Abstract
The Biomolecular Interaction Network Database (BIND) (http://bind.ca) archives biomolecular interaction, reaction, complex and pathway information. Our aim is to curate the details about molecular interactions that arise from published experimental research and to provide this information, as well as tools to enable data analysis, freely to researchers worldwide. BIND data are curated into a comprehensive machine-readable archive of computable information and provides users with methods to discover interactions and molecular mechanisms. BIND has worked to develop new methods for visualization that amplify the underlying annotation of genes and proteins to facilitate the study of molecular interaction networks. BIND has maintained an open database policy since its inception in 1999. Data growth has proceeded at a tremendous rate, approaching over 100 000 records. New services provided include a new BIND Query and Submission interface, a Standard Object Access Protocol service and the Small Molecule Interaction Database (http://smid.blueprint.org) that allows users to determine probable small molecule binding sites of new sequences and examine conserved binding residues.
INTRODUCTION
In light of the vast scientific resources made available through genomics, the science of deciphering molecular mechanisms is expanding rapidly. Scientists who once hunted for disease genes or sought to distinguish key concepts in evolution are now turning their attention to the details of molecular assembly and mechanism to further understand medicine and the key concepts underlying biology. The Biomolecular Interaction Network Database (BIND) was designed to store complete information about molecular assembly through a database structure in order to archive interactions and reactions arising from biopolymers (protein, RNA and DNA), as well as small molecules, lipids and carbohydrates. Detailed information about molecular mechanism, such as the chemical product(s) of an enzymatic reaction, can be encoded in BIND. The underlying ontology of the BIND database is chemistry, and as such, BIND is capable of storing information about molecular interactions to atomic resolution. The taxonomic scope of BIND is also very broad, such that any organism that has a taxon identifier in the NCBI/EMBL/DDBJ taxonomy can be represented in BIND. One of the long-term goals of BIND is to arrive at a sufficiently complete set of interaction and reaction records for each major model organism such that the underlying computable data can act as feedstock for complete cellular simulations. BIND's short-term goals include making data and analysis tools freely available to the community of researchers who strive to discover new molecular mechanisms and function. The Blueprint Initiative seeks to make BIND a global interaction resource that proactively supports third-party tool developers, model organism databases and bioinformatics researchers to achieve their goals. To that end, Blueprint has secured initial large-scale funding for database curation operations in Toronto (Blueprint North America) and Singapore (Blueprint Asia).
DATA GROWTH
The number of interactions in BIND has increased ∼10-fold since our previous Nucleic Acids Research article (1) by the addition of approximately 80 000 interactions to a current total as of September 2004 of over 100 000 interaction records. Approximately 71% of BIND records arise from high-throughput experiments. There are 58 266 protein–protein interactions and 4225 genetic interactions in BIND. There are also 874 protein–small-molecule interactions in BIND, but it should be noted that we have not yet undertaken any deliberate metabolic pathway annotation, and that small molecules from the Protein Data Bank (PDB) are not counted in this number. A total of 19 348 BIND biopolymer–biopolymer interaction records are derived from the PDB structures with full annotation of atomic contacts, after discarding crystal symmetry artifacts and grouping redundant structure interfaces (2). About half of these data represents biological oligomer interactions. Another 25 857 BIND records are protein–DNA interactions, with 23 865 of these originating from high-throughput chromatin-immunoprecipitation-style transcription-factor binding experiments, representing a very fast growing experimental trend. In total, 31 972 protein sequences, as well as 4560 DNA sequences and 759 RNA sequences are represented in BIND, and all of these records reflect the content of 11 649 unique publications. Organisms represented in BIND include Saccharomyces cerevisiae (48 151 records), Drosophila melanogaster (21 309), Homo sapiens (13 902), Caenorhabditis elegans (5266), Mus musculus (3823), Helicobacter pylori (1470), Bos taurus (1064), human immunodeficiency virus 1 (442), Gallus gallus (318) and Arabidopsis thaliana (180) with over 10 000 BIND records arising from other taxonomies. A total of 901 taxa are represented in BIND. Blueprint's Small Molecule Curation Database, the Molecular Object Database (MOD) has a total of 1450 small molecules fully curated, a database that grows as BIND curators find small molecules to add. Blueprint's Small Molecule Interaction Database (SMID) contains 114 305 small molecule–protein interactions extracted from the PDB records and annotated on 22 215 domains as described by the National Center for Biotechnology Information (NCBI) Conserved Domain Database (CDD) spanning 3806 small molecules from the three-dimensional (3D) structure dataset. Integration of small molecule binding specificity information is a challenge we will be pursuing in the coming year.
