Abstract
Phenobarbital was the first therapeutic drug to be characterized for its induction of hepatic drug metabolism. Essentially at the same time, cytochrome P450, an enzyme that metabolizes drugs, was discovered. After nearly 50 years of investigation, the molecular target of phenobarbital induction has now been delineated to phosphorylation at threonine 38 of the constitutive androstane receptor (NR1I3), a member of the nuclear receptor superfamily. Determining this mechanism has provided us with the molecular basis to understand drug induction of drug metabolism and disposition. Threonine 38 is conserved as a phosphorylation motif in the majority of both mouse and human nuclear receptors, providing us with an opportunity to integrate diverse functions of nuclear receptors. Here, I review the works and accomplishments of my laboratory at the National Institutes of Health National Institute of Environmental Health Sciences and the future research directions of where our study of the constitutive androstane receptor might take us.
Phenobarbital Induction Mechanism
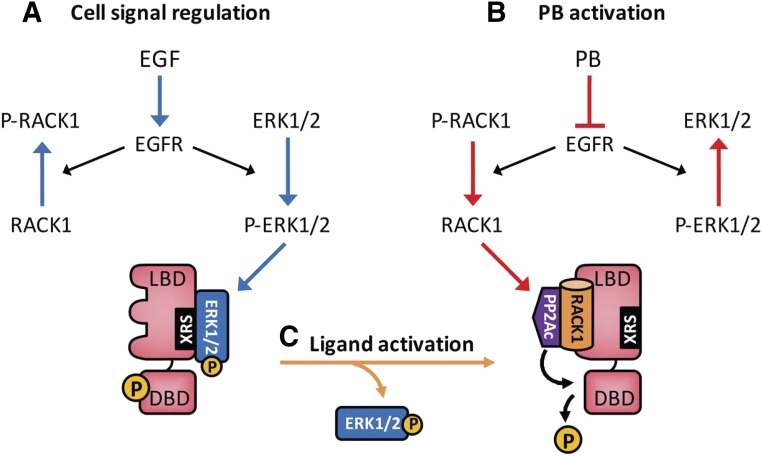
To begin, the current cell signal/molecular mechanism of phenobarbital (PB) induction/constitutive androstane receptor (CAR) activation is depicted in Fig. 1. The underlying mechanism of CAR regulation is dephosphorylation of threonine 38, which can be inhibited by epidermal growth factor (EGF) to inactivate CAR (Fig. 1A) or induced by PB to activate CAR (Fig. 1B). Ligands directly bind and enable CAR to be dephosphorylated for its activation (Fig. 1C). Here, I will detail these regulatory processes.
Fig. 1.
The molecular mechanism of PB induction. (A) EGF activates EGFR, preventing CAR from being dephosphorylated and keeping CAR inactivated. (B) PB binds EGFR and represses its signaling, dissociating ERK1/2 from XRS and allowing RACK1/PP2Ac to dephosphorylate threonine 38 for CAR activation. (C) Ligands such as CITCO directly bind phosphorylated CAR, dissociating P-ERK1/2 from XRS. Blue arrows indicate signals to inactivate CAR, and red and orange arrows indicate signals to activate CAR. XRS, xenochemical response signal.
Phenobarbital-Responsive Enhancer Module.
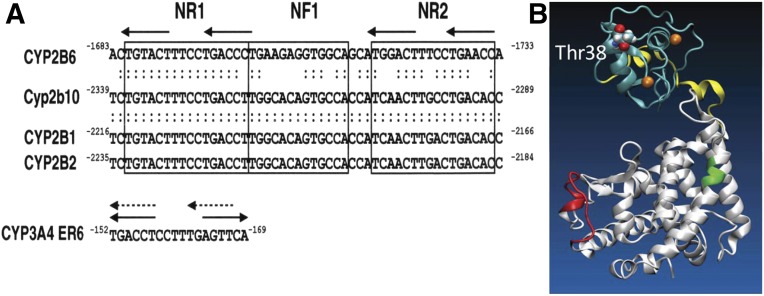
There have been long and intensive investigations to identify a regulatory DNA element in PB-induced cytochrome P450 (P450) genes (e.g., CYP2B) (Omura and Sato, 1962; Remmer and Merker, 1963; Honkakoski and Negishi, 1998; Mutoh et al., 2009). A DNA sequence that responds to PB was first identified within a distal region of the rat CYP2B2 promoter (Trottier et al., 1995) and characterized as the 51-bp phenobarbital-responsive enhancer module (PBREM) in the mouse Cyp2b10 promoter (Honkakoski et al., 1998b). PBREM consists of two nuclear receptor binding DR4 motifs (NR1 and NR2) that flank a nuclear factor-1 binding site. In mouse primary hepatocytes, PBREM was activated by drugs such as clotrimazole and chlorpromazine in addition to PB and by numerous chemicals, including a potent PB-like inducer 1,4-Bis-[2-(3,5-dichloropyridyloxy)]benzene, 3,3′,5,5′-Tetrachloro-1,4-bis(pyridyloxy)benzene (TCPOBOP) and polychlorinated biphenyls 2,2′,4,4′-tetrachlorobiphenyl, 2,2′,5,5′-tetrachlorobiphenyl, and 2,3,3′,5,6-hexachlorobiphenyl, but not by aryl hydrocarbon receptor ligands 3-methylcholanthrene and1,4-bis[2-(3-chloro-pyridyloxy)]benzene (Honkakoski et al., 1998a). The PBREM is conserved in the CYP2B genes from rodents to humans, providing us with a cross-species consensus enhancer for activation by PB and PB-type inducers as the experimental basis for subsequent investigations (Fig. 2A).
Fig. 2.
(A) PBREM sequences are conserved in the mouse (Cyp2b10), rat (CYP2B1 and CYP2B2), and human (CYP2B6) genes (Sueyoshi et al., 1999). NR1 and NR2 denote nuclear receptor binding sites 1 and 2, respectively, and NF1 denotes nuclear factor 1. A response element in the human CYP3A4 promoter was reported as ER6, which can also be DR4. (B) A three-dimensional model of the CAR structure is depicted from our previous publication (Mutoh et al., 2013). PB indirectly activates the CAR by inhibition of EGFR signaling (from Mutoh et al., 2013; reprinted with permission from AAAS). DBD is shown in light blue, the hinge in yellow, and the LBD in white. Green and red indicate the XRS and the RACK1/PP2Ac binding site, respectively. XRS, xenochemical response signal.
Nuclear Receptor CAR.
Given the caveat that PB induction is a liver-predominant event, various hepatocyte-enriched nuclear receptors were screened. Among them, nuclear receptor CAR activated PBREM (Honkakoski et al., 1998b). In its short history, the cDNA MB67 was cloned as a nuclear receptor that activates an empirical set of retinoic acid response elements (Baes et al., 1994). Because its activation occurs spontaneously, the term constitutive activator of retinoic acid response replaced MB67 and was subsequently shortened to “constitutive active receptor” or CAR. Later, androstanes were found to repress constitutive activity of CAR in cell-based assays, which became the basis of “CAR” to mean constitutive androstane receptor (Forman et al., 1998). This trivial name gave rise to the notion that nuclear receptors are ligand-activated transcription factors and should affiliate with so-called endogenous ligands. Although androstanes are useful to identify CAR activators in cell-based reactivation assays (Sueyoshi et al., 1999), there is no biologic significance to CAR being repressed by androstanes. In addition, an androstane also activates the pregnane X receptor (Moore et al., 2000). The true characteristic of CAR resides in its constitutively activated nature that has become the biologic basis to understand CAR functions.
