Abstract
DNA methylation in transcriptional regulatory regions is crucial for gene expression. The DNA methylation status of the edges of CpG islands, called CpG island shore, is involved in tissue/cell-type-specific gene expression. Haploinsufficiency diseases are caused by inheritance of one mutated null allele and are classified as autosomal dominant. However, in the same pedigree, phenotypic variances are observed despite the inheritance of the identical mutated null allele, including Fibrillin1 (FBN1), which is responsible for development of the haploinsufficient Marfan disease. In this study, we examined the relationship between gene expression and DNA methylation patterns of the FBN1 CpG island shore focusing on transcriptionally active hypomethylated alleles (Hypo-alleles). No difference in the DNA methylation level of FBN1 CpG island shore was observed in porcine fetal fibroblast (PFF) and the liver, whereas FBN1 expression was higher in PFF than in the liver. However, Hypo-allele ratio of the FBN1 CpG island shore in PFF was higher than that in the liver, indicating that Hypo-allele ratio of the FBN1 CpG island shore likely correlated with FBN1 expression level. In addition, oocyte-derived DNA hypermethylation in preimplantation embryos was erased until the blastocyst stage, and re-methylation of the FBN1 CpG island shore was observed with prolonged in vitro culture of blastocysts. These results suggest that the establishment of the DNA methylation pattern within the FBN1 CpG island shore occurs after the blastocyst stage, likely during organogenesis. In conclusion, Hypo-allele ratios of the FBN1 CpG island shore correlated with FBN1 expression levels in porcine tissues.
Keywords: CpG island shore, DNA methylation, Fibrillin1 (FBN1), Hypo-allele
DNA methylation in transcriptional regulatory regions is crucial for cell type-specific gene expression [1,2,3]. In mammalian genomes, cytosines in cytosine-guanine (CpG) sites of repetitive sequences are stably methylated, whereas the DNA methylation status of genes change dynamically during embryonic development by denovo methylation and demethylation related to cell fate determination [4, 5]. In preimplantation development, genome-wide demethylation occurs after fertilization until the blastocyst stage [6, 7], and cell-type specific DNA methylation patterns are established during organogenesis. Thus, DNA methylation plays an important role for cellular differentiation and mammalian development.
In mammalian genomes, frequency of CpG dinucleotides is about one-fourth of the expected value [8, 9], whereas relatively CG-rich regions, the so-called CpG islands, can be found in the genome. Many CpG islands tend to be located around the transcriptional start site [10, 11], overlapping with promoter regions of most housekeeping genes and half of cell/tissue-specifically expressed genes [12, 13]. Previously, CpG islands were considered unmethylated regardless of the expression status of the corresponding genes. However, a part of or the entire CpG island is methylated in cell-type-specifically expressed genes [14, 15]. The edges of CpG islands are called CpG island shore and are involved in transcriptional regulation [16]. Our and others’ previous findings indicated that these regions are subject to tissue/cell-type-dependent DNA methylation in association with their expression patterns [14,15,16]. In addition, our previous data indicated that abnormal hypomethylation within the CpG island shore of estrogen receptor alpha (ERa) was associated with uterine leiomyoma [17]. Thus, deciphering DNA methylation of CpG island shores will provide new insights into molecular mechanisms of disease onset and pathogenesis.
Haploinsufficiency is caused by inheritance of one mutated null allele together with the other normal allele as heterozygote, which can result in the amount of functional protein being less than half that of the diploid level [18]. Haploinsufficiency diseases are classified as autosomal dominant and often exhibit phenotypic variances despite the inheritance of the identical mutated null allele in the same pedigree. By definition in genetics, a certain autosomal dominant trait that appears only in some of the affected individuals is termed incomplete penetrance, and the severity of a phenotype that varies among affected individuals is named variable expressivity. However, incomplete penetrance and variable expressivity in autosomal dominant diseases, including haploinsufficiency, cannot be explained only by the genetic mutation of a single responsible gene. In addition, although haploinsufficiency diseases are genetically autosomal dominant, some carriers do not show any symptoms, leading to non-penetrance in the same pedigree. Since these phenotypic variances in haploinsufficiency cannot be explained by the simple inheritance of one mutated null allele and dealt only as a statistical probability in classical genetics, molecular mechanisms underlying these phenomena remain to be elucidated.
Fibrillin1 (FBN1) encoding a microfibril protein, Fibrillin, is expressed in various tissues and cell types [19]. Many Marfan syndrome pedigrees inherit mutated null alleles of FBN1, some of which likely result in haploinsufficiency in an autosomal dominant fashion [20, 21]. As mentioned above, Marfan syndrome caused by haploinsufficiency exhibits phenotypic variations, including non-penetrance even in the same Marfan pedigree inheriting the identical loss-of-function null allele [20, 22, 23]. In general, null alleles result from a premature termination codon (PTC) by nonsense or frameshift mutations. These PTCs are recognized by nonsense-mediated mRNA decay system, and the pre-mRNAs containing PTCs are degraded during the RNA splicing process. Thus, heterozygous individuals with one normal allele and the other PTC-containing null allele exhibit reduced mRNA levels translated to the functional protein [24]. Analysis of fibroblast cells derived from Marfan’s patients containing one null allele with PTC indicated that the FBN1 mRNA level varied among patients [25, 26]. It has also been reported that patients with a low level of the FBN1 mRNA from the normal allele suffer an increased risk of ectopia lentis, pectus excavatum, and aortic abnormalities [26]. Taken together, these findings strongly suggest that skewed FBN1 expression from the normal allele, which is transcribed and translated into the functional Fibrillin protein, is associated with disease onset and severity of Marfan syndrome.
DNA methylation analysis by sodium bisulfite polymerase chain reaction (PCR) followed by sequencing of cloned PCR fragments enabled us to identify DNA methylation patterns of each sequenced fragment that can be recognized as one allele/cell in a tissue. Based on this concept, we previously focused on transcriptionally active DNA hypomethylated alleles (Hypo-alleles), which can be recognized by the high ratio of unmethylated CpGs in each sequenced fragment (allele), and indicated that Hypo-allele ratio of a non-imprinted and autosomal gene can be used to estimate the proportion of the cell type expressing the corresponding gene in the tissue [27]. Therefore, the Hypo-allele concept can be applied to the detection of transcriptional regulation in each allele involved in certain tissues or cell populations.
