Abstract
The natural environment serves as a reservoir of opportunistic pathogens. A well-established method for studying the epidemiology of such opportunists is multilocus sequence typing, which in many cases has defined strains predisposed to causing infection. Burkholderia multivorans is an important pathogen in people with cystic fibrosis (CF) and its epidemiology suggests that strains are acquired from non-human sources such as the natural environment. This raises the central question of whether the isolation source (CF or environment) or the multilocus sequence type (ST) of B. multivorans better predicts their genomic content and functionality. We identified four pairs of B. multivorans isolates, representing distinct STs and consisting of one CF and one environmental isolate each. All genomes were sequenced using the PacBio SMRT sequencing technology, which resulted in eight high-quality B. multivorans genome assemblies. The present study demonstrated that the genomic structure of the examined B. multivorans STs is highly conserved and that the B. multivorans genomic lineages are defined by their ST. Orthologous protein families were not uniformly distributed among chromosomes, with core orthologs being enriched on the primary chromosome and ST-specific orthologs being enriched on the second and third chromosome. The ST-specific orthologs were enriched in genes involved in defense mechanisms and secondary metabolism, corroborating the strain-specificity of these virulence characteristics. Finally, the same B. multivorans genomic lineages occur in both CF and environmental samples and on different continents, demonstrating their ubiquity and evolutionary persistence.
Introduction
Burkholderia cepacia complex (Bcc) bacteria are rare but potentially virulent pathogens in cystic fibrosis (CF) patients [1]. Epidemiological surveys revealed that B. multivorans is the most prevalent Bcc CF pathogen in many countries [1–7]. The continued emergence of unique B. multivorans strains in CF patients suggests acquisition from non-human sources, such as the natural environment [8]. Environmental conditions or non-human hosts in which virulence factors might be adaptive can select for traits that confer virulence and natural environments could therefore serve as reservoirs of opportunistic pathogens [9, 10].
Multilocus sequence typing (MLST) is a well-established method for studying the epidemiology and population structure of Bcc organisms [11, 12]. The Bcc MLST scheme takes into account the allelic variation of seven housekeeping genes and each strain is defined by its unique allelic profile and multilocus sequence type (ST) [13]. Baldwin et al. [14] demonstrated that roughly one fifth of the clinical isolates in the Bcc PubMLST database had the same ST as environmental isolates, suggesting these isolates represent the same strain. A follow-up study demonstrated that several B. multivorans STs were globally distributed and that the natural environment (e.g. water and soil) may be an important reservoir for infection with this species [8].
Burkholderia genomes vary in size from 2.4 Mb (Ca. Burkholderia schumannianae UZHbot8) [15] to 11.5 Mb (Burkholderia terrae BS001) [16], are characterized by a high G+C content (62-68 mol%) and consist of multiple replicons [17, 18]. To gain insight into the overall genome biology of B. multivorans, we sequenced the genomes of eight isolates representing four distinct STs. For each ST, a CF and an environmental isolate were sequenced using the PacBio Single-Molecule Real-Time (SMRT) sequencing technology. The present study provides the first comprehensive comparative genome analysis of B. multivorans and assesses to which extent isolates with the same ST but from different origin (CF versus environmental) differ in genetic potential.
Materials and methods
Studied isolates
We searched the Bcc PubMLST database (http://pubmlst.org/bcc/) [13], identified four B. multivorans STs that included both CF and environmental (ENV) isolates and selected eight isolates for whole-genome sequencing (Table 1). Strains were grown aerobically on Tryptone Soya Agar (Oxoid) and incubated at 28°C. Cultures were preserved in MicroBank™ vials at -80°C.
Table 1. B. multivorans isolates included in the present study.
| Isolate | Strain number | ST | Isolation source | Depositor |
|---|---|---|---|---|
| ST180-ENV | LMG 29305, J2943 | 180 | Rhizosphere soil (United Kingdom, 2000) | J.R.W. Govan |
| ST180-CF | LMG 29313, 8335 | 180 | CF sputum (Czech republic, 2011) | P. Drevinek |
| ST189-ENV | LMG 29309 | 189 | Succulent soil (Belgium, 2003) | Own isolate |
| ST189-CF | LMG 29312, BCC0208 | 189 | CF patient (Canada, 1999) | E. Mahenthiralingam |
| ST287-ENV | LMG 29306, J2947 | 287 | Rhizosphere soil (United Kingdom, 2000) | J.R.W. Govan |
| ST287-CF | LMG 29311, BCC0059 | 287 | CF patient (Canada, 1995) | E. Mahenthiralingam |
| ST650-ENV | LMG 29308 | 650 | Pond water (Belgium, 2003) | Own isolate |
| ST650-CF | LMG 29310, Q113 | 650 | CF patient (Germany, 2010) | B. Kahl |
LMG, BCCM/LMG Bacteria Collection, Laboratory of Microbiology, Ghent University, Ghent, Belgium. CF, cystic fibrosis; ENV, environmental; ST, multilocus sequence type.
Genome sequencing, assembly and annotation
High-quality DNA was prepared using Qiagen Genomic tips (20/G) and genomes were sequenced using the P5-C3 chemistry on the PacBio SMRT II platform of the Department of Genetics and Genomic Sciences of the Icahn School of Medicine at Mount Sinai (New York, USA). One SMRT cell per isolate was sequenced, except for isolates ST189-CF and ST287-ENV for which a second SMRT cell was run to increase the quality of the raw data. PacBio reads were assembled in the sequencing center using the SMRT analysis software (including HGAP3 and Quiver) and contigs were ordered against the complete reference genome of B. multivorans strain ATCC 17616 (PRJNA17407) using Mauve [19]. We further polished the assemblies in five steps. The first step consisted of removing spurious contigs that were small in size, had a low coverage and resulted in a highest BLAST hit with the primary chromosome of its own genome [20]. Reads were mapped using pbalign and QC reports were created based on the resulting BAM files using Qualimap [21]. Contigs smaller than 20 kb and with less than 20x coverage or a high variation (SD) in coverage were discarded. In step two, read mappings were used to further polish the contigs using Pilon [22] with default parameters. In step three, contigs with overlapping ends were merged using Gap5 [23] to exclude artificially duplicated regions, often including many frameshifts and fragmented open reading frames. In step four, the duplicated ends of circular contigs were trimmed using Gepard [24] and Gap5 as these duplications were a consequence of the circular nature of the replicons in combination with the long-read sequencing technology [25]. Importantly, this artificial duplication of contig ends not only resulted in a highly variable rRNA copy number, but also falsely excluded genes from the ortholog dataset because they were artificially duplicated. Since the merging of overlapping ends by Gap5 might be imperfect, we ran Quiver in a final polishing step. The PacBio sequencing reads of one SMRT cell resulted in a coverage ranging from 76x to 119x.
Annotation was performed using Prokka v1.11 [26] with a genus-specific database based on reference genomes from the Burkholderia Genome database (http://beta.burkholderia.com/, accessed March 2015) [27]. The annotated genome assemblies were submitted to the European Nucleotide Archive and are publicly available through the GenBank/EMBL/DDBJ accession numbers FKJT01000000, FKJS01000000, FKJU01000000, FKJP01000000, FKJV01000000, FKJW01000000, FKJX01000000 and FKJY01000000. The original PacBio sequencing data were submitted to the Sequence Read Archive and are publicly available through the accession numbers ERX1955257, ERX1955260, ERX1955324, ERX1955331, ERX1955371, ERX1955980, ERX1955987 and ERX1955995.
The genome sequence of B. multivorans strain ATCC 17616 (PRJNA17407) was included as a reference in all further analyses. A multiple genome alignment was performed using Mauve [28] to assess the basic genome structure.
Analysis of protein-coding genes and ortholog identification
We mapped for each protein-coding gene (CDS) on which chromosome it was located and to which cluster of orthologous groups (COG) [29] it belonged (S5 Table). COGs were assigned by a reversed position-specific BLAST (RPSBLAST v2.2.29+) with an e-value cut-off of 1E-3 against the NCBI conserved domain database (CDD v3.14). Orthologous genes were identified in the eight B. multivorans genomes of the present study and the ATCC 17616 reference strain using custom perl scripts (https://github.com/hatcherunh/GeneFamilyAnalysis) as described previously [30, 31]. In short, homologs were identified as reciprocal best BLAST hits with a normalized bit score (bit score of hit/bit score of self-hit, see [31]), providing an empirically determined taxon-specific threshold. A CDS was defined to be non-orthologous if no orthologs were found in the dataset. The putative panorthologs (i.e. single-copy orthologous genes conserved in all genomes) were computed while varying the bit score threshold from 0.1 to 0.9 in 0.1 increments and the largest set of panorthologs was selected. For each orthologous protein family, the consensus chromosome location and COG were determined (S6 Table). Conflicts in COG mapping were resolved by the majority rule.