While characterizing CAR as a PBREM-activating transcription factor in cell-based assays, we performed an independent study in which we used NR1-affinity chromatography to purify a nuclear protein that binds to the PBREM within the Cyp2b10 promoter in the mouse liver after PB treatment. Western blot and peptide sequence analyses of purified fractions determined this binding protein to be CAR. The results obtained from both cell-based assays and purification implicated CAR as the PB-activated transcription factor (Honkakoski et al., 1998b). Shortly thereafter, as expected, the Car gene was knocked out in mice, confirming that CAR is a nuclear receptor that mediates PB activation of the CYP2B genes in the liver (Wei et al., 2000). Subsequently, studies with transgenic mice that express either human CAR or pregnane X receptor in the absence of their mouse counterpart determined CAR as the PB-activated nuclear receptor in the mouse liver (Scheer et al., 2008). At that time, CAR (a relatively obscure nuclear receptor for 5 years after it was cloned) became a target for intense investigations in the research field of drug metabolism and toxicity as well as drug–drug interactions.
Phosphorylation of Threonine 38.
CAR is constitutively activated and this activity must be repressed to acquire response capability to PB. Western blot analysis of the mouse liver revealed that CAR translocates from the cytoplasm into the nucleus after PB treatment (Kawamoto et al., 1999). Moreover, this nuclear translocation correlated with a PB-induced increase of CYP2B10 mRNA. This observation led us to hypothesize that cytoplasmic retention is the mechanism by which CAR represses its constitutive activity. In mouse primary hepatocytes, treatment with okadaic acid, a protein phosphatase (PP) inhibitor, inhibited this nuclear translocation (Kawamoto et al., 1999). In 2009, the target of this dephosphorylation was delineated to threonine 38 of CAR; threonine 38 was phosphorylated and dephosphorylated before and after PB treatment in the mouse liver and primary hepatocytes (Mutoh et al., 2009). The topology of threonine 38 and the two functional features, which will be discussed later, is defined in the simulated three-dimensional structure of CAR as including both the DNA binding domain (DBD) and ligand binding domain (LBD) (Fig. 2B). Subsequently, phosphorylation and dephosphorylation were shown to occur in human primary hepatocytes (Yang et al., 2014), confirming that PB-induced dephosphorylation at threonine 38 is conserved in both mice and humans. In fact, the phosphomimetic CAR T38D mutant was unable to bind NR1 in gel-shift assays and was retained in the cytoplasm of human hepatoma-derived HepG2 cells. In the mouse liver, the CAR T38D mutant was retained in the cytoplasm even after PB treatment, whereas the nonphosphomimetic CAR T38A mutant spontaneously accumulated in the nucleus. Although threonine 38 constitutes a protein kinase C (PKC) phosphorylation motif and is phosphorylated by PKC in in vitro kinase assays, the kinase that phosphorylates threonine 38 in the liver in vivo and where it is phosphorylated in the cytoplasm and/or in the nucleus remain vague. Our recent study showed that threonine 38 can be phosphorylated by p38 mitogen-activated protein kinase (MAPK) in the nucleus of liver cells. p38 MAPK can be the kinase that phosphorylates threonine 38 in the nucleus, exporting phosphorylated CAR into the cytoplasm (Hori et al., 2016). Nevertheless, it is now accepted that the dephosphorylation of threonine 38 is the underlying mechanism of CAR activation.
Cell Signal Regulation.
CAR is activated by dephosphorylation of threonine 38. How is this dephosphorylation regulated? First, investigators found that growth factors such as EGF, hepatocyte growth factor, and insulin repress CAR activation and nuclear translocation in rat and/or mouse primary hepatocytes (Bauer et al., 2004; Koike et al., 2007; Yasujima et al., 2016). Therefore, CAR is, in principle, a cell signal–regulated nuclear receptor (Fig. 1A). The enzyme that dephosphorylates threonine 38 is PP2A, with which CAR forms a complex in the mouse liver in response to PB treatment (Yoshinari et al., 2003). In fact, in in vitro assays, PP2Ac used the receptor for protein kinase C (RACK) 1 as a regulatory subunit to dephosphorylate threonine 38. Moreover, small hairpin RNA knockdown of either RACK1 or PP2Ac nearly abrogated PB-induced dephosphorylation of CAR and an increase of CYP2B10 mRNA levels in mouse primary hepatocytes (Mutoh et al., 2013). When the epidermal growth factor receptor (EGFR) signal is activated, dephosphorylation of CAR at threonine 38 is repressed. Upon activation, EGFR signaling phosphorylates extracellular signal–regulated kinase ERK1/2, enabling phospho-ERK1/2 to bind a previously characterized peptide motif called the xenochemical response signal of CAR (Zelko et al., 2001), preventing threonine 38 from being dephosphorylated. In addition, EGFR signaling also phosphorylates RACK1 at tyrosine 52, preventing RACK1 from interacting with CAR and activating PP2Ac to dephosphorylate threonine 38 (Mutoh et al., 2013). Thus, EGF coordinates two different downstream signals (ERK1/2 and Src1) to repress dephosphorylation and activation of CAR. It was found that inhibition of ERK1/2 phosphorylation by U0126 (1,4-diamino-2,3-dicyano-1,4-bis[2-aminophenylthio]butadiene), a mitogen-activated protein kinase kinase MEK1/2 inhibitor is sufficient to activate CAR and activate the Cyp2b10 gene in the absence of exogenous CAR activators in mouse primary hepatocytes (Koike et al., 2007; Osabe and Negishi, 2011). These findings indicate that CAR is truly a cell signal–regulated nuclear receptor and ERK1/2, one of the two signals, may dominate CAR regulation.
Indirect Activation Mechanism.
Our work using isothermal titration calorimetry, competitive binding assays, and dynamic computer simulations of binding corroborated the findings that PB directly binds EGFR. Through this binding, PB repressed EGFR’s downstream signals ERK1/2 and Src1 in mouse primary hepatocytes (Mutoh et al., 2013). These observations provided the underlying molecular basis for PB activation of CAR (Fig. 1B). By reversing EGFR activation by EGF, PB reverses EGFR signaling. ERK1/2 is dephosphorylated and dissociates from the xenochemical response signal of phosphorylated CAR. The Src1 signal is attenuated, resulting in dephosphorylation of RACK1 at tyrosine 52. Dissociation of ERK1/12 allows dephosphorylated RACK1 to bind CAR and PP2Ac, dephosphorylating threonine 38 of CAR for nuclear translocation and activation.
