Abstract
The Anopheles gambiae complex contains a number of highly anthropophilic mosquito species that have acquired exceptional ability to thrive in complex human habitats. Thus, examining the evolutionary history of this Afrotropical mosquito may yield vital information on the selective processes that occurred during the adaptation to human-dominated environments. We performed reduced representation sequencing on 941 mosquitoes of the Anopheles gambiae complex collected across four ecogeographic zones in Cameroon. We find evidence for genetic and geographic subdivision within An. coluzzii and An. gambiae sensu stricto—the two most significant malaria vectors in the region. Importantly, in both species, rural and urban populations are genetically differentiated. Genome scans reveal pervasive signatures of selection centered on genes involved in xenobiotic resistance. Notably, a selective sweep containing detoxification enzymes is prominent in urban mosquitoes that exploit polluted breeding sites. Overall, our study suggests that recent anthropogenic environmental modifications and widespread use of insecticides are driving population differentiation and local adaptation in vectors with potentially significant consequences for malaria epidemiology.
Keywords: Anopheles gambiae, selective sweep, insecticides, pollution, adaptive divergence
Introduction
Humans and their actions on ecosystems can profoundly alter the evolutionary trajectory of other organisms in various ways (Palumbi 2001; Bull and Maron 2016; Hendry et al. 2017). Signatures of human-driven evolutionary changes are particularly evident in cities and urbanizing regions, which often necessitate rapid adaptation to complex environments (Alberti 2015; Alberti et al. 2017; Hendry et al. 2017). A wealth of data exists on genetic and phenotypic signals associated with adaptation to anthropogenic changes in multiple taxa, but the extent of signatures across genomes remain largely unexplored. The widespread use of xenobiotics in agriculture and public health also increases selective pressures on pathogens and their vectors, prompting significant evolutionary adjustments that have yet to be fully elucidated (Georghiou 1972; Mallet 1989; Palumbi 2001). The application of powerful genomic tools now available will help identify the genes and genomic regions underlying adaptation to human-induced selective pressures.
The most effective malaria mosquitoes of the Afrotropical region are remarkably anthropophilic and closely associated with human habitats (White 1974; Coluzzi et al. 1979; Antonio-Nkondjio et al. 2006; Sinka et al. 2010; Dia et al. 2013). Therefore, dissecting the genome of contemporary populations of these species may provide significant insights into the genetic targets of recent selection driven by anthropogenic disturbance. Adaptation to human-dominated environments is challenging and very selective even among highly specialized mosquitoes that feed almost exclusively on human hosts. For example, tens of Anopheles species transmit human malaria parasites across the continent, but only four members of the Anopheles gambiae complex thrive in dense urban areas (Antonio-Nkondjio et al. 2005; Oyewole and Awolola 2006; Mourou et al. 2010; Dabiré et al. 2012; Kamdem et al. 2012). The recent scaling up of measures using insecticides and insecticide-treated materials to combat malaria exposes mosquitoes to an even more disruptive adaptive challenge (Sokhna et al. 2013; WHO 2013). Although the genetic basis remains unknown, studies suggest that both physiological and ecological limits and species ranges are profoundly altered in Anopheles mosquitoes as a result of rapid urbanization and widespread use of insecticides (Jones et al. 2012; Kamdem et al. 2012; Mwangangi et al. 2013; Tene Fossog et al. 2013; Antonio-Nkondjio et al. 2015).
The Anopheles gambiae complex is a group of at least nine isomorphic mosquito species exhibiting varying degrees of geographic and reproductive isolation (Riehle et al. 2011; Coetzee et al. 2013; Fontaine et al. 2015). Owing to ongoing speciation, the principal vectors of the complex have an amazing plasticity in adjusting to diverse ecological conditions, allowing them to track humans across most of tropical Africa (Davidson 1964; Coluzzi et al. 1979, 2002). This adaptive flexibility and the underlying behavioral complexity have undermined at least one major effort at vector control (Molineaux and Gramiccia 1980). Importantly, the epidemiological consequences of adaptive radiation will be even more challenging to tackle if a part of the divergence recently described within the particularly efficient vectors An. gambiae sensu stricto (hereafter An. gambiae) and An. coluzzii is driven by human-induced changes (Touré et al. 1998; Wondji et al. 2005; Slotman et al. 2007; Pinto et al. 2013; Caputo et al. 2014). This hypothesis has been formulated based on ecological observations and population structure, but the genetic underpinnings still need to be clearly assessed by examining patterns of variation across the genome (Coluzzi et al. 2002; Kamdem et al. 2012; Caputo et al. 2014).
To delineate the population structure and the role of recent processes—mainly urbanization and the rapid scaling up of insecticide-treated bed net coverage—in genetic differentiation, we genotyped 941 mosquitoes of the An. gambiae complex collected from diverse environments in Cameroon at >8,000 single nucleotide polymorphisms (SNPs). We found strong evidence for complex geographic and genetic structuring giving rise to seven subpopulations distributed along a continuum of genomic differentiation within both An. gambiae and An. coluzzii. By combining genomic evidence and ecological knowledge, we revealed that the strongest correlates of genetic differentiation in this region are intense suburban agriculture, urbanization, insecticides, and forest-savannah subdivisions. These findings add to a growing body of data indicating that the inevitable rise of insecticide resistance together with ongoing local adaptation of Anopheles species in urban areas are the major challenges for current vector control strategies.
Results
Population Structure of An. gambiae SensuLatoSiblingSpecies
We performed extensive sampling of human-associated Anopheles across the main ecological zones in Cameroon (supplementary table S1 and fig. S1, Supplementary Material online) to collect diverse populations belonging to the four species of the An. gambiae complex that are known to occur in the country (Simard et al. 2009). Certain subpopulations or species can be overlooked when sampling is biased toward one type of population (Riehle et al. 2011). To maximize the chances that our samples best represent the genetic diversity within each species, we used several sampling methods (Service 1993) to collect both larvae and adult populations. In addition, populations of An. gambiae and An. coluzzii segregate along urbanization gradients, which seem to be the most important driver of ecological divergence in the forest zone (Kamdem et al. 2012). To validate this hypothesis and to investigate the genomic targets of local adaptation in urban environments, we surveyed several neighborhoods representing the urban and suburban ecosystems in the two largest cities of the forest area: Douala and Yaoundé (supplementary fig. S1, Supplementary Material online).
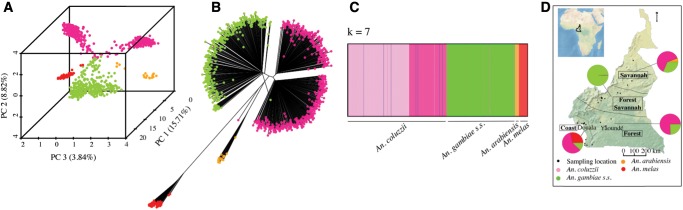
To assess the genetic relatedness among individuals, we subjected all 941 mosquitoes that were morphologically identified as An. gambiae sensu lato (s.l.) to population genomic analysis. Individual mosquitoes were genotyped in parallel at a dense panel of markers using double-digest restriction-associated DNA sequencing (ddRADseq), which enriches for a representative and reproducible fraction of the genome that can be sequenced on the Illumina platform (Peterson et al. 2012). After aligning ddRADseq reads to the An. gambiae reference genome, we used STACKS (Catchen et al. 2011, 2013) to identify 8,476 SNPs (∼1 SNP every 30 kb across the genome) within consensus RAD loci present in more than 80% of individuals, and we inferred the population structure based on these variants. First, we performed principal component analysis (PCA) across all 941 individuals (fig. 1A). The top three components explain 28.4% of the total variance and group individuals into five main clusters. Likewise, a neighbor-joining (NJ) tree, based on Euclidian distance of allele frequencies, shows five distinct clades of mosquitoes (fig. 1B). We hypothesized that these groups at least partially correspond to the four sibling species—An. gambiae, An. coluzzii, An. arabiensis, and An. melas—known to occur in Cameroon. To confirm, we typed a subset of 288 specimens using species identification polymerase chain reactions (PCRs) (Fanello et al. 2002; Santolamazza et al. 2004) and found that each cluster comprised a single species. In agreement with previous surveys (Wondji et al. 2005; Simard et al. 2009), our collections indicate that the brackish water breeding An. melas is limited to coastal regions, whereas the arid-adapted An. arabiensis is restricted to the savannah. In contrast, An. gambiae and An. coluzzii are distributed across the four ecogeographic zones of Cameroon (fig. 1D). Lee et al. (2013) recently reported frequent bouts of hybridization between An. gambiae and An. coluzzii in Cameroon. Although both the PCA and NJ trees clearly separate the two species, the PCA does show the overlap between some rare individuals suggestive of semipermeable species boundaries.
Fig. 1.
Anopheles gambiae complex sibling species are genetically distinct. (A) PCA, (B) NJ tree, and (C) fastSTRUCTURE analyses clearly separate the four An. gambiae complex species that occur in Cameroon into discrete genetic clusters. Additional subdivision below the species level is apparent within An. coluzzii and An. gambiae. (D) Species composition varies strongly between ecogeographic regions. Sampling sites are denoted by black dots.
