and water molecules in a bilayer collectively increases the dipole density at the membrane-water interface and is directly reported by the helix, which is also homogeneously distributed throughout the bilayer.
At high sterol concentration, 40 mol%, the distribution of phospholipids, sterols, and the helix are different for cholesterol and 6-kc. In the case of cholesterol-containing bilayers, cholesterol forms small clusters distributed throughout the bilayer, shown schematically in Figure 11(c). The multiple prominent peaks in the RDF of sterol-sterol interactions at 40 mol% cholesterol (shown in Figure 9(b)) suggest a long-range distribution of such clusters. The nitrile-containing helix is distributed homogeneously among the phospholipids and these cholesterol-rich clusters. This hypothesis is supported by two of our results: (1) the RDF plot of helix-sterol at 40 mol% cholesterol in Figure 8; and (2) the experimental fwhm of the absorption peak of the nitrile in αLAX(16) in Figure 5, which is significantly larger at 40 mol% cholesterol. The greater fwhm values can be explained if the nitrile is positioned in two chemically heterogeneous domains: one enriched in phospholipids with ordered tails, and the other enriched in cholesterol. In this environment, the relative concentration of cholesterol increases in comparison to phospholipids. As cholesterol molecules displace phospholipids, the water molecules associated with those phospholipids through hydrogen bonding are displaced as well. The intrinsic dipole moment of cholesterol is 2.01 D, which is significantly smaller than the molecular dipole moment of a DMPC phospholipid (~14 D), particularly when it is decorated with hydrogen-bound water molecules.18 The nitrile, which is distributed among the phospholipids and cholesterol clusters, therefore directly reports the net reduction in the dipole density by the small magnitude of the absorption energy shift and thus the smaller electric field.
In the case of membranes containing high concentrations of 6-kc, the sterol segregates into larger clusters of 6-kc compared to cholesterol. The single prominent peak in the RDF plot of 40 mol% 6-kc in Figure 9(b) provides strong evidence of such cluster formation. The RDF plot of helix-sterol in Figure 8 clearly suggests greater separation between helix and the 6-kc clusters. Furthermore, in contrast to cholesterol, we do not observe significant differences in the fwhm of the nitrile in αLAX(16) at 40 mol% 6-kc versus 10 mol% 6-kc. This implies that the nitrile is continuously placed within the same phospholipid tail region at all concentrations of 6-kc. As the concentration of 6-kc increases, so do the magnitudes of the contributed dipole moments from the sterol (~4 D) and associated water molecules. This effect is larger than that seen for cholesterol because of the extra hydrogen bond acceptor oxygen atom on 6-kc. This is verified by the quantitative hydrogen bond analysis between sterol and water molecules (Figure 10), in which the number of hydrogen bonds associated with 6-kc is larger than with cholesterol. This leaves the helix surrounded by phospholipids, but sequestered from 6-kc.
At mid range concentrations we show an opposite behavior of the electric field for 6-kc and cholesterol in both experiment and simulations. The electric field decreases as the concentration of cholesterol increases while it increases for 6-kc. This surprising difference between two similar sterols can be rationalized by their relative positions in the membrane. Cholesterol is placed deeper in the membrane and is unlikely to orient the water dipoles successfully. This task is conducted effectively by 6-kc, whose capacity for an additional hydrogen bond places it closer to the interface between the aqueous solution and the membrane (the electric field contribution of water is similar between the two sterols). Our results shed light on sometimes conflicting results about the effect of sterols, particularly cholesterol, on membrane electrostatics. We show that the effect of cholesterol on the electric field is divided into two regimes based on its concentration. At low concentration, the simulations results are too noisy to obtain significant signal, but experiments suggest that the electric field is increasing. At high concentration, cholesterol reduces the polar environment near the helical probe and therefore reduces the electric field as shown in Figure 12. 6-kc is susceptible to forming a greater number of hydrogen bonds with water molecules compared to cholesterol, and the dipole density at the membrane-water interface increases with the increasing 6-kc mole fraction due to its larger intrinsic dipole moment.
