Abstract
Nuclear factor erythroid 2-related factor 2 (Nrf2) is a stress-activated transcription factor activated by stimuli such as electrophilic compounds and other reactive xenobiotics. Previously, we have shown that the commonly used food additive and Nrf2 activator tert-butylhydroquinone (tBHQ) suppresses interleukin-2 (IL-2) production, CD25 expression, and NFκB activity in human Jurkat T cells. The purpose of the current studies was to determine whether these effects were dependent upon Nrf2 by developing a human Nrf2-null T cell model using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 technology. The current studies show that suppression of CD25 expression by tBHQ is partially dependent on Nrf2, whereas inhibition of IL-2 secretion is largely Nrf2-independent. Interestingly, tBHQ inhibited NFκB activation in an Nrf2-independent manner. This was an unexpected finding since Nrf2 inhibits NFκB activation in other models. These results led us to investigate another more potent Nrf2 activator, the synthetic triterpenoid 1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO-Im). Treatment of wild-type and Nrf2-null Jurkat T cells with CDDO-Im resulted in an Nrf2-dependent suppression of IL-2. Furthermore, susceptibility to reactive oxygen species was significantly enhanced in the Nrf2-null clones as determined by decreased mitochondrial membrane potential and cell viability. Importantly, this study is the first to describe the generation of a human Nrf2-null model, which is likely to have multiple applications in immunology and cancer biology. Collectively, this study demonstrates a role for Nrf2 in the effects of CDDO-Im on CD25 and IL-2 expression, whereas the effect of tBHQ on these parameters is complex and likely involves modulation of multiple stress-activated transcription factors, including NFκB and Nrf2.
Introduction
Nuclear factor erythroid 2-related factor 2 (Nrf2) is a transcription factor that is activated by cell stress, including the presence of reactive oxygen species, electrophilic compounds, and other reactive xenobiotics (Chan et al., 1996; Dinkova-Kostova et al., 2002; Kensler et al., 2007). In the absence of cell stress, Nrf2 is tethered within the cytosol by its repressor protein Kelch ECH associated protein 1 (Keap1), which facilitates the continual degradation of Nrf2 by the 26S proteasome (Itoh et al., 1999; McMahon et al., 2003). Upon activation, Nrf2 accumulates in the nucleus and binds to its consensus sequence in the promoters of target genes, inducing transcription (Venugopal and Jaiswal, 1996). Genes regulated by Nrf2 are often cytoprotective in function and include genes involved in glutathione homeostasis, detoxification, and metabolism. Within leukocytes, Nrf2 has been shown to regulate transcription of NADPH quinone oxidoreductase 1 (Nqo1), making it a good target to assess Nrf2 activity (Itoh et al., 1997; Kensler et al., 2007).
Nrf2 is activated by numerous chemicals, including environmental contaminants, such as heavy metals; synthetic compounds, such as 1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO-Im); and food preservatives, such as tert-butylhydroquinone (tBHQ) and butylated hydroxyanisole. The present studies focused on tBHQ, as it activates Nrf2, is a widely used food preservative, and has robust immunomodulatory effects in T cells at low concentrations (Zagorski et al., 2013; Turley et al., 2015). In the food industry, tBHQ is often added to oils (vegetable, soybean, etc.) to increase shelf life, but it has numerous applications and is found in a variety of foods (Shahidi, 2000). Previous studies have shown that human volunteers who ingested between 100 and 150 mg of tBHQ achieved a range of serum concentrations in some cases in excess of 200 µM. Thus, in addition to serving as a useful tool for activating Nrf2, tBHQ is also relevant to human exposure at the concentrations used in the present studies (WHO, 1975; Zagorski et al., 2013; Turley et al., 2015).
The role of Nrf2 in antioxidant and metabolic responses to reactive toxicants is well described, but it also has a strong anti-inflammatory role. For instance, Nrf2-null mice are more sensitive to insults in a host of inflammatory models, including traumatic brain injury, models of multiple sclerosis, and models of sepsis, among others (Thimmulappa et al., 2006; Johnson et al., 2010; Saykally et al., 2012). Nrf2 has also been implicated in the development of autoimmunity, as female Nrf2-null mice develop a disease similar to systemic lupus erythematosus in humans (Yoh et al., 2001; Li et al., 2004; Rangasamy et al., 2005; Ma et al., 2006; Thimmulappa et al., 2006; Johnson et al., 2010). In humans, the role of Nrf2 is less clear, but our previous studies demonstrate that a known Nrf2 activator, tBHQ, inhibits several events of T cell activation in both primary human T cells as well as in human Jurkat T cells, which suggests Nrf2 may play a role in regulating human T cell activation (Zagorski et al., 2013; Turley et al., 2015).
T cell activation requires two signals. The first is stimulation and signaling through the T cell receptor complex, one member of which is CD3. The second signal is propagated through a costimulatory receptor, such as CD28 (Cantrell, 1996). Experimentally, we activate T cells with antibodies targeting both CD3 and CD28, resulting in a signaling cascade and the activation of NFκB and other transcription factors, induction of the early cytokine interleukin-2 (IL-2), as well as the expression of early activation markers CD25 and CD69.
Use of clustered regularly interspaced short palindromic repeats (CRISPR) in conjunction with the endonuclease CRISPR-associated protein 9 (Cas9), isolated from Streptococcus pyogenes, to mediate targeted gene disruption has greatly increased over the last several years due to the high efficiency of CRISPR/Cas9 over other methods of gene editing. This technology relies on Watson-Crick base pairing and complementarity of a guide RNA to a specific site within the genome to target the endonuclease Cas9 to the gene of interest and facilitate a double-strand DNA break. The cell then initiates nonhomologous end joining, which can be error prone. Subsequent insertions or deletions (INDEL) at the site of DNA repair can result in a frameshift mutation, early stop codon, and/or a gene knockout (Cong et al., 2013; Mali et al., 2013a,b; Wang et al., 2013).
The characterization of the physiologic and pathophysiologic effects of Nrf2 activators on human cells is vital given the ubiquitous nature of such chemicals in the human environment. However, these studies are hampered by lack of adequate tools for mechanistic research, including availability of Nrf2-null human cells. The purpose of the present studies was to address this problem by generating Nrf2-null Jurkat T cell clones as well as to use these clones to determine the role of Nrf2 on T cell function when stimulated with tBHQ, CDDO-Im, and H2O2.
Materials and Methods
Materials.
Unless specified, reagents, including tBHQ, were purchased from Sigma-Aldrich (St. Louis, MO).
Plasmids.
The NFκB luciferase reporter was purchased from Promega (Madison, WI). The CRISPR/Cas9 plasmid (PX458) was purchased from Addgene (Cambridge, MA).
Cell Culture.
Human Jurkat E6-1 cells were purchased from American Type Culture Collection (Manassas, VA). Cells were then cultured in RPMI 1640 medium with the addition of 10% fetal bovine serum, 25 mM HEPES, 1 mM sodium pyruvate, 1× nonessential amino acids, 100 U/ml penicillin, and 100 U/ml streptomycin. Fetal bovine serum was purchased from BioWest (Riverside, MO). The protocols for tBHQ treatment of Jurkat T cells are described in the corresponding figure legends.
