Abstract
Background
Powdery mildew (PM) causing fungi are well-known pathogens, infecting over 10.000 plant species, including the economically important crop cucumber (Cucumis sativus L.). Loss-of-function mutations in clade V MLO genes have previously been shown to lead to recessively inherited broad-spectrum resistance to PM in several species. In cucumber, one clade V MLO homolog (CsaMLO8) was previously identified as being a susceptibility factor to PM. Two other closely related homologs (CsaMLO1 and CsaMLO11) were found, but their function was not yet unravelled.
Methods
CsaMLO1 and CsaMLO11 were cloned from cucumber and overexpressed in a tomato mlo mutant. The transcript abundances of all three CsaMLO genes in different cucumber tissues were quantified using qRT-PCR and RNA-seq, with and without inoculation with the cucumber PM fungus Podosphaera xanthii. Allelic variation of CsaMLO1 and CsaMLO11 was screened in silico in sequenced cucumber germplasm.
Results
Heterologous overexpression of all three CsaMLO genes in the tomato mlo mutant restored susceptibility to PM caused by Oidium neolycopersici, albeit to a different extent: whereas overexpression of CsaMLO1 or CsaMLO8 completely restored susceptibility, overexpression of CsaMLO11 was only partially able to restore PM susceptibility. Furthermore, it was observed by qRT-PCR and RNA-seq that CsaMLO8 was significantly higher expressed in non-inoculated cucumber compared to the other two MLO genes. However, inoculation with P. xanthii led to upregulation of CsaMLO1, but not to upregulation of CsaMLO8 or CsaMLO11.
Conclusions
Both CsaMLO1 and CsaMLO11 are functional susceptibility genes, although we conclude that based on the transcript abundance CsaMLO8 is probably the major clade V MLO gene in cucumber regarding providing susceptibility to PM. Potential loss-of-function mutations in CsaMLO1 and CsaMLO11 have not been identified. The generation and analysis of such mutants are interesting subjects for further investigation.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-017-1029-z) contains supplementary material, which is available to authorized users.
Keywords: Cucumber (Cucumis sativus L.), Powdery mildew, MLO, Susceptibility genes, Gene expression
Background
Powdery mildew (PM), caused by ascomycete fungi of the order Erysiphales, is one of the most well-known plant diseases [1]. PM fungi are able to cause disease on leaves, stems, flowers and fruits of nearly 10.000 different angiosperm plant species, including various economically important plants, such as cucumber (Cucumis sativus L.). PM in cucumber can be caused by two different species, Golovinomyces orontii and Podosphaera xanthii. In greenhouses and open field cultivation in warm regions, P. xanthii appears to be the most occurring agent of PM in cucurbits, whereas G. orontii is the major PM species on cucurbits grown in the open field in colder regions [2].
According to the gene-for-gene concept, plants have dominantly inherited resistance genes (R-genes), encoding R proteins which recognize the products of corresponding avirulence genes (Avr-genes) of pathogens, triggering defence responses [3, 4]. Even though this can give a strong, complete resistance, the pathogen can mutate, lose or silence the recognized Avr gene to break the resistance, leading to a new virulent race of the pathogen, often within a few years after the commercial introduction of a new R gene. The cloning of R genes in several plant species has led to the finding that they typically encode receptor proteins of various classes with leucine-rich repeat (LRR) domains [3]. An exception to R-gene mediated resistance was discovered in X-ray irradiated summer barley populations in the 1940’s [5]. A recessively inherited monogenic resistance was observed, which was active against all known isolates of barley PM (caused by Blumeria graminis f. sp. hordei). Later, other alleles at the same genetic locus were obtained in various barley genotypes, including a naturally occurring mutant allele that was found in resistant Ethiopian barley landraces [6]. The durability of this so-called mlo (Mildew Locus O) resistance is exemplified by the fact that cultivars with this type of resistance have been extensively cultivated since the 1980s without new races of the pathogen breaking the resistance [7]. By positional cloning, the causal gene for mlo based resistance was isolated [8]. It was found to encode a plasma membrane-anchored protein with seven transmembrane helices, reminiscent of animal G-protein-coupled receptors [9].
After the barley MLO gene was cloned, it was found that mutations in homologs of this gene in other plant species can also lead to recessively inherited resistance to different PM causing fungi. In the model species Arabidopsis thaliana T-DNA insertion mutations in three MLO homologs contribute to PM resistance, although a mutation in one of the three genes (AtMLO2) has a larger effect compared to mutations in the other two genes (AtMLO6 and AtMLO12). The effect of loss of function of AtMLO6 or AtMLO12 is only additive in Atmlo2 background, but not detectable when AtMLO2 is intact [10]. In several crop species, i.e. tomato [11], pea [12], cucumber [13] and tobacco [14], recessively inherited PM resistance with similar characteristics compared to barley mlo resistance was indeed found to be caused by naturally occurring mutations in MLO homologs. In other species, e.g. pepper, wheat, apple and grapevine, knockdown of MLO homologs by virus induced gene silencing (VIGS) or RNA interference (RNAi) led to PM resistance [15–18]. This indicates that mlo-based resistance is very common in plants, rather than a particular oddity occurring in barley. It has been shown that mlo-based resistance depends on the formation of cell wall depositions (papillae) by the plant cell directly beneath PM penetration attempts [19]. The molecular basis of mlo-based resistance is however yet poorly understood, although it has been shown that it depends on the function of several molecular components, such as the BAX-inhibitor protein (BI-1) which plays a role in control of programmed cell death [20]; on an intact actin cytoskeleton [21] and on the t-SNARE proteins PEN1 (Arabidopsis) and ROR2 (barley) involved in targeted exocytosis [22].
Since the year 2000, when the first plant genome sequence was published, i.e. that of the model plant Arabidopsis thaliana [23], an increasing number of plant genomes has been sequenced. In all available sequenced plant genomes MLO homologs occur as medium sized gene families with seven to thirty-nine MLO genes per plant species [24]. In phylogenetic analyses of the MLO gene family, it has been found that MLO genes can be divided into seven phylogenetic clades, although not all plant species harbour representatives of all clades [25]. The mosses, representing the most basal lineages of land plants, have MLO homologs only one of the clades, i.e. clade I. In other lineages of plants, especially in angiosperm species, the MLO gene family has diversified. However, several plant species apparently lost genes in some of the MLO clades during evolution, such as the monocotyledonous family of the Poaceae which has no MLO homologs in clades V and VI, or several dicotyledonous species such as Arabidopsis thaliana and tomato which have lost clade IV MLO homologs, even though a basal angiosperm species, Amborella trichopoda, has homologs of clade I to VI [25].
Not all the MLO genes found so far have been characterised as being required for susceptibility towards PM fungi. For most MLO genes mutant phenotypes have not been described yet, although there are examples of clade I mlo mutants with a defect in root formation [26] and of clade III mlo mutants with defects in pollen tube perception [27] and pollen hydration [28]. So far, all MLO genes involved in PM susceptibility (i.e. susceptibility genes, S-genes) have been found to group either in clade IV (for monocotyledonous species) or clade V (for dicotyledonous species). It has been shown that heterologous overexpression of the barley clade IV MLO gene can functionally complement loss of function mutations in clade V MLO genes in tomato [29], exemplifying that although there are significant differences in amino acid sequence between clade IV and V MLO proteins, they are functionally conserved.
In cucumber, the genome sequence of which was published in 2009 [30], thirteen MLO homologs have previously been identified. Of these thirteen homologs, three genes were found to group phylogenetically in clade V. These three genes, named CsaMLO1, CsaMLO8 and CsaMLO11, should therefore be considered as potential PM S-genes in cucumber [31]. In other cucurbit crops, i.e. melon (Cucumis melo), watermelon (Citrullus lanatus) and pumpkin (Cucurbita pepo), similar numbers of clade V MLO genes have been identified, although pumpkin has four rather than three clade V MLO genes [32]. Phylogenetic analysis reveals that the last common ancestor of these cucurbit crops already had at least three clade V MLO genes.
Of the three cucumber clade V MLO genes, CsaMLO8 has previously been proven to be a susceptibility gene for PM caused by P. xanthii. From cucumber genotypes with recessively inherited PM resistance a natural Csamlo8 mutant allele was cloned [13, 33]. While heterologous overexpression of the wild-type CsaMLO8 was able to functionally complement mlo loss-of-function mutants in both tomato [13] and Arabidopsis [33], the mutant allele failed to restore susceptibility. The mutation in Csamlo8 has been caused by the integration of a retrotransposable element in the coding sequence of the gene [13, 34]. This mutant allele was found to occur frequently in cultivated cucumber germplasm [13], and additionally two other Csamlo8 loss-of-function mutations have been found in resistant genotypes due to either a frameshift indel leading to an early stop codon, or a SNP in an intron-exon junction causing aberrant splicing of the pre-mRNA [33].
In a review of co-localization of cucumber MLO genes with QTLs for PM resistance, Schouten et al. [31] mentioned that two previously described QTLs for PM resistance co-localized with the other two cucumber clade V MLO genes, CsaMLO1 and CsaMLO11. Fukino et al. [35] performed QTL analysis in a RIL population derived from a cross between the PM resistant genotype CS-PMR1 (an inbred line derived from the PM resistant wild cucumber accession PI 197088) and the moderately susceptible genotype Santou, a native Japanese cultivar. Of the nine detected QTLs for PM resistance, one QTL (pm1.1) co-localized with CsaMLO1, whereas another QTL (pm6.1) co-localized with CsaMLO11. The resistance associated with pm1.1 was contributed by the allele from CS-PMR1, while the Santou allele contributed to resistance at the pm6.1 locus.
Here we report the functional characterization of two cucumber clade V MLO genes, CsaMLO1 and CsaMLO11. We show that heterologous overexpression of either of the genes in a tomato mlo mutant led to restoration of susceptibility to PM. Furthermore, we investigated the transcription profile of the three cucumber clade V MLO genes in various tissues, both prior to and after inoculation with the PM causing fungus P. xanthii. Also, we screened a set of 115 resequenced cucumber accessions in silico for potential loss of function mutations in either of the clade V MLO genes, and resequenced two additional cucumber genotypes with reported PM resistance QTLs in regions containing either CsaMLO1 or CsaMLO11.
Results
Functional analysis of cucumber clade V MLO genes by complementation of a tomato mlo mutant
We amplified the clade V MLO genes CsaMLO1 [Csa1M085890] and CsaMLO11 [Csa6M292430] from cDNA of a cucumber inbred line. PCR products were of the expected sizes (1.749 bp and 1.782 bp, respectively) and sequences were identical to the reference genome of the PM susceptible genotype ‘Chinese Long 9930’ [30]. To test whether these genes are susceptibility genes, CsaMLO1 and CsaMLO11 were overexpressed in a tomato mlo mutant, which is resistant to PM due to a mutation in the SlMLO1 gene [11]. It was expected that if CsaMLO1 and/or CsaMLO11 are susceptibility genes, overexpression in the tomato mlo mutant would lead to restoration of susceptibility to PM. After transformation, cuttings from eight (CsaMLO1) or seven (CsaMLO11) individual transformants were obtained and inoculated with Oidium neolycopersici, the causal agent of PM in tomato. Sporulation was observed on five of the eight CsaMLO1 transformants and on two of the seven CsaMLO11 transformants (Table 1).
