ABSTRACT
It is no longer necessary to demonstrate that ribosome is the central machinery of protein synthesis. But it is less known that it is also key player of the protein folding process through another conserved function: the protein folding activity of the ribosome (PFAR). This ribozyme activity, discovered more than 2 decades ago, depends upon the domain V of the large rRNA within the large subunit of the ribosome. Surprisingly, we discovered that anti-prion compounds are also potent PFAR inhibitors, highlighting an unexpected link between PFAR and prion propagation.
In this review, we discuss the ancestral origin of PFAR in the light of the ancient RNA world hypothesis. We also consider how this ribosomal activity fits into the landscape of cellular protein chaperones involved in the appearance and propagation of prions and other amyloids in mammals. Finally, we examine how drugs targeting the protein folding activity of the ribosome could be active against mammalian prion and other protein aggregation-based diseases, making PFAR a promising therapeutic target for various human protein misfolding diseases.
KEYWORDS: chaperon, PFAR, prion, protein folding, ribosome, RNA
Present in every cell and mitochondria, ribosomes are composed of 2 subunits, which themselves are made up of ribosomal RNA (rRNA) and proteins. Broadly, the small subunit initiates the translational process and ensures the correct decoding of genetic information, while the large subunit catalyzes the peptidyl transferase activity that covalently links amino acids together. While the central role of the ribosome in protein synthesis is well appreciated, little is known about the second ribozyme activity of the ribosome: its protein folding activity (PFAR).
Since its discovery, the existence of PFAR has been controversial and difficult to gain acceptance within the scientific community. Despite PFAR being first identified in 1994 by the group of C. Das Gupta1 and this activity being corroborated by several other teams,2 PFAR is often overlooked, as illustrated by its absence from reviews on non-coding RNAs. However, once getting over the initial surprise of the existence of such an activity, it seems self-evident that the biological entity that has evolved to synthesize proteins must also aid them achieve a proper 3-dimensional functional state before their release. In addition, this is fully consistent with the estimation that, apart from spontaneously folded proteins, only a fraction of the total cellular proteins can be folded by classical protein chaperones.3
Initially, PFAR was described in vitro for bacterial ribosomes and was quickly shown to be a conserved function of any ribosome, whether from bacteria, eubacteria, eukaryotes or even mitochondria.2 The conservation of the protein folding capacity of the ribosome among species is not a surprise when considering the high rate of conservation of rRNA in the ribosome core throughout evolution.4 As one may expect, PFAR is a versatile process that has been demonstrated to be able to refold any protein challenged so far in vitro2 and in vivo.5-8 This folding activity appears to be inherent to the conserved RNA domain that also harbors the peptidyl transferase activity: domain V of the rRNA of the large subunit of the ribosome (23S for bacteria, 25S for yeast and 28S for mammals). However, the nucleotides of domain V involved in its protein folding activity are different from those involved in its peptidyl transferase activity 9,10 (Fig. 1).
FIGURE 1.
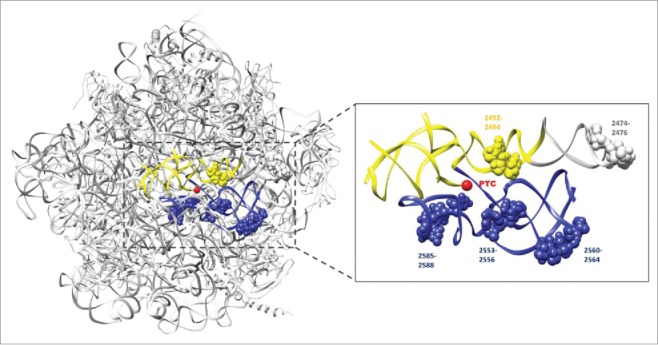
The sites of major importance for PFAR activity are included within the PTC region. The ribosomal large subunit is shown in light gray (Protein Data Bank code 4V6F). The yellow and blue helices indicate the 2 helices part of the symmetrical region forming the PTC cavity. The nucleotides involved in PFAR activity are numbered and represented as spheres (Thermus thermophilus numbering). The core of the PTC is indicated by a red sphere (Adapted from ref. 9). Yeast nucleotide U28926 corresponds to position 2492.