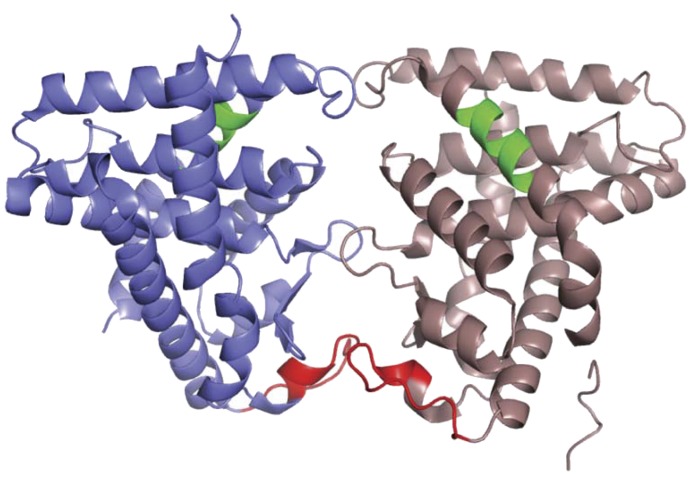
The binding site for RACK1/PP2Ac is previously delineated to a loop (from residues 140 to 152) of the CAR LBD (Mutoh et al., 2013). CAR utilizes this binding site as a molecular basis to integrate EGFR signaling into its activation mechanism. Figure 3 depicts the reported homodimer configuration in a CAR LBD crystal structure (Shun et al., 2004). Three loops form the dimer interface, the bottom loop of which bears the RACK1/PP2Ac binding site. Thus, the CAR homodimer buries the RACK1/PP2Ac binding site within this interface. EGF promotes dimerization of CAR to hide the binding site, while PB stimulates monomerization to expose the binding site for RACK1/PP2Ac to dephosphorylate CAR (Shizu et al., 2017). The homodimer-monomer conversion is the molecular base that regulates CAR activation.
Fig. 3.
Novel configuration of the CAR homodimer. The mouse CAR dimer from the X-ray crystal structure (Protein Data Bank identifier 1XNX) is shown. The RACK1 binding motif (residues 150–162) is in red and the XRS motif (ERK1/2 binding site; residues 312–319) is in green.
Direct Activation Mechanism.
Various chemicals directly activate CAR in cell-based assays, among which TCPOBOP and 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO) activate mouse and human CARs, respectively (Sueyoshi et al., 1998; Maglich et al., 2003). Chemicals that activate or repress CAR in cell-based assays are generally considered ligands. They were found to bind the LBD of CAR in X-ray crystal structures (Suino et al., 2004; Xu et al., 2004). Although EGF repressed ligand activation of CAR (e.g., by ligands such as TCPOBOP and CITCO), unlike PB, these ligands did not repress EGF signaling at the EGFR (Koike et al., 2007; Shizu et al., 2017). Thus, ligands should crosstalk with EGF signaling to activate CAR, but not at the EGFR. Recent studies with human CAR and CITCO have demonstrated that CITCO directly dissociates the CAR homodimer into its monomer for RACK1/PP2Ac to dephosphorylate threonine 38 (Fig. 1C) (Shizu et al., 2017). Thus, the phosphorylated CAR homodimer is the common target of indirect CAR activators such as PB and ligands such as CITCO. Although CAR activation has been investigated from two different angles (the so-called “direct versus indirect activation” approach), it is now clear that both direct and indirect activation share the same mechanism. The CAR homodimer can be the target of PB induction, which we have been looking for over the last 50 years.
Regulation in the Nucleus.
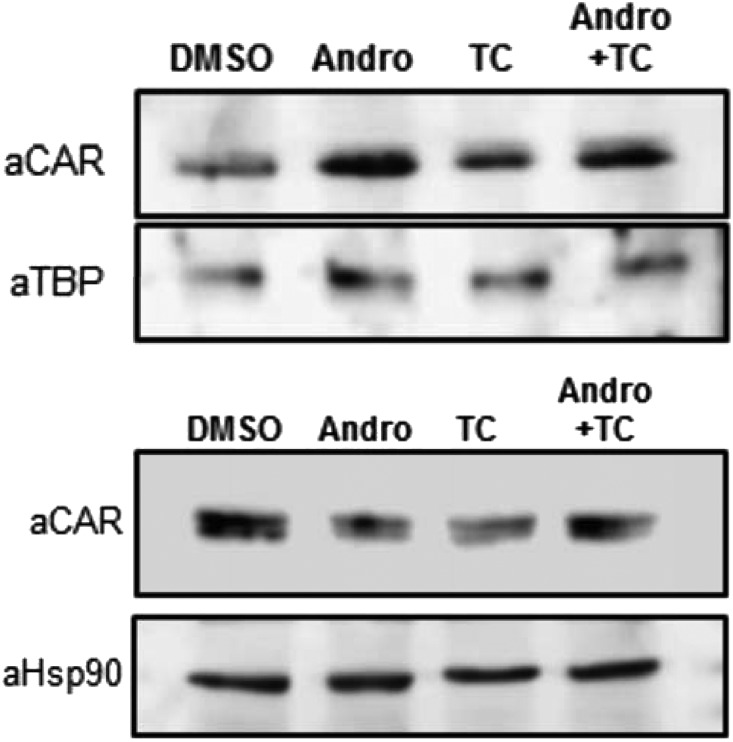
Nuclear translocation of dephosphorylated CAR is now accepted to be a key process of CAR activation. Nuclear CAR was once thought to elicit constitutive activity to activate transcription. However, it is now known that CAR activation is more complex and tightly regulated in the nucleus. The most unexpected finding was that CAR accumulates in the nucleus in mouse primary hepatocytes after treatment with androstanol, a mouse CAR antagonist (Fig. 4). Apparently, nuclear translocation is essential but not sufficient for CAR to activate transcription and CAR activation requires distinct processes in the nucleus (Ohno et al., 2014). Recent studies with cytoplasmic CAR retention protein (CCRP) knockout (KO) mice showed that CCRP does not regulate basal intracellular localization of CAR (Ohno et al., 2014). However, CCRP appeared to constrain PB-induced nuclear translocation, as indicated by excess nuclear accumulation of CAR in CCRP KO compared with CCRP wild-type mice. In response to PB treatment, the Cyp2b10 promoter recruited CAR but not RNA polymerase II in the liver of CCRP KO mice, resulting in an attenuation of PB induction of CYP2B10 mRNA. In addition, CCRP also regulated the CAR-independent but PB-induced demethylation of the Cyp2b10 promoter. Contrary to the Cyp2b10 gene, the other CAR target Cyp2c55 gene was not under CCRP regulation.
Fig. 4.
CAR antagonist accumulates CAR in the nucleus. Mouse hepatocytes were prepared from C3H mice, precultured for 3 hours, and treated with 10 µM androstenol and/or 250 nM TCPOBOP for 2 hours. Hepatocytes were fractionated by using NE-PER (Thermo Fisher Scientific, Rockford, IL), a nuclear and cytoplasmic extraction kit, subjected to Western blot analysis with an anti-CAR TATA-box binding protein or Hsp90 antibody. Andro, androstenol; DMSO, dimethylsulfoxide; TBP, TATA-box binding protein; TC, TCPOBOP.