Table 1.
Cuttings of five out of eight CsaMLO1 transformants and two out of seven CsaMLO11 transformants were found to be susceptible to Oidium neolycopersici
| Gene | Transformant | Transgene expression | PM symptoms |
|---|---|---|---|
| CsaMLO1 | 35S::CsaMLO1-A | 2.3 | + |
| 35S::CsaMLO1-B | 1.3 | + | |
| 35S::CsaMLO1-C | 1.2 | + | |
| 35S::CsaMLO1-D | 0.8 | − | |
| 35S::CsaMLO1-E | 0.7 | − | |
| 35S::CsaMLO1-F | 0.4 | − | |
| 35S::CsaMLO1-G | 0.4 | + | |
| 35S::CsaMLO1-H | 0.1 | + | |
| CsaMLO11 | 35S::CsaMLO11-A | 0.3 | − |
| 35S::CsaMLO11-B | 0.3 | − | |
| 35S::CsaMLO11-C | 0.2 | + | |
| 35S::CsaMLO11-D | 0.2 | − | |
| 35S::CsaMLO11-E | 0.1 | − | |
| 35S::CsaMLO11-F | 0.1 | − | |
| 35S::CsaMLO11-G | 0.1 | + |
The tomato Slmlo1 mutant, with a frameshift deletion in the SlMLO1 gene [11], was transformed with either a 35S::CsaMLO1 construct or a 35S::CsaMLO11 construct. CsaMLO1 or CsaMLO11 expression in each of the primary transformants (one sample per individual transformant) was quantified relatively to housekeeping gene SlEF-α using qRT-PCR. Two cuttings per individual transformant were inoculated with Oidium neolycopersici, the causal agent of powdery mildew (PM) in tomato. Disease phenotypes were scored based on whether or not PM symptoms were visible on the leaves at 10 days post inoculation
In order to confirm the ability of CsaMLO1 and CsaMLO11 to restore PM susceptibility, primary transformants were self-pollinated to obtain T2 families. T2 families were obtained from two individual transformants per gene. Additionally, two T2 families were obtained from previously described CsaMLO8 [Csa5M623470] overexpressing transformants [13]. From each T2 family, 22 to 30 plants were sown. Plants were screened by PCR for the presence of the overexpression construct. It was found that the T2 populations obtained from the CsaMLO1 and CsaMLO11 transformants segregated for the presence of the transgene in ratios close to 3:1, suggesting one transgene insertion site per individual primary transformant. The T2 populations obtained from (both) the CsaMLO8 transformants showed skewed segregation patterns with either one out of 22 (T2-A) or one out of 30 (T2-B) individuals not having the transgene, suggesting multiple inserts per individual transformant.
T2 families were inoculated with O. neolycopersici and PM symptoms were scored based on a 0–3 scale, with 0 being completely free of PM symptoms and 3 being fully infected. The non-transformed slmlo1 mutant did not show any PM symptoms (i.e. all plants scored a disease index of 0), whereas in the susceptible control (Moneymaker) 75% of the plants scored the maximum disease index of 3, and 25% of the plants scored a disease index of 2 (Fig. 1). An overall analysis showed significant differences between the groups (Kruskal-Wallis test, P < 0.05). Stepdown post hoc analysis revealed that overexpression of CsaMLO8 or CsaMLO1 restored susceptibility completely, leading to a susceptibility level not significantly different from Moneymaker (P > 0.05). However, overexpression of CsaMLO11 restored susceptibility only partially, giving disease indices between 0 and 1, which is significantly higher than the resistant control slmlo1 (P < 0.05) but significantly lower than the susceptible control Moneymaker (P < 0.05).
Fig. 1.
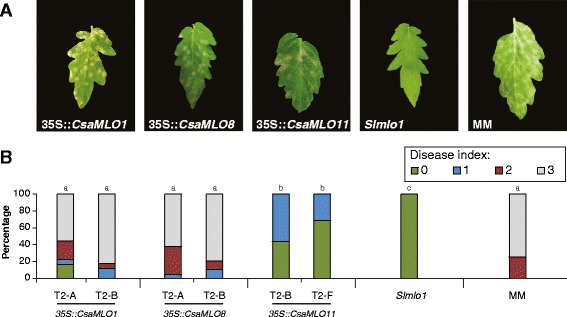
Complementation of a tomato mlo mutant with either CsaMLO1, CsaMLO8 or CsaMLO11. The tomato mlo mutant, with a frameshift deletion in the SlMLO1 gene [11], was transformed with either a 35S::CsaMLO1 construct, a 35S::CsaMLO8 construct or a 35S::CsaMLO11 construct (Table 1). Two individual transformants per transgene were self-pollinated to obtain T2 populations. T2 plants were screened for the presence of the overexpression construct. Plants carrying an overexpression construct were inoculated with O. neolycopersici. a Representative individuals from T2 families expressing either CsaMLO1, CsaMLO8 or CsaMLO11 in the tomato mlo mutant, showing powdery mildew (PM) symptoms. Non-transformed tomato slmlo1 mutant (PM resistant) and cv. Moneymaker (MM, PM susceptible) are shown as controls. b Disease indices were scored visually on a scale from 0 (completely resistant) to 3 (completely susceptible), as described in [11]. T2 families consisted of 16 to 29 individuals positive for the presence of the overexpression construct. Resistant and susceptible controls consisted of 12 individuals. Bars represent percentages of plants within each disease index class. Different letters above bars indicate significant differences between populations (Kruskal-Wallis test with Stepwise-Stepdown Multiple Comparisons, P < 0.05)
Per family, five plants (positive for the presence of the overexpression construct) were randomly chosen to measure the expression of the transgene using qRT-PCR (Additional file 1). It was found that the transgene expression in both CsaMLO8 T2 families was significantly higher than in the CsaMLO1 T2-A family and both of the CsaMLO11 T2 families (ANOVA with Bonferroni post hoc test, P < 0.05), whereas there was no significant difference in transgene expression between CsaMLO1 and CsaMLO11 T2 families (ANOVA with Bonferroni post hoc test,P > 0.05). The CsaMLO1 T2-B family did not have a significantly different transgene expression compared to either of the other T2 families (ANOVA with Bonferroni post hoc tests,P > 0.05). Transgene expression was not detectable in (untransformed) susceptible Moneymaker or resistant mlo control plants.
Transcript abundance profiling of cucumber clade V MLO genes
The relative transcript abundance of the cucumber clade V MLO genes CsaMLO1, CsaMLO8 and CsaMLO11 was determined in three different tissues (hypocotyl, cotyledon and leaf) of the susceptible cucumber cultivar Sheila, using qRT-PCR. It was found that in each of the three tissues the transcript abundance of CsaMLO8 was several orders of magnitude higher than that of CsaMLO1 and CsaMLO11 (Fig. 2a), a difference which was found to be statistically significant (ANOVA with Bonferroni post hoc tests, P < 0.05). To confirm this, we re-examined a previously obtained dataset consisting of RNA-seq data obtained from a variety of cucumber tissues from the reference cultivar ‘Chinese Long 9930’. This showed that in all examined aerial tissues (hypocotyl, stem, cotyledon, leaf and fruit tissue) CsaMLO8 was higher expressed (on average ca. tenfold) compared to either CsaMLO1 or CsaMLO11, although in stem tissue CsaMLO1 also appeared to be rather highly expressed. For the aerial tissues for which data on more than one biological replicate was available (leaf and fruit tissue), the observed difference in expression was found to be statistically significantly (ANOVA with Bonferroni post hoc tests, P < 0.05). In root and root tip tissue however, both CsaMLO1 and CsaMLO11 were highly expressed, whereas CsaMLO8 was lowly expressed (Fig. 2b), although this difference was not found to be statistically significant (ANOVA, P > 0.05). For comparison we also examined the expression of the A. thaliana Clade V MLO genes (AtMLO2, AtMLO6 and AtMLO12) in a publicly available RNA-seq dataset of A. thaliana tissues [36]. We found that AtMLO2 expression was much higher than AtMLO6 or AtMLO12 expression in all four sampled tissues (root, leaf, flower and fruit), although the difference in roots was smaller than in the other tissues (Additional file 2).
Fig. 2.
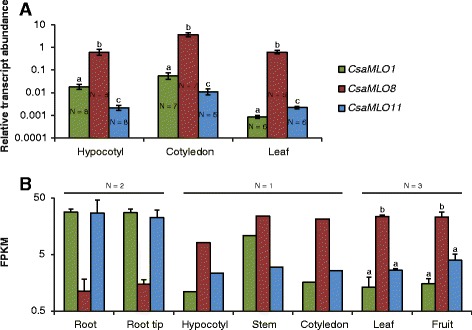
Expression profile of clade V cucumber MLO genes in different tissues. a The relative transcript abundances of CsaMLO1, CsaMLO8 and CsaMLO11 in three tissues of the PM-susceptible cucumber cultivar ‘Sheila’ (hypocotyl, cotyledon and leaf) were determined using qRT-PCR. Data were normalized, using the geometric average of the Ct values of reference genes Ef-α and TIP41. Relative transcript abundances were calculated as 2-dCt. Each bar shows the average transcript abundance of five to eight biological replicates on a logarithmic scale. The number of independent biological replicates per gene/tissue combination is given in the respective bars. Error bars indicate standard error of the mean. Different letters above bars indicate significant differences between genes (ANOVA with Bonferroni post hoc tests, P < 0.05). b The transcript abundances in seven tissues of cucumber (‘Chinese long’ inbred line 9930) were determined using RNA-seq. The FPKM values (Fragments Per Kilobase of transcript per Million mapped fragments) for CsaMLO1, CsaMLO8 and CsaMLO11 in each of these tissues are shown on a logarithmic scale. The amount of independent biological replicates per tissue was either one (hypocotyl, stem and cotyledon), two (root and root tip) or three (leaf and fruit). If applicable, error bars indicate the standard error of the mean. Different letters above bars indicate significant differences between genes (ANOVA with Bonferroni (for fruit tissue) or Dunnet T3 (for leaf tissue) post hoc tests, P < 0.05)
Subsequently we investigated the expression profile in cucumber tissues inoculated with P. xanthii. Samples were taken of cucumber hypocotyl, cotyledon and leaf tissue prior to and at four, six, eight and 24 h post inoculation. The relative expression of CsaMLO1, CsaMLO8 and CsaMLO11 in those samples was determined using qRT-PCR (Fig. 3). We found that in leaf tissue there were significant differences in CsaMLO1 transcript abundance between time points (ANOVA, P < 0.05). At four, 6 and 8 h post inoculation CsaMLO1 transcript abundance was significantly higher compared to the transcript abundance prior to inoculation (Bonferroni post hoc tests, P < 0.05). This induction of CsaMLO1 was not significant anymore at 24 h after inoculation (Bonferroni post hoc test, P > 0.05). CsaMLO11 was significantly downregulated at 4 h post inoculation compared to the transcript abundance prior to inoculation (ANOVA with Bonferroni post hoc test, P < 0.05). In cotyledon or hypocotyl tissue there were no significant differences of Clade V MLO gene transcript abundance between any of the time-points (ANOVA, P > 0.05).
