ABSTRACT
A pair of Arabidopsis thaliana resistance proteins, RPS4 and RRS1, recognizes the cognate Avr effector from the bacterial pathogens Pseudomonas syringae pv. tomato expressing avrRps4 (Pst-avrRps4), Ralstonia solanacearum, and the fungal pathogen Colletotrichum higginsianum and leads to defense signaling activation against the pathogens. In the present study, we analyzed 14 A. thaliana accessions for natural variation in Pst-avrRps4 and C. higginsianum susceptibility, and found new compatible and incompatible Arabidopsis–pathogen interactions. We first found that A. thaliana accession Cvi-0 is susceptible to Pst-avrRps4. Interestingly, the genome sequence assembly indicated that Cvi-0 lost both RPS4 and RRS1, but not RPS4B and RRS1B, compared to the reference genome sequence from A. thaliana accession Col-0. On the other hand, the natural variation analysis of RPS4 alleles from various Arabidopsis accessions revealed that one amino-acid change, Y950H, is responsible for the loss of resistance to Pst-avrRps4 and C. higginsianum in RLD-0. Our data indicate that the amino acid change, Y950H, in RPS4 resulted in the loss of both RPS4 and RRS1 functions and resistance to pathogens.
KEYWORDS: Arabidopsis, colletotrichum higginsianum, pseudomonas syringae, R gene, RPS4, RRS1
Introduction
Plant disease resistance, known as gene-for-gene relationship, requires a resistance (R) gene in the host plant and a cognate avirulence (Avr) gene in the insect, pest, or pathogen.1 The R-gene product detects the corresponding Avr gene product and initiates signal transduction to confer resistance.
Many R-genes encode NB-LRR proteins, also known as NLRs, which consist of a central NB-ARC domain (a nucleotide-binding adaptor shared with Apaf-1, plant resistance proteins, and CED-4) and a C-terminal leucine-rich repeat (LRR) domain. NLRs possess either a Toll/interleukin 1 receptor (TIR) domain or a coiled-coil (CC) domain in their N-terminal structures.2
In most cases, a single plant NLR recognizes the cognate Avr effector and results in the activation of defense signaling against the pathogen.3,4 Moreover, some pairs of NLRs are reported to be required for both recognizing the cognate Avr effectors and conferring resistance to pathogens.5 Our recent studies showed that a pair of Arabidopsis thaliana NLRs, RPS4 (Resistance to Pseudomonas syringae 4) and RRS1 (Resistance to Ralstonia solanacearum 1), mediate recognition of multiple pathogens, such as the fungal pathogen Colletotrichum higginsianum (anthracnose) and bacterial pathogens Pseudomonas syringae pv. tomato expressing avrRps4 (Pst-avrRps4; bacterial speck) and Ralstonia solanacearum (bacterial wilt).6 In addition, Debieu et al.7 recently reported that RPS4/RRS1 pair was required to confer resistance to Xanthomonas campestris pv. campestris (black rot). Thus, the paired NLRs are required for recognition of at least three bacterial Avr effectors, AvrRps4 from Pst-avrRps4, PopP2 from R. solanacearum, and a putative bacterial Avr effector from X. campestris pv. campestris, and a putative fungal Avr effector from C. higginsianum.6,8-10 Our previous studies also showed that introduction of the NLR gene pair, RPS4 and RRS1, into several crops provided disease resistance to different classes of pathogens.11,12
The genes, RPS4 and RRS1, constituting a pair are localized near each other and are encoded in opposite directions. Although both RPS4 and RRS1 are TIR-type NLRs, RRS1 contains a leucine zipper (LZ) motif and a WRKY domain at the C-terminus. The paired NLRs interact with each other physically to form a hetero-complex.6,13-14
RPS4 and RRS1 play different roles in effector-triggered immunity in plants. According to the “integrated decoy” model, acetylation of the C-terminal WRKY domain of RRS1 protein triggers activation of NLR complex and thus, initiates the immune response.15-16 On the other hand, the precise mechanism of how RPS4 interacts with acetylated RRS1 and how RPS4/RRS1 complex triggers defense activation is unclear.
A previous analysis of natural variation in RPS4 alleles from Col-0, Ws-2, Ler, and RLD-0 accessions revealed that two amino-acid changes, N195D and Y950H, might be responsible for the loss of resistance to Pst-avrRps4 in RLD.8,17 The objectives of this study were to investigate whether: 1) the above-mentioned amino acid changes also cause susceptibility to C. higginsianum in RLD-0 and Ws-2 background, 2) these amino acid polymorphisms account for the non-functionality of RPS4-RLD, and 3) specific amino acid polymorphisms in RPS4 play a role in the RPS4/RRS1 complex.
