Abstract
In recent years, there have been increasing numbers of transcripts identified that do not encode proteins, many of which are developmentally regulated and appear to have regulatory functions. Here, we describe the construction of a comprehensive mammalian noncoding RNA database (RNAdb) which contains over 800 unique experimentally studied non-coding RNAs (ncRNAs), including many associated with diseases and/or developmental processes. The database is available at http://research.imb.uq.edu.au/RNAdb and is searchable by many criteria. It includes microRNAs and snoRNAs, but not infrastructural RNAs, such as rRNAs and tRNAs, which are catalogued elsewhere. The database also includes over 1100 putative antisense ncRNAs and almost 20 000 putative ncRNAs identified in high-quality murine and human cDNA libraries, with more to be added in the near future. Many of these RNAs are large, and many are spliced, some alternatively. The database will be useful as a foundation for the emerging field of RNomics and the characterization of the roles of ncRNAs in mammalian gene expression and regulation.
INTRODUCTION
The traditional view of the genomic programming of cells and organisms is predicated on the belief that genetic information normally flows from DNA to RNA to protein. As a consequence, genes are generally considered to be synonymous with proteins, which carry out the majority of the structural, catalytic and regulatory transactions in living cells, with RNA functioning primarily as an intermediate coding template for protein synthesis, aided by infrastructural RNAs that are central to the process (tRNAs and rRNAs). This view of the structure of molecular genetic systems is essentially correct in prokaryotes, whose genomes consist almost entirely of closely spaced protein-coding sequences flanked by cis-acting regulatory elements that act to control transcription and translation, although it has recently become clear that prokaryotes also express limited numbers of small regulatory RNAs. However, these occupy <1% of the genome sequence in prokaryotes (1). In contrast, non-protein-coding RNA (ncRNAs) sequences are abundant in the genomes of the eukaryotes, especially the developmentally complex multicellular eukaryotes, where they dominate transcriptional output (2). These RNAs are composed of transcribed introns from protein-coding genes, and an increasing but, as yet, undetermined number of transcripts that do not encode proteins and may or may not themselves contain introns, collectively termed ncRNAs (2–5).
The evidence for large numbers of ncRNA transcripts in eukaryotes is increasing. There are large numbers of sequences that do not contain a significant open reading frame (ORF) in well-constructed mammalian cDNA collections (6,7), many of which show variation in their expression in different cells and developmental stages. The number of detectable RNAs derived from human chromosomes 21 and 22 has been shown to be at least an order of magnitude higher than that expected from known protein-coding sequences (8,9). All well-studied loci (including β-globin, bithorax and various imprinted loci) show a predominance of non-coding transcripts, many of which are developmentally regulated and appear themselves to have regulatory functions (10–12). Careful analysis of human chromosome 7 has identified 213 ncRNA genes, indicating by extrapolation that the number of ncRNA genes in the human genome is more than 3500 (13) while other estimates based on the integration of data from a wide variety of sources suggests that the majority of all human genes encode ncRNAs (3,5). Indeed, while protein-coding sequences occupy <1.5% of the human genome, it is clear that at least 50% of the genome is transcribed (2).
ncRNAs have been identified in various ways. Early examples such as lin-4, Xist, IPW, NTT and BC200 were discovered and characterized experimentally on an ad hoc basis, and their lack of protein-coding capacity was unexpected (references for individual ncRNAs are listed in Supplementary Material 1). Many of these RNAs, expressed in particular tissues and/or developmental stages, are associated with particular diseases including various cancers (14–20), schizophrenia (21), ataxia (22), cartilage-hair hypoplasia (19), DiGeorge syndrome (23) and autism (13), and/or are involved in complex genetic phenomena such as imprinting and other forms of epigenetic control of gene expression (12,24). More recently, there have been targeted experimental approaches to the large-scale discovery of ncRNA genes. The presence of hundreds of microRNAs (miRNAs) and small nucleolar RNAs (snoRNAs) has been established experimentally by specifically screening for tiny RNA species (25–27), at least some of which have been implicated in the control of development (28–30) and in the etiology of cancer (31–33). As noted above, thousands of larger ncRNA transcripts have also been putatively identified via the systematic sequencing and annotation of tens of thousands of full-length cDNAs (6,7). Recent advances in computational genomics are also helping to identify ncRNAs of particular types. New algorithms have been used to search the genomic sequence databases for members that share secondary structure motifs with existing ncRNA families (34,35), although there remain large numbers of ncRNAs which do not yet appear to share recognizable primary or secondary structural motifs and hence cannot be identified by these methods.
