Abstract
Polyploidy is an example of instantaneous speciation when it involves the formation of a new cytotype that is incompatible with the parental species. Because new polyploid individuals are likely to be rare, establishment of a new species is unlikely unless polyploids are able to reproduce through self-fertilization (selfing), or asexually. Conversely, selfing (or asexuality) makes it possible for polyploid species to originate from a single individual—a bona fide speciation event. The extent to which this happens is not known. Here, we consider the origin of Arabidopsis suecica, a selfing allopolyploid between Arabidopsis thaliana and Arabidopsis arenosa, which has hitherto been considered to be an example of a unique origin. Based on whole-genome re-sequencing of 15 natural A. suecica accessions, we identify ubiquitous shared polymorphism with the parental species, and hence conclusively reject a unique origin in favor of multiple founding individuals. We further estimate that the species originated after the last glacial maximum in Eastern Europe or central Eurasia (rather than Sweden, as the name might suggest). Finally, annotation of the self-incompatibility loci in A. suecica revealed that both loci carry non-functional alleles. The locus inherited from the selfing A. thaliana is fixed for an ancestral non-functional allele, whereas the locus inherited from the outcrossing A. arenosa is fixed for a novel loss-of-function allele. Furthermore, the allele inherited from A. thaliana is predicted to transcriptionally silence the allele inherited from A. arenosa, suggesting that loss of self-incompatibility may have been instantaneous.
Keywords: polyploidy, Arabidopsis suecica, Arabidopsis thaliana, Arabidopsis arenosa, shared polymorphism, speciation, hybridization
Introduction
Polyploidy requires a series of unlikely events: the formation of unreduced gametes, hybridization, and the establishment of a new polyploid population (Ramsey and Schemske 1998; Soltis et al. 2015). Nevertheless, whole-genome duplication events have occurred throughout evolutionary history, and have been frequent in plants (Vision et al. 2000; Jiao et al. 2011; Vanneste, Baele, et al. 2014).
The genus Arabidopsis includes two relatively young allotetraploid species: Arabidopsiskamchatica and Arabidopsissuecica (Hylander 1957; Shimizu et al. 2005; Shimizu-Inatsugi et al. 2009). The former is a hybrid between A. lyrata and A. halleri and is limited to East Asia and North America (Shimizu et al. 2005); the latter is a hybrid between A. thaliana and A. arenosa and is limited to the Fennoscandinavian region (O’Kane et al. 1996). Previous studies have suggested that A. suecica originated from a single hybridization event between 12 and 300 Kya (Jakobsson et al. 2006) with A. thaliana as the maternal parent (Price et al. 1994; Hurka 1995; Comai et al. 2000; Säll et al. 2003). The latter conclusion is based partly on sequences from maternally inherited chloroplast genomes, partly on the fact that “synthetic” allotetraploids can be generated by fertilizing autotetraploid A. thaliana (which occur rarely in nature, but can readily be generated in the laboratory) with pollen from naturally autotetraploid A. arenosa (which are common), whereas the reciprocal cross cannot be made (Comai et al. 2000). Thus, the most likely scenario for the formation of A. suecica is that in which a normal (diploid) pollen from tetraploid A. arenosa fertilizes an unreduced gamete of diploid A. thaliana (Jakobsson et al. 2006). In support of this scenario, the A. arenosa complement of A. suecica is more closely related to tetraploid rather than diploid A. arenosa (Novikova et al. 2016). The alternative scenario of mating between diploid parents followed by whole-genome duplication seems less likely.
Whether inheritance in an allotetraploid will be disomic or tetrasomic (or meiosis will fail and lead to aneuploidy) depends largely on the divergence between the parental species (because it allows to prevent homologous pairing), but can also be controlled by specific molecular mechanisms (Griffiths et al. 2006). Cytological studies revealed a diploid-like, homologous chromosomal pairing in A. suecica (Comai et al. 2003). Interestingly, synthetic lines appear to be much less stable (Comai et al. 2000; Madlung et al. 2005; Henry et al. 2014). It has been suggested that such meiotic regularity has a genetic basis and is under selection in polyploids (Henry et al. 2014).
Arabidopsis suecica is currently widely used as a model for studying allotetraploidy in terms of the evolutionary retention of homologs (Chang et al. 2010), the epigenetic regulation of nucleolar dominance (Chen et al. 1998; Pikaard 1999; Pontes et al. 2007; Costa-Nunes et al. 2010; Pontvianne et al. 2012), overall gene expression (Wang et al. 2006; Ha et al. 2009; Ng, Miller, et al. 2014; Ng, Shi, et al. 2014; Tian et al. 2014; Miller et al. 2015), and heterosis (Solhaug et al. 2016). One of the main advantages of A. suecica as a model (in addition to the fact that one of the parents is the model plant A. thaliana), is the possibility to “re-run evolution” by creating synthetic hybrids (Chen et al. 1998; Comai et al. 2000). However, to fully capitalize on this, it is important to understand the history and origin of the natural species better: hence this article. Using whole genome sequencing data of multiple natural A. suecica accessions that cover most of its geographic distribution, we aim to describe the population history of this allotetraploid species: the location and timing of its origin and also the evolution of its ability to self-fertilize which ultimately led to the establishment of A. suecica as a new species.
Results and Discussion
We sequenced (using Illumina 100-bp paired-end reads) 15 natural accessions of A. suecica sampled at different locations throughout the species distribution (supplementary table S1 and fig. S1, Supplementary Material online). We mapped A. suecica reads to the A. thaliana and A. lyrata reference genomes simultaneously, and obtained variant calls from the A. thaliana and A. arenosa components of A. suecica, respectively (see “Materials and Methods” section). Our approach was greatly facilitated by the fact that A. suecica accessions are natural inbred lines as a result of selfing: by only retaining homozygous calls, we avoid many spurious polymorphisms that would have arisen from the misalignment of reads to the wrong parental genome. We mapped 77% of the raw reads on average (supplementary table S1, Supplementary Material online), identifying 167,283 polymorphic sites in 15 A. suecica accessions on the A. thaliana portion of the reference and 416,898 sites on the A. lyrata portion. Throughout the study, results generated for A. suecica are compared with data from the parental species A. thaliana (1001 Genomes Consortium 2016) and A. arenosa (Novikova et al. 2016).
Previous results have suggested that A. suecica had a unique origin, undergoing an extreme bottleneck that completely wiped out ancestral polymorphism, at least in the A. thaliana portion of the genome (Jakobsson et al. 2006). We were thus very surprised to find that 89% of identified polymorphisms for the A. thaliana portion of A. suecica are shared with contemporary A. thaliana. A similar result was obtained for the A. arenosa portion of the genome: 91% of polymorphic sites are shared with A. arenosa (fig. 1, supplementary fig. S2, Supplementary Material online). This amount of shared or, rather, retained ancestral variation clearly contradicts the previously suggested unique origin of A. suecica (Jakobsson et al. 2006), especially since A. thaliana was already selfing when it contributed to A. suecica (see below, supplementary fig. S3, Supplementary Material online), and thus is unlikely to have contributed more than one allele at each locus. Most A. thaliana individuals are almost completely homozygous, and although outcrossing occurs, even a single generation of selfing renders half the genome homozygous (furthermore, as we shall see below, many regions of the genome harbor more than two ancestral haplotypes and must therefore have more than a single ancestor).