Results
Natural variation in the susceptibility to P. syringae pv. tomato strain DC3000 expressing avrRps4 among A. thaliana accessions
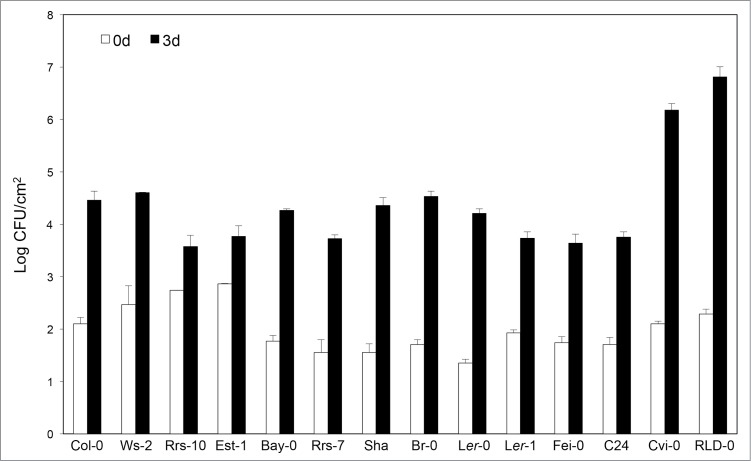
To investigate natural variations in Pst-avrRps4 susceptibility, we tested 14 A. thaliana accessions (Col-0, Ws-2, Ler-0, Ler-1, Rrs-7, Rrs-10, Est-1, Bay-0, Sha, Br-0, Fei-0, C24, Cvi-0, and RLD-0). We evaluated the disease reactions of A. thaliana accessions to Pst-avrRps4 based on certain aspects, such as the assays of bacterial growth by colony formation in plate culture of samples from infected leaves. Most of the interactions observed with these accessions were incompatible (Fig. 1). However, a compatible phenotype was found following the inoculation of accessions RLD-0 and Cvi-0 (Fig. 1). A. thaliana plants that appeared to be susceptible, developed chlorotic lesions at the inoculation sites, 3–4 day post inoculation (dpi), which expanded further. It is the first report that Cvi-0 is compatible to Pst-avrRps4.
Figure 1.
Growth of Pst-avrRps4 in Arabidopsis thaliana accessions after inoculation with Pst-avrRps4. Leaves of 5-week-old plants were infiltrated with bacterial suspensions (5 × 104 cfu ml−1). The leaves were harvested at 0 (white columns) and 3 days (black columns) after inoculation. Bacterial growth (cfu cm−2) was assessed using five leaf disks by cell counting. Bars indicate SE. This experiment was repeated three times with similar results.
Comparison of the nucleotide sequences of RPS4 and RRS1 alleles between Cvi-0 and Col-0
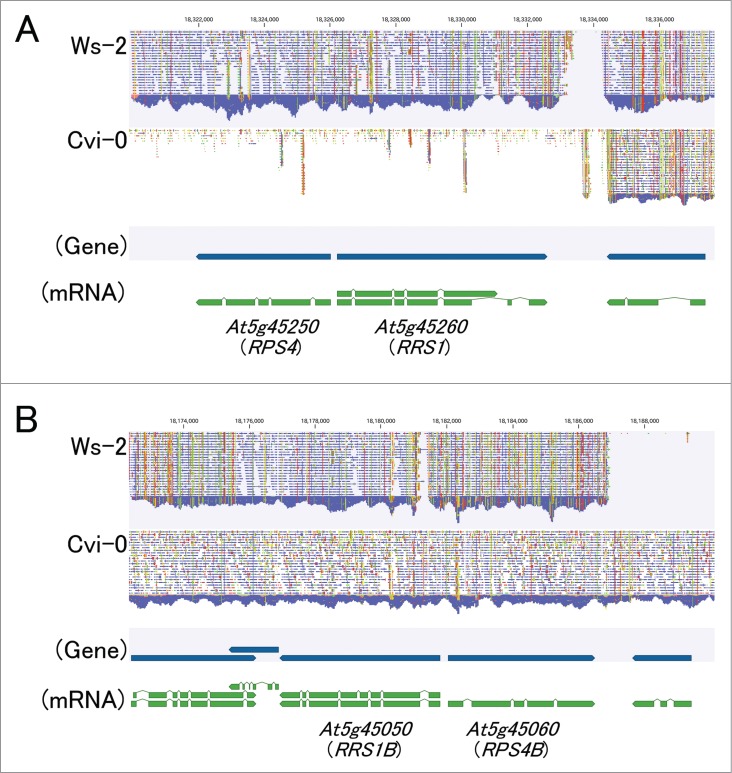
The evolutionary conservation of RPS4/RRS1 gene pair localized near each other in a head to head arrangement indicates their cooperative function in disease resistance. To understand whether the putative RPS4-Cvi and RRS1-Cvi are responsible for susceptibility to C. higginsianum and Pst-avrRps4, we analyzed DNA sequences of the RPS4-Cvi and RRS1-Cvi alleles. Surprisingly, these genes from Arabidopsis 1001 genome project18 could not be assembled against the corresponding region of Col-0 reference sequence. Although Clark et al.19 reported single-nucleotide polymorphisms (SNPs) in 20 wild accessions of A. thaliana using high-density oligonucleotide arrays containing putative RPS4-Cvi and RRS1-Cvi alleles, the data from Arabidopsis 1001 genome project indicate that the putative RPS4-Cvi and RRS1-Cvi genes are less homologous to the corresponding regions of other Arabidopsis RPS4/RRS1 genes (Fig. 2A). On the other hand, Saucet et al.20 reported that RPS4B (At5g45060)/RRS1B (At5g45050) is paralogous and functionally similar to RPS4/RRS1. RPS4B and RRS1B recognize AvrRps4 but not PopP2. Therefore, we analyzed DNA sequences of RPS4B-Cvi and RRS1B-Cvi alleles. These genes from Arabidopsis 1001 genome project assembled against the corresponding region of Col-0 reference sequence (Fig. 2B).