Fig. 3.
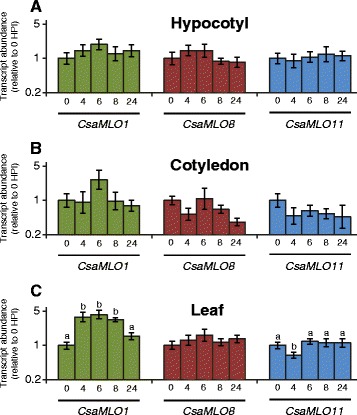
Relative expression level of clade V MLO genes in different cucumber tissues in response to Podosphaera xanthii inoculation. Relative transcript abundances in three tissues (a) hypocotyl, b cotyledon and c leaf) of PM-susceptible cucumber genotype ‘Sheila’ before and at 4, 6, 8 and 24 h post inoculation with P. xanthii were determined using qRT-PCR. Data were normalized relative to the geometric average of the Ct values of reference genes Ef-α and TIP41, and subsequently normalized relative to the average dCt value at 0 hpi for each gene/tissue combination. Relative transcript abundances were calculated as 2-ddCt. Each bar shows the relative expression of four to eight biological replicates on a logarithmic scale. Error bars indicate standard error of the mean. Different letters above bars indicate significant differences between time points (ANOVA with Bonferroni post hoc tests, P < 0.05)
As these results were in conflict with the finding in [13] that CsaMLO8 transcription was upregulated upon PM inoculation, the experiment was repeated independently, with samples of cucumber hypocotyl and leaf tissue prior to and at 4, 6, 8, 12 and 24 h post inoculation. The relative expression of CsaMLO8 in those samples was determined using qRT-PCR (Additional file 3). No significant differences in CsaMLO8 transcript abundance were observed in any of the time points compared to the transcript abundance prior to inoculation (ANOVA, P > 0.05).
Screening of sequenced cucumber germplasm for potential CsaMLO1 or CsaMLO11 mutants
We anticipated that if CsaMLO1 and CsaMLO11 are susceptibility genes towards PM in cucumber, loss-of-function mutations in those genes, leading to resistance, would have been selected for in cucumber germplasm. As Qi et al. published resequencing data of a collection of 115 divergent cucumber genotypes [37], we decided to screen these data for potential loss-of-function mutations in either CsaMLO1 or CsaMLO11. Complete lists of detected SNPs and indels in the 115 lines were downloaded from the Cucurbit Genomics Database [38], and filtered for the chromosomal locations of CsaMLO1 and CsaMLO11. Additional files 4 and 5 give an overview of all the detected SNPs and indels in CsaMLO1 and CsaMLO11 regions, respectively. The lists of SNPs/indels were manually curated to obtain variants in coding regions of the genes, with an effect on the predicted amino acid sequence. For CsaMLO11 we observed no SNPs or indels with an effect on the predicted protein. For CsaMLO1 three SNPs (and no indels) were found with an effect on the predicted amino acid sequence (Table 2). The first substitution (V170G) was in an amino acid residue conserved to be either a Valine, an Isoleucine or a Leucine in other clade V MLO proteins with a proven function as susceptibility gene (Additional file 6). The other two detected substitutions (V472I and V557I) were in non-conserved regions.
Table 2.
Non-synonymous SNPs detected in the CsaMLO1 coding sequence of 115 resequenced cucumber accessions
| Chromosome | Chromosome position | Exon number | Nucleotide substitution | Amino acid substitution |
|---|---|---|---|---|
| Chr1 | 8,163,552 | Exon 4 | T - > G | V170G |
| Chr1 | 8,159,763 | Exon 14 | G - > A | V472I |
| Chr1 | 8,159,508 | Exon 14 | G - > A | V557I |
Resequencing of cucumber genotypes CS-PMR1 and Santou to find potential CsaMLO1 or CsaMLO11 mutants
As it was mentioned that two previously reported QTLs for PM resistance co-localize with the genomic positions of CsaMLO1 and CsaMLO11 [31], we hypothesized that the sources of these resistances might have mutant alleles of CsaMLO1 and/or CsaMLO11, causal for the observed resistance. In order to test this hypothesis, we isolated DNA from leaves of genotypes CS-PMR1 and cv. Santou, the parental genotypes used for the QTL analysis [35], and performed whole genome sequencing (WGS). We aligned the obtained reads to the reference genome (Chinese long 9930 [30]) and identified SNPs and small indels in the genomic regions of CsaMLO1 and CsaMLO11 (Additional files 7, 8 and 9). In the genomic region of CsaMLO1 we identified 23 SNPs and indels in CS-PMR1 and four SNPs and indels in Santou, but none of these had an effect on the predicted encoded protein. Similarly we identified 23 SNPs and indels in the genomic region of CsaMLO11 in CS-PMR1, neither of which had an effect on the predicted encoded protein. The CsaMLO11 sequence of Santou was found to be identical to that of the reference genome.
In addition to calling SNPs and indels we identified two regions in genotype CS-PMR1, in intron 6 of CsaMLO1 and intron 12 of CsaMLO11 respectively, where we observed a local low read coverage combined with flanking read pairs with insert sizes deviating from the average or for which one of the mates in the read pair could not be mapped (Additional files 7 and 8). As this can be indicative for larger structural variations that are harder to characterize with short read sequencing, we amplified these regions from DNA of CS-PMR1 and sequenced the PCR products by Sanger sequencing. Compared to the reference genome, a 10 bp deletion and a 23 bp insertion in intron 6 of CsaMLO1 (Additional file 10A), and a 231 bp deletion in intron 12 of CsaMLO11 (Additional file 10D) were found in genotype CS-PMR1. To verify whether these large indels in intron 6 of CsaMLO1 and intron 12 of CsaMLO11 have any effect on the splicing of the genes, we also amplified and sequenced the corresponding regions from cDNA of CS-PMR1 and Santou by PCR, and found that there was no observable difference in PCR product size or sequence for either CsaMLO1 (Additional file 10B-C) or CsaMLO11 (Additional file 10E-F).
As we anticipated that the resistances of genotypes Santou and CS-PMR1 could also be caused by a difference in CsaMLO1 or CsaMLO11 expression rather than a difference in encoded protein sequence, we determined the relative transcript abundances of CsaMLO1 and CsaMLO11 in leaf tissue of both genotypes by qRT-PCR, using the susceptible genotype Sheila as a control (Additional file 11). Although the relative transcript abundance of both genes was found to be slightly higher in CS-PMR1 compared to the other two genotypes, these differences were found to be not statistically significant for either CsaMLO1 (ANOVA, P = 0.156) or CsaMLO11 (ANOVA, P = 0.239).
Discussion
Overexpression of all three cucumber clade V MLO genes restores PM susceptibility in a tomato mlo mutant
Previously, it has been shown that loss of susceptibility towards PM causing fungi due to mutations in MLO genes can be restored by overexpression of functional MLO genes, both by cloning an MLO gene from a susceptible individual from the same plant species [10–12] as well as by heterologous expression of clade V MLO genes from other dicot species [13, 15, 39], and even by heterologous expression of clade IV MLO genes from a monocot species [29]. This shows that even though there are considerable differences in amino acid sequence between MLO proteins, they are functionally conserved between plant species. This enabled us to study the function of cucumber clade V MLO genes by heterologous expression in the tomato slmlo1 mutant background. We have shown that overexpression of either of the three clade V CsaMLO genes restored susceptibility to O. neolycopersici, the PM causing fungus in tomato, albeit to a different extent. Overexpression of either CsaMLO1 or CsaMLO8 resulted in full restoration of PM susceptibility towards wild-type levels. Overexpression of CsaMLO11 on the other hand only partially complemented loss of SlMLO1 function (Fig. 1). This shows that, at least to some extent, all three genes are functionally conserved.
Overexpression of CsaMLO11 appeared to be less efficient in restoring PM susceptibility compared to the other CsaMLO genes (Fig. 1). Although the proteins encoded by each of the three genes are not identical, they are highly similar to one another and to clade V MLO genes of other species (Additional file 6). It appears difficult to attribute the lower efficiency of CsaMLO11 compared to the other CsaMLO genes in restoring PM susceptibility to a particular difference in amino acid sequence.
CsaMLO8 is the major clade V MLO gene in aerial tissues, whereas CsaMLO1 and CsaMLO11 are the major clade V genes in roots
Susceptibility genes are defined as those genes that facilitate infection and support compatibility to plant pathogens [40]. Within the MLO gene family, homologs from clade IV (in monocotyledonous species) and V (in dicotyledonous species) have been found to be susceptibility genes [24]. However, not all clade V MLO genes in all dicotyledonous plant species have been found to be S-genes. For instance, in grapevine (Vitis vinifera) it was found that there are four clade V MLO genes. Silencing of one of them (VvMLO7) by transformation with RNAi constructs led to gain of PM resistance, whereas silencing of two other homologs only increases resistance when VvMLO7 was already silenced. Silencing of the fourth homolog did not contribute to resistance at all [16]. This unequal genetic redundancy was previously also observed in Arabidopsis with one major MLO S-gene (AtMLO2) and two minor MLO S-genes [10]; and in tomato, with one major MLO S-gene (SlMLO1), two minor MLO S-genes and one clade V MLO homolog which does not seem to play a role in PM susceptibility [41].
We have shown, using qRT-PCR data (Fig. 2a) and RNA-seq data (Fig. 2b), that in aerial cucumber tissues (hypocotyl, stem, cotyledon, leaf and fruit tissue) CsaMLO8 is several folds higher expressed than the other cucumber clade V MLO genes, CsaMLO1 and CsaMLO11. This is reminiscent of the findings in tomato by Zheng et al. [41], who showed that the major clade V MLO gene, SlMLO1, is much higher expressed than the other clade V homologs in case of absence of PM. Interestingly, they showed that silencing of SlMLO1 by transformation with an RNAi construct led to gain of resistance, whereas silencing of the other MLO homologs did not. Indeed, a natural slmlo1 loss-of-function mutant had previously been characterised to be resistant to PM [11], even though the other clade V MLO genes were presumably still intact. Additionally, we have shown that in a publicly available Arabidopsis RNA-seq dataset one clade V MLO homolog, AtMLO2, is much higher expressed than the other clade V homologs, AtMLO6 and AtMLO12 (Additional file 2). Previously it has been shown that loss-of-function mutations in AtMLO2, but not in AtMLO6 or AtMLO12 lead to (partial) resistance against PM, although double AtMLO2/6 or AtMLO2/12 and triple AtMLO2/6/12 mutants showed even higher levels of resistance [10].