Generation of CRISPR/Cas9 Plasmids.
A CRISPR/Cas9 plasmid (PX458) lacking a guide RNA (gRNA) sequence was purchased from Addgene. gRNA sequences targeting different locations within Nfe2l2 (the Nrf2 gene) were chosen from http://arep.med.harvard.edu/human_crispr/, and Nrf2 specificity was verified using http://blast.ncbi.nlm.nih.gov/Blast.cgi. Oligos for the identified gRNA sequences were generated by Integrated DNA Technologies (Coralville, IA), and restriction digest cloning was performed as described by the Zhang laboratory (Massachusetts Institute of Technology) (Ran et al., 2013). After CRISPR/Cas9-transformed bacterial clones were identified, the plasmids were isolated by QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA). Plasmids were sent to Genewiz (South Plainfield, NJ) using the primer sequence 5′-GGGCCTATTTCCCATGATTC-3′ to ensure proper cloning of the gRNA sequence into the PX458 vector.
Transient Transfection of CRISPR/Cas9 Plasmids.
Human Jurkat T cells were reverse transfected with 1.5 μg of plasmid and 3 μl of Lipofectamine 2000 (Life Technologies, Grand Island, NY) per 5 × 105 cells for 12 hours in complete medium. After 12 hours, the cells were washed and resuspended at a concentration of 5 × 105 cells/ml in complete medium. After resuspension, the cells were seeded in a 48-well plate at a volume of 1 ml per well. The cells were then incubated at 37°C for 36 hours. After incubation, the cells were sorted by Green Fluorescent Protein (GFP) expression, and a single GFP-positive cell was deposited per well in a 96-well tissue culture plate using a BD Influx flow cytometer (BD Biosciences, San Jose, CA) and allowed to clonally expand.
Identification of Nrf2 Mutants by Sanger Sequencing.
Genomic DNA was extracted from Jurkat clones using a DNeasy Blood and Tissue kit from Qiagen. The region of Nrf2 containing the targeted cut site was then amplified using the high fidelity Herculase II fusion DNA polymerase kit (Agilent Genomics, Santa Clara, CA). The primers used were as follows: exon 2 of Nrf2 forward primer: 5′- GAGGCTGAGGTTGGAAAGTAG-3′, reverse primer: 5′-CAGCGACGGAAAGAGTATGAG-3′; exon 5 of Nrf2 forward primer: 5′-TCTACAGGGAATGGGATATGGA-3′, reverse primer 5′-CTACTTGGCCTCAGTGATTCTG -3. DNA extracts from individual clones were then sent to Genewiz for Sanger sequencing using the forward primer from the polymerase chain reaction (PCR) for sequencing.
mRNA Quantification by Real-Time PCR.
Total RNA was isolated from 2 × 106 wild-type and Nrf2-null Jurkat T cells using TRIzol Reagent, per the manufacturer’s protocol (Life Technologies). After isolation, reverse transcription was performed prior to Sybr Green real-time PCR analysis. Relative mRNA expression was calculated by Delta-Delta-Cycle Threshold, normalized to the housekeeping gene ribosomal protein RPL13a, which has previously been shown to be a stable housekeeping gene in T cell analysis (Mane et al., 2008), using Ct values quantified by Life Technologies/Applied Biosystems Sequence Detection System 7500. All primer sequences were retrieved from Primer Depot (http://primerdepot.nci.nih.gov/) and synthesized by Integrated DNA Technologies. The primer sequences were as follows: RPL13A forward primer: 5′GTTGATGCCTTCACAGCGTA-3′, RPL13A reverse primer: 5′-AGATGGCGGAGGTGCAG-3′; NQO1 forward primer: 5′-TCCTTTCTTCAAAGCCG-3′, NQO1 reverse primer: 5′-GGACTGCACCAGAGCCAT-3′.
IL-2 Quantification by Enzyme-Linked Immunosorbent Assay.
Wild-type and Nrf2-null Jurkat T cells were cultured in 96-well plates (1 × 105 cells/well), treated as described, and cell supernatants were harvested 24 hours after activation with anti-CD3/anti-CD28. IL-2 was quantified from cell supernatants by the use of a commercially available enzyme-linked immunosorbent assay kit (BioLegend, San Diego, CA) per the manufacturer’s protocol. The relative light intensity was quantified at 450 nm using an m1000 Pro microplate reader (Tecan, Männedorf, Switzerland).
Measurement of Extracellular Markers by Flow Cytometry.
Wild-type and Nrf2-null Jurkat T cells were labeled with anti-CD69/PECy7 (BioLegend) and anti-CD25/APC (eBioscience, San Diego, CA). After labeling, cells were washed and resuspended in Fluorescence-activated cell sorting (FACS) buffer. After resuspension, cells were analyzed by flow cytometry using the BD Accuri C6 (BD Biosciences). Fluorescence was quantified using CFlow software (BD Biosciences).
Quantification of Mitochondrial Membrane Potential and Viability by Flow Cytometry.
2 × 105 cells were stained with MitoProbe DilC1(5) per the manufacturer’s protocol (Invitrogen, Eugene, OR). Immediately prior to analysis by flow cytometry, 5 µl of propidium iodide (BioLegend) was added to each sample. All samples were analyzed by flow cytometry using the BD Accuri C6 (BD Biosciences). Fluorescence was quantified using CFlow software (BD Biosciences).
Nuclear Protein Isolation.
Wild-type and Nrf2-null Jurkat T cells (1 × 107 cells) were collected 3 hours after activation by anti-CD3/anti-CD28. Cells were lysed using a solution containing 10 mM HEPES, 100 mM KCl, 1.5 mM MgCl2, 0.1 mM EGTA, 0.5 mM Dithiothreitol, 0.5% NP40 substitute, and 1× Halt protease inhibitor cocktail (Thermo Scientific, Waltham, MA). After lysis, lysates were spun down and resuspended in nuclear extraction buffer containing 10 mM HEPES, 420 mM NaCl, 1.5 mM MgCl2, 0.1 mM EGTA, 0.5 mM Dithiothreitol, 5% glycerol, and 1× Halt protease inhibitor cocktail. Samples were kept in nuclear extraction buffer for 1 hour and were vortexed intermittently. After incubation, samples were spun down, and supernatants containing the nuclear fraction of the protein extract were collected. Nuclear protein was quantified via Bradford assay (Bio-Rad, Hercules, CA).
Western Analysis.
After quantification, nuclear protein was diluted in Laemmli Sample Buffer (Bio-Rad) supplemented with 2-mercaptoethanol so that each well contained 10 µg of nuclear protein. Samples were subjected to SDS-PAGE and subsequently transferred to a polyvinylidene difluoride membrane. Membranes were blocked with 5% nonfat dehydrated milk (NFDM) in phosphate-buffered saline containing 0.05% Tween 20 (PBST). Histone H3 (7074S) and Nrf2 (H-300) primary antibodies were diluted 1:1000 in PBST containing 5% NFDM. The H3 primary antibody was purchased from Cell Signaling Technology (Danvers, MA). The Nrf2 primary antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The secondary antibody was horseradish peroxidase–linked and was diluted 1:2000 in 0.05% PBST containing 2% NFDM. The anti-rabbit IgG secondary antibody was obtained from Cell Signaling Technology. All blots were developed via Pierce ECL Western Blotting Substrate (Thermo Scientific, Rockford, IL) per the manufacturer’s protocol. The bands were visualized by the LI-COR Odyssey FC infrared imaging system (Lincoln, NE).