Figure 2.
A comparison of the nucleotide sequences of the RPS4/RRS1 and RPS4B/RRS1B alleles between Cvi-0 and Col-0. RPS4/RRS1 (A) and RPS4B/RRS1B (B) from genome sequence data sets for Arabidopsis thaliana accession Ws-2 (SRR492407) and Cvi-0 (SRR492239) were compared with the corresponding region of Col-0 reference sequence.
Responses of A. thaliana accession RLD-0 to inoculation with C. higginsianum
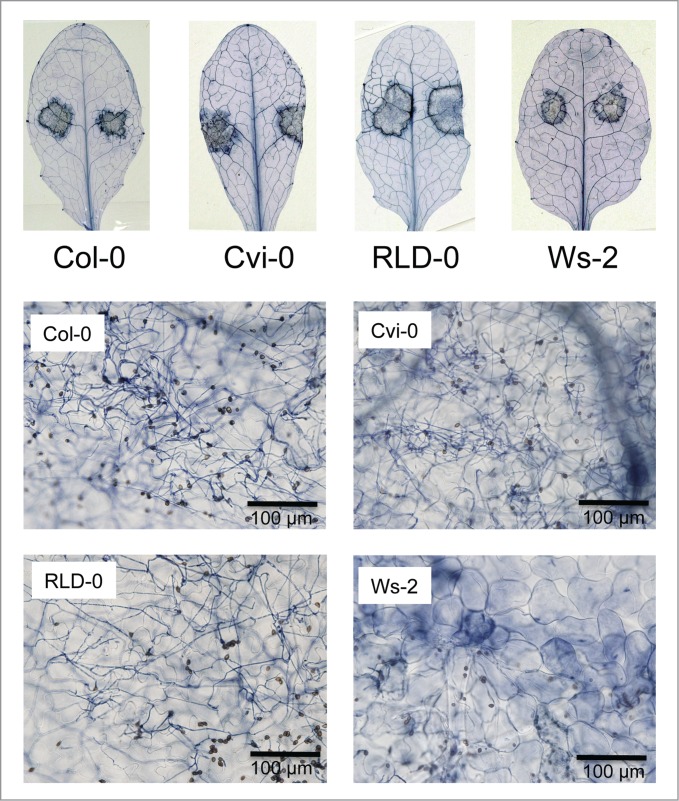
In this study, A. thaliana accession RLD-0 and Cvi-0 appeared to be compatible to C. higginsianum. Light microscopy revealed that infection hyphae developed in the invaded epidermal cells of the susceptible accession Cvi-0 and RLD-0 but not in Ws-2 (Fig. 3).
Figure 3.
Infection phenotypes of leaves inoculated with C. higginsianum. Mature leaves of 28-day-old plants were inoculated by placing 5 µl of spore suspension of C. higginsianum (5 × 105 spores ml−1) on each side of the leaf. The leaves were harvested at 6 dpi and stained with trypan blue. Each picture shows a representative of three independent experiments.
Susceptibility to C. higginsianum and Pst avrRps4: Which is responsible—RPS4-RLD or RRS1-RLD ?