Recently, researchers investigated the expression pattern of MLO genes in a large number of tissues from Arabidopsis and rice, based on microarray data [42]. The data presented there for the Arabidopsis MLO genes are in agreement with our findings (Additional file 2), as they report that in most tissues the major MLO S-gene AtMLO2 is much higher expressed than the other two clade V MLO genes [42]. Rice has two clade IV MLO genes, OsMLO3 and OsMLO6 [25], it was found that in most rice tissues OsMLO3 transcription was much higher than OsMLO6 transcription [42]. Based on this finding, the authors conclude that OsMLO3 is likely to be the major clade IV MLO in rice, rather than OsMLO6.
Taken together, this suggests that loss-of-function mutations in the most abundantly expressed MLO gene have a large effect compared to loss-of-function mutations in the less abundantly expressed genes. This would imply that in cucumber, CsaMLO8 would be the major clade V MLO gene concerning PM susceptibility, comparable in function to e.g. SlMLO1 in tomato and AtMLO2 in Arabidopsis. We postulate that differences in transcription efficiency between different clade V MLO genes in a species are the main reason for the observed unequal genetic redundancy, and that characterization of the relative transcript abundances of clade V MLO genes in a species may help identify the major MLO gene regarding susceptibility. To our knowledge, it has not been attempted in either Arabidopsis or tomato to express the minor clade V MLO genes under a strong constitutive promoter. On the basis of our results, we would expect that overexpression of e.g. AtMLO6 or AtMLO12 in Atmlo2 background would be sufficient for restoration of susceptibility, if it were to be true that transcript abundance rather than differences in amino acid sequence determine which clade V MLO gene is the major S-gene.
Contrastingly, RNA-seq results show that CsaMLO1 and CsaMLO11 are highly expressed in root tissue, whereas CsaMLO8 is not (Fig. 1b). Interestingly, all three Arabidopsis Clade V MLO genes were also found to be highly expressed in root tissue (Additional file 2). As PM causing fungi are foliar pathogens, which do not infect roots, the finding that several clade V MLO genes in cucumber and Arabidopsis are highly expressed in roots will probably not have much consequence on the interaction of plants with PM causing fungi. However, we should note that it is likely that CsaMLO1 and CsaMLO11 fulfil an important, yet unknown role in cucumber roots.
Loss-of-function mutations in HvMLO in barley and AtMLO2/AtMLO6 in Arabidopsis lead to increased susceptibility to necrotrophic and hemibiotrophic pathogens such as leaf spot blotch disease caused by Bipolaris sorokiniana [43], rice blast on barley caused by Magnaporthe grisea [44], leaf spot disease caused by Alternaria spp. and late blight caused by Phytophthora infestans [10]. This suggests that MLO-genes can in some cases contribute to resistance to necrotrophic and hemibiotrophic pathogens, in contrast to their role as susceptibility gene for (biotrophic) PM causing fungi. Therefore, it might be interesting to study the effect of loss-of-function mutations in the root-expressed CsaMLO1 and CsaMLO11 on the interaction with necrotrophic, soil-borne cucumber pathogens such as vascular wilt caused by the necrotrophic fungus Fusarium oxysporum f. sp. cucumerinum or root rot caused by the necrotrophic oomycete Pythium spp. Furthermore, as it is known that barley mlo mutants are less efficiently colonized by mutualistic arbuscular mycorrhiza fungi [45], it could also be interesting to see the effect of CsaMLO1 and CsaMLO11 loss-of-function mutations on the mutualistic interaction with arbuscular mycorrhiza fungi in cucumber.
CsaMLO1 expression is induced upon PM inoculation, whereas CsaMLO8 and CsaMLO11 are not
For several plant species it has been shown that the expression of MLO susceptibility genes is induced upon inoculation with PM causing fungi (e.g. [46–48]), potentially due to the fungus actively upregulating the expression of those genes to induce susceptibility. It was previously reported based on RNA-seq data of P. xanthii inoculated cucumber leaves that CsaMLO1, but not CsaMLO8 or CsaMLO11 was upregulated in leaves in response to the inoculation, showing a ca. 3.5 fold upregulation in expression 8 h post inoculation [31]. Figure 3c confirms this finding, since we have found significant (ca. four-fold) upregulation of CsaMLO1 expression in leaves at 4, 6 and 8 h after inoculation with P. xanthii. As upregulation of MLO gene expression due to inoculation with PM causing fungi is often regarded as putative evidence for a role as susceptibility gene, one might argue that this suggests that CsaMLO1 is a functional susceptibility gene, and CsaMLO8 or CsaMLO11 are not. However we would like to point out that even though CsaMLO1 expression in leaves is significantly induced upon PM inoculation relative to the expression before inoculation (Fig. 3c), the expression of CsaMLO1 before inoculation is much lower than that of CsaMLO8 which is constitutively higher expressed (Fig. 2), so consequently even after inoculation the transcript abundance of CsaMLO8 is still higher than that of CsaMLO1.
Previously we observed that inoculation of cucumber with P. xanthii led to upregulation of CsaMLO8 in hypocotyl tissue, but not in leaf or cotyledon [13]. To our surprise we could not reproduce this result in our experiments described here, even though using the same cucumber genotype, P. xanthii isolate, climatic conditions and inoculation protocol, and using the same qRT-PCR primers and protocol (Fig. 3a). Even though we did observe a small induction of CsaMLO8 expression in hypocotyl at 4 and 6 h post inoculation, differences in CsaMLO8 transcript abundance in hypocotyl tissue between time points were far from significant (ANOVA, P = 0.389). It should be noted that the variation in transcript abundances between different biological replicates is quite high, both in our experiments described here and in our previously published results. As the current experiment has a larger sample size (four to eight independent biological replicates per time point instead of three independent biological replicates in [13]) and was repeated with similar results (Additional file 3), we conclude that upregulation of CsaMLO8 in hypocotyl tissue [13] was probably an artefact caused by a low number of biological replicates.
We previously described that a loss-of-function mutant allele of CsaMLO8 leads to hypocotyl-specific resistance towards PM, with partial resistance in leaf tissue, and attributed this tissue specificity to the supposed tissue-specific upregulation of CsaMLO8 [13]. Now that we have shown that CsaMLO8 is in fact not upregulated in hypocotyl tissue, we have to come up with a different explanation for the observed tissue specificity of Csamlo8-based resistance. It is in this sense interesting to note that CsaMLO1 basal expression is very low (Fig. 2) whereas it is induced upon PM inoculation in leaf tissue but not in hypocotyl tissue (Fig. 3). Assuming that both CsaMLO1 and CsaMLO8 are functional susceptibility genes, we can hypothesize that in a Csamlo8 loss-of-function mutant, which expresses functional CsaMLO1, there is hardly any expression of a functional MLO susceptibility gene in hypocotyl tissue, whereas there is induced expression of CsaMLO1 in leaf tissue, resulting in complete resistance in hypocotyl tissue and only partial resistance in leaf tissue. If this would be true, a double Csamlo1/Csamlo8 loss-of-function mutant would be expected to have complete resistance in both leaf and hypocotyl tissue.
No putative loss-of-function CsaMLO1 or CsaMLO11 mutants could be identified in resequenced cucumber germplasm
As loss-of-function mutations in functional susceptibility genes can lead to durable resistance [40], it would be worthwhile to obtain cucumber lines with mutations in clade V MLO genes. It has previously been described that a natural mutant allele in CsaMLO8, caused by insertion of a retrotransposable element, leads to partial resistance to P. xanthii. This mutant allele has a rather high frequency in breeding material, probably because of its beneficial effect on PM resistance [13]. Furthermore, several other mutant alleles of CsaMLO8 were identified in resistant cucumber genotypes [33]. It is therefore reasonable to assume that if loss-of-function mutations in CsaMLO1 and/or CsaMLO11 would contribute to PM resistance, they also would have been selected for during cucumber breeding. Therefore we decided to screen a publicly available dataset of SNPs and indels in a collection of 115 cucumber genotypes [37] for putative loss-of-function alleles in the coding regions of CsaMLO1 and CsaMLO11 (Additional files 4 and 5). We did not find any evidence for variant alleles with a large effect on the amino acid sequence in either of the genes (e.g. a SNP leading to gain of an early stop codon or a frameshift indel), although we observed three SNPs in CsaMLO1 leading to amino acid substitutions (Table 2). Of these three SNPs, two were predicted to cause an amino acid substitution from valine to isoleucine, two amino acid residues with very similar physiochemical properties. Furthermore, those amino acid residues were in the C-terminal domain of the CsaMLO1 protein, a region which is not conserved compared to other clade V MLO proteins (Additional file 6). A third SNP was predicted to lead to a substitution of the 170th amino acid residue, a valine, into a glycine residue, at a location conserved to be either a valine, a leucine or an isoleucine in other clade V MLO proteins (Additional file 6). As glycine and valine are both relatively small, aliphatic, non-polar amino acids, this substitution can be considered a rather conservative mutation. Without further evidence it does not seem very likely that this SNP represents a loss-of-function allele of CsaMLO1. In conclusion, we did not find strong evidence in this dataset for possible loss-of-function alleles of CsaMLO1 and/or CsaMLO11, although it should be noted that by focussing on SNPs and indels we could have overlooked mutant alleles that are harder to find by short-read resequencing, such as the transposable element characterised in CsaMLO8, which can have a profound effect on the function of the genes.
In another approach to try to identify possible mutant alleles of CsaMLO1 or CsaMLO11 we resequenced two additional cucumber genotypes, CS-PMR1 and cv. Santou, which were previously mentioned to have QTLs for PM resistance colocalizing with the genomic positions of CsaMLO1 and CsaMLO11 [31, 35]. Although we identified several SNPs, indels and structural variations at the CsaMLO1 and CsaMLO11 loci, especially in the more PM resistant genotype CS-PMR1, none of these are predicted to lead to any change in the encoded CsaMLO1 or CsaMLO11 proteins (Additional files 7, 8, 9 and 10). In addition, we verified whether the transcript abundances of CsaMLO1 and CsaMLO11 were different between these cucumber genotypes (Additional file 11), but we concluded that there were no significant differences among them. Therefore, we concluded that the observed resistance by Fukino et al. [35] is likely caused by other genes rather than CsaMLO1 or CsaMLO11.
The fact that we could not find convincing loss-of-function alleles of CsaMLO1 or CsaMLO11 in a diverse panel of cucumber genotypes implies that loss-of-function mutations in either of these genes have apparently not been selected for in cucumber breeding. An explanation for this finding could be that Csamlo1 and Csamlo11 knockout mutations could have only a small, additive effect on PM resistance in Csamlo8 mutant background, and not have any effect in CsaMLO8 background, comparable to the situation in Arabidopsis [10]. Furthermore it could be possible that loss-of-function mutations in CsaMLO1 and CsaMLO11 have pleiotropic effects on plant fitness, and are therefore selected against.
