Abstract
Viruses infect host cells releasing their genome (DNA or RNA) containing all information needed to replicate themselves. The viral genome takes control of the cells and helps the virus to evade the host immune system. Some viruses alter the functions of infected cells without killing them. In some cases infected cells lose control over normal cell proliferation and becomes cancerous. Viruses, such as HCMV and HIV-1, may leave their viral genome in the host cells for a certain period (latency) and begin to replicate when the cells are stressed causing diseases. HCMV and HIV-1 have developed multiple strategies to avoid recognition and elimination by the host’s immune system. These strategies rely on viral products that mimic specific components of the host cells to prevent immune recognition of virally infected cells. In addition to viral proteins, viruses encode short non-coding RNAs (vmiRNAs) that regulate both viral and host cellular transcripts to favor viral infection and actively curtail the host’s antiviral immune response. In this review, we will give an overview of the general functions of microRNAs generated by HCMV and HIV-1, their processing and interaction with the host’s immune system.
Keywords: microRNA, HCMV, HIV, immune system, cancer
Introduction
MicroRNAs (miRNAs) are short (∼22 nucleotides) single-stranded non-coding RNA molecules that negatively regulate gene expression at post-transcriptional level. The miRNA maturation machinery involves several steps and multiple proteins in both nucleus and cytoplasm. miRNAs can be transcribed as part of independent primary transcripts (pri-miRNAs), which are mainly generated by RNA polymerase II (Lee et al., 2004; Ozsolak et al., 2008), and display all features commonly associated with Pol II-mediated transcription, such as histone marks, CpG islands, transcription factor binding sites (Cai et al., 2004). In the canonical miRNA biogenesis pathway, the Microprocessor complex, a multiprotein complex with Drosha and Di George Syndrome critical region gene 8 (DGCR8), cleaves the double-stranded pri-miRNA generating a hairpin-shaped RNA molecule (pre-miRNA) of about 70–100 bp (Denli et al., 2004; Han et al., 2006). This process can be modulated by different proteins (Suzuki et al., 2009; Trabucchi et al., 2009). Once generated, nuclear pre-miRNAs are exported to the cytoplasm by the exportin-5/Ran-GTP complex to be further processed by the RNase III enzyme Dicer (Grishok et al., 2001). The pre-miRNA processing is finely regulated and its inhibition affects miRNA-mediated differentiation in embryonic stem cells, embryonal carcinoma cells and certain primary tumors. An example is provided by Lin-28, an developing regulated RNA binding protein, which promotes cell proliferation and tumorigenesis of embryonic cells by affecting let-7 maturation (Viswanathan et al., 2008). The activity of Dicer can be either enhancing or inhibiting by A-to-I RNA editing mediated by ADARs enzymes which convert Adenosine to Inosine (Kawahara et al., 2008; Nishikura, 2010; Tomaselli et al., 2013). ADAR1 has been shown to directly bind Dicer, thus increasing the maximum rate of pre-microRNA cleavage by Dicer (Ota et al., 2013). In human adult brain, it has been estimated that approximately 20% of pri-miRNAs can undergo A-to-I RNA editing (Nishikura, 2010; Tomaselli et al., 2013) by affecting miRNAs maturation at different steps (Luciano et al., 2004; Chawla and Sokol, 2014; Tomaselli et al., 2015).
The result of Dicer cleavage is the formation of a double stranded RNA of 22 nt in length whose strand with the less stability is normally chosen as guide strand and transferred to the RNA-induced silencing complex (RISC) for the annealing of miRNAs to the target mRNA, whereas the other strand [the star (∗)-strand] is usually degraded. The RISC complex contains Dicer and many associated proteins such as Argonaute (Ago) protein family and the RNA-binding protein TRBP [human immunodeficiency virus transactivating response RNA (TAR) binding protein] (Chendrimada et al., 2005).
Mature miRNAs recognize their target mRNAs through 6–8 nucleotides (the seed region) at the 5′ end of the miRNA. Complete complementarity between the miRNA and target mRNA sequence directs mRNA degradation, while absent of perfect complementarity will silence the gene target by preventing its translation (Lim et al., 2005). A given miRNA may have hundreds of different mRNA targets, and a given target mRNA might be regulated by several miRNAs. These evidences suggest that the biogenesis of miRNAs is extremely complex and regulated at different levels, thus highlighting the importance of these short RNA molecules in crucial cell programs including viral infection.