The protein folding mediated by domain V is a 2-step post-translational process: the neo-synthesized polypeptide is first folded by the central loop of domain V (RNA1) and remains associated to it until the intervention of a second part of domain V (RNA2) which is responsible for the release of the folded protein.11 The nucleotides involved in PFAR are localized at the interface of the ribosome's small and large subunits.9 Ribosomal subunits dissociate in the presence of unfolded polypeptides, making them more accessible to PFAR-involved nucleotides and thus enhancing their folding ability.2 This is in good agreement with the fact that protein synthesis and protein folding are synchronized: as long as the peptidyl transferase activity continues, the protein folding activity of the ribosome is silenced and only becomes operational when translation is completed.12
In the midst of identifying new anti-prion compounds, we identified anti-prion drugs such as 6-aminophenanthridine (6AP13), guanabenz (GA14) and imiquimod (IQ15) which are active in vivo against yeast and mammalian prions and that were amazingly identified as PFAR inhibitors. The discovery that 6AP and GA anti-prion drugs are also anti-PFAR drugs led us naturally to suggest that PFAR may be linked to prion propagation. Although the primary role of protein chaperones is to prevent protein misfolding and aggregation, involvement of protein chaperones like PFAR in the propagation of prion conformation is self-evident, as amyloid-forming proteins exist in several conformations and replication corresponds to propagation of differentially folded states.16 This hypothesis has strong support as Hsp104p, a yeast heat shock protein, has been shown for a long time to be involved in yeast prion propagation, together with Hsp70 and Hsp40. Similarly to Hsp104p, PFAR misregulation leads to a defect in the propagation of the yeast prion [PSI+]6 (Fig. 2). The anti-PFAR drugs we identified were the first of that kind: all the previous anti-PFAR drugs described so far are antibiotics that also bind to domain V and inhibit its peptidyl transferase activity.5,7 Of note, the fact that some antibiotics specifically target PFAR is a further indication of the central function of this ribozyme activity.
FIGURE 2.
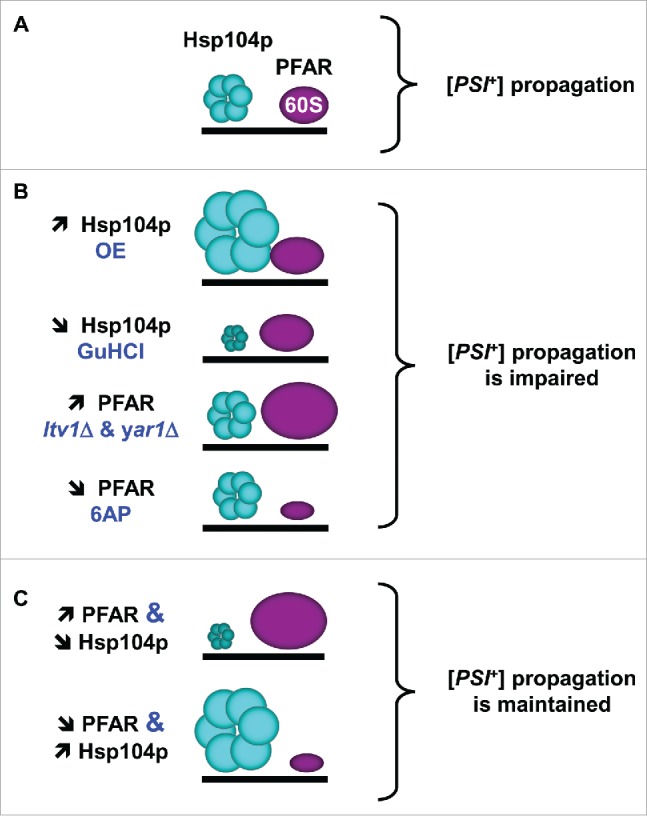
Interplay between protein folding activities of Hsp104p and ribosome in modulating [PSI+] propagation (adapted from ref. 6). (A)- Hsp104p and PFAR both participate in [PSI+] propagation. (B)- [PSI+] propagation is affected by enrichment or inhibition of Hsp104p and PFAR. (C)- Hsp104p and PFAR interplay results in the compensation of the increase of one by the reduction of the other, and vice versa. OE, overexpression. GuHCl, Hsp104p inhibitor guanidine hydrochloride. ltv1Δ and yar1Δ, PFAR-enriched yeast strains deleted for LTV1 or YAR1 genes.6
In this review, we consider the ancestral origin of PFAR and the relationship of this emerging ribosomal activity with the global protein folding capacity of the cell. We also discuss PFAR involvement in the propagation of prions and other amyloids in mammals.
