Abstract
In photosynthetic organisms the accumulation of harmful photodynamic chlorophyll precursors is prevented because of the tight regulation of the tetrapyrrole pathway. FLU is one of the regulatory factors involved in this process in land plants. We have examined the function of a Flu-like gene (FLP) from Chlamydomonas that gives rise to two FLP transcripts through alternative splicing. These transcripts are translated into a short and a long protein that differ by only 12 amino acids but that interact differently with glutamyl-tRNA reductase, an enzyme involved in an early step of the chlorophyll biosynthetic pathway. Expression of FLPs is light-regulated at the level of RNA accumulation and splicing and is altered by mutations affecting the pathway. The relative levels of the long and short forms of FLP can be correlated with the accumulation of specific porphyrin intermediates, some of which have been implicated in a signaling chain from the chloroplast to the nucleus. Reciprocally, reduction of the FLP proteins by RNA interference leads to the accumulation of several porphyrin intermediates and to photobleaching when cells are transferred from the dark to the light. Thus the FLP proteins act as regulators of chlorophyll synthesis, and their expression is controlled by light and plastid signals.
Keywords: Chlorophyll synthesis, plastid signal, light regulation, alternative splicing, Chlamydomonas
Coordination between the activities of organelles and the nucleus requires the exchange of factors and signals. Genetic analysis of numerous mutants deficient in photosynthesis in Chlamydomonas reinhardtii and land plants revealed a large number of nucleus-encoded factors that are imported into the chloroplast, where they are required for several post-transcriptional steps of plastid gene expression (Barkan and Goldschmidt-Clermont 2000). In recent years it has become clear that these interactions operate bidirectionally from the nucleus to the chloroplast and from the chloroplast to the nucleus. The existence of a plastid signal was originally proposed based on studies of carotenoid-deficient plants that photobleach when they are exposed to high intensities of light (Taylor 1989). Under these conditions transcription of nuclear genes encoding proteins of the photosynthetic apparatus is specifically repressed.
Tetrapyrrole molecules like chlorophylls, heme, and their precursors play an important role in the process of light absorption and energy metabolism in photosynthesis and respiration (Timko 1998; Cornah et al. 2003). Moreover, genetic and physiological studies in Arabidopsis and Chlamydomonas revealed that one of the retrograde signaling pathways from chloroplasts to nucleus is mediated by chlorophyll precursors (Gray 2003). In plants and algae, chlorophyll biosynthesis takes place exclusively in the chloroplast and most of the enzymes involved in this pathway are encoded by nuclear genes. Chlorophyll biosynthesis needs to be tightly controlled in order to optimize the photosynthetic yield in response to metabolic and environmental changes. Moreover, chlorophyll and chlorophyll precursors in free form are highly reactive molecules in the light and can produce reactive oxygen species (Reinbothe et al. 1996a; Timko 1998). In photosynthetic eukaryotes, all tetrapyrroles are synthesized from a common precursor, 5-aminolaevulinic acid (ALA) (see Fig. 6A, below), which is formed from glutamate through the activity of three enzymes (glutamyl-tRNA synthetase, glutamyl-tRNA reductase [GluTR], and glutamate-1-semialdehyde aminotransferase [GSA]). ALA is subsequently converted in several steps to protoporphyrin IX (Proto-IX), through a highly conserved pathway (Timko 1998). Proto-IX is the last common intermediate before the separation of the Fe2+ and the Mg2+ branches that lead to the formation of heme and chlorophyll, respectively. Mg-chelatase is the first committed enzyme in the Mg branch that converts PROTO-IX into Mg-PROTO-IX.
Figure 6.