Transient Transfection and Luciferase Assay.
Wild-type and Nrf2-null human Jurkat T cells were reverse transfected with 1.5 μg of plasmid and 3 μl of Lipofectamine 2000 (Life Technologies) per 5 × 105 cells for 12 hours in complete medium. After 12 hours, the cells were washed and resuspended at a concentration of 5 × 105 cells/ml in complete medium. After resuspension, the cells were seeded in a 96-well plate at a volume of 200μl per well. The cells were then treated as described in the figure legends and incubated for 12 hours. After incubation, luciferase activity was measured via a commercially available luciferase assay system kit, using the manufacturer’s protocol (Promega, Madison, WI). Luciferase luminescence was quantified by the Tecan Infinite M1000 Pro Microplate Reader (Tecan, San Jose, CA).
Statistical Analysis.
The mean ± standard error was determined for each treatment group in the individual experiments. Data were determined to be homogeneous upon passing both the Shapiro-Wilk normality test and the equal variance test. Homogeneous data were subsequently evaluated by two-way parametric analysis of variance (in which genotype was one factor and cell treatment was a second factor). When significant differences were observed, the Holm-Sidak post-hoc test was used to compare treatment groups to the vehicle control using SigmaPlot 12.3 software (Systat Software, Inc., Chicago, IL).
Results
CRISPR/Cas9 Gene Editing Causes INDEL Formation in Nrf2.
Previously, we have shown that the Nrf2 activator tBHQ causes a significant suppression in the secretion of IL-2, cell-surface expression of CD25, and transactivation activity of NFκB in human Jurkat T cells (Zagorski et al., 2013). The purpose of the present studies was to determine if these effects are dependent upon Nrf2. Toward that end, CRISPR/Cas9-mediated gene editing was implemented. Guide RNA sequences were selected from a database listing the optimal guide RNA sequences for 40% of the human exome (Mali et al., 2013b). The selected sequences were cloned into empty Cas9 vectors coexpressing GFP, as described previously (Ran et al., 2013). Sanger sequencing of the resulting Jurkat clones revealed that both guide RNAs were successful in generating INDELs at the Cas9 cut site (Fig. 1). Four clones with frameshift mutations from each guide RNA were then selected to begin screening for Nrf2 disruption.
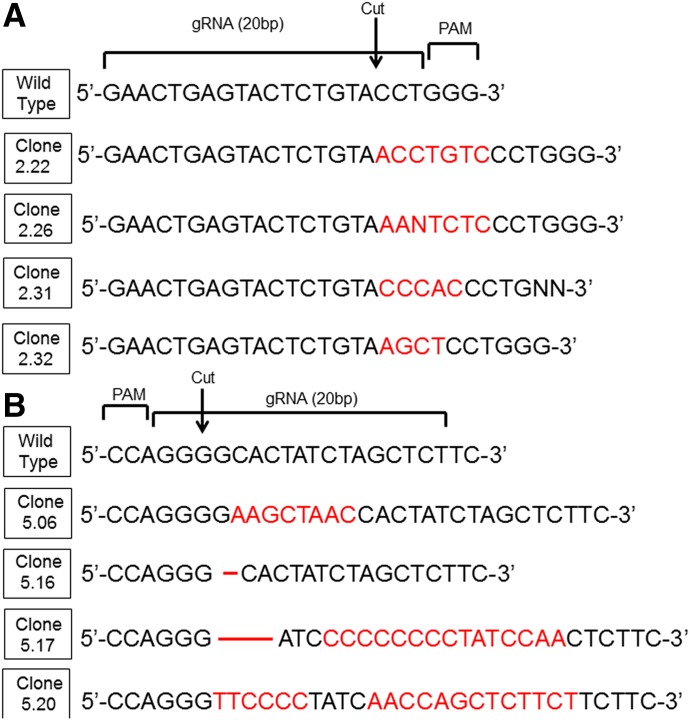
Fig. 1.
Wild-type Jurkat E6-1 cells were transiently transfected with one of two CRISPR/Cas9 plasmids coexpressing GFP, targeting different sites within the genome. The two plasmids (A and B) contained different guide RNA sequences and targeted different exons on the plus and minus strands of Nrf2, respectively. After sorting positive transfectants, cells were clonally expanded and analyzed by Sanger sequencing. INDEL mutations are listed in red font. gRNA, guide RNA; PAM, pattern adjacent motif.
CRISPR/Cas9-Mediated Gene Editing Results in Nrf2 Deficiency.
Jurkat clones identified as having frameshift mutations were then screened for Nrf2 activity, as ascertained by Nrf2 translocation and induction of Nrf2 target genes when treated with tBHQ. Wild-type Jurkat T cells displayed robust nuclear accumulation 3 hours after treatment with 10 µM tBHQ. In contrast, none of the CRISPR/Cas9 clones showed nuclear accumulation under the same conditions (Fig. 2A). Furthermore, tBHQ potently induced expression of the Nrf2 target gene Nqo1 in wild-type cells, but not pooled Nrf2-null clones (Fig. 2B). Likewise, basal expression of Nqo1 was decreased in the Nrf2-null Jurkat T cells as compared with wild-type cells. Taken together, the data strongly suggest that targeting Nrf2 by CRISPR/Cas9 successfully generated Nrf2-null clones.
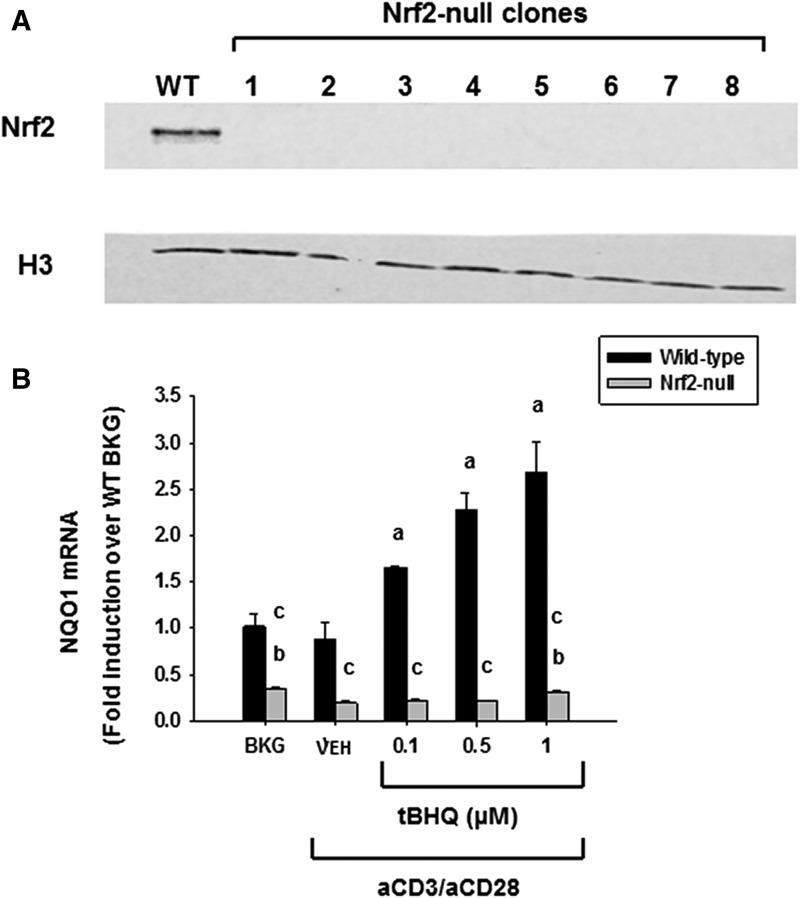
Fig. 2.