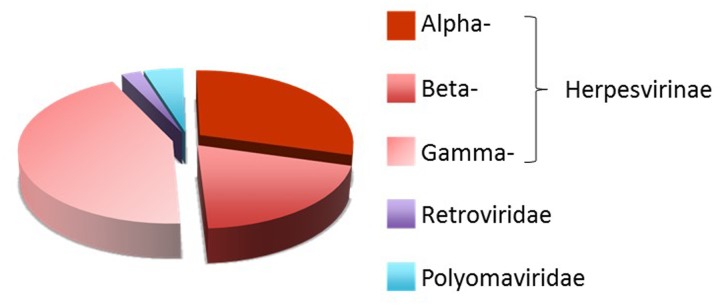
Viruses have developed multiple strategies to avoid recognition and elimination by the host’s immune system. These strategies rely on viral products that mimic specific components of the host cells to prevent immune recognition of virally infected cells. In addition to viral proteins, viruses encode miRNAs (vmiRNA) that regulate both viral and host cellular transcripts during viral infection. The first vmiRNAs was identified in a cell line latently infected with Epstein–Barr virus (EBV) (Pfeffer et al., 2004), a member of the Herpesviridae, that in humans is associated with Burkitt’s lymphoma, Hodgkin’s disease and nasopharyngeal carcinoma (Raab-Traub, 2007). Currently, different members of Herpesviridae, Polyomaviridae and Adenoviridae families are known to express vmiRNAs (Skalsky and Cullen, 2010). To date, 172 viral-encoded mature miRNAs are listed in the miRBase collection1 (Kozomara and Griffiths-Jones, 2014), the majority of which (93%) belongs to the three subfamilies (Alphaherpesvirinae, Betaherpesvirinae and Gammaherpesvirinae) of Herpesviridae family (Figure 1).
FIGURE 1.
Distribution of human viral miRNAs. Distribution of human viral-encoded miRNAs listed in the miRBase collection (http://www.mirbase.org/) (Kozomara and Griffiths-Jones, 2014).
Biogenesis of vmiRNAs largely relies on host-derived machineries. The common pathway of vmiRNA maturation involves the transcription from host RNA polymerase II into a long transcript precursor known as viral pri-miRNA (vpri-miRNAs), which is trimmed by the host RNase III endonuclease Drosha microprocessor into approximately 80 nt long hairpin structures, known as viral pre-miRNAs (vpre-miRNAs). Vpre-miRNAs are rapidly exported to the cytoplasm where a specialized multi-domain ribonuclease III enzyme, known as Dicer, removes the loop structure leaving the vmiRNA duplex. Some functional vmiRNAs, such as murine γ-herpesvirus 68 (MHV68) miRNAs, are produced by host RNA polymerase III and tRNase Z, independently from the microprocessor-Drosha component (Bogerd et al., 2010), whereas others are generated by different additional non-canonical pathways (Pfeffer et al., 2005; Diebel et al., 2010, 2014; Kincaid et al., 2012; Burke et al., 2014; Feldman et al., 2014; Kincaid et al., 2014; Whisnant et al., 2014).
In general, virus and the host-cell’s miRNAs use different mechanisms to interfere each other. Viruses can either block or impair the host cells miRNA pathway by interacting with key proteins (Lu and Cullen, 2004; Bennasser et al., 2006), synthesize and regulate their own miRNAs (Grundhoff and Sullivan, 2011; Harwig et al., 2014), exploit cellular miRNAs to complete their replication cycle (Luna et al., 2015). Conversely, host cells can target vmiRNAs with endogenous miRNAs (Lecellier et al., 2005; Delorme-Axford et al., 2013; Bai and Nicot, 2015). The complex interplay between viruses and host cells usually favors viral infection by either reducing immune recognition or promoting cell growth and lytic or latent infection. One of the best examples of virus-host-miRNA interplay important for human infection and diseases come from HCMV. In this review, we summarize the general functions of miRNAs generated by HCMV and their interactions with immune system and we discuss new finding regarding miRNA from HIV-1.
HCMV MicroRNAs
HCMV is a ubiquitous and highly specific herpesvirus that establishes lifelong latent infections, coexisting asymptomatically with its host in a healthy immune system, with periodic and spontaneous phases of reactivation, lytic replication and virus shedding (Stern-Ginossar et al., 2012). In immunocompromised individuals, such as transplant recipients, HIV-infected patients and individuals with an immunological immaturity, as the fetus in utero, HCMV can cause different clinical syndromes the severity of which depends on the degree of immunosuppression (Varani and Landini, 2011). Of note, during pregnancy HCMV primary infections can lead to mental retardation and severe neonatal pathologies.
To persist indefinitely within the host, HCMV has elaborated several strategies that act to subvert host cellular immune responses (Noriega et al., 2012). Many HCMV proteins and vmiRNAs are known to target cellular and viral transcripts to establish and maintain latency (Murphy et al., 2008). The complex interplay between virus and host is further enhanced by post-transcriptional events, such as RNA editing. Indeed, during HCMV infection, the expression of the short form of the RNA editing enzyme ADAR1 (ADAR1-p110) is enhanced and the host miR-376a precursor undergo editing at specific sites. The increased edited-miR-376a during HCMV infection, downregulates HLA-E transcript leading the infected cells more visible by NK cells (Nachmani et al., 2014).