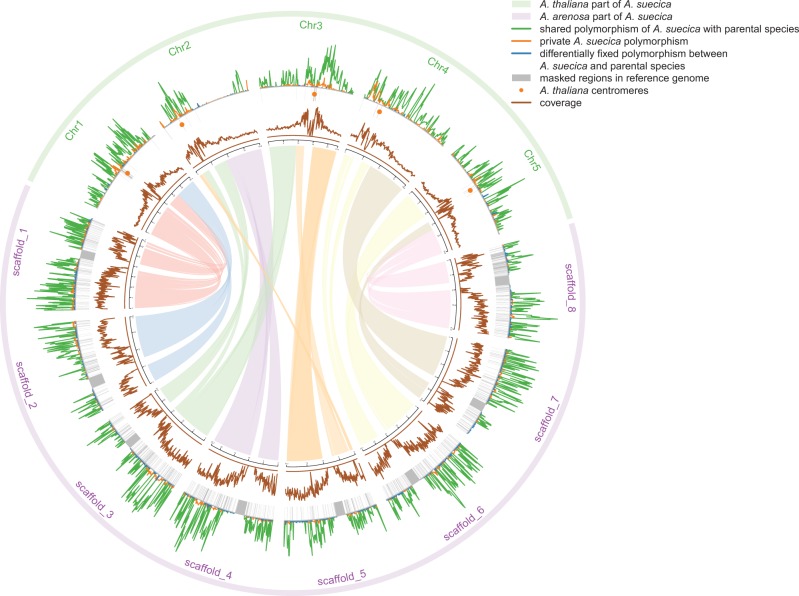
Fig. 1.
Polymorphism density (outer graph) and sequencing coverage (inner, brown graph) along the chromosomes of A. suecica (shown on the outer rim, with the five A. thaliana chromosomes indicated in green and the eight A. lyrata reference genome scaffolds indicated in purple). The polymorphism density (number of SNPs per aligned site) along the genome is shown separately for shared (green), private (orange), and differentially fixed polymorphism (blue). A large non-polymorphic region is located between 7.8 and 16.2 Mbp on chromosome 2 of the A. thaliana portion of A. suecica. Links between the A. thaliana and A. lyrata reference genomes (center) are adapted from (Hu et al. 2011).
Nevertheless, there are clear traces of a major bottleneck, presumably associated with the origin of the new polyploid species from a relatively small number of founders. Not only is the overall level of polymorphism strongly reduced (to roughly 30 and 12% of that of A. thaliana and A. arenosa, respectively), but also non-synonymous and putatively deleterious alleles are present at higher frequencies than in the parental species (supplementary fig. S4A and B, Supplementary Material online)—as expected as a consequence of drift during a bottleneck. We note, however, that purifying selection appears to have been operating after the establishment of the species: among polymorphisms private to A. suecica (i.e., polymorphisms that must have arisen in the species), non-synonymous ones are again biased toward rare alleles (supplementary fig. S4C and D, Supplementary Material online).
There are also large chromosomal regions almost devoid of variation (fig. 1) in the A. suecica genome. While some of these may reflect selective sweeps in the new species, a simpler explanation is that they are a consequence of the foundation bottleneck. Indeed, the ubiquity and size of these regions will make it very difficult to find any genuine selective sweeps. The largest region, on the second chromosome of the A. thaliana portion of the genome, covers most of the long arm (∼8 Mb). We can use this to estimate how old A. suecica is. Under the assumption that the small amount of polymorphism that does exist in this region has been generated solely by new (i.e., non-ancestral) mutations (only 4.5% of polymorphism in this genomic region is shared with A. thaliana, which should be compared with the genome-wide average of 89%, see above), we estimate that the bottleneck occurred ∼16 Kya (95% CI [14.1–18.4 Kya]: other bottlenecked regions give similar results; see “Materials and Methods” section). Consistent with this, estimates of the effective population size over time (using MSMC; Schiffels and Durbin 2014) based on the full data point to a sharp decline following the last glacial maximum roughly 22 Kya (supplementary fig. S5, Supplementary Material online). The decline is particularly noticeable in the A. arenosa portion of the genome, which is expected given that the ancestral species was an obligate outcrosser, implying that this portion underwent a transition to selfing as well.
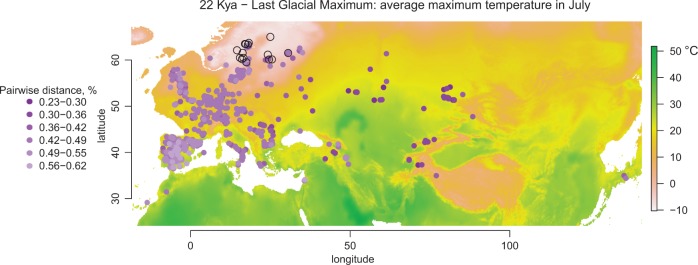
The next question is where A. suecica originated. We sought the ancestral A. thaliana population in the worldwide collection of sequenced A. thaliana genomes (TG Consortium 2016). Based on pairwise sequence divergence, the most closely related A. thaliana accessions appear to be found around the Ural Mountains, and in northern and central Eurasia (fig. 2). Clustering accessions using ADMIXTURE (Alexander et al. 2009), similarly groups A. thaliana accessions from northern and central Eurasia with A. suecica, suggesting a shared past (supplementary fig. S6, Supplementary Material online). Thus A. suecica is not most closely related to the A. thaliana with which it currently coexists, and the ancestral population must have been elsewhere. Indeed, because the Fennoscandinavian region was covered by ice until ∼6 Kya (fig. 2; http://www.worldclim.org/; last accessed January 2017; Hijmans et al. 2005), it is obvious that both species must be recent immigrants. Given that Swedish A. thaliana (unlike Swedish A. suecica) do not appear to be particularly closely related to Russian A. thaliana (1001 Genomes Consortium 2016), a plausible scenario is that A. thaliana mainly reached Scandinavia from the south, via present-day Denmark (1001 Genomes Consortium 2016), while A. suecica took the northern route, via present-day Finland.
Fig. 2.
The A. thaliana accessions most closely related to A. suecica are found in northern and central Eurasia (indicated by the dark violet circles). The background color of the map indicates the average maximum temperature in July during the last glacial maximum (Hijmans et al. 2005). The present distribution of A. suecica (Fennoscandinavia; A. suecica sampling locations are indicated with open black circles) was covered by ice during this time, experiencing temperatures below 0 °C in July.
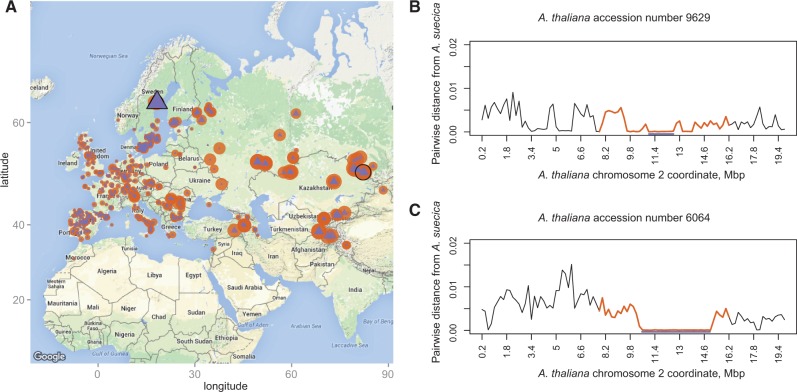
Interestingly, although A. suecica is clearly most closely related to Russian A. thaliana (fig. 2), the largest shared haplotype was found in A. thaliana from northern Sweden. An almost ∼4 Mb segment of the ∼8 Mb bottlenecked region on chromosome 2 (fig. 1) appears to be shared (i.e., identical-by-descent) with an A. thaliana accession from northern Sweden (fig. 3). However, this accession is not particularly closely related to A. suecica in any other sense, and the bottlenecked region does not show a pattern of relatedness different from the genome-wide pattern (fig. 3A–C). Since recent admixture is extremely unlikely (A. thaliana and A. suecica do not produce fertile offspring), one explanation is that this is a case of ancestral haplotype sharing, with the extreme length in northern Sweden being another facet of the generally much more extensive haplotype sharing and linkage disequilibrium in A. thaliana in this part of the world (Long et al. 2013). Simply put, there has effectively been less recombination in northern Swedish A. thaliana since the last glaciation, and this has led to greater haplotype sharing with ancient A. thaliana, and hence with A. suecica. Consistent with this interpretation, there is extensive haplotype sharing in this chromosomal region throughout Europe (fig. 3A).
