Abstract
As database management systems expand their array of analytical functionality, they become powerful research engines for biomedical data analysis and drug discovery. Databases can hold most of the data types commonly required in life sciences and consequently can be used as flexible platforms for the implementation of knowledgebases. Performing data analysis in the database simplifies data management by minimizing the movement of data from disks to memory, allowing pre-filtering and post-processing of datasets, and enabling data to remain in a secure, highly available environment. This article describes the Oracle Database 10g implementation of BLAST and Regular Expression Searches and provides case studies of their usage in bioinformatics. http://www.oracle.com/technology/software/index.html
INTRODUCTION
The complexity of experimental and computational methods used in biological discovery has increased as a result of new biotechnologies and the wealth of molecular biological data available today. Holistic views of cells and organisms are opening the way to the development of more sophisticated models and promise to increase the throughput in biomedical discovery. As a consequence of these advancements there are unprecedented opportunities to discover new drugs, but at the expense of escalating costs.
The primary reason for the rising cost of biological discovery is the challenge of managing and analyzing the increasing volumes of heterogeneous biological data (http://www.bcg.com/publications/files/eng_genomicsgenetics_rep_11_01.pdf). Scientists need to draw together data from many diverse sources, including public databanks, proprietary data providers, and internal wet-lab experiments. Data from these sources belong to a wide range of data types, including relational tables, three-dimensional (3D) biochemical structures, images, web pages and flat files. Most of these data, which have been developed in individual research laboratories worldwide over several decades, often lack common data formats, common vocabulary and the common record identifiers that are needed for interoperability (1). In addition, there is a high rate of development of new scientific algorithms in academia, which further increases the complexity of data management.
Researchers facing the biological data management challenge have been benefiting from enhancements that have been made to database systems for the life sciences. Oracle Database 10g, for instance, has a range of improved data management and data sharing features that can support the biology domain. Its distributed data architecture allows users to write queries that span over relational databases located locally and remotely, and over data in flat files. It is also possible to store several data types such as images, XML documents, biomedical publications and chemical structures. These features, along with new database functionalities expanded into areas such as online analytical processing (OLAP), data mining, statistics, regular expression searches, text mining and most recently BLAST (2–5), take database systems far beyond simple data repositories. Incorporating sophisticated analytics and enhancing the inferential capabilities of databases has been a topic of discussion by academics for several years (6–8); however, it is only recently that commercial enterprise databases have truly begun to take that step.
Embedding analytics in the database is an attractive approach because it minimizes data movement. Embedding not only allows the data to stay in the database, but also enables analytical tasks to be run automatically, asynchronously and independently. This tight integration with the database provides a scalable and automated environment, which is required for the development and deployment of sophisticated analytics. Integrating analytics into a database enables users to routinely run complex analysis queries. Application developers can also integrate the analytical functions into their software products.
In this article, we discuss two database features in Oracle Database 10g: Oracle Data Mining (ODM) BLAST and Regular Expression Searches. Scientists can use ODM BLAST to perform sequence homology searches and Regular Expression Searches to perform pattern matching, inside the database. These new database features enable scientists to take advantage of a new analytical paradigm to simplify data management in research.
BLAST
BLAST overview
BLAST is a family of heuristic algorithms for identifying local alignments between genome sequences (9). More specifically, the BLAST family of algorithms can be used to search nucleotide and amino acid query sequences against databases of nucleotide and amino acid sequences. The discovery of sequence homology can help scientists to establish the evolutionary origin of particular genes, or help to make predictions about protein structure and function (10). In addition to being a fast algorithm, an important advantage of BLAST is that it provides a measure of the statistical significance of the alignment scores.
Oracle Data Mining BLAST implementation
A version of BLAST, like NCBI BLAST 2.0, has been implemented in Oracle Database 10g (http://download-west.oracle.com/docs/cd/B14117_01/datamine.101/b10699/6blast.htm#76938) as a part of ODM. The implementation includes the five core variants of BLAST (BLASTP, BLASTN, BLASTX, TBLASTN and TBLASTX). ODM BLAST provides a MATCH function that can be used to return the sequence ID, expect value and score; and an ALIGN function that can be used to return the sequence ID, expect value, score and full alignment information.