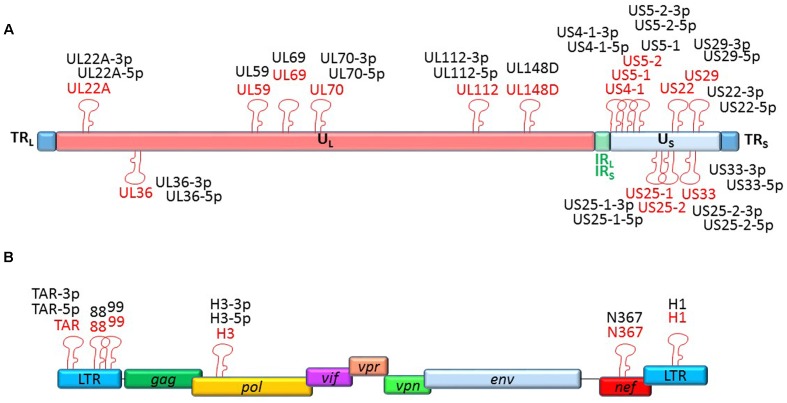
To date, 15 stem-loop precursors and 26 mature HCMV miRNAs are deposited in the miRBase (Figure 2). HCMV miRNAs are differently expressed during latent and lytic infection. Of note, only a subset of vmiRNAs are produced during HCMV latency, most of which originate from the unique long (UL) region of the HCMV genome (Meshesha et al., 2016). Interestingly, during reactivation of the virus from latency all known vmiRNAs, including those that were absent during latency, restored their expression. An alternative expression of the two stands of miR-US29 was detected in the lytic and latent infection. Specifically, miR-US29-5p prevailed during lytic infection, whereas miR-US29-3p dominated viral latency, suggesting the presence of a specific mechanism that regulates expression of the two arms of the vmiRNA hairpins during the viral life cycle (Meshesha et al., 2016). It is possible that expression of vmiRNAs during latency is required to manipulate host-signaling pathways and make the latently infected cell ready for reactivation.
FIGURE 2.
Genomic organization of the miRNAs encoded by HCMV and HIV-1. Location of precursors (red) and mature (black) miRNAs is shown by hairpin loop structures along the linear representation of the HCMV (A) and HIV-1 (B) genomes. miRNAs shown above the line representing viral double-strand DNA are transcribed in sense direction (from right to left), while those shown below are transcribed in the opposite direction.
HCMV miRNAs are known to target several cellular genes to evade immune system, control cell cycle and vesicle trafficking (Table 1). Since this subject has been recently reviewed by Piedade and Azevedo-Pereira (2016), in this review we will deepen the HCMV miRNAs with proved implications in viral evasion from innate and adaptive immune responses.
Table 1.
HCMV and HIV-1 microRNAs with potential role in viral infection and pathogenesis.
| Targets | Predicted role | Reference | |
|---|---|---|---|
| CMV-encoded miRNA | |||
| miR-UL36 | UL138* | Latent infection | Huang et al., 2013 |
| miR-UL36-5p | ANT3 | Cell survival | Guo et al., 2015 |
| miR-UL112 | MICB | Immune evasion | Stern-Ginossar et al., 2007 |
| IE72 (UL123, IE1), UL112/113, UL120/121 | Viral infection | Grey et al., 2007 | |
| IL-32 | Immune evasion | Huang et al., 2013 | |
| type I IFN signaling | Immune evasion | Huang et al., 2015 | |
| VAMP3, RAB5C, RAB11A, SNAP23, CDC42 | Vesicle pathway | Hook et al., 2014 | |
| TLR2 | Immune evasion | Landais et al., 2015 | |
| IKKα, IKKβ | Immune evasion | Hancock et al., 2017 | |
| miR-UL148D | CCL5 | Immune evasion | Kim et al., 2012 |
| IEX-1 | Cell survival | Wang et al., 2013 | |
| CDC25B | Latent infection | Pan et al., 2016 | |
| ACVR1B | Immune evasion | Lau et al., 2016 | |
| miR-US4 | ERAP1 | Immune evasion | Kim et al., 2011 |
| QARS | Cell survival | Shao et al., 2016 | |
| miR-US5-1 | US7 | Viral infection | Tirabassi et al., 2011 |
| VAMP3, RAB5C, RAB11A, SNAP23, CDC42 | Vesicle pathway | Hook et al., 2014 | |
| IKKα, IKKβ | Immune evasion | Hancock et al., 2017 | |
| miR-US5-2 | US7 | Viral infection | Tirabassi et al., 2011 |
| VAMP3, RAB5C, RAB11A, SNAP23, CDC42 | Vesicle pathway | Hook et al., 2014 | |
| miR-US25-1-5p | VAMP3, RAB5C, RAB11A, SNAP23, CDC42 | Vesicle pathway | Hook et al., 2014 |
| miR-US25-1 | E2, BRCC3∗, EID1, MAPRE2, CD147 | Cell survival | Grey et al., 2010; Fan et al., 2014 |
| miR-US25-2-3p | eIF4A1 | Viral infection | Qi et al., 2013 |
| miR-US33-5p | STX3 | Vesicle pathway | Guo et al., 2015 |
| HIV-1-encoded miRNA | |||
| miR-TAR | ERCC1, IER3 NPM/B23, Caspase 8, Aiolos, Ikaros | Apoptosis | Klase et al., 2009; Ouellet et al., 2013 |
| miR-88 | Immune evasion | Bernard et al., 2014 | |
| miR-99 | Immune evasion | Bernard et al., 2014 | |
| miR-H3 | HIV-1 5′ LTR | Viral replication | Zhang et al., 2014 |
| miR-N367 | NEF§ | Viral replication | Omoto et al., 2004 |
| miR-H1 | AATF | Apoptosis | Kaul et al., 2009 |
* In italic viral targets.