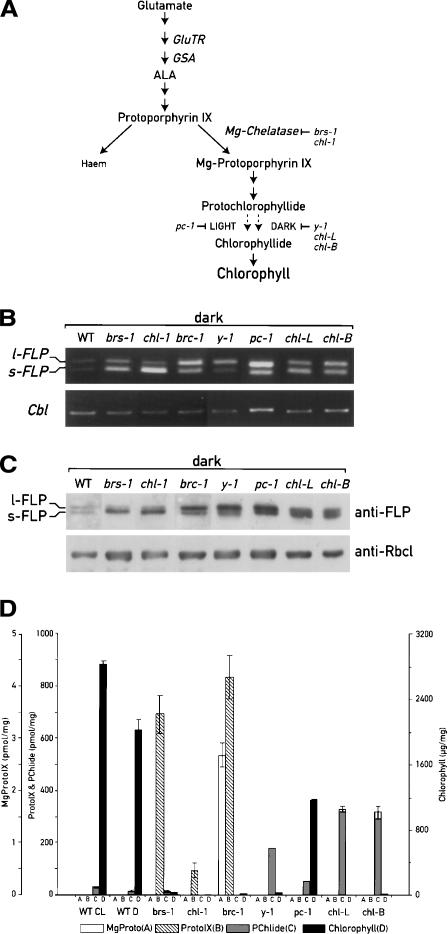
Analysis of expression of FLPs and pigment accumulation in the wild-type strain and chlorophyll biosynthesis mutants. (A) Schematic representation of chlorophyll biosynthesis pathway and the steps blocked in the mutants analyzed. (B) RT–PCR analysis performed with oligonucleotides specific for the FLPs, as indicated in Figure 1, and for the G protein (Cbl) as control. (C) Immunoblot analysis using FLP antiserum and anti-RbcL as control. (D) Quantification of the pigment content by HPLC. All the pigments were normalized relative to the dry weight of cell pellets.
Some chlorophyll precursors were proposed to act as negative regulators of Lhc gene expression and were also shown to increase the expression of the Hsp70 gene that is normally induced by heat stress and light (see Kropat et al. 1997; Gray 2003). Moreover, addition of the precursors Mg-PROTO-IX and its methyl esters to Chlamydomonas cells in the dark can substitute for the light requirement for induction of the nuclear Hsp70 gene, which encodes a chloroplast heat-shock protein (Kropat et al. 1997). Important information about tetrapyrrole signaling derives from the identification and characterization of five gun (genomes uncoupled) mutants of Arabidopsis, which continue to transcribe nuclear genes involved in photosynthesis under conditions of photo-oxidation when they are normally not expressed (see Gray 2003). These mutants are thus deficient in a chloroplast–nuclear signaling pathway. Three of them have defects in enzymes involved in tetrapyrrole synthesis. Of particular interest is gun5, affected in the H subunit of Mg chelatase (Mochizuki et al. 2001). Another mutant, gun4, is deficient in a factor that is required for the regulation of Mg-chelatase activity (Larkin et al. 2003). The analysis of these mutants indicated that accumulation of Mg-ProtoIX is both necessary and sufficient for regulating the expression of many nuclear genes involved in photosynthesis (Strand et al. 2003).
An important step in the Mg2+ branch is the reduction of protochlorophyllide (PChlide) to chlorophyllide (Chlide). In angiosperms this process is catalyzed by the enzyme NADPH:PChlide oxidoreductase (POR) in a light-dependent reaction (Reinbothe et al. 1996b; Timko 1998). Thus in these plants the chlorophyll synthesis pathway is blocked at the level of PChlide in the dark (see Fig. 6A, below). Once a critical level of this intermediate has been reached in the dark, an early step of the pathway is inhibited at the level of ALA synthesis, preventing the accumulation of PChlide, which would cause photobleaching when the plants are illuminated. The FLU protein of Arabidopsis was identified as a key component of this negative feedback loop through its interaction with GluTR (Meskauskiene et al. 2001; Meskauskiene and Apel 2002). Etiolated seedlings of flu mutants accumulate 15-fold higher levels of PChlide than the wild type. After transfer from the dark to the light, the mutant plants rapidly bleach and die. Since the flu seedlings do not overaccumulate heme, FLU appears to regulate specifically the Mg2+ branch of the tetrapyrrole pathway and operates independently of heme. Recently it was shown that the gene affected in the barley tigrina d mutant, which overaccumulates PChlide in the dark, is an ortholog of the Arabidopsis FLU protein (Lee et al. 2003).
Anoxygenic bacteria, cyanobacteria, algae, nonvascular plants, ferns, and gymnosperms are able to convert PChlide to Chlide through a light-independent reaction, and are thus capable to produce chlorophyll in the dark (Timko 1998). Because these organisms are not expected to accumulate PChlide in darkness, our finding of a FLU-like gene (FLP) in Chlamydomonas came as a surprise and raised new questions on the role(s) of this protein in algae and plants. In this study we show that two FLP proteins are present in Chlamydomonas, whereas only one form was detected in angiosperms. These proteins are encoded by a single nuclear gene, generated by alternative splicing, and their expression is controlled both at the level of RNA accumulation and splicing by light and plastid signals. We used RNA interference to show that the FLPs play a major role in the control of chlorophyll biosynthesis.
Results
Two FLP transcripts are generated from a single-copy gene by alternative splicing in Chlamydomonas