Phylogenomic analysis
The whole-genome phylogeny (of the eight B. multivorans genomes of the present study and the ATCC 17616 reference strain) was calculated based on the sequences of the panorthologs as described previously [15]. In short, amino acid sequences were aligned using MUSCLE [32] and translated back to the respective nucleotide sequences using T-Coffee [33]. Nucleotide alignments were trimmed using trimAl [34] by removing positions with gaps in more than 50% of the sequences, and were subsequently concatenated to construct a maximum likelihood tree using RaXML v7.4.2 [35] with the GTRGAMMA substitution model and 1000 rapid bootstrap analyses.
In a second approach, the presence/absence matrix of all orthologs was used in a discrete character-state parsimony analysis using pars from the PHYLIP package [36] to assess the relatedness of the genomes in terms of gene content.
Comparison of B. multivorans and B. cenocepacia COG profiles
Complete genome sequences of Burkholderia cenocepacia strains J2315, H111, K56-2Valvano, AU1054, HI2424 and MC0-3 were downloaded from the Burkholderia Genome database (http://beta.burkholderia.com/) [27]. COG mapping of B. cenocepacia CDS was performed as described above for B. multivorans. The number of CDS per COG category for each species (B. multivorans versus B. cenocepacia) was counted and the distributions were compared using Pearson chi-square analysis.
Data visualization and statistical analyses
Data visualization and statistical analyses were performed using RStudio with R v3.2.3. Pearson’s chi-square analyses were used to test the association between different sets of categorical variables. When a significant relationship was found between two variables, we further examined the standardized Pearson residuals. Standardized Pearson residuals with high absolute values indicate a lack of fit of the null hypothesis of independence in each cell [37] and thus indicate observed cell frequencies in the contingency table that are significantly higher or lower than expected based on coincidence. In case multiple COG categories were registered for the same COG, each COG category was counted separately for Pearson chi-square analysis on COG categories. For the 198 CDS that were involved in the translocation within the ST650-CF isolate from the primary to the secondary chromosome, the consensus chromosome mapping was set to the primary chromosome for Pearson chi-square analysis on chromosome distribution.
Results
The genomic structure of B. multivorans is highly conserved
The final assemblies produced closed genomes for five of the eight sequenced B. multivorans isolates. The genomes were 6.2-6.9 Mb in size with a G+C content of ˜67 mol% and the number of predicted CDS ranged from 5,415 to 6,155 CDS per genome (Table 2). No clustered regularly interspaced short palindromic repeats (CRISPRs) were identified. Each of the genomes contained one tmRNA and 75-79 tRNAs.
Table 2. B. multivorans genome characteristics.
| Isolate | Contigs | Size (bp) | %GC | C1 | C2 | C3 | Plasmid | Total CDS | Orthologous CDS | Non-orthologous CDS |
|---|---|---|---|---|---|---|---|---|---|---|
| ST180-ENV | 4 | 6,464,081 | 67.1 | 1 | 2 | 3 | 4 | 5,794 | 5,271 | 523 |
| ST180-CF | 4 | 6,296,736 | 67.3 | 1 | 2 | 3 | 4 | 5,551 | 5,266 | 285 |
| ST189-ENV | 3 | 6,223,431 | 67.3 | 1 | 2 | 3 | - | 5,467 | 5,144 | 323 |
| ST189-CF | 13 | 6,157,395 | 67.3 | 1-5 | 6-9 | 10-13 | - | 5,415 | 5,132 | 283 |
| ST287-ENV | 4 | 6,559,547 | 67.2 | 1-2 | 3 | 4 | - | 5,800 | 5,494 | 306 |
| ST287-CF | 6 | 6,857,684 | 67.0 | 1-4 | 5 | 6 | - | 6,155 | 5,505 | 650 |
| ST650-ENV | 3 | 6,322,929 | 67.2 | 1 | 2 | 3 | - | 5,594 | 5,275 | 319 |
| ST650-CF | 3 | 6,308,820 | 67.2 | 1 | 2 | 3 | - | 5,599 | 5,250 | 349 |
| ATCC 17616 | 4 | 7,008,622 | 66.7 | 1 | 2 | 3 | 4 | 6,258 | 4,893 | 1,365 |
C1, C2 and C3: contigs mapping to chromosome 1, 2 and 3 of B. multivorans strain ATCC 17616, respectively; bp, base pairs.
The multiple genome alignment of the examined B. multivorans STs (S3 Fig) revealed a highly conserved genomic structure with three chromosomes (from here on referred to as C1, C2 and C3). C1, C2 and C3 were on average 3.4 Mb, 2.4 Mb and 0.6 Mb in size. Both ST180 isolates harbored one contig that did not map onto the reference genome of ATCC 17616. These contigs were 22,339 and 28,809 bp in size, did not contain any rRNA genes, had a G+C content of 58 mol% and were therefore considered plasmids. Both plasmids contained genes for an initiator repB protein, an AsnC transcriptional regulator, a cobyrinic acid a,c-diamide synthase (parA homologue), multiple integrases and several hypothetical proteins. The multiple genome alignment also revealed a fairly large translocation (207 genes, 198 CDS) within the ST650-CF isolate from C1 to C2 that was delimited by rRNA operons at both ends. All isolates except ST650-CF contained 3, 1 and 1 rRNA copies on C1, C2 and C3, respectively. As a result of the translocation, isolate ST650-CF contained 2, 2 and 1 rRNA copies on C1, C2 and C3, respectively.
ST predicts genomic lineage
Orthologous genes were identified to determine the conserved genome content of B. multivorans. The ortholog analysis identified 6,254 homologous protein families (S6 Table) comprising 47,230 CDS in total (Table 2). The largest set of panorthologs, i.e. orthologs conserved in all nine B. multivorans genomes and present as single copies, was found at a reciprocal best bit score threshold of 0.7 (see Methods section) and comprised 4,503 ortholog families.
The frequency of orthologous versus non-orthologous CDS (i.e. CDS with versus without orthologs in the dataset) varied significantly per isolate (X2(8) = 1829.6, p<0.001) and ST (X2(3) = 67.3, p<0.001), but not isolation source (p>0.05). The genomes of isolates ST287-CF and ATCC 17616 were significantly enriched with non-orthologous CDS, while those of ST180-CF, ST287-ENV, ST650-ENV and both ST189 isolates were significantly deprived in non-orthologous CDS (Table 2 and S1 Table). Analysis of the relationship between orthologous versus non-orthologous CDS and ST showed that the ST287 genomes were significantly enriched with non-orthologous CDS, while the ST189 and ST650 genomes were significantly deprived in non-orthologous CDS (S2 Table).
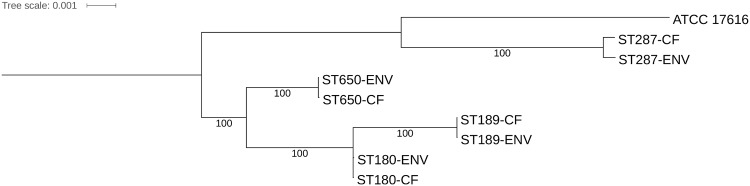
The ortholog dataset enabled two subsequent analyses of strain phylogeny. In the first approach, a whole-genome phylogeny was obtained based on nucleotide sequence divergence of the panorthologs (Fig 1). In the second approach, the presence/absence matrix of the ortholog families was used to assess the relatedness of the genomes in terms of gene content using parsimony (S1 Fig). These analyses both demonstrated that the ST, and not the isolation source, of the B. multivorans isolates predicted their phylogeny and gene content. This finding demonstrated that isolates with the same ST represent the same genomic lineage, irrespective of their isolation source.
Fig 1. Phylogenomic analysis showing the relatedness of the genomes in terms of sequence divergence of the panorthologs.