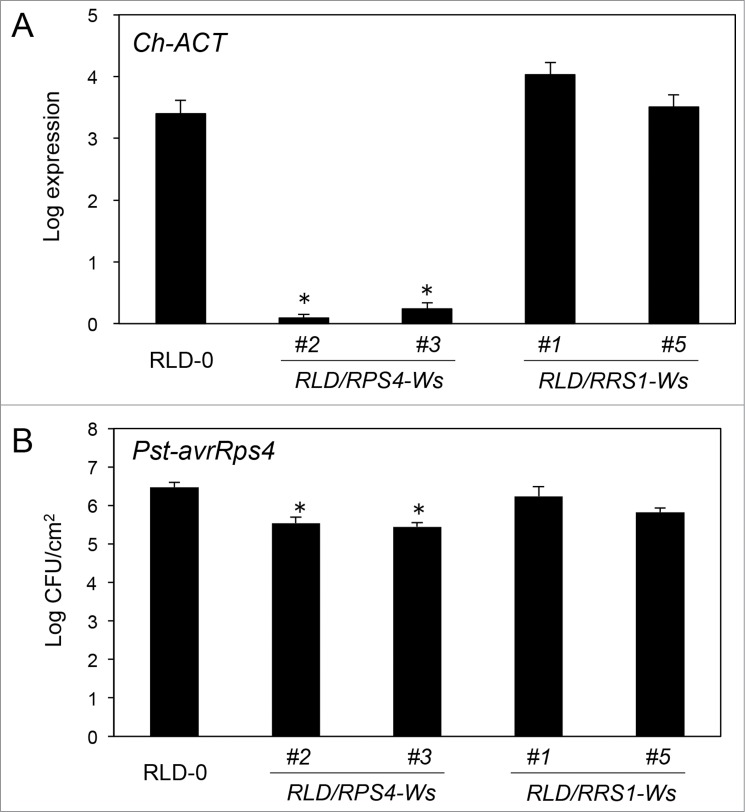
To confirm whether RPS4-RLD or RRS1-RLD is responsible for susceptibility to C. higginsianum and Pst-avrRps4, we introduced either 6.3-kbp genomic RRS1-Ws fragments, including approximately 1.7-kbp upstream and 176-bp downstream regions (Ws-2 background),6 or the 6-kbp genomic RPS4-Ws fragments, including approximately 2-kbp upstream and 109-bp downstream regions (Ws-2 background)6 into the susceptible accession RLD-0. RPS4-Ws transgenic RLD-0 plants conferred resistance to C. higginsianum and Pst-avrRps4, but RRS1-Ws transgenic RLD-0 plants were susceptible to the pathogen (Fig. 4). We concluded that RPS4-RLD is responsible for resistance to C. higginsianum and Pst-avrRps4.
Figure 4.
C. higginsianum and Pst-avrRps4 resistance analysis in the transgenic lines. RLD/RPS4-Ws-#2 and -#3 lines represent independent transgenic RLD-0 plants harboring the genomic RPS4-Ws fragment. RLD/RRS1-Ws-#1 and -#5 lines represent independent RLD-0 transgenic plants harboring the genomic RRS1-Ws fragment. (A) Quantification of C. higginsianum in planta by qRT-PCR. Twenty eight-day-old plants were spray-inoculated with C. higginsianum. The inoculated leaves were harvested at 5 dpi and total RNA was isolated. QRT-PCR was performed with Ch-ACT primers for each sample. (B) Quantification of Pst-avrRps4 in planta. Leaves of 5-week-old plants were infiltrated with bacterial suspensions (5 × 104 cfu ml−1). The leaves were harvested 3 days after inoculation. Bacterial growth (cfu cm−2) was assessed using five leaf disks by cell counting. Bars indicate SE. The asterisks indicate statistical significance from the RLD-0 WT controls (Dunnett's method,28 P < 0.01). This experiment was repeated at least two times with similar results.
Complementation of mutated RPS4 (N195D or Y950H) to rps4-21 mutants
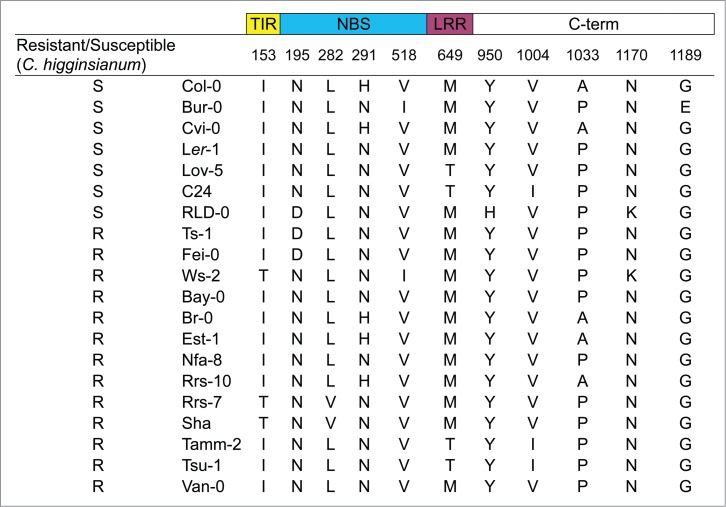
Amino acid sequence variations in RPS4 proteins among 20 Arabidopsis accessions are shown in Fig. 5. Only one amino-acid substitution, Y950H, observed in RPS4-RLD is not shared with RPS4 from any of the other accessions. In addition, Gassmann et al. reported that N195D and/or Y950H were responsible for the loss of resistance to Pst-avrRps4 in RLD-08,17. Therefore, the mutated RPS4-Ws clones (Ws-2 background) with the amino acid changes, N195D and Y950H, were designated as RPS4-WsN195D and RPS4-WsY950H, respectively. Subsequently, these clones were complemented into rps4-21 mutants (Ws-2 background).6
Figure 5.