DATABASE STRUCTURE
BIND originated from object-relational database architecture in use at the NCBI. BIND was the first database structure to define biomolecular interactions, reactions and pathways in a united schema, and the first of its kind to base its underlying ontology on chemistry, with a US patent awarded (6 745 204) on June 1, 2004 from provisional filing date of February 12, 1999. Particular innovations originally made in BIND (3) have found their way into many other interaction and pathway databases, such as chemical state transitions (4,5) and the ability to represent fragmented or incomplete pathways (6). The unique use of a chemical ontology has allowed BIND to uniquely represent 3D molecular interactions arising from structural studies (2), and allows BIND users to explore this information with the visualization tool Cn3D (7), available from the NCBI. Although originally provided in ASN.1, the BIND dataset has been available in the XML format since 2001 (8), making it the first openly available XML system for interaction and pathway data interchange. Other derivative representations include the HUPO-PSI (9) format and the BioPAX format (www.biopax.org), which are considered as subsets of the BIND schema. BIND is split into non-overlapping divisions according to taxonomic lines, with separate branches for highly represented organisms. The divisions of BIND till date include BIND-Metazoa, BIND-Fungi and the remainder of records in BIND-Taxroot. Additional divisions arise from data extracted from third-party databases, which till date includes BIND-3DBP (2) the 3D biopolymer interaction data from the PDB.
DATABASE CURATION
A variety of approaches to collect information from the literature have been proposed. We have chosen to curate BIND records and make them fulfill documented standards of quality for a wide variety of use-cases. BIND validation and quality assurance programs have been established to ensure a high fidelity of capture of the underlying experimental information. BIND curation is organized into two tracks, low-throughput (LTP) and high-throughput (HTP), where HTP records are defined as papers that have more than 40 interaction results arising from the same experimental design and methodology. LTP BIND curators are selected for M.Sc.- or Ph.D.-level experience in a laboratory setting having carried out interaction research on the bench, and are further trained through the Canadian Bioinformatics Workshops, with course material provided online at www.bioinformatics.ca, and in-house at Blueprint using the BIND Curation Training Manual found at http://www.blueprint.org/bind/bind_documentation.html.
HTP curators are selected for training and experience in bioinformatics programming. HTP curators are responsible for collecting and archiving experimental data, often in the form of supplementary data stored separate from a research publication. HTP curators create scripts to fulfill the curation of BIND records from each publication. The HTP data and scripts to create BIND records are archived in a versioning system so that the database records may be updated.
The large metadata space of BIND spans over 2000 fields representing an extensive space in which to curate experimental information. It is not intended that any one experiment fill this entire metadata space, but rather that an accumulation of experimental evidence will provide a portfolio of information for each molecular interaction and reaction. The methods by which BIND curators find and transcribe experimental data are documented in the BIND Curation Reference Manual available at http://www.blueprint.org/bind/bind_documentation.html.
The emphasis on documented standards and controlled use-cases results in homogeneous database records, and without them, we note that the personal ‘style’ of the record curator becomes evident. The BIND Curation Reference Manual also describes the process used for BIND record validation, a process that has been built into the BIND software system ‘B*S’ used to track submissions and internal BIND curation workflow.
Small-molecule chemistry data are curated separately from interaction data by curation staff trained in chemistry. Small-molecule curators use standard chemoinformatics tools and a wide variety of chemistry resources to curate these records. When a BIND curator encounters a research article that discusses a small molecule, the article is passed onto a chemistry curator, who creates a MOD record for each unique small molecule found in BIND. A Small Molecule Curation Reference Manual outlining the process followed and software used is available at http://www.blueprint.org/bind/bind_documentation.html.