The term ‘non-coding RNA’ in its broadest sense includes all RNAs that do not code for protein (i.e. non-messenger RNAs) and encompasses transfer RNAs (tRNAs), ribosomal RNAs (rRNAs) and spliceosomal RNAs, which largely have basic housekeeping functions in cells. Many other ncRNAs, however, have been shown to perform regulatory functions within the cell including phenomena such as the temporal suppression of mRNA translation, RNA interference, imprinting, DNA methylation and X chromosome dosage compensation (3,5). So, while precise biological roles for the vast majority of these non-messenger, non-infrastructural RNAs are still to be elucidated, such ncRNAs have been proposed to serve as a diverse and hitherto hidden regulatory network in eukaryotic cells (2,3).
At present, there is no comprehensive database of ncRNAs, although there are several existing databases that cover aspects of the field. tRNAs and rRNAs are listed within multiple databases (35–37). The miRNA registry provides a searchable database of published miRNA sequences (38). The Rfam database contains thousands of mammalian RNAs, the majority of which are infrastructural RNAs (tRNAs, etc.) and predicted using co-variance models from multiple-sequence alignments of genomic datasets with little direct experimental support for their transcription (35). Notably, many well-documented regulatory ncRNAs such as Xist and NTT are not listed in Rfam, reflecting a bias to include only those entries that are members of particular structure-based RNA families. A ‘regulatory’ ncRNA database has been published previously (39) but the total number of unique mammalian ncRNAs in this database is <40 excluding homologs and miRNAs.
AIMS OF THE DATABASE
Here, we describe a new ncRNA database, RNAdb. The broad aims of RNAdb are 2-fold. First, to provide a searchable sequence repository for mammalian ncRNAs whose function may be regulatory and whose existence is supported by experimental evidence. Being sequence-oriented, the database permits not only bioinformatic analyses but also the rational design of experimental tools such as microarray chips to characterize the roles of ncRNAs in mammalian gene expression and regulation. The second aim is to highlight to the scientific community the presence of thousands of potentially regulatory ncRNAs.
RNAdb contains a comprehensive listing of mammalian ncRNAs, but excludes tRNAs, rRNAs and spliceosomal RNAs. More than 800 unique known mammalian ncRNAs are included in the database, around two-thirds of which are miRNAs and snoRNAs. The rest are largely of unknown function, but some are known to be developmentally regulated, disease-associated, imprinted, expressed pseudogenes or antisense transcripts. As well as sequence data, additional information—including GenBank accession numbers, species, references, chromosomal location, transcript length, splicing status, conservation notes, function, disease associations, antisense relationships, imprinting status and tissue expression patterns—is provided wherever possible in separate searchable fields. The database also includes almost 20 000 putative ncRNAs identified in high-quality cDNA libraries (6,7), with more to be added in the near future. At this stage, we have restricted the database to mammalian ncRNAs, as this group has the best information and is most relevant to humans and disease, but we expect that many of these ncRNAs will have paralogs in other vertebrates, and that the database will eventually be extended to include ncRNAs from other vertebrates and from invertebrates.
DATABASE DEVELOPMENT AND DESIGN
ncRNAs were included in RNAdb provided that (i) there was experimental (i.e. EST, cDNA, RT–PCR and/or northern blot) evidence to support their existence as RNAs; (ii) they did not contain a significant ORF (i.e. <100 amino acids); (iii) they were not annotated as rRNAs, tRNAs and spliceosomal RNAs; and (iv) they were mammalian.
ncRNAs that satisfied these requirements were identified from the following sources: a comprehensive literature review; the Functional Annotation of Mouse (FANTOM) and the H-Invitational databases, which contain over 60 000 and 20 000 annotated gene candidates identified from mouse and human full-length cDNA libraries, respectively; the human chromosome 7 annotation project; the miRNA registry; and public sequence databases at NCBI, Ensembl and UCSC. A novel pipeline was designed to identify putative antisense ncRNAs from the public databases (Supplementary Material 2).