tBHQ increases Nrf2 nuclear accumulation and target gene expression in wild-type cells, but not Nrf2-null clones. (A) Wild-type Jurkat T cells or Nrf2-null Jurkat clones were treated with 10 µM tBHQ for 3 hours. Nuclear protein was then isolated, and expression of Nrf2 and the housekeeper histone H3 was probed by western analysis. (B) Wild-type (WT) and Nrf2-null Jurkat T cells were left untreated as background (BKG) or pretreated with vehicle (VEH) control (0.001% ethanol) or tBHQ (0.1–1 μM) prior to activation by anti-CD3/anti-CD28. Six hours post activation, cells were harvested, and total RNA was collected. Quantification of NQO1 was performed by SYBR Green real-time PCR. The data (n = 3) are presented as the mean ± S.E. aP < 0.05 versus wild-type VEH; bP < 0.05 versus Nrf2-null VEH; cP < 0.05 between the wild-type and Nrf2-null genotypes.
The Effect of tBHQ on IL-2 Is Largely Independent of Nrf2.
We have previously shown that the secretion of IL-2 by activated Jurkat T cells is suppressed by the Nrf2 activator tBHQ (Zagorski et al., 2013). To test whether this effect was dependent upon Nrf2, we pretreated wild-type and Nrf2-null Jurkat T cells with tBHQ (0.1–1 µM) 30 minutes prior to T cell stimulation. IL-2 secretion was evaluated 24 hours later. Our data show that suppression of IL-2 by tBHQ is largely independent of Nrf2, as tBHQ inhibits IL-2 production in both genotypes, although the Nrf2-null Jurkat cells were slightly less sensitive (Fig. 3). In the present study, only noncytotoxic concentrations of tBHQ were used. To determine whether the effect of tBHQ on T cells is similar at physiologic oxygen concentrations, we treated Jurkat cells with tBHQ and cultured the cells at 5% oxygen to better mimic physiologic oxygen concentrations in blood. However, the effect of tBHQ on IL-2 production was similar at 5% oxygen concentration (data not shown).
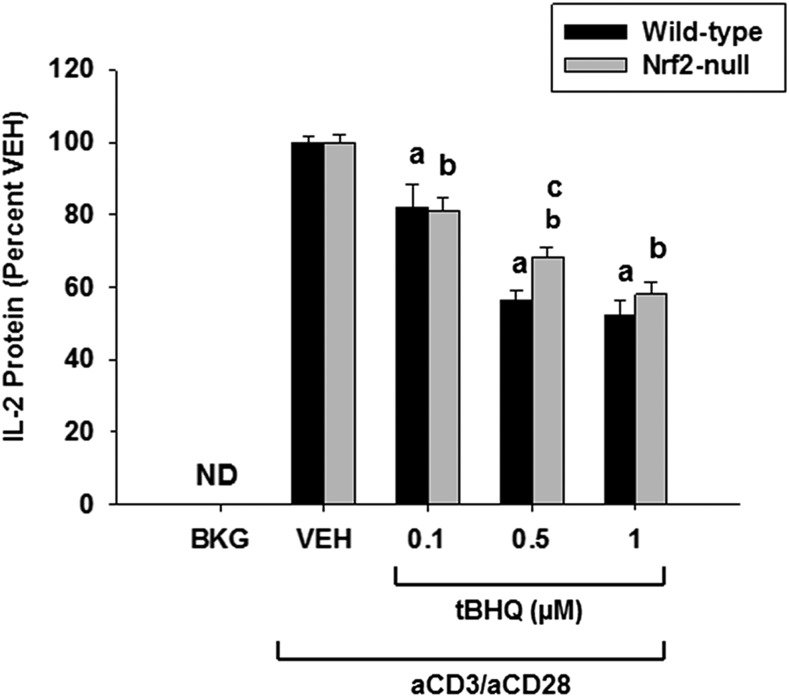
Fig. 3.
Suppression of IL-2 production by tBHQ is largely independent of Nrf2. Human Jurkat T cells were left untreated as background (BKG) or pretreated with vehicle (VEH) control (0.001% ethanol) or tBHQ (0.1–1 µM) for 30 minutes prior to activation with anti-CD3/anti-CD28. After 24 hours, the supernatants were collected, and IL-2 was detected and quantified via enzyme-linked immunosorbent assay. The data (n = 3) are presented as the mean ± S.E. aP < 0.05 versus wild-type VEH; bP < 0.05 versus Nrf2-null VEH; cP < 0.05 between the wild-type and Nrf2-null genotypes. ND, not detectable.
Suppression of CD25 by tBHQ Is Partially Nrf2-Dependent.
In addition to the suppression of IL-2, we have previously shown that tBHQ suppresses the cell-surface expression of CD25, the high-affinity chain of the IL-2 receptor (Zagorski et al., 2013). To determine the role of Nrf2 in the suppression of CD25 by tBHQ, wild-type and Nrf2-null Jurkat T cells were pretreated with tBHQ prior to T cell stimulation with anti-CD3/anti-CD28. Twenty-four hours post stimulation, the expression of CD25 and CD69 was quantified by flow cytometry. CD25 and CD69 are cell-surface molecules that are rapidly upregulated after T cell activation and thus serve as early markers of T cell activation. The data indicate that the inhibition of CD25 expression by tBHQ is partially dependent on the expression of Nrf2, as Nrf2-null Jurkat T cells are more sensitive than wild-type cells to tBHQ treatment. Additionally, tBHQ had little effect on CD69 expression, which is consistent with previous studies (Fig. 4).