Conclusion
It is well known that the non-covalent intercalation of cholesterol into biological membranes has a great impact on membrane fluidity, self-association, and function. Despite extensive experimental and theoretical investigations, there is limited agreement on how and to what extent cholesterol and other sterols such as 6-kc alter membrane electrostatics, largely the result of limited techniques for studying a property contained entirely within membrane interior. By addressing this with VSE spectroscopy and MD simulations, we measured changes in electric field as a function of sterol concentration and identity. We used MD simulations to identify distinct patterns of lateral organization of sterols inside the lipid bilayer especially at higher sterol concentration. Future work in this laboratory will focus on how this electrostatic field regulates significantly more complex lipid membrane mechanisms such as binding of membrane-proteins and ion channel formations.
Supplementary Material
Acknowledgments
This research was supported by the Welch Foundation (Grant No. F-1722) and the Burroughs Wellcome Fund (Grant No. 1007207.01) to L.J.W, and the Welch Foundation (Grant No. F- 1785 and F-1896) and the NIH (Grant No. GM059796 and GM111364) to R.E. We acknowledge The Targeted Therapeutic Drug Discovery & Development Program (TTDDDP) at the University of Texas at Austin for the use of their circular dichroic spectrometer and the Texas Materials Institute (TMI) for the use of the Zetasizer Nano ZS DLS.
Footnotes
Circular Dichromism data of nitrile-containing peptides intercalated in vesicles, and simulation data on the orientation of the peptide and nitrile functional group within the vesicle are given in Supporting Information.
References
- 1.van Meer G. Lipid Traffic in Animal Cells. Annu Rev Cell Biol. 1989;5:247–275. doi: 10.1146/annurev.cb.05.110189.001335. [DOI] [PubMed] [Google Scholar]
- 2.Jacob RF, Cenedella RJ, Mason RP. Direct Evidence for Immiscible Cholesterol Domains in Human Ocular Lens Fiber Cell Plasma Membranes. J Biol Chem. 1999;274:31613–31618. doi: 10.1074/jbc.274.44.31613. [DOI] [PubMed] [Google Scholar]
- 3.Léonard A, Dufourc EJ. Interactions of Cholesterol with the Membrane Lipid Matrix. A Solid State NMR Approach. Biochimie. 1991;73:1295–1302. doi: 10.1016/0300-9084(91)90092-f. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs R, Oldfield E. Deuterium Nuclear Magnetic Resonance Investigation of Dimyristoyllecithin-Dipalmitoyllecithin and Dimyristoyllecithin-Cholesterol Mixtures. Biochemistry. 1979;18:3280–3285. doi: 10.1021/bi00582a013. [DOI] [PubMed] [Google Scholar]
- 5.Veatch SL, Polozov IV, Gawrisch K, Keller SL. Liquid Domain in Vesicles Investigated by NMR and Fluorescence Microscopy. Biophys J. 2004;86:2910–2922. doi: 10.1016/S0006-3495(04)74342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bin X, Lipkowski J. Electrochemical and PM-IRRAS Studies of the Effect of Cholesterol on the Properties of the Headgroup Region of a DMPC Bilayer Supported at a Au(111) Electrode. J Phys Chem B. 2006;110:26430–26441. doi: 10.1021/jp0660132. [DOI] [PubMed] [Google Scholar]
- 7.Shimshick EJ, McConnel HM. Lateral Phase Separations in Binary-Mixtures of Cholesterol and Phospholipids. Biochem Biophys Res Commun. 1973;53:446–451. doi: 10.1016/0006-291x(73)90682-7. [DOI] [PubMed] [Google Scholar]
- 8.Oldfield E, Chapman D. Effects of Cholesterol and Cholesterol Derivatives on Hydrocarbon Chain Mobility in Lipids. Biochem Biophys Res Commun. 1971;43:610–616. doi: 10.1016/0006-291x(71)90658-9. [DOI] [PubMed] [Google Scholar]