For individual ncRNAs with direct support in the literature, the details (sequence, accession number, organism, chromosomal location, etc.) were manually curated where possible. For ncRNAs identified from large-scale discovery projects such as FANTOM and H-Invitational, information was restricted to that already publicly available. Further information will be obtained and incorporated in the near future.
All the information is currently stored in a Microsoft SQL2000 relational database which runs on a Microsoft.NET platform.
DATABASE ACCESS AND INTERFACE
RNAdb is hosted at the Institute for Molecular Bioscience and is freely available at http://research.imb.uq.edu.au/RNAdb. The entire database is available for download in XML format via the website.
The web interface allows users to browse the entire collection or to search for specific ncRNAs of interest. Searches are executed either by performing keyword searches or by applying filters to nominated fields including organism, chromosome number, ncRNA category and ncRNA feature (e.g. antisense, disease-associated, etc.) either singly or in combination (Figure 1A). Once an individual ncRNA is selected, the sequence and relevant annotations are displayed in a tabular format (Figure 1B).
Figure 1.
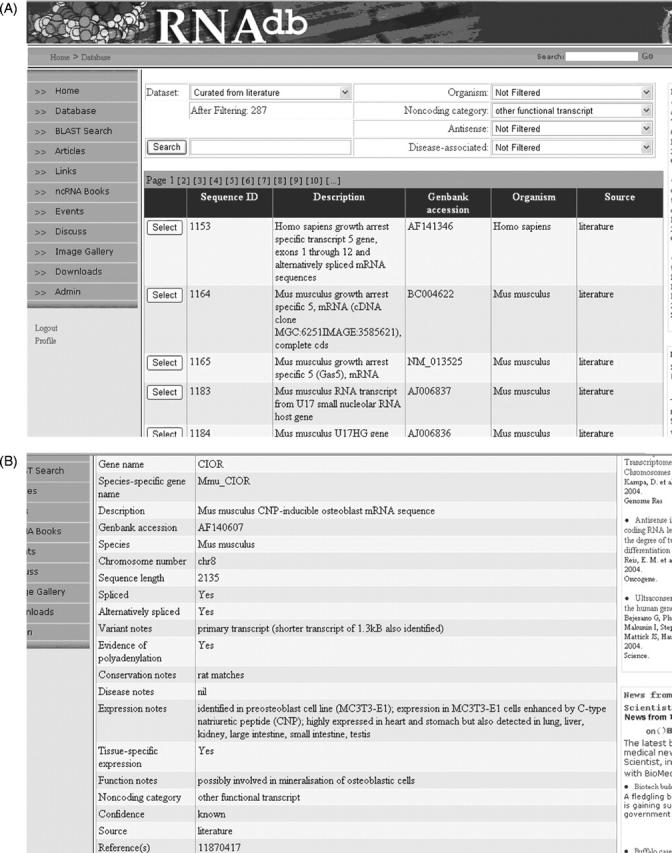
(A) RNAdb search interface. Users can look for ncRNAs of interest either using a keyword search or by applying filters. (B) Example of RNAdb entry listing. When an individual ncRNA is selected, the annotated information is displayed in a tabular format.
A BLAST tool allows users to query sequences of interest against those present in the database.
Questions, feedback and submissions of new mammalian ncRNAs are welcomed, and should be directed via email to RNAdb@imb.uq.edu.au.