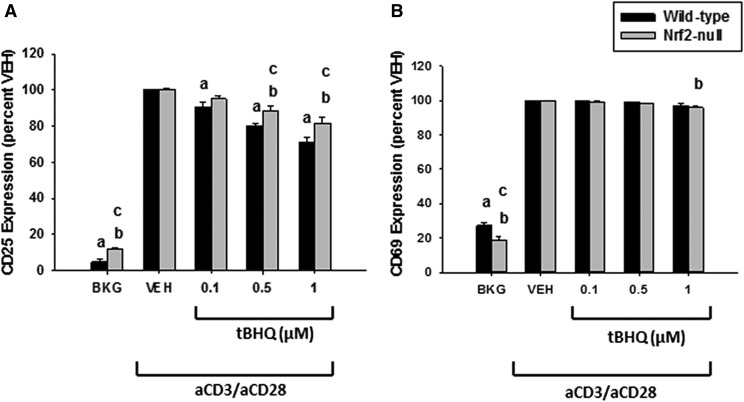
Fig. 4.
Treatment of Jurkat T cells with tBHQ suppresses CD25 in a partly Nrf2-dependent manner. Human Jurkat T cells were left untreated as background (BKG) or pretreated with vehicle (VEH) control (0.001% ethanol) or tBHQ (0.1–1 µM) for 30 minutes prior to activation with anti-CD3/anti-CD28. After 24 hours, the percentage of cells expressing CD25 (A) and CD69 (B) was quantified by flow cytometry. The data (n = 3) are presented as the mean ± S.E. aP < 0.05 versus wild-type VEH; bP < 0.05 versus Nrf2-null VEH; cP < 0.05 between the wild-type and Nrf2-null genotypes.
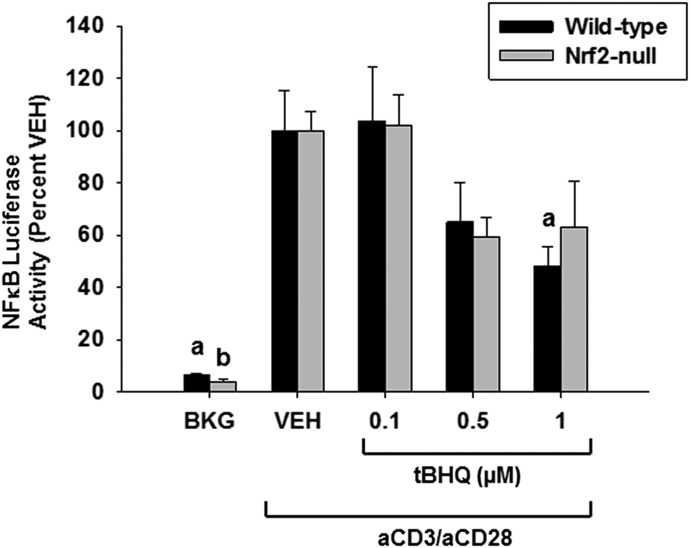
NFκB Luciferase Activity Is Inhibited by tBHQ in an Nrf2-Independent Manner.
Previous studies have established there is cross-talk between the Nrf2 and NFκB signaling pathways, where activation of Nrf2 inhibits the phosphorylation of inhibitor of κB kinase, resulting in a loss of NFκB activity (Thimmulappa et al., 2006; Liby et al., 2008). We have previously shown that tBHQ suppresses NFκB transcriptional activity in Jurkat T cells (Zagorski et al., 2013). To assess the role of Nrf2 in tBHQ-induced suppression of NFκB, we transiently transfected an NFκB luciferase reporter into wild-type and Nrf2-null Jurkat T cells 12 hours prior to treatment with tBHQ and T cell activation. Genotypes were normalized to their own vehicle control to control for potential differences in transfection efficiency. These data demonstrate that the inhibition of NFκB activity by tBHQ occurs independently of Nrf2, as both genotypes display similar inhibition of NFκB (Fig. 5).
Fig. 5.
NFκB transcriptional activity is decreased by tBHQ treatment independently of Nrf2. Human Jurkat T cells were transfected with luciferase reporter plasmids driven by NFκB 12 hours prior to tBHQ treatment and activation. Jurkat T cells were left untreated as background (BKG) or pretreated with vehicle (VEH) control (0.001% ethanol) or tBHQ (0.1–1 µM) for 30 minutes prior to activation with anti-CD3/anti-CD28. Twelve hours post stimulation, luciferase activity was measured by luminescence, using a Tecan microplate reader. The data (n = 3) are presented as the mean ± S.E. aP < 0.05 versus wild-type VEH; bP < 0.05 versus Nrf2-null VEH; cP < 0.05 between the wild-type and Nrf2-null genotypes.
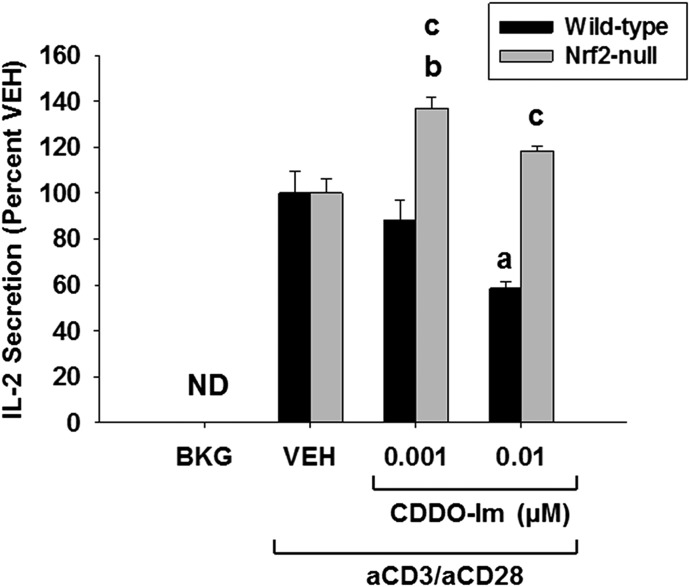
CDDO-Im Inhibits IL-2 Secretion in an Nrf2-Dependent Manner.
The current data show that the inhibition of IL-2 secretion and NFκB activity by tBHQ is largely independent of the transcription factor Nrf2. To further investigate the role of Nrf2 in the early events following T cell activation, we investigated the effects of another, more potent, Nrf2 activator, CDDO-Im (Liby and Sporn, 2012). Wild-type and Nrf2-null T cells were treated with CDDO-Im (0.001–0.01 µM) for 30 minutes prior to stimulation of T cells with anti-CD3/anti-CD28. After 24 hours, analysis of cell supernatants demonstrated an Nrf2-dependent suppression of IL-2 secretion in wild-type Jurkat T cells (Fig. 6). Interestingly, treatment of Nrf2-null Jurkat T cells resulted in an Nrf2-independent increase in IL-2 secretion (Fig. 6). In the present study, only noncytotoxic concentrations of CDDO-Im were used. Overall, these data demonstrate that the inhibition of IL-2 secretion by CDDO-Im in activated Jurkat T cells occurs through Nrf2.
Fig. 6.
Suppression of IL-2 production by CDDO-Im is Nrf2-dependent. Human Jurkat T cells were left untreated as background (BKG) or pretreated with vehicle (VEH) control (0.001% dimethylsulfoxide) or CDDO-Im (0.001–0.01 µM) for 30 minutes prior to activation with anti-CD3/anti-CD28. After 24 hours, the supernatants were collected, and IL-2 was detected and quantified via enzyme-linked immunosorbent assay. The data (n = 3) are presented as the mean ± S.E. aP < 0.05 versus wild-type VEH; bP < 0.05 versus Nrf2-null VEH; cP < 0.05 between the wild-type and Nrf2-null genotypes. ND, = not detectable.