In support of population structuring below the species level, Bayesian clustering analysis with fastSTRUCTURE (Raj et al. 2014) finds that seven population clusters (k) best explain the genetic variance present in our data (fig. 1C, supplementary fig. S2, Supplementary Material online). Indeed, grouping of samples within An. gambiae and An. coluzzii clades suggests that additional subdivision may exist within each species (fig. 1A and B). Ancestry plots further support inference from the PCA and NJ tree: Although An. arabiensis and An. melas are panmictic, An. coluzzii and An. gambiae display composite ancestries indicative of within-species population structure (fig. 1C, supplementary fig. S2, Supplementary Material online). A cryptic subgroup of An. gambiae has been discovered recently by comparing indoor and outdoor fauna from the same village in Burkina Faso (Riehle et al. 2011). Visual inspections of our PCA, NJ, and fastSTRUCTURE clustering results do not indicate any genetic subdivision based on the collection methods or the developmental stage. To explicitly test for the effects of the sampling methods and the geographic origin of samples on the genetic variance among individuals, we applied a hierarchical analysis of molecular variance (AMOVA) (Excoffier et al. 1992), which confirmed the absence of genetic structuring based on microhabitats or temporal segregations in both species. The large majority of the genetic variation is attributable to differences among individuals. Although the geographic origin accounts for 13.2% (P < 0.001) and 9.4% (P < 0.001) of the variance in An. coluzzii and An. gambiae, respectively, the effect of the types of sample is marginal and not significant.
Population Structure within An. gambiae
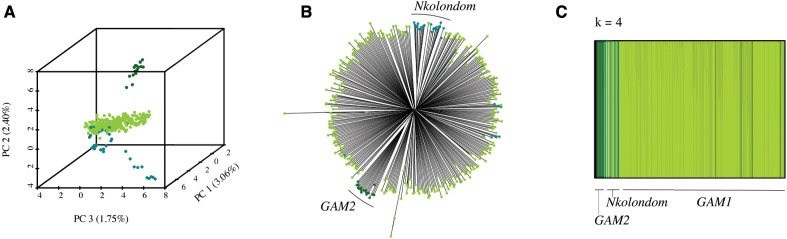
To further resolve the population structure within 357 An. gambiae specimens, we performed population genetic analysis with a set of 9,345 filtered SNPs. Using a combination of PCA, NJ trees, and ancestry assignment, we consistently identify three distinct subgroups within An. gambiae (fig. 2, supplementary fig. S2, Supplementary Material online). The first and largest subgroup (termed GAM1) comprises the vast majority of all An. gambiae specimens including individuals collected in all four ecogeographic regions (supplementary table S1, Supplementary Material online). A total of 17 individuals make up a second small subgroup (termed GAM2). Interestingly, individuals assigned to this cluster include both larvae and adults collected in three different villages spread across two ecogeographic regions. In the absence of any obvious evidence of niche differentiation between GAM1 and GAM2, it is unclear what is driving and/or maintaining divergence between the two sympatric subgroups. Specimens collected from Nkolondom, a suburban neighborhood of Yaoundé where larval sites associated with small-scale agricultural irrigation are common (Fossog Tene et al. 2013; Nwane et al. 2013), form a genetically distinct third subgroup (termed Nkolondom) that appears to be a locally adapted ecotype.
Fig. 2.
Anopheles gambiae is divided into three subpopulations. (A) PCA, (B) NJ tree, and (C) fastSTRUCTURE analyses reveal subdivisions within An. gambiae. We term the most abundant group GAM1, whereas a second small, but widely distributed group, is termed GAM2. Finally, most individuals from the village of Nkolondom are genetically distinct from other An. gambiae suggestive of local adaptation.
Population Structure within An. coluzzii
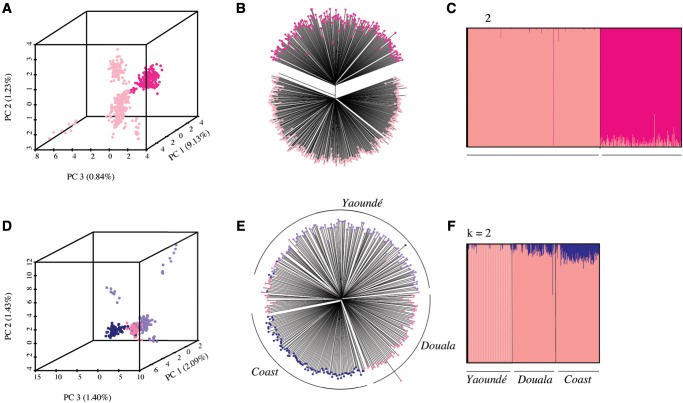
To examine population structure within 521 An. coluzzii specimens, we utilized 9,822 SNPs that passed stringent filtration. All analyses show a clear split between individuals from the northern savannah region and the southern three forested regions of Cameroon (coastal, forest, forest-savannah) (fig. 3A–C). In principle, the north–south structuring could be caused solely by differences in chromosome 2 inversion frequencies, which form a cline from near absence in the humid south to fixation in the arid north (Simard et al. 2009; Fouet et al. 2012). However, we find SNPs from all five chromosomal arms consistently separate northern and southern mosquitoes, indicating a substantial genome-wide divergence between the two populations (supplementary fig. S3, Supplementary Material online).
Fig. 3.
Anopheles coluzzii is divided into four subgroups. (A) PCA, (B) NJ tree, and (C) fastSTRUCTURE reveal major population structuring between An. coluzzii from the northern Savannah ecogeographic region and An. coluzzii from the southern three forested regions. Within the south, (D) PCA, (E) NJ tree, and (F) fastSTRUCTURE analyses separate mosquitoes based on geographic origin, although clustering is not fully discrete indicating a dynamic interplay between local adaptation and migration.
Southern populations of An. coluzzii were collected from three different areas: Douala (the largest city of Cameroon), Yaoundé (the second largest city), and the rural coastal region. PCA, NJ trees, and fastSTRUCTURE show clear clustering of southern samples by collection site (fig. 3D–F). Mosquitoes from Douala, situated on the coastal border, contain a mixture of urban and coastal polymorphisms as illustrated by their intermediate position along PC3 (fig. 3D). Despite considerable geographic segregation, clusters are not fully discrete, likely owing to substantial migration between the three sites. Taken together, the data suggest a dynamic and ongoing process of local adaptation within southern An. coluzzii. In contrast, no similar geographic clustering is observed in northern populations (supplementary figs. S2 and S4, Supplementary Material online). Slight variations can be observed between genetic clustering patterns suggested by different methods. We have only considered subdivisions that were consistent across the three methods we employed. For example, individuals from Yaoundé have been treated as a single subgroup, although PCA showed that a few specimens were relatively detached from the main cluster. This putative subdivision was not supported by the NJ tree and fastSTRUCTURE analyses (fig. 3D–F). All populations described as “urban” were collected from the most urbanized areas of the city, which are characterized by a high frequency of built environments as described in Kamdem et al. (2012).
Relationships between Species and Subgroups
Population genomic analysis identified four different An. gambiae s.l. species present within our samples. Within An. gambiae and An. coluzzii we identified seven potential subgroups with apparently varying levels of divergence. To further explore the relationships between different populations, we built an unrooted NJ tree using pairwise levels of genomic divergence (FST) between all species and subgroups (fig. 4, supplementary table S2, Supplementary Material online). As previously observed in a phylogeny based on whole-genome sequencing (Fontaine et al. 2015), we find that An. melas is highly divergent from all other species (FST ∼ 0.8), whereas An. arabiensis shows intermediate levels of divergence (FST ∼ 0.4) from An. gambiae and An. coluzzii. As expected, the sister species An. gambiae and An. coluzzii are more closely related to each other (FST ∼ 0.2) than any other species. When examining differentiation between subgroups within An. coluzzii, we find that the southern and northern subgroups are relatively divergent (FST > 0.1), whereas differentiation between local ecotypes within the south is much lower (FST < 0.04), corroborating patterns of population structure described within this species (fig. 3D–F). Similarly, as suggested by the genetic structuring, the level of differentiation between An. gambiae subgroups GAM1 and GAM2 is relatively significant (FST ∼ 0.1), whereas the suburban ecotype from Nkolondom shows a low level of divergence from GAM1 (FST ∼ 0.05). To further examine the degree of isolation of subgroups within species, we assessed the reductions in observed heterozygosity with respect to that expected under Hardy–Weinberg Equilibrium among An. coluzzii and An. gambiae populations, by computing the average Wright’s inbreeding coefficient, FIS, across genome-wide SNPs. Values of FIS close to 1 indicate a deviation from Hardy–Weinberg Equilibrium and the existence of cryptic subdivisions, whereas FIS close to 0 suggest that there are no barriers to gene flow. In spite of the strong population genetic structure observed within An. coluzzii and An. gambiae, we found surprisingly low genome-wide FIS values (less than 0.0003) in both species, suggesting a lack of assortative mating. Overall, there is no evidence for reproductive isolation within An. gambiae and An. coluzzii despite ongoing local adaptation and significant population differentiation.
Fig. 4.