CONTENT OF THE DATABASE
RNAdb contains more than 800 unique, known mammalian ncRNAs. Including homologs, splice variants and precursor forms, there are more than 2000 individual sequences listed. Of the uniquely known ncRNAs in the database, around two-thirds comprise miRNAs and snoRNAs. The rest are generally much longer and while some of these have documented biological roles, most are transcripts of unknown function. Altogether 36 mammalian organisms are represented within the dataset but the majority of unique ncRNAs are either murine and/or human.
miRNAs
Over 200 unique mammalian miRNAs are found within RNAdb and were chiefly obtained from the miRNA registry (38). While it is not our aim to duplicate this valuable resource, we have included miRNAs here because they represent an important population of potentially regulatory ncRNAs and consequently fall within the scope of this database. In addition, we have manually annotated the miRNAs to reflect recent published results (30,40–43).
snoRNAs
More than 300 snoRNAs have been described. They fall into two general classes, C/D box and H/ACA snoRNAs, which guide ribose methylation and pseudo-uridylation of rRNAs, respectively. Some snoRNAs are expressed in a tissue-specific manner and show complementarity to mRNAs rather than rRNAs (44), pointing to possible roles in post-transcriptional modification and regulation.
Disease-associated ncRNAs
Over 40 unique ncRNAs have been linked to diseases ranging from malignancies to psychiatric illnesses and neuro-developmental disorders. These include: miR-15 and 16, BCMS (B cell chronic lymphocytic leukemia), TTY1 and 2 (gonadoblastoma), NCRMS (rhabdomyosarcoma), BIC (ALV-induced B cell lymphoma), H19 (breast/colon/bladder/Wilm's tumor), MALAT-1 (non-small cell lung cancer), DISC2 (schizophrenia), SCA8 (spinocerebellar ataxia), ST7OT1-4 (autism), IPW (Prader–Willi syndrome), LIT-1 (Beckwith Wiedemann syndrome), RMRP (cartilage hair hypoplasia) and UBE3A antisense (Angelman syndrome).
Developmentally regulated ncRNAs
More than 40 ncRNAs show tissue-specific expression and/or regulation during development. Recent examples include multiple miRNAs differentially expressed during brain development and hematopoiesis (30,40), and others include Xist, 7H4, BC1, BC200, BORG, Bsr, CIOR, MBII-13/52 and 85, Ntab, NTT, tncb and Zim3.
Natural antisense ncRNAs
Natural antisense transcription is now recognized as being much more common than previously thought, with recent estimates indicating that there may be thousands of sense–antisense pairs in the human genome (45). RNAdb includes over 30 previously described antisense ncRNAs, a few examples of which are aHIF, Air, DISC2, EMX2OS, FGF antisense, HFE antisense, HOXA11S, LIT1, Msx1 antisense and Nespas.
Expressed pseudogenes
The number of pseudogenes in the human genome has been estimated at ∼20 000 (46), only a minority of which are thought to be transcribed into ncRNAs (47). Here, we catalog over 50 expressed pseudogenes. These include Makorin1-p1, the disruption of which was recently shown to play a role in the pathogenesis of a mouse mutant exhibiting polycystic kidneys and bone deformity.
Imprinted ncRNAs
We have also found over 40 ncRNAs that are imprinted. Several of these cluster together including IPW, PWCR1, UBE3A antisense, PAR1 and PEG13 on human chromosome 15q11-12, and PEG-11 antisense and MEG8 (on ovine chromosome 18). Many are natural antisense transcripts, such as (apart from those above) LIT-1, Air, GNAS1 antisense, PEG8, Copg2IT1, PEG1 antisense and Zim3.
Alternatively spliced ncRNAs
Over 20 ncRNAs are known to be alternatively spliced. Some of these include BCMS, CIOR, DD3, DGCR5, Enox, G.B7, Gas5, GTL2, NCRMS, Nespas, SCA8 and Tmevpg1.
Putative ncRNAs
In addition to the 800 unique known ncRNAs, RNAdb also contains almost 20 000 putative ncRNAs. Approximately 15 000 of these were originally identified from over 60 000 high-quality largely full-length mouse cDNA clones annotated by the FANTOM consortium (6). A further 2000 putative ncRNAs were obtained from the H-Invitational database, which contains over 20 000 validated gene candidates derived from analysis of high-quality largely full-length human cDNA clones (7). The human chromosome 7 annotation project has described over 350 putative ncRNAs derived from computer-based annotation in conjunction with extensive laboratory experimentation, and these are also included among our putative ncRNA dataset. Finally, using a computational pipeline, we identified over 1100 putative antisense ncRNAs (Supplementary Material 2), and these are also contained in the database.