It would in our opinion be interesting to study the effect of knock-out mutants of CsaMLO1 and CsaMLO11, for instance by targeted mutation using the increasingly popular CRISPR-Cas9 technology [49], such as was already done in the bread wheat TaMLO-A1 gene [50]. This might lead to a new, durable source of PM resistance in cucumber, especially when combined with the already existing Csamlo8 partial resistance [13].
Conclusions
In this study we analysed the role of cucumber clade V MLO genes in susceptibility to PM. We showed by means of heterologous overexpression of the cucumber MLO genes CsaMLO1, CsaMLO8 and CsaMLO11 that all three genes are able to restore susceptibility in mlo tomato, although the effect of CsaMLO11 overexpression was weaker compared to CsaMLO1 or CsaMLO8 overexpression. Additionally, we studied the transcription levels of these genes in different tissues of cucumber, both with and without inoculation with a PM causing fungus, P. xanthii, showing that CsaMLO8 is higher expressed compared to CsaMLO1 and CsaMLO11 in all aerial tissues, although CsaMLO1 expression in leaves is induced by inoculation with P. xanthii. We discuss that CsaMLO8 is therefore likely to be the major clade V MLO gene in cucumber concerning PM susceptibility, with a potential minor role for CsaMLO1 and CsaMLO11, comparable to earlier findings for mlo genes in Arabidopsis and tomato. In roots, however, CsaMLO1 and CsaMLO11 appeared to be much higher expressed, which might have implications on the interactions of cucumber with root pathogens or beneficial microbes. No potential natural loss-of-function mutations in either CsaMLO1 or CsaMLO11 have been found so far. It would be interesting to generate Csamlo1 and Csamlo11 mutants, for instance by CRISPR-Cas9 technology, to investigate whether such mutations have an added effect on top of PM resistance caused by Csamlo8.
Methods
Cloning of CsaMLO1 and CsaMLO11
A homozygous cucumber breeding line derived from a parental line of cv. Anaxo was grown in a greenhouse in Wageningen, the Netherlands. Growing conditions were 20 °C (day) and 16 °C (night), with a 16 h/8 h day/night cycle, and a relative humidity of 70%. RNA isolation and cDNA synthesis were performed as previously described [13].
The coding sequence of CsaMLO1 was amplified from cDNA with primers 5′-caccTTCCTTCCACACCCCTAAGA-3′ (Forward) and 5′-TGAATGGTGTAAACGAGATTGC-3′ (Reverse). As template 50 ng cDNA was used in a 50 μl reaction using Advantage 2 polymerase (Takara Bio, U.S.A.). Cycling conditions were: 1 min initial denaturation at 95 °C, followed by 35 cycles of 30 s denaturation at 95 °C and 3 min annealing and extension at 68 °C. Reactions were finished by 3 min incubation at 68 °C. The PCR product was subsequently diluted 100 times, and used as template for a 50 μl reaction using Phusion high-fidelity polymerase (ThermoFisher Scientific, U.S.A.). Cycling conditions were: 30 s initial denaturation at 98 °C, followed by 25 cycles of 20 s denaturation at 98 °C, 30 s annealing at 55 °C, and 30 s extension at 72 °C. Reactions were finished by 10 min incubation at 72 °C.
The coding sequence of CsaMLO11 was amplified from cDNA as previously described [13], using primers 5′-caccTTTGTTTCCCTACGCGTTCT-3′ (Forward) and 5′-TATACCAACCCCCAACCTCA-3′ (Reverse).
Cloning of CsaMLO1 and CsaMLO11 PCR products, through the Gateway-compatible vector pENTR/D-TOPO (ThermoFisher Scientific, U.S.A.) to binary vector pK7WG2, which harbours the constitutively active 35S Cauliflower Mosaic Virus promotor and the nptII selectable marker gene for kanamycin resistance [51], was done as previously described [13].
Complementation of ol-2 tomato with cucumber MLO genes
Cotyledon explants of ol-2 mutant tomato seedlings were transformed with CsaMLO1 and CsaMLO11 overexpression constructs as previously described [13]. The ol-2 mutant carries a loss-of-function mutation in the SlMLO1 gene [11]. Obtained tomato transformants were assessed for presence of the transgenes by PCR using the same primers as used for the cloning of CsaMLO1 and CsaMLO11 (see above) and for presence of the nptII marker gene with primers 5′-GAAGGGACTGGCTGCTATTG-3′ (nptII forward) and 5′-AATATCACGGGTAGCCAACG-3′ (nptII reverse). For each of the two transformations with a different construct, seven (CsaMLO11) or eight (CsaMLO1) independent transgenic plants were selected, and were assessed for transgene expression by qRT-PCR using primer pairs specific for CsaMLO1: 5′-TGAAAGTTTCCGGCGGAGTT-3′ (CsaMLO1-Forward) and 5′-AGGAAGCTTTACCCTTGGCG-3′ (CsaMLO1-Reverse) or specific for CsaMLO11: 5′-GCGACGGCGTTGAGAAATTG-3′ (CsaMLO11-Forward) and 5′-GGGTGACAAGTGGTGGGAGG-3′ (CsaMLO11-Reverse). As housekeeping gene for normalization of CsaMLO1 or CsaMLO11 expression in tomato, SlEF-α was used, with primer pair 5′-ATTGGAAACGGATATGCCCCT-3′ (SlEF-α forward) and 5′-TCCTTACCTGAACGCCTGTCA-3′ (SlEF-α reverse). qRT-PCR was performed as previously described [13].
Evaluation of PM resistance of CsaMLO overexpressing ol-2 tomato
From each of the individual transformants of both CsaMLO1 and CsaMLO11 overexpressing ol-2 plants, two cuttings were inoculated with an isolate of Oidium neolycopersici maintained on susceptible tomato plants in a climate chamber in Wageningen, the Netherlands, as previously described [13].
For two individual T1 transformants per overexpression construct, seeds of self-pollinated plants were harvested. In principle the T1 transformants with highest transgene expression were chosen for generation of T2 families, although as the CsaMLO11 transformant with the highest expression did not give viable seeds another T1 transformant with enough viable seeds was randomly selected. In addition, seeds of two self-pollinated CsaMLO8 overexpressing plants described in [13] were harvested. Twenty-two to thirty seeds of each of the six T2 families were sown on soil, plantlets were assayed for presence of the nptII marker gene by PCR as described above. As susceptible control, 12 seeds of cultivar Moneymaker (MM) were sown. As resistant control, 12 seeds of resistant mutant line ol-2, the background used for the complementation, were sown. For five plants per T2 family and for five plants of the susceptible and resistant controls, leaf samples were harvested and immediately frozen in liquid nitrogen. Transgene expression efficiency in these plants was determined by means of qRT-PCR using primers and conditions as described above. To analyse differences in transgene expression efficiency between the T2 families a one-way ANOVA test was performed on the dCt values, followed by Dunnet’s T3 post hoc tests. Homogeneity of the variances was tested using Levene’s test. All statistical analyses were performed using SPSS v23 software (IBM). T2 families and controls were inoculated with O. neolycopersici as described above. The susceptibility/resistance of plants was scored after two weeks on a 0–3 scale as described earlier [11]. Differences in PM susceptibility between the T2 families and controls were analysed with a nonparametric Kruskal-Wallis test followed by Stepwise Stepdown Multiple Comparison post hoc tests to identify homogeneous subsets, using SPSS v23 software (IBM).
Expression analysis of MLO genes in cucumber using qRT-PCR
For expression analysis of MLO genes in PM-inoculated cucumber tissues, PM susceptible cucumber cultivar ‘Sheila’ was grown and inoculated with a P. xanthii isolate as previously described [13]. Prior to inoculation and at 4, 6, 8 and 24 h post inoculation (hpi), from eight individual plants per time point hypocotyl, cotyledon and (first) true leaf samples were harvested separately, and were immediately frozen in liquid nitrogen. RNA isolation, cDNA synthesis and qRT-PCR were performed as previously described [13]. For quantification of CsaMLO1 and CsaMLO11 expression, respectively, we used the primer sequences CsaMLO1-Forward and CsaMLO1-Reverse and CsaMLO11-Forward and CsaMLO11-Reverse, as described above. Primer pairs specific for the cucumber housekeeping genes TIP41 and EF-α, as described by Warzybok et al. [52], were used for normalization of expression.
Samples for which the difference in Ct value between the technical replicates was larger than 1.0, or for which one or both of the technical replicates did not reach the detection threshold were excluded from the analysis. Ct values per sample were normalised by subtracting the geometric mean of the Ct values for the two housekeeping genes, giving deltaCt, abbreviated as dCt. In the time series of inoculated cucumber tissues, dCt values were subsequently normalized by the average dCt value for each gene/tissue combination at 0 hpi, giving ddCt. Averages and standard errors of ddCt values were calculated over four to eight biological replicates per gene/tissue/time point combination. Normality of ddCt distributions was tested using Shapiro-Wilk tests (P > 0.05). Differences in ddCt value between time points were analysed with ANOVA tests. Homogeneity of variances were tested using Levene’s test. If ANOVA tests showed a significant effect of time points (P < 0.05), Bonferroni post hoc tests were performed to analyse which time points were significantly different from one another. All statistical analyses were performed using SPSS v23 software (IBM). Relative transcript abundances were calculated as 2-ddCt.
In a second experiment, PM susceptible cucumber cultivar ‘Sheila’ was grown and inoculated with P. xanthii under the same conditions as described above. Samples of leaf and hypocotyl tissue were harvested prior to inoculation and at 4, 6, 8, 12 and 24 h post inoculation (hpi), from eight individual plants per time point. Total RNA was isolated using a phenol-based protocol described by [53]. cDNA synthesis and qRT-PCR were performed as described above.
In another experiment, cucumber genotypes ‘Sheila’, ‘Santou’ and ‘CS-PMR1’ were grown in a greenhouse in Wageningen, the Netherlands. Five weeks post seeding, leaf samples were harvested and were immediately frozen in liquid nitrogen. Total RNA was isolated using the RNeasy plant mini kit (Qiagen, Germany). cDNA was synthesised using 500 ng of RNA samples with the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, U.S.A.). Before use in qRT-PCR, cDNA samples were diluted 2-fold. To quantify the expression of CsaMLO1 and CsaMLO11, qRT-PCR was performed with conditions and subsequent data analysis as described above, normalizing dCt values using the average dCt value of the ‘Sheila’ samples.