Fig. 3.
The relationship between A. suecica and A. thaliana accessions in the bottlenecked region corresponding to A. thaliana chromosome 2 (fig. 1). (A) The size of the orange circles is inversely proportional to the average pairwise distance between A. thaliana and A. suecica w.r.t. the bottlenecked region of interest; this distribution is similar to the genome-wide pattern shown in figure 2. The closest accession (9,629) is highlighted with a black circle. The size of the violet triangles is proportional to the length of a haplotype that is shared with A. suecica. The accession with the longest haplotype (6,064) is highlighted with a black triangle. (B,C) The average pairwise distance between 9,629 and 6,064, respectively, and A. suecica. The violet line shows the position of the longest haplotype that is shared with A. suecica; the orange lines show the region inherited from one founder in A. suecica and used to calculate the pairwise distance on A.
Alternatively, the extensive haplotype sharing in northern accessions is a consequence of some A. thaliana having migrated to Sweden together with A. suecica via a northern route, and since admixed with A. thaliana coming from the south. Interestingly, the A. thaliana accession closest to A. suecica (by average pairwise divergence) is from the Altai mountain range in Central Asia (fig. 3B), a region which served as a refugium for many species (including humans; Reich et al. 2010; Prufer et al. 2014) during the glacial interchanges (Tarasov et al. 2000; Pavelkova Ricankova et al. 2014). One can thus imagine a scenario wherein a local A. thaliana population, which contributed to the formation of A. suecica, occupied new territories following the retraction of the ice sheets alongside its hybrid progeny. Where this would have taken place is far from clear. While the closest relatives on the A. thaliana side are currently found in Central Asia, there is no evidence for the other parent of A. suecica—A. arenosa—in this region. Thus, the most likely scenario may be that A. suecica originated somewhere in Eastern Europe and migrated to Fennoscandinavia following the retracting ice, while its parental A. thaliana population additionally spread into Central Asia.
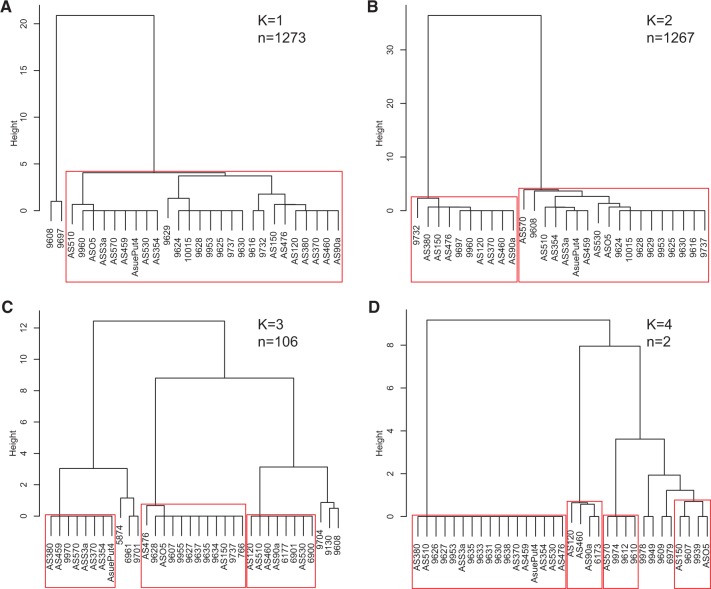
Next, we considered the number of A. thaliana individuals that contributed to the founding of A. suecica. As we have seen, the number of founding haplotypes is one for several regions of the genome, most noticeably on chromosome 2 (figs. 1 and3). As it happens, another example (as in fig. 4A) is a region located at the top of chromosome 4, which harbors the four loci previously used to conclude that A. suecica likely had a unique origin (Jakobsson et al. 2006). This conclusion was thus correct for this region, but is clearly incorrect for most of the genome. To gain further insight into the number of founders, we searched the genome for ancestral haplotype blocks shared between A. suecica and A. thaliana (using PLINK; see “Materials and Methods” section). We identified 1,273 haplotype blocks for which all A. suecica accessions fell into a single cluster of almost identical haplotypes, which we interpret as sharing a single founder haplotype (fig. 4A). Similarly, we found 1,267 blocks for which the accessions can be clearly divided into 2 haplotype clusters; 106 for which they can be divided into 3 haplotype clusters, and 2 for which there were 4 clusters (fig. 4B–D). As a consistency check, we estimated the divergence time for haplotypes belonging to the same founder haplotype and obtained a very similar estimate to that reported above for the chromosome 2 region (95% CI [15.1–16.6 Kya], see “Materials and Methods” section).
Fig. 4.
Number of founder haplotypes in A. suecica. (A–D) Examples of the different number of founder haplotypes in A. suecica. A. suecica accessions are divided into clusters (red boxes) which also include accessions of A. thaliana and are strongly supported by P-values following multiscale bootstrap resampling (“Materials and Methods” section).
The observed numbers of founder haplotypes do not directly correspond to the number of A. thaliana founders, as lineages may have been lost through drift (Nordborg 1998). Of course the number of haplotypes provides a lower bound, and we can therefore conclude that at least four founding individuals contributed (under the assumption that the founders were inbred, which is likely). To see if it is possible to be more precise, we simulated gene genealogies under linear growth models with varying numbers of founding individuals at the estimated time of origin (∼16 Kya). These simulations showed that the observed distribution of founder lineages is compatible with a wide range of parameters (supplementary fig. S7, Supplementary Material online), and it is therefore unlikely that we will be able to refine our estimate of the number of founders further.
Finally, we considered the transition to selfing in A. suecica. A. suecica is self-compatible (Säll et al. 2004), which agrees with the general association between polyploidy and selfing in plants (Barringer 2007). Selfing often evolves through the loss of the self-incompatibility system, which is controlled by the S-locus in flowering plants (Barrett 2002). In the genus Arabidopsis, the tightly linked male SCR (S-locus cysteine-rich protein—present in the pollen coat) and female SRK (S-locus receptor kinase—expressed on the surface of the stigma) determine the specificities of the self-recognition system: the male gene coding for the ligand and the female gene coding for the receptor (Takayama and Isogai 2005). Recognition of SCR by the SRK protein triggers a downstream signaling pathway that prevents pollen tube growth (Comai et al. 2000; Takayama and Isogai 2005; Chapman and Goring 2010). In the predominantly selfing A. thaliana, the S-locus is nonfunctional due to several loss-of-function mutations (Nasrallah et al. 2002; Liu et al. 2007; Sherman-Broyles et al. 2007; Tang et al. 2007; Shimizu et al. 2008; Boggs et al. 2009; Tsuchimatsu et al. 2010).
“Synthetic A. suecica” F1 hybrids, produced by fertilizing colchicine-induced tetraploids of A. thaliana with pollen from naturally tetraploid A. arenosa, were not immediately selfing, and exhibited many abnormal phenotypes compared to natural A. suecica (Chen et al. 1998; Comai et al. 2000), however, they became increasing self-compatible after several rounds of forced self-pollination (Z.J. Chen, personal communication). However, a single A. arenosa collect (Care-1) was used in these crosses, and it is thus not known if other combinations of parents produce fully self-compatible hybrids.