BIND CURATION PRIORITIES
BIND has been focusing its curation priorities on low-throughput curation, so that we can collect information about molecular interactions as it arises from journals. In early 2004, BIND surveyed 110 journals, each over a 3-month period to determine the rate of publication of data that could be curated in BIND. This survey found that a total of 1963 interactions per month are published in 79 journals, a number that rivals high-throughput interactions arriving, which we estimate at an average of 2600 per month (with wide variations). The top 20 journals with interaction data are listed in Table 1. Blueprint is seeking prepublication relationships with journal publishers, such as those used by GenBank and PDB for sequence and structure information, respectively, to capture this impressive influx of low-throughput molecular interaction data, and we are happy to note early success with this approach. In addition, a network of collaborating interaction databases has been organized, the International Molecular-Interaction Exchange (IMEx) consortium which seeks to achieve similar goals, comprising the DIP (10), MINT (11), IntAct (12), MIPS (13) and BIND database organizations.
Table 1. Top-ranked list of surveyed journals with low-throughput interaction data suitable for BIND curation.
| Journal | Interactions per month | |
|---|---|---|
| 1 | Journal of Biological Chemistry | 492 |
| 2 | The EMBO Journal | 300 |
| 3 | Biochemistry | 148 |
| 4 | PNAS | 90 |
| 5 | Cell | 80 |
| 5 | European Journal of Biochemistry | 80 |
| 5 | Molecular Cell | 80 |
| 6 | Journal of Virology | 48 |
| 7 | Journal of Molecular Biology | 44 |
| 8 | Nature | 40 |
| 8 | Science | 40 |
| 9 | Molecular and Cellular Biology | 32 |
| 10 | Virology | 30 |
| 11 | FEBS Letters | 28 |
| 12 | Journal of Cell Biology | 26 |
| 12 | Molecular Microbiology | 26 |
| 13 | Oncogene | 24 |
| 14 | Molecular Biology of the Cell | 23 |
| 15 | Journal of Bacteriology | 22 |
| 16 | Archives of Biochemistry and Biophysics | 20 |
| 17 | Journal of Immunology | 16 |
| 18 | EMBO Reports | 15 |
| 19 | Biochemical Pharmacology | 14 |
| 20 | Developmental Biology | 12 |
| 20 | Nature Genetics | 12 |
DATABASE QUERY AND RETRIEVAL
New BIND 3.5 software, released in September 2004, offers significant new methods for the query and retrieval of information from the BIND database. Most notably, the user-interface has been refined thanks to feedback from users and our Scientific Advisory Group to streamline the information retrieval process. BIND supports a broad range of query mechanisms, including browsing the database and database identifier searching (BIND ID, GI, PMID, Taxon ID, LocusLink, PDB, Entrez Gene, MMDB ID, GO, PFAM, CDD, SGD, FlyBase, WormBase, Interpro, MGI, RGD, OMIM, SMART, Swiss-Prot, TrEMBL and AfCS, with others to be added). Advanced field-specific queries can be constructed using a wizard-like tool that highlights and explains the myriad of BIND fields that can be queried in a precisely controlled manner.
BIND BLAST is provided for users who wish to find interactions with a protein similar to one specified as a query, and BLASTable BIND databases are now provided as BIND-ALL-NR, BIND-METAZOA-NR, BIND-FUNGI-NR and BIND-TAXROOT-NR, each reflecting the BIND divisions as defined previously. At query time, the BIND user is offered check-boxes that allow the user to exclude or include high-throughput BIND records, interactions, pathways and complexes. Additional query fine-tuning is being added to reflect user feedback as the BQS 3.5 system is further refined. For example, Adobe Acrobat PDF format reports containing BIND interactions may be retrieved when a paper record of an interaction is desired.