- 9.Mainali L, Raguz M, Subczynski WK. Formation of Cholesterol Bilayer Domains Precedes Formation of Cholesterol Crystals in Cholesterol/Dimyristoylphosphatidylcholine Membranes: EPR and DSC Studies. J Phys Chem B. 2013;117:8994–9003. doi: 10.1021/jp402394m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mabrey S, Sturtevant JM. Investigation of Phase Transitions of Lipids and Lipid Mixtures by High Sensitivity Differential Scanning Calorimetry. Proc Natl Acad Sci. 1976;73:3862–3866. doi: 10.1073/pnas.73.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blandamer MJ, Briggs B, Cullis PM, Rawlings BJ, Engberts JBFN. Vesicle-Cholesterol Interactions: Effects of Added Cholesterol on Gel-to-Liquid Crystal Transitions in a Phospholipid Membrane and Five Dialkyl-based Vesicles as Monitored Using DSC. Phys Chem Chem Phys. 2003;5:5309–5312. [Google Scholar]
- 12.McMullen TPW, McElhaney RN. New Aspects of the Interaction of Cholesterol with Dipalmitoylphosphatidylcholine Bilayers as Revealed by High-Sensitivity Differential Scanning Calorimetry. Biochim Biophys Acta. 1995;1234:90–98. doi: 10.1016/0005-2736(94)00266-r. [DOI] [PubMed] [Google Scholar]
- 13.McMullen TPW, Lewis RNAH, McElhaney RN. Cholesterol–Phospholipid Interactions, the Liquid-Ordered Phase and Lipid Rafts in Model and Biological Membranes. Curr Opin Colloid Interface Sci. 2004;8:459–468. [Google Scholar]
- 14.Carquin M, D’Auria L, Pollet H, Bongarzone ER, Tyteca D. Recent Progress on Lipid Lateral Heterogeneity in Plasma Membranes: From Rafts to Submicrometric Domains. Prog Lipid Res. 2016;62:1–24. doi: 10.1016/j.plipres.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lingwood D, Simons K. Lipid Rafts As a Membrane-Organizing Principle. Science. 2009;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 16.Levental I, Grzybek M, Simons K. Raft Domains of Variable Properties in Plasma Membrane Vesicles. Proc Natl Acad Sci. 2011;108:11411–11416. doi: 10.1073/pnas.1105996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Efimova SS, Ostroumova OS. Effect of Dipole modifiers on the Magnitude of the Dipole Potential of Sterol-Containing Bilayers. Langmuir. 2012;28:9908–9914. doi: 10.1021/la301653s. [DOI] [PubMed] [Google Scholar]
- 18.Haldar S, Kanaparthi RK, Samanta A, Chattopadhyay A. Differential Effect of Cholesterol and Its Biosynthetic Precursors on Membrane Dipole Potential. Biophys J. 2012;102:1561–1569. doi: 10.1016/j.bpj.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starke-Peterkovic T, Turner N, Vitha MF, Waller MP, Hibbs DE, Clarke RJ. Cholesterol Effect on the Dipole Potential of Lipid Membranes. Biophys J. 2006;90:4060–4070. doi: 10.1529/biophysj.105.074666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honig BH, Hubbell WL, Flewelling RF. Electrostatic Interactions in Membranes and Proteins. Annu Rev Biophys Biophys Chem. 1986;15:163–193. doi: 10.1146/annurev.bb.15.060186.001115. [DOI] [PubMed] [Google Scholar]
- 21.Cevc G. Membrane Electrostatics. Biochim Biophys Acta. 1990;1031:311–382. doi: 10.1016/0304-4157(90)90015-5. [DOI] [PubMed] [Google Scholar]
- 22.Schamberger J, Clarke RJ. Hydrophobic Ion Hydration and the Magnitude of the Dipole Potential. Biophys J. 2002;82:3081–3088. doi: 10.1016/S0006-3495(02)75649-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duffin RL, Garrett MP, Flake KB, Durrant JD, Busath DD. Modulation of Lipid Bilayer Interfacial Dipole Potential by Phloretin, RH421, and 6-ketocholestanol as Probed by Gramicidin Channel Conductance. Langmuir. 2003;19:1439–1442. [Google Scholar]