Amino acid sequence variations in RPS4 proteins among 20 Arabidopsis accessions. Only amino acid differences from 20 Arabidopsis accessions are shown, without identical residues. Resistance and susceptibility to C. higginsianum are indicated by ‘R’ and ‘S’, respectively.
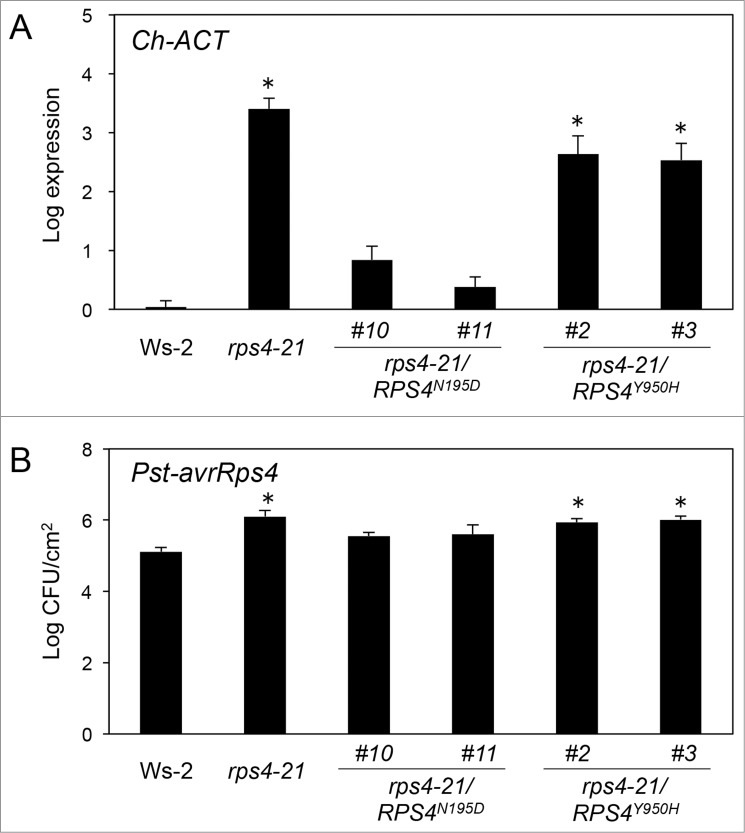
To determine whether N195 and Y950 are required for resistance to C. higginsianum, fungal infection levels were monitored in Ws-2 and in the mutants, 5 days post-inoculation (Fig. 6A). The RPS4-WsY950H complemented rps4-21 plants were susceptible to C. higginsianum. On the contrary, RPS4-WsN195D complemented rps4-21 plants were resistant to C. higginsianum.
Figure 6.
Complementation of mutated RPS4 (N195D or Y950H) to Arabidopsis rps4-21 mutants. The rps4-21/RPS4-WsN195D -#10, and -#11, and rps4-21/RPS4-WsY950H -#2, and -#3 lines represent independent transgenic rps4-21 plants harboring the genomic RPS4-WsN195D and RPS4-WsY950H fragments, respectively. (A) Quantification of C. higginsianum in planta by qRT-PCR. Twenty eight-day-old plants were spray-inoculated with C. higginsianum. The inoculated leaves were harvested at 5 dpi and total RNA was isolated. QRT-PCR was performed with Ch-ACT primers for each sample. (B) Quantification of Pst-avrRps4 in planta. The leaves of 5-week-old plants were infiltrated with bacterial suspensions (5 × 104 cfu ml−1). The leaves were harvested at 3 days after inoculation. Bacterial growth (cfu cm−2) was assessed using five leaf disks by cell counting. Bars indicate SE. The asterisks indicate statistical significance from the Ws-2 WT controls (Dunnett's method,28 P < 0.01). This experiment was repeated at least two times with similar results.
To determine whether N195 and Y950 are required for resistance to Pst-avrRps4, the bacterial infection levels were monitored in Ws-2 and in the mutants, 3 days post-inoculation (Fig. 6B). Pst-avrRps4 count was about three- or eight-times higher in RPS4-WsN195D or RPS4-WsY950H complemented rps4-21 plants, respectively, than in wild-type Ws-2.