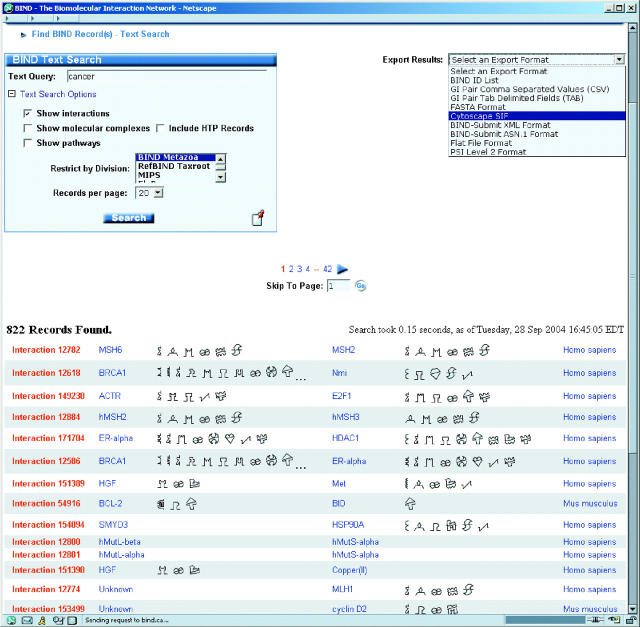
BIND now has a new look for query results retrived featuring OntoGlyphs as shown in Figure 1, a series of symbolic characters representing a high-level summary of Gene Ontology (GO) information (14). This helps users by concentrating a large amount of biological annotation information into a small space, and providing links back to the original GO annotation, as well as links to the sources for that annotation and the appropriate evidence codes.
Figure 1.
One view from the BIND interface showing a ‘cancer’ text query, restricting results to the BIND-Metazoa division and excluding high-throughput interactions. At the bottom, the list of pairwise interactions shows the OntoGlyph symbols which have click-through links for GO annotation details. At the top right is a list of export formats for the entire set of query results including Microsoft Excel (CSV), Cytoscape (highlighted) and PSI-Level II data files.
Beginning with BIND v3.5, the details of a large set of query results retrieved using the BIND web interface can be captured in a variety of formats for further processing by the BIND end-user, including Cytoscape SIF (15), Comma Separated Values (CSV), PSI level 2, GI pair list, FASTA sequences, BIND ID list, BIND FlatFile, BIND XML, BIND ASN.1 and Summary XML formats. We anticipate that the two most useful formats will be the Cytoscape SIF (as noted below) and CSV (for Microsoft Excel), from the perspective of the research biologist user of BIND. PreBIND (16) is currently a separate information system with interactions derived from text mining of MEDLINE abstracts (17). When the user cannot find BIND records, we suggest that they next search through the PreBIND database. PreBIND is, at time of writing, a separate query interface, but will be integrated into the BIND query interface in the future.
BIND is now also complemented by a new database called SMID and the new tool SMID-BLAST. SMID was built to help scientists answer the question ‘to what small molecule might this protein bind?’, a direct query supporting the search for ‘druggable’ targets. SMID's underlying data originates from the PDB data processed into the BIND-3DSM division that contains the small-molecule–protein interactions. SMID allows the user to query by CDD/SMART or Pfam domain to find instances where a member of that domain family is found in the PDB interacting with a small molecule. Users may see a list of small molecules found to bind that protein domain, and see a consensus sequence originating from the curated CDD domain set with the specific small-molecule binding sites highlighted in the view. SMID-BLAST is a version of RPS-BLAST (7). SMID-BLAST matches a protein sequence query to a CDD domain (18), thereby returning the set of small-molecule interaction partners to other members of that domain family. The resulting set of small molecules are candidate small-molecule interaction partners for the query sequence.
VISUALIZATION TOOLS
Visualization plays an important role in interaction database research and discovery. Interactions can be viewed from the BIND data model with the molecular entities represented as nodes, and the interaction or reaction depicted as an edge in a number of tools, including those we provide and third-party tools. A new wave of visualization tools has replaced earlier systems like Pajek (19) with biology-specific information including GO annotation and better support for the multidimensional mapping of annotation onto interaction nodes. Cytoscape (www.cytoscape.org) is a third-party interaction network visualization tool supported by good documentation and tutorials. Cytoscape also supports a number of plug-in algorithm components for exploring interaction and microarray data (15), including our own MCODE (Molecular Complex Detection) algorithm (20), which finds dense regions in interaction networks corresponding to molecular complexes. For example, a BIND query result can be saved directly from the BIND web interface to the user's local hard disk in Cystoscape SIF format. This file can then be loaded and viewed in Cytoscape as an interaction network and explored using a variety of built-in and plug-in Cytoscape features.