PFAR Activity: Another Relic of the Ancient RNA World into Modern Ribosomes?
In current living organisms, the newly synthesized polypeptides undergo several maturation mechanisms to be correctly processed and translocated to achieve their functional folded state. These maturation processes are often linked to protein synthesis. They depend on translational speed and on interactions of the nascent polypeptide chain with the peptide exit tunnel and with the nascent chain-interacting protein factors, which are located on and around the ribosomal tunnel exit area. Beyond, a network of other cooperating chaperones acting post-translationally in a coordinated ballet are also involved during the latter stages of the protein folding process.17 However, this system brings the endless chicken-and-egg paradox back into play: to get mature and functional proteins, the cell needs… mature and functional chaperoning factors! To address this paradox, one has to consider the evolution of the protein folding process and how the ancient foundations may have developed. In the ancient RNA world hypothesis, earlier forms of life may rely on the dual function of RNA that simultaneously catalyzes enzymatic reactions and store genetic information.18 This suggests that RNA could be the main actor that performed the chemical reactions allowing proteins to gradually emerge and become more complex due to the development of more and more sophisticated protein folding capabilities. Many molecular fossils of the ancient RNA world are still present and sometimes still active in modern organisms. Candidates must be catalytic, ubiquitous, and/or central to some aspect of metabolism.19 Accordingly, one of the most significant piece of evidence to support that the first peptide bonding machines emerged in an RNA world is the fact that the modern ribosome is a ribozyme, or in other words that only RNA performs its key role in peptide bond formation.20 The peptidyl-transferase center (PTC) of the modern ribosome is a universal and highly conserved RNA-only structure located in the domain V of 23S rRNA in bacteria or its equivalent parts in eukaryotic ribosome. It forms a symmetrical pocket in the heart of the large ribosomal subunit and probably originated from the dimerization of 2 stem–elbow–stem motifs.21 The PTC is considered to be the first proto-ribosome dating from the ancient RNA world since it was certainly capable of triggering catalytic reactions by itself. Strikingly, as supported by the binding sites of its protein substrates or inhibitors, PFAR activity is clustered within the PTC binding pocket, surrounding the core where the catalytic reaction takes place9,22 (Fig. 1). Therefore, PFAR encompasses most criteria for making it a perfect vestige of the ancient RNA world as it is: i) part of another ribozyme, the PTC, ii) as ancient as the PTC, iii) supported by RNA only, iv) conserved from bacteria to eukaryotes and v) capable of playing a key role in modern protein synthesis. In this scenario, the first PTCs (proto-PTCs) were formed by the dimerization of short helical structures that created the first random but fragile peptides aleatory using amino acids. In a process that might be assimilated as “molecular Darwinism,”23 these proto-PTCs would then have evolved into a stable PTC, able to better favor protection, stability and activity of the peptides they synthesized. While one can assume that the earliest peptides were short, single domain and rapidly selected for robust chaperone-independent folding, the emergence of a primitive PFAR certainly conferred a selective advantage to the peptides. In turn, and resulting from the more and more efficient peptides and proteins being produced, the proto-PTC slowly co-evolved into a proto-ribosome and finally into the modern ribosome we recognize today, likely by progressively integrating multiple small ribonucleoprotein complexes into a much more complex machinery.24 The traces of these ancient times remain active RNAs such as domain V harboring PTC and PFAR, still embedded in contemporary ribosomes, and may continue to play a role as part of the global protein folding capability of the cell.
Which Place PFAR Could Take in the Array of Protein Chaperones?
Prion propagation in yeast has been shown to be intimately linked to the expression and activity of a plethora of protein chaperones. We recently showed that PFAR and Hsp104p partially compensate each other for [PSI+] propagation in yeast (Fig. 2). We indeed observed that PFAR up-modulation can compensate for the partial loss of Hsp104p activity and that PFAR partial down-modulation is also compensated by Hsp104p increase.6 These results clearly indicate that PFAR is linked to the cellular array of protein chaperones, at least through Hsp104p. To this respect, PFAR participates to the very complex network required for cell proteostasis, i.e. the global protein quality control and the maintenance of proteome homeostasis. Of note, polynucleotides, and more particularly RNAs, have been shown to exhibit potent chaperone activity in vitro.25 Proteostasis involves hundreds of proteins constituting a highly regulated and integrated network.