In our sample of natural A. suecica, we found that the S-locus inherited from A. thaliana is fixed for the 213-bp inversion in the SCR gene (supplementary fig. S3, Supplementary Material online) that is suggested to have led to loss of self-incompatibility in A. thaliana (Tsuchimatsu et al. 2010). Therefore, A. thaliana was almost certainly already self-compatible when it contributed to A. suecica, supporting the notion that the transition to selfing is more ancient (Bechsgaard et al. 2006; Tang et al. 2007; Hu et al. 2011).
A rabidopsis arenosa is an obligate outcrosser (Säll et al. 2004), hence A. suecica should have inherited fully functional S-alleles from A. arenosa, and these must somehow have been rendered nonfunctional or silenced. Arabidopsis S-alleles have a complex dominance hierarchy, determined by a small RNA regulatory network (Tarutani et al. 2010; Durand et al. 2014) in which small RNAs from some S-alleles can silence expression of SCR on other S-alleles. The S-locus is a classic example of a long-term balancing selection, with suppressed recombination between SRK and SCR leading to highly diverged S-allele shared across species (Mable et al. 2003, 2004; Castric and Vekemans 2004; Castric et al. 2008; Llaurens et al. 2008). In other words, an S-allele from A. arenosa may be more closely related to, for example, an S-alleles from A. halleri, than to other A. arenosa alleles. The A. thaliana S-allele found in A. suecica is predicted (based on cross-species alignments; see fig. 5 and “Materials and Methods” section) to be an ortholog of the A. halleri S-haplogroup 4, whereas the A. arenosa S-allele found in A. suecica is orthologous to S-haplogroup 2. Based on crosses in A. halleri, the former has been shown to be dominant over the latter (Llaurens et al. 2008), meaning that in A. suecica the S-allele at the locus inherited from A. arenosa is predicted to be transcriptionally silenced by the S-allele at the locus inherited from A. thaliana. Further investigation showed that the main players ensuring such a dominance mechanism (miRNA-producing loci and target sites) are most probably functional in A. suecica (“Materials and Methods” section, supplementary fig. S8, Supplementary Material online).
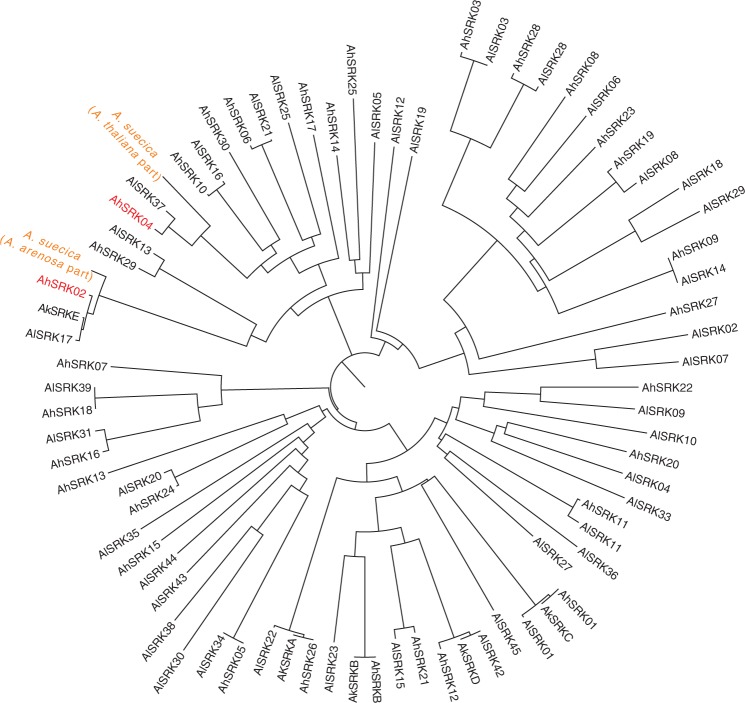
Fig. 5.
The phylogeny of the SRK sequences belonging to A. lyrata (AlSRK), A. halleri (AhSRK), A. kamchatica (AkSRK), and A. suecica. The alignment is adapted from Tsuchimatsu et al. (2012) and the phylogeny was generated using a neighbor-joining algorithm.
Moreover, mapping A. suecica to a complete BAC-sequence of A. halleri S-allele 2 (“Materials and Methods” section) revealed that the S-locus inherited from A. arenosa is fixed for a single S-allele, which exhibited a frame-shift mutation in SCR that is predicted to lead to loss of function (supplementary fig. S9, Supplementary Material online). It is therefore possible that A. suecica was immediately at least partly self-compatible due to the non-functional allele from A. thaliana being dominant over the functional allele from A. arenosa, and that the loss-of-function mutation was fixed later—it could either have been fixed by drift, or by selection. Independent of explanation, this mutation rendered the species fully self-compatible. In support of the former explanation, however, there is no sign of a selective sweep in the S-allele region from A. arenosa (supplementary fig. S10, Supplementary Material online).
In conclusion, whole-genome sequencing of 15 natural A. suecica accessions from the entire species range revealed that the species was almost certainly founded by multiple hybridizations between A. thaliana (as the mother) and tetraploid A. arenosa (as the father), which is in line with generalizations made for the origins of the other well-established allotetraploid species from multiple founders (Soltis et al. 2004; Shimizu-Inatsugi et al. 2009; Vallejo-Marin et al. 2015). Any scenario involving a single origin would have to invoke a completely outcrossed parent on the A. thaliana side (to explain genome-wide allele sharing), and subsequent gene flow between species with dramatically different karyotypes (to explain the regions of the genome that harbor more than two ancestral haplotypes). The species appears to have originated somewhere in central Eurasia in conjunction with the last glacial maximum, and subsequently migrated to Fennoscandinavia. A possibility is that hybridization was facilitated by the production of an unusually high frequency of unreduced A. thaliana ovules in a population containing both parental species, perhaps as a result of environmental stress (Madlung 2013; Vanneste, Maere, et al. 2014; Mason and Pires 2015; Zhou et al. 2015). The transition to self-fertilization may have been facilitated by the dominance of the non-functional S-allele from A. thaliana over the S-allele from A. arenosa, followed by inactivation of the latter.
Materials and Methods
DNA Extraction, Library Preparation, and Sequencing
Whole genomic DNA was extracted from fresh leaf material using DNEasy Plant Mini Kit (Qiagen). Total genomic DNA libraries were prepared using a slightly modified Illumina Genomic DNA Sample Prep protocol. In brief, 100–200 ng DNA was fragmented by sonication with Bioruptor (Diagenode); the peak of fragment sizes was ∼400 bp. End-repair of sheared DNA fragments, A-tailing and adapter ligation were done with the NEXTflex DNA Sequencing Kit (Bio Scientific). Adaptor-modified DNA was resolved on 1.5% low melt agarose (Peqlab) gel. For size selection of the library, DNA was excised from the gel with the size range from 300 to 500 bp. The paired-end DNA libraries were amplified for 10–12 cycles by PCR with VeraSeq PCR Mix (Biozym). Libraries were sequenced in 100-bp paired-end mode on Illumina HiSeq 2000 Analyzers using manufacturer’s standard cluster generation and sequencing protocols.
Raw sequencing data for 12 new A. suecica accessions were uploaded to NCBI SRA under BioProject ID PRJNA309929; data for 3 additional accessions were already published (Novikova et al. 2016) and are available under BioProject ID PRJNA284572 (supplementary table S1, Supplementary Material online).