BIND's own visualization tool v3.1 continues to offer new features, including support for OntoGlyphs. In total, there are 83 OntoGlyph characters, which represent three types of molecule attributes: function, binding, and cellular localization. Ontoglyphs are derived from a combination of the US NCBI's Cluster of Orthologous Groups (COGs) functional categories (21) and GO terms (14), and are based on grouping the nearly 17 000 GO terms in the categories used most frequently by biologists in describing genes and protein function. The 34 functional OntoGlyphs cover molecule attributes ranging from cell physiology to ion transport to signal transduction. Similarly, the 25 binding OntoGlyphs divide molecules into ligand-binding categories such as ATP binding, DNA binding or transition metal ion binding. The 24 localization OntoGlyphs visually inform researchers about a molecule's location within the cell, anywhere from the nucleus to the cytoskeleton to the cell surface. With just a few mouse clicks in the BIND Interaction Viewer, individual OntoGlyphs can be selected, highlighted and manipulated, allowing researchers to hide all of the molecules involved in a certain pathway or not found within a particular cellular compartment, such as the nucleus. This mechanism helps researchers to make better sense of complex interaction networks by allowing them to focus on specific subsets of the data, without the distraction of secondary or tertiary partners. Similarly, through visual pattern recognition, researchers are more likely to see linkages through common interacting partners between different pathways that have not yet been identified in the literature. This has the potential to open new doors of scientific inquiry.
The Cn3D viewer (13) available from the NCBI offers the utility of an interaction-specific view on protein structure which is immediately seen with large molecular complexes like the ribosome. These can be very difficult to study from an interaction perspective with conventional structure-visualization tools. BIND's annotation of the intermolecular interfaces between RNA and protein molecules allows a user to select a BIND record with only the interface between two specific molecules within the complex (e.g. BIND ID 109757 from 1FFK, showing the interaction between the large ribosomal subunit rRNA and 50S ribosomal protein L30P from Haloarcula marismortui). One can further specifically limit the complexity of the returned data to a backbone-only model (e.g. choose ‘virtual bond model’ before launching Cn3D) to improve the responsiveness of systems with insufficient memory to display such large structures.
SUPPORT FOR THIRD-PARTY SOFTWARE
BIND software is available under terms that begin with the GNU Public License, although other licenses are available upon request. BIND data distributions and file formats support a variety of third-party software packages and ships by default with a number of commercial interaction network tools. Users should ensure that they have up-to-date versions of the BIND database as it is supplemented on a daily basis. The BIND web services v3.5 offers a SOAP (Standard Object Access Protocol) interface for developers who wish to access the data from third-party software. In addition, the SeqHound data warehouse system (22) supports the BIND interface, as well as high-throughput access to a variety of up-to-date databases including sequences, 3D protein structures, sequence redundancies, pre-computed BLAST neighbors, taxonomy information, complete genome sequences, conserved protein domains, GO terms and PubMed links from a central repository hosted by Blueprint, or alternatively in a format that can be hosted locally on the users' own servers. SeqHound is used as supporting data warehouse infrastructure for the BioMoby (23) bioinformatics middleware and Taverna (24) bioinformatics workflow projects and is being further supported in BioPerl (25) for automating bioinformatics analyses.
DATA DISTRIBUTION VIA FTP SITE
BIND data are available on the ftp.blueprint.org/pub/BIND/ FTP site in a variety of formats for users with a variety of bioinformatics skill sets. The BIND FTP site includes a simplified relational-table format view of the BIND data called the BIND Index (ftp://ftp.blueprint.org/pub/BIND/data/bindflatfiles/bindindex/). The BIND Index is recommended for researchers who prefer to work directly in SQL and contains the core information in BIND records including primary database identifiers, publications, non-redundant interactions, matrix and spoke models of BIND complexes (26), taxonomies, short labels and experimental methods.
Users who are able to work with more complex data grammars, such as the XML and ASN.1 versions of BIND, should refer to the BIND divisions to obtain a complete BIND database: ftp://ftp.blueprint.org/pub/BIND/data/divisions/. Daily non-cumulative (nc) updates to BIND are provided in the daily-nc directory for those who write scripts but do not wish to download all of BIND on a daily basis. Likewise, users who are looking for specific subsets of BIND data should refer to the BIND datasets (ftp://ftp.blueprint.org/pub/BIND/data/datasets/). The file with all of the BIND sequences in the FASTA format is found at ftp://ftp.blueprint.org/pub/BIND/data/divisions/fasta/bindall.fsa.gz BIND BLAST databases are found throughout the BIND FTP site as *.fsa files in the Divisions and Dataset directories and are updated in the daily-nc directory.