In the present study, we determined the relationship between the DNA methylation pattern of the FBN1 CpG island shore, focusing on Hypo-alleles, and FBN1 expression, together with the epigenetic reprogramming and re-establishment of FBN1 during early embryonic development using embryos and tissues from pigs, whose physiological similarity to humans is very useful to biomedical research.
Materials and Methods
Porcine fetal fibroblast (PFF) culture
Primary culture of PFF was performed as previously described [28]. Briefly, PFF was cultured in Minimum Essential Medium Eagle (MEM) Alpha Modifications (Invitrogen, Carlsbad, CA, USA) supplemented with 15% fetal bovine serum (Biowest, Nuaillè, France) and Penicillin-Streptomycin (Invitrogen) with type I collagen-coated dish in a humidified atmosphere of 5% CO2 at 37°C. In addition, inhibition of DNA methylation in PFF was performed by 5-aza-2′-deoxycytidine (5-aza-dC, Sigma-Aldrich, Tokyo, Japan) treatment of 1 or 5 µM for four days.
In vitro fertilization (IVF) and parthenogenetic porcine embryos
The IVF method has been described previously [29]. Briefly, porcine ovaries were collected at local abattoirs, and in vitro matured oocytes with expanded cumulus cells were used for IVF. Frozen sperm collected from an LWD pig was thawed in warm water and suspended in DPBS supplemented with 0.1% bovine serum albumin. After washing, sperm was resuspended in porcine fertilization medium (PFM; Research Institute for the Functional Peptides, Yamagata, Japan) at a concentration of 1 × 107 cells/ml. For insemination, sperm and in vitro matured oocytes were incubated for 8 h in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2 at 38.5°C. After insemination, the eggs were treated with Tyrode’s lactose medium containing 10 mM HEPES and 0.3% (w/v) polyvinylpyrrolidone (HEPES-TL-PVP), and cumulus cells and excess sperm were removed by gentle pipetting. Eggs that released one or more polar bodies were cultured in porcine zygote medium-5 (PZM-5; Research Institute for the Functional Peptides) under paraffin oil (Kanto Chemical, Tokyo, Japan) in the same humidified atmosphere as insemination, and morula or blastocyst stage embryos were collected for DNA methylation analysis.
Generation of porcine parthenogenetic embryos has been previously described [30]. Briefly, in vitro matured oocytes with expanded cumulus cells were treated with 1 mg/ml of hyaluronidase dissolved in HEPES-TL-PVP followed by separation from the cumulus cells by gentle pipetting. Oocytes with evenly granulated ooplasm and an extruded first polar body were selected for subsequent experiments. Oocytes were transferred to an activation solution containing 0.3 M mannitol (Nacalai Tesque, Kyoto, Japan), 50 µM CaCl2, 100 µM MgCl2, and 0.01% (w/v) PVA and activation was performed by applying a single direct current pulse (150 V/mm, 100 µsec) using an electrical pulsing machine (LF201, Nepa Gene, Chiba, Japan). Activated oocytes were treated with 5 µg/mL cytochalasin B for 3 h to suppress extrusion of the second polar body followed by in vitro culture for up to 6 days to obtain parthenogenetic embryos at 2 to 4-cell-stages (day 2) and blastocyst stage (days 5 and 6). In vitro culture of parthenogenetic embryos was performed in PZM-5 under paraffin oil in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2 at 38.5°C.
Genomic DNA extraction and sodium bisulfite reaction
Genomic DNA extraction and bisulfite conversion were carried out as described previously [31]. Genomic DNA was purified from PFF, the liver, the sperm, and the aorta by Proteinase K digestion and PCI (phenol, chloroform, and isoamyl alcohol: 50/49/1, v/v) extraction followed by ethanol precipitation. In the case of genomic DNA extraction from sperm, mercaptoethanol was added to a final concentration of 0.144 M with Proteinase K. Tissues (liver and aorta) and PFF were collected from LWD pigs. Purified genomic DNA digested with HindIII (TaKaRa, Kyoto, Japan) was purified by ethanol precipitation. After denaturing HindIII-digested genomic DNA with 0.3 M NaOH, sodium metabisulfite (pH 5.0) and hydroquinone were added to final concentrations of 2.0 M (4.0 M equivalent of sodium bisulfite) and 0.5 mM, respectively. The bisulfite reaction was performed using a thermocycler as follows: 20 cycles of 95°C for 30 sec and 55°C for 15 min, followed by 55°C for 10 h. Bisulfite-treated genomic DNA was purified with a QIAquick Gel Extraction Kit (Qiagen GmbH, Hilden, Germany), desulfonated with 0.3 M NaOH at 37°C for 15 min, and ethanol-precipitated. For the bisulfite reactions of porcine IVF-derived and parthenogenetic early embryos, EZ DNA Methylation-Direct Kit (Zymo Research, Irvine, CA, USA) was used following the manufacturer’s protocol. Briefly, 11 to 80 early embryos or oocytes (appropriately 80 cells) were pooled and digested with Proteinase K for 20 min at 50°C followed by the bisulfite reaction with CT Conversion Reagent under the following conditions: 98°C for 8 min and then 64°C for 3.5 h. Purified bisulfite-treated DNA was amplified with BioTaq HS DNA polymerase (BIOLINE, London, UK) using specific primers for porcine FBN1 regions (FBN1_Bis_F: 5′-GAGGTAATGGGAAGAAGTGGAGTAG-3′, FBN1_Bis_R: 5′-CCAAACTAACATTCACAAACCTCTAA-3′). PCR was performed under the following conditions: 95°C for 10 min; 43 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min; final extension 72°C for 2 min. PCR products were subcloned into pGEM-T Easy vector (Promega, Madison, WI, USA) for sodium bisulfite sequencing.