Read Mapping and Variants Discovery
We mapped A. suecica reads to the A. thaliana (TAIR10) and the A. lyrata (v1.0) references simultaneously using the BWA-MEM algorithm from BWA (Li and Durbin 2009) (version 0.7.8) with an increased penalty of 15 for unpaired read pairs. The A. lyrata reference genome is the closest available reference to A. arenosa, and the successful mapping of A. suecica to a combination of the A. thaliana and A. lyrata genomes has been reported before (Henry et al. 2014). We used Samtools (Li et al. 2009) (version 0.1.19) to sort, index and remove potential duplications from the PCR amplification step of library preparation. We then performed a local realignment with IndelRealigner from Genome Analysis Toolkit (McKenna et al. 2010; DePristo et al. 2011) (version 3.3.0). After filtering for uniquely aligned reads with Samtools, we called sites and variants using GATK UnifiedGenotyper with default parameters. We combined called sites from all A. suecica samples with CombineVariants from GATK and annotated called sites using SNPeff (Cingolani et al. 2012). In order to decrease the number of variant calls from any misaligned homoeologous regions, we filtered out all the heterozygous calls, which were present in the population sample more than once.
We used previously published raw data from A. arenosa clade samples (Novikova et al. 2016). A. arenosa reads were mapped to the A. lyrata (v1.0) reference, using the pipeline described above. All confident A. arenosa calls, including variants, were combined with the A. suecica calls for the A. lyrata component of the reference used for the mapping of A. suecica. The A. thaliana-derived portion of A. suecica was compared throughout the study with variant calls of Eurasian accessions taken from the 1001 Arabidopsis genome project (1001 Genomes Consortium 2016).
Ancestral A. thaliana Population(s) for A. suecica
Shared, private, and differentially fixed polymorphism in A. suecica were compared with the parental species (fig. 1) and were calculated for all sites, for which data were available for at least 80% of individuals in both populations for each comparison. Polymorphism density in figure 1 represents the number of polymorphic sites per aligned site in 200-kb windows along the genome. The R library ‘circlize’ was used to visualize data along the chromosomes of A. suecica.
Pairwise divergence between A. thaliana and A. suecica accessions was calculated with custom scripts as a percentage of diverged sites from all aligned sites, excluding indels. Figure 2 shows minimal divergence between each A. thaliana accession and all A. suecica accessions. Maximum likelihood estimates of individual ancestries (with K = 9 for the number of clusters) were done via ADMIXTURE (Alexander et al. 2009), allowing for missing data. The R library ‘ggmap’ was used to visualize the data points on Google Maps. The R library ‘raster’ was used for visualizing the climate data from http://www.worldclim.org/ with a resolution of 10 min.
In figure 3B and C, the average pairwise distance between 9,629 and 6,064 was calculated in 200-kb windows along the genome. The violet lines in figure 3B and C show the position of the longest haplotype that is shared with A. suecica and is defined by using a threshold of a 0.1% error rate along the region of interest (indicated in orange).
Number of Founder Haplotypes in A. suecica
We divided the A. thaliana portion of the A. suecica genome into 200-kb windows, and estimated pairwise divergence between all A. thaliana and A. suecica accessions. We choose the five closest A. thaliana accessions for each A. suecica accession, which resulted in a range of 5–43 unique A. thaliana accessions, depending on the examined interval. For each interval, we calculated haplotype blocks and all possible haplotype phases for A. suecica together with the selected A. thaliana accessions, using PLINK (Purcell et al. 2007) with default parameters. Using consensus sequences of the most likely haplotype phases for each individual, we performed hierarchical clustering and calculated an uncertainty level for each cluster with the R package ‘pvclust’ (Suzuki and Shimodaira 2006) (method.hclust=“ward.D2”). The uncertainty of each cluster was assessed using an ‘AU’ (Approximately Unbiased) P-value, which is computed by multiscale bootstrap resampling. In order to estimate the number of founder haplotypes in A. suecica, we counted, for each haplotype, the number of clusters with an AU greater than 99%, where A. suecica and A. thaliana accessions are present in the same cluster.
We ran coalescent simulations using the R library ‘scrm’ (analogous to ms; Hudson 2002) under a linear growth model with a varying θ (measured in 4N0 generations) and population size at time t: Nt = n*N0, where N0 is the contemporary population size of A. suecica and Nt is the number of founders for A. suecica. These parameters were uniformly distributed (N0 varied from 1,000 to 100,000) and (Nt varied from 1 to 1,000 for each N0) and result in a total of 100,000 MS runs. For each MS run, 2,648 gene genealogies were generated, where we calculated the number of ancestral lineages for time t = 16 Kya, which match our estimate for the origin time of A. suecica (supplementary fig. S7, Supplementary Material online). A comparison with the observed number of ancestral lineages for 2,648 loci (fig. 4) was conducted via a least squared distance method. An additional 1,000,000 simulations were run with a fixed N0 of 5,000 and from this we chose 1,000 simulations that were close to the observed data, allowing a posterior distribution of the Nt parameter to be inferred.
Dating the Origin of A.suecica
Our estimation of the time of origin of A. suecica was based on the assumption that at loci inherited from one founder, the population diversity within A. suecica is generated solely by new mutations. Therefore, we can estimate the origin time of A. suecica simply as π/2 μ, where π is nucleotide diversity within A. suecica at the single founder region, μ is mutation rate and the generation time is 1 year (A. suecica is an annual plant). Here, we take the mutation rate of A. suecica to equal that of A. thaliana: 7 × 10−9 base substitutions per site per generation (Ossowski et al. 2010). We calculated the origin time of A. suecica as the expected coalescence time within accessions at loci which belong to the same founder haplotype: 95% CI [15.1–16.6 Kya]. Confidence intervals for the median of the distribution were calculated using the basic bootstrap method in the R package ‘boot’. We obtained a similar result for the estimated origin time of A. suecica (95% CI [14.1–18.4 Kya]) by applying the same logic at the largest single founder region on the second chromosome of the A. thaliana portion of the A. suecica genome (between 7.8 and 16.2 Mbp). Nucleotide diversity was calculated in 200-kb windows along this region.
Coalescent rates and the scaled population size over time were inferred using MSMC (Schiffels and Durbin 2014) for six combinations of four randomly chosen A. suecica accessions separately for mapping to A. thaliana and A. lyrata (representing the A. arenosa portion of the A. suecica genome) chromosomes of the combined reference genome. Only intervals with a continuous coverage over 10 kb were chosen for the analysis.
Mechanism of Self-Compatibility in A. suecica
Combining de novo assembly and alignment tactics, we assigned S-haplogroups to A. thaliana- and A. arenosa-derived S-alleles in A. suecica using partial SRK sequences (see below). A. thaliana-derived and A. arenosa-derived S-alleles of A. suecica appear to be orthologous to corresponding A. halleri S-alleles 4 (AhS04) and 2 (AhS02) (fig. 5).
AhS02 and AhS04 are members of the second most recessive class of Arabidopsis S-haplogroups (Durand et al. 2014). However, the inferred pollen phenotype of A. halleri plants with a heterozygous AhS02/AhS04 genotype was that of AhS04 (Llaurens et al. 2008). This suggests that the A. thaliana-derived S-haplogroup (an ortholog of AhS04) could be partially dominant over the A. arenosa-derived S-haplogroup (an ortholog of AhS02) in A. suecica; and that the A. suecica pollen phenotype should correspond to the A. thaliana-derived SCR, which is truncated and allows for selfing. Such a combination of S-haplogroups also appears to be present in all the analyzed A. suecica accessions and, most probably, fixed in the A. suecica species.