The ODM BLAST API is a table function which can be used in the FROM clause of an SQL query. This implementation allows ODM BLAST to be invoked either by embedding the functionality into applications or by ad hoc SQL queries.
The ODM BLAST table function accepts as input a query sequence, a reference cursor that specifies the sequences that the query sequence needs to be searched against, and other input parameters that control the search. Once the query sequence has been specified, it is passed onto the underlying server side program code as a Character Large OBject (CLOB). The reference cursor, which specifies the target sequences, must contain a sequence identifier of the data type VARCHAR and a sequence data string of the data type CLOB. The programming code takes the input, performs the search and sends the results as a virtual table to the invoking ODM BLAST table function. Users can specify the substitution matrix used to assign a score for aligning any possible pair of residues. The different options include PAM30, PAM70, BLOSUM45, BLOSUM62 and BLOSUM80.
To take advantage of the ODM BLAST functionality, the sequence data must be accessible from inside Oracle Database 10g. The optimal way, therefore, for using the ODM BLAST functionality is to store the sequence information in the database as a CLOB. However, Oracle does provide features such as Generic Connectivity, Transparent Gateways and External Tables that allow users to query data that is held in non-Oracle databases and in external flat files.
ODM BLAST can be freely downloaded for non-commercial and non-production use with Oracle Database 10g (http://www.oracle.com/technology/software/products/database/oracle10g/index.html).
ODM BLAST case study
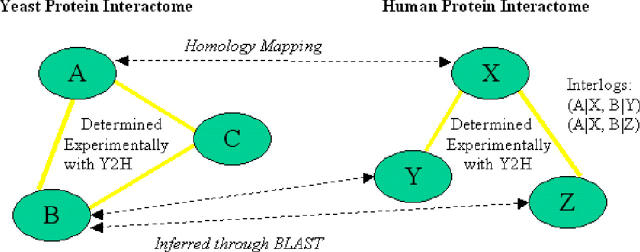
Research was undertaken to identify human protein–protein interactions using a yeast two-hybrid (Y2H) approach (11). Over 100 000 protein interactions were identified, but as the Y2H technique is known to generate a large number of false positive results due to sticky proteins and self interactions (12), we wanted to verify which protein–protein interactions were genuine. ODM BLAST was used to infer true-positive interactions by examining whether similar interactions occurred in yeast. Figure 1 depicts the experimental approach.
Figure 1.
Experimental approach for the identification of protein–protein interactions using ODM BLAST.
All three reading frames of chromosomes 1–16 of the yeast proteome were downloaded from the Web (www.yeastgenome.org), as was yeast protein–protein interaction data (13). The data files were converted into CSV format and loaded into Oracle Database 10g using SQL*Loader. A PL/SQL script was then written that allowed ODM BLAST to perform a homology search where all interacting human protein sequences were queried against the yeast proteome in an iterative fashion. The script is available in Figure 1 of the Supplementary Material. Query performance was optimized to get parallel throughput by running multiple ODM BLAST threads, each of which was iterating through a subset of human proteins.
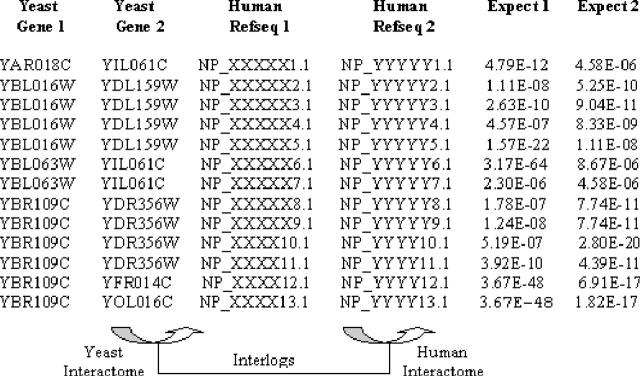
As a result of the analysis, the human protein–protein interactions that are statistically most likely to be genuine were identified (Figure 2). This has enabled discovery efforts to be focused on the most promising targets.
Figure 2.
A sample of the protein–protein interaction results gained from ODM BLAST analysis.
ODM BLAST offers a high-performance automated solution that minimizes the typical data management challenges. This approach simplified an otherwise complex sequence homology search.