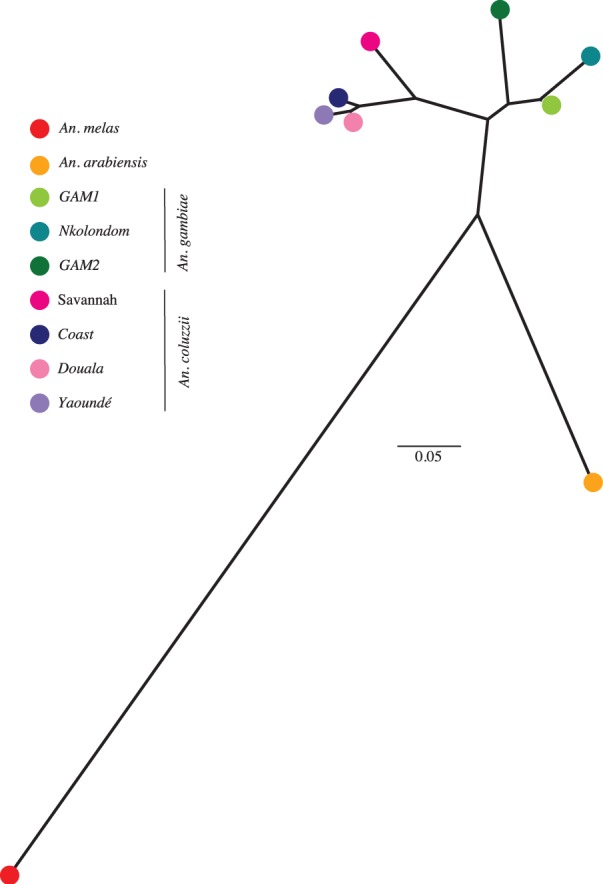
The genetic distance (FST) between populations shows recent radiation within An. gambiae and An. coluzzii clades. In an unrooted, FST-based NJ tree, An. melas is most distant from all other species, whereas An. gambiae and An. coluzzii are sister species. Southern populations of An. coluzzii are more closely related to each other than to the northern savannah population. In contrast to geographic distance, the Douala subpopulation is genetically closer to Yaoundé rather than Coastal mosquitoes. Within An. gambiae, a relatively deep split is present between GAM2 and GAM1, whereas Nkolondom appears to have recently diverged from GAM1.
Signatures of Selection
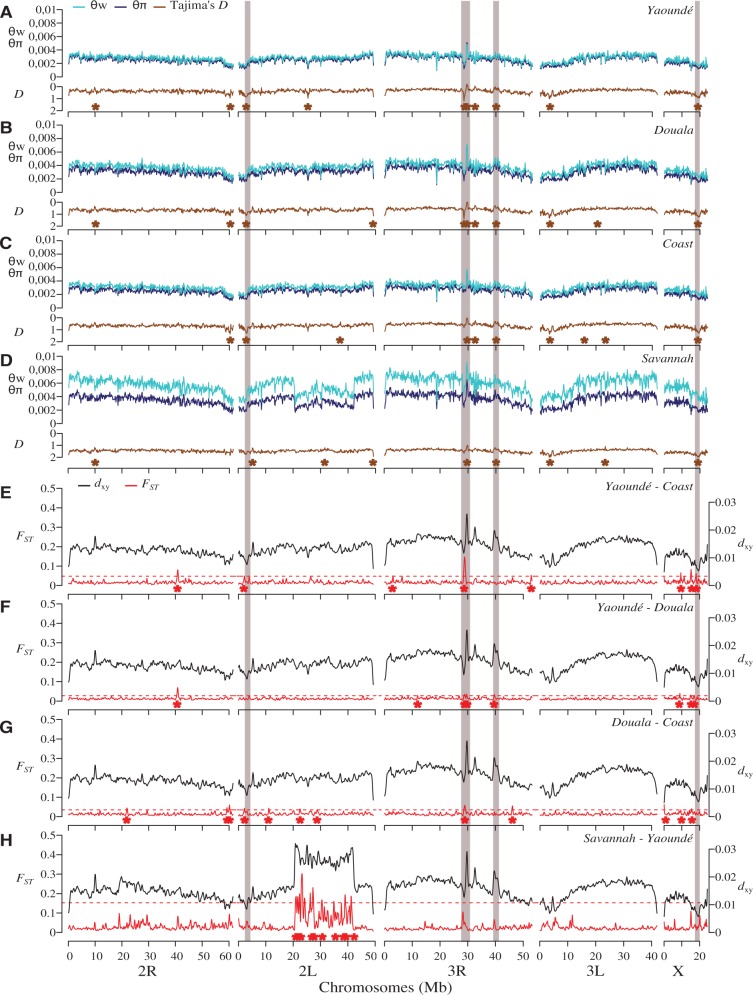
To find potential targets of selection within subgroups, we performed scans of nucleotide diversity (θw, θπ) and allele frequency spectrum (Tajima’s D) in nonoverlapping 150-kb windows across the genome. Scans of θw, θπ, and Tajima’s D were conducted by importing aligned, but otherwise unfiltered, reads directly into ANGSD, which uses genotype likelihoods to calculate summary statistics (Korneliussen et al. 2014). Natural selection can increase the frequency of an adaptive variant within a population, leading to localized reductions in genetic diversity as the haplotype containing the adaptive variant(s) sweeps toward fixation (Maynard Smith and Haigh 1974; Tajima 1989). Selective processes can also promote the coexistence of multiple alleles in the gene pool of a population (balancing selection). Thus, genomic regions harboring targets of selection should exhibit extreme values of diversity and allele frequency spectra relative to genome-wide averages (Storz 2005).
We also performed genome scans using both a relative (FST) and absolute (dxy) measure of divergence calculated with STACKS and ngsTools (Fumagalli et al. 2014), respectively. For both diversity and divergence scans we used a maximum of 40 mosquitoes per population, prioritizing individuals with the highest coverage in populations when sample size exceeded 40. In contrast to Tajima’s D and FST, the genome-wide distribution of dxy and nucleotide diversity in 150-kb sliding windows yielded relatively noisy patterns due to a large variance (Wakeley 1996) (figs. 5 and 6). As a result, the identification of signatures of selection was based primarily on outliers of Tajima’s D and FST. Estimates of dxy and nucleotide diversity were used to confirm genomic locations that were pinpointed as candidate selective sweep on the basis of values of Tajima’s D and FST. Precisely, genomic regions were considered as targets of selection if they mapped to significant peaks or depressions of diversity and dxy, and their values of FST and Tajima’s D were among the top 1% of the empirical distribution in at least one population. Negative Tajima’s D suggests an excess of low-frequency mutations due to population expansion, background selection or selective sweeps, whereas positive Tajima’s D indicates balancing selection.
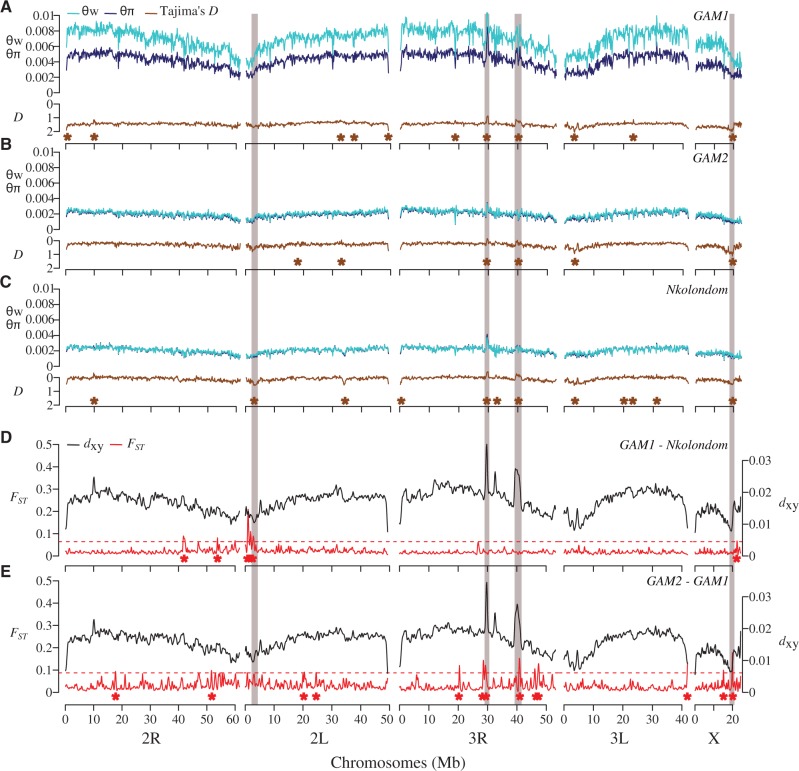
Fig. 5.
Genome scans reveal footprints of global and local adaptation in An. gambiae subpopulations. (A–C) Diversity and Tajima's D are plotted for each of the three subpopulations. Brown asterisks denote windows above the 99th percentile or below the 1st percentile of empirical distribution of Tajima’s D. (D, E) Both absolute (dxy) and relative (FST) divergence between populations are plotted across 150-kb windows. Red asterisks indicate windows above the 99th percentile of empirical distribution of FST. In all populations, concordant dips in diversity and Tajima’s D are evident near the pericentromeric region of 2L where the para sodium channel gene is located. Three other selective sweeps located on the X and 3R chromosomes are highlighted (gray boxes).
Targets of Selection within An. gambiae Subgroups
Our estimates of genome-wide diversity levels (supplementary table S3, Supplementary Material online) within An. gambiae subgroups are comparable to previous values based on RAD sequencing of East African An. gambiae s.l. populations (O’Loughlin et al. 2014). As expected, the large GAM1 population harbors more genetic diversity than the apparently rare GAM2 population or the geographically restricted Nkolondom ecotype (supplementary table S3, Supplementary Material online, fig. 5A–C). Tajima’s D is consistently negative across the entire genome of all three subgroups, indicating an excess of low-frequency variants that are likely the result of recent population expansion (fig. 5A–C). Indeed, demographic models infer relatively recent bouts of population expansion in all three subgroups (supplementary table S4, Supplementary Material online).