FUTURE DIRECTIONS
To date, we have cataloged hundreds of known non-infrastructural mammalian ncRNAs. This number quickly stretches to thousands once the putative ncRNAs are taken into account. While it has been proposed that many of these ncRNAs function as components in a regulatory network (3,5), in the majority of cases, virtually nothing is known about their precise biological roles, tissue expression patterns and significance. Elucidating the functional significance of these transcripts must, therefore, become a priority. Until this is done, we acknowledge the important caveats that some of the RNAs classified here as ncRNAs may not have intrinsic function, as in the case of SRG1 in yeast (48), and that some may encode small proteins as recently described for the steroid receptor RNA activator (49). Nonetheless, having a searchable database such as RNAdb dedicated to non-infrastructural ncRNAs will be a vital foundation for the emerging field of RNomics as the future knowledge base grows. We are, therefore, setting up a regular database update schedule, and invite researchers to submit new ncRNAs to RNAdb as they are published. In the near future, we will be adding thousands of novel putative ncRNAs that have arisen from the latest round of analysis of the FANTOM cDNA libraries, as they become publicly available.
POSTSCRIPT
At the time of submission, it came to our attention that a new noncoding RNA database (NONCODE) had been released online in the previous month. This database appears to share a similar interest with ours but RNAdb has the following advantages: (i) over 1100 putative antisense ncRNAs and almost 20 000 putative ncRNAs identified in high-quality murine and human cDNA libraries are included and annotated; (ii) BLAST searches against the non-coding dataset can be performed; and (iii) the database is available for download to permit more thorough local bioinformatic analyses.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jeff McDonald and Steve Scherer for providing us with information on ncRNAs identified by the chromosome 7 annotation project, as well as Carole Charlier for providing us with information on callipyge locus ncRNAs. K.C.P. is supported by the NHMRC with a Postgraduate Medical Research Scholarship (234711). W.C. is supported by a Wellcome Trust International Senior Research Fellow Fellowship (066646Z01Z). J.S.M. is supported by the Australian Research Council and the Queensland State Government.
REFERENCES
- 1.Vogel J., Bartels,V., Tang,T.H., Churakov,G., Slagter-Jager,J.G., Huttenhofer,A. and Wagner,E.G. (2003) RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res., 31, 6435–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattick J.S. (2001) Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep., 2, 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattick J.S. and Gagen,M.J. (2001) The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol. Biol. Evol., 18, 1611–1630. [DOI] [PubMed] [Google Scholar]
- 4.Ruvkun G. (2001) Molecular biology. Glimpses of a tiny RNA world. Science, 294, 797–799. [DOI] [PubMed] [Google Scholar]
- 5.Mattick J.S. (2003) Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays, 25, 930–939. [DOI] [PubMed] [Google Scholar]
- 6.Okazaki Y., Furuno,M., Kasukawa,T., Adachi,J., Bono,H., Kondo,S., Nikaido,I., Osato,N., Saito,R., Suzuki,H. et al. (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs [comment]. Nature, 420, 563–573. [DOI] [PubMed] [Google Scholar]
- 7.Imanishi T., Itoh,T., Suzuki,Y., O'Donovan,C., Fukuchi,S., Koyanagi,K.O., Barrero,R.A., Tamura,T., Yamaguchi-Kabata,Y., Tanino,M. et al. (2004) Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol., 2, E162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapranov P., Cawley,S.E., Drenkow,J., Bekiranov,S., Strausberg,R.L., Fodor,S.P. and Gingeras,T.R. (2002) Large-scale transcriptional activity in chromosomes 21 and 22. Science, 296, 916–919. [DOI] [PubMed] [Google Scholar]