REGULAR EXPRESSION SEARCHES
Regular Expression Searches overview
A Regular Expression is a sequence of characters that describe a pattern in text (14,15). Metacharacters are used so that matches can be performed when only the general pattern of text is known. The functionality is useful in solving many different tasks involving text searching and pattern recognition.
Oracle Regular Expression Searches implementation
While middle-tier technologies have long performed regular expression searching, support in the backend database is a valuable and often overlooked consideration. Oracle Regular Expressions provide a simple yet powerful mechanism for rapidly describing patterns and greatly simplifies the way in which users can search, extract, format and otherwise manipulate text in the database (http://www.oracle.com/technology/products/database/application_development/pdf/TWP_Regular_Expressions.pdf).
The Oracle implementation of Regular Expressions conforms to the IEEE Portable Operating System Interface (POSIX) standard draft 1003.2/D11.2 and to the Unicode Regular Expression Guidelines of the Unicode Consortium. The Oracle Database follows the exact syntax and matching semantics for these operators as defined in the POSIX standard for matching ASCII (English language) data (http://www.opengroup.org/onlinepubs/007908799/xbd/re.html).
The Oracle Database enhances Regular Expression support by extending the matching capabilities for multilingual data beyond what is specified in the POSIX standard, adds support for the common Perl Regular Expression extensions that are not included in the POSIX standard but do not conflict with it and provides built-in support for some of the most heavily used Perl Regular Expression operators, e.g. character class shortcuts and the non-greedy modifier (16).
The implementation of Regular Expressions comes in the form of a range of SQL functions and a WHERE clause operator. Oracle Regular Expressions are implemented by interfaces that are available in both SQL and PL/SQL (Table 1).
Table 1. SQL and PL/SQL interfaces for Oracle Regular Expressions.
| SQL function | Description |
|---|---|
| REGEXP_LIKE | Determine whether pattern matches |
| REGEXP_SUBSTR | Determine what string matches the pattern |
| REGEXP_INSTR | Determine where the match occurred in the string |
| REGEXP_REPLACE | Search and replace a pattern |
Regular Expression Searches case study
The goal of the case study was to identify locally conserved regions within protein sequences that exhibit a predictable pattern and to annotate a proprietary protein–protein interaction database with this information by using Oracle Regular Expressions.
The PROSITE protein motif repository was downloaded from the Web (http://www.expasy.org/prosite/). Standard AWK and SED routines were used to convert the PROSITE data file into CSV format, and to extract motif patterns and convert them into Oracle Regular Expressions. The Oracle Regular Expressions were then loaded into the Oracle Database 10g using the External Tables functionality.
Once the data were loaded in the database, it was possible to iterate through all of the Oracle Regular Expressions to identify their presence in both the proprietary protein–protein interaction database and in RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq/).
Oracle Regular Expressions are very similar in format to PROSITE Patterns. For example, the Tyrosine Kinase Phosphorylation (TKP) site can be represented as ‘[RK].{2,3}[DE].{2,3}[Y]’ with Oracle Regular Expressions, which is similar to the ‘[RK]-x(2,3)-[DE]-x(2,3)-Y’ expression with PROSITE. The SQL statement for identifying the first four instances of the TKP site in each protein is shown as Figure 2 in the Supplementary Material, and the result of the query is displayed in Figure 3.
Figure 3.
Depicts a segment of the results from the Regular Expressions Search. The sequence for protein ‘NP_003983’ is shown with the Regular Expressions highlighted in boldface.
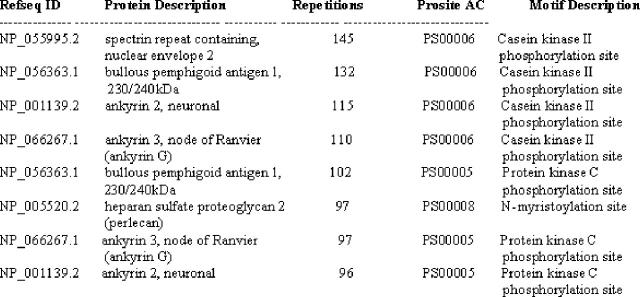
It was discovered that TKP sites were found in 56% of proteins in the protein-protein interaction database, which appeared to be significantly higher than the occurrence of TKP sites in RefSeq proteins. Using Oracle Regular Expressions, it was trivial to extend the case study to writing queries that were able to identify the most frequently occurring protein motifs in a protein database (Figure 4) and to identify the most frequently occurring motifs in individual proteins (Figure 5).