- 24.Phillips LR, Cole CD, Hendershot RJ, Cotten M, Cross TA, Busath DD. Noncontact Dipole Effects on Channel Permeation. III. Anomalous Proton Conductance Effects in Gramicidin. Biophys J. 1999;77:2492–2501. doi: 10.1016/S0006-3495(99)77085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hladky SB. The Energy Barriers to Ion Transport by Nonactin Across Thin Lipid Membranes. Biochim Biophys Acta. 1974;352:71–85. doi: 10.1016/0005-2736(74)90180-1. [DOI] [PubMed] [Google Scholar]
- 26.Bala S, Kombrabail MH, Prabhananda BS. Effect of Phloretin on Ionophore Mediated Electroneutral Transmembrane Translocations of H(+), K(+) and Na(+) in Phospholipid Vesicles. Biochim Biophys Acta. 2001;1510:258–269. doi: 10.1016/s0005-2736(00)00357-6. [DOI] [PubMed] [Google Scholar]
- 27.Cladera J, Martin I, Ruysschaert JM, O’Shea P. Characterization of the Sequence of Interactions of the Fusion Domain of the Simian Immunodeficiency Virus With Membranes. J Biol Chem. 1999;274:29951–29959. doi: 10.1074/jbc.274.42.29951. [DOI] [PubMed] [Google Scholar]
- 28.Alakoskela JMI, Kinnunen PKJ. Control of a Redox Reaction on Lipid Bilayer Surfaces by Membrane Dipole Potential. Biophys J. 2001;80:294–304. doi: 10.1016/S0006-3495(01)76014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alakoskela JMI, Soderlund T, Holopainen JM, Kinnunen PKJ. Dipole Potential and Head-group Spacing are Determinants for the Membrane Partitioning of Pregnanolone. Mol Pharmacol. 2004;66:161–168. doi: 10.1124/mol.104.000075. [DOI] [PubMed] [Google Scholar]
- 30.Cladera J, O’Shea P. Intramembrane Molecular Dipoles Affect the Membrane Insertion and Folding of a Model Amphiphilic Peptide. Biophys J. 1998;74:2434–2442. doi: 10.1016/S0006-3495(98)77951-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starke-Peterkovic T, Turner N, Else PL, Clarke RJ. Electric Field Strength of Membrane Lipids From Vertebrate Species: Membrane Lipid Composition and Na1,K1-ATPase molecular activity. Am J Physiol Regul Integr Comp Physiol. 2005;288:R663–R670. doi: 10.1152/ajpregu.00434.2004. [DOI] [PubMed] [Google Scholar]
- 32.Maggio B. Modulation of Phospholipase A2 by Electrostatic Fields and Dipole Potential of Glycosphingolipids in Monolayers. J Lipid Res. 1999;40:930–939. [PubMed] [Google Scholar]
- 33.Buzon V, Cladera J. Effect of Cholesterol on the Interaction of the HIV GP41 Fusion Peptide with Model Membranes. Importance of the Membrane Dipole Potential. Biochemistry. 2006;45:15768–15775. doi: 10.1021/bi060622i. [DOI] [PubMed] [Google Scholar]
- 34.McIntosh TJ, Magid AD, Simon SA. Cholesterol Modifies the Short-range Repulsive Interactions Between Phosphatidyl-choline Membranes. Biochemistry. 1989;28:17–25. doi: 10.1021/bi00427a004. [DOI] [PubMed] [Google Scholar]
- 35.Szabo G. Dual Mechanism for the Action of Cholesterol on Membrane Permeability. Nature. 1974;252:47–49. doi: 10.1038/252047a0. [DOI] [PubMed] [Google Scholar]
- 36.Hofsäβ C, Lindahl E, Edholm O. Molecular Dynamics Simulations of Phospholipid Bilayers with Cholesterol. Biophys J. 2003;84:2192–2206. doi: 10.1016/S0006-3495(03)75025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smondyrev AM, Berkowitz ML. Structure of Dipalmitoylphosphatidylcholine/ Cholesterol Bilayer at Low and High Cholesterol Concentrations: Molecular Dynamics Simulation. Biophys J. 1999;77:2075–2089. doi: 10.1016/S0006-3495(99)77049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiu SW, Jakobsson E, Scott HL. Combined Monte Carlo and Molecular Dynamics Simulation of Hydrated Dipalmitoyl–phosphatidylcholine–Cholesterol Lipid Bilayers. J Chem Phys. 2001;114:5432–5443. [Google Scholar]