The maximum likelihood tree was inferred using the GTRGAMMA substitution model and is based on a concatenated nucleotide alignment of 4,503 CDS (4,457,847 positions). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap analyses (1,000 replicates) are shown next to the branches. Scale bar represents number of substitutions per site. The tree was rooted on the branch with the largest branch length.
Orthologous genes are enriched on C2 and are involved in carbohydrate metabolism and transport
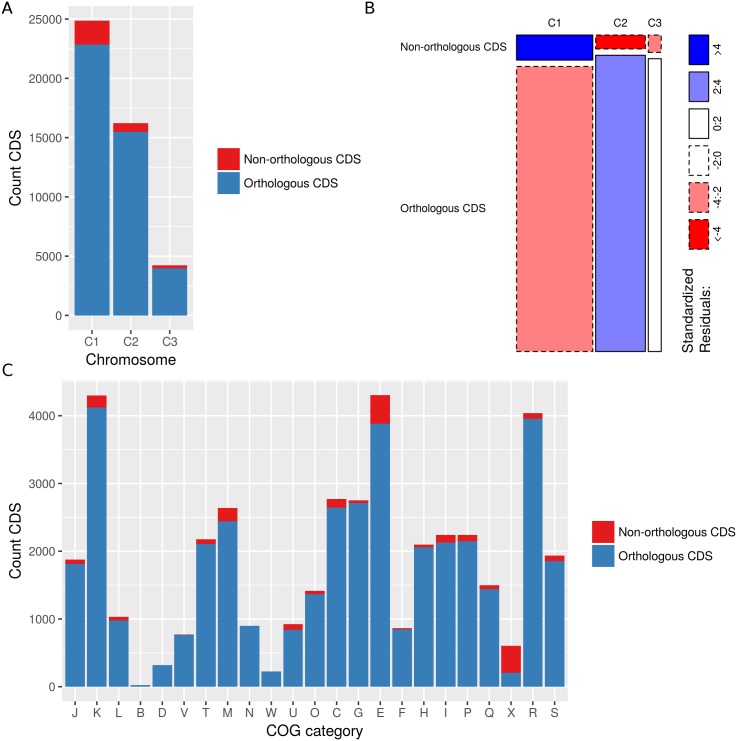
Because the fraction of genes that are involved in housekeeping functions varies among the chromosomes [17], we mapped the chromosome location of each CDS (S5 Table). Consistent with the average chromosome size, the total number of CDS was highest on C1 (27,813 CDS), followed by C2 (18,565 CDS) and C3 (5,047 CDS). The plasmid of the ST180 isolates harbored 208 CDS, of which 206 were non-orthologous CDS. The translocation within the ST650-CF isolate from C1 to C2 comprised 198 CDS, of which 13 were non-orthologous CDS. The frequency of orthologous versus non-orthologous CDS varied significantly among the different chromosomes (X2(2) = 213.4, p<0.001) (Fig 2A). C1 was significantly enriched with non-orthologous CDS, while C2 was significantly enriched with orthologous CDS (Fig 2B).
Fig 2. The frequency of orthologous versus non-orthologous CDS varies among chromosomes and COG categories.
Bar plots show the number of orthologous and non-orthologous CDS per chromosome (X2(2) = 213.4, p<0.001) (a) and COG category (X2(22) = 5101.2, p<0.001) (c). Mosaic plots show the standardized residuals of the Pearson chi-square analysis for the number of orthologous and non-orthologous CDS per chromosome (b). Solid and dashed boundaries represent positive and negative residuals, respectively. Rectangles are colored only if the standardized residual is significant at p<0.05 (outside ±1.96). COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
To identify biological functions that were over- or underrepresented, we assigned each CDS to a COG (S5 Table). Roughly 80% of the CDS (41,520 CDS in total) could be assigned to a COG and its associated COG functional category. The frequency of orthologous versus non-orthologous CDS varied significantly among the different COG categories (X2(22) = 5101.2, p<0.001) (Fig 2C). The non-orthologous CDS were significantly enriched in the COG categories cell wall/membrane/envelope biogenesis (M), intracellular trafficking, secretion and vesicular transport (U), amino acid transport and metabolism (E) and mobilome (X), while the orthologous CDS were significantly enriched in the COG categories carbohydrate transport and metabolism (G) and general function prediction only (R) (Table 3).
Table 3. The frequency of orthologous versus non-orthologous CDS varies among the COG categories.
| Orthologous CDS | Non-orthologous CDS | ||||
|---|---|---|---|---|---|
| Information storage and processing | |||||
| J | Translation, ribosomal structure and biogenesis | 1814 (0.789) | − | 65 (-3.359) | |
| K | Transcription | 4122 (0.766) | − | 176 (-3.258) | |
| L | Replication, recombination and repair | 977 (-0.032) | 55 (0.137) | ||
| B | Chromatin structure and dynamics | 24 (0.263) | 0 (-1.121) | ||
| Cellular processes and signaling | |||||
| D | Cell cycle control, cell division, chromosome partitioning | 319 (0.848) | − | 2 (-3.610) | |
| V | Defense mechanisms | 765 (1.199) | − | 8 (-5.101) | |
| T | Signal transduction mechanisms | 2108 (0.988) | − | 69 (-4.207) | |
| M | Cell wall/membrane/envelope biogenesis | 2441 (-1.161) | + | 196 (4.941) | |
| N | Cell motility | 898 (1.510) | − | 3 (-6.429) | |
| W | Extracellular structures | 225 (0.806) | − | 0 (-3.431) | |
| U | Intracellular trafficking, secretion, and vesicular transport | 838 (-1.241) | + | 85 (5.283) | |
| O | Posttranslational modification, protein turnover, chaperones | 1367 (0.711) | − | 48 (-3.025) | |
| Metabolism | |||||
| C | Energy production and conversion | 2641 (0.329) | 128 (-1.401) | ||
| G | Carbohydrate transport and metabolism | + | 2710 (2.016) | − | 41 (-8.579) |
| E | Amino acid transport and metabolism | − | 3880 (-3.142) | + | 426 (13.373) |
| F | Nucleotide transport and metabolism | 845 (0.916) | − | 19 (-3.897) | |
| H | Coenzyme transport and metabolism | 2058 (1.521) | − | 42 (-6.475) | |
| I | Lipid transport and metabolism | 2127 (0.091) | 113 (-0.387) | ||
| P | Inorganic ion transport and metabolism | 2148 (0.567) | − | 91 (-2.415) | |
| Q | Secondary metabolites biosynthesis, transport and catabolism | 1442 (0.543) | − | 58 (-2.311) | |
| Poorly characterized | |||||
| R | General function prediction only | + | 3954 (2.026) | − | 86 (-8.623) |
| S | Function unknown | 1851 (0.358) | 86 (-1.524) | ||
| Mobile elements | |||||
| X | Mobilome: prophages, transposons | − | 207 (-15.300) | + | 398 (65.117) |
Pearson chi-square analysis testing the independence of gene conservation (orthologous vs. non-orthologous CDS) and COG category (X2(22) = 5101.2, p<0.001). Each cell in the contingency represents the observed frequency and standardized residual (in between brackets) and is preceded by + or − if the standardized residual is >1.96 or <-1.96, respectively, and significant at p<0.05.
Each ST harbors unique orthologs
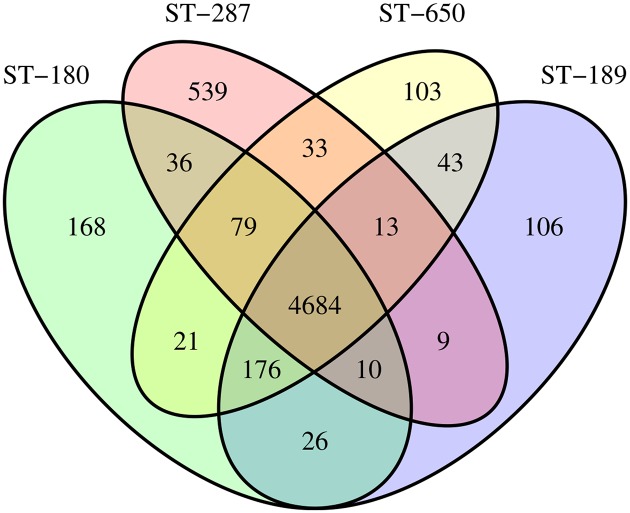
For each ortholog family we examined whether it was present in all eight isolates (i.e. showed core specificity), specific for isolates of one or more STs, specific for isolates of a specific source or randomly present (S6 Table). None of the ortholog families was present in all four isolates of a specific source (CF vs. ENV), but a small number of ortholog families were present in only one, two or three isolates from the same source, thus leaving five relevant specificity groups: core (n = 4,684), ST (n = 1,362), CF-only (n = 38), ENV-only (n = 51) and random (n = 119). The Venn diagram (Fig 3) visualizes the number of ortholog families in the core and ST specificity groups (n = 6,046) and shows that each ST harbors 103-539 orthologs that were not present in any other ST.