Figure 4.
Most frequently occurring motifs in a protein–protein interaction database.
Figure 5.
Frequency of motifs within proteins.
CONCLUSIONS
In this article, we provided case studies to show how a scientist can perform BLAST sequence homology searches and Regular Expression Searches for protein motifs without leaving the Oracle Database Management System. In both instances, the work was relatively easy to perform and interesting results were gained.
Databases are traditionally known for their capabilities in managing large volumes of data, storing a variety of data types and being able to access distributed data. However, scientists are less aware of the analytical functionality embedded in databases, e.g. statistical analyses, supervised and unsupervised data-mining capabilities and the ability to perform BLAST and Regular Expression Searches.
There are many advantages to performing analytics in the database. Examples include not needing to take data out of a highly available, secure and reliable environment; taking advantage of the database's inherent strengths which include the ability to perform queries in parallel and in batch; and users can pre-filter and post-process queries to get rapidly to the subset of data of interest.
As advances in science and technology are resulting in rapidly expanding data volumes, it becomes increasingly important for scientists to embrace in silico technology for both managing and analyzing data. As databases already manage much life sciences data, they provide a strong analytical platform for drug discovery.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.Clark T., Martin,S. and Liefeld,T. (2004) Globally distributed object identification for biological knowledgebases. Brief. Bioinformatics, 5, 59–70. [DOI] [PubMed] [Google Scholar]
- 2.Stephens S., Chen,J.Y. and Thomas,S. (2004) ODM BLAST: sequence homology search in the RDBMS. IEEE Data Eng. Bull., 27, 20–23. [Google Scholar]
- 3.Gali R. and Stephens,S. (2004) Oracle Database 10g functionality for bioinformatics. Brief. Bioinformatics, 5, 294–299. [Google Scholar]
- 4.Buckingham S. (2004) Bioinformatics: data's future shock. Nature, 428, 774–777. [DOI] [PubMed] [Google Scholar]
- 5.Stephens S. and Tamayo,P. (2003) Supervised and unsupervised data mining techniques for the life sciences. Curr. Drug Discov., 34–36. [Google Scholar]
- 6.Flach P. (1990) Inductive characterisation of database relations. In Ras,Z.W., Zemankova,M. and Emrich,M.L. (eds), Proceedings of the 4th International Symposium on Methodologies for Intelligent Systems. North-Holland Press, Amsterdam, pp. 371–378. [Google Scholar]
- 7.Mannila H. (1997) Inductive database and condensed representations for data mining. In Maluszynski,J. (ed.), Proceedings of the International Logic Programming Symposium. MIT Press, MA, pp. 21–30. [Google Scholar]
- 8.Chen J.C. and Carlis,J.V. (2003) Similar_Join: extending DBMS with a bio-specific operator. In Proceedings of the 18th ACM Symposium on Applied Computing. ACM Press, NY, pp. 109–114. [Google Scholar]
- 9.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 10.Searls D.B. (2003) Pharmacophylogenomics: genes, evolution and drug targets. Nature Rev. Drug Discov., 2, 613–623. [DOI] [PubMed] [Google Scholar]
- 11.Fields S. and Song,O. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- 12.Ito T., Ota,K., Kubota,H., Yamaguchi,Y., Chiba,T., Sakuraba,K. and Yoshida,M. (2002) Roles for the two-hybrid system in exploration of the yeast protein interactome. Mol. Cell. Proteomics, 1, 561–566. [DOI] [PubMed] [Google Scholar]
- 13.Bader G.D., Betel,D. and Hogue,C.W. (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res., 31, 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stubbletine T. (2003) Regular Expression Pocket Reference. O'Reilly Media, Sebastopol, CA. [Google Scholar]
- 15.Friedl J.E.F. (1997) Mastering Regular Expressions. O'Reilly Media, Sebastopol, CA. [Google Scholar]
- 16.Gennick J. and Linsley,P. (2003) Oracle Regular Expressions Pocket Reference. O'Reilly Media, Sebastopol, CA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.