One of the main challenges of the cellular response to stress is to synthesize, under adverse conditions, correctly folded proteins essential for survival. Since the PTC and PFAR are intimately linked, PFAR activity directly depends upon the synthesis of proteins, thus allowing the cell to establish a proper equilibrium between protein production and folding activities. In this context, the link between the PTC and PFAR can be seen as a strategy to avoid the synthesis of a polypeptide in the absence of efficient cellular folding activities. Remarkably, in situations of cellular stress, translation is strongly reduced, which consequently increases the population of non-translating ribosomes becoming available for protein folding. As a consequence, the corresponding misfolded proteins emerging from the deficient ribosome will be more prone to proteolysis. Thus, PFAR could also participate to proteostasis as a potential proofreading process of the PTC activity.
While Hsp104, and now PFAR, are key players in dictating the appearance and propagation of naturally occurring amyloid-based yeast prions, members of the cytosolic Hsp70 chaperone machinery, and associated co-chaperones, are important modulators of prion propagation, and in some instances de novo prion formation.16,26 The cytosolic Hsp70-Ssa [Stress Seventy Sub-Family A] family members are well characterized as playing an integral role in modulating the propagation of a variety of yeast prions,27,28 and the ribosome-associated Hsp70-Ssb [Stress Seventy Sub-Family B] family members are anti-prion in their action since they repress the spontaneous appearance of [PSI+].29 Of note, contrary to Hsp104p and Hsp70 which expression is induced by stress, PFAR is a ready-to-use protein folding source immediately available in case of heatshock to supplement Hsp104 or to compensate for its absence, similarly to mammals where there is no HSP104 homolog.
Where could PFAR “sit” among these other cellular factors and how may they interact to influence prion propagation, or other protein folding events in vivo? One possibility is that some prion phenotypes consecutive to alterations in PFAR activity could result from indirect effects through alteration of chaperone systems such as the cytosolic Hsp70 machinery. However, such phenotypes are not due to indirect effects on Hsp70 abundance6 and it is unlikely that alterations in PFAR activity could significantly disturb Hsp70 specific functions, unless through indirect effect on Hsp70 co-chaperone activities. PFAR could possibly work in conjunction or in tandem with Hsp70s or other chaperones and the association of Hsp70-Ssb and the interaction with RAC [Ribosome Associated Complex] in prion formation and propagation30 provides the basis for such interactions to evolve. Chaperones often function as multi-complex machines and it is conceivable that Hsp70 or co-chaperones could be involved into the delivery or processing of substrates acted upon by PFAR, or that PFAR could process substrates acted on by downstream chaperone machinery. The construction and characterization of yeast cells modified to alter PFAR activity now provides the basis for a thorough genetic assessment of interaction between PFAR and an array of protein chaperone pathways. The involvement of PFAR in protein homeostasis could extend well beyond prion propagation and should it be the case, the integration with other chaperone networks is even more likely.
PFAR: Mammalian Prion Propagation and Beyond?