- 9.Kampa D., Cheng,J., Kapranov,P., Yamanaka,M., Brubaker,S., Cawley,S., Drenkow,J., Piccolboni,A., Bekiranov,S., Helt,G. et al. (2004) Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res., 14, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashe H.L., Monks,J., Wijgerde,M., Fraser,P. and Proudfoot,N.J. (1997) Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev., 11, 2494–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drewell R.A., Bae,E., Burr,J. and Lewis,E.B. (2002) Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc. Natl Acad. Sci. USA, 99, 16853–16858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes R., Williamson,C., Peters,J., Denny,P. and Wells,C. (2003) A comprehensive transcript map of the mouse Gnas imprinted complex. Genome Res., 13, 1410–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scherer S.W., Cheung,J., MacDonald,J.R., Osborne,L.R., Nakabayashi,K., Herbrick,J.A., Carson,A.R., Parker-Katiraee,L., Skaug,J., Khaja,R. et al. (2003) Human chromosome 7: DNA sequence and biology. Science, 300, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wrana J.L. (1994) H19, a tumour suppressing RNA? Bioessays, 16, 89–90. [DOI] [PubMed] [Google Scholar]
- 15.Bussemakers M.J., van Bokhoven,A., Verhaegh,G.W., Smit,F.P., Karthaus,H.F., Schalken,J.A., Debruyne,F.M., Ru,N. and Isaacs,W.B. (1999) DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res., 59, 5975–5979. [PubMed] [Google Scholar]
- 16.Tam W., Ben-Yehuda,D. and Hayward,W.S. (1997) bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol. Cell Biol., 17, 1490–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tam W. (2001) Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene, 274, 157–167. [DOI] [PubMed] [Google Scholar]
- 18.van den Berg A., Kroesen,B.J., Kooistra,K., de Jong,D., Briggs,J., Blokzijl,T., Jacobs,S., Kluiver,J., Diepstra,A., Maggio,E. et al. (2003) High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer, 37, 20–28. [DOI] [PubMed] [Google Scholar]
- 19.Ridanpaa M., van Eenennaam,H., Pelin,K., Chadwick,R., Johnson,C., Yuan,B., vanVenrooij,W., Pruijn,G., Salmela,R., Rockas,S. et al. (2001) Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell, 104, 195–203. [DOI] [PubMed] [Google Scholar]
- 20.Ji P., Diederichs,S., Wang,W., Boing,S., Metzger,R., Schneider,P.M., Tidow,N., Brandt,B., Buerger,H., Bulk,E. et al. (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene, 22, 6087–6097. [DOI] [PubMed] [Google Scholar]
- 21.Millar J.K., Wilson-Annan,J.C., Anderson,S., Christie,S., Taylor,M.S., Semple,C.A., Devon,R.S., Clair,D.M., Muir,W.J., Blackwood,D.H. et al. (2000) Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet., 9, 1415–1423. [DOI] [PubMed] [Google Scholar]
- 22.Nemes J.P., Benzow,K.A., Moseley,M.L., Ranum,L.P. and Koob,M.D. (2000) The SCA8 transcript is an antisense RNA to a brain-specific transcript encoding a novel actin-binding protein (KLHL1). Hum. Mol. Genet., 9, 1543–1551. [DOI] [PubMed] [Google Scholar]
- 23.Sutherland H.F., Wadey,R., McKie,J.M., Taylor,C., Atif,U., Johnstone,K.A., Halford,S., Kim,U.J., Goodship,J., Baldini,A. et al. (1996) Identification of a novel transcript disrupted by a balanced translocation associated with DiGeorge syndrome. Am. J. Hum. Genet., 59, 23–31. [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston C.M., Newall,A.E., Brockdorff,N. and Nesterova,T.B. (2002) Enox, a novel gene that maps 10 kb upstream of Xist and partially escapes X inactivation. Genomics, 80, 236–244. [DOI] [PubMed] [Google Scholar]
- 25.Lagos-Quintana M., Rauhut,R., Lendeckel,W. and Tuschl,T. (2001) Identification of novel genes coding for small expressed RNAs. Science, 294, 853–858. [DOI] [PubMed] [Google Scholar]
- 26.Lee R.C. and Ambros,V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science, 294, 862–864. [DOI] [PubMed] [Google Scholar]
- 27.Lau N.C., Lim,L.P., Weinstein,E.G. and Bartel,D.P. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, 294, 858–862. [DOI] [PubMed] [Google Scholar]