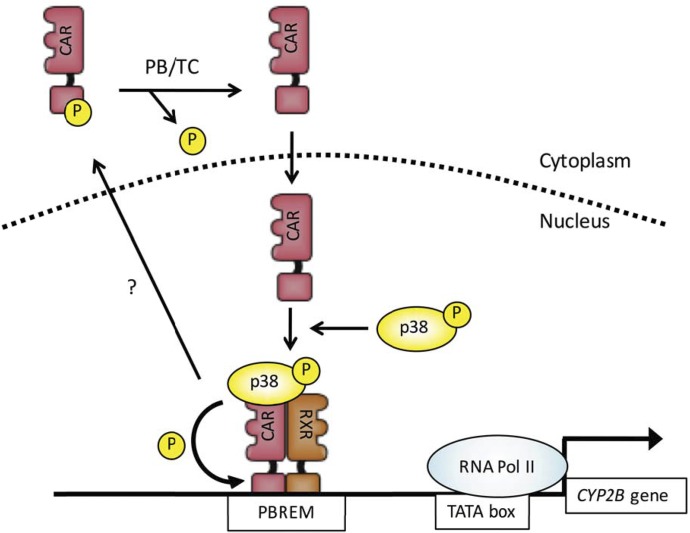
CAR requires p38 MAPK activity to activate the CYP2B6 gene in HepG2 cells (Saito et al., 2013). Our recent study with mouse CAR-expressing HepG2 cells and mouse primary hepatocytes demonstrated that, in the nucleus, p38 MAPK recruits CAR to the PBREM of the CYP2B promoter for activation and subsequently phosphorylates threonine 38 for inactivation (Hori et al., 2016). Through these sequential reactions, p38 MAPK appears to link CAR activation and inactivation as shown in Fig. 5.
Fig. 5.
p38 MAPK phosphorylates CAR at threonine 38 to link PB activation, inactivation, and intracellular localization, as shown in our previous publication (Hori et al., 2016). PB/TC is used to indicate dephosphorylation in response to PB or TCPOBOP. TC, TCPOBOP.
Experiments with growth arrest and DNA damage induce 45B (GADD45B) KO mice showed that CAR also needs GADD45B to fully activate the Cyp2b10 gene, but not the Cyp2c55 gene (Yamamoto and Negishi, 2008). Although CCRP (the cochaperone DNAJC7) was first characterized for accumulating ectopic CAR in the cytoplasm of HepG2 cells (Kobayashi et al., 2003), CCRP is now known to regulate CAR in the nucleus (Ohno et al., 2014). CAR-mediated transcription is diverse but tightly regulated in the nucleus. It will take novel ideas and efforts to understand PPARγ coactivator-1 complex mechanisms that involve many protein–protein interactions.
Beyond PB and Drug Metabolism.
The National Institutes of Health National Institute of Environmental Health Sciences (NIEHS) was developing its own cDNA microarray system around 2000. This array allowed us to analyze PB-induced and CAR-dependent genes using CAR KO mice, although it was far smaller than currently available arrays. Our array experiments generated two significant findings: one is that CAR regulates PB induction of not only P450 but also various enzymes that orchestrate drug metabolism, and the other is that CAR mediates PB-induced repression of hepatic gluconeogenesis (Ueda et al., 2002). Regarding the mechanism, activated CAR directly interacts with a forkhead transcription factor FOXO1, preventing it from activating gluconeogenic genes (Kodama et al., 2004). Since insulin stimulates phosphorylation of FOXO1 to inactivate it, PB and insulin share the same target to repress gluconeogenesis. Recently, it was also suggested that CAR forms a complex with PPARγ coactivator-1 (PGC-1) to stimulate its degradation, thereby repressing gluconeogenesis (Gao et al., 2015). CAR activation may benefit humans to repress hepatic gluconeogenesis as well as associated diseases such as type 2 diabetes and steatosis.
CAR as a Signal Transducer.
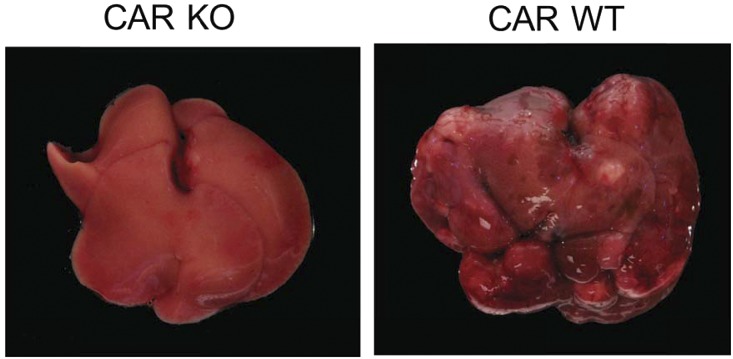
CAR directly interacts with and regulates cell signal molecules to determine cell fate, such as cell growth and death and inflammation. For example, CAR represses stress-induced signaling. CAR forms a complex with a growth arrest and DNA damage-inducible β GADD45B to repress c-Jun N-terminal kinase JNK1 signaling in mouse primary hepatocytes, attenuating apoptosis (Yamamoto et al., 2010). Recently, it was also found that the CAR-GADD45B complex suppresses MKK6-catalized phosphorylation of p38 MAPK and its downstream signaling in the mouse liver (unpublished observations). CAR activation is essential for PB to promote nongenomic development of hepatocellular carcinoma (HCC) in rodents as shown in Fig. 6 (Yamamoto et al., 2004), although this promotion mechanism remains undetermined. Repression of these JNK1 and p38 MAPK signal pathways may be the foundation of the molecular mechanism by which PB promotes HCC. An epidemiologic study with patients with epilepsy suggested no direct link between PB treatment with HCC development (Clemmesen and Hjalgrim-Jensen, 1978). However, defining the underlying process of cell signaling that leads to HCC development in rodents is critically important for us to investigate and understand HCC in humans.
Fig. 6.
No PB-promoted HCC in CAR KO mice. Car−/− (CAR KO) and Car+/+ (CAR WT) mice with a C3H/HeNCrlBR background were treated with a single dose of diethylnitrosamine for tumor initiation and chronically with PB for tumor promotion, according to a standard protocol of two-step hepatocarcinogenesis (Yamamoto et al., 2014). After 32-week PB treatment, 8 of 20 Car+/+ mice developed carcinoma, whereas none of the 20 Car−/− mice did. DEN, diethylnitrosamine; WT, wild type.
Integrating Nuclear Receptors
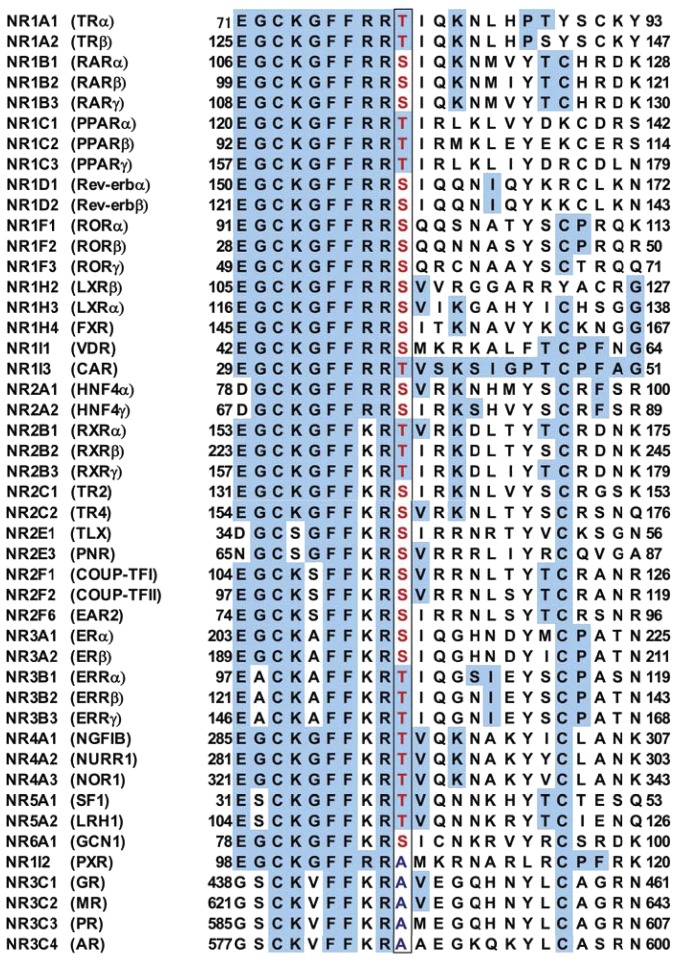
Threonine 38 Conserved in Nuclear Receptors.