§ Hypothetical target.
The first host cellular mRNA target reported for a HCMV vmiRNA encodes MICB, a stress-induced ligand for the NK cell activating receptor NKG2D critical for the NK cell killing of virus-infected and tumor cells. VmiR-UL112 specifically binds to MICB-3′ untranslated regions (3′UTR) and downregulates MICB expression during viral infection, therefore, leading to decreased binding of NKG2D and reduced killing by NK cells (Stern-Ginossar et al., 2007). This miRNA-mediated MICB inhibition is not exclusive to HCMV, but conserved in the herpesviruses family. Other viral miRNAs encoded by EBV and HHV-8 (miR-BART2-5p and miR-k12-7, respectively), target MICB to escape NK cell recognition (Nachmani et al., 2009). These vmiRNAs exhibit poor sequence homology with HCMV miR-UL112 and target MICB at different binding sites. HCMV miR-UL112 attenuates NK cell activity also by targeting others transcripts, such as IL-32, type I IFN and the toll-like receptor 2 (TLR2) signaling (Landais et al., 2015).
In addition, the HCMV miR-UL148D, one of the most highly expressed vmiRNAs during latent infection, contribute to immune evasion by directly targeting the chemokine (C-C Motif) ligand 5 (CCL5), a chemokine known to attract immune cells to sites of inflammation and tissue damage (Kim et al., 2012). This downregulation was reverted by treatment with a miR-UL148D-specific inhibitor, supporting the role of this agent as therapeutic tool against HCMV infection. VmiR-UL148D also targets the activating receptor type-1B (ACVR1B) in monocytes, resulting in a reduced secretion of IL-6 (Lau et al., 2016). More recently, vmiR-US5-1 and vmiR-UL112-3p have been shown to play important role in modulating NF-κB signaling at late time of infection by reducing the expression of the IKK complex and induce the release of proinflammatory cytokines (Hancock et al., 2017). The limited or delayed secretion of proinflammatory cytokines is thought to be one of the mechanisms of immune evasion exploited by vmiRNAs to limit the recruitment of immune cells and killing of infected cells.
Finally, the HCMV miR-US4-1 was shown to directly target the endoplasmic reticulum aminopeptidase 1 (ERAP1), a key peptidase that trims peptide precursors to their optimal length to bind MHC class I molecules (Kim et al., 2011). The reduced trimming due to HCMV-specific action resulted in an immuno-evasion of HCMV-infected cells (Kim et al., 2011).
All these mechanisms focus on the possibility to enhance viral replication by hindering viral clearance by NK cells and T cells for the period of time necessary to virus to replicate.
HIV-1 miRNAs
HIV-1 infection cause progressive CD4+ T-cell loss making individuals susceptible to get infections and develop a wide range of immunological abnormalities until oncological complications. Although HIV infections are able to induce vigorous antiviral immune responses, HIV-1 replication is not fully controlled by the innate and adaptive immune system. Like many other viruses, HIV-1 has evolved a number of strategies to evade host immune responses, most notably by using viral accessory proteins and RNA.