Fig 3. Venn diagram showing the number of core and ST-specific ortholog families.
ST, multilocus sequence type.
ST-specific orthologs are enriched on C2 and C3 and are involved in defense mechanisms and secondary metabolism
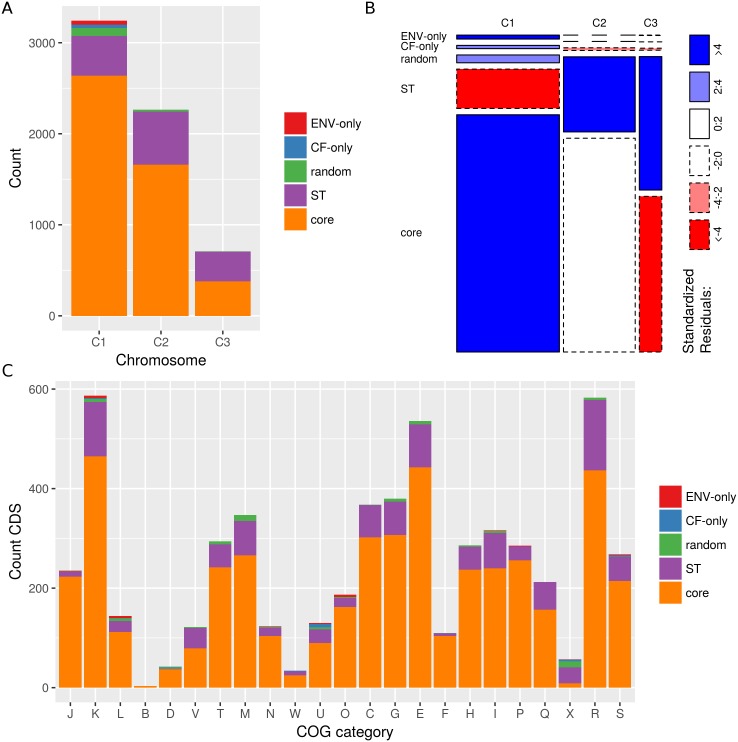
Based on the chromosome and COG mapping for the individual CDS, we mapped the consensus chromosome location and COG category for each ortholog family (S6 Table). Consistent with the average chromosome size, the number of orthologs was highest on C1 (3,242), followed by C2 (2,264) and C3 (710). For 37 ortholog families there was a conflict in chromosome mapping, and 1 ortholog was located on the plasmid of the ST180 isolates. COGs and their associated COG functional category could be assigned to 4,896 of the 6,254 ortholog families.
The specificity of the ortholog families varied significantly among the chromosomes (X2(8) = 469.8, p<0.001) (Fig 4A) and COG categories (X2(88) = 649.8, p<0.001) (Fig 4C). C2 and C3 were significantly enriched with ST-specific orthologs, while C1 was significantly enriched with orthologs belonging to the specificity groups core, random, CF-only and ENV-only (Fig 4B). The ST-specific orthologs were significantly enriched in the COG categories defense mechanisms (V), secondary metabolites biosynthesis, transport and catabolism (Q), mobilome (X) and general function prediction only (R) (Table 4).
Fig 4. Ortholog specificity varies among chromosomes and COG categories.
Bar plots show the number of orthologs per specificity group for different chromosomes (X2(8) = 469.8, p<0.001) (a) and COG categories (X2(88) = 649.8, p<0.001) (c). Mosaic plots show the standardized residuals of the Pearson chi-square analysis on the number of orthologs per specificity group per chromosome (b). Solid and dashed boundaries represent positive and negative residuals, respectively. Rectangles are colored only if the standardized residual is significant at p<0.05 (outside ±1.96). COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
Table 4. Ortholog specificity varies among the COG categories.
| Core | ST | CF-only | ENV-only | Random | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Information storage and processing | |||||||||||
| J | Translation, ribosomal structure and biogenesis | + | 223 (2.570) | − | 10 (-4.947) | 0 (-0.865) | 1 (0.044) | 1 (-1.306) | |||
| K | Transcription | 465 (-0.187) | 109 (0.375) | 2 (0.095) | 5 (1.688) | 6 (-0.864) | |||||
| L | Replication, recombination and repair | 112 (-0.286) | 22 (-0.748) | 1 (0.799) | + | 4 (4.458) | + | 5 (2.013) | |||
| B | Chromatin structure and dynamics | 3 (0.389) | 0 (-0.733) | 0 (-0.098) | 0 (-0.111) | 0 (-0.209) | |||||
| Cellular processes and signaling | |||||||||||
| D | Cell cycle control, cell division, chromosome partitioning | 37 (0.594) | − | 1 (-2.378) | + | 1 (2.368) | 0 (-0.414) | + | 3 (3.061) | ||
| V | Defense mechanisms | 79 (-1.872) | + | 41 (4.095) | 0 (-0.623) | 0 (-0.705) | 2 (0.172) | ||||
| T | Signal transduction mechanisms | 242 (0.461) | 46 (-0.919) | 0 (-0.968) | 0 (-1.094) | 6 (0.839) | |||||
| M | Cell wall/membrane/envelope biogenesis | 266 (-0.677) | 69 (0.867) | 0 (-1.052) | 0 (-1.189) | + | 12 (3.102) | ||||
| N | Cell motility | 104 (0.576) | 16 (-1.286) | 1 (0.971) | 1 (0.705) | 1 (-0.588) | |||||
| W | Extracellular structures | 25 (-0.416) | 8 (0.774) | + | 1 (2.709) | 0 (-0.372) | 0 (-0.703) | ||||
| U | Intracellular trafficking, secretion, and vesicular transport | 90 (-1.362) | 27 (0.769) | + | 7 (10.233) | + | 2 (2.022) | 4 (1.538) | |||
| O | Posttranslational modification, protein turnover, chaperones | 162 (1.028) | − | 18 (-2.678) | 0 (-0.772) | + | 5 (4.858) | 2 (-0.434) | |||
| Metabolism | |||||||||||
| C | Energy production and conversion | 302 (0.463) | 65 (-0.114) | 0 (-1.083) | 0 (-1.224) | 1 (-1.879) | |||||
| G | Carbohydrate transport and metabolism | 307 (0.192) | 67 (-0.130) | 0 (-1.100) | 0 (-1.244) | 6 (0.206) | |||||
| E | Amino acid transport and metabolism | 443 (0.710) | 86 (-1.023) | 0 (-1.307) | 0 (-1.477) | 7 (-0.280) | |||||
| F | Nucleotide transport and metabolism | 104 (1.717) | − | 5 (-3.313) | 0 (-0.592) | 0 (-0.669) | 1 (-0.472) | ||||
| H | Coenzyme transport and metabolism | 237 (0.560) | 46 (-0.731) | 0 (-0.955) | 0 (-1.079) | 3 (-0.565) | |||||
| I | Lipid transport and metabolism | 240 (-0.787) | 71 (1.913) | 0 (-1.003) | 1 (-0.253) | 4 (-0.274) | |||||
| P | Inorganic ion transport and metabolism | 256 (1.873) | − | 28 (-3.227) | 0 (-0.953) | 1 (-0.149) | − | 0 (-2.034) | |||
| Q | Secondary metabolites biosynthesis, transport and catabolism | 157 (-0.953) | + | 55 (2.762) | 0 (-0.822) | 0 (-0.929) | 0 (-1.754) | ||||
| Poorly characterized | |||||||||||
| R | General function prediction only | 437 (-1.337) | + | 141 (3.577) | 0 (-1.363) | 0 (-1.541) | 5 (-1.190) | ||||
| S | Function unknown | 214 (-0.010) | 49 (0.143) | 2 (1.240) | 2 (0.870) | 1 (-1.465) | |||||
| Mobile elements | |||||||||||
| X | Mobilome: prophages, transposons | − | 9 (-5.415) | + | 32 (6.818) | + | 3 (6.613) | 1 (1.594) | + | 12 (12.283) | |
Pearson chi-square analysis testing the independence of ortholog specificity and COG category (X2(88) = 469.8, p<0.001). Each cell in the contingency represents the observed frequency and standardized residual (in between brackets) and is preceded by + or − if the standardized residual is >1.96 or <-1.96, respectively, and significant at p<0.05.