Discussion
In the present study, we analyzed 14 A. thaliana accessions for understanding the natural variation in C. higginsianum and Pst-avrRps4 susceptibility, and found new compatible and incompatible Arabidopsis-pathogens interactions. We first found that A. thaliana accession Cvi-0 is susceptible to Pst-avrRps4. RPS4 and RRS1 together recognize bacterial effector AvrRps4 in Pst-avrRps4 and subsequently induce resistance to the pathogen. However, the genome sequence assembly indicated that Cvi-0 lacks both RPS4 and RRS1; however, RPS4B and RRS1B, which are closely linked to RPS4 and RRS1, respectively are present in the genome. It suggests that RRS1B and RPS4B in Cvi-0 might recognize AvrRps4 from Pst-avrRps4, as susceptibility to Pst-avrRps4 in Cvi-0 was slightly lower than that in RLD-0, which is super susceptible to the pathogen. Clark et al.19 reported that most of the NLR genes harbor at least one ‘major-effect change’, i.e. SNP with large-effects on gene integrity and/or polymorphic region prediction. One hypothesis is that the major-effect change arose in putative RPS4-Cvi and RRS1-Cvi alleles resulting in trade-offs between plant growth and defense against the pathogen.21 A. thaliana Cvi-0 accession might evolve in an environment without exposure to the pathogens that could be recognized by the RPS4 and RRS1 pair. As we do not understand which of the two R-gene pairs, RPS4/RRS1 or RPS4B/RRS1B, is an ancestor, it is also interesting for understanding the branching process in the evolution of RRS4 and RRS1 among the A. thaliana accessions.
Previously, Gassmann et al.8 also reported that RLD-0 was susceptible to Pst-avrRps4 and Col-0 showed resistance against the pathogen. A previous study indicated that the two amino-acid changes, N195D and Y950H, in RLD-0 are sufficient for the non-functionality of RPS4-RLD and for the loss of resistance to Pst-avrRps417. Similarly, we found that RLD-0 accession is also susceptible to C. higginsianum. The C. higginsianum strain MAFF305635 causes anthracnose disease symptoms in Col-0 and Cvi-0 plants.6,22 In contrast, incompatible accession, Ws-2 (moderate resistance), formed restricted brown necrotic lesions at the inoculation sites, which did not expand.6,22 Moreover, we also found that two accessions, Fei-0 and Ts-1, which contain RPS4 with N195D but not Y950H, are resistant to C. higginsianum. In addition, Fei-0 showed resistance to Pst-avrRps4. The analyses of natural variation showed that one amino acid change, Y950H, in RLD-0 is unique among all the accessions used here. As the RPS4-WsY950H complemented rps4-21 plants were susceptible to Pst-avrRps4, Y950H in RPS4 is likely to be responsible for susceptibility to the pathogen and also possibly to Pst-avrRps4 in RLD-0.
We also showed that RPS4-WsY950H transgenic plants were susceptible to C. higginsianum; therefore, one amino acid change compromised the RPS4 function. Y950 is located in the C-terminal end after LRR; the consequence of the amino acid change on the protein activity and stability is unknown. It is known that proteins can bind to one another by using phosphorylated tyrosines.23,24 The replacement of Y950 may cause an unstable LRR domain structure. Our recent report showed that the C-termini of RPS4 and RRS1 are responsible for resistance signaling against C. higginsianum.13 Therefore, the C-terminal region of RPS4, containing Y950, likely plays an important role in the activation of RPS4/RRS1-dependent defense responses. On the other hand, Saucet et al.20 suggested that A. thaliana accession RLD-0 lacks function of both RPS4/RRS1 and RPS4B/RRS1B. In this study, we showed that RPS4-Ws transferred RLD-0 plants, containing nonfunctional RPS4B/RRS1B, were resistant to the pathogens and RPS4-WsY950H complemented rps4-21 plants (Ws-2 background, containing functional RPS4B/RRS1B) were compatible to the pathogen. Therefore, some structural domains in RPS4 and RRS1 are essential for disease resistance and contribute to the interaction of RPS4 with RRS1. On the other hand, we found that a pair of RRS1B and RPS4B in RLD-0 and Cvi-0 was not required for resistance to C. higginsianum.
In conclusion, our data indicated that the amino acid change, Y950H, in RPS4 resulted in the loss of both the RPS4 and RRS1 functions and resistance to pathogens. Further experiments will be required to address the evolutionary genetic and functional studies in RPS4 and RRS1 pair using Cvi-0 as an important tool.
Material and methods
Plant materials and growth
Arabidopsis ecotypes Bay-0, Br-0, Bur-0, C24, Cvi-0, Est-1, Fei-0, Ler-1, Lov-5, Nfa-8, Rrs-7, Rrs-10, Sha, Tamm-2, Ts-1, Tsu-1, and Van-0 were obtained from the Arabidopsis Biological Resource Center (ABRC; USA). Col-0, Ler-0, RLD-0, and Ws-2 were obtained from the RIKEN BRC, Japan. The rps4-21 mutant has been described previously.6 Arabidopsis plants were grown in soil mix (Sakata Seed Corp.) and expanded vermiculite (2–5 mm granules) at a 1:1 ratio for 28 days in a growth chamber at 22°C under a 12-h light/12-h dark cycle.