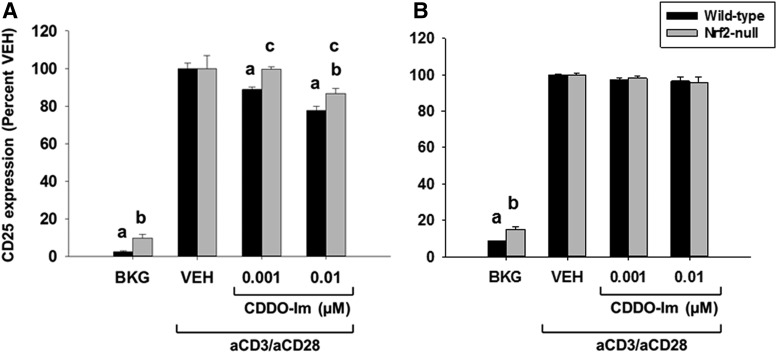
Suppression of CD25 by CDDO-Im Is Partially Nrf2-Dependent.
To further characterize CDDO-Im in activated Jurkat T cells, we quantified expression of CD25 and CD69. Wild-type and Nrf2-null T cells were treated with CDDO-Im (0.001–0.01 µM) for 30 minutes prior to T cell stimulation. After a 24-hour incubation, the expression of CD25 and CD69 was quantified by flow cytometry. Similar to tBHQ (Fig. 4A), CDDO-Im suppressed CD25 expression in a partially Nrf2-dependent manner (Fig. 7A) but had little effect on CD69 expression (Fig. 7B). Taken together, these results demonstrate that CDDO-Im has both Nrf2-dependent and -independent effects upon early events following activation of Jurkat T cells.
Fig. 7.
Treatment of Jurkat T cells with CDDO-Im suppresses CD25 in a partly Nrf2-dependent manner. Human Jurkat T cells were left untreated as background (BKG) or pretreated with vehicle (VEH) control (0.001% dimethylsulfoxide) or CDDO-Im (0.001–0.01 µM) for 30 minutes prior to activation with anti-CD3/anti-CD28. After 24 hours, the percentage of cells expressing CD25 (A) and CD69 (B) was quantified by flow cytometry. The data (n = 3) are presented as the mean ± S.E. aP < 0.05 versus wild-type VEH; bP < 0.05 versus Nrf2-null VEH; cP < 0.05 between the wild-type and Nrf2-null genotypes.
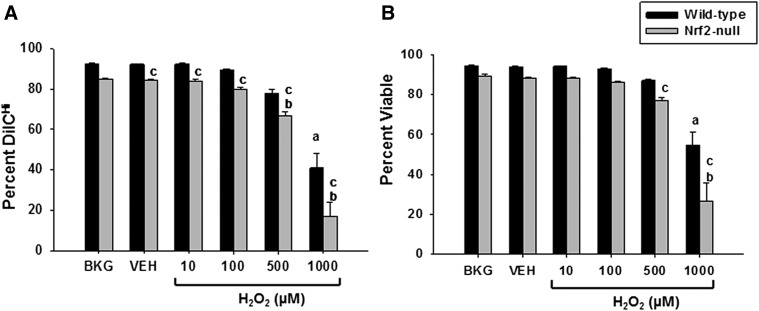
Nrf2-Null Jurkat T Cells Are More Sensitive to H2O2.
Reactive oxygen species play an important role in the immune response, both as cytotoxic agents against pathogens as well as signaling molecules. Accordingly, such reactive moieties contribute to and are frequently present during the course of an immune response and thus represent potential endogenous activators of Nrf2 in immune cells. Although important contributors to host defense, reactive oxygen species produced by classically activated macrophages and other leukocytes can also damage immune cells in the vicinity of the pathogens. To determine the role of Nrf2 in providing protection against oxidative insult, Jurkat T cells were treated with cytotoxic levels of hydrogen peroxide (10–1000 μM). Mitochondrial membrane potential was assessed with the cell-permeant dye DilC, which accumulates in mitochondria with normal membrane potential. A reduction in DilC fluorescence indicates a disruption of mitochondrial membrane potential and mitochondrial stress. Hydrogen peroxide caused a significant decrease in mitochondrial membrane potential in Jurkat T cells that was more pronounced in the Nrf2-null clones (Fig. 8A). The mitochondrial stress correlated with a decrease in cell viability as assessed by propidium iodide exclusion. The loss of cell viability by hydrogen peroxide was significantly greater in the Nrf2-null clones (Fig. 8B). These data demonstrate that Nrf2-null Jurkat T cells are more sensitive to oxidative stress induced by hydrogen peroxide than wild-type cells, which suggests that Nrf2 may provide protection against endogenous reactive oxygen species produced during the course of an immune response.
Fig. 8.
Nrf2-null Jurkat clones are more sensitive to exposure to hydrogen peroxide. Human Jurkat T cells were left untreated as background (BKG) or treated with vehicle (VEH) control (0.01% water in RPMI) or H2O2 (1–1000 µM) for 16 hours prior to flow cytometry analysis for mitochondrial membrane potential as measured using the cell permeable dye DilC1(5) (A) and cell viability as quantified by propidium iodide exclusion (B). The data (n = 3) are presented as the mean ± S.E. aP < 0.05 versus wild-type VEH; bP < 0.05 versus Nrf2-null VEH; cP < 0.05 between the wild-type and Nrf2-null genotypes.
Discussion
Nrf2 has emerged as a critical signaling pathway in detoxification, cell survival, and immune regulation (Kensler et al., 2007; Ma, 2013). There is also a growing body of evidence to suggest Nrf2 may play an important role in the promotion of tumorigenesis in some models (DeNicola et al., 2011). Accordingly, investigation into the multiple and complex roles of Nrf2 has become a very active area of research. We have developed the first human Nrf2-null model using CRISPR/Cas9 technology. As such, we believe this model will be a useful tool for a variety of studies. In addition, the model may help to distinguish between therapeutically beneficial and adverse effects of Nrf2 activation in human immune response and in human cancer, which has been difficult to parse apart thus far.
In the literature, the role of Nrf2 in human T cell function has been unclear. We have previously shown in both cell lines and primary human T cells that the Nrf2 activator tBHQ suppresses cytokine production, expression of surface markers of activation, and activity of NFκB(Zagorski et al., 2013; Turley et al., 2015). Other groups have used other Nrf2 activators, including benzo[a]pyrene and arsenic compounds, to both implicate and absolve Nrf2 in immune modulation (Katika et al., 2011; Murugaiyan et al., 2013; Morzadec et al., 2014; Kaufmann et al., 2016). In addition, the Jurkat cell line has been used to observe the role of Nrf2 in cellular function apart from T cell activation, including measurement of glutathione induction by Nrf2, as well as antioxidant response (Kataoka et al., 2001; Ogasawara et al., 2014). As pharmacological tools specifically modulating Nrf2 are not readily available, the field as a whole has been reliant upon techniques such as small interfering RNA and small hairpin RNA, which result in less than total knockout and thus make the data difficult to interpret. Ultimately, these technical problems have left the question of Nrf2-specific effects in human T cells largely unanswered.