Threonine 38 is in the loop between two zinc fingers within the DBD and conserved as a PKC phosphorylation motif in both mouse and human nuclear receptors and in 41 of the 46 total human nuclear receptors as shown in Fig. 7 (Hashiguchi et al., 2016). Although this phosphorylation motif is the most conserved in nuclear receptors, it has been ignored in nuclear receptor research. This scorn may have arisen from the idea that the DBD was only considered for DNA binding and as the entity regulated by the LBD and/or N-terminal domain [Activation Function 1 (AF1)]. In addition, the fact that phosphorylation of this motif was not confirmed in endogenous nuclear receptors in tissues in vivo may have perpetuated this ignorance. In addition to CAR, three more nuclear receptors [estrogen receptor (ER) α at serine 216, farnesoid X receptor (FXR) at serine 154, and retinoid X receptor (RXR) at threonine 162] are phosphorylated in mice in vivo (Shindo et al., 2013 unpublished; Hashiguchi et al., 2016; unpublished observations). Further investigations may establish this phosphorylation in numerous nuclear receptors, shedding light on the conserved motif of the DBD and providing us with an opportunity to integrate nuclear receptors.
Fig. 7.
Threonine 38 is conserved as a PKC phosphorylation motif in nuclear receptors. Only four members of the subfamily 3C and NR1I2 (PXR) do not conserve this motif. Some data are from a previous publication (Hashiguchi et al., 2016). These conserved phosphorylation residues are in red and boxed. Letters shaded in blue are amino acid residues that are conserved among nuclear receptors.
Phosphorylated DBD as a Nuclear Receptor Degradation Signal.
This conserved phosphorylation motif has only sporadically been the subject of investigations with a limited number of nuclear receptors, such as the vitamin D receptor (VDR), hepatocyte-enriched nuclear factor (HNF) 4α, and FXR (Hsieh et al., 1991; Sun et al., 2007; Gineste et al., 2008). Our current work systematically examined 11 different human nuclear receptors by expressing them in COS-1 cells (Hashiguchi et al., 2016). Among these 11 nuclear receptors, 8 (FXR, CAR, VDR, thyroid hormone receptorα, Rev-erb α, RAR-related receptor α, HNF4α, and peroxisome proliferator–activated receptor α) degraded through ubiquitination and proteasomes in COS-1 cells upon a phosphomimetic mutation at their conserved motifs. These observations have now defined the phosphorylated DBD as a minimum structural signal for nuclear receptors to degradation. Interestingly, whether and how nuclear receptors use this degradation signal appeared to be determined by other subdomains such as AF1 and/or the LBD. For example, AF1 enabled FXR to degrade through ubiquitination and proteasomes, whereas the LBD enabled RAR-related receptor α to evade degradation and accumulate. Thus, nuclear receptors can diverge their functions and regulations by utilizing this conserved phosphorylation differently.
Control Constitutive Activity.
One major question that has persisted around many nuclear receptors is how their constitutive activity is controlled. The idea of so-called endogenous ligands does not appear to have helped us much in answering this question, as indicated by the following: “Estrogen-related receptors (ERRs) are founding members of the orphan nuclear receptor (ONR) subgroup of the nuclear receptor superfamily: Twenty-seven years of study have yet to identify cognate ligands for the ERRs, though they have firmly placed ERRα (ESRRA) and ERRγ (ESRRG) at the intersection of cellular metabolism and oncogenesis” (Divekar et al., 2016). In contrast with the hypothesis of “endogenous ligands,” the conserved phosphorylation of DBD presents an experimental basis for us to determine the underlying mechanism by which nuclear receptors control their constitutive activities.
Threonine 38, directing its sidechain to be opposite from the surface of CAR binding DNA, does not directly interact with DNA. Phosphorylation of threonine 38 distorts the C-terminal side helix of the two helices flanking the loop bearing threonine 38, disabling CAR from binding and activating PBREM (Mutoh et al., 2009). Phosphorylation of threonine 87 of HNF4α is suggested to alter interactions between its DBD and LBD to inactivate DNA binding (Chandra et al., 2013). In general, phosphomimetic mutations at the conserved motif inactivate nuclear receptors in both constitutive and ligand-based activities in cell-based reporter assays. Thus, this conserved phosphorylation motif promotes nuclear receptors’ ability to regulate both constitutive and response activities. Moreover, this motif enables nuclear receptors to coordinate their activation and inactivation. For example, threonine 38 is dephosphorylated by PP2A in the cytoplasm to translocate CAR into the nucleus. In the nucleus, p38 MAPK forms a complex with CAR, enabling it to bind to the PBREM and activate the Cyp2b10 promoter. Subsequently, p38 MAPK phosphorylates threonine 38 and dissociates CAR from the promoter, thereby exporting it back to the cytoplasm (Hori et al., 2016). Not only PB but ligands such as CITCO and TCPOBOP also use this phosphorylation to coordinate CAR activation and inactivation. For this purpose, threonine 38 links CAR activation and inactivation as well as nuclear import and export through phosphorylation and dephosphorylation, probably for recycling CAR. Further investigations with additional nuclear receptors may conceptualize phosphorylation of the conserved motif of the DBD as a common regulatory mechanism for nuclear receptors.
Nuclear Receptor Crosstalk via Phosphorylation.