How could drugs targeting the protein folding activity of the ribosome, which mainly lies in the cytoplasm, be active against mammalian prion PrPSc, which is believed to be modified mostly at the cell surface or on endocytic compartments? Addressing this question emphasizes our lack of precise knowledge regarding the cellular compartments where prion conversion and subsequent accumulation occur. This is partly due to the fact that direct and dynamic visualization in cells remains a highly challenging issue, notably because tagging the cellular form of the prion protein (PrPC) most often prevents its conversion into the disease-specific isoform PrPSc. In many differentiated cell types, a major proportion of PrPC is detected in lipid rafts at the cell surface, being anchored by a GPI moiety (for review31). PrPC expression at the cell surface is required for conversion into misfolded PrPSc,32,33 and this process may occur rapidly.32 Several reports also indicate that a substantial fraction of PrPC cycles constitutively between the plasma membrane and endocytic compartments and that conversion can also occur in several intracellular compartments all along the endocytic pathway.34-36 A minor proportion of PrPC can be found in the cytosol or in contact with the cytosol, due to retrotranslocation from the endoplasmic reticulum (or escape before translocation) or transmembrane anchoring, respectively (for review37). In physiological conditions, these forms are rapidly cleared by quality control processes (ERAD (Endoplasmic-reticulum-associated protein degradation), aggresomes). However, in the prion disease state, or with mutated PrP mimicking familial forms of prion diseases, the levels of cytosolic PrP are increased.37,38 The exact contribution of these variants to prion pathology remains unknown. PrPC is also involved in many cellular processes and signaling pathways. These processes include response to oxidative stress, ER stress, apoptosis, proliferation and differentiation (reviewed in39-41). Certain stress can even trigger unusual localization of PrPC into the nucleus.42 Late stages of prion pathogenesis involve over-activation of the PERK (protein kinase RNA-like endoplasmic reticulum kinase) pathway of the unfolded protein response in experimental mouse models.43 Such pathways and the deregulation of the PrP-dependent physiological pathways, or a yet to discover PFAR-dependent proteostatic pathway in mammals,44 provide plenty of scope for direct or indirect interactions with PFAR to occur.
PFAR involvement goes most probably further than prion-based diseases such as Creutzfeldt-Jakob disease. There is indeed growing evidence that amyloid-based diseases like OPMD (Oculopharyngeal muscular dystrophy), Alzheimer's, Parkinson's and Huntington's diseases share key biophysical and biochemical characteristics with prionopathies: they involve accumulation of aggregates of misfolded host-encoded proteins through prion-like processes of seeded polymerisation.45-47 As metazoa have no Hsp104p ortholog, the protein folding activity of domain V of the ribosome might thus correspond to the Hsp104-like activity responsible for amyloid handling that is absent in these organisms. This original hypothesis makes sense in light of our recent findings that some anti-prion compounds with anti-PFAR activity, like 6AP, GA and IQ, are also active in models of other protein aggregation-based diseases like Huntington's disease, Parkinson's disease (unpublished data) and OPMD.48 Our data thus indicate that PFAR involvement in amyloid handling may be shared by several human protein misfolding-based diseases, hence making PFAR a potential therapeutic target for human protein misfolding diseases. Despite the central role of PFAR in the proteostasis of the cell, it is still a promising therapeutic target to treat this class of diseases, either as a sole target or as part of a collaborative strategy targeting more than one cellular chaperone system. Indeed, its mild inhibition, together with other therapeutics, may help overcome the progression of these invariably fatal pathologies.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors declare no competing financial interests in relation to the work described.