ST287 harbors extra orthologous and non-orthologous genes
Both ST287 genomes were considerably larger and contained a higher number of CDS (Table 2), suggesting that this genomic lineage contains extra genes. ST287 was not only enriched with non-orthologous CDS (S2 Table) but also harbored 539 orthologs that were not present in the other three STs (Fig 3), showing that the extra genes in ST287 are both orthologous and non-orthologous CDS. A similar trend was observed for the ST287-specific orthologs as compared to the ST-specific orthologs in general (Table 4), as they were enriched in the same COG categories.
C1 is enriched with orthologs showing CF and ENV specificity
C1 was enriched with orthologs that were present in only one, two or three isolates from the same source (CF-only or ENV-only) (Fig 4A and 4B). CF-only orthologs were significantly enriched in the COG categories cell cycle control, cell division and chromosome partitioning (D), extracellular structures (W), intracellular trafficking, secretion and vesicular transport (U) and mobilome (X), while ENV-only orthologs were significantly enriched in replication, recombination and repair (L), intracellular trafficking, secretion and vesicular transport (U) and posttranslational modification, protein turnover and chaperones (O) (Fig 4C and Table 4).
Additionally, C1 was also enriched with orthologs showing random specificity (Fig 4A and 4B) and these orthologs with random specificity were significantly enriched in the COG categories replication, recombination and repair (L), cell cycle control, cell division and chromosome partitioning (D), cell wall/membrane/envelope biogenesis (M) and mobilome (X) (Fig 4C and Table 4).
Comparison of B. multivorans and B. cenocepacia average COG profiles
During the past two decades, B. multivorans and B. cenocepacia have been the most prevalent Bcc pathogens in CF. Historically, B. cenocepacia strains have been responsible for large epidemics within the CF community and are often extremely virulent [38]. In contrast, only a limited number of B. multivorans outbreak strains were described and B. multivorans is generally considered a less virulent Bcc pathogen as compared to B. cenocepacia [17]. To examine the species-specific genome content of B. multivorans, we compared its average COG profile to that of B. cenocepacia. Generally, B. cenocepacia genomes contained more genes (6,477-7,116 CDS per genome) (S3 Table) than B. multivorans genomes (5,415-6,155 CDS per genome) (Table 2) and more CDS per COG category (S2 Fig). We compared the average COG profile of the two species by calculating the average number of CDS per genome in each COG category and by comparing these distributions. The distribution of CDS among COG categories varied significantly between the two species (X2(22) = 102.9, p<0.001) (S2 Fig). B. cenocepacia genomes harbor significantly more CDS in the COG categories transcription (K), defense mechanisms (V) and general function prediction only (R) and significantly less in translation, ribosomal structure and biogenesis (J) and replication, recombination and repair (L). Conversely, B. multivorans genomes harbor significantly more CDS in the COG categories replication, recombination and repair (L) and less in transcription (K) (Table 5).
Table 5. The distribution of B. multivorans versus B. cenocepacia CDS varies among COG categories.
| B. cenocepacia | B. multivorans | ||||
|---|---|---|---|---|---|
| Information storage and processing | |||||
| J | Translation, ribosomal structure and biogenesis | − | 1528 (-2.166) | 2118 (1.931) | |
| K | Transcription | + | 4314 (3.883) | − | 4866 (-3.462) |
| L | Replication, recombination and repair | − | 809 (-2.797) | + | 1206 (2.494) |
| B | Chromatin structure and dynamics | 21 (-0.057) | 27 (0.051) | ||
| Cellular processes and signaling | |||||
| D | Cell cycle control, cell division, chromosome partitioning | 261 (-1.082) | 369 (0.964) | ||
| V | Defense mechanisms | + | 800 (2.046) | 880 (-1.825) | |
| T | Signal transduction mechanisms | 2032 (0.726) | 2482 (-0.647) | ||
| M | Cell wall/membrane/envelope biogenesis | 2438 (0.658) | 2993 (-0.587) | ||
| N | Cell motility | 715 (-1.808) | 1012 (1.612) | ||
| W | Extracellular structures | 226 (0.949) | 253 (-0.846) | ||
| U | Intracellular trafficking, secretion, and vesicular transport | 750 (-1.798) | 1058 (1.603) | ||
| O | Posttranslational modification, protein turnover, chaperones | 1251 (-0.421) | 1607 (0.376) | ||
| Metabolism | |||||
| C | Energy production and conversion | 2357 (-1.677) | 3151 (1.496) | ||
| G | Carbohydrate transport and metabolism | 2519 (0.619) | 3098 (-0.552) | ||
| E | Amino acid transport and metabolism | 3694 (-1.403) | 4840 (1.251) | ||
| F | Nucleotide transport and metabolism | 747 (-0.603) | 977 (0.538) | ||
| H | Coenzyme transport and metabolism | 1854 (-0.344) | 2364 (0.297) | ||
| I | Lipid transport and metabolism | 2090 (1.130) | 2513 (-1.008) | ||
| P | Inorganic ion transport and metabolism | 1883 (-1.820) | 2550 (1.623) | ||
| Q | Secondary metabolites biosynthesis, transport and catabolism | 1367 (0.495) | 1678 (-0.441) | ||
| Poorly characterized | |||||
| R | General function prediction only | + | 3843 (1.965) | 4562 (-1.752) | |
| S | Function unknown | 1685 (-0.887) | 2202 (0.791) | ||
| Mobile elements | |||||
| X | Mobilome: prophages, transposons | 630 (0.830) | 746 (-0.740) | ||
Pearson chi-square analysis testing the independence of species and COG category (X2(22) = 102.9, p<0.001). Each cell in the contingency represents the observed frequency and standardized residual (in between brackets) and is preceded by + or − if the standardized residual is >1.96 or <-1.96, respectively, and significant at p<0.05.
Finally, we searched for COGs that were exclusively present in either B. multivorans or B. cenocepacia genomes. In total, 124 COGs were exclusively present in one or more B. multivorans genomes, but only 21 COGs were uniquely present in all nine of the B. multivorans genomes (Table 6). Conversely, 204 COGs were exclusively present in one or more B. cenocepacia genomes, but only 72 COGs were uniquely present in all six of the B. cenocepacia genomes (S4 Table).
Table 6. B. multivorans-specific COGs.
| COG | COG name | COG category |
|---|---|---|
| COG0062 | NAD(P)H-hydrate repair enzyme Nnr, NAD(P)H-hydrate epimerase domain | F |
| COG0645 | Predicted kinase | R |
| COG1585 | Membrane protein implicated in regulation of membrane protease activity | O |
| COG2312 | Erythromycin esterase homolog | Q |
| COG4121 | tRNA U34 5-methylaminomethyl-2-thiouridine-forming methyltransferase MnmC | J |
| COG5567 | Predicted small periplasmic lipoprotein YifL (function unknown) | S |
| COG5615 | Uncharacterized membrane protein | S |
| COG2519 | tRNA A58 N-methylase Trm61 | J |
| COG2905 | Signal-transduction protein containing cAMP-binding, CBS, and nucleotidyltransferase domains | T |
| COG3059 | Uncharacterized membrane protein YkgB | S |
| COG3095 | Chromosome condensin MukBEF, MukE localization factor | D |
| COG3220 | Uncharacterized conserved protein, UPF0276 family | S |
| COG4823 | Abortive infection bacteriophage resistance protein | V |
| COG5453 | Uncharacterized protein | S |
| COG1107 | Archaea-specific RecJ-like exonuclease, contains DnaJ-type Zn finger domain | L |
| COG1140 | Nitrate reductase beta subunit | CP |
| COG2180 | Nitrate reductase assembly protein NarJ, required for insertion of molybdenum cofactor | CPO |
| COG2181 | Nitrate reductase gamma subunit | CP |
| COG2202 | PAS domain | T |
| COG2427 | Uncharacterized conserved protein YjgD, DUF1641 family | S |
| COG5013 | Nitrate reductase alpha subunit | CP |
COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
Discussion
Of the 20 formally named species within the Bcc, B. multivorans and B. cenocepacia are generally the most prevalent Bcc species in CF [1]. Historically, B. cenocepacia strains have been responsible for large epidemics within the CF community and are often extremely virulent [38]. While infection control measures reduced patient-to-patient transmission and thereby the prevalence of B. cenocepacia, B. multivorans is characterized by a limited person-to-person transmission and subsequently emerged as the most prevalent Bcc pathogen in many countries [1–7]. The low number of outbreaks caused by B. multivorans [39–41] and the fact that isolates from CF patients commonly represent unique strains suggest that strains are acquired from non-human sources such as the natural environment [8]. To examine the extent to which the ST of B. multivorans isolates from CF versus environmental samples explains their genomic content and functionality, we selected four pairs of B. multivorans isolates for whole-genome sequencing, representing distinct STs and consisting of one CF and one environmental isolate each.