Analysis of MLO data in RNA-seq datasets
Cucumber genotype ‘Chinese long’ inbred line 9930 was cultivated under standard greenhouse conditions (20 °C (day) and 16 °C (night), with a 16 h/8 h day/night cycle, and a relative humidity of 70%). Separate samples of roots, root tips, hypocotyls, cotyledons, stems, leaves and fruit were harvested and immediately frozen in liquid nitrogen, using one (hypocotyls, cotyledons and stems), two (roots and root tips) or three (leafs and fruit) individual samples per tissue. Material was sent to KeyGene B.V., The Netherlands, for RNA-seq. Total RNA from each sample was isolated using the RNeasy plant mini kit (Qiagen, Germany). Subsequently, RNA-seq libraries were made following the TruSeq RNA Sample Preparation v2 Guide protocol. After concentration measurement by qPCR (LightCycler 480; Roche), PhiX (~0.6%) was spiked as a control according to the manufacturer’s recommendations. The libraries were pooled, and sequenced using a Illumina HiSeq 2000 sequencer. The resulting reads were sorted into single fasta files per sample based on the sample tag sequences. The obtained read length was approximately 100 nt at a minimal read length of 36 nt. Reads were aligned to the reference genome (‘Chinese long’ inbred line 9930, version 2 [30]). The transcript abundance per sample was assessed on the basis of the number of sequenced fragments, normalised by the length of the coding sequence of the gene, per million of total reads sequenced (fragments per kilobase of transcript per million sequenced reads, FPKM). We extracted FPKM values per sample from the total dataset for each of the three clade V MLO genes, using Excel. For samples with more than one biological replicate, differences in FPKM values were analysed with ANOVA tests, followed by Bonferroni post hoc tests if variance was homogeneous, or Dunnet T3 post hoc tests if variance was not homogeneous. Homogeneity of variances was tested using Levene’s test. All statistical analyses were performed using SPSS v23 software (IBM).
RNA-seq data for Arabidopsis were analysed using the Gene Expression Atlas from EMBL-EBI [54]. Baseline expression values in Arabidopsis tissues as quantified by [36] were filtered for clade V MLO genes AtMLO2 [AT1G11310], AtMLO6 [AT1G61560] and AtMLO12 [AT2G39200] and downloaded as a tabular file.
In silico screening of MLO sequence variants in 115 lines
Total lists of SNPs and indels identified in 115 cucumber genotypes by [37] were downloaded from the Cucurbit Genomics Database [38]. SNPs and indels were filtered by Excel based on the genomic positions of CsaMLO1 (Chr1: 8,159,427.8,165,253, negative strand) or CsaMLO11 (Chr6: 14,120,024.14,125,039, positive strand). Sequence variants were manually annotated based on their genomic location to see whether they were located in introns or exons, when they were located in exons the effect on the coding sequence was scored using CLC Main Workbench v. 7.6.4.
Resequencing of CS-PMR1 and Santou
Seeds of cucumber genotypes CS-PMR1 and Santou were ordered from the Genetic Resources Center, NARO (National Agriculture and Food Research Organization), Japan. Young leaves of both genotypes were harvested and immediately frozen in liquid nitrogen. DNA was isolated from leaves as described by [55]. Total DNA was send for library preparation and whole genome sequencing with an average coverage of 25 reads per base pair using Illumina Hiseq PE150 technology with insert size of 350 bp, by Novogene Company Limited, Hong Kong, People’s Republic of China. Resulting reads were aligned to the cucumber reference genome (‘Chinese long’ inbred line 9930, version 2 [30]) using Bowtie, version 2.2.6 [56]. Filtering the reads for the genomic location of CsaMLO1 and CsaMLO11 and SNP/indel calling were performed using the SAMtools software package, version 0.1.18 [57].
To inspect a region with low read coverage in intron 6 of CsaMLO1, this region was amplified by PCR from DNA of genotype CS-PMR1 using primers 5′-CCTGCCTTGATGTGGATCGT-3′ (Forward) and 5′-AGTGCCTTCTTCTGACCGTT-3′ (Reverse). To inspect a region with low read coverage in intron 12 of CsaMLO11, this region was amplified by PCR from DNA of genotype CS-PMR1 using primers 5′- AGCACACAGAGGATTTGGTCA-3′ (Forward) and 5′-TGAACGAGAACCCTGATGCA-3′ (Reverse). For both PCR reactions, as template 2 μl DNA was used in a 50 μl reaction using DreamTaq DNA polymerase (ThermoFisher Scientific, U.S.A.). Cycling conditions were: 1 min initial denaturation at 95 °C, followed by 40 cycles of 30 s denaturation at 95 °C, 30 s annealing at 60 °C and 1 min annealing at 72 °C. Reactions were finished by 7 min incubation at 72 °C. PCR products were visualised by staining with GelRed and electrophoresis on agarose gels. Sequencing reactions were performed in triplicate, using the same primers used for amplification (GATC Biotech, Germany). Obtained sequences were aligned using CLC Main Workbench v. 7.6.4. The consensus sequence for the amplified region was extracted from the alignment. This consensus sequences were then aligned to the genomic reference sequences of CsaMLO1 and CsaMLO11, respectively.
To verify whether the observed sequence variants in intron 6 of CsaMLO1 and intron 12 of CsaMLO11 in genotype CS-PMR1 cause any effect on splicing, the corresponding regions were amplified from cDNA. RNA was isolated from young leaves of cucumber genotypes CS-PMR1 and Santou using the RNeasy Kit (Qiagen, Germany). Possible DNA contamination of RNA samples was removed by treatment with DNase I, Amp. Grade (Invitrogen life technologies, U.S.A.). cDNA was synthesised using 2 μg of RNA samples with an iScript cDNA Synthesis Kit (Bio-Rad Laboratories, U.S.A.). A CsaMLO1 coding sequence fragment was amplified from cDNA using primers CsaMLO1-Forward (as above) and 5′-ATGGCAGCCATAGATACGCC (Reverse). A CsaMLO11 coding sequence fragment was amplified from cDNA using primers 5′-GTGGTGGTCAGTATCAGCCC-3′ (Forward) and 5′-CGTCGAACCCATCTGTGTGA-3′ (Reverse). PCR reactions were performed using DreamTaq DNA polymerase (ThermoFisher Scientific, U.S.A.), with cycling conditions as described above. PCR products were visualised by staining with GelRed and electrophoresis on agarose gels. Sequencing reactions were performed in triplicate, using the same primers used for amplification (GATC Biotech, Germany). Obtained sequences were aligned using CLC Main Workbench v. 7.6.4. The consensus sequence for the amplified region was extracted from the alignment. These consensus sequences were then aligned to the cDNA reference sequences of CsaMLO1 and CsaMLO11, respectively.
Additional files
The relative transcript abundances of CsaMLO1, CsaMLO8 and CsaMLO11 in T2 families of a tomato mlo mutant overexpressing CsaMLO1, CsaMLO8 and CsaMLO11, were determined by qRT-PCR. Data were normalised relatively to the reference gene SlEF-α. Average transcript abundances of four or five randomly selected individuals are shown on a logarithmic scale. Error bars represent standard error of the mean. Different letters above the bars indicate statistical significance of Ct-values (One-way ANOVA with Dunnet’s T3 post hoc test, P < 0.05). CsaMLO1, CsaMLO8 or CsaMLO11 expression was not detectable in non-transformed tomato mlo mutant or Moneymaker. (PDF 341 kb)
Data on the transcript abundance in four tissues of Arabidopsis thaliana, determined using RNA-seq was investigated and downloaded using the Expression Atlas of EMBL-EBI (https://www.ebi.ac.uk/gxa/home). The FPKM values (Fragments Per Kilobase of transcript per Million mapped fragments) for AtMLO2, AtMLO6 and AtMLO12 in each of the tissues is shown on a logarithmic scale. (PDF 350 kb)
Relative transcript abundances in two tissues of PM susceptible cucumber cultivar ‘Sheila’ (A) hypocotyl and B) leaf, before and at 4, 6, 8, 12 and 24 h post inoculation with P. xanthii were determined using qRT-PCR. Data were normalized relative to the geometric average of the Ct values of reference genes Ef-α and CACS, and subsequently normalized relative to the average dCt value at 0 hpi for both tissues. Each bar shows the relative expression of five to eight biological replicates, as indicated above the bars, on a logarithmic scale. Error bars indicate standard error of the mean. (PDF 374 kb)
The list of SNPs and indels in 115 resequenced cucumber accessions, available from the Cucurbit Genomics Database [38], was filtered for the genomic region of CsaMLO1 (Chr1:8,159,428.8165253). The genic location of each SNP/Indel was determined manually (exon/intron), for exonic SNPs the effect on the predicted amino acid sequence was scored (synonymous/nonsynonymous). (XLSX 57 kb)
The list of SNPs and indels in 115 resequenced cucumber accessions, available from the Cucurbit Genomics Database [38], was filtered for the genomic region of CsaMLO11 (Chr6:14,120,024.14,125,039). The genic location of each SNP/Indel was determined manually (exon/intron), for exonic SNPs the effect on the predicted amino acid sequence was scored (synonymous/nonsynonymous). (XLSX 50 kb)
Protein alignment of clade V MLO proteins of Arabidopsis thaliana (AtMLO2, 6 and 12), Medicago trunculata (MtMLO1), Pisum sativum (PsMLO1), Lotus japonicus (LjMLO1), Cucumis sativus (CsaMLO1, 8 and 11), Solanum lycopersicum (SlMLO1), Capsicum annuum (CaMLO2) and Nicotiana tabacum (NtMLO1). A bar graph shows the conservation of the individual residues. Colours indicate amino acid residues with similar physiochemical properties according to the RasMol colour scheme. Locations of amino acid substitutions in CsaMLO1 due to SNPs are indicated by a red arrow. (PDF 3224 kb)
Resequencing data of the genomic region of CsaMLO1 in cucumber genotypes CS-PMR1 and Santou. The location of the gene on the chromosome is indicated by a red cursor. For each of the two genotypes, the total reads mapping to the location and the coverage per base pair are given. SNPs are indicated by coloured stripes (green for A, red for T, blue for C, brown for G), indels are indicated by black stripes. Read pairs with a small (smallest 0.5%) or large (largest 0.5%) insert size are coloured dark blue or dark red, respectively. Reads for which the other mate in the mate pair was not mapped are indicated by a bright red outline. (PDF 356 kb)
Resequencing data of the genomic region of CsaMLO11 in cucumber genotypes CS-PMR1 and Santou. The location of the gene on the chromosome is indicated by a red cursor. For each of the two genotypes, the total reads mapping to the location and the coverage per base pair are given. SNPs are indicated by coloured stripes (green for A, red for T, blue for C, brown for G), indels are indicated by black stripes. Read pairs with a small (smallest 0.5%) or large (largest 0.5%) insert size are coloured dark blue or dark red, respectively. Reads for which the other mate in the mate pair was not mapped are indicated by a bright red outline. (PDF 356 kb)
The list of SNPs and indels identified in the genomic sequence of CsaMLO1 and CsaMLO11 in the resequenced cucumber genotypes CS-PMR1 and Santou. The reference allele (in cucumber reference genome Chinese Long 9930) and the alternative allele are given. Furthermore, the genic location of each SNP/Indel was determined (exon/intron), for exonic SNPs the effect on the predicted amino acid sequence was scored (synonymous/nonsynonymous). (XLSX 12 kb)
Intron 6 of CsaMLO1 (A) and intron 12 of CsaMLO11 (D) were amplified from genomic DNA isolated from the cucumber genotype CS-PMR1, and subsequently sequenced in triplicate by Sanger sequencing. The obtained sequences were aligned to the reference cucumber genome (Chinese long inbred 9930, v2). Numbers above the alignment are relative to the start codon of the respective genes. The region surrounding intron 6 of CsaMLO1 (B and C) and the region surrounding intron 12 of CsaMLO11 (E and F) were amplified from cDNA of cucumber genotypes CS-PMR1 and Santou, and subsequently sequenced in triplicate by Sanger sequencing. Amplified products were analysed on 1.25% agarose gels. It was found that for both amplified regions, the products amplified from CS-PMR1 and Santou were of similar sizes. Sequences of cDNA were identical to one another and to the reference cDNA sequence (Chinese long inbred 9930, v2). (PDF 2940 kb)
Relative transcript abundances of CsaMLO1 and CsaMLO11 in leaf tissues of cucumber genotypes Sheila, Santou and CS-PMR1 were determined using qRT-PCR. Data were normalized relative to the geometric average of the Ct values of reference genes Ef-α, TIP41 and CACS, and subsequently normalized relative to the average dCt value of Sheila. Each bar shows the relative expression of three biological replicates, on a logarithmic scale. Error bars indicate standard error of the mean. (PDF 349 kb)
Acknowledgements
We are grateful to foundation TKI Starting Materials, the Netherlands, for funding the majority of this work.