Genome sequence
Genome sequence data sets for Arabidopsis thaliana accession Ws-2 (SRR492407) and Cvi-0 (SRR492239) were downloaded from the sequence read archive of DDBJ (DNA Data Bank of Japan, https://trace.ddbj.nig.ac.jp/index_e.htm). These sequence data, originally open to public from the 1,001 Arabidopsis Genomes project, were contributed by the Salk Institute (http://1001genomes.org/, http://signal.salk.edu/atg1001/accessions.php). The reference genome sequence and gene annotation data of A. thaliana (TAIR10) were obtained using the download function at the sequence analysis software, CLC genomic workbench (QIAGEN Bioinformatics). These genome sequence data from Ws-2 and Cvi-0 were independently mapped to the reference genome sequence of Arabidopsis thaliana by the CLC genomic workbench.
Pst-avrRps4 infections
Arabidopsis plants were inoculated as described previously.6 The quantification of Pst-avrRps4 was performed as described previously.6
C. higginsianum inoculation
C. higginsianum Saccardo isolates (MAFF305635) were obtained from the Ministry of Agriculture, Forestry and Fisheries (MAFF) Genebank, Japan. The Arabidopsis plants were inoculated as described previously,6,25 and harvested at 5 dpi for qRT-PCR analysis. The quantification of C. higginsianum was performed as described previously.25 Fungal hyphae within the resulting lesions and dead cells were stained with lactophenol-trypan blue, as described previously.26
Construction of the R-gene plasmid
All the DNA fragments containing RRS1 and/or RPS4 used in this study were derived from the genome of A. thaliana Ws-2 accession. The plasmids used in this study have been described in a previous study.6 All the clones were verified by DNA sequencing. The 6.3-kbp genomic RPS4 fragment, including approximately 2.1-kbp upstream and 109-bp downstream regions, was cloned into pBI101-SK+.6 Site-directed mutagenesis of genomic RPS4 was performed by a custom cloning service (Takara Bio Inc.) to generate RPS4-WsN195D and RPS4-WsY950H, carrying Asn to Asp at 195 a.a. and Tyr to His at 950 a.a. mutations, respectively.
Arabidopsis transformation
Arabidopsis transformation was carried out by the floral inoculation method using Agrobacterium tumefaciens strain GV3101 (pMP90).27 T3 homozygous lines were used for further analysis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Yukiko Kurosaki, Yasuyo Katayama, Masami Miyamoto, Shoko Nieda, Aya Okada of RIBS, and Atsuko Iuchi, Yukie Aso of RIKEN BRC for their excellent technical assistance.
Funding
This work was supported by the Science and Technology Research Promotion Program for the Agriculture, Forestry, Fisheries, and Food industry awarded to Y.N. and by Grant-in-Aid for Scientific Research (KAKENHI) (15K07321 to Y.N. and 16K08152 to M.N.).
References
- 1.Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol 1971; 9:275-96; http://dx.doi.org/ 10.1146/annurev.py.09.090171.001423 [DOI] [Google Scholar]
- 2.Van der Biezen EA, Jones JD. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci 1998; 23(12):454-6; PMID:9868361; http://dx.doi.org/ 10.1016/S0968-0004(98)01311-5 [DOI] [PubMed] [Google Scholar]
- 3.Jones JD, Dangl JL. The plant immune system. Nature 2006; 444(7117):323-9; PMID:17108957; http://dx.doi.org/ 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- 4.Bernoux M, Ellis JG, Dodds PN. New insights in plant immunity signaling activation. Curr Opin Plant Biol 2011; 14(5):512-8; PMID:21723182; http://dx.doi.org/ 10.1016/j.pbi.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eitas TK, Dangl JL. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 2010; 13(4):472-7; PMID:20483655; http://dx.doi.org/ 10.1016/j.pbi.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J 2009; 60(2):218-26; PMID:19519800; http://dx.doi.org/ 10.1111/j.1365-313X.2009.03949.x [DOI] [PubMed] [Google Scholar]
- 7.Debieu M, Huard-Chauveau C, Genissel A, Roux F, Roby D. Quantitative disease resistance to the bacterial pathogen Xanthomonas campestris involves an Arabidopsis immune receptor pair and a gene of unknown function. Mol Plant Pathol 2016; 17(4):510-20; PMID:26212639; http://dx.doi.org/ 10.1111/mpp.12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gassmann W, Hinsch ME, Staskawicz BJ. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J 1999; 20(3):265-77; PMID:10571887; http://dx.doi.org/ 10.1046/j.1365-313X.1999.t01-1-00600.x [DOI] [PubMed] [Google Scholar]
- 9.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A 2003; 100(13):8024-9; PMID:12788974; http://dx.doi.org/ 10.1073/pnas.1230660100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birker D, Heidrich K, Takahara H, Narusaka M, Deslandes L, Narusaka Y, Reymond M, Parker JE, O'Connell R. A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J 2009; 60(4):602-13; PMID:19686535; http://dx.doi.org/ 10.1111/j.1365-313X.2009.03984.x [DOI] [PubMed] [Google Scholar]