Recent reports have demonstrated the existence of HIV-1-derived miRNAs from coding and non-coding regions of the viral genome, which regulate both viral and host gene expression (Table 1). Despite so, the presence of HIV-1 derived vmiRNAs has been highly controversial and further studies are necessary to better understand their potential role in viral infection and pathogenesis. The importance of viral and cell host miRNAs in the context of HIV-1 infection was first suggested by silencing of Drosha and Dicer leading to significant enhancement of HIV-1 replication (Triboulet et al., 2007). The first description of HIV-1-derived miRNAs came in 2004 from a group that identified a Nef-derived miRNA, named miR-N367 (Omoto et al., 2004). MiR-N367 reduces HIV-1 transcription by blocking Nef expression and long terminal repeat (LTR) transcription (Omoto et al., 2004; Omoto and Fujii, 2005). The HIV-1 transactivation RNA (TAR), which regulates viral translation, also encodes for a vmiRNA (called TAR-miR-5p and -3p) (Ouellet et al., 2008; Klase et al., 2009). TAR-miR has been shown to downregulate host genes (such as ERCC1 and IER3) important for apoptosis and cell survival, thus giving HIV-1-infected cells a survival advantage by preventing host cell death (Klase et al., 2009). Recent studies demonstrated that additional host substrates, including Caspase 8, Aiolos, Ikaros and Nucleophosmin (NPM)/B23, are modulated by TAR-miR (Ouellet et al., 2013). VmiR-H1, located in the LTR has been reported to downregulate the apoptosis-antagonizing transcription factor (AATF) gene product and act as an antagonist of the anti-apoptotic effect mediated by TAR-miR. Additionally, hiv1-mir-H1 can downregulate the host miR149 expression recognized to target HIV-1 Vpr transcript (Kaul et al., 2009). Recently, Zhang et al. (2014) have reported the existence of a novel HIV-1-encoded vmiRNA called miR-H3 located in the region of the HIV-1 RNA genome that encodes for reverse transcriptase (RT). Overexpression of miR-H3 increases viral production and mutations within miR-H3 sequence significantly impair the viral replication of wild-type HIV-1 viruses by targeting HIV-1 5′-LTR (TATA box) (Zhang et al., 2014). Two additional vmiRNAs (called as vmiR88 and vmiR99) were identified in overlapping regions of HIV LTR from viral infected human macrophages (Bernard et al., 2014). They were able to directly stimulate TNFα release by human macrophages through TLR8 activation by a mechanism that is partially dependent on vmiRNA sequence motifs. Of note, vmiR88 and vmiR99 were detected in sera of HIV infected individuals suggesting their role in stimulating recipient macrophages in vivo and contributing to chronic immune activation.
Future Outlook and Conclusion
vmiRNAs have overall a protecting role for the viral infection. By interfering with host cellular proteins and mechanisms of immune evasion, they represent a potential target for future specific therapies. Studies on the complex interplay between viral miRNAs and host genes has been only recently started and further investigations are required to identify more efficient and less toxic therapeutic strategies. The identification of differentially expressed miRNAs during viral infection (for example in HIV-1 infection) may provide a new approach to control disease progression. Finally, understanding the mechanism of vmiRNA-mediated immune evasion will allow us to develop novel strategies to prevent and cure viral infections. Targeting vmiRNAs with molecules such as antagomirs may represent a novel therapeutic strategy to limit chronic immune activation and the progression of viral infection.
Author Contributions
DF and AG analyzed the literature, wrote the manuscript, and approved the final version for publication. RR analyzed the literature, revised the manuscript and approved the final version for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Italian Ministry of Health (Rome, Italy) grant PE-2011-02351866 (DF) and Associazione Italiana per la Ricerca sul Cancro (AIRC, Milan, Italy) grants # 18495 (DF), # 17615 (AG) and # 15312 (RR).
Footnotes
References
- Bai X. T., Nicot C. (2015). miR-28-3p is a cellular restriction factor that inhibits human T cell leukemia virus, type 1 (HTLV-1) replication and virus infection. J. Biol. Chem. 290 5381–5390. 10.1074/jbc.M114.626325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y., Yeung M. L., Jeang K. T. (2006). HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J. Biol. Chem. 281 27674–27678. 10.1074/jbc.C600072200 [DOI] [PubMed] [Google Scholar]