Also we would like to thank the Technological Top Institute Green Genetics, the Netherlands (TTI-GG project INTCFD040RP), together with Hazera Seeds B.V., Bayer Vegetable Seeds B.V., Takii Europe B.V., and Rijk Zwaan B.V. for providing funding for the RNA-seq data.
Availability of data and materials
All relevant data are within this article and its supporting information files.
Authors’ contributions
JAB, HJS and YB designed the experiments. JAB, MA and GB performed the experiments. JAB drafted the manuscript. MA, RGFV, HJS and YB critically revised the manuscript. All authors read and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jeroen A. Berg, Email: jeroen.berg@wur.nl
Michela Appiano, Email: michela.appiano@wur.nl.
Gerard Bijsterbosch, Email: gerard.bijsterbosch@wur.nl.
Richard G. F. Visser, Email: richard.visser@wur.nl
Henk J. Schouten, Email: henk.schouten@wur.nl
Yuling Bai, Email: bai.yuling@wur.nl.
References
- 1.Glawe DA. The powdery mildews: a review of the world ’ s most familiar (yet poorly known) plant pathogens. Annu Rev Phytopathol. 2008;46:27–51. doi: 10.1146/annurev.phyto.46.081407.104740. [DOI] [PubMed] [Google Scholar]
- 2.Kristkova E, Lebeda A, Sedlakova B. Species spectra, distribution and host range of cucurbit powdery mildews in the Czech Republic, and in some other European and middle eastern countries. Phytoparasitica. 2009;37:337–350. doi: 10.1007/s12600-009-0045-4. [DOI] [Google Scholar]
- 3.Jones JDG, Dangl JL. The plant immune system. Nat Rev. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 4.Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. doi: 10.1146/annurev.py.09.090171.001423. [DOI] [Google Scholar]
- 5.Freisleben R, Lein A. Über die Auffindung einer mehltauresistenten Mutante nach Röntgenbestrahlung einer anfälligen reinen Linie von Sommergerste. Naturwissenschaften. 1942;30:608. doi: 10.1007/BF01488231. [DOI] [Google Scholar]
- 6.Piffanelli P, Ramsay L, Waugh R, Benabdelmouna A, D’Hont A, Hollricher K, Jørgensen JH, Schulze-Lefert P, Panstruga R. A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature. 2004;430:887–891. doi: 10.1038/nature02781. [DOI] [PubMed] [Google Scholar]
- 7.Jørgensen JH. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica. 1992;63:141–152. doi: 10.1007/BF00023919. [DOI] [Google Scholar]
- 8.Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, Töpsch S, Vos P, Salamini F, Schulze-Lefert P. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/S0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 9.Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, Von Heijne G, Schulze-Lefert P. Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem. 1999;274:34993–35004. doi: 10.1074/jbc.274.49.34993. [DOI] [PubMed] [Google Scholar]
- 10.Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, Vogel J, Lipka V, Kemmerling B, Schulze-Lefert P, Somerville SC, Panstruga R. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet. 2006;38:716–720. doi: 10.1038/ng1806. [DOI] [PubMed] [Google Scholar]
- 11.Bai Y, Pavan S, Zheng Z, Zappel NF, Reinstädler A, Lotti C, De Giovanni C, Ricciardi L, Lindhout P, Visser R, Theres K, Panstruga R. Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of mlo function. Mol Plant-Microbe Interact. 2008;21:30–39. doi: 10.1094/MPMI-21-1-0030. [DOI] [PubMed] [Google Scholar]
- 12.Humphry M, Reinstädler A, Ivanov S, Bisseling T, Panstruga R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol Plant Pathol. 2011;12:866–878. doi: 10.1111/j.1364-3703.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg JA, Appiano M, Santillán Martínez M, Hermans FW, Vriezen WH, Visser RG, Bai Y, Schouten HJ. A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biol. 2015;15:243. doi: 10.1186/s12870-015-0635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimura T, Sato S, Tajima T, Arai M. Powdery mildew resistance in the Japanese domestic tobacco cultivar Kokubu is associated with aberrant splicing of MLO orthologs. Plant Pathol. 2016;65:1358–1365. doi: 10.1111/ppa.12498. [DOI] [Google Scholar]
- 15.Zheng Z, Nonomura T, Appiano M, Pavan S, Matsuda Y, Toyoda H, Wolters A-MA, Visser RGF, Bai Y. Loss of function in Mlo Orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica. PLoS One. 2013;8:e70723. doi: 10.1371/journal.pone.0070723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pessina S, Lenzi L, Perazzolli M, Campa M, Costa LD, Urso S, Valè G, Salamini F, Velasco R, Malnoy M. Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine. Hortic Res. 2016, 3. doi:10.1038/hortres.2016.16. [DOI] [PMC free article] [PubMed]
- 17.Pessina S, Angeli D, Martens S, Visser R, Bai Y, Salamini F, Velasco R, Schouten H, Malnoy M. The knock-down of the expression of MdMLO19 reduces susceptibility to powdery mildew (Podosphaera leucotricha) in apple (Malus domestica). Plant Biotechnol J. 2016;14(10):2033–44. [DOI] [PMC free article] [PubMed]
- 18.Varallyay E, Giczey G, Burgyan J. Virus-induced gene silencing of Mlo genes induces powdery mildew resistance in Triticum aestivum. Arch Virol. 2012;157:1345–1350. doi: 10.1007/s00705-012-1286-y. [DOI] [PubMed] [Google Scholar]
- 19.Wolter M, Hollricher K, Salamini F, Schulze-Lefert P. The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol Gen Genet. 1993;239:122–128. doi: 10.1007/BF00281610. [DOI] [PubMed] [Google Scholar]
- 20.Hückelhoven R, Dechert C, Kogel K-H. Overexpression of barley BAX inhibitor 1 induces breakdown of mlo-mediated penetration resistance to Blumeria graminis. PNAS. 2003;100:5555–5560. doi: 10.1073/pnas.0931464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miklis M, Consonni C, Bhat RA, Lipka V, Schulze-Lefert P, Panstruga R. Barley MLO modulates Actin-dependent and Actin-independent antifungal defense pathways at the cell periphery. Plant Physiol. 2007;144:1132–1143. doi: 10.1104/pp.107.098897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu J-LL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, Schulze-Lefert P. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 23.The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 24.Acevedo-Garcia J, Kusch S, Panstruga R. Magical mystery tour: MLO proteins in plant immunity and beyond. J Physiol. 2014;204:273–281. doi: 10.1111/nph.12889. [DOI] [PubMed] [Google Scholar]
- 25.Kusch S, Pesch L, Panstruga R. Comprehensive Phylogenetic analysis sheds light on the diversity and origin of the MLO family of integral membrane proteins. Genome Biol Evol. 2016;8:878–895. doi: 10.1093/gbe/evw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Z, Noir S, Kwaaitaal M, Hartmann HA, Wu M, Mudgil Y, Sukumar P, Muday G, Panstruga R, Jones AM. Two seven-Transmembrane domain MILDEW RESISTANCE LOCUS O proteins Cofunction in Arabidopsis root Thigmomorphogenesis. Plant Cell. 2009;21:1972–1991. doi: 10.1105/tpc.108.062653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U. Conserved molecular components for pollen tube reception and fungal invasion. Science. 2010;330:968–971. doi: 10.1126/science.1195211. [DOI] [PubMed] [Google Scholar]
- 28.Yi J, An S, An G. OsMLO12, encoding seven transmembrane proteins, is involved with pollen hydration in rice. Plant Reprod. 2014;27:169–180. doi: 10.1007/s00497-014-0249-8. [DOI] [PubMed] [Google Scholar]
- 29.Appiano M, Catalano D, Santillán Martínez M, Lotti C, Zheng Z, Visser RGF, Ricciardi L, Bai Y, Pavan S. Monocot and dicot MLO powdery mildew susceptibility factors are functionally conserved in spite of the evolution of class-specific molecular features. BMC Plant Biol. 2015;15:257. doi: 10.1186/s12870-015-0639-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P, Ren Y, Zhu H, Li J, Lin K, Jin W, Fei Z, Li G, Staub J, Kilian A, van der Vossen EAG, Wu Y, Guo J, He J, Jia Z, Ren Y, Tian G, Lu Y, Ruan J, Qian W, Wang M, et al. The genome of the cucumber, Cucumis sativus L. Nat Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 31.Schouten HJ, Krauskopf J, Visser RGF, Bai Y. Identification of candidate genes required for susceptibility to powdery or downy mildew in cucumber. Euphytica. 2014;200:475–486. doi: 10.1007/s10681-014-1216-z. [DOI] [Google Scholar]
- 32.Iovieno P, Andolfo G, Schiavulli A, Catalano D, Ricciardi L, Frusciante L, Ercolano MR, Pavan S. Structure, evolution and functional inference on the Mildew Locus O (MLO) gene family in three cultivated Cucurbitaceae spp. BMC Genomics. 2015;16:1112. doi: 10.1186/s12864-015-2325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie J, Wang Y, He H, Guo C, Zhu W, Pan J, Li D, Lian H, Pan J, Cai R. Loss-of-Function Mutations in CsMLO1 Confer Durable Powdery Mildew Resistance in Cucumber (Cucumis sativus L.) Front Plant Sci. 2015;6:1155. doi: 10.3389/fpls.2015.01155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nie J, He H, Peng J, Yang X, Bie B, Zhao J, Wang Y, Si L, Pan J-S, Cai R: Identification and fine mapping of pm5.1: a recessive gene for powdery mildew resistance in cucumber (Cucumis sativus L.). Mol Breed 2015, 35:7.