RNA extraction and cDNA synthesis
Total RNA was extracted using TRIzol reagent (Invitrogen). After treatment of the total RNAs with RNase-free DNase I (Invitrogen), first-strand cDNA synthesis was performed using a Superscript III First-strand Synthesis System (Invitrogen) with random hexamers. RT-PCR was performed using TaKaRa LA Taq (TaKaRa) with specific primer pairs for FBN1 (FBN1_RT_F; 5′-TGCTTACTGTTGCCCTGGAT-3′, FBN1_RT_R; 5’-CTGGAGCCACAGGAAGGAG-3′) and GAPDH (GAPDH_RT_F; 5′-ACCACAGTCCATGCCATCAC-3′, GAPDH_RT_R; 5′-TCCACCACCCTGTTGCTGTA-3′) mRNAs under the following conditions: 95°C for 5 min; 30 cycles (FBN1) or 25 cycles (GAPDH) of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min; final extension 72°C for 10 min. Statistical comparisons of FBN1 mRNA levels were performed using the Student’s t-test, and for the multiple comparisons Student’s t-test with a Bonferroni correction was applied.
Homology search
Homology search and sequence alignment of the FBN1 CpG island shore among pig, human, and mouse were performed using YASS program [32] and ApE (http://biologylabs.utah.edu/jorgensen/wayned/ape/), respectively. Genomic sequence data of the human (chr15:48,935,985-48,939,984 GRCh37/hg19), pig (chr1:137,101,509-137,105,508 Sscrofa10.2/susScr3), and mouse (chr2:125,330,175-125,334,174 NCBI37/mm9) FBN1 loci were obtained from the University of California, Santa Cruz genome browser (https://genome.ucsc.edu/).
Results
Cell/tissue-dependent DNA methylation patterns within the FBN1 CpG island shore
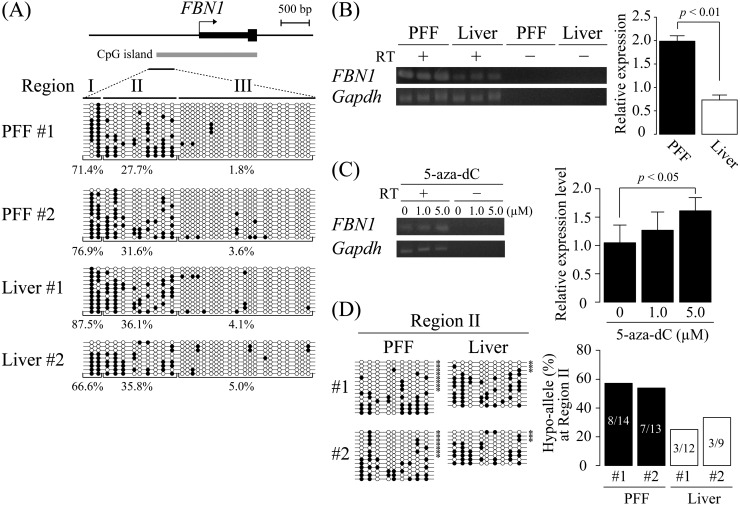
To examine DNA methylation patterns of the FBN1 promoter region, including CpG island, sodium bisulfite sequencing was performed using a bisulfite PCR primer pair that can amplify the FBN1 CpG island and its upstream island shore region (Fig. 1A). Genomic DNA from PFF and the liver were subjected to bisulfite sequencing. Judging from the methylation levels, the analyzed region could be divided into three sub-regions (Region I: outside of the CpG island, Region II: CpG island shore, Region III: inside the CpG island). Regions I and III were hypermethylated (66.6−87.5%) and hypomethylated (1.8−5.0%), respectively, in PFF and the liver. On the other hand, Region II exhibited a moderate level of DNA methylation (27.7−36.1%) in PFF and the liver. The FBN1 expression was analyzed by RT-PCR (Fig. 1B), and PFF isolated from a connective tissue, the skin, showed a higher expression level of FBN1 than the liver (P < 0.01). This indicated that simple DNA methylation level of the FBN1 promoter region did not correlate with mRNA expression levels. However, treatment of PFF with 5 µM 5-aza-dC, a DNA methylation inhibitor, resulted in a statistically significant increase of the FBN1 mRNA level compared with non-treated control cells (Fig. 1C). This strongly suggested that DNA methylation is at least in part related to transcriptional repression of FBN1. To further examine the correlation between DNA methylation and FBN1 expression, the previously proposed Hypo-allele concept [27] was applied to FBN1. CpG island shores are involved in the tissue/cell-type-dependent gene expression [14, 15]. Consistent with this finding, Hypo-allele ratios of FBN1 CpG island shore (Region II) in PFF were relatively high at 57.1% (8/14 fragments) and 53.8% (7/13 fragments), whereas those in the liver were 25.0% (3/12 fragments) and 33.3% (3/9 fragments), indicating that PFF contain more alleles/cells that are prone to express FBN1 than the liver (Fig. 1D). In contrast, Hypo-allele ratios of Regions I and III (outside and inside of CpG islands, respectively) did not show significant difference between PFF and the liver (Supplementary Fig. 1A: online only). In addition, 5-aza-dC treatment caused an increase in the Hypo-allele ratios of Region II in PFF (Supplementary Fig. 1B). Taken together, the DNA methylation pattern of the Region II, focusing on Hypo-alleles, is likely correlated with the expression of mRNA at the FBN1 locus. FBN1 is highly expressed in the aorta, whose abnormal phenotype has been reported in patients with Marfan syndrome [33]. DNA methylation analysis focusing on each sequenced allele also exhibited relatively high Hypo-allele ratios of 42.9 and 57.1% in two aorta samples (Supplementary Fig. 1C). Therefore, Hypo-allele ratios in the Region II (CpG island shore) correlated with FBN1 mRNA expression levels.
Fig. 1.