Genome scans of each subgroup reveal genomic regions that show concordant reductions in diversity and allele frequency spectrum consistent with selective sweep (highlighted in fig. 5A–C). Based on the 1% cutoff of Tajima’s D (upper and lower bounds) and FST, we identified four candidate regions exhibiting strong signatures of selection. It should be noted that due to the reduced representation sequencing approach we used, our analysis may be highlighting only clear instances of selection, which are likely both recent and strong (Tiffin and Ross-Ibarra 2014). An apparent selective event on the left arm of chromosome 2 near the centromere is found in all populations. This selective sweep is characterized by a prominent depression of Tajima’s D (∼1.5 Mb in width) and contains ∼80 genes including the pyrethroid knockdown resistance gene (kdr). Although it is difficult to precisely identify the specific gene that has been influenced by selection in this region, the voltage-gated sodium channel gene (kdr), which is a pervasive target of selection in Anopheles mosquitoes (Clarkson et al. 2014; Norris et al. 2015), represents the strongest candidate in our populations. Three mutations of the para-type sodium channel conferring knockdown resistance (1014F, 1014L, and 1014S) are widespread in Cameroon (Nwane et al. 2011). The drop in Tajima’s D in the putative kdr sweep is sharpest in the Nkolondom population. Both 1014F and 1014L kdr alleles coexist at high frequencies in mosquitoes from this suburban neighborhood of Yaoundé where larvae can be readily collected from irrigated garden plots that likely contain elevated levels of pesticides directed at agricultural pests (Nwane et al. 2011, 2013; Fossog Tene et al. 2013).
Another region exhibiting consistent signatures of selection in all populations is found around the centromere on the X chromosome. Functional analyses of gene ontology (GO) terms (supplementary table S5, Supplementary Material online) revealed a significant representation of chitin binding proteins in this sweep (P = 1.91e-4). A strong genetic divergence among the three subgroups of An. gambiae characterized by significant FST and dxy peaks is also observed at ∼30 and ∼40 Mb on chromosome 3R. Positive outliers of both Tajima’s D and nucleotide diversity suggest that this genetic divergence is due to balancing selection on multiple alleles among populations. Functional analyses of GO terms indicate that the region at ∼40 Mb on 3R is enriched in cell membrane proteins, genes involved in olfaction and epidermal growth factor genes (supplementary table S5, Supplementary Material online). Finally, a striking depression in Tajima’s D supported by a marked dip in nucleotide diversity occurs on chromosome 2L from ∼33 to 35 Mb in the Nkolondom population exclusively (fig. 5A–C). Despite the lack of genetic differentiation, this region—enriched in six epidermal growth factor genes (supplementary table S5, Supplementary Material online)—is probably a recent selective sweep, which could facilitate larval development of this subgroup in pesticide-laced agricultural water.
Targets of Selection within An. coluzzii Subpopulations
As above, we used diversity, allele frequency spectra, and genetic differentiation metrics to scan for targets of selection in the four subgroups of An. coluzzii. Overall, genetic diversity is higher in the northern savannah population than either of three southern populations, which all exhibit similar levels of diversity (supplementary table S3, Supplementary Material online). Just as in An. gambiae, all subgroups have consistently negative Tajima’s D values confirming demographic models of population expansion (supplementary table S4, Supplementary Material online).
We detected 33 regions where either FST or Tajima’s D fell in the 1% cutoff in at least one population. This included at least ten hotspots of FST clustered within the 2La inversion, which segregates between forest and savannah populations, as well as a region centered on the resistance to dieldrin (rdl) locus at ∼25 Mb on 2L (fig. 6H). However, based on the 1% threshold of both Tajima’s D and FST, we found only five candidate selective sweeps including the kdr region, the sweep on the X chromosome, and the two hot spots of balancing selection detected on the chromosome 3R in An. gambiae (fig. 6A–D). The fifth putative selective sweep characterized by a sharp drop in both diversity and Tajima’s D occurs on 3R from ∼28.5–29.0 Mb, with the decline being more significant in urban populations. The particularly reduced diversity at this sweep in urban mosquitoes strongly suggests it may contain variant(s) that confer adaptation to human-induced changes. Indeed, this genomic region harbors a cluster of both Glutathione S-transferase (GSTE1–GSTE7) and cytochrome P450 (CYP4C27, CYP4C35, CYP4C36) genes, and functional analyses of GO terms reveals an overrepresentation of terms containing “Glutathione S-transferase” (supplementary table S5, Supplementary Material online, P = 5.14e-10). Both the GSTE and cytochrome P450 gene families are known to confer metabolic resistance to insecticides and pollutants in mosquitoes (Enayati et al. 2005; David et al. 2013). In particular, GSTE5 and GSTE6 are intriguing candidate targets of selection as each is upregulated in highly insecticide resistant An. arabiensis populations that recently colonized urban areas of Bobo-Dioulasso, Burkina Faso (Jones et al. 2012).
Fig. 6.
Strong signatures of selection are centered on xenobiotic resistance loci in subpopulations of An. coluzzii. (A–H) Gray boxes highlight five selective sweeps characterized by extreme values of FST and Tajima’s D. As in An. gambiae, sharp declines in diversity and allele frequency spectrum at the para sodium channel gene are present in all populations. A sweep encompassing a cluster of detoxification genes on 3R is sharpest in urban mosquitoes.
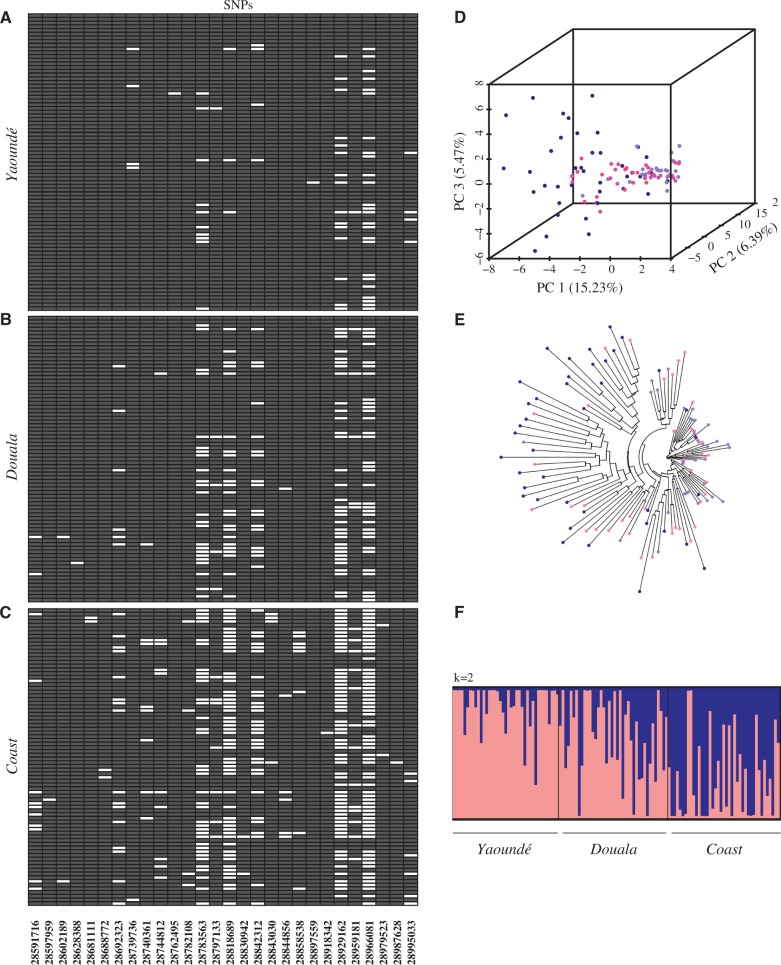
Both relative and absolute divergences between populations at the two selective sweeps involved in xenobiotic resistance probably reflect spatial variation in selection along gradients of anthropogenic disturbance. In particular, the kdr locus exhibits minimal divergence in all pairwise comparisons, suggesting that the same resistance haplotype is under selection in each population (fig. 6E–H). This hypothesis is corroborated by data on the distribution of kdr mutations in Cameroon (Nwane et al. 2011). As opposed to An. gambiae populations, which are broadly polymorphic for three kdr alleles, the 1014L mutation is the major allele present at very high frequencies in An. coluzzii throughout the country. The GSTE/CYP450 sweep on 3R shows a peak in FST, but surprisingly low absolute divergence between Yaoundé and Coastal mosquitoes—a pattern consistent with genomic regions that underwent a selective sweep over a broad geographic area followed by episodes of within-population selection leading to reduced diversity across certain locations (Irwin et al. 2016). Comparisons between Douala and Coastal populations show a more moderate increase in FST, presumably due to high rates of migration between these nearby sites. To further explore the 3R GSTE/CYP450 sweep, we reconstructed haplotypes for all 240 An. coluzzii southern chromosomes across the 28 SNPs found within the sweep. In the Yaoundé population, a single haplotype is present on 44 out of 80 (55%) chromosomes (all gray SNPs), while an additional 11 haplotypes are within one mutational step of this common haplotype (fig. 7A). In Douala, the same haplotype is the most common, but present at a lower frequency (31%) than in Yaoundé (fig. 7B). This haplotype is found on only 6/80 (7.5%) coastal chromosomes (fig. 7C). Population genomic analyses of the same 28 SNPs (fig. 7D–F) mirror results of the haplotype analysis, confirming that haplotype inference did not bias the results.