Mapping A. suecica to a complete BAC-sequence of A. halleri S-allele 2 (see below) revealed that a loss-of-function mutation at the SCR gene (supplementary fig. S9, Supplementary Material online) is fixed in A. suecica, while functionally and structurally important residues (supplementary fig. S11, Supplementary Material online) are conserved in the SRK gene. It is therefore possible that A. suecica became a selfer following the frame-shift mutation in SCR. However, it is also possible that silencing of the A. arenosa-derived S-allele by the A. thaliana-derived S-allele provided an immediate selfing opportunity for A. suecica, followed by the subsequent pseudogenization of the SCR gene, making A. suecica an irrevocable selfer. In line with this hypothesis, we found that mir867 expressed in the A. thaliana haplotype A of Col-0 as well as the corresponding A. halleri haplotype 4, which is predicted to be able to target the A. halleri haplotype 2 is fully identical to the mature miRNA-producing portion in A. suecica (supplementary fig. S8, Supplementary Material online). Its target sequence in the orthologous A. suecica SCR02 is also fully identical, suggesting that the silencing mechanism could have been active at the speciation time of A. suecica. Target sites of sRNA reads produced by mir867 of AhS04 and the A. thaliana haplotype A were predicted by mapping to the AhSCR02 genomic sequence using a modified Smith-Waterman algorithm and a threshold of 18 (Durand et al. 2014). sRNA sequencing data were from (Durand et al. 2014) for Ah04 in A. halleri (GSM1378105) and from (Montgomery et al. 2008) (ago1-25, GSE13605), (Mi et al. 2008) (AGO4-IP, GSE10036), and (Zheng et al. 2010) (rdr6, GSE23439) for haplotype A in A. thaliana.
Assembly of the A.suecica Accession ASS3a and the Assignment of S-Alleles
We assembled one A. suecica accession (ASS3a) that possessed the highest number of Illumina reads, using SOAPdenovo2 (Luo et al. 2012) (127mer version 1.4.10) with a kmer length equal to 73. We used all the reads, both at the contig and scaffold assembly level (asm_flags = 3). The resulting assembly had an N50 length of scaffolds equal to 38,763 bp.
Using the SCR sequence from A. thaliana (Col-0) as a query (Shimizu et al. 2008), we searched in our de novo A. suecica assembly for the scaffold containing SCR-like sequences using Blast (v. 2.2.28) and applying penalties for the opening and extension of gaps equal to 2 and 1, respectively. With this, we identified scaffold3258 as the scaffold that contains the A. suecica SCR sequence for the A. thaliana-derived portion of the genome.
We obtained available SRK sequences for A. thaliana, A. halleri, and A. lyrata (Schierup et al. 2001; Charlesworth et al. 2003; Bechsgaard et al. 2006; Castric and Vekemans 2007; Tang et al. 2007; Castric et al. 2008, 2010; Tsuchimatsu et al. 2012). Using those SRK sequences as a query, we searched for the scaffolds containing SRK-like sequences in our A. suecica assembly using Blast with the same parameters. Combining the percent of identity with the bit score, we identified C3267705 and scaffold11240 as scaffolds containing the SRK sequences for the A. thaliana and A. arenosa portions of A. suecica, respectively. In order to assign the S-haplogroups in the ASS3a A. suecica accession, we incorporated the obtained SRK sequences from C3267705 and scaffold11240 scaffolds into the SRK alignment (Tsuchimatsu et al. 2012). We used CLC Main Workbench v7.0.2 (CLC bio, Aarhus, Denmark) to align the SRK sequences and applied default parameters for a ‘slow’ alignment: with gap open and extension cost being 10.0 and 1.0, respectively. A neighbor-joining tree from the SRK alignment was constructed using the same software, applying the Jukes-Cantor nucleotide distance as a measure.
In order to check whether all A. suecica accessions carry the same S-haplogroups, we included the partially assembled A. arenosa S-locus sequence from our A. suecica assembly (scaffold11240) to the combined reference genome of A. thaliana (TAIR10) and A. lyrata (v1.0) and mapped all the A. suecica accessions to this novel reference. Mapping was conducted using the pipeline described above, however, we did not filter for ‘primary’ aligned reads. The same pipeline was used for mapping of A. suecica accessions to the BAC-sequence of A. halleri S-allele 2, together with the A. thaliana and A. lyrata reference genomes. Consensus sequences of SCR and SRK genes were obtained with GATK FastaAlternateReferenceMaker (McKenna et al. 2010; DePristo et al. 2011), aligned with MAFFT (Katoh and Standley 2013) (version 7) and visualized with JalView (Waterhouse et al. 2009).
Construction of BAC Libraries
High Molecular Weight (HMW) DNA was prepared from young leaves of Arabidopsis halleri var. P21M53. For the extraction, 20 g of frozen leaf tissue was grounded to a powder in liquid nitrogen with a mortar and pestle in order to prepare megabase-size DNA embedded in agarose plugs. HMW DNA was prepared as described by Peterson et al. (2000) and modified as described by Gonthier et al. (2010). Embedded HMW DNA was partially digested with HindIII (New England Biolabs, Ipswich, MA), and subjected to two size selection steps by pulsed-field electrophoresis, using a BioRad CHEF Mapper system (Bio-Rad Laboratories, Hercules, CA), and ligated to pIndigoBAC-5 HindIII-Cloning Ready vector (EpicentreBiotecnologies, Madison, WI). Pulsed-field migration programs, electrophoresis buffer, and ligation desalting conditions were performed according to Chalhoub et al. (2004). The BAC library is composed by 18,432 clones with a mean insert size of 110 kb and represents 6 genome equivalents.
PacBio RS II Sequencing and Assembly of the S-Locus
2 µg of BAC clone Aha_P21M53_40F08 were pooled with 11 other BAC clones DNA to obtain a total amount of 24 µg. One library was generated using the standard Pacific Biosciences library preparation protocol for 8–12 kb libraries. This library was sequenced in one PacBio RS II SMRT Cell using the P4 polymerase in combination with the C2 chemistry (sequencing service following the standard operating procedures was provided by IGM Genomic Center).
Assembly of the PacBio RS II reads was performed following the HGAP workflow. The SMRT Analysis (v2.2.0) software suite was used for HGAP implementation (https://github.com/PacificBiosciences/Bioinformatics-Training/wiki/HGAP; last accessed January 2017). Reads were first aligned using BLASR (“Blasr on Pacific Biosciences repository website,” n.d.; Chaisson and Tesler 2012) against “Escherichia coli str. K12 substr. DH10B, complete genome”. Identified E. coli reads and low quality reads (read quality <0.80 and read length <500 bp) were removed from data used for the BAC clone sequences assembly. Vector sequences were trimmed as part of the assembly process. Each BAC assembly was individualized by matching its BES to the ends of assembled sequences using BLAST. Annotation of the SRK and SCR genes followed (Goubet et al. 2012) and annotation of small RNA precursors followed (Durand et al. 2014).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Next Generation Sequencing Facility at Vienna Biocenter Core Facilities (VBCF) for obtaining the sequence data. This project received funding, in part, from: the German Research Foundation DFG SPP 1529 to MN; the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (NOVEL project, grant agreement No. 648321) to VC; and a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science to TT.