ACKNOWLEDGMENT
We thank E. Giudice for generously helping preparing Fig. 1.
FUNDING
This work was supported by FA29 and défi organization to CV and MB, Agence Nationale pour la Recherche and Direction Générale de l'Armement (ANR-14-ASTR-0001) and Institut Universitaire de France to RG.
REFERENCES
- [1].Chattopadhyay S, Das B, Bera AK, Dasgupta D, Dasgupta C. Refolding of denatured lactate dehydrogenase by Escherichia coli ribosomes. Biochem J 1994; 300 (Pt 3):717-21; PMID:8010952; http://dx.doi.org/ 10.1042/bj3000717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Das D, Das A, Samanta D, Ghosh J, Dasgupta S, Bhattacharya A, Basu A, Sanyal S, Das Gupta C. Role of the ribosome in protein folding. Biotechnol J 2008; 3:999-1009; PMID:18702035; http://dx.doi.org/ 10.1002/biot.200800098 [DOI] [PubMed] [Google Scholar]
- [3].Pechmann S, Willmund F, Frydman J. The ribosome as a hub for protein quality control. Mol Cell 2013; 49:411-21; PMID:23395271; http://dx.doi.org/ 10.1016/j.molcel.2013.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol 2012; 19:560-7; PMID:22664983; http://dx.doi.org/ 10.1038/nsmb.2313 [DOI] [PubMed] [Google Scholar]
- [5].Basu A, Samanta D, Bhattacharya A, Das A, Das D, Dasgupta C. Protein folding following synthesis in vitro and in vivo: association of newly synthesized protein with 50S subunit of E. coli ribosome. Biochem Biophys Res Commun 2008; 366:592-7; http://dx.doi.org/ 10.1016/j.bbrc.2007.11.142 [DOI] [PubMed] [Google Scholar]
- [6].Blondel M, Soubigou F, Evrard J, Nguyen PH, Hasin N, Chedin S, Gillet R, Contesse MA, Friocourt G, Stahl G, et al.. Protein Folding Activity of the Ribosome is involved in Yeast Prion Propagation. Sci Rep 2016; 6:32117; PMID:27633137; http://dx.doi.org/ 10.1038/srep32117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chattopadhyay S, Pal S, Pal D, Sarkar D, Chandra S, Das Gupta C. Protein folding in Escherichia coli: role of 23S ribosomal RNA. Biochim Biophys Acta 1999; 1429:293-8; PMID:9989214; http://dx.doi.org/ 10.1016/S0167-4838(98)00179-4 [DOI] [PubMed] [Google Scholar]
- [8].Tribouillard-Tanvier D, Dos Reis S, Gug F, Voisset C, Beringue V, Sabate R, Kikovska E, Talarek N, Bach S, Huang C, et al.. Protein folding activity of ribosomal RNA is a selective target of two unrelated antiprion drugs. PLoS One 2008; 3:e2174; PMID:18478094; http://dx.doi.org/ 10.1371/journal.pone.0002174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pang Y, Kurella S, Voisset C, Samanta D, Banerjee D, Schabe A, Das Gupta C, Galons H, Blondel M, Sanyal S. The antiprion compound 6-aminophenanthridine inhibits the protein folding activity of the ribosome by direct competition. J Biol Chem 2013; 288:19081-9; PMID:23673663; http://dx.doi.org/ 10.1074/jbc.M113.466748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Samanta D, Mukhopadhyay D, Chowdhury S, Ghosh J, Pal S, Basu A, Bhattacharya A, Das A, Das D, DasGupta C. Protein folding by domain V of Escherichia coli 23S rRNA: specificity of RNA-protein interactions. J Bacteriol 2008; 190:3344-52; PMID:18310328; http://dx.doi.org/ 10.1128/JB.01800-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pal S, Chandra S, Chowdhury S, Sarkar D, Ghosh AN, Gupta CD. Complementary role of two fragments of domain V of 23 S ribosomal RNA in protein folding. J Biol Chem 1999; 274:32771-7; PMID:10551837; http://dx.doi.org/ 10.1074/jbc.274.46.32771 [DOI] [PubMed] [Google Scholar]
- [12].Mondal S, Pathak BK, Ray S, Barat C. Impact of P-Site tRNA and antibiotics on ribosome mediated protein folding: studies using the Escherichia coli ribosome. PLoS One 2014; 9:e101293; PMID:25000563; http://dx.doi.org/ 10.1371/journal.pone.0101293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bach S, Talarek N, Andrieu T, Vierfond JM, Mettey Y, Galons H, Dormont D, Meijer L, Cullin C, Blondel M. Isolation of drugs active against mammalian prions using a yeast-based screening assay. Nat Biotechnol 2003; 21:1075-81; PMID:12910243; http://dx.doi.org/ 10.1038/nbt855 [DOI] [PubMed] [Google Scholar]