BIND datasets are collections of BIND data based on fixed queries of taxonomy, experimental system or publications, and files are available in BIND XML, BIND Asn.1 and FASTA sequence formats. BIND datasets arising from experimental-system annotation are referred to by name on the FTP site (e.g. two_hybrid_test.1.xml.gz). Datasets organized by taxonomy can be used to collect all interactions in BIND that are known for each organism in BIND. To access the appropriate file, first use the NCBI Taxonomy browser to find the taxon identifier number of the organism of interest (e.g. 9606–Homo sapiens), then find the corresponding file with the taxon identifier in its name (e.g. taxid9606.1.xml.gz). In a similar fashion, dataset files arising from unique publications are organized on the FTP site according to the PubMed identifier of the paper, which can be obtained from an NCBI PubMed query. Publication datasets can be used to assemble specific sets of interactions from cited publications without having to search across multiple websites to collect a variety of supplemental data stored at publisher websites in ad hoc formats.
MMDBBIND data are supplemented on the FTP site with sequence files that convey the specific pairwise residue–residue interaction between 3D biopolymer molecules using uppercase sequence characters to indicate interacting residues, as well as the redundant groupings of structure interaction data. The PDB sequences have also been matched to other databases like RefSeq with high confidence via BLAST and customized alignment tools. These data are also available in the same sequence representation with interacting residues mapped onto the typically longer versions of the sequences found in RefSeq. BIND's MOD data, containing curated, validated small molecules can be downloaded in *.mol or *.sdf file format at ftp://ftp.blueprint.org/pub/BIND/data/MOD/. Curated MOD records will be provided through the NCBI PubChem web interface, with links back to BIND in the near future.
Acknowledgments
ACKNOWLEDGEMENTS
BIND is funded in Canada by a consortium that includes Genome Canada through the Ontario Genomics Institute, the Ontario R&D Challenge Fund, the Canadian Institutes of Health Research in partnership with IT providers Sun Microsystems and Foundry Networks. BIND activity in Asia is funded by an investment of the Economic Development Board of Singapore. C.W.V.H. wrote this manuscript. All other authors contributed to database and software products mentioned herein and are listed alphabetically.
REFERENCES
- 1.Bader G.D., Betel,D. and Hogue,C.W.V. (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res., 31, 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salama J.J., Donaldson,I. and Hogue,C.W. (2002) Automatic annotation of BIND molecular interactions from three-dimensional structures. Biopolymers, 61, 111–120. [DOI] [PubMed] [Google Scholar]
- 3.Bader G.D. and Hogue,C.W. (2000) BIND—a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics, 16, 465–477. [DOI] [PubMed] [Google Scholar]
- 4.Duan X.J., Xenarios,I. and Eisenberg,D. (2002) Describing biological protein interactions in terms of protein states and state transitions: the LiveDIP database. Mol. Cell Proteomics, 2, 104–116. [DOI] [PubMed] [Google Scholar]
- 5.Papin J. and Subramaniam,S. (2004) Bioinformatics and cellular signaling. Curr. Opin. Biotechnol., 15, 78–81. [DOI] [PubMed] [Google Scholar]
- 6.Demir E., Babur,O., Dogrusoz,U., Gursoy,A., Ayaz,A., Gulesir,G., Nisanci,G. and Cetin-Atalay,R. (2004) An ontology for collaborative construction and analysis of cellular pathways. Bioinformatics, 20, 349–356. [DOI] [PubMed] [Google Scholar]
- 7.Marchler-Bauer A., Panchenko,A.R., Shoemaker,B.A., Thiessen,P.A., Geer,L.Y. and Bryant,S.H. (2002) CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res., 30, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bader G.D., Donaldson,I., Wolting,C., Ouellette,B.F., Pawson,T. and Hogue,C.W. (2001) BIND—the biomolecular interaction network database. Nucleic Acids Res., 29, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermjakob H., Montecchi-Palazzi,L., Bader,G., Wojcik,J., Salwinski,L., Ceol,A., Moore,S., Orchard,S., Sarkans,U., von Mering,C. et al. (2004) The HUPO PSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol., 22, 177–183. [DOI] [PubMed] [Google Scholar]