References
- 1001 Genomes Consortium. 2016. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166:481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SC. 2002. The evolution of plant sexual diversity. Nat Rev Genet. 3:274–284. [DOI] [PubMed] [Google Scholar]
- Barringer BC. 2007. Polyploidy and self-fertilization in flowering plants. Am J Bot. 94:1527–1533. [DOI] [PubMed] [Google Scholar]
- Bechsgaard JS, Castric V, Charlesworth D, Vekemans X, Schierup MH.. 2006. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol Biol Evol. 23:1741–1750. [DOI] [PubMed] [Google Scholar]
- Boggs NA, Nasrallah JB, Nasrallah ME.. 2009. Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet. 5:e1000426.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric V, Bechsgaard J, Schierup MH, Vekemans X.. 2008. Repeated adaptive introgression at a gene under multiallelic balancing selection. PLoS Genet. 4:e1000168.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric V, Bechsgaard JS, Grenier S, Noureddine R, Schierup MH, Vekemans X.. 2010. Molecular evolution within and between self-incompatibility specificities. Mol Biol Evol. 27:11–20. [DOI] [PubMed] [Google Scholar]
- Castric V, Vekemans X.. 2004. Plant self-incompatibility in natural populations: a critical assessment of recent theoretical and empirical advances. Mol Ecol. 13:2873–2889. [DOI] [PubMed] [Google Scholar]
- Castric V, Vekemans X.. 2007. Evolution under strong balancing selection: how many codons determine specificity at the female self-incompatibility gene SRK in Brassicaceae?. BMC Evol Biol. 7:132.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Tesler G.. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13:238.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B, Belcram H, Caboche M.. 2004. Efficient cloning of plant genomes into bacterial artificial chromosome (BAC) libraries with larger and more uniform insert size. Plant Biotechnol J. 2:181–188. [DOI] [PubMed] [Google Scholar]
- Chang PL, Dilkes BP, McMahon M, Comai L, Nuzhdin SV.. 2010. Homoeolog-specific retention and use in allotetraploid Arabidopsis suecica depends on parent of origin and network partners. Genome Biol. 11:R125.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman LA, Goring DR.. 2010. Pollen-pistil interactions regulating successful fertilization in the Brassicaceae. J Exp Bot. 61:1987–1999. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Mable BK, Schierup MH, Bartolomé C, Awadalla P.. 2003. Diversity and linkage of genes in the self-incompatibility gene family in Arabidopsis lyrata. Genetics 164:1519–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Comai L, Pikaard CS.. 1998. Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc Natl Acad Sci U S A. 95:14891–14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Tyagi AP, Lysak MA.. 2003. FISH analysis of meiosis in Arabidopsis allopolyploids. Chromosome Res. 11:217–226. [DOI] [PubMed] [Google Scholar]
- Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y, Byers B.. 2000. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12:1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Nunes P, Pontes O, Preuss SB, Pikaard CS.. 2010. Extra views on RNA-dependent DNA methylation and MBD6-dependent heterochromatin formation in nucleolar dominance. Nucleus 1:254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E, Meheust R, Soucaze M, Goubet PM, Gallina S, Poux C, Fobis-Loisy I, Guillon E, Gaude T, Sarazin A, et al. 2014. Dominance hierarchy arising from the evolution of a complex small RNA regulatory network. Science 346:1200–1205. [DOI] [PubMed] [Google Scholar]
- Gonthier L, Bellec A, Blassiau C, Prat E, Helmstetter N, Rambaud C, Huss B, Hendriks T, Berges H, Quillet MC.. 2010. Construction and characterization of two BAC libraries representing a deep-coverage of the genome of chicory (Cichorium intybus L., Asteraceae). BMC Res Notes 3:225.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet PM, Berges H, Bellec A, Prat E, Helmstetter N, Mangenot S, Gallina S, Holl AC, Fobis-Loisy I, Vekemans X, et al. 2012. Contrasted patterns of molecular evolution in dominant and recessive self-incompatibility haplotypes in Arabidopsis. PLoS Genet. 8:e1002495.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, Colas I, Moore G.. 2006. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439:749–752. [DOI] [PubMed] [Google Scholar]
- Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chen X, Wang X-J, Chen ZJ.. 2009. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci U S A. 106:17835–17840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Dilkes BP, Tyagi A, Gao J, Christensen B, Comai L.. 2014. The BOY NAMED SUE quantitative trait locus confers increased meiotic stability to an adapted natural allopolyploid of Arabidopsis. Plant Cell 26:181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A.. 2005. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 25:1965–1978. [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng J-F, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 43:476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18:337–338. [DOI] [PubMed] [Google Scholar]
- Hurka KMA. 1995. Allopolyploid origin of Arabidopsis suecica (Fries) Norrlin: evidence from chloroplast and nuclear genome markers. Botanica Acta 108:449–456. [Google Scholar]
- Hylander N. 1957. Cardaminopsis suecica (Fr.) Hiit., A northern amphidiploid species. Bull Jard Bot État Bruxelles 271:591–604. [Google Scholar]
- Jakobsson M, Hagenblad J, Tavaré S, Säll T, Halldén C, Lind-Halldén C, Nordborg M.. 2006. A unique recent origin of the allotetraploid species Arabidopsis suecica: evidence from nuclear DNA markers. Mol Biol Evol. 23:1217–1231. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. Subgroup GPDP. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Sherman-Broyles S, Nasrallah ME, Nasrallah JB.. 2007. A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr Biol. 17:734–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llaurens V, Billiard S, Leducq J-B, Castric V, Klein EK, Vekemans X.. 2008. Does frequency-dependent selection with complex dominance interactions accurately predict allelic frequencies at the self-incompatibility locus in Arabidopsis halleri? Evolution 62:2545–2557. [DOI] [PubMed] [Google Scholar]
- Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, Platzer A, Zhang Q, Vilhjalmsson BJ, Korte A, Nizhynska V, et al. 2013. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 45:884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable BK, Beland J, Di Berardo C.. 2004. Inheritance and dominance of self-incompatibility alleles in polyploid Arabidopsis lyrata. Heredity 93:476–486. [DOI] [PubMed] [Google Scholar]
- Mable BK, Schierup MH, Charlesworth D.. 2003. Estimating the number, frequency, and dominance of S-alleles in a natural population of Arabidopsis lyrata (Brassicaceae) with sporophytic control of self-incompatibility. Heredity 90:422–431. [DOI] [PubMed] [Google Scholar]
- Madlung A. 2013. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity 110:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A, Tyagi AP, Watson B, Jiang H, Kagochi T, Doerge RW, Martienssen R, Comai L.. 2005. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 41:221–230. [DOI] [PubMed] [Google Scholar]
- Mason AS, Pires JC.. 2015. Unreduced gametes: meiotic mishap or evolutionary mechanism? Trends Genet. 31:5–10. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Song Q, Shi X, Juenger TE, Chen ZJ.. 2015. Natural variation in timing of stress-responsive gene expression predicts heterosis in intraspecific hybrids of Arabidopsis. Nat Commun. 6:7453.. [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Yoo SJ, Fahlgren N, Gilbert SD, Howell MD, Sullivan CM, Alexander A, Nguyen G, Allen E, Ahn JH, et al. 2008. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci U S A. 105:20055–20062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah ME, Liu P, Nasrallah JB.. 2002. Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297:247–249. [DOI] [PubMed] [Google Scholar]