MLST is a well-established method for studying the population structure of Bcc organisms [11, 12] and Baldwin et al. [8] previously reported the occurrence of B. multivorans STs that were globally distributed. Recently, whole-genome sequencing of B. pseudomallei revealed that the unexpected occurrence of two B. pseudomallei STs on two continents was due to homoplasy [42]. In contrast, the present study demonstrated that the ST predicted both phylogeny and gene content of B. multivorans isolates (Fig 1 and S1 Fig) and hence corroborates the use of MLST for epidemiological surveillance of Bcc bacteria.
The clinical isolates of ST189 and ST287 were obtained from samples of Canadian CF patients, but the environmental isolates of these STs were soil isolates from Belgium and the United Kingdom, respectively (Table 1). Yet, our analyses showed that, despite this transatlantic barrier, each B. multivorans genomic lineage was defined by its ST, harboring a highly conserved set of genes (S1 Fig). Moreover, isolates belonging to the same ST were isolated up to eleven years apart (Table 1). Finally, searching the Bcc PubMLST database (http://pubmlst.org/bcc/) [13] for additional isolates of the studied STs (Table 1) revealed yet another ST189 isolate that was isolated in 2000 from an Australian CF patient. Altogether, these findings underscore the ubiquity of B. multivorans strains in different niches and on different continents.
To gain insight into the genome biology of B. multivorans, we analyzed all protein-coding genes in terms of homology, specificity, chromosome location and predicted function (i.e. COG category). Firstly, we identified orthologous genes because the conservation of genes may hold clues about which genes are essential for the species-specific lifestyle of B. multivorans. Secondly, we mapped the chromosome location of each CDS because the different chromosomes are associated with different gene copy numbers, mutation rates and expression levels and because the chromosomal location of a gene has an influence on its evolutionary course [31, 43]. Finally, we assigned each CDS to a COG to assess which biological functions were over- or underrepresented [29]. A large fraction of the orthologs (72%) was present in all nine B. multivorans genomes (Fig 3), showing that the B. multivorans isolates possessed a large set of genes regardless of their isolation source. Accordingly, Wolfgang et al. [44] compared clinical and environmental isolates of Pseudomonas aeruginosa, which is also a significant CF pathogen, and demonstrated that most strains, regardless of source, possess the basic pathogenic mechanisms necessary to cause a wide variety of human infections.
The highly conserved multireplicon genomic structure found in the present study was in agreement with the general genome architecture of Bcc organisms [45]. Since primary chromosomes contain generally more core genes [17] it was not surprising to find that C1 was enriched with core orthologs, while C2 and C3 were enriched with ST-specific orthologs (Fig 4B). These ST-specific orthologs were enriched in genes involved in defense mechanisms and secondary metabolism (Table 4), two functional categories that are generally characterized by a large degree of strain-specificity. As shown by Cooper et al. [31], multiple replicons allow for long-term segregation of genes by expression rates and dispensability. This way, secondary chromosomes might serve as evolutionary test beds and the ST-specific orthologs located on C2 and C3 are expected to evolve faster.
C1 was not only enriched with core orthologs but also with orthologs showing random, CF-only and ENV-only specificity (Fig 4B) and non-orthologous CDS (Fig 2B). The enrichment of C1 with random specificity orthologs may be explained by stochastic gene loss or the fact that primarily C1 suffered from unclosed assemblies (Table 2) and annotations could therefore be missing at contig ends. Nevertheless, these findings suggest that, in contrast with the overall highly conserved nature of the largest Bcc chromosome [17], C1 harbors a rather large number of non-orthologous CDS and orthologous CDS that are found only in a smaller subset (CF-only, ENV-only) of the B. multivorans genomes in the present study.
Because the absence or presence of specific genes may hold clues about how B. multivorans differs in lifestyle and epidemiology from B. cenocepacia, we compared the average COG profiles of these two Bcc species. In comparison with B. cenocepacia, the genome of B. multivorans was enriched in COGs involved in translation (J) and replication (L) and deprived in COGs involved in transcription (K). Since COG category K contains many transcriptional regulators, the deprivation in this category may indicate a lower adaptability of B. multivorans to varying environments. This different distribution in COGs involved in information storage and processing may also reflect the overall difference in genome size between these two Bcc species (Table 2 and S3 Table). Indeed, several studies [46, 47] previously demonstrated that the categories translation (J) and replication (L) showed a strong negative correlation with genome size, while transcription (K) showed a strong positive correlation with genome size. Similarly, Carlier et al. [48, 49] showed that the genomes of the obligate leaf nodule endosymbionts Candidatus Burkholderia crenata and Candidatus Burkholderia kirkii were smaller, enriched in COG categories J and L and deprived in COG category K when compared to free-living, facultative endophytic Burkholderia species. Consequently, we may expect that larger genomes require greater regulatory capacity to control their versatile metabolic capacity, as reflected by the higher number of transcriptional regulators.
Next to the differences in average COG profile related to information storage and processing, our comparison also revealed that, as compared to B. cenocepacia, B. multivorans genomes contained less COGs involved in defense mechanisms (V) (Table 5). This finding correlates with B. multivorans generally being less virulent than B. cenocepacia [50, 51]. Similarly, Bartell et al. [52] recently showed that B. cenocepacia produces a wider array of virulence factors compared to B. multivorans. This difference in average COG profile was also reflected by the fact that B. cenocepacia genomes harbored several COGs involved in resistance to antimicrobial compounds (S4 Table). B. multivorans on the other hand harbored COGs that encode a nitrate reductase (Table 6), which corresponds to B. multivorans showing nitrate reduction activity [53, 54]. Although dissimilatory nitrate reduction could enable anaerobic growth, Schwab et al. [54] previously showed that Bcc bacteria are incapable of anaerobic respiration and use fermentation rather than anaerobic respiration to gain energy. Altogether, the present study did not reveal any difference in the average COG profile between B. multivorans and B. cenocepacia that could explain their difference in CF epidemiology.
In this high-throughput sequencing era it is relatively straightforward to obtain draft genome sequences to study the molecular epidemiology of bacterial pathogens [55]. While short-read sequencing platforms yield draft genome assemblies at a low cost, Burkholderia genomes can only be fully resolved using long-read sequencing technologies such as PacBio SMRT sequencing [56, 57]. The present study provides high-quality genome assemblies for eight B. multivorans isolates and the final assemblies produced closed genomes for five of the eight isolates (Table 2). Although the SMRT analysis software already produced high-quality assemblies there was still a need to further polish the resulting assemblies manually (see Methods section). The circular nature of the replicons in combination with the long-read sequencing technology resulted in artificial duplications, as exemplified by the fact that the B. multivorans genomes initially harbored five to seven rRNA operons, while they all harbored five copies after the manual curation. The rRNA copy number is generally quite stable within a species [58] and is thus an easy quality checkpoint when evaluating the status of PacBio assemblies.
In conclusion, the present study demonstrates that the genomic structure of B. multivorans is highly conserved and that the ST predicts the genomic lineage. The high-quality genome assemblies provided in the present study may serve as reference genomes for future studies using transcriptomics and proteomics to try to further elucidate the epidemiology and pathogenicity of this CF pathogen.
Supporting information
Pearson chi-square analysis testing the independence of gene conservation (orthologous vs. non-orthologous CDS) and isolate (X2(8) = 1829.6, p<0.001). Each cell in the contingency represents the observed frequency and standardized residual (in between brackets) and is preceded by + or − if the standardized residual is >1.96 or <-1.96, respectively, and significant at p<0.05.