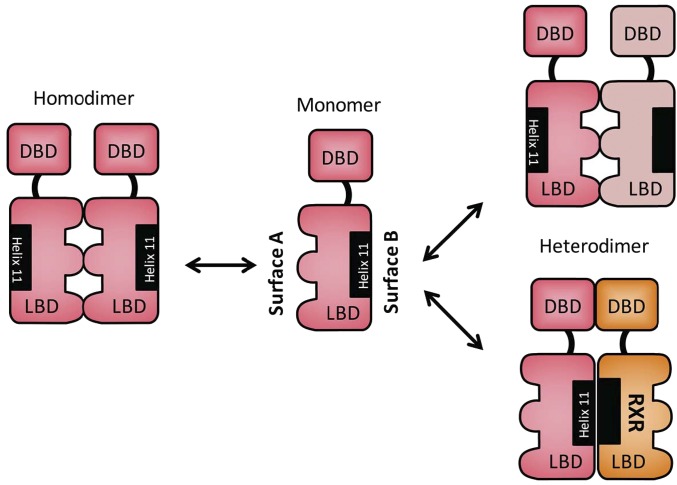
Nuclear receptors crosstalk to coregulate P450 expression and drug metabolism (Gotoh et al., 2015), the molecular mechanism of which remains nebulous. The conserved phosphorylation motif within the DBD can be used as a foundation for nuclear receptors to crosstalk with each other. As described, CAR forms its homodimer through the interactions between two LBDs by utilizing a surface (surface A) opposite from the surface (surface B) that forms a heterodimer with RXR (Fig. 8). Nuclear receptors should be able to form their homodimers in the same configuration as observed with the CAR homodimer. In addition to homodimerization, nuclear receptors should also heterodimerize with another nuclear receptor through surface A in the cytoplasm in the absence of RXR. In the presence of RXR, it may be possible for two nuclear receptors to form a tetramer (RXR-NR1-NR2-RXR) in cases such as crosstalk between CAR and VDR to repress the CYP24A1 gene by locking the corepressor SMRT (silencing mediator for retinoid and thyroid hormone receptor) onto its promoter in the nucleus (Konno et al., 2009). Although this crosstalk can be a molecular basis for investigating metabolic bone diseases caused by chronic drug therapy with PB and/or phenytoin, which activate CAR, the mechanism of this locking is not yet understood. Phosphorylation of the conserved motif could determine functionality of nuclear receptor crosstalk by altering formations of their complexes. When it becomes available in the future, a knock-in (KI) mouse library bearing a nonphosphomimetic mutation in nuclear receptors will help us to investigate nuclear receptor crosstalk as well as correlate regulation of phosphorylation and dimerization and indicate their pharmacological significance.
Fig. 8.
Two different dimer surfaces of nuclear receptors. Surfaces A and B are on the opposite sides of the CAR molecule. Three loops shaped with half circles comprise surface A for homodimerization and may also for heterodimerization of two different nuclear receptors, such as CAR and VDR. Surface B is well established as heterodimerizing with RXR. ERα was suggested to form its homodimer through surface B.
Biology of Conserved Phosphorylation.
In addition to CAR, two nuclear receptors (FXR and ERα) are now confirmed to be phosphorylated at their conserved motifs in vivo in mice. FXR becomes phosphorylated at serine 154 in the nucleus of the liver of ligand-treated mice, in which nuclear kinase vaccinia-related kinase 1 appears to phosphorylate serine 154 (Hashiguchi et al., 2016). As suggested by the FXR S154D mutant, phosphorylated FXR does not bind or activate FXRE and it is degraded by proteasomes either in the nucleus or cytoplasm. Thus, FXR utilizes the conserved phosphorylation motif to link its activation, inactivation, and degradation. This FXR linking mechanism resembles that observed with CAR and p38 MAPK (Fig. 5). The lack of this phosphorylation may prolong the activated status of FXR, dysregulating liver functions such as bile acid, fatty acid, and glucose metabolism and contributing to the development of liver disease.
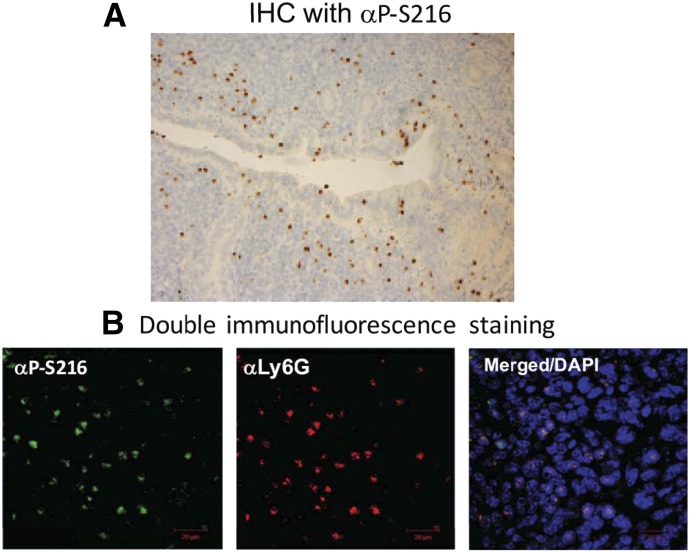
ERα, which conserves this phosphorylation residue at serine 212 in humans and serine 216 in mice, was found to be phosphorylated in mouse immune cells (Shindo et al., 2013). Immunohistochemistry with an anti–phospho-S216 peptide antibody (αP-S216) first revealed staining of certain cells but not uterine cells of the mouse uterus (Fig. 9A). Double fluorescence immunostaining confirmed that these cells were infiltrating neutrophils. Neutrophils infiltrate the uterus from the blood stream (Fig. 5B), about 20% of which express phosphorylated ERα. Only blood neutrophils expressing phosphorylated ERα migrated in in vitro migration assays and infiltrated the uterus of female mice, as shown in Fig. 9 (Shindo et al., 2013). Phosphorylation of the conserved motif enables neutrophils to migrate and infiltrate. ERα is also phosphorylated in mouse microglia, the resident macrophages, and only immune cells in the brain (Shindo et al., 2016). KI mice bearing a single mutation of serine 216 to alanine (Esr1S216A) were generated and began to reveal anti-inflammatory and antiapoptotic functions of phosphorylated ERα in immune cells. Esr1S216A mice, obese but fertile in both sexes, can be an excellent animal model for the study of inflammation-related diseases (including obesity) and neuronal degenerative diseases (e.g., Alzheimer and Parkinson diseases). Again, the majority of nuclear receptors contain this conserved phosphorylation motif within their DBDs in both humans and mice. To determine the biologic roles of the conserved phosphorylation of nuclear receptors, it is important to generate a library of KI mouse lines that bear the nonphosphomimetic mutation of this motif in each nuclear receptor.
Fig. 9.
ERα phosphorylated at serine 216 in neutrophils infiltrating the mouse uterus. (A) An anti-Ser216 peptide antibody (αP-S216) was used to stain sections of the mouse uterus in immunohistochemistry studies. (B) Double immunostaining with an anti–P-S216 (green) and anti-Ly6G, a neutrophil marker (red) was performed. DAPI-stained nuclei are also shown. These pictures are assembled from our previous publication (Shindo et al., 2013). DAPI, 4′,6-diamidino-2-phenylindole; IHC, immunohistochemistry.
Epilogue
A Single Residue Matters.
Mammalian P450s consist of about 500 amino acid residues. Mouse CYP2A4 and CYP2A5 specifically catalyze testosterone 15α-hydroxylation and coumarin 7-hydrosylation, yet they differ by only 11 amino acid residues from their total 494 residues (Negishi et al., 1989). Later, it was found that CYP2A6, the human homolog of mouse CYP2A5, also catalyzes coumarin 7-hydroxylation. The residue at position 209 is leucine in CYP2A4 and phenylalanine in CYP2A5. A single mutation of phenylalanine 209 to leucine converted the catalytic activity of CYP2A5 from coumarin to testosterone hydroxylation (Lindberg and Negishi, 1989). Furthermore, mutation of phenylalanine 209 to asparagine conferred corticosterone 15α hydroxylase activity to CYP2A5 (Iwasaki et al., 1993). Subsequently, using a dynamic simulation study, with the only available crystal structure of bacterial P450cam (now CYP101) at that time, F and G helices and the loop between them were suggested to be structurally flexible, enabling CYP2A5 to determine substrate specificity (Iwasaki et al., 1993). In addition, our mutagenesis work suggested that CYP2A5 may have two different substrate binding sites (Lindberg and Negishi, 1989). The idea of two substrates in one substrate binding pocket was first proposed more than 30 years ago by Alan Conney’s group: flavonoids stimulate benzo(a)pyrene hydroxylation activity of rat liver microsomes (Huang et al., 1981). Since the first mammalian P450 crystal structure was solved with CYP2C5 (Johnson, 2002), a large number of the crystal structures have been determined, which confirm our early predictions in the 1990s that a single amino acid along with these helices, loop, and the two substrate binding sites enable P450s to be flexible and diverse in their catalytic activities.