DNA methylation analysis of the porcine FBN1 CpG island shore. (A) DNA methylation status of the FBN1 CpG island shore within the FBN1 promoter region was analyzed by sodium bisulfite sequencing using two samples of porcine fetal fibroblast (PFF #1 and #2) and liver (Liver #1 and #2). The FBN1 CpG island shore was divided into three regions (Regions I, II, and III), and their DNA methylation levels (methylated CpGs/all CpGs) were calculated for each region. Open and closed circles indicate unmethylated and methylated CpGs, respectively. (B) FBN1 mRNA expression. FBN1 mRNA expression levels in PFF and the liver were assessed by RT-PCR. The relative expression levels were normalized to those of GAPDH. The expression levels are shown as mean ± SD (n = 3). Statistical comparisons of the expression levels were performed using the Student’s t-test. (C) FBN1 mRNA expression level was increased by 5-aza-dC treatment. FBN1 mRNA expression in PFF treated with 5-aza-dC (0, 1.0, or 5.0 µM) for 4 days was assessed by RT-PCR. The relative expression levels were normalized to that of GAPDH. The 5-aza-dC treatment with each dose was performed twice independently, and PCR for each sample was performed in triplicate independently. The expression levels are shown as mean ± SD, and statistical comparisons of the expression levels among the samples of 0, 1.0, 5.0 µM treatment were performed using the Student’s t-test with a Bonferroni correction. (D) Hypo-allele ratio of the Region II in the FBN1 CpG island shore. Based on the bisulfite sequencing data of the Region II in Fig. 1A, sequenced fragments that contain ≥ 75% unmethylated CpG sites (7–9 out of the 9 CpG sites in the Region II) were defined as Hypo-alleles (left panel), and the Hypo-allele ratio (Hypo-allele fragments/all sequenced fragments) was calculated (right panel). * Hypo-alleles in the sequenced fragments.
Establishment of the DNA methylation pattern in the FBN1 CpG island shore during early porcine embryonic development
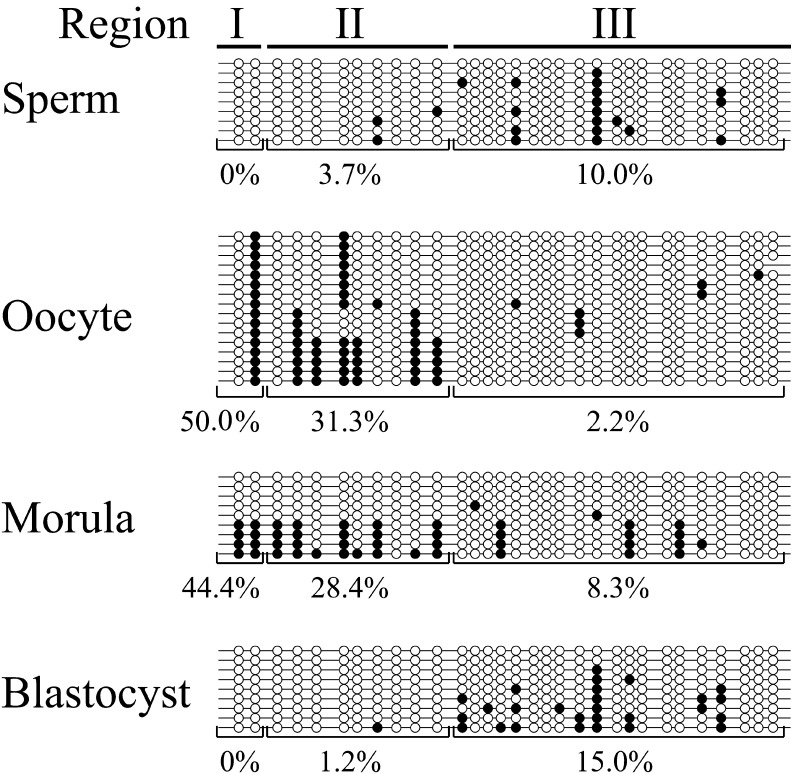
It is well-known that after fertilization of hypermethylated sperm and hypomethylated oocytes, genome-wide DNA demethylation occurs during preimplantation development followed by genome-wide DNA re-methylation in later stage embryos/fetuses in association with cellular differentiation events [6, 7]. To determine whether DNA methylation pattern of FBN1 changes during preimplantation embryos through the blastocyst stage, germ cells (sperm and oocytes), and early stage embryos were analyzed by sodium bisulfite sequencing (Fig. 2). In contrast to genome-wide DNA methylation level, CpG island and its upstream shore were unexpectedly hypomethylated in all three regions (I–III) of sperm (0−10.0%), but hypermethylated especially in Regions I and II of the oocyte (50.0 and 31.3%, respectively). On the other hand, the other FBN1 CpG island shore in intron 1 was almost completely methylated in sperm and oocytes (Supplementary Fig. 2: online only). After fertilization, DNA demethylation within the CpG island and shore regions occurred, and DNA methylation level was the lowest in the blastocyst stage embryos.
Fig. 2.
DNA methylation patterns were reprogrammed in the promoter region, including the FBN1 CpG island shore during preimplantation development. In sperm, oocytes, IVF-produced morula, and blastocyst stage embryos, DNA methylation status was analyzed by sodium bisulfite sequencing. In morula and blastocyst stage embryos, genomic DNA extracted from about ten embryos was used for DNA methylation analysis. DNA methylation level of each region (Regions I, II, and III) was calculated based on the methylated CpGs/all CpGs. Open and closed circles indicate unmethylated and methylated CpGs, respectively.