- Bernard M. A., Zhao H., Yue S. C., Anandaiah A., Koziel H., Tachado S. D. (2014). Novel HIV-1 miRNAs stimulate TNFalpha release in human macrophages via TLR8 signaling pathway. PLoS ONE 9:e106006 10.1371/journal.pone.0106006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H. P., Karnowski H. W., Cai X., Shin J., Pohlers M., Cullen B. R. (2010). A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol. Cell. 37 135–142. 10.1016/j.molcel.2009.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Bass C. R., Kincaid R. P., Sullivan C. S. (2014). Identification of tri-phosphatase activity in the biogenesis of retroviral microRNAs and RNAP III-generated shRNAs. Nucleic Acids Res. 42 13949–13962. 10.1093/nar/gku1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Hagedorn C. H., Cullen B. R. (2004). Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10 1957–1966. 10.1261/rna.7135204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla G., Sokol N. S. (2014). ADAR mediates differential expression of polycistronic microRNAs. Nucleic Acids Res. 42 5245–5255. 10.1093/nar/gku145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., et al. (2005). TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436 740–744. 10.1038/nature03868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme-Axford E., Donker R. B., Mouillet J. F., Chu T., Bayer A., Ouyang Y., et al. (2013). Human placental trophoblasts confer viral resistance to recipient cells. Proc. Natl. Acad. Sci. U.S.A. 110 12048–12053. 10.1073/pnas.1304718110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli A. M., Tops B. B., Plasterk R. H., Ketting R. F., Hannon G. J. (2004). Processing of primary microRNAs by the Microprocessor complex. Nature 432 231–235. 10.1038/nature03049 [DOI] [PubMed] [Google Scholar]
- Diebel K. W., Claypool D. J., Van Dyk L. F. (2014). A conserved RNA polymerase III promoter required for gammaherpesvirus TMER transcription and microRNA processing. Gene 544 8–18. 10.1016/j.gene.2014.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebel K. W., Smith A. L., Van Dyk L. F. (2010). Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA 16 170–185. 10.1261/rna.1873910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Zhang W., Liu Q. (2014). Human cytomegalovirus-encoded miR-US25-1 aggravates the oxidised low density lipoprotein-induced apoptosis of endothelial cells. Biomed. Res. Int. 2014:531979 10.1155/2014/531979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman E. R., Kara M., Coleman C. B., Grau K. R., Oko L. M., Krueger B. J., et al. (2014). Virus-encoded microRNAs facilitate gammaherpesvirus latency and pathogenesis in vivo. MBio 5:e981-14 10.1128/mBio.00981-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F., Meyers H., White E. A., Spector D. H., Nelson J. (2007). A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 3:e163 10.1371/journal.ppat.0030163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F., Tirabassi R., Meyers H., Wu G., Mcweeney S., Hook L., et al. (2010). A viral microRNA down-regulates multiple cell cycle genes through mRNA 5’UTRs. PLoS Pathog. 6:e1000967 10.1371/journal.ppat.1000967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., Ha I., et al. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106 23–34. 10.1016/S0092-8674(01)00431-7 [DOI] [PubMed] [Google Scholar]
- Grundhoff A., Sullivan C. S. (2011). Virus-encoded microRNAs. Virology 411 325–343. 10.1016/j.virol.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Huang Y., Qi Y., Liu Z., Ma Y., Shao Y., et al. (2015). Human cytomegalovirus miR-UL36-5p inhibits apoptosis via downregulation of adenine nucleotide translocator 3 in cultured cells. Arch. Virol. 160 2483–2490. 10.1007/s00705-015-2498-8 [DOI] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K. H., Nam J. W., Heo I., Rhee J. K., et al. (2006). Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125 887–901. 10.1016/j.cell.2006.03.043 [DOI] [PubMed] [Google Scholar]
- Hancock M. H., Hook L. M., Mitchell J., Nelson J. A. (2017). Human cytomegalovirus microRNAs miR-US5-1 and miR-UL112-3p block proinflammatory cytokine production in response to NF-kappaB-activating factors through direct downregulation of IKKalpha and IKKbeta. mBio 8:e00109-17. 10.1128/mBio.00109-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwig A., Das A. T., Berkhout B. (2014). Retroviral microRNAs. Curr. Opin. Virol. 7 47–54. 10.1016/j.coviro.2014.03.013 [DOI] [PubMed] [Google Scholar]
- Hook L. M., Grey F., Grabski R., Tirabassi R., Doyle T., Hancock M., et al. (2014). Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe 15 363–373. 10.1016/j.chom.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Chen D., He J., Cai J., Shen K., Liu X., et al. (2015). Hcmv-miR-UL112 attenuates NK cell activity by inhibition type I interferon secretion. Immunol. Lett. 163 151–156. 10.1016/j.imlet.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Huang Y., Qi Y., Ma Y., He R., Ji Y., Sun Z., et al. (2013). The expression of interleukin-32 is activated by human cytomegalovirus infection and down regulated by hcmv-miR-UL112-1. Virol. J. 10:51 10.1186/1743-422X-10-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul D., Ahlawat A., Gupta S. D. (2009). HIV-1 genome-encoded hiv1-mir-H1 impairs cellular responses to infection. Mol. Cell. Biochem. 323 143–148. 10.1007/s11010-008-9973-4 [DOI] [PubMed] [Google Scholar]