- 28.Pasquinelli A.E. and Ruvkun,G. (2002) Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol., 18, 495–513. [DOI] [PubMed] [Google Scholar]
- 29.Ambros V. (2003) MicroRNA pathways in flies and worms. Growth, death, fat, stress and timing. Cell, 114, 269. [DOI] [PubMed] [Google Scholar]
- 30.Chen C.Z., Li,L., Lodish,H.F. and Bartel,D.P. (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science, 303, 83–86. [DOI] [PubMed] [Google Scholar]
- 31.McManus M.T. (2003) MicroRNAs and cancer. Semin. Cancer Biol., 13, 253–258. [DOI] [PubMed] [Google Scholar]
- 32.Metzler M., Wilda,M., Busch,K., Viehmann,S. and Borkhardt,A. (2004) High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer, 39, 167–169. [DOI] [PubMed] [Google Scholar]
- 33.Michael M.Z., O'Connor,S.M., van Holst Pellekaan,N.G., Young,G.P. and James,R.J. (2003) Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res., 1, 882–891. [PubMed] [Google Scholar]
- 34.Eddy S.R. (2002) Computational genomics of noncoding RNA genes. Cell, 109, 137–140. [DOI] [PubMed] [Google Scholar]
- 35.Griffiths-Jones S., Bateman,A., Marshall,M., Khanna,A. and Eddy,S.R. (2003) Rfam: an RNA family database. Nucleic Acids Res., 31, 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wuyts J., Perriere,G. and Van De Peer,Y. (2004) The European ribosomal RNA database. Nucleic Acids Res., 32, D101–D103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffiths-Jones S. (2004) The microRNA Registry. Nucleic Acids Res., 32, D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szymanski M., Erdmann,V.A. and Barciszewski,J. (2003) Noncoding regulatory RNAs database. Nucleic Acids Res., 31, 429–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krichevsky A.M., King,K.S., Donahue,C.P., Khrapko,K. and Kosik,K.S. (2003) A microRNA array reveals extensive regulation of microRNAs during brain development. RNA, 9, 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seitz H., Youngson,N., Lin,S.P., Dalbert,S., Paulsen,M., Bachellerie,J.P., Ferguson-Smith,A.C. and Cavaille,J. (2003) Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nature Genet., 34, 261–262. [DOI] [PubMed] [Google Scholar]
- 42.Calin G.A., Dumitru,C.D., Shimizu,M., Bichi,R., Zupo,S., Noch,E., Aldler,H., Rattan,S., Keating,M., Rai,K. et al. (2002) Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA, 99, 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calin G.A., Sevignani,C., Dumitru,C.D., Hyslop,T., Noch,E., Yendamuri,S., Shimizu,M., Rattan,S., Bullrich,F., Negrini,M. et al. (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA, 101, 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cavaille J., Buiting,K., Kiefmann,M., Lalande,M., Brannan,C.I., Horsthemke,B., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization [comment]. Proc. Natl Acad. Sci. USA, 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yelin R., Dahary,D., Sorek,R., Levanon,E.Y., Goldstein,O., Shoshan,A., Diber,A., Biton,S., Tamir,Y., Khosravi,R. et al. (2003) Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol., 21, 379–386. [DOI] [PubMed] [Google Scholar]
- 46.Harrison P.M., Hegyi,H., Balasubramanian,S., Luscombe,N.M., Bertone,P., Echols,N., Johnson,T. and Gerstein,M. (2002) Molecular fossils in the human genome: identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res., 12, 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mighell A.J., Smith,N.R., Robinson,P.A. and Markham,A.F. (2000) Vertebrate pseudogenes. FEBS Lett., 468, 109–114. [DOI] [PubMed] [Google Scholar]
- 48.Martens J.A., Laprade,L. and Winston,F. (2004) Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature, 429, 571–574. [DOI] [PubMed] [Google Scholar]
- 49.Chooniedass-Kothari S., Emberley,E., Hamedani,M.K., Troup,S., Wang,X., Czosnek,A., Hube,F., Mutawe,M., Watson,P.H. and Leygue,E. (2004) The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett., 566, 43–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