For the current studies, we used this model to address a long-standing question in our laboratory, and the Nrf2 community, concerning the role of Nrf2 in the early events following T cell activation. Our results suggest that the activation of Nrf2 by tBHQ in this setting is complex; whereas inhibition of IL-2 production by tBHQ appears to be largely Nrf2-independent, the suppression of CD25 expression by tBHQ appears to be partially Nrf2-dependent. The decrease in NFκB activity by tBHQ appears to be Nrf2-independent. With respect to cell stress caused by an Nrf2 activator that is endogenously produced (hydrogen peroxide), we found that absence of Nrf2 increased the susceptibility of Jurkat T cells to mitochondrial stress and loss of viability.
Although the effects of tBHQ upon the early events of human T cell activation were determined to be independent of Nrf2, the overarching goal of these studies was to determine the role of Nrf2 in human T cell activation. Toward this end, we used the synthetic triterpenoid CDDO-Im, which is widely used as a potent Nrf2 activator. Treatment of wild-type and Nrf2-null human Jurkat T cells with CDDO-Im resulted in an Nrf2-dependent suppression of IL-2 secretion. Importantly, to our knowledge, this is the first time in which activation of Nrf2 has been shown to modulate a human immune response, which is consistent with a role for Nrf2 as an important regulator of human immune function. The concentrations of CDDO-Im used in the present studies were markedly lower than those of tBHQ because CDDO-Im is a more potent activator of Nrf2 (Dinkova-Kostova et al., 2005; Liby and Sporn, 2012). Furthermore, higher concentrations of CDDO-Im were cytotoxic to both wild-type and Nrf2- null cells (data not shown).
One unexpected finding of the current studies was that the inhibition of NFκB activation by tBHQ seems to be independent of Nrf2. This was surprising since tBHQ is a strong Nrf2 activator, and activation of Nrf2 has been previously shown to inhibit NFκB activation in immune cells (Thimmulappa et al., 2006). The regulation of NFκB is quite complex. Classic activation of NFκB involves a series of phosphorylation events that culminate in the phosphorylation of inhibitor of κBα, which is subsequently degraded by the proteasome and thus liberates the p65/p50 homodimer, resulting in activation (Flohe et al., 1997). However, in addition to this component of NFκB activation, it is also well established that reactive oxygen intermediates also play an essential role. Accordingly, a number of structurally disparate antioxidants have been shown to inhibit NFκB activation in immune cells, including butylated hydroxyanisole, an antioxidant that is structurally similar to tBHQ (Schulze-Osthoff et al., 1993). Thus, it seems plausible that the inhibition of NFκB transactivation activity by tBHQ may instead be due to direct antioxidant effects.
Recent evidence has led some researchers to suggest that Nrf2 may be a proto-oncogene because it functions endogenously to resolve cell stress to xenobiotics and reactive oxygen species, but the system appears to be hijacked in certain types of cancer (Shelton and Jaiswal, 2013). One of the hallmarks of cancer is immune cell evasion, and this occurs by several mechanisms, such as the upregulation of the T cell inhibitory marker PD-L1. Notably, we have previously shown that tBHQ inhibits Th1 differentiation in an Nrf2-dependent manner (Rockwell et al., 2012). This potentially adds another mechanism by which Nrf2 may promote the survival of tumor cells, as the Th1 response is known to be crucial for tumor cell surveillance. Our current model is unique in that it has applicability beyond immune endpoints. The Jurkat E6-1 cell line is a T cell leukemia. Until recently, determining the role of Nrf2 in human cancer biology has been difficult in that it has relied upon either overexpression or knockdown of Nrf2 mRNA, both of which may result in the generation of artifacts (Sanchez-Rivera and Jacks, 2015). The implementation of CRISPR/Cas9 methodology has, for the first time, provided a model in which the role of Nrf2 can be studied, without the addition of such confounding factors. Furthermore, our data demonstrate that, in the absence of Nrf2, Jurkat T cells are significantly more sensitive to oxidative stress induced by hydrogen peroxide, further supporting the hypothesis that Nrf2 promotes tumor cell survival.
Overall, this study is novel because, to our knowledge, it is the first to generate a human Nrf2-null model using a genome-editing approach, which is a significant improvement over RNA interference previously used by our laboratory and others. In most model systems and particularly in immune cells, targeting Nrf2 using small hairpin RNA or small interfering RNA has fallen far short of full gene knockout. Our data demonstrate that targeting Nrf2 by CRISPR/Cas9 genome editing essentially abrogated Nrf2 expression and function. Using this model, we determined that the inhibition of IL-2 and its receptor, CD25, by the widely used food additive tBHQ is complex, with Nrf2-dependent and Nrf2-independent components. The data also suggest a role for NFκB in this process, which likely occurs through direct antioxidant effects of tBHQ. Furthermore, utilization of a second, more potent, Nrf2 activator, CDDO-Im, resulted in an Nrf2-dependent suppression of IL-2 secretion, as well as a partially Nrf2-dependent suppression of the early activation marker CD25. Collectively, these studies demonstrate that low concentrations of tBHQ that are relevant to human exposure modulate multiple stress-activated transcription factors, which ultimately impairs early T cell response following activation. Furthermore, these studies also showed that Nrf2 activation by CDDO-Im suppresses IL-2 secretion after T cell stimulation, demonstrating that Nrf2 is a regulator of early events following activation of human Jurkat T cells.
Acknowledgments
The authors thank Dr. Bryan Copple and his laboratory for the use of some of the reagents and instrumentation used in this study. The authors also thank Dr. Louis King and the Michigan State flow cytometry core for assistance in Fluorescence-activated cell sorting of Jurkat clones.
Abbreviations
- Cas9
CRISPR-associated protein 9
- CDDO-Im
1[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole
- CRISPR
clustered regularly interspaced short palindromic repeats
- GFP
Green Florescent Protein
- gRNA
guide RNA
- IL-2
interleukin-2
- INDEL
insertions or deletions
- NFDM
nonfat dehydrated milk
- Nqo1
NADPH quinone oxidoreductase 1
- Nrf2
nuclear factor erythroid 2-related factor 2
- PBST
phosphate-buffered saline containing Tween 20
- PCR
polymerase chain reaction
- tBHQ
tert-butylhydroquinone
Authorship Contributions
Participated in research design: Zagorski, Rockwell.
Conducted experiments: Zagorski, Maser.
Contributed new reagents or analytic tools: Zagorski, Liby.
Performed data analysis: Zagorski, Rockwell.
Wrote or contributed to the writing of the manuscript: Zagorski, Maser, Liby, Rockwell.
Footnotes
These studies were supported by the National Institutes of Health [Grants ES018885 and ES024966 to C.E.R. and GM092715 to J.W.Z.].