- Kawahara Y., Megraw M., Kreider E., Iizasa H., Valente L., Hatzigeorgiou A. G., et al. (2008). Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 36 5270–5280. 10.1093/nar/gkn479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Lee S., Shin J., Kim Y., Evnouchidou I., Kim D., et al. (2011). Human cytomegalovirus microRNA miR-US4-1 inhibits CD8(+) T cell responses by targeting the aminopeptidase ERAP1. Nat. Immunol. 12 984–991. 10.1038/ni.2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lee S., Kim S., Kim D., Ahn J. H., Ahn K. (2012). Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog. 8:e1002577 10.1371/journal.ppat.1002577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid R. P., Burke J. M., Sullivan C. S. (2012). RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci. U.S.A. 109 3077–3082. 10.1073/pnas.1116107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid R. P., Chen Y., Cox J. E., Rethwilm A., Sullivan C. S. (2014). Noncanonical microRNA (miRNA) biogenesis gives rise to retroviral mimics of lymphoproliferative and immunosuppressive host miRNAs. MBio 5:e00074-14 10.1128/mBio.00074-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klase Z., Winograd R., Davis J., Carpio L., Hildreth R., Heydarian M., et al. (2009). HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology 6:18 10.1186/1742-4690-6-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42 D68–D73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landais I., Pelton C., Streblow D., Defilippis V., Mcweeney S., Nelson J. A. (2015). Human cytomegalovirus miR-UL112-3p targets TLR2 and modulates the TLR2/IRAK1/NFkappaB signaling pathway. PLoS Pathog. 11:e1004881 10.1371/journal.ppat.1004881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau B., Poole E., Krishna B., Sellart I., Wills M. R., Murphy E., et al. (2016). The expression of human cytomegalovirus microRNA MiR-UL148D during latent infection in primary myeloid cells inhibits activin A-triggered secretion of IL-6. Sci. Rep. 6:31205 10.1038/srep31205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier C. H., Dunoyer P., Arar K., Lehmann-Che J., Eyquem S., Himber C., et al. (2005). A cellular microRNA mediates antiviral defense in human cells. Science 308 557–560. 10.1126/science.1108784 [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K. H., Lee S., Baek S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23 4051–4060. 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., et al. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433 769–773. 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Lu S., Cullen B. R. (2004). Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J. Virol. 78 12868–12876. 10.1128/JVI.78.23.12868-12876.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano D. J., Mirsky H., Vendetti N. J., Maas S. (2004). RNA editing of a miRNA precursor. RNA 10 1174–1177. 10.1261/rna.7350304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna J. M., Scheel T. K., Danino T., Shaw K. S., Mele A., Fak J. J., et al. (2015). Hepatitis C virus RNA functionally sequesters miR-122. Cell 160 1099–1110. 10.1016/j.cell.2015.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshesha M. K., Bentwich Z., Solomon S. A., Avni Y. S. (2016). In vivo expression of human cytomegalovirus (HCMV) microRNAs during latency. Gene 575 101–107. 10.1016/j.gene.2015.08.040 [DOI] [PubMed] [Google Scholar]
- Murphy E., Vanicek J., Robins H., Shenk T., Levine A. J. (2008). Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. U.S.A. 105 5453–5458. 10.1073/pnas.0711910105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachmani D., Stern-Ginossar N., Sarid R., Mandelboim O. (2009). Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5 376–385. 10.1016/j.chom.2009.03.003 [DOI] [PubMed] [Google Scholar]
- Nachmani D., Zimmermann A., Oiknine Djian E., Weisblum Y., Livneh Y., Khanh Le V. T., et al. (2014). MicroRNA editing facilitates immune elimination of HCMV infected cells. PLoS Pathog. 10:e1003963 10.1371/journal.ppat.1003963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. (2010). Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79 321–349. 10.1146/annurev-biochem-060208-105251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriega V., Redmann V., Gardner T., Tortorella D. (2012). Diverse immune evasion strategies by human cytomegalovirus. Immunol. Res. 54 140–151. 10.1007/s12026-012-8304-8 [DOI] [PubMed] [Google Scholar]
- Omoto S., Fujii Y. R. (2005). Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J. Gen. Virol. 86 751–755. 10.1099/vir.0.80449-0 [DOI] [PubMed] [Google Scholar]
- Omoto S., Ito M., Tsutsumi Y., Ichikawa Y., Okuyama H., Brisibe E. A., et al. (2004). HIV-1 nef suppression by virally encoded microRNA. Retrovirology 1:44 10.1186/1742-4690-1-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota H., Sakurai M., Gupta R., Valente L., Wulff B. E., Ariyoshi K., et al. (2013). ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 153 575–589. 10.1016/j.cell.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet D. L., Plante I., Landry P., Barat C., Janelle M. E., Flamand L., et al. (2008). Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 36 2353–2365. 10.1093/nar/gkn076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet D. L., Vigneault-Edwards J., Letourneau K., Gobeil L. A., Plante I., Burnett J. C., et al. (2013). Regulation of host gene expression by HIV-1 TAR microRNAs. Retrovirology 10:86 10.1186/1742-4690-10-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F., Poling L. L., Wang Z., Liu H., Liu X. S., Roeder R. G., et al. (2008). Chromatin structure analyses identify miRNA promoters. Genes Dev. 22 3172–3183. 