Fig. 7.
Spatially varying selection between urban and coastal populations. For each of the three southern An. coluzzii subpopulations, 80 reconstructed haplotypes are visualized by color-coding 28 biallelic SNPs in the 3R GSTE/CYP450 sweep either gray or white. A single invariant haplotype—all gray SNPs—is common in (A) Yaoundé, less so in (B) Douala, and very rare in (C) coastal populations. (D, E) Similarly, in PCA and NJ tree analysis of the same 28 SNPs, coastal individuals (navy blue) are diffuse across genotypic space, whereas Yaoundé mosquitoes (purple) are tightly clustered. As expected, Douala (pink) exhibits an intermediate degree of variation. (F) STRUCTURE analysis based solely on the 28 SNPs within the sweep shows clear distinctions between the three populations.
Discussion
Genetic differentiation within species of the An. gambiae Complex
The An. gambiae complex is a model of adaptive radiation with a puzzling evolutionary history (Coluzzi et al. 2002; Ayala and Coluzzi 2005). Due to the lack of observable phenotypes that segregate between populations, it is almost impossible to tackle the genetic basis of divergence between and within species of the complex using traditional genetic mapping methods. Consequently, the examination of sequence divergence in the wild with high-throughput sequencing and genotyping approaches is particularly adapted to address the environmental and genomic targets of ecological divergence in An. gambiae s.l. In that respect, patterns of genomic divergence have started to be dissected and significant insights have been gained into the signatures of selection among continental populations (Lawniczak et al. 2010; Neafsey et al. 2010; White et al. 2011). However, ecological divergence and local adaptation within species of the An. gambiae complex sometimes occur at smaller spatial scales (often a few kilometers) (Sogoba et al. 2008; Kamdem et al. 2012) that are ignored by continental-scale analyses. Here we have applied a population genomic approach to investigate the genomic architecture of selection across a more restricted geographic area characterized by great environmental diversity. We first showed that reduced representation sequencing of 941 An. gambiae s.l. collected in or near human settlements in 33 sites scattered across Cameroon facilitated rapid identification of known sibling species and revealed a complex hierarchical population structure within An. gambiae and An. coluzzii. This result is opposite to that found in East Africa, where RADseq markers detected no population differentiation within species of the An. gambiae complex (O’Loughlin et al. 2014). Indeed, contrary to their East African counterparts, West African populations have a long history of genetic differentiation that is still unresolved despite the application of several generations of genetic markers (Coluzzi et al. 1979, 2002; Touré et al. 1998; Wondji et al. 2002). Polymorphic chromosomal inversions and microsatellite markers have suggested the presence of additional subdivision within An. coluzzii in Cameroon (Wondji et al. 2005; Slotman et al. 2007). Using genome-wide SNPs, we have shown that this subdivision results in at least three genetically distinct clusters including populations from the savannah, coastal, and urban areas. Little is known about the phenotypic differentiation of these populations, but recent studies suggest that mosquitoes from the coastal region tolerate higher concentrations of salt (Tene Fossog et al. 2015) while urban populations are highly resistant to insecticides and environmental pollutants (Antonio-Nkondjio et al. 2011, 2015; Fossog Tene et al. 2013; Tene Fossog et al. 2013). Models of ecological niches of An. gambiae s.l. in Cameroon predict that favorable habitats of An. coluzzii populations are fragmented and marginal in contrast to An. gambiae, which occupies a broad range across the country (Simard et al. 2009). In line with this prediction, we have found a weak differentiation and higher diversity in An. gambiae over significant geographic areas despite the presence of an emerging suburban cluster in Nkolondom.
Genomic Signatures of Selection
When approached with powerful tools, sensitive enough to capture shallow and recent processes, populations depicting increasing levels of genetic differentiation along a speciation continuum may provide insights into the targets of selection at early stages of ecological divergence (Savolainen et al. 2013). We presumed that the genomic divergence will be low given the weak genetic differentiation between the subgroups we described (Nosil and Feder 2012; Andrew and Rieseberg 2013). Indeed, we found signatures of differentiation consistent with selection only across a handful of loci in structured populations of An. gambiae and An. coluzzii. Although some targets of selection are likely missing due to the limited genomic coverage of the RADseq approach we used (Arnold et al. 2013; Tiffin and Ross-Ibarra 2014), the few selective sweeps we found are relevant based on the genes they contain. Specifically, the five main targets of selection in our populations are enriched in genes whose functions include insecticide resistance and detoxification, epidermal growth, cuticle formation and olfaction. The cuticle plays a major role at the interface between several biological functions in insects and recent data support the hypothesis that the coevolution of multiple mechanisms, including cuticular barriers, has occurred in highly pyrethroid-resistant An. gambiae (Balabanidou et al. 2016). Olfaction also mediates a wide range of both adult and larval behaviors including feeding, host preference, and mate selection in blood-feeding insects (Bowen 1991; Carey et al. 2010; Takken and Verhulst 2013). Finally, due to the extreme association between the most effective vectors of the An. gambiae complex and human habitats, a long-standing hypothesis has suggested that human-driven selection is the main modulator of ecological divergence between and within species (Coluzzi et al. 1979, 2002; Kamdem et al. 2012; Caputo et al. 2014). Consistent with this hypothesis, we have found that urban and suburban mosquitoes are genetically differentiated from rural populations and exhibit strong signatures of recent selection at loci containing xenobiotic resistance genes.
Anopheles mosquitoes currently experience strong selective pressures across the African continent due to the increased use of pesticides/insecticides in agriculture and vector control over the last few decades (WHO 2012; Reid and McKenzie 2016). The idea that insecticide resistance can affect the population structure of mosquitoes as a result of a limited gene flow and/or local adaptation between resistant and sensitive populations was already evoked in the 1990s (Silvestrini et al. 1998; Lenormand et al. 1999). In addition to increased resistance, large-scale exposure of vector populations to insecticide-treated bed nets in Africa is also correlated with changes in species distribution and behavioral shifts (Bøgh et al. 1998; Derua et al. 2012; Moiroux et al. 2012; Mwangangi et al. 2013; Sokhna et al. 2013). However, so far, despite ongoing efforts, there is no evidence that these changes can lead to genetic clustering or the creation of cryptic populations within species (e.g., Main et al. 2016). Our data show that the genetic differentiation within the An. gambiae complex in Cameroon is due primarily to regional-scale subdivisions that do not correspond to known distributional or behavioral shifts. Previous studies have documented significant reductions in the population size of An. gambiae s.l. after introduction of long-lasting insecticide-treated nets (Athrey et al. 2012). The demographic history of subgroups of An. gambiae and An. coluzzii does not indicate signals of recent population bottlenecks that can be associated with insecticide exposure (supplementary table S4, Supplementary Material online).
Insecticide resistance, involving both multiple alleles of kdr and significant levels of metabolic resistance, is widespread among An. gambiae and An. coluzzii populations in Cameroon especially in urban areas and locations where intensive farming practices are common (Ndjemaï et al. 2009; Antonio-Nkondjio et al. 2011, 2015; Nwane et al. 2011, 2013; Fossog Tene et al. 2013). Alongside a mass distribution of insecticide-treated nets throughout the country that was held in 2011, indoor residual insecticide spraying has been tested in a few pilot locations, but this measure is not yet applied on a large scale (Etang et al. 2011; Bowen 2013). We hypothesized that regional variations in insecticide exposure exist across the country and contribute to the patterns observed at the two selective sweeps associated with xenobiotic resistance. As expected, urban and suburban populations that are exposed to higher levels of insecticides/pollutants exhibit stronger signals of selection (Kamdem et al. 2012; Fossog Tene et al. 2013; Tene Fossog et al. 2013). Indeed, the synergistic effects of the two types of xenobiotics could be exerting intense selection pressure for pleiotropic resistance in urban mosquitoes (Nwane et al. 2013). Further analysis of the 3R GSTE/CYP450 sweep using a combination of whole-genome resequencing and functional genomic approaches should help resolve the specific genes that are targets of local adaptation in this region of the genome. Additional studies are also needed to further characterize the spatial and temporal variations as well as the haplotypes correlated with insecticide-induced selection within the sweeps.
Effective strategies for managing insecticide resistance that has reached a critical tipping point among malaria vectors on the continent are urgently needed (Hemingway et al. 2016; Ranson and Lissenden 2016). In addition, considerable evidence suggests that insecticide resistance has much deeper consequences on the mosquito biology by interfering with other key biological functions such as the immune system, which in turn profoundly affects the fitness of the vector and its efficiency to transmit the pathogen (vectorial capacity) (Alout et al. 2013, 2014, 2016). As a result, detailed knowledge of genomic signatures of insecticide resistance and their pleiotropic effects on vectorial capacity is necessary to design effective resistance management. Surprisingly, despite intense research on insecticide resistance, this information is lacking in all malaria vectors partly because efforts have so far focused on a few resistance genes and have failed to capture the widespread effects of insecticide resistance across the genome. The population genomic approaches we have used are among the new tools that are being applied to investigate the genomic targets of insecticide resistance at a fine scale in mosquitoes and will facilitate the development of rational resistance management strategies.