- 39.Gabdoulline RR, Vanderkooi G, Zheng C. Comparison of the Structures of Dimyristoylphosphatidylcholine in the Presence and Absence of Cholesterol by Molecular Dynamics Simulations. J Phys Chem. 1996;100:15942–15946. [Google Scholar]
- 40.Yang Y, Mayer KM, Wickremasignhs NS, Hafner JH. Probing the Lipid Membrane Dipole Potential by Atomic Force Microscopy. Biophys J. 2008;95:5193–5199. doi: 10.1529/biophysj.108.136507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franklin JC, Cafiso DS. Internal Electrostatic Potentials in Bilayers: Measuring and Controlling Dipole Potentials in Lipid Vesicles. Biophys J. 1993;65:289–299. doi: 10.1016/S0006-3495(93)81051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross E, Bedlack RS, Loew LM. Dual-Wavelength Ratiometric Fluorescence Measurement of the Membrane Dipole Potential. Biophys J. 1994;67:208–216. doi: 10.1016/S0006-3495(94)80471-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haldar S, Kanaparthi RK, Samanta A, Chattopadhyay A. Differential Effect of Cholesterol and Its Biosynthetic Precursors on Membrane Dipole Potential. Biophys J. 2012;102:1561–1569. doi: 10.1016/j.bpj.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Efimova SS, Ostroumova OS. Effect of Dipole Modifiers on the Magnitude of the Dipole Potential of Sterol-Containing Bilayers. Langmuir. 2012;28:9908–9914. doi: 10.1021/la301653s. [DOI] [PubMed] [Google Scholar]
- 45.Smondyrev AM, Berkowitz ML. Effects of Oxygenated Sterol on Phospholipid Bilayer Properties: a Molecular Dynamics Simulation. Chem Phys Lipids. 2001;112:31–39. doi: 10.1016/s0009-3084(01)00160-8. [DOI] [PubMed] [Google Scholar]
- 46.Simon SA, McIntosh TJ, Magid AD, Needham D. Modulation of the Interbilayer Hydration Pressure by the Addition of Dipoles at the Hydrocarbon/Water Interface. Biophys J. 1992;61:786–799. doi: 10.1016/S0006-3495(92)81883-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chattopadhyay A, Boxer SG. Vibrational Stark Effect Spectroscopy. J Am Chem Soc. 1995;117:1449–1450. [Google Scholar]
- 48.Andrews SS, Boxer SG. Vibrational Stark Effects of Nitriles I. Methods and Experimental Results. J Phys Chem A. 2000;104:11853–11863. [Google Scholar]
- 49.Andrews SS, Boxer SG. Vibrational Stark Effects of Nitriles II. Physical Origins of Stark Effects from Experiment and Perturbation Models. J Phys Chem A. 2002;106:469–477. [Google Scholar]
- 50.Hu W, Webb LJ. Direct Measurement of the Membrane Dipole Field in Bicelles Using Vibrational Stark Effect Spectroscopy. J Phys Chem Lett. 2011;2:1925–1930. [Google Scholar]
- 51.Shrestha R, Cardenas AE, Elber R, Webb LJ. Measurement of the Membrane Dipole Electric Field in DMPC Vesicles Using Vibrational Shifts of p-Cyanophenylalanine and Molecular Dynamics Simulations. J Phys Chem B. 2015;119:2869–2876. doi: 10.1021/jp511677j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johansson ACV, Lindahl E. Amino-Acid Solvation Structure in Transmembrane Helices from Molecular Dynamics Simulations. Biophys J. 2006;91:4450–4463. doi: 10.1529/biophysj.106.092767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morrissey JH. Morrisey Lab Protocol for Preparing Phospholipid Vesicles (SUV) by Sonication. [accessed March 15, 2012];Trigger Website. http://tf7.org/suv.pdf.