- [14].Tribouillard-Tanvier D, Beringue V, Desban N, Gug F, Bach S, Voisset C, Galons H, Laude H, Vilette D, Blondel M. Antihypertensive drug guanabenz is active in vivo against both yeast and mammalian prions. PLoS One 2008; 3:e1981; PMID:18431471; http://dx.doi.org/ 10.1371/journal.pone.0001981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Oumata N, Nguyen PH, Beringue V, Soubigou F, Pang Y, Desban N, Massacrier C, Morel Y, Paturel C, Contesse MA, et al.. The toll-like receptor agonist imiquimod is active against prions. PLoS One 2013; 8:e72112; PMID:23977222; http://dx.doi.org/ 10.1371/journal.pone.0072112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jones GW, Tuite MF. Chaperoning prions: the cellular machinery for propagating an infectious protein? Bioessays 2005; 27:823-32; PMID:16015602; http://dx.doi.org/ 10.1002/bies.20267 [DOI] [PubMed] [Google Scholar]
- [17].Gloge F, Becker AH, Kramer G, Bukau B. Co-translational mechanisms of protein maturation. Curr Opin Struct Biol 2014; 24:24-33; PMID:24721450; http://dx.doi.org/ 10.1016/j.sbi.2013.11.004 [DOI] [PubMed] [Google Scholar]
- [18].Atkins JF, Gesteland RF, Cech TR (eds.). The RNA worlds: From life's origins to diversity in gene regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2011. [Google Scholar]
- [19].Jeffares DC, Poole AM, Penny D. Relics from the RNA world. J Mol Evol 1998; 46:18-36; PMID: 9419222; http://dx.doi.org/ 10.1007/PL00006280 [DOI] [PubMed] [Google Scholar]
- [20].Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science 2000; 289:920-30; PMID: 10937990; http://dx.doi.org/ 10.1126/science.289.5481.920 [DOI] [PubMed] [Google Scholar]
- [21].Krupkin M, Matzov D, Tang H, Metz M, Kalaora R, Belousoff MJ, Zimmerman E, Bashan A, Yonath A. A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philos Trans R Soc Lond B Biol Sci 2011; 366:2972-8; PMID:21930590; http://dx.doi.org/ 10.1098/rstb.2011.0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Reis SD, Pang Y, Vishnu N, Voisset C, Galons H, Blondel M, Sanyal S. Mode of action of the antiprion drugs 6AP and GA on ribosome assisted protein folding. Biochimie 2011; 93:1047-54; PMID:21396977; http://dx.doi.org/ 10.1016/j.biochi.2011.03.002 [DOI] [PubMed] [Google Scholar]
- [23].Mace K, Gillet R. Origins of tmRNA: the missing link in the birth of protein synthesis? Nucleic Acids Res 2016; 44:8041-51; PMID:27484476; http://dx.doi.org/ 10.1093/nar/gkw693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Harish A, Caetano-Anolles G. Ribosomal history reveals origins of modern protein synthesis. PLoS One 2012; 7:e32776; PMID:22427882; http://dx.doi.org/ 10.1371/journal.pone.0032776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Docter BE, Horowitz S, Gray MJ, Jakob U, Bardwell JC. Do nucleic acids moonlight as molecular chaperones? Nucleic Acids Res 2016; 44:4835-45; PMID:27105849; http://dx.doi.org/ 10.1093/nar/gkw291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Reidy M, Masison DC. Yeast prions help identify and define chaperone interaction networks. Curr Pharm Biotechnol 2014; 15:1008-18; PMID:25373385; http://dx.doi.org/ 10.2174/1389201015666141103021035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hines JK, Li X, Du Z, Higurashi T, Li L, Craig EA. [SWI], the prion formed by the chromatin remodeling factor Swi1, is highly sensitive to alterations in Hsp70 chaperone system activity. PLoS Genet 2011; 7:e1001309; PMID:21379326; http://dx.doi.org/ 10.1371/journal.pgen.1001309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol 2002; 22:3590-8; PMID:11997496; http://dx.doi.org/ 10.1128/MCB.22.11.3590-3598.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol 1999; 19:8103-12; PMID:10567536; http://dx.doi.org/ 10.1128/MCB.19.12.8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kiktev DA, Melomed MM, Lu CD, Newnam GP, Chernoff YO. Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol Microbiol 2015; 96:621-32; PMID:25649498; http://dx.doi.org/ 10.1111/mmi.12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Linden R, Martins VR, Prado MA, Cammarota M, Izquierdo I, Brentani RR. Physiology of the prion protein. Physiol Rev 2008; 88:673-728; PMID: 18391177; http://dx.doi.org/ 10.1152/physrev.00007.2007 [DOI] [PubMed] [Google Scholar]