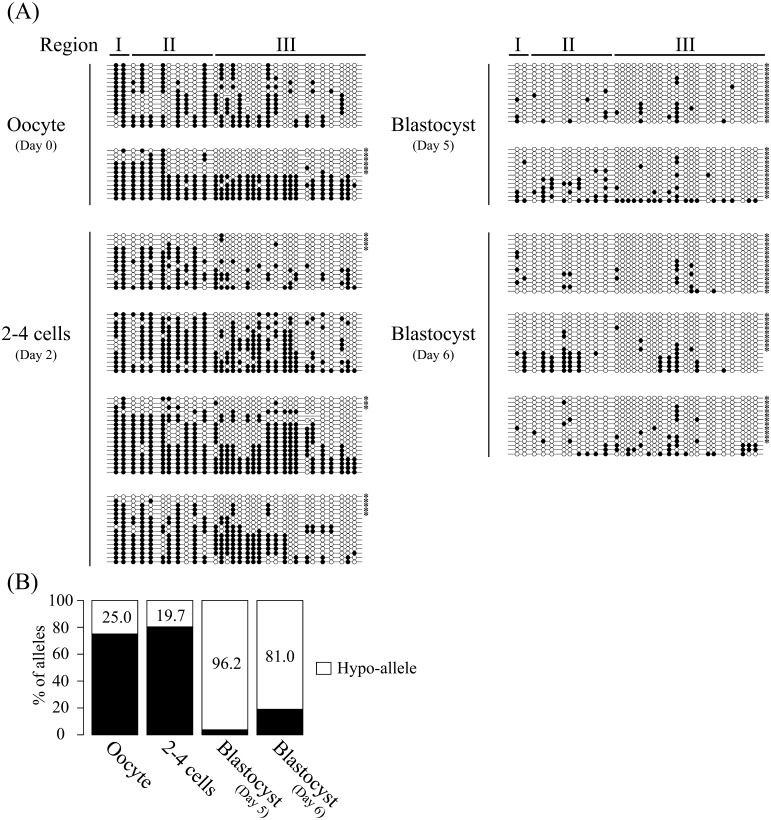
As shown in Fig. 2, the FBN1 CpG island shore in the oocyte was obviously hypermethylated compared with sperm, and the blastocyst stage embryos exhibited a hypomethylated status similar to that of sperm. These data suggested that oocyte-derived alleles of the FBN1 CpG island shore are subjected to demethylation during early embryonic development until blastocyst stage. Therefore, DNA methylation reprogramming of the FBN1 CpG island shore in early porcine embryonic development was further analyzed using parthenogenetic embryos that contained only oocyte-derived genomic DNA with the hypermethylated FBN1 allele (Fig. 3A). In the developing parthenogenetic embryos, oocytes and 2–4 cell embryos were hypermethylated in the whole analyzed region, and gradual demethylation occurred, as expected, until the blastocyst stage, indicating that the DNA methylation pattern of oocytes was erased during early embryonic development. By analyzing the Hypo-allele ratio of the whole sequenced region, the onset of DNA re-methylation in an allele-dependent manner was observed along with a reduced Hypo-allele ratio in blastocyst embryos with a prolonged culture period through day 6 (81.0%) compared with that of day 5 blastocyst (96.2%) (Fig. 3B). However, Region II-specific hypomethylated alleles together with Region I hypermethylation and Region III hypomethylation were not observed in the embryos until the blastocyst stage, suggesting that Hypo-alleles in Region II detected in PFF and the aorta would be established in later stage embryos/fetuses.
Fig. 3.
De novo methylation occurred in vitro cultured embryos with a prolonged culture period. (A) DNA methylation status of FBN1 CpG island shore was analyzed in the oocyte (day 0), parthenogenetic 2–4 cell stage embryos (day 2), and blastocyst stage-embryos (days 5 and 6). Each sample contains genomic DNA extracted from about 80 cells, and the experiments were performed 2–4 times independently for each sample. DNA methylation rates (calculated by methyl CpG/total CpG) were 44.8% in the oocyte, 46.3% in the 2−4 cell embryos, 10.5% in the day 5 blastocyst, and 9.8% in the day 6 blastocyst. (B) Hypo-allele ratios at each developmental stage. Hypo-allele ratios were calculated from the 27, 61, 27, and 42 sequenced clones amplified by PCR from bisulfite-modified genomic DNAs of oocyte (day 0), 2–4 cells (day 2), blastocyst (day 5), and blastocyst (day 6), respectively. In the oocyte and preimplantation stage embryos, DNA methylation status was clearly different among the sequenced fragments across the whole sequenced region. Thus, the sequenced fragments that contain ≥75% unmethylated CpG sites (more than 24 of the 31 CpG sites in the amplified fragments) in the whole analyzed region were defined as Hypo-alleles (white bar).
Higher sequence conservation of the FBN1 CpG island shore between humans and pigs than between humans and mice
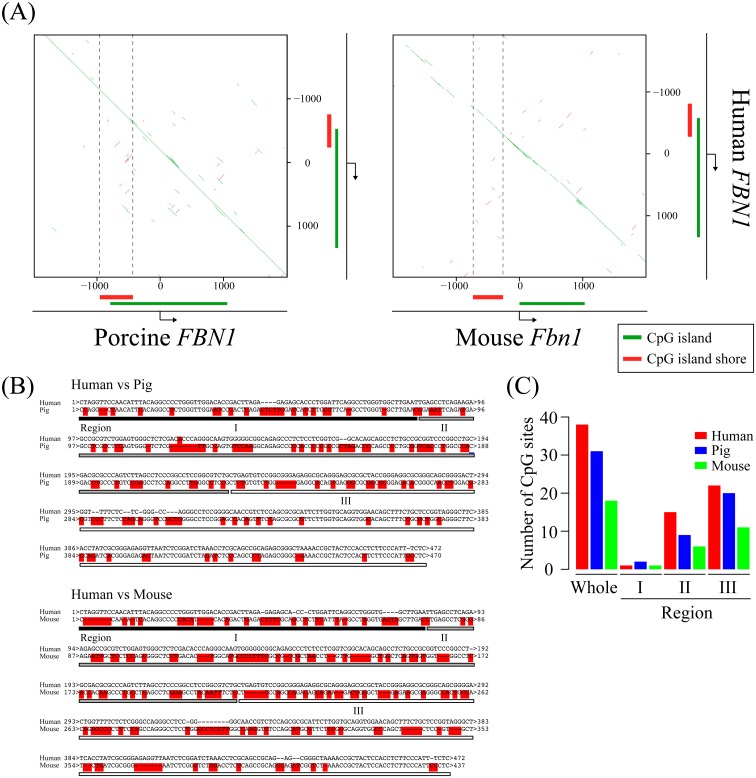
The pig is increasingly recognized as a good animal model to study human disease owing to its physiological and anatomical similarity to humans [34,35,36]. Our present data indicated that FBN1 expression level is at least in part regulated by DNA methylation of the FBN1 CpG island shore. To apply our finding to Marfan disease research in humans, we sought to determine whether the genomic DNA sequence of the porcine FBN1 CpG island shore was conserved in the equivalent region of human FBN1. For this purpose, the porcine genomic sequence of the FBN1 CpG island shore and its surrounding regions were compared with those of human and mouse equivalent regions. A comparison of pig and human sequences (transcription start site ± 2 kb of FBN1) with YASS program exhibited high homology between these two mammalian species (Fig. 4A, left panel). On the other hand, the equivalent sequence of mouse Fbn1 analyzed by the same method showed less conservation between humans and mice, especially for the CpG island shore region (Fig. 4A, right panel). Since our present data suggested that the DNA methylation of CpGs in the FBN1 CpG island shore was involved in the transcriptional regulation of FBN1, we focused on the number of CpGs in the FBN1 CpG island shore regions of pig, human, and mouse using ApE (Fig. 4B). As shown in Fig. 4C, human FBN1 CpG island shore contained 38 CpGs, similar to that in pigs (31 CpGs). In contrast, mouse Fbn1 CpG island shore contained only 18 CpGs, which is less than half the number of CpGs in the equivalent human region. Taken together, both the genomic sequence and the number of CpG are more highly conserved between pig and human than between mouse and human. Therefore, the pig is likely more advantageous for translational research of Marfan syndrome caused by haploinsufficiency of FBN1 than the mouse not only because of the physiological and anatomical similarity to human, but also because of the significant conservation of genomic sequence and the number of CpG sites that can be methylated.