- 54.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 55.Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elber R, Roitberg A, Simmerling C, Goldstein R, Li HY, Verkhivker G, Keasar C, Zhang J, Ulitsky A. Moil: A Program for Simulations of Macrmolecules. Comput Phys Commun. 1995;91:159–189. [Google Scholar]
- 57.Ruymgaart AP, Cardenas AE, Elber R. MOIL-OPT: Energy-Conserving Molecular Dynamics on a GPU/CPU System. J Chem Theory Comput. 2011;7:3072–3082. doi: 10.1021/ct200360f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berger O, Edholm O, Jahnig F. Molecular Dynamics Simulations of a Fluid Bilayer of Dipalmitoylphosphatidylcholine at Full Hydration, Constant Pressure, and Constant Temperature. Biophys J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jorgensen WL, Tiradorives J. The OPLS Potential Functions for Proteins - Energy Minimizations for Crystals of Cyclic-Peptides and Crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 60.Pasenkiewicz-Gierula M, Rog T, Kitamura K, Kusumi A. Cholesterol Effects on the Phosphatidylcholine Bilayer Polar Region: A Molecular Simulation study. Biophys J. 2000;78:1376–1389. doi: 10.1016/S0006-3495(00)76691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price MLP, Ostrovsky D, Jorgensen WL. Gas-Phase and Liquid-State Properties of Esters, Nitriles, and Nitro Compounds with the OPLS-AA Force Field. J Comput Chem. 2001;22:1340–1352. [Google Scholar]
- 62.Berendsen HJC, Grigera JR, Straatsma TP. The Missing Term in Effective Pair Potentials. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 63.Di Pierro M, Elber R, Leimkuhler B. A Stochastic Algorithm for the Isobaric-Isothermal Ensemble with Ewald Summations for All Long Range Forces. J Chem Theory Comput. 2015;11:5624–5637. doi: 10.1021/acs.jctc.5b00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 65.Weinbach Y, Elber R. Revisiting and Parallelizing SHAKE. J Comp Phys. 2005;209:193–206. [Google Scholar]
- 66.Tuckerman M, Berne BJ, Martyna GJ. Reversible Multiple Time Scale Molecular-Dynamics. J Chem Phys. 1992;97:1990–2001. [Google Scholar]
- 67.Kabsh W. A Discussion of the Best Solution For the Best Rotation to Relate Two Sets of Vectors. Acta Cryst. 1978;A34:827–828. [Google Scholar]
- 68.Frohlich M, Brecht V, Peschka-Suss R. Parameters Influencing the Determination of Liposome Lamellarity by 31P-NMR. Chem Phys Lipids. 2001;109:103–112. doi: 10.1016/s0009-3084(00)00220-6. [DOI] [PubMed] [Google Scholar]
- 69.Bystrov VF, Shapiro YE, Viktorov AV, Barsukov LI, Bergelson LD. 31P-NMR Signals From Inner and Outer Surfaces of Phospholipid Membranes. FEBS Lett. 1972;25:337–338. doi: 10.1016/0014-5793(72)80518-0. [DOI] [PubMed] [Google Scholar]