- 35.Fukino N, Yoshioka Y, Sugiyama M, Sakata Y, Matsumoto S. Identification and validation of powdery mildew (Podosphaera xanthii)-resistant loci in recombinant inbred lines of cucumber (Cucumis sativus L.) Mol Breed. 2013;32:267–277. doi: 10.1007/s11032-013-9867-3. [DOI] [Google Scholar]
- 36.Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua N-H. Genome-wide analysis uncovers regulation of long Intergenic Noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–4345. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi J, Liu X, Shen D, Miao H, Xie B, Li X, Zeng P, Wang S, Shang Y, Gu X, Du Y, Li Y, Lin T, Yuan J, Yang X, Chen J, Chen H, Xiong X, Huang K, Fei Z, Mao L, Tian L, Städler T, Renner SS, Kamoun S, Lucas WJ, Zhang Z, Huang S. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat Genet. 2013;45:1510–1515. doi: 10.1038/ng.2801. [DOI] [PubMed] [Google Scholar]
- 38.The Cucurbit Genomics Database. http://www.icugi.org/. Accessed 13 June 2016.
- 39.Appiano M, Pavan S, Catalano D, Zheng Z, Bracuto V, Lotti C, Visser RGF, Ricciardi L, Bai Y. Identification of candidate MLO powdery mildew susceptibility genes in cultivated Solanaceae and functional characterization of tobacco NtMLO1. Transgenic Res. 2015;24:847–858. doi: 10.1007/s11248-015-9878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Schie CCN, Takken FLW. Susceptibility genes 101: how to be a good host. Annu Rev Phytopathol. 2014;52:551–581. doi: 10.1146/annurev-phyto-102313-045854. [DOI] [PubMed] [Google Scholar]
- 41.Zheng Z, Appiano M, Pavan S, Bracuto V, Ricciardi L, Visser RG, Wolters A-MA, Bai Y. Genome-wide study of the tomato SlMLO gene family and its functional characterization in response to the powdery mildew fungus Oidium neolycopersici. Front Plant Sci. 2016;7:380. doi: 10.3389/fpls.2016.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen VN, Vo KTX, Park H, Jeon J-S, Jung K-H. A systematic view of the MLO family in rice suggests their novel roles in morphological development, diurnal responses, the light-signaling pathway, and various stress responses. Front Plant Sci. 2016;7:1413. doi: 10.3389/fpls.2016.01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar J, Hückelhoven R, Beckhove U, Nagarajan S, Kogel K. A compromised Mlo pathway affects the response of barley to the Necrotrophic fungus Bipolaris sorokiniana (Teleomorph: Cochliobolus sativus) and its toxins. Phytopathology. 2001;91:127–133. doi: 10.1094/PHYTO.2001.91.2.127. [DOI] [PubMed] [Google Scholar]
- 44.Jarosch B, Kogel K, Schaffrath U. The ambivalence of the barley Mlo locus: mutations conferring resistance against powdery mildew (Blumeria graminis f. Sp. hordei) enhance susceptibility to the Rice blast fungus Magnaporthe grisea. Mol Plant-Microbe Interact. 1999;12:508–514. doi: 10.1094/MPMI.1999.12.6.508. [DOI] [Google Scholar]
- 45.Ruiz-Lozano JM, Gianinazzi S, Gianinazzi-Pearson V. Genes involved in resistance to powdery mildew in barley differentially modulate root colonization by the mycorrhizal fungus Glomus mosseae. Mycorrhiza. 1999;9:237–240. doi: 10.1007/s005720050273. [DOI] [Google Scholar]
- 46.Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, Collins NC, Panstruga R, Schulze-Lefert P. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129:1076–1085. doi: 10.1104/pp.010954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z, Hartmann HA, Wu M-J, Friedman EJ, Chen J-G, Pulley M, Schulze-Lefert P, Panstruga R, Jones AM. Expression analysis of the AtMLO gene family encoding plant-specific seven-transmembrane domain proteins. Plant Mol Biol. 2006;60:583–597. doi: 10.1007/s11103-005-5082-x. [DOI] [PubMed] [Google Scholar]
- 48.Pessina S, Pavan S, Catalano D, Gallotta A, Visser RGF, Bai Y, Malnoy M, Schouten HJ. Characterization of the MLO gene family in Rosaceae and gene expression analysis in Malus domestica. BMC Genomics. 2014;15:618. doi: 10.1186/1471-2164-15-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346 [DOI] [PubMed]
- 50.Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu J-L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32:947–952. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 51.Karimi M, Inzé D, Depicker A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 52.Warzybok A, Migocka M. Reliable reference genes for normalization of Gene expression in cucumber grown under different nitrogen nutrition. PLoS One. 2013;8:e72887. doi: 10.1371/journal.pone.0072887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Box MS, Coustham V, Dean C, Mylne JS. Protocol: a simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods. 2011;7:7. doi: 10.1186/1746-4811-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.EMBL-EBI Expression Atlas. http://www.ebi.ac.uk/gxa. Accessed 23 June 2016.
- 55.Healey A, Furtado A, Cooper T, Henry RJ. Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods. 2014;10:21. doi: 10.1186/1746-4811-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 genome project data processing subgroup: the sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The relative transcript abundances of CsaMLO1, CsaMLO8 and CsaMLO11 in T2 families of a tomato mlo mutant overexpressing CsaMLO1, CsaMLO8 and CsaMLO11, were determined by qRT-PCR. Data were normalised relatively to the reference gene SlEF-α. Average transcript abundances of four or five randomly selected individuals are shown on a logarithmic scale. Error bars represent standard error of the mean. Different letters above the bars indicate statistical significance of Ct-values (One-way ANOVA with Dunnet’s T3 post hoc test, P < 0.05). CsaMLO1, CsaMLO8 or CsaMLO11 expression was not detectable in non-transformed tomato mlo mutant or Moneymaker. (PDF 341 kb)
Data on the transcript abundance in four tissues of Arabidopsis thaliana, determined using RNA-seq was investigated and downloaded using the Expression Atlas of EMBL-EBI (https://www.ebi.ac.uk/gxa/home). The FPKM values (Fragments Per Kilobase of transcript per Million mapped fragments) for AtMLO2, AtMLO6 and AtMLO12 in each of the tissues is shown on a logarithmic scale. (PDF 350 kb)
Relative transcript abundances in two tissues of PM susceptible cucumber cultivar ‘Sheila’ (A) hypocotyl and B) leaf, before and at 4, 6, 8, 12 and 24 h post inoculation with P. xanthii were determined using qRT-PCR. Data were normalized relative to the geometric average of the Ct values of reference genes Ef-α and CACS, and subsequently normalized relative to the average dCt value at 0 hpi for both tissues. Each bar shows the relative expression of five to eight biological replicates, as indicated above the bars, on a logarithmic scale. Error bars indicate standard error of the mean. (PDF 374 kb)
The list of SNPs and indels in 115 resequenced cucumber accessions, available from the Cucurbit Genomics Database [38], was filtered for the genomic region of CsaMLO1 (Chr1:8,159,428.8165253). The genic location of each SNP/Indel was determined manually (exon/intron), for exonic SNPs the effect on the predicted amino acid sequence was scored (synonymous/nonsynonymous). (XLSX 57 kb)
The list of SNPs and indels in 115 resequenced cucumber accessions, available from the Cucurbit Genomics Database [38], was filtered for the genomic region of CsaMLO11 (Chr6:14,120,024.14,125,039). The genic location of each SNP/Indel was determined manually (exon/intron), for exonic SNPs the effect on the predicted amino acid sequence was scored (synonymous/nonsynonymous). (XLSX 50 kb)
Protein alignment of clade V MLO proteins of Arabidopsis thaliana (AtMLO2, 6 and 12), Medicago trunculata (MtMLO1), Pisum sativum (PsMLO1), Lotus japonicus (LjMLO1), Cucumis sativus (CsaMLO1, 8 and 11), Solanum lycopersicum (SlMLO1), Capsicum annuum (CaMLO2) and Nicotiana tabacum (NtMLO1). A bar graph shows the conservation of the individual residues. Colours indicate amino acid residues with similar physiochemical properties according to the RasMol colour scheme. Locations of amino acid substitutions in CsaMLO1 due to SNPs are indicated by a red arrow. (PDF 3224 kb)
Resequencing data of the genomic region of CsaMLO1 in cucumber genotypes CS-PMR1 and Santou. The location of the gene on the chromosome is indicated by a red cursor. For each of the two genotypes, the total reads mapping to the location and the coverage per base pair are given. SNPs are indicated by coloured stripes (green for A, red for T, blue for C, brown for G), indels are indicated by black stripes. Read pairs with a small (smallest 0.5%) or large (largest 0.5%) insert size are coloured dark blue or dark red, respectively. Reads for which the other mate in the mate pair was not mapped are indicated by a bright red outline. (PDF 356 kb)
Resequencing data of the genomic region of CsaMLO11 in cucumber genotypes CS-PMR1 and Santou. The location of the gene on the chromosome is indicated by a red cursor. For each of the two genotypes, the total reads mapping to the location and the coverage per base pair are given. SNPs are indicated by coloured stripes (green for A, red for T, blue for C, brown for G), indels are indicated by black stripes. Read pairs with a small (smallest 0.5%) or large (largest 0.5%) insert size are coloured dark blue or dark red, respectively. Reads for which the other mate in the mate pair was not mapped are indicated by a bright red outline. (PDF 356 kb)
The list of SNPs and indels identified in the genomic sequence of CsaMLO1 and CsaMLO11 in the resequenced cucumber genotypes CS-PMR1 and Santou. The reference allele (in cucumber reference genome Chinese Long 9930) and the alternative allele are given. Furthermore, the genic location of each SNP/Indel was determined (exon/intron), for exonic SNPs the effect on the predicted amino acid sequence was scored (synonymous/nonsynonymous). (XLSX 12 kb)
Intron 6 of CsaMLO1 (A) and intron 12 of CsaMLO11 (D) were amplified from genomic DNA isolated from the cucumber genotype CS-PMR1, and subsequently sequenced in triplicate by Sanger sequencing. The obtained sequences were aligned to the reference cucumber genome (Chinese long inbred 9930, v2). Numbers above the alignment are relative to the start codon of the respective genes. The region surrounding intron 6 of CsaMLO1 (B and C) and the region surrounding intron 12 of CsaMLO11 (E and F) were amplified from cDNA of cucumber genotypes CS-PMR1 and Santou, and subsequently sequenced in triplicate by Sanger sequencing. Amplified products were analysed on 1.25% agarose gels. It was found that for both amplified regions, the products amplified from CS-PMR1 and Santou were of similar sizes. Sequences of cDNA were identical to one another and to the reference cDNA sequence (Chinese long inbred 9930, v2). (PDF 2940 kb)
Relative transcript abundances of CsaMLO1 and CsaMLO11 in leaf tissues of cucumber genotypes Sheila, Santou and CS-PMR1 were determined using qRT-PCR. Data were normalized relative to the geometric average of the Ct values of reference genes Ef-α, TIP41 and CACS, and subsequently normalized relative to the average dCt value of Sheila. Each bar shows the relative expression of three biological replicates, on a logarithmic scale. Error bars indicate standard error of the mean. (PDF 349 kb)
Data Availability Statement
All relevant data are within this article and its supporting information files.