Materials and Methods
Mosquito Collections
We collected Anopheles mosquitoes from 33 locations spread across the four major ecogeographic regions of Cameroon from August to November 2013 (supplementary table S1, Supplementary Material online). All collections were performed by researchers who were given malaria prophylaxis before and after the collection period. Indoor resting adult mosquitoes were collected by pyrethrum spray catch, whereas host-seeking adults were obtained via indoor/outdoor human-baited landing catch. Larvae were sampled using standard dipping procedures (Service 1993). Individual mosquitoes belonging to the An. gambiae complex were identified using morphological keys (Gillies and De Meillon 1968; Gillies and Coetzee 1987).
ddRADseq Library Construction
Genomic DNA was extracted from adults using the ZR-96 Quick-gDNA kit (Zymo Research) and from larvae using the DNeasy Extraction kit (Qiagen). A subset of individuals were assigned to sibling species using PCR-RFLP (restriction fragment length polymorphism) assays that type fixed SNP differences in the rDNA (Fanello et al. 2002; Santolamazza et al. 2004). Preparation of ddRAD libraries largely followed (Turissini et al. 2014). Briefly, approximately one-third of the DNA extracted from an individual mosquito (10 µl) was digested with MluCI and NlaIII (New England Biolabs). Barcoded adapters (1 of 48) were ligated to overhangs and 400-bp fragments were selected using 1.5% gels on a BluePippin (Sage Science). One of six indices was added during PCR amplification. Each library contained 288 individuals and was subjected to single-end, 100-bp sequencing across one or two flow cells lanes run on an Illumina HiSeq2500.
Raw sequence reads were demultiplexed and quality filtered using the STACKS v 1.29 process_radtags pipeline (Catchen et al. 2011, 2013). After removal of reads with ambiguous barcodes, incorrect restriction sites, and low sequencing quality (mean Phred < 33), GSNAP (Wu and Nacu 2010) was used to align reads to the An. gambiae PEST reference genome (AgamP3) allowing up to five mismatches per read. The STACKS pipeline was then used to identify unique RAD tags and construct consensus assemblies for each and to call individual SNP genotypes using a maximum-likelihood statistical model.
Population Genomic Analysis
Population genetic structure was assessed using the SNP data set output by the “populations” program of STACKS. We used PLINK v 1.19 to retrieve subsets of genome-wide SNPs as needed (Purcell et al. 2007). PCA, NJ tree analyses, and Bayesian information criterion were implemented using the packages adegenet and ape in R (Paradis et al. 2004; Jombart 2008; R Development Core Team 2014). Ancestry analyses were conducted in fastSTRUCTURE v 1.0 (Raj et al. 2014) using the logistic method. The choosek.py script was used to find the appropriate number of populations (k); in cases where a range of k was suggested, the Bayesian information criterion-inferred number of clusters was chosen. CLUMPP v1.1.2 (Jakobsson and Rosenberg 2007) was used to summarize assignment results across independent runs and DISTRUCT v1.1 (Rosenberg 2004) was used to visualize ancestry assignment of individual mosquitoes. We used a subset of 1,000 randomly chosen SNPs to calculate average pairwise FST between populations in GENODIVE v 2.0 using up to 40 individuals—prioritized by coverage—per population (Meirmans and Van Tienderen 2004). Using this same subset of 1,000 SNPs, we conducted an AMOVA (Excoffier et al. 1992) to quantify the effect of the sampling method and the geographic origin on the genetic variance among individuals in GENODIVE. We used 10,000 permutations to assess significance of FST values and AMOVA. We input pairwise FST values into the program Fitch from the Phylip (Felsenstein 1989) suite to create the population-level NJ tree. FIS values were computed with the “populations” program in STACKS.
Genome Scans for Selection
We used ANGSD v 0.612 (Korneliussen et al. 2014) to calculate nucleotide diversity (θw and θπ) and Tajima’s D in 150-kb nonoverlapping windows. Unlike most genotyping algorithms, ANGSD does not perform hard SNP calls, instead taking genotyping uncertainty into account when calculating summary statistics. Similarly, absolute divergence (dxy) was calculated using ngsTools (Fumagalli et al. 2014) based on genotype likelihoods generated by ANGSD. Kernel-smoothed values for 150-kb windows for all four metrics (θw, θπ,D, dxy) were obtained with the R package KernSmooth. FST (based on AMOVA) was calculated with the “populations” program in STACKS using only loci present in 80% of individuals. A Kernel smoothing procedure implemented in STACKS was used to obtain FST values across 150-kb windows. Because regions with unusually high or low read depth can yield unreliable estimates of diversity and divergence parameters due to the likelihood of repeats and local misassembly, we checked that the average per-locus sequencing coverage was consistent throughout the genome (supplementary fig. S5, Supplementary Material online). To determine whether selective sweeps were enriched for specific functional annotation classes, we used the program DAVID 6.7 with default settings (Huang et al. 2009). We delimitated the physical limits of the selective sweep as the region corresponding to the base of the peak or the depression of Tajima’s D. Haplotypes across the GSTE/CYP450 sweep were reconstructed by PHASE v 2.1.1 using the default recombination model (Stephens et al. 2001; Stephens and Scheet 2005).
Data Accessibility
Raw data (fastq files) for 309 Anopheles gambiae s.l. individuals with the best sequencing coverage are available at http://dx.doi.org/10.5061/dryad.jf402.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
C.K. and B.J.W. conceived and designed the experiments; C.K., B.J.W., and S.G. performed the experiments; C.K., C.F., and B.J.W. analyzed the data and wrote the article.
Supplementary Material
Acknowledgments
This work was supported by the University of California Riverside and National Institutes of Health (1R01AI113248, 1R21AI115271 to B.J.W.). We thank Elysée Nchoutpouem and Raymond Fokom for assistance collecting mosquitoes and Sina Hananian for assisting in DNA extraction. We also thank the editor and three anonymous reviewers for their constructive comments, which helped us to improve the manuscript. This work would not have been possible without the collaboration of inhabitants and administrative authorities of all sampled sites and we want to underscore their generosity and patience.
References
- Alberti M. 2015. Eco-evolutionary dynamics in an urbanizing planet. Trends Ecol Evol. 30:114–126. [DOI] [PubMed] [Google Scholar]
- Alberti M, Marzluff J, Hunt VM.. 2017. Urban driven phenotypic changes: empirical observations and theoretical implications for eco-evolutionary feedback. Philos Trans R Soc B Biol Sci. 372:20160029.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alout H, Dabiré RK, Djogbénou L, Abate L, Corbel V, Chandre F, Cohuet A.. 2016. Interactive cost of Plasmodium infection and insecticide resistance in the malaria vector Anopheles gambiae. Sci Rep. 6:29755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alout H, Djègbè I, Chandre F, Djogbénou LS, Dabiré RK, Corbel V, Cohuet A.. 2014. Insecticide exposure impacts vector–parasite interactions in insecticide-resistant malaria vectors. Proc R Soc B Biol Sci. 281:20140389.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alout H, Ndam NT, Sandeu MM, Djégbe I, Chandre F, Dabiré RK, Djogbénou LS, Corbel V, Cohuet A.. 2013. Insecticide resistance alleles affect vector competence of Anopheles gambiae s.s. for Plasmodium falciparum field isolates. PLoS One 85:e63849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew RL, Rieseberg LH.. 2013. Divergence is focused on few genomic regions early in speciation: incipient speciation of sunflower ecotypes. Evolution 67:2468–2482. [DOI] [PubMed] [Google Scholar]
- Antonio-Nkondjio C, Fossog BT, Ndo C, Djantio BM, Togouet SZ, Awono-Ambene P, Costantini C, Wondji CS, Ranson H.. 2011. Anopheles gambiae distribution and insecticide resistance in the cities of Douala and Yaoundé (Cameroon): influence of urban agriculture and pollution. Malar J. 10:154.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio-Nkondjio C, Kerah CH, Simard F, Awono-Ambene P, Chouaibou M, Tchuinkam T, Fontenille D.. 2006. Complexity of the malaria vectorial system in Cameroon: contribution of secondary vectors to malaria transmission. J Med Entomol. 43:1215–1221. [DOI] [PubMed] [Google Scholar]
- Antonio-Nkondjio C, Simard F, Awono-Ambene P, Ngassam P, Toto JC, Tchuinkam T, Fontenille D.. 2005. Malaria vectors and urbanization in the equatorial forest region of south Cameroon. Trans R Soc Trop Med Hyg. 99:347–354. [DOI] [PubMed] [Google Scholar]
- Antonio-Nkondjio C, Tene Fossog B, Kopya E, Poumachu Y, Menze Djantio B, Ndo C, Tchuinkam T, Awono-Ambene P, Wondji CS.. 2015. Rapid evolution of pyrethroid resistance prevalence in Anopheles gambiae populations from the cities of Douala and Yaoundé (Cameroon). Malar J. 14:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold B, Corbett-Detig RB, Hartl D, Bomblies K.. 2013. RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Mol Ecol. 22:3179–3190. [DOI] [PubMed] [Google Scholar]