- 70.Kučerka N, Nieh MP, Katsaras J. Fluid Phase Lipid Areas and Bilayer Thicknesses of Commonly Used Phosphatidylcholines as a Function of Temperature. Biochim Biophys Acta. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 71.Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. J Molec Graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 72.Cardenas AE, Shrestha R, Webb LJ, Elber R. Membrane Permeation of a Peptide: It is Better to be Positive. J Phys Chem B. 2015;119:6412–6420. doi: 10.1021/acs.jpcb.5b02122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khelashvili G, Pabst G, Harries D. Cholesterol Orientation and Tilt Modulus in DMPC Bilayers. J Phys Chem B. 2010;114:7524–7534. doi: 10.1021/jp101889k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rog T, Pasenkiewicz-Gierula M. Cholesterol Effects on the Phosphatidylcholine Bilayer Nonpolar Region: A Molecular Simulation Study. Biophys J. 2001;81:2190–2202. doi: 10.1016/S0006-3495(01)75867-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jedlovszky P, Mezei M. Effect of Cholesterol on the Properties of Phospholipid Membranes. 1. Structural Features. J Phys Chem B. 2003;107:5311–5321. doi: 10.1021/jp051832s. [DOI] [PubMed] [Google Scholar]
- 76.Henriksen J, Rowat AC, Brief E, Hsueh YW, Thewalt JL, Zuckermann MJ, Ipsen JH. Universal Behaviour of Membranes with Sterols. Biophys J. 2006;90:1639–1649. doi: 10.1529/biophysj.105.067652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smaby JM, Momsen M, Brockman HL, Brown RE. Phosphatidylcholine Acyl Unsaturation Modulates the Decrease in Interfacial Elasticity Induced by Cholesterol. Biophys J. 1997;73:1492–1505. doi: 10.1016/S0006-3495(97)78181-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marsh D, Smith ICP. Interacting Spin Labels as Probes of Molecular Separation within Phospholipid Bilayers. Biochem Biophys Res Commun. 1972;49:916–922. doi: 10.1016/0006-291x(72)90299-9. [DOI] [PubMed] [Google Scholar]
- 79.Kusumi A, Tsuda M, Akino T, Ohnishi S, Terayama Y. Protein-Phospholipid-Cholesterol Interaction in the Photolysis Invertebrate Rhodopsin. Biochemistry. 1983;22:1165–1170. doi: 10.1021/bi00274a027. [DOI] [PubMed] [Google Scholar]
- 80.Mouritsen OG, Jørgensen K. Dynamical Order and Disorder in Lipid Bilayers. Chem Phys Lipids. 1994;73:3–25. doi: 10.1016/0009-3084(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 81.McConnell HM, Radhakrishnan A. Condensed Complexes of Cholesterol and Phospholipids. Biochim Biophys Acta. 2003;1610:159–173. doi: 10.1016/s0005-2736(03)00015-4. [DOI] [PubMed] [Google Scholar]
- 82.Ohvo-Rekila H, Ramstedt B, Leppimaki P, Slotte JP. Cholesterol Interactions with Phospholipids in Membranes. Prog Lipid Res. 2002;41:66–97. doi: 10.1016/s0163-7827(01)00020-0. [DOI] [PubMed] [Google Scholar]
- 83.Rheinstadter MC, Mouritsen OG. Small-Scale Structure in Fluid Cholesterol-Lipid Bilayers. Curr Opin Colloid Interface Sci. 2013;18:440–447. [Google Scholar]
- 84.Auner BG, O’Neill MAA, Valenta C, Hadgraft J. Interaction of Phloretin and 6-Ketocholestanol With DPPC-Liposomes as Phospholipid Model Membranes. Int J Pharm. 2005;294:149–155. doi: 10.1016/j.ijpharm.2005.01.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.