Fig. 4.
Homology search of the FBN1 CpG island shore among human, pig, and mouse. (A) Homology search of the FBN1 transcriptional start site ± 2 kilobase pair (kbp), including the FBN1 CpG island shore between human and pig/mouse was performed using YASS program. Green lines indicate highly conserved regions between the two species, and vertical dotted black lines indicate the FBN1 CpG island shore. (B) Sequence alignment between human and pig/mouse in the FBN1 CpG island shore. Nucleotides with red highlights mean mismatch between human and pig or mouse. The lengths of the conserved FBN1 CpG island shore in human, pig and mouse are 472, 470, and 437 base pairs (bp), respectively. (C) The number of CpG sites in the FBN1 CpG island shore (whole) and sub-regions (Regions I, II, and III) is represented as a bar graph.
Discussion
In the present study, the role of FBN1 epigenetic regulation, especially by DNA methylation, was analyzed in porcine tissues focusing on DNA methylation patterns of each allele in the CpG island shore. Our DNA methylation analysis indicated that DNA methylation levels of the classical definition that is calculated by methylated CpGs out of all CpGs did not exhibit clear correlation with FBN1 expression levels. In contrast, based on the previously proposed Hypo-alleles [27], the Hypo-allele ratio of the CpG island shore within the FBN1 promoter region correlated with FBN1 mRNA expression levels. An increased expression of FBN1 with a DNA methylation inhibitor, 5-aza-dC, further supports the involvement of DNA methylation on the transcriptional repression of FBN1. Therefore, these results strongly suggest that hypomethylation of the FBN1 CpG island shore is required to increase FBN1 mRNA expression.
DNA methylation analysis of porcine early embryos exhibited progressive demethylation in the promoter region of FBN1, including the CpG island shore, until the blastocyst stage, indicating that the DNA methylation patterns of germ cells are erased during early embryonic development. Genome-wide DNA demethylation occurs during early embryonic development [6, 7], and the FBN1 CpG island shore in the promoter region should be subjected to this demethylation wave. Although sperm and oocytes are generally hypermethylated and hypomethylated, respectively, the CpG island shore in the FBN1 promoter exhibited unexpected hypomethylation in sperm and hypermethylation in the oocyte. This indicates that certain germ-cell-specific regulation of DNA methylation different from genome-wide changes in DNA methylation levels are involved in the formation of DNA methylation patterns in the FBN1 promoter region, including the CpG island shore, in germ cells. The Hypo-allele ratio was highest in the day 5 blastocysts and decreased in day 6 blastocysts resulting in further development of the embryos by prolongation of the culture period. Thus, FBN1 promoter methylation starts after the blastocyst stage, whereas the DNA methylation patterns specific to the FBN1 CpG island shore in the aorta, PFF, and the liver are likely established later in embryonic/fetal development.
The pig FBN1 CpG island shore shared a higher homology with the equivalent human region than with the mouse equivalent in terms of genomic sequence and the number of CpG. Our previous data indicated that tissue/cell-type-dependent differentially methylated regions tend to be in CpG island shore regions that are the boundary between hypomethylated CpG islands and hypermethylated outside regions [14, 15]. Consistent with this finding, the FBN1 CpG island shore presented a Hypo-allelic region correlated with gene expression at the boundary between the hypomethylated CpG island and the hypermethylated outside region. Thus, these data strongly suggest that pigs are a more advantageous model than mice to study epigenetic regulation of FBN1 expression that would be crucial for the onset and/or manifestation of Marfan disease caused by FBN1 haploinsufficiency.
FBN1 heterozygous knockout pigs (containing one null allele with PTC) produced by our group exhibited a broad spectrum of abnormal individual phenotypes, including pectus excavatum, cleft palate, and aortic abnormality as well as non-penetrance, although the heterozygous knockout pigs present the syngeneic background produced by somatic cell nuclear transfer [37]. These symptoms are similar to those observed in patients with Marfan syndrome, including phenotypic variations [20, 22]. A cause of Marfan syndrome is haploinsufficiency by inheriting one PTC-containing null allele, and our porcine model for Marfan syndrome generated by genome editing also has a PTC-containing null allele [37], which can result in shortage of functional Fibrillin protein level despite having the other normal allele. In humans, the genetic background of individuals even in the same Marfan pedigree varies, except for the inheritance of the PTC-containing null allele of FBN1, which, in turn, would lead us to hypothesize the existence of certain genetic modifiers of FBN1-related genes. However, each individual Marfan model pig produced by somatic cell nuclear transfer contains the same genetic background, and, thus, modifier loci/genes are unlikely to be involved in the phenotypic variations among individual pigs. Therefore, phenotypic variations of Marfan syndrome could be explained solely by haploinsufficiency, resulting variable decreases in the functional Fibrillin protein level in FBN1-expressing connective tissues of each individual without modifier loci/genes.