- Ng DW, Miller M, Yu HH, Huang TY, Kim ED, Lu J, Xie Q, McClung CR, Chen ZJ.. 2014. A role for CHH methylation in the parent-of-origin effect on altered circadian rhythms and biomass heterosis in Arabidopsis intraspecific hybrids. Plant Cell 26:2430–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DW, Shi X, Nah G, Chen ZJ.. 2014. High-throughput RNA-seq for allelic or locus-specific expression analysis in Arabidopsis-related species, hybrids, and allotetraploids. Methods Mol Biol. 1112:33–48. [DOI] [PubMed] [Google Scholar]
- Nordborg M. 1998. On the probability of Neanderthal ancestry. Am J Hum Genet. 63:1237–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova PY, Hohmann N, Nizhynska V, Tsuchimatsu T, Ali J, Muir G, Guggisberg A, Paape T, Schmid K, Fedorenko OM, et al. 2016. Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat Genet. 48:1077–1082. [DOI] [PubMed] [Google Scholar]
- O’Kane SL, Schaal BA, Al-Shehbaz IA.. 1996. The origins of Arabidopsis suecica (Brassicaceae) as indicated by nuclear rDNA sequences. Syst Bot. 21:559. [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledo JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M.. 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327:92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelkova Ricankova V, Robovsky J, Riegert J.. 2014. Ecological structure of recent and last glacial mammalian faunas in northern Eurasia: the case of Altai-Sayan refugium. PLoS One 9:e85056.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DG, Tomkins JP, Frisch DA, Wing RA, Paterson AH.. 2000. Construction of plant bacterial artificial chromosome (BAC) libraries: an illustrated guide. J Agric Genom. 5:1–3. [Google Scholar]
- Pikaard CS. 1999. Nucleolar dominance and silencing of transcription. Trends Plant Sci. 4:478–483. [DOI] [PubMed] [Google Scholar]
- Pontes O, Lawrence RJ, Silva M, Preuss S, Costa-Nunes P, Earley K, Neves N, Viegas W, Pikaard CS.. 2007. Postembryonic establishment of megabase-scale gene silencing in nucleolar dominance. PLoS One 2:e1157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Chandrasekhara C, Feng W, Stroud H, Jacobsen SE, Michaels SD, Pikaard CS.. 2012. Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes Dev. 26:945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RA, Al-Shebaz IA, Palmer JD.. 1994. Systematic relationships of Arabidopsis: a molecular and morphological perspective In:. Meyerowitz E,, Somerville C, editors. Arabidopsis. New York: Cold Spring Harbour Laboratory Press; p. 7–19. [Google Scholar]
- Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey JS, Schemske WD.. 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst. 29:467–501. [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468:1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Säll T, Jakobsson M, Lind-Hallden C, Hallden C.. 2003. Chloroplast DNA indicates a single origin of the allotetraploid Arabidopsis suecica. J Evol Biol. 16:1019–1029. [DOI] [PubMed] [Google Scholar]
- Säll T, Lind-Halldén C, Jakobsson M, Halldén C.. 2004. Mode of reproduction in Arabidopsis suecica. Hereditas 141:313–317. [DOI] [PubMed] [Google Scholar]
- Schierup MH, Mable BK, Awadalla P, Charlesworth D.. 2001. Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158:387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Durbin R.. 2014. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 46:919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman-Broyles S, Boggs N, Farkas A, Liu P, Vrebalov J, Nasrallah ME, Nasrallah JB.. 2007. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell 19:94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu KK, Fujii S, Marhold K, Watanabe K, Kudoh H.. 2005. Arabidopsis kamchatica (Fisch. ex DC.) К. Shimizu & Kudoh and A. kamchatica subsp. kawasakiana (Makino) K. Shimizu & Kudoh, new combinations. Acta Phytotaxonomica Et Geobotanica 56:163–172. [Google Scholar]
- Shimizu KK, Shimizu-Inatsugi R, Tsuchimatsu T, Purugganan MD.. 2008. Independent origins of self-compatibility in Arabidopsis thaliana. Mol Ecol. 17:704–714. [DOI] [PubMed] [Google Scholar]
- Shimizu-Inatsugi R, Lihova J, Iwanaga H, Kudoh H, Marhold K, Savolainen O, Watanabe K, Yakubov VV, Shimizu KK.. 2009. The allopolyploid Arabidopsis kamchatica originated from multiple individuals of Arabidopsis lyrata and Arabidopsis halleri. Mol Ecol. 18:4024–4048. [DOI] [PubMed] [Google Scholar]
- Solhaug EM, Ihinger J, Jost M, Gamboa V, Marchant B, Bradford D, Doerge RW, Tyagi A, Replogle A, Madlung A.. 2016. Environmental regulation of heterosis in the allopolyploid Arabidopsis suecica. Plant Physiol. 170:2251–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE,, Soltis PS, Pires JC, Kovarik A, Tate JA, Mavrodiev E.. 2004. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biol J Linn Soc. 485–501. [Google Scholar]
- Soltis PS, Marchant DB, Van de Peer Y, Soltis DE.. 2015. Polyploidy and genome evolution in plants. Curr Opin Genet Dev. 35:119–125. [DOI] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H.. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542. [DOI] [PubMed] [Google Scholar]
- Takayama S, Isogai A.. 2005. Self-incompatibility in plants. Annu Rev Plant Biol. 56:467–489. [DOI] [PubMed] [Google Scholar]
- Tang C, Toomajian C, Sherman-Broyles S, Plagnol V, Guo Y-L, Hu TT, Clark RM, Nasrallah JB, Weigel D, Nordborg M.. 2007. The evolution of selfing in Arabidopsis thaliana. Science 317:1070–1072. [DOI] [PubMed] [Google Scholar]
- Tarasov PE, Volkova VS, Webb T, Guiot J, Andreev AA, Bezusko LG, Bezusko TV, Bykova GV, Dorofeyuk NI, Kvavadze EV, et al. 2000. Last glacial maximum biomes reconstructed from pollen and plant macrofossil data from northern Eurasia. J Biogeogr. 27:609–620. [Google Scholar]
- Tarutani Y, Shiba H, Iwano M, Kakizaki T, Suzuki G, Watanabe M, Isogai A, Takayama S.. 2010. Trans-acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature 466:983–986. [DOI] [PubMed] [Google Scholar]
- Tian L, Li X, Ha M, Zhang C, Chen ZJ.. 2014. Genetic and epigenetic changes in a genomic region containing MIR172 in Arabidopsis allopolyploids and their progenitors. Heredity 112:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimatsu T, Kaiser P, Yew C-L, Bachelier JB, Shimizu KK.. 2012. Recent loss of self-incompatibility by degradation of the male component in Allotetraploid Arabidopsis kamchatica. PLoS Genet. 8:e1002838.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimatsu T, Suwabe K, Shimizu-Inatsugi R, Isokawa S, Pavlidis P, Städler T, Suzuki G, Takayama S, Watanabe M, Shimizu KK.. 2010. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464:1342–1346. [DOI] [PubMed] [Google Scholar]
- Vallejo-Marin M, Buggs RJ, Cooley AM, Puzey JR.. 2015. Speciation by genome duplication: repeated origins and genomic composition of the recently formed allopolyploid species Mimulus peregrinus. Evolution 69:1487–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K, Baele G, Maere S, Van de Peer Y.. 2014. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 24:1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K, Maere S, Van de Peer Y.. 2014. Tangled up in two: a burst of genome duplications at the end of the Cretaceous and the consequences for plant evolution. Philos Trans R Soc Lond B Biol Sci. 369:20130353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD.. 2000. The origins of genomic duplications in Arabidopsis. Science 290:2114–2117. [DOI] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee HS, Wei NE, Jiang H, Watson B, Madlung A, Osborn TC, Doerge RW, Comai L, et al. 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172:507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ.. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Ryvkin P, Li F, Dragomir I, Valladares O, Yang J, Cao K, Wang LS, Gregory BD.. 2010. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 6:e1001141.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Mo X, Gui M, Wu X, Jiang Y, Ma L, Shi Z, Luo Y, Tang W.. 2015. Cytological, molecular mechanisms and temperature stress regulating production of diploid male gametes in Dianthus caryophyllus L. Plant Physiol Biochem. 97:255–263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.