10.1101/gad.1706508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Zhu D., Wang Y., Li L., Li D., Liu F., et al. (2016). Human cytomegalovirus miR-UL148D facilitates latent viral infection by targeting host cell immediate early response gene 5. PLoS Pathog. 12:e1006007 10.1371/journal.ppat.1006007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S., Sewer A., Lagos-Quintana M., Sheridan R., Sander C., Grasser F. A., et al. (2005). Identification of microRNAs of the herpesvirus family. Nat. Methods 2 269–276. 10.1038/nmeth746 [DOI] [PubMed] [Google Scholar]
- Pfeffer S., Zavolan M., Grasser F. A., Chien M., Russo J. J., Ju J., et al. (2004). Identification of virus-encoded microRNAs. Science 304 734–736. 10.1126/science.1096781 [DOI] [PubMed] [Google Scholar]
- Piedade D., Azevedo-Pereira J. M. (2016). The role of microRNAs in the pathogenesis of herpesvirus infection. Viruses 8:E156 10.3390/v8060156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M., Qi Y., Ma Y., He R., Ji Y., Sun Z., et al. (2013). Over-expression of human cytomegalovirus miR-US25-2-3p downregulates eIF4A1 and inhibits HCMV replication. FEBS Lett. 587 2266–2271. 10.1016/j.febslet.2013.05.057 [DOI] [PubMed] [Google Scholar]
- Raab-Traub N. (2007). “EBV-induced oncogenesis,” in Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis eds Arvin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roizman B., Whitley R.et al. (Cambridge: Cambridge University Press; ). [PubMed] [Google Scholar]
- Shao Y., Qi Y., Huang Y., Liu Z., Ma Y., Guo X., et al. (2016). Human cytomegalovirus-encoded miR-US4-1 promotes cell apoptosis and benefits discharge of infectious virus particles by targeting QARS. J. Biosci. 41 183–192. 10.1007/s12038-016-9605-1 [DOI] [PubMed] [Google Scholar]
- Skalsky R. L., Cullen B. R. (2010). Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64 123–141. 10.1146/annurev.micro.112408.134243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N., Elefant N., Zimmermann A., Wolf D. G., Saleh N., Biton M., et al. (2007). Host immune system gene targeting by a viral miRNA. Science 317 376–381. 10.1126/science.1140956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N., Weisburd B., Michalski A., Le V. T., Hein M. Y., Huang S. X., et al. (2012). Decoding human cytomegalovirus. Science 338 1088–1093. 10.1126/science.1227919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H. I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. (2009). Modulation of microRNA processing by p53. Nature 460 529–533. 10.1038/nature08199 [DOI] [PubMed] [Google Scholar]
- Tirabassi R., Hook L., Landais I., Grey F., Meyers H., Hewitt H., et al. (2011). Human cytomegalovirus US7 is regulated synergistically by two virally encoded microRNAs and by two distinct mechanisms. J. Virol. 85 11938–11944. 10.1128/JVI.05443-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaselli S., Bonamassa B., Alisi A., Nobili V., Locatelli F., Gallo A. (2013). ADAR enzyme and miRNA story: a nucleotide that can make the difference. Int. J. Mol. Sci. 14 22796–22816. 10.3390/ijms141122796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaselli S., Galeano F., Alon S., Raho S., Galardi S., Polito V. A., et al. (2015). Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 16:5 10.1186/s13059-014-0575-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M., Briata P., Garcia-Mayoral M., Haase A. D., Filipowicz W., Ramos A., et al. (2009). The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459 1010–1014. 10.1038/nature08025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R., Mari B., Lin Y. L., Chable-Bessia C., Bennasser Y., Lebrigand K., et al. (2007). Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315 1579–1582. 10.1126/science.1136319 [DOI] [PubMed] [Google Scholar]
- Varani S., Landini M. P. (2011). Cytomegalovirus-induced immunopathology and its clinical consequences. Herpesviridae 2:6 10.1186/2042-4280-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S. R., Daley G. Q., Gregory R. I. (2008). Selective blockade of microRNA processing by Lin28. Science 320 97–100. 10.1126/science.1154040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. P., Qi Y., Huang Y. J., Qi M. L., Ma Y. P., He R., et al. (2013). Identification of immediate early gene X-1 as a cellular target gene of hcmv-mir-UL148D. Int. J. Mol. Med. 31 959–966. 10.3892/ijmm.2013.1271 [DOI] [PubMed] [Google Scholar]
- Whisnant A. W., Kehl T., Bao Q., Materniak M., Kuzmak J., Lochelt M., et al. (2014). Identification of novel, highly expressed retroviral microRNAs in cells infected by bovine foamy virus. J. Virol. 88 4679–4686. 10.1128/JVI.03587-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Fan M., Geng G., Liu B., Huang Z., Luo H., et al. (2014). A novel HIV-1-encoded microRNA enhances its viral replication by targeting the TATA box region. Retrovirology 11:23 10.1186/1742-4690-11-23 [DOI] [PMC free article] [PubMed] [Google Scholar]