References
- Cantrell DA. (1996) T cell antigen receptor signal transduction pathways. Cancer Surv 27:165–175. [PubMed] [Google Scholar]
- Chan K, Lu R, Chang JC, Kan YW. (1996) NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci USA 93:13943–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. (2011) Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475:106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. (2002) Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA 99:11908–11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Liby KT, Stephenson KK, Holtzclaw WD, Gao X, Suh N, Williams C, Risingsong R, Honda T, Gribble GW, et al. (2005) Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proc Natl Acad Sci USA 102:4584–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flohé L, Brigelius-Flohé R, Saliou C, Traber MG, Packer L. (1997) Redox regulation of NF-kappa B activation. Free Radic Biol Med 22:1115–1126. [DOI] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–322. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. (1999) Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DA, Amirahmadi S, Ward C, Fabry Z, Johnson JA. (2010) The absence of the pro-antioxidant transcription factor Nrf2 exacerbates experimental autoimmune encephalomyelitis. Toxicol Sci 114:237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K, Handa H, Nishizawa M. (2001) Induction of cellular antioxidative stress genes through heterodimeric transcription factor Nrf2/small Maf by antirheumatic gold(I) compounds. J Biol Chem 276:34074–34081. [DOI] [PubMed] [Google Scholar]
- Katika MR, Hendriksen PJ, van Loveren H, Peijnenburg A. (2011) Exposure of Jurkat cells to bis (tri-n-butyltin) oxide (TBTO) induces transcriptomics changes indicative for ER- and oxidative stress, T cell activation and apoptosis. Toxicol Appl Pharmacol 254:311–322. [DOI] [PubMed] [Google Scholar]
- Kaufmann KB, Gothwal M, Schallner N, Ulbrich F, Rücker H, Amslinger S, Goebel U. (2016) The anti-inflammatory effects of E-α-(p-methoxyphenyl)-2′,3,4,4′-tetramethoxychalcone are mediated via HO-1 induction. Int Immunopharmacol 35:99–110. [DOI] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. (2007) Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47:89–116. [DOI] [PubMed] [Google Scholar]
- Li J, Stein TD, Johnson JA. (2004) Genetic dissection of systemic autoimmune disease in Nrf2-deficient mice. Physiol Genomics 18:261–272. [DOI] [PubMed] [Google Scholar]
- Liby K, Risingsong R, Royce DB, Williams CR, Yore MM, Honda T, Gribble GW, Lamph WW, Vannini N, Sogno I, et al. (2008) Prevention and treatment of experimental estrogen receptor-negative mammary carcinogenesis by the synthetic triterpenoid CDDO-methyl Ester and the rexinoid LG100268. Clin Cancer Res 14:4556–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liby KT, Sporn MB. (2012) Synthetic oleanane triterpenoids: multifunctional drugs with a broad range of applications for prevention and treatment of chronic disease. Pharmacol Rev 64:972–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. (2013) Role of Nrf2 in oxidative stress and toxicity. Ann Rev Pharmacol Toxicol 53:401–426.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Battelli L, Hubbs AF. (2006) Multiorgan autoimmune inflammation, enhanced lymphoproliferation, and impaired homeostasis of reactive oxygen species in mice lacking the antioxidant-activated transcription factor Nrf2. Am J Pathol 168:1960–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. (2013a) Cas9 as a versatile tool for engineering biology. Nat Methods 10:957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. (2013b) RNA-guided human genome engineering via Cas9. Science 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mane VP, Heuer MA, Hillyer P, Navarro MB, Rabin RL. (2008) Systematic method for determining an ideal housekeeping gene for real-time PCR analysis. J Biomol Tech 19:342–347. [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Hayes JD. (2003) Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem 278:21592–21600. [DOI] [PubMed] [Google Scholar]
- Morzadec C, Macoch M, Sparfel L, Kerdine-Römer S, Fardel O, Vernhet L. (2014) Nrf2 expression and activity in human T lymphocytes: stimulation by T cell receptor activation and priming by inorganic arsenic and tert-butylhydroquinone. Free Radic Biol Med 71:133–145. [DOI] [PubMed] [Google Scholar]
- Murugaiyan J, Rockstroh M, Wagner J, Baumann S, Schorsch K, Trump S, Lehmann I, Bergen Mv, Tomm JM. (2013) Benzo[a]pyrene affects Jurkat T cells in the activated state via the antioxidant response element dependent Nrf2 pathway leading to decreased IL-2 secretion and redirecting glutamine metabolism. Toxicol Appl Pharmacol 269:307–316. [DOI] [PubMed] [Google Scholar]
- Ogasawara Y, Takeda Y, Takayama H, Nishimoto S, Ichikawa K, Ueki M, Suzuki T, Ishii K. (2014) Significance of the rapid increase in GSH levels in the protective response to cadmium exposure through phosphorylated Nrf2 signaling in Jurkat T-cells. Free Radic Biol Med 69:58–66. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, Fryer AD, Yamamoto M, Kensler TW, Tuder RM, Georas SN, et al. (2005) Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med 202:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell CE, Zhang M, Fields PE, Klaassen CD. (2012) Th2 skewing by activation of Nrf2 in CD4(+) T cells. J Immunol 188:1630–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rivera FJ, Jacks T. (2015) Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer 15:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykally JN, Rachmany L, Hatic H, Shaer A, Rubovitch V, Pick CG, Citron BA. (2012) The nuclear factor erythroid 2-like 2 activator, tert-butylhydroquinone, improves cognitive performance in mice after mild traumatic brain injury. Neuroscience 223:305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Osthoff K, Beyaert R, Vandevoorde V, Haegeman G, Fiers W. (1993) Depletion of the mitochondrial electron transport abrogates the cytotoxic and gene-inductive effects of TNF. EMBO J 12:3095–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahidi F. (2000) Antioxidants in food and food antioxidants. Nahrung 44:158–163. [DOI] [PubMed] [Google Scholar]
- Shelton P, Jaiswal AK. (2013) The transcription factor NF-E2-related factor 2 (Nrf2): a protooncogene? FASEB J 27:414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmulappa RK, Lee H, Rangasamy T, Reddy SP, Yamamoto M, Kensler TW, Biswal S. (2006) Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest 116:984–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley AE, Zagorski JW, Rockwell CE. (2015) The Nrf2 activator tBHQ inhibits T cell activation of primary human CD4 T cells. Cytokine 71:289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK. (1996) Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci USA 93:14960–14965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (1975) Toxicological evaluation of some food colours, thickening agents, and certain other substances: tertiary butyl hydroquinone, in WHO Food Additives Series, World Health Organization, Geneva, Switzerland. [Google Scholar]
- Yoh K, Itoh K, Enomoto A, Hirayama A, Yamaguchi N, Kobayashi M, Morito N, Koyama A, Yamamoto M, Takahashi S. (2001) Nrf2-deficient female mice develop lupus-like autoimmune nephritis. Kidney Int 60:1343–1353. [DOI] [PubMed] [Google Scholar]
- Zagorski JW, Turley AE, Dover HE, VanDenBerg KR, Compton JR, Rockwell CE. (2013) The Nrf2 activator, tBHQ, differentially affects early events following stimulation of Jurkat cells. Toxicol Sci 136:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]