- Athrey G, Hodges TK, Reddy MR, Overgaard HJ, Matias A, Ridl FC, Kleinschmidt I, Caccone A, Slotman MA.. 2012. The effective population size of malaria mosquitoes: large impact of vector control. PLoS Genet. 8:e1003097.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala FJ, Coluzzi M.. 2005. Chromosome speciation: humans, Drosophila, and mosquitoes. Proc Natl Acad Sci USA. 102(Suppl.):6535–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabanidou V, Kampouraki A, MacLean M, Blomquist GJ, Tittiger C, Juárez MP, Mijailovsky SJ, Chalepakis G, Anthousi A, Lynd A, et al. 2016. Cytochrome P450 associated with insecticide resistance catalyzes cuticular hydrocarbon production in Anopheles gambiae. Proc Natl Acad Sci USA. 113:201608295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bøgh C, Pedersen EM, Mukoko DA, Ouma JH.. 1998. Permethrin-impregnated bednet effects on resting and feeding behaviour of lymphatic filariasis vector mosquitoes in Kenya. Med Vet Entomol. 12:52–59. [DOI] [PubMed] [Google Scholar]
- Bowen HL. 2013. Impact of a mass media campaign on bed net use in Cameroon. Malar J. 12:36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen MF. 1991. The sensory physiology of host-seeking behavior in mosquitoes. Annu Rev Entomol. 36:139–158. [DOI] [PubMed] [Google Scholar]
- Bull JW, Maron M.. 2016. How humans drive speciation as well as extinction. Proc R Soc B. 283:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo B, Nwakanma D, Caputo FP, Jawara M, Oriero EC, Hamid-Adiamoh M, Dia I, Konate L, Petrarca V, Pinto J, et al. 2014. Prominent intraspecific genetic divergence within Anopheles gambiae sibling species triggered by habitat discontinuities across a riverine landscape. Mol Ecol. 23:4574–4589. [DOI] [PubMed] [Google Scholar]
- Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR.. 2010. Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko W.. 2013. Stacks: an analysis tool set for population genomics. Mol Ecol. 22:3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH.. 2011. Stacks: building and genotyping loci de novo from short-read sequences. G3 (Bethesda) 1:171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson CS, Weetman D, Essandoh J, Yawson AE, Maslen G, Manske M, Field SG, Webster M, Antão T, MacInnis B, et al. 2014. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat Commun. 5:4248.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee M, Hunt RH, Wilkerson R, Della TA, Coulibaly MB, Besansky NJ.. 2013. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 3619:246–274. [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Petrarca V, Di Deco M.. 1979. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 73:483–497. [DOI] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Torre A, Angela M, Deco D, Petrarca V.. 2002. A polytene chromosome analysis of the Anopheles gambiae species complex. Science (80-.) 298:1415–1419. [DOI] [PubMed] [Google Scholar]
- Dabiré RK, Namountougou M, Sawadogo SP, Yaro LB, Toé HK, Ouari A, Gouagna L-C, Simard F, Chandre F, Baldet T, et al. 2012. Population dynamics of Anopheles gambiae s.l. in Bobo-Dioulasso city : bionomics, infection rate and susceptibility to insecticides. Parasit Vectors. 5:127.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David J, Ismail HM, Chandor-Proust A, Ingraham MJ, John M, Paine I.. 2013. Role of cytochrome P450s in insecticide resistance : impact on the control of mosquito-borne diseases and use of insecticides on Earth. Philos Trans R Soc Lond B Biol Sci. 3681612:20120429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson G. 1964. The five mating types of the Anopheles gambiae complex. Riv Malariol. 13:167–183. [PubMed] [Google Scholar]
- Derua YA, Alifrangis M, Hosea KM, Meyrowitsch DW, Magesa SM, Pedersen EM, Simonsen PE.. 2012. Change in composition of the Anopheles gambiae complex and its possible implications for the transmission of malaria and lymphatic filariasis in north-eastern Tanzania. Malar J. 11:188.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dia I, Guelbeogo M, Ayala D.. 2013. Advances and perspectives in the study of the malaria mosquito Anopheles funestus. Anopheles mosquitoes - New insights into malaria vectors, Prof. Sylvie Manguin, editor, InTech, doi: 10.5772/55389.
- Enayati AA, Ranson H, Hemingway J.. 2005. Insect glutathione transferases and insecticide resistance. Insect Mol Biol. 14:3–8. [DOI] [PubMed] [Google Scholar]
- Etang J, Nwane P, Mbida JA, Piameu M, Manga B, Souop D, Awono-Ambene P, Ahmed AM, Braimah N, Drakeley C, et al. 2011. Variations of insecticide residual bio-efficacy on different types of walls: results from a community-based trial in south Cameroon. Malar J. 10:333.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM.. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanello C, Santolamazza F, DellaTorre A.. 2002. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol. 16:461–464. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1989. PHYLIP - Phylogeny Inference Package (Version 3.2). Cladistics 5:164–166. [Google Scholar]
- Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, Jiang X, Hall AB, Catteruccia F, Kakani E, et al. 2015. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science (80-.) 347:1258524–1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossog Tene B, Poupardin R, Costantini C, Awono-Ambene P, Wondji CS, Ranson H, Antonio-Nkondjio C.. 2013. Resistance to DDT in an urban setting: common mechanisms implicated in both M and S forms of Anopheles gambiae in the city of Yaoundé Cameroon. PLoS One 84:e61408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet C, Gray E, Besansky NJ, Costantini C.. 2012. Adaptation to aridity in the malaria mosquito Anopheles gambiae: chromosomal inversion polymorphism and body size influence resistance to desiccation. PLoS One 7:e34841.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Vieira FG, Linderoth T.. 2014. ngsTools : methods for population genetics analyses from Next-Generation Sequencing data. Bioinformatics. 3010:1486–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georghiou GP. 1972. The evolution of resistance to pesticides. Annu Rev Ecol Syst. 3:133–168. [Google Scholar]
- Gillies MT, Coetzee M.. 1987. A supplement to the Anophelinae of Africa south of the Sahara. Johannesburg (South Africa: ): The South African Institute for Medical Research. [Google Scholar]
- Gillies MT, De Meillon B.. 1968. The Anophelinae of Africa South of the Sahara. 2nd ed. Johannesburg(South Africa): Publications of the South African Institute for Medical Research.
- Hemingway J, Ranson H, Magill A, Kolaczinski J, Fornadel C, Gimnig J, Coetzee M, Simard F, Roch DK, Hinzoumbe CK, et al. 2016. Averting a malaria disaster: will insecticide resistance derail malaria control? Lancet 387:1785–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry AP, Gotanda KM, Svensson EI.. 2017. Human influences on evolution, and the ecological and societal consequences. Philos Trans R Soc Lond B Biol Sci. 3721712:20160028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Irwin DE, Alcaide M, Delmore KE, Irwin JH, Owens GL.. 2016. Recurrent selection explains genomic regions of high relative but low absolute differentiation in the greenish warbler ring species. Mol Ecol. 25:4488–4507. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg N.. 2007. CLUMPP: a cluster matching and permutation program for dealing with multimodality in analysis of population structure. Bioinformatics 23:1801–1806. [DOI] [PubMed] [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. [DOI] [PubMed] [Google Scholar]
- Jones CM, Toé HK, Sanou A, Namountougou M, Hughes A, Diabaté A, Dabiré R, Simard F, Ranson H.. 2012. Additional selection for insecticide resistance in urban malaria vectors: DDT resistance in Anopheles arabiensis from Bobo-Dioulasso, Burkina Faso. PLoS One 7:e45995.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdem C, Tene Fossog B, Simard F, Etouna J, Ndo C, Kengne P, Boussès P, Etoa F-X, Awono-Ambene P, Fontenille D, et al. 2012. Anthropogenic habitat disturbance and ecological divergence between incipient species of the malaria mosquito Anopheles gambiae. PLoS One 7:e39453.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen T, Albrechtsen A, Nielsen R.. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15:356.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak MKN, Emrich SJ, Holloway AK, Regier AP, Olson M, White B, Redmond S, Fulton L, Appelbaum E, Godfrey J, et al. 2010. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science 330:512–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Marsden CD, Norris LC, Collier TC, Main BJ, Fofana A, Cornel AJ, Lanzaro GC.. 2013. Spatiotemporal dynamics of gene flow and hybrid fitness between the M and S forms of the malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci USA. 110:19854–19859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T, Bourguet D, Guillemaud T, Raymond M.. 1999. Tracking the evolution of insecticide resistance in the mosquito Culex pipiens. Nature 400:861–864. [DOI] [PubMed] [Google Scholar]
- Main BJ, Lee Y, Ferguson HM, Kreppel KS, Kihonda A, Govella NJ, Collier TC, Cornel AJ, Eskin E, Kang EY, et al. 2016. The genetic basis of host preference and resting behavior in the major African malaria vector, Anopheles arabiensis. PLoS Genet. 12:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. 1989. The evolution of insecticide resistance: have the insects won? Trends Ecol Evol. 4:336–340. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J.. 1974. The hitch-hiking effect of a favorable gene. Genet Res. 23:22–35. [PubMed] [Google Scholar]
- Meirmans P, Van Tienderen P.. 2004. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes. 4:792–794. [Google Scholar]
- Moiroux N, Gomez MB, Pennetier C, Elanga E, Djènontin A, Chandre F, Djègbé I, Guis H, Corbel V.. 2012. Changes in Anopheles funestus biting behavior following universal coverage of long-lasting insecticidal nets in Benin. J Infect Dis. 206:1622–1629. [DOI] [PubMed] [Google Scholar]
- Molineaux L, Gramiccia G.. 1980. The Garki Project. Research on the epidemiology and control of malaria in the Sudan Savanna of West Africa. Geneva: World Health Organization. [Google Scholar]