(PDF)
Pearson chi-square analysis testing the independence of gene conservation (orthologous vs. non-orthologous CDS) and ST (X2(3) = 67.3, p<0.001). Each cell in the contingency represents the observed frequency and standardized residual (in between brackets) and is preceded by + or − if the standardized residual is >1.96 or <-1.96, respectively, and significant at p<0.05.
(PDF)
CF, cystic fibrosis; ENV, environmental.
(PDF)
COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
(PDF)
Ortholog, orthologous CDS; Unique, non-orthologous CDS. C1, chromosome 1; C2, chromosome 2; C3, chromosome 3. TL4B, translocation from C1 to C2 in the ST650-CF isolate. COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
(XLSX)
Core, present in all eight (spec-profile) or nine (spec-profile-atcc, including ATCC 17616) isolates; ST, specific for isolates of one or more STs; ENV-only, only occurring in environmental isolates; CF-only, only occurring in CF isolates; Random, randomly present. C1, chromosome 1; C2, chromosome 2; C3, chromosome 3. TL4B, translocation from C1 to C2 in the ST650-CF isolate. COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
(XLSX)
Scale bar represents number of changes of state required in each character. The tree was rooted on the branch with the largest branch length.
(PDF)
Bar plot showing the average number of CDS per genome per COG category. COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
(PDF)
The eight sequenced B. multivorans isolates from this study were aligned using Mauve [28] against B. multivorans strain ATCC 17616 (PRJNA17407) as a reference.
(TIF)
Acknowledgments
We thank all strain depositors listed in Table 1 to make this study possible. We thank Robert Sebra from the Department of Genetics and Genomic Sciences of the Icahn School of Medicine at Mount Sinai for the PacBio sequencing service. CP is indebted to the Special Research Council of Ghent University.
Data Availability
The annotated genome assemblies were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena) and are publicly available through the GenBank/EMBL/DDBJ accession numbers FKJT01000000, FKJS01000000, FKJU01000000, FKJP01000000, FKJV01000000, FKJW01000000, FKJX01000000 and FKJY01000000.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1. LiPuma JJ. The changing microbial epidemiology in cystic fibrosis. Clinical Microbiology Reviews. 2010;23(2):299–323. 10.1128/CMR.00068-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brisse S, Cordevant C, Vandamme P, Bidet P, Loukil C, Chabanon G, et al. Species distribution and ribotype diversity of Burkholderia cepacia complex isolates from French patients with cystic fibrosis. Journal of Clinical Microbiology. 2004;42(10):4824–4827. 10.1128/JCM.42.10.4824-4827.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. De Boeck K, Malfroot A, Van Schil L, Lebecque P, Knoop C, Govan JRW, et al. Epidemiology of Burkholderia cepacia complex colonisation in cystic fibrosis patients. European Respiratory Journal. 2004;23(6):851–856. 10.1183/09031936.04.00118804 [DOI] [PubMed] [Google Scholar]
- 4. Govan JRW, Brown AR, Jones AM. Evolving epidemiology of Pseudomonas aeruginosa and the Burkholderia cepacia complex in cystic fibrosis lung infection. Future Microbiology. 2007;2(2):153–164. 10.2217/17460913.2.2.153 [DOI] [PubMed] [Google Scholar]
- 5. Norskov-Lauritsen N, Johansen HK, Fenger MG, Nielsen XC, Pressler T, Olesen HV, et al. Unusual distribution of Burkholderia cepacia complex species in Danish cystic fibrosis clinics may stem from restricted transmission between patients. Journal of Clinical Microbiology. 2010;48(8):2981–2983. 10.1128/JCM.00383-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pope CE, Short P, Carter PE. Species distribution of Burkholderia cepacia complex isolates in cystic fibrosis and non-cystic fibrosis patients in New Zealand. Journal of Cystic Fibrosis. 2010;9(6):442–446. 10.1016/j.jcf.2010.08.011 [DOI] [PubMed] [Google Scholar]
- 7. Medina-Pascual MJ, Valdezate S, Villalon P, Garrido N, Rubio V, Saez-Nieto JA. Identification, molecular characterisation and antimicrobial susceptibility of genomovars of the Burkholderia cepacia complex in Spain. European Journal of Clinical Microbiology and Infectious Diseases. 2012;31(12):3385–3396. 10.1007/s10096-012-1707-6 [DOI] [PubMed] [Google Scholar]
- 8. Baldwin A, Mahenthiralingam E, Drevinek P, Pope C, Waine DJ, Henry DA, et al. Elucidating global epidemiology of Burkholderia multivorans in cases of cystic fibrosis by multilocus sequence typing. Journal of Clinical Microbiology. 2008;46(1):290–295. 10.1128/JCM.01818-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coenye T, Vandamme P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environmental Microbiology. 2003;5(9):719–729. 10.1046/j.1462-2920.2003.00471.x [DOI] [PubMed] [Google Scholar]
- 10. Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environmental Microbiology. 2005;7(11):1673–1685. 10.1111/j.1462-2920.2005.00891.x [DOI] [PubMed] [Google Scholar]
- 11. Baldwin A, Mahenthiralingam E, Thickett KM, Honeybourne D, Maiden MCJ, Govan JR, et al. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. Journal of Clinical Microbiology. 2005;43(9):4665–4673. 10.1128/JCM.43.9.4665-4673.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spilker T, Baldwin A, Bumford A, Dowson CG, Mahenthiralingam E, LiPuma JJ. Expanded multilocus sequence typing for Burkholderia species. Journal of Clinical Microbiology. 2009;47(8):2607–2610. 10.1128/JCM.00770-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11(595). 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baldwin A, Mahenthiralingam E, Drevinek P, Vandamme P, Govan JR, Waine DJ, et al. Environmental Burkholderia cepacia complex isolates in human infections. Emerging Infectious Diseases. 2007;13(3):458–461. 10.3201/eid1303.060403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pinto-Carbo M, Sieber S, Dessein S, Wicker T, Verstraete B, Gademann K, et al. Evidence of horizontal gene transfer between obligate leaf nodule symbionts. The ISME Journal. 2016;10(9):2092–2105. 10.1038/ismej.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nazir R, Hansen MA, Sorensen S, van Elsas JD. Draft genome sequence of the soil bacterium Burkholderia terrae strain BS001, which interacts with fungal surface structures. Journal of Bacteriology. 2012;194(16):4480–4481. 10.1128/JB.00725-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nature Reviews Microbiology. 2005;3(2):144–156. 10.1038/nrmicro1085 [DOI] [PubMed] [Google Scholar]
- 18. Ussery DW, Kiil K, Lagesen K, Sicheritz-Ponten T, Bohlin J, Wassenaar TM. The genus Burkholderia: analysis of 56 genomic sequences. Microbial Pathogenomics. 2009;6:140–157. 10.1159/000235768 [DOI] [PubMed] [Google Scholar]
- 19. Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. Reordering contigs of draft genomes using the Mauve Aligner. Bioinformatics. 2009;25(16):2071–2073. 10.1093/bioinformatics/btp356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liao YC, Lin SH, Lin HH. Completing bacterial genome assemblies: strategy and performance comparisons. Scientific Reports. 2015;5:8747 10.1038/srep08747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garcia-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Gotz S, Tarazona S, et al. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 2012;28(20):2678–2679. 10.1093/bioinformatics/bts503 [DOI] [PubMed] [Google Scholar]
- 22. Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9(11):e112963 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bonfield JK, Whitwham A. Gap5—editing the billion fragment sequence assembly. Bioinformatics. 2010;26(14):1699–1703. 10.1093/bioinformatics/btq268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krumsiek J, Arnold R, Rattei T. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23(8):1026–1028. 10.1093/bioinformatics/btm039 [DOI] [PubMed] [Google Scholar]
- 25. Scott D, Ely B. Comparison of genome sequencing technology and assembly methods for the analysis of a GC-rich bacterial genome. Current Microbiology. 2015;70(3):338–344. 10.1007/s00284-014-0721-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 27. Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FSL. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics. 2008;24(23):2803–2804. 10.1093/bioinformatics/btn524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5(6):e11147 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Research. 2015;43(D1):D261–D269. 10.1093/nar/gku1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lerat E, Daubin V, Moran NA. From gene trees to organismal phylogeny in prokaryotes: the case of the γ-Proteobacteria. PLoS Biology. 2003;1(1):e19 10.1371/journal.pbio.0000019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cooper VS, Vohr SH, Wrocklage SC, Hatcher PJ. Why genes evolve faster on secondary chromosomes in bacteria. PLoS Computational Biology. 2010;6(4):e1000732 10.1371/journal.pcbi.1000732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5(1):113 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Notredame C, Higgins DG, Heringa J. T-coffee: a novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology. 2000;302(1):205–217. 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- 34. Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6; 2005.