- [32].Goold R, Rabbanian S, Sutton L, Andre R, Arora P, Moonga J, Clarke AR, Schiavo G, Jat P, Collinge J, et al.. Rapid cell-surface prion protein conversion revealed using a novel cell system. Nat Commun 2011; 2:281; PMID:21505437; http://dx.doi.org/ 10.1038/ncomms1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Paquet S, Sabuncu E, Delaunay JL, Laude H, Vilette D. Prion infection of epithelial Rov cells is a polarized event. J Virol 2004; 78:7148-52; PMID:15194791; http://dx.doi.org/ 10.1128/JVI.78.13.7148-7152.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Godsave SF, Wille H, Kujala P, Latawiec D, DeArmond SJ, Serban A, Prusiner SB, Peters PJ. Cryo-immunogold electron microscopy for prions: toward identification of a conversion site. J Neurosci 2008; 28:12489-99; PMID:19020041; http://dx.doi.org/ 10.1523/JNEUROSCI.4474-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Marijanovic Z, Caputo A, Campana V, Zurzolo C. Identification of an intracellular site of prion conversion. PLoS Pathog 2009; 5:e1000426; PMID:19424437; http://dx.doi.org/ 10.1371/journal.ppat.1000426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yim YI, Park BC, Yadavalli R, Zhao X, Eisenberg E, Greene LE. The multivesicular body is the major internal site of prion conversion. J Cell Sci 2015; 128:1434-43; PMID:25663703; http://dx.doi.org/ 10.1242/jcs.165472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Miesbauer M, Rambold AS, Winklhofer KF, Tatzelt J. Targeting of the prion protein to the cytosol: mechanisms and consequences. Curr Issues Mol Biol 2010; 12:109-18; PMID:19767654 [PubMed] [Google Scholar]
- [38].Hegde RS, Tremblay P, Groth D, DeArmond SJ, Prusiner SB, Lingappa VR. Transmissible and genetic prion diseases share a common pathway of neurodegeneration. Nature 1999; 402:822-6; PMID:10617204; http://dx.doi.org/ 10.1038/45574 [DOI] [PubMed] [Google Scholar]
- [39].Martin-Lanneree S, Hirsch TZ, Hernandez-Rapp J, Halliez S, Vilotte JL, Launay JM, Mouillet-Richard S. PrP(C) from stem cells to cancer. Front Cell Dev Biol 2014; 2:55; PMID:25364760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Martins VR, Beraldo FH, Hajj GN, Lopes MH, Lee KS, Prado MA, Linden R. Prion protein: orchestrating neurotrophic activities. Curr Issues Mol Biol 2010; 12:63-86; PMID:19767651 [PubMed] [Google Scholar]
- [41].Halliez S, Passet B, Martin-Lanneree S, Hernandez-Rapp J, Laude H, Mouillet-Richard S, Vilotte JL, Béringue V. To develop with or without the prion protein. Front Cell Dev Biol 2014; 2:58; PMID:25364763; http://dx.doi.org/ 10.3389/fcell.2014.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bravard A, Auvre F, Fantini D, Bernardino-Sgherri J, Sissoeff L, Daynac M, Xu Z, Etienne O, Dehen C, Comoy E, et al.. The prion protein is critical for DNA repair and cell survival after genotoxic stress. Nucleic Acids Res 2015; 43:904-16; PMID:25539913; http://dx.doi.org/ 10.1093/nar/gku1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Smith HL, Mallucci GR. The unfolded protein response: mechanisms and therapy of neurodegeneration. Brain 2016; 139:2113-21; PMID:27190028; http://dx.doi.org/ 10.1093/brain/aww101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mays CE, Kim C, Haldiman T, van der Merwe J, Lau A, Yang J, Grams J, Di Bari MA, Nonno R, Telling GC, et al.. Prion disease tempo determined by host-dependent substrate reduction. J Clin Invest 2014; 124:847-58; PMID:24430187; http://dx.doi.org/ 10.1172/JCI72241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Brundin P, Melki R, Kopito R. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol 2010; 11:301-7; PMID:20308987; http://dx.doi.org/ 10.1038/nrm2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci 2010; 11:155-9; PMID:20029438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Goedert M, Clavaguera F, Tolnay M. The propagation of prion-like protein inclusions in neurodegenerative diseases. Trends Neurosci 2010; 33:317-25; PMID:20493564; http://dx.doi.org/ 10.1016/j.tins.2010.04.003 [DOI] [PubMed] [Google Scholar]
- [48].Barbezier N, Chartier A, Bidet Y, Buttstedt A, Voisset C, Galons H, Blondel M, Schwarz E, Simonelig M. Antiprion drugs 6-aminophenanthridine and guanabenz reduce PABPN1 toxicity and aggregation in oculopharyngeal muscular dystrophy. EMBO Mol Med 2011; 3:35-49; PMID:21204267; http://dx.doi.org/ 10.1002/emmm.201000109 [DOI] [PMC free article] [PubMed] [Google Scholar]