In retrospect, this belief that a single residue matters sustained my laboratory throughout our ordeal to discover phosphorylation of threonine 38 of CAR in PB induction and has been extended beyond CAR by us. It is still thought that phosphorylation of a single residue cannot and should not regulate nuclear receptors, since there are many potential phosphorylation sites in any given nuclear receptor. As it turns out, threonine 38 is not one of these sites; it is the most conserved phosphorylation site of nuclear receptors. Moreover, this conserved phosphorylation motif has renewed research interests in the DBD as a regulatory determinant of nuclear receptor functions beyond being the simple DNA binding entity of a nuclear receptor.
Many Thanks.
I started my career in science as a graduate student in Professor Ryo Sato’s laboratory at Institute for Protein Research at Osaka University, under the supervision of Professor Tsuneo Omura in 1968. From the beginning to the present, Omura sensei (mentor) has never stopped helping me develop my career, although I was his worst student in many ways. For 5 years after I obtained my degree, I worked at Kansai Medical University under Professor Yutaka Tashiro, who provided me with an opportunity to work in the United States in 1976. Since 1983, I have been a group leader at NIEHS, and it has been my privilege to work with more than 100 postdoctoral fellows, graduate students, and guest researchers (Fig. 10). I am sincerely thankful for all of these mentors and colleagues who made my life happy and fruitful. Now I finish my review by “entrusting the next 50 years to the next generation of young scientists and students.”
Fig. 10.
Thank you to all laboratory members past, present, and constant.
Acknowledgments
The author extends deepest appreciation to Dr. Joyce Goldstein, who has been a friend and collaborator for the last 33 years at NIEHS. The author also thanks Drs. Lalith Perera, Lars Pedersen, Ryo Shizu, Takeshi Hori, and Mack Sobhany for help with writing this review.
Abbreviations
- CAR
constitutive active/androstane receptor
- CCRP
cytoplasmic CAR retention protein
- CITCO
6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime
- DBD
DNA binding domain
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- ERα
estrogen receptor α
- ERK
extracellular signal–regulated kinase
- ERR
estrogen receptor–related receptor
- FXR
farnesoid X receptor
- HCC
hepatocellular carcinoma
- HNF
hepatocyte-enriched nuclear factor
- KI
knock in
- KO
knockout
- LBD
ligand binding domain
- MAPK
mitogen-activated protein kinase
- P450
cytochrome P450
- PBREM
phenobarbital-responsive enhancer module
- PKC
protein kinase C
- PP
protein phosphatase
- RACK
receptor for protein kinase C
- RXR
retinoid X receptor
- TCPOBOP
1,4-bis[2-(3,5-dichloropyridyloxy)]benzene
- U0126
1,4-diamino-2,3-dicyano-1,4-bis[2-aminophenylthio]butadiene
- VDR
vitamin D receptor
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Negishi.
Footnotes
This research was supported by the Intramural Research Program of the National Institutes of Health National Institute of Environmental Health Sciences [Grant Z01ES71005-01].
References
- Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. (1994) A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol 14:1544–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D, Wolfram N, Kahl GF, Hirsch-Ernst KI. (2004) Transcriptional regulation of CYP2B1 induction in primary rat hepatocyte cultures: repression by epidermal growth factor is mediated via a distal enhancer region. Mol Pharmacol 65:172–180. [DOI] [PubMed] [Google Scholar]
- Chandra V, Huang P, Potluri N, Wu D, Kim Y, Rastinejad F. (2013) Multidomain integration in the structure of the HNF-4α nuclear receptor complex. Nature 495:394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmesen J, Hjalgrim-Jensen S. (1978) Is phenobarbital carcinogenic? A follow-up of 8078 epileptics. Ecotoxicol Environ Saf 1:457–470. [DOI] [PubMed] [Google Scholar]
- Divekar SD, Tiek DM, Fernandez A, Riggins RB. (2016) Estrogen-related receptor β (ERRβ) - renaissance receptor or receptor renaissance? Nucl Recept Signal 14:e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, Moore DD. (1998) Androstane metabolites bind to and deactivate the nuclear receptor CAR-β. Nature 395:612–615. [DOI] [PubMed] [Google Scholar]
- Gao J, Yan J, Xu M, Ren S, Xie W. (2015) CAR suppresses hepatic gluconeogenesis by facilitating the ubiquitination and degradation of PGC1α. Mol Endocrinol 29:1558–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gineste R, Sirvent A, Paumelle R, Helleboid S, Aquilina A, Darteil R, Hum DW, Fruchart JC, Staels B. (2008) Phosphorylation of farnesoid X receptor by protein kinase C promotes its transcriptional activity. Mol Endocrinol 22:2433–2447. [DOI] [PubMed] [Google Scholar]
- Gotoh S, Ohno M, Yoshinari K, Negishi M, and Honkakoski P (2015) Nuclear receptor-mediated regulation of cytochrome P450 genes, in Cytochrome P450: Structure, Mechanism, and Biochemistry (Ortiz de Montellano PR ed) 5th ed, pp 787–812, Springer, New York. [Google Scholar]
- Hashiguchi T, Arakawa S, Takahashi S, Gonzalez FJ, Sueyoshi T, Negishi M. (2016) Phosphorylation of farnesoid X receptor at serine 154 links ligand activation with degradation. Mol Endocrinol 30:1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P, Moore R, Washburn KA, Negishi M. (1998a) Activation by diverse xenochemicals of the 51-base pair phenobarbital-responsive enhancer module in the CYP2B10 gene. Mol Pharmacol 53:597–601. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Negishi M. (1998) Regulatory DNA elements of phenobarbital-responsive cytochrome P450 CYP2B genes. J Biochem Mol Toxicol 12:3–9. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Zelko I, Sueyoshi T, Negishi M. (1998b) The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol 18:5652–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Moore R, Negishi M. (2016) p38 MAP kinase links CAR activation and inactivation in the nucleus via phosphorylation at threonine 38. Drug Metab Dispos 44:871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JC, Jurutka PW, Galligan MA, Terpening CM, Haussler CA, Samuels DS, Shimizu Y, Shimizu N, Haussler MR. (1991) Human vitamin D receptor is selectively phosphorylated by protein kinase C on serine 51, a residue crucial to its trans-activation function. Proc Natl Acad Sci USA 88:9315–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MT, Chang RL, Fortner JG, Conney AH. (1981) Studies on the mechanism of activation of microsomal benzo[a]pyrene hydroxylation by flavonoids. J Biol Chem 256:6829–6836. [PubMed] [Google Scholar]
- Iwasaki M, Darden TA, Pedersen LG, Davis DG, Juvonen RO, Sueyoshi T, Negishi M. (1993) Engineering mouse P450coh to a novel corticosterone 15 α-hydroxylase and modeling steroid-binding orientation in the substrate pocket. J Biol Chem 268:759–762. [PubMed] [Google Scholar]