- 11.Narusaka M, Kubo Y, Hatakeyama K, Imamura J, Ezura H, Nanasato Y, Tabei Y, Takano Y, Shirasu K, Narusaka Y. Interfamily transfer of dual NB-LRR genes confers resistance to multiple pathogens. PLoS One 2013; 8(2):e55954; PMID:23437080; http://dx.doi.org/ 10.1371/journal.pone.0055954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narusaka M, Hatakeyama K, Shirasu K, Narusaka Y. Arabidopsis dual resistance proteins, both RPS4 and RRS1, are required for resistance to bacterial wilt in transgenic Brassica crops. Plant Signal Behav 2014; 9(7):e29130. PMID:25763492; http://dx.doi.org/ 10.4161/psb.29130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narusaka M, Toyoda K, Shiraishi T, Iuchi S, Takano Y, Shirasu K, Narusaka Y. Leucine zipper motif in RRS1 is crucial for the regulation of Arabidopsis dual resistance protein complex RPS4/RRS1. Sci Rep 2016; 6:18702; PMID:26750751; http://dx.doi.org/ 10.1038/srep18702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, Segonzac C, Ve T, Ma Y, Saucet SB, Ericsson DJ, et al.. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 2014; 344(6181):299-303. PMID:24744375; http://dx.doi.org/ 10.1126/science.1247357 [DOI] [PubMed] [Google Scholar]
- 15.Le Roux C, Huet G, Jauneau A, Camborde L, Trémousaygue D, Kraut A, Zhou B, Levaillant M, Adachi H, Yoshioka H, et al.. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 2015;161(5):1074-88. PMID:26000483; http://dx.doi.org/ 10.1016/j.cell.2015.04.025 [DOI] [PubMed] [Google Scholar]
- 16.Sarris PF, Duxbury Z, Huh SU, Ma Y, Segonzac C, Sklenar J, Derbyshire P, Cevik V, Rallapalli G, Saucet SB, et al.. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 2015; 161(5):1089-100. PMID:26000484; http://dx.doi.org/ 10.1016/j.cell.2015.04.024 [DOI] [PubMed] [Google Scholar]
- 17.Zhang XC, Gassmann W. RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell 2003; 15(10):2333-42; PMID:14523247; http://dx.doi.org/ 10.1105/tpc.013474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weigel D, Mott R. The 1001 genomes project for Arabidopsis thaliana. Genome Biol 2009; 10(5):107; PMID:19519932; http://dx.doi.org/ 10.1186/gb-2009-10-5-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al.. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 2007; 317(5836):338-42; PMID:17641193; http://dx.doi.org/ 10.1126/science.1138632 [DOI] [PubMed] [Google Scholar]
- 20.Saucet SB, Ma Y, Sarris PF, Furzer OJ, Sohn KH, Jones JD. Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat Commun 2015; 6:6338; PMID:25744164; http://dx.doi.org/ 10.1038/ncomms7338 [DOI] [PubMed] [Google Scholar]
- 21.Huot B, Yao J, Montgomery BL, He SY. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant 2014; 7(8):1267-87; PMID:24777989; http://dx.doi.org/ 10.1093/mp/ssu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narusaka Y, Narusaka M, Park P, Kubo Y, Hirayama T, Seki M, Shiraishi T, Ishida J, Nakashima M, Enju A, et al.. RCH1, a locus in Arabidopsis that confers resistance to the hemibiotrophic fungal pathogen Colletotrichum higginsianum. Mol Plant Microbe Interact 2004; 17(7):749-62; PMID:15242169; http://dx.doi.org/ 10.1094/MPMI.2004.17.7.749 [DOI] [PubMed] [Google Scholar]
- 23.Yaffe MB. Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol 2002; 3(3):177-86; PMID:11994738; http://dx.doi.org/ 10.1038/nrm759 [DOI] [PubMed] [Google Scholar]
- 24.Grossmann A, Benlasfer N, Birth P, Hegele A, Wachsmuth F, Apelt L, Stelzl U. Phospho-tyrosine dependent protein-protein interaction network. Mol Syst Biol 2015; 11(3):794; PMID:25814554; http://dx.doi.org/ 10.15252/msb.20145968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narusaka M, Shiraishi T, Iwabuchi M, Narusaka Y. Monitoring fungal viability and development in plants infected with Colletotrichum higginsianum by quantitative reverse transcription-polymerase chain reaction. J Gen Plant Pathol 2010; 76(1):1-6; http://dx.doi.org/ 10.1007/s10327-009-0211-z [DOI] [Google Scholar]
- 26.Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 1997; 9(9):1573-84; PMID:9338960; http://dx.doi.org/ 10.1105/tpc.9.9.1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narusaka M, Shiraishi T, Iwabuchi M, Narusaka Y. The floral inoculating protocol: a simplified Arabidopsis thaliana transformation method modified from floral dipping. Plant Biotech 2010; 27:349-51; http://dx.doi.org/ 10.5511/plantbiotechnology.27.349 [DOI] [Google Scholar]
- 28.Dunnett CW. A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc 1955; 50(272):1096-211; http://dx.doi.org/ 10.1080/01621459.1955.10501294 [DOI] [Google Scholar]