- Mourou J-R, Coffinet T, Jarjaval F, Pradines B, Amalvict R, Rogier C, Kombila M, Pagès F.. 2010. Malaria transmission and insecticide resistance of Anopheles gambiae in Libreville and Port-Gentil, Gabon. Malar J. 9:321.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangangi JM, Mbogo CM, Orindi BO, Muturi EJ, Midega JT, Nzovu J, Gatakaa H, Githure J, Borgemeister C, Keating J, et al. 2013. Shifts in malaria vector species composition and transmission dynamics along the Kenyan coast over the past 20 years. Malar J. 12:13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndjemaï HNM, Patchoké S, Atangana J, Etang J, Simard F, Bilong CFB, Reimer L, Cornel A, Lanzaro GC, Fondjo E.. 2009. The distribution of insecticide resistance in Anopheles gambiae s.l. populations from Cameroon: an update. Trans R Soc Trop Med Hyg. 103:1127–1138. [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Lawniczak MKN, Park DJ, Redmond SN, Coulibaly MB, Traoré SF, Sagnon N, Costantini C, Johnson C, Wiegand RC, et al. 2010. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science 330:514–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris LC, Main BJ, Lee Y, Collier TC, Fofana A, Cornel AJ, Lanzaro GC.. 2015. Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proc Natl Acad Sci USA. 1123:815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Feder JL.. 2012. Widespread yet heterogeneous genomic divergence. Mol Ecol. 21:2829–2832. [DOI] [PubMed] [Google Scholar]
- Nwane P, Etang J, Chouaïbou M, Toto J, Mimpfoundi R, Simard F.. 2011. Kdr-based insecticide resistance in Anopheles gambiae s.s populations in. BMC Res Notes. 4:463.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwane P, Etang J, Chouaïbou M, Toto JC, Koffi A, Mimpfoundi R, Simard F.. 2013. Multiple insecticide resistance mechanisms in Anopheles gambiae s.l. populations from Cameroon, Central Africa. Parasit Vectors. 6:41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loughlin SM, Magesa S, Mbogo C, Mosha F, Midega J, Lomas S, Burt A.. 2014. Genomic analyses of three malaria vectors reveals extensive shared polymorphism but contrasting population histories. Mol Biol Evol. 314:889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyewole IO, Awolola TS.. 2006. Impact of urbanisation on bionomics and distribution of malaria vectors in Lagos, southwestern Nigeria. J Vector Borne Dis. 43:173–178. [PubMed] [Google Scholar]
- Palumbi SR. 2001. Evolution—humans as the world’s greatest evolutionary force. Science (80-.) 293:1786–1790. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. Analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. [DOI] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE.. 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7:e37135.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto J, Egyir-Yawson A, Vicente J, Gomes B, Santolamazza F, Moreno M, Charlwood J, Simard F, Elissa N, Weetman D, et al. 2013. Geographic population structure of the African malaria vector Anopheles gambiae suggests a role for the forest-savannah biome transition as a barrier to gene flow. Evol Appl. 6:910–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, Maller J, Sklar P, de Bakker P, Daly M, et al. 2007. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 813:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2014. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing. [Google Scholar]
- Raj A, Stephens M, Pritchard JK.. 2014. FastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197:573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson H, Lissenden N.. 2016. Insecticide resistance in African Anopheles mosquitoes: a worsening situation that needs urgent action to maintain malaria control. Trends Parasitol. 32:187–196. [DOI] [PubMed] [Google Scholar]
- Reid MC, McKenzie FE.. 2016. The contribution of agricultural insecticide use to increasing insecticide resistance in African malaria vectors. Malar J. 15:107.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehle MM, Guelbeogo WM, Gneme A, Eiglmeier K, Holm I, Bischoff E, Garnier T, Snyder GM, Li X, Markianos K, et al. 2011. A cryptic subgroup of Anopheles gambiae is highly susceptible to human malaria parasites. Science 331:596–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. 2004. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Resour. 4:137–138. [Google Scholar]
- Santolamazza F, Della TA, Caccone A.. 2004. Short report : a new polymerase chain reaction—restriction fragment length polymorphism method to identify Anopheles arabiensis from An. gambiae and its two molecular forms from degraded DNA templates or museum samples. Trop Med. 70:604–606. [PubMed] [Google Scholar]
- Savolainen O, Lascoux M, Merilä J.. 2013. Ecological genomics of local adaptation. Nat Rev Genet. 14:807–820. [DOI] [PubMed] [Google Scholar]
- Service MW. 1993. Mosquito ecology: field sampling methods. London: Elsevier Applied Science; (editor). [Google Scholar]
- Silvestrini F, Severini C, di Pardo V, Romi R, de Matthaeis E, Raymond M.. 1998. Population structure and dynamics of insecticide resistance genes in Culex pipiens populations from Italy. Heredity 81:342–348. [Google Scholar]
- Simard F, Ayala D, Kamdem GC, Pombi M, Etouna J, Ose K, Fotsing J-M, Fontenille D, Besansky NJ, Costantini C.. 2009. Ecological niche partitioning between Anopheles gambiae molecular forms in Cameroon: the ecological side of speciation. BMC Ecol. 9:17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinka ME, Bangs MJ, Manguin S, Coetzee M, Mbogo CM, Hemingway J, Patil AP, Temperley WH, Gething PW, Kabaria CW, et al. 2010. The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic précis. Parasit Vectors. 3:117.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman MA, Tripet F, Cornel AJ, Meneses CR, Lee Y, Reimer LJ, Thiemann TC, Fondjo E, Fofana A, Traoré SF, et al. 2007. Evidence for subdivision within the M molecular form of Anopheles gambiae. Mol Ecol. 16:639–649. [DOI] [PubMed] [Google Scholar]
- Sogoba N, Vounatsou P, Bagayoko MM, Doumbia S, Dolo G, Gosoniu L, Traoré SF, Smith TA, Touré YT.. 2008. Spatial distribution of the chromosomal forms of Anopheles gambiae in Mali. Malar J. 7:205.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokhna C, Ndiath MO, Rogier C.. 2013. The changes in mosquito vector behaviour and the emerging resistance to insecticides will challenge the decline of malaria. Clin Microbiol Infect. 19:902–907. [DOI] [PubMed] [Google Scholar]
- Stephens M, Scheet P.. 2005. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 76:449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P.. 2001. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 68:978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF. 2005. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol. 14:671–688. [DOI] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken W, Verhulst NO.. 2013. Host preferences of blood-feeding mosquitoes. Annu Rev Entomol. 58:433–453. [DOI] [PubMed] [Google Scholar]
- Tene Fossog B, Antonio-Nkondjio C, Kengne P, Njiokou F, Besansky NJ, Costantini C.. 2013. Physiological correlates of ecological divergence along an urbanization gradient: differential tolerance to ammonia among molecular forms of the malaria mosquito Anopheles gambiae. BMC Ecol. 13:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tene Fossog B, Ayala D, Acevedo P, Kengne P, Ngomo Abeso Mebuy I, Makanga B, Magnus J, Awono-Ambene P, Njiokou F, Pombi M, et al. 2015. Habitat segregation and ecological character displacement in cryptic African malaria mosquitoes. Evol Appl. 8:326–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin P, Ross-Ibarra J.. 2014. Advances and limits of using population genetics to understand local adaptation. Trends Ecol Evol. 29:673–680. [DOI] [PubMed] [Google Scholar]
- Touré Y, Petrarca V, Traoré S, Coulibaly A, Maiga H, Sankaré O, Sow M, Di Deco M, Coluzzi M.. 1998. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia 40:477–511. [PubMed] [Google Scholar]
- Turissini D, Gamez S, White BJ.. 2014. Genome-wide patterns of polymorphism in an inbred line of the African malaria mosquito Anopheles gambiae. Genome Biol Evol. 6:3094–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J. 1996. The variance of pairwise nucleotide differences in two populations with migration. Theor Popul Biol. 49:39–57. [DOI] [PubMed] [Google Scholar]
- White B, Lawniczak M, Cheng C, Coulibaly MB, Wilson MD, Sagnon N, Costantini C, Simard F, Christophides GK, Besansky NJ.. 2011. Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium. Proc Natl Acad Sci USA. 108:244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White GB. 1974. Anopheles gambiae complex and disease transmission in Africa. Trans R Soc Trop Med Hyg. 68:278–298. [DOI] [PubMed] [Google Scholar]
- WHO (World Health Organization). 2012. Global plan for insecticide resistance management in malaria vectors (GPIRM). 132.
- WHO (World Health Organization). 2013. World Malaria Report 2013. Geneva, Switzerland, 284.
- Wondji C, Simard F, Fontenille D.. 2002. Evidence for genetic differentiation between the molecular forms M and S within the Forest chromosomal form of Anopheles gambiae in an area of sympatry. Insect Mol Biol. 11:11–19. [DOI] [PubMed] [Google Scholar]
- Wondji C, Simard F, Petrarca V.. 2005. Species and populations of the Anopheles gambiae complex in Cameroon with special emphasis on chromosomal and molecular forms of Anopheles gambiae ss. J Med Entomol. 426:998–1005. [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S.. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26:873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data (fastq files) for 309 Anopheles gambiae s.l. individuals with the best sequencing coverage are available at http://dx.doi.org/10.5061/dryad.jf402.