- Johnson EF. (2002) The 2002 Bernard B. Brodie Award lecture: deciphering substrate recognition by drug-metabolizing cytochromes P450. Drug Metab Dispos 31:1532–1540. [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. (1999) Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol Cell Biol 19:6318–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Sueyoshi T, Inoue K, Moore R, Negishi M. (2003) Cytoplasmic accumulation of the nuclear receptor CAR by a tetratricopeptide repeat protein in HepG2 cells. Mol Pharmacol 64:1069–1075. [DOI] [PubMed] [Google Scholar]
- Kodama S, Koike C, Negishi M, Yamamoto Y. (2004) Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol 24:7931–7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike C, Moore R, Negishi M. (2007) Extracellular signal-regulated kinase is an endogenous signal retaining the nuclear constitutive active/androstane receptor (CAR) in the cytoplasm of mouse primary hepatocytes. Mol Pharmacol 71:1217–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno Y, Kodama S, Moore R, Kamiya N, Negishi M. (2009) Nuclear xenobiotic receptor pregnane X receptor locks corepressor silencing mediator for retinoid and thyroid hormone receptors (SMRT) onto the CYP24A1 promoter to attenuate vitamin D3 activation. Mol Pharmacol 75:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg RL, Negishi M. (1989) Alteration of mouse cytochrome P450coh substrate specificity by mutation of a single amino-acid residue. Nature 339:632–634. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, et al. (2003) Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem 278:17277–17283. [DOI] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, et al. (2000) Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem 275:15122–15127. [DOI] [PubMed] [Google Scholar]
- Mutoh S, Osabe M, Inoue K, Moore R, Pedersen L, Perera L, Rebolloso Y, Sueyoshi T, Negishi M. (2009) Dephosphorylation of threonine 38 is required for nuclear translocation and activation of human xenobiotic receptor CAR (NR1I3). J Biol Chem 284:34785–34792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh S, Sobhany M, Moore R, Perera L, Pedersen L, Sueyoshi T, Negishi M. (2013) Phenobarbital indirectly activates the constitutive active androstane receptor (CAR) by inhibition of epidermal growth factor receptor signaling. Sci Signal 6:ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi M, Lindberg R, Burkhart B, Ichikawa T, Honkakoski P, Lang M. (1989) Mouse steroid 15 α-hydroxylase gene family: identification of type II P-450(15)α as coumarin 7-hydroxylase. Biochemistry 28:4169–4172. [DOI] [PubMed] [Google Scholar]
- Ohno M, Kanayama T, Moore R, Ray M, Negishi M. (2014) The roles of co-chaperone CCRP/DNAJC7 in Cyp2b10 gene activation and steatosis development in mouse livers. PLoS One 9:e115663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T, Sato R. (1962) A new cytochrome in liver microsomes. J Biol Chem 237:1375–1376. [PubMed] [Google Scholar]
- Osabe M, Negishi M. (2011) Active ERK1/2 protein interacts with the phosphorylated nuclear constitutive active/androstane receptor (CAR; NR1I3), repressing dephosphorylation and sequestering CAR in the cytoplasm. J Biol Chem 286:35763–35769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmer H and Merker HJ (1963) Drug-induced changes in the liver endoplasmic reticulum: association with drug-metabolizing enzymes. Science 142:1657–1658. [DOI] [PubMed] [Google Scholar]
- Saito K, Moore R, Negishi M. (2013) p38 Mitogen-activated protein kinase regulates nuclear receptor CAR that activates the CYP2B6 gene. Drug Metab Dispos 41:1170–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer N, Ross J, Rode A, Zevnik B, Niehaves S, Faust N, Wolf CR. (2008) A novel panel of mouse models to evaluate the role of human pregnane X receptor and constitutive androstane receptor in drug response. J Clin Invest 118:3228–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo S, Moore R, Flake G, Negishi M. (2013) Serine 216 phosphorylation of estrogen receptor α in neutrophils: migration and infiltration into the mouse uterus. PLoS One 8:e84462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo S, Sakuma T, Negishi M, Squires J. (2012) Phosphorylation of serine 212 confers novel activity to human estrogen receptor α. Steroids 77:448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizu R, Osabe M, Perera L, Moore R, Sueyoshi T, Negishi M. (2017) Phosphorylated nuclear receptor CAR forms a homodimer to repress its constitutive activity for ligand activation. Mol Cell Biol DOI: 10.1128/MCB.00649-16 [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. (1999) The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem 274:6043–6046. [DOI] [PubMed] [Google Scholar]
- Suino K, Peng L, Reynolds R, Li Y, Cha JY, Repa JJ, Kliewer SA, Xu HE. (2004) The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol Cell 16:893–905. [DOI] [PubMed] [Google Scholar]
- Trottier E, Belzil A, Stoltz C, Anderson A. (1995) Localization of a phenobarbital-responsive element (PBRE) in the 5′-flanking region of the rat CYP2B2 gene. Gene 158:263–268. [DOI] [PubMed] [Google Scholar]
- Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. (2002) Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol Pharmacol 61:1–6. [DOI] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. (2000) The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature 407:920–923. [DOI] [PubMed] [Google Scholar]
- Xu RX, Lambert MH, Wisely BB, Warren EN, Weinert EE, Waitt GM, Williams JD, Collins JL, Moore LB, Willson TM, et al. (2004) A structural basis for constitutive activity in the human CAR/RXRalpha heterodimer. Mol Cell 16:919–928. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Moore R, Flavell RA, Lu B, and Negishi M (2010) Nuclear receptor CAR represses TNFalpha-induced cell death by interacting with the anti-apoptotic GADD45B. PLoS One 5:e10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Moore R, Goldsworthy TL, Negishi M, Maronpot RR. (2004) The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res 64:7197–7200. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Negishi M. (2008) The antiapoptotic factor growth arrest and DNA-damage-inducible 45 β regulates the nuclear receptor constitutive active/androstane receptor-mediated transcription. Drug Metab Dispos 36:1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Garzel B, Heyward S, Moeller T, Shapiro P, Wang H. (2014) Metformin represses drug-induced expression of CYP2B6 by modulating the constitutive androstane receptor signaling. Mol Pharmacol 85:249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasujima T, Saito K, Moore R, Negishi M. (2016) Phenobarbital and insulin reciprocate activation of the nuclear constitutive androstane receptor through the insulin receptor. J Pharmacol Exp Ther 357:367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari K, Kobayashi K, Moore R, Kawamoto T, Negishi M. (2003) Identification of the nuclear receptor CAR:HSP90 complex in mouse liver and recruitment of protein phosphatase 2A in response to phenobarbital. FEBS Lett 548:17–20. [DOI] [PubMed] [Google Scholar]
- Zelko I, Sueyoshi T, Kawamoto T, Moore R, Negishi M. (2001) The peptide near the C terminus regulates receptor CAR nuclear translocation induced by xenochemicals in mouse liver. Mol Cell Biol 21:2838–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]