The DNA methylation pattern of the FBN1 CpG island shore varied depending on sequenced fragments (alleles) in PFF and the aorta exhibiting high expression levels of FBN1, and DNA methylation inhibition by 5-aza-dC in PFF resulted in a further increase of FBN1 expression in the present study. These data suggest that the DNA methylation levels of the CpG island shore on each allele correlates with the FBN1 expression. Our previous data indicated that DNA methylation levels in CpG island shores are important for tissue/cell-type-dependent expression of CpG-island-containing genes [14, 15]. Our previous data also indicate that aberrant regulation of DNA methylation in CpG island shores of ERa and spalt like transcription factor 3 (Sall3) are associated with uterine leiomyomas in humans and placentomegaly in cloned mice, respectively [17, 38]. Since FBN1 is a non-imprinted autosomal gene exhibiting about 40–60% Hypo-alleles in the CpG island shore in PFF and the aorta, about half of total alleles in the analyzed cell populations can be thought as DNA hypomethylated (Hypo-alleles) in the CpG island shore and prone to express FBN1. Taken together, in a FBN1 heterozygous situation with one normal allele and the other PTC-containing null allele as observed in patients with Marfan syndrome, about 50% Hypo-alleles in FBN1-expressing tissues should include both normal and null alleles, whose ratio is likely random because of the non-imprinted autosomal gene. Our present data indicated that the oocyte-derived DNA hypermethylation is reprogrammed to the hypomethylated status around the FBN1 CpG island shore during early embryonic development until blastocyst stage followed by initiation of DNA re-methylation. Thus, in early embryonic development, reprogramming of the DNA methylation pattern of the FBN1 CpG island shore is likely required for the establishment of the DNA methylation patterns in FBN1-expressing tissue/cell types. These data strongly suggest that in FBN1 heterozygous individuals, DNA re-methylation of about 50% of the alleles in the expressing tissues would occur both in normal and PTC-containing null allele in a random manner, which affects the functional FBN1 protein level from the normal alleles. When normal alleles tend to be hypermethylated by chance within the CpG island shore in FBN1-expressing tissues or cell populations, the tissues or cell populations should exhibit a non-negligible decrease of the functional Fibrillin protein level, which may meet the criteria of haploinsufficiency. Consistent with this, reduced expression from the normal FBN1 allele has been reported to correlate with the severity of symptoms in patients with Marfan syndrome [26]. This leads us to hypothesize that stochastic hypermethylation of the normal alleles, resulting in decreased functional Fibrillin protein levels, can be a cause of the onset and/or manifestation of Marfan’s symptoms together with phenotypic variations. Therefore, allelic variations of DNA methylation levels themselves can be an epigenetic modifier of haploinsufficiency, but not a genetic modifier of unknown other gene loci. Thus, the next issue to be addressed should be to distinguish between the normal and null alleles of FBN1 and determine whether Hypo-allele ratios of the FBN1 CpG island shore in the normal alleles correlates with the expression level of the functional FBN1 mRNA together with the disease onset and/or manifestation of Marfan’s symptoms in FBN1 heterozygous pigs.
The present study indicates that hypomethylated status (Hypo-allele ratio) of the FBN1 CpG island shore correlates with FBN1 mRNA expression level. Establishment of the DNA methylation pattern in the FBN1 CpG island shore is suggested to start after the blastocyst stage during organogenesis. As a perspective, the Hypo-allele concept proposed as the stochastic regulation of FBN1 expression might provide useful information for the onset and manifestation of haploinsufficiency dominant diseases, whose molecular mechanisms cannot be explained by simple genetic differences of responsible genes in classical genetics.
Supplementary
Acknowledgments
Financial support for this study was provided by a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to HN and JO), Research Project Grant (B) by Institute of Science and Technology Meiji University (to JO), and Grant-in-Aid for Young Scientists (B) (to YA). This work is supported in part by a Research Funding for Computational Software Supporting Program from Meiji University, Meiji University International Institute for Bio-Resource Research, and Leading Advanced Project for medical innovation (LEAP), AMED. JO and YA thank Dr Koichiro Nishino for his kind cooperation with this project. The authors have no competing financial interest to declare.
References
- 1.Shiota K. DNA methylation profiles of CpG islands for cellular differentiation and development in mammals. Cytogenet Genome Res 2004; 105: 325–334. [DOI] [PubMed] [Google Scholar]
- 2.Lieb JD, Beck S, Bulyk ML, Farnham P, Hattori N, Henikoff S, Liu XS, Okumura K, Shiota K, Ushijima T, Greally JM. Applying whole-genome studies of epigenetic regulation to study human disease. Cytogenet Genome Res 2006; 114: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohgane J, Yagi S, Shiota K. Epigenetics: the DNA methylation profile of tissue-dependent and differentially methylated regions in cells. Placenta 2008; 29(Suppl A): S29–S35. [DOI] [PubMed] [Google Scholar]
- 4.Shiota K, Kogo Y, Ohgane J, Imamura T, Urano A, Nishino K, Tanaka S, Hattori N. Epigenetic marks by DNA methylation specific to stem, germ and somatic cells in mice. Genes Cells 2002; 7: 961–969. [DOI] [PubMed] [Google Scholar]
- 5.Sakamoto H, Suzuki M, Abe T, Hosoyama T, Himeno E, Tanaka S, Greally JM, Hattori N, Yagi S, Shiota K. Cell type-specific methylation profiles occurring disproportionately in CpG-less regions that delineate developmental similarity. Genes Cells 2007; 12: 1123–1132. [DOI] [PubMed] [Google Scholar]
- 6.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science 2001; 293: 1089–1093. [DOI] [PubMed] [Google Scholar]
- 7.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 2002; 241: 172–182. [DOI] [PubMed] [Google Scholar]
- 8.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol 1987; 196: 261–282. [DOI] [PubMed] [Google Scholar]
- 9.Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet 2009; 10: 285–311. [DOI] [PubMed] [Google Scholar]
- 10.Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci 2003; 60: 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat Rev Genet 2004; 5: 446–455. [DOI] [PubMed] [Google Scholar]