- 10.Salwinski L., Miller,C.S., Smith,A.J., Pettit,F.K., Bowie,J.U. and Eisenberg,D. (2004) The Database of Interacting Proteins: 2004 update. Nucleic Acids Res., 32, D449–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanzoni A., Montecchi-Palazzi,L., Quondam,M., Ausiello,G., Helmer-Citterich,M. and Cesareni,G. (2002) MINT: a Molecular INTeraction database. FEBS lett., 513, 135–140. [DOI] [PubMed] [Google Scholar]
- 12.Hermjakob H., Montecchi-Palazzi,L., Lewington,C., Mudali,S., Kerrien,S., Orchard,S., Vingron,M., Roechert,B., Roepstorff,P., Valencia,A. et al. (2004) IntAct: an open source molecular interaction database. Nucleic Acids Res., 32, 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mewes H.W., Amid,C., Arnold,R., Frishman,D., Guldener,U., Mannhaupt,G., Munsterkotter,M., Pagel,P., Strack,N., Stumpflen,V. et al. (2004) MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res., 32, D41–D44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris M.A., Clark,J., Ireland,A., Lomax,J., Ashburner,M., Foulger,R., Eilbeck,K., Lewis,S., Marshall,B., Mungall,C. et al. (2004) The Gene Ontology (GO) database and informatics resource. The Gene Ontology Consortium. Nucleic Acids Res., 32, D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shannon P., Markiel,A., Ozier,O., Baliga,N.S., Wang,J.T., Ramage,D., Amin,N., Schwikowski,B. and Ideker,T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donaldson I., Martin,J., de Bruijn,B., Wolting,C., Lay,V., Tuekam,B., Zhang,S., Baskin,B., Bader,G.D., Michalickova,K. et al. (2003) PreBIND and Textomy—mining the biomedical literature for protein–protein interactions using a support vector machine. BMC Bioinformatics, 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis K.N., Robinson,M.D., Hughes,T.R. and Hogue,C.W.V. (2004) MyMED: an internal XML relational database implementation of MEDLINE citations in DB2. IBM Syst. J., 43, 756–757. [Google Scholar]
- 18.Marchler-Bauer A., Anderson,J.B., DeWeese-Scott,C., Fedorova,N.D., Geer,L.Y., He,S., Hurwitz,D.I., Jackson,J.D., Jacobs,A.R. et al. (2003) CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res., 31, 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batagelj V. and Mrvar,A. (1998) Pajek—program for large network analysis. Connections, 2, 47–57. [Google Scholar]
- 20.Bader G.D. and Hogue,C.W. (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics, 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatusov R.L., Fedorova,N.D., Jackson,J.D., Jacobs,A.R., Kiryutin,B., Koonin,E.V., Krylov,D.M., Mazumder,R., Mekhedov,S.L., Nikolskaya,A.N. et al. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics, 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michalickova K., Bader,G.D., Dumontier,M., Lieu,H., Betel,D., Isserlin,R. and Hogue,C.W. (2002) SeqHound: biological sequence and structure database as a platform for bioinformatics research. BMC Bioinformatics, 3, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson M.D. and Links,M. (2002) BioMOBY: an open source biological web services proposal. Brief Bioinformatics, 3, 331–341. [DOI] [PubMed] [Google Scholar]
- 24.Oinn T., Addis,M., Ferris,J., Marvin,D., Greenwood,M., Carver,T., Pocock,M.R., Wipat,A. and Li,P. (2004) Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics, 20, 3045–3054. [DOI] [PubMed] [Google Scholar]
- 25.Stajich J.E., Block,D., Boulez,K., Brenner,S.E., Chervitz,S.A., Dagdigian,C., Fuellen,G., Gilbert,J.G., Korf,I., Lapp,H. et al. (2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res., 12, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bader G.D. and Hogue,C.W. (2002) Analyzing yeast protein–protein interaction data obtained from different sources. Nat. Biotechnol., 20, 991–997. [DOI] [PubMed] [Google Scholar]