- 37. Agresti A. Inference for contingency tables In: Categorical Data Analysis. 2nd ed John Wiley & Sons, Inc.; 2002. p. 70–114. Available from: 10.1002/0471249688.ch3 [DOI] [Google Scholar]
- 38. Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clinical Microbiology and Infection. 2010;16(7):821–830. 10.1111/j.1469-0691.2010.03237.x [DOI] [PubMed] [Google Scholar]
- 39. Whiteford ML, Wilkinson JD, McColl JH, Conlon FM, Michie JR, Evans TJ, et al. Outcome of Burkholderia (Pseudomonas) cepacia colonisation in children with cystic fibrosis following a hospital outbreak. Thorax. 1995;50(11):1194–1198. 10.1136/thx.50.11.1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Segonds C, Heulin T, Marty N, Chabanon G. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. Journal of Clinical Microbiology. 1999;37(7):2201–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Biddick R, Spilker T, Martin A, LiPuma JJ. Evidence of transmission of Burkholderia cepacia, Burkholderia multivorans and Burkholderia dolosa among persons with cystic fibrosis. FEMS Microbiology Letters. 2003;228(1):57–62. 10.1016/S0378-1097(03)00724-9 [DOI] [PubMed] [Google Scholar]
- 42. De Smet B, Sarovich DS, Price EP, Mayo M, Theobald V, Kham C, et al. Whole-genome sequencing confirms that Burkholderia pseudomallei multilocus sequence types common to both Cambodia and Australia are due to homoplasy. Journal of Clinical Microbiology. 2015;53(1):323–326. 10.1128/JCM.02574-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morrow JD, Cooper VS. Evolutionary effects of translocations in bacterial genomes. Genome Biology and Evolution. 2012;4(12):1256–1262. 10.1093/gbe/evs099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wolfgang MC, Kulasekara BR, Liang X, Boyd D, Wu K, Yang Q, et al. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences. 2003;100(14):8484–8489. 10.1073/pnas.0832438100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Agnoli K, Schwager S, Uehlinger S, Vergunst A, Viteri DF, Nguyen DT, et al. Exposing the third chromosome of Burkholderia cepacia complex strains as a virulence plasmid. Molecular Microbiology. 2012;83(2):362–378. 10.1111/j.1365-2958.2011.07937.x [DOI] [PubMed] [Google Scholar]
- 46. Cases I, de Lorenzo V, Ouzounis CA. Transcription regulation and environmental adaptation in bacteria. Trends in Microbiology. 2003;11(6):248–253. 10.1016/S0966-842X(03)00103-3 [DOI] [PubMed] [Google Scholar]
- 47. Konstantinidis KT, Tiedje JM. Trends between gene content and genome size in prokaryotic species with larger genomes. Proceedings of the National Academy of Sciences. 2004;101(9):3160–3165. 10.1073/pnas.0308653100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Carlier AL, Eberl L. The eroded genome of a Psychotria leaf symbiont: hypotheses about lifestyle and interactions with its plant host. Environmental Microbiology. 2012;14(10):2757–2769. 10.1111/j.1462-2920.2012.02763.x [DOI] [PubMed] [Google Scholar]
- 49. Carlier A, Fehr L, Pinto-Carbo M, Schaberle T, Reher R, Dessein S, et al. The genome analysis of Candidatus Burkholderia crenata reveals that secondary metabolism may be a key function of the Ardisia crenata leaf nodule symbiosis. Environmental Microbiology. 2016;18(8):2507–2522. 10.1111/1462-2920.13184 [DOI] [PubMed] [Google Scholar]
- 50. Aris RM, Routh JC, LiPuma JJ, Heath DG, Gilligan PH. Lung transplantation for cystic fibrosis patients with Burkholderia cepacia complex. American Journal of Respiratory and Critical Care Medicine. 2001;164(11):2102–2106. 10.1164/ajrccm.164.11.2107022 [DOI] [PubMed] [Google Scholar]
- 51. Jones AM, Dodd ME, Govan JRW, Barcus V, Doherty CJ, Morris J, et al. Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax. 2004;59(11):948–951. 10.1136/thx.2003.017210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bartell JA, Yen P, Varga JJ, Goldberg JB, Papin JA. Comparative metabolic systems analysis of pathogenic Burkholderia. Journal of Bacteriology. 2014;196(2):210–226. 10.1128/JB.00997-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, et al. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. International Journal of Systematic Bacteriology. 1997;47(4):1188–1200. 10.1099/00207713-47-4-1188 [DOI] [PubMed] [Google Scholar]
- 54. Schwab U, Abdullah LH, Perlmutt OS, Albert D, Davis CW, Arnold RR, et al. Localization of Burkholderia cepacia complex bacteria in cystic fibrosis lungs and interactions with Pseudomonas aeruginosa in hypoxic mucus. Infection and Immunity. 2014;82(11):4729–4745. 10.1128/IAI.01876-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McAdam PR, Richardson EJ, Fitzgerald JR. High-throughput sequencing for the study of bacterial pathogen biology. Current Opinion in Microbiology. 2014;19:106–113. 10.1016/j.mib.2014.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shin SC, Ahn DH, Kim SJ, Lee H, Oh TJ, Lee JE, et al. Advantages of single-molecule real-time sequencing in high-GC content genomes. PLoS ONE. 2013;8(7):e68824 10.1371/journal.pone.0068824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koren S, Phillippy AM. One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Current Opinion in Microbiology. 2015;23:110–120. 10.1016/j.mib.2014.11.014 [DOI] [PubMed] [Google Scholar]
- 58. Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Research. 2015;43(D1):D593–D598. 10.1093/nar/gku1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pearson chi-square analysis testing the independence of gene conservation (orthologous vs. non-orthologous CDS) and isolate (X2(8) = 1829.6, p<0.001). Each cell in the contingency represents the observed frequency and standardized residual (in between brackets) and is preceded by + or − if the standardized residual is >1.96 or <-1.96, respectively, and significant at p<0.05.
(PDF)
Pearson chi-square analysis testing the independence of gene conservation (orthologous vs. non-orthologous CDS) and ST (X2(3) = 67.3, p<0.001). Each cell in the contingency represents the observed frequency and standardized residual (in between brackets) and is preceded by + or − if the standardized residual is >1.96 or <-1.96, respectively, and significant at p<0.05.
(PDF)
CF, cystic fibrosis; ENV, environmental.
(PDF)
COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
(PDF)
Ortholog, orthologous CDS; Unique, non-orthologous CDS. C1, chromosome 1; C2, chromosome 2; C3, chromosome 3. TL4B, translocation from C1 to C2 in the ST650-CF isolate. COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
(XLSX)
Core, present in all eight (spec-profile) or nine (spec-profile-atcc, including ATCC 17616) isolates; ST, specific for isolates of one or more STs; ENV-only, only occurring in environmental isolates; CF-only, only occurring in CF isolates; Random, randomly present. C1, chromosome 1; C2, chromosome 2; C3, chromosome 3. TL4B, translocation from C1 to C2 in the ST650-CF isolate. COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
(XLSX)
Scale bar represents number of changes of state required in each character. The tree was rooted on the branch with the largest branch length.
(PDF)
Bar plot showing the average number of CDS per genome per COG category. COG categories: J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
(PDF)
The eight sequenced B. multivorans isolates from this study were aligned using Mauve [28] against B. multivorans strain ATCC 17616 (PRJNA17407) as a reference.
(TIF)
Data Availability Statement
The annotated genome assemblies were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena) and are publicly available through the GenBank/EMBL/DDBJ accession numbers FKJT01000000, FKJS01000000, FKJU01000000, FKJP01000000, FKJV01000000, FKJW01000000, FKJX01000000 and FKJY01000000.