- 12.Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics 1992; 13: 1095–1107. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki Y, Tsunoda T, Sese J, Taira H, Mizushima-Sugano J, Hata H, Ota T, Isogai T, Tanaka T, Nakamura Y, Suyama A, Sakaki Y, Morishita S, Okubo K, Sugano S. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res 2001; 11: 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imamura T, Ohgane J, Ito S, Ogawa T, Hattori N, Tanaka S, Shiota K. CpG island of rat sphingosine kinase-1 gene: tissue-dependent DNA methylation status and multiple alternative first exons. Genomics 2001; 76: 117–125. [DOI] [PubMed] [Google Scholar]
- 15.Yagi S, Hirabayashi K, Sato S, Li W, Takahashi Y, Hirakawa T, Wu G, Hattori N, Hattori N, Ohgane J, Tanaka S, Liu XS, Shiota K. DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res 2008; 18: 1969–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 2009; 41: 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asada H, Yamagata Y, Taketani T, Matsuoka A, Tamura H, Hattori N, Ohgane J, Hattori N, Shiota K, Sugino N. Potential link between estrogen receptor-alpha gene hypomethylation and uterine fibroid formation. Mol Hum Reprod 2008; 14: 539–545. [DOI] [PubMed] [Google Scholar]
- 18.Cook DL, Gerber AN, Tapscott SJ. Modeling stochastic gene expression: implications for haploinsufficiency. Proc Natl Acad Sci USA 1998; 95: 15641–15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakai LY, Keene DR, Engvall E. Fibrillin, a new 350-kD glycoprotein, is a component of extracellular microfibrils. J Cell Biol 1986; 103: 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Judge DP, Biery NJ, Keene DR, Geubtner J, Myers L, Huso DL, Sakai LY, Dietz HC. Evidence for a critical contribution of haploinsufficiency in the complex pathogenesis of Marfan syndrome. J Clin Invest 2004; 114: 172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jondeau G, Michel JB, Boileau C. The translational science of Marfan syndrome. Heart 2011; 97: 1206–1214. [DOI] [PubMed] [Google Scholar]
- 22.Robinson PN, Booms P, Katzke S, Ladewig M, Neumann L, Palz M, Pregla R, Tiecke F, Rosenberg T. Mutations of FBN1 and genotype-phenotype correlations in Marfan syndrome and related fibrillinopathies. Hum Mutat 2002; 20: 153–161. [DOI] [PubMed] [Google Scholar]
- 23.Potter KJ, Creighton S, Armstrong L, Eydoux P, Duncan W, Penny DJ, Fan Y, Gibson WT. The c.7409G>A (p.Cys2470Tyr) Variant of FBN1: Phenotypic Variability across Three Generations. Mol Syndromol 2013; 4: 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrier L, Schlossarek S, Willis MS, Eschenhagen T. The ubiquitin-proteasome system and nonsense-mediated mRNA decay in hypertrophic cardiomyopathy. Cardiovasc Res 2010; 85: 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchinson S, Furger A, Halliday D, Judge DP, Jefferson A, Dietz HC, Firth H, Handford PA. Allelic variation in normal human FBN1 expression in a family with Marfan syndrome: a potential modifier of phenotype? Hum Mol Genet 2003; 12: 2269–2276. [DOI] [PubMed] [Google Scholar]
- 26.Aubart M, Gross MS, Hanna N, Zabot MT, Sznajder M, Detaint D, Gouya L, Jondeau G, Boileau C, Stheneur C. The clinical presentation of Marfan syndrome is modulated by expression of wild-type FBN1 allele. Hum Mol Genet 2015; 24: 2764–2770. [DOI] [PubMed] [Google Scholar]
- 27.Arai Y, Fukukawa H, Atozi T, Matsumoto S, Hanazono Y, Nagashima H, Ohgane J. Ultra-Deep Bisulfite Sequencing to Detect Specific DNA Methylation Patterns of Minor Cell Types in Heterogeneous Cell Populations: An Example of the Pituitary Tissue. PLoS ONE 2016; 11: e0146498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurome M, Hisatomi H, Matsumoto S, Tomii R, Ueno S, Hiruma K, Saito H, Nakamura K, Okumura K, Matsumoto M, Kaji Y, Endo F, Nagashima H. Production efficiency and telomere length of the cloned pigs following serial somatic cell nuclear transfer. J Reprod Dev 2008; 54: 254–258. [DOI] [PubMed] [Google Scholar]
- 29.Umeyama K, Honda K, Matsunari H, Nakano K, Hidaka T, Sekiguchi K, Mochizuki H, Takeuchi Y, Fujiwara T, Watanabe M, Nagaya M, Nagashima H. Production of diabetic offspring using cryopreserved epididymal sperm by in vitro fertilization and intrafallopian insemination techniques in transgenic pigs. J Reprod Dev 2013; 59: 599–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsunari H, Onodera M, Tada N, Mochizuki H, Karasawa S, Haruyama E, Nakayama N, Saito H, Ueno S, Kurome M, Miyawaki A, Nagashima H. Transgenic-cloned pigs systemically expressing red fluorescent protein, Kusabira-Orange. Cloning Stem Cells 2008; 10: 313–323. [DOI] [PubMed] [Google Scholar]
- 31.Arai Y, Ohgane J, Yagi S, Ito R, Iwasaki Y, Saito K, Akutsu K, Takatori S, Ishii R, Hayashi R, Izumi S, Sugino N, Kondo F, Horie M, Nakazawa H, Makino T, Shiota K. Epigenetic assessment of environmental chemicals detected in maternal peripheral and cord blood samples. J Reprod Dev 2011; 57: 507–517. [DOI] [PubMed] [Google Scholar]
- 32.Noé L, Kucherov G. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res 2005; 33: W540-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Judge DP, Dietz HC. Marfans syndrome. Lancet 2005; 366: 1965–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Spoel TI, Jansen of Lorkeers SJ, Agostoni P, van Belle E, Gyöngyösi M, Sluijter JP, Cramer MJ, Doevendans PA, Chamuleau SA. Human relevance of pre-clinical studies in stem cell therapy: systematic review and meta-analysis of large animal models of ischaemic heart disease. Cardiovasc Res 2011; 91: 649–658. [DOI] [PubMed] [Google Scholar]
- 35.Zhao MT, Prather RS. The multi-potentiality of skin-derived stem cells in pigs. Theriogenology 2011; 75: 1372–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsunari H, Nagashima H. Application of genetically modified and cloned pigs in translational research. J Reprod Dev 2009; 55: 225–230. [DOI] [PubMed] [Google Scholar]
- 37.Umeyama K, Watanabe K, Watanabe M, Horiuchi K, Nakano K, Kitashiro M, Matsunari H, Kimura T, Arima Y, Sampetrean O, Nagaya M, Saito M, Saya H, Kosaki K, Nagashima H, Matsumoto M. Generation of heterozygous fibrillin-1 mutant cloned pigs from genome-edited foetal fibroblasts. Sci Rep 2016; 6: 24413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohgane J, Wakayama T, Senda S, Yamazaki Y, Inoue K, Ogura A, Marh J, Tanaka S, Yanagimachi R, Shiota K. The Sall3 locus is an epigenetic hotspot of aberrant DNA methylation associated with placentomegaly of cloned mice. Genes Cells 2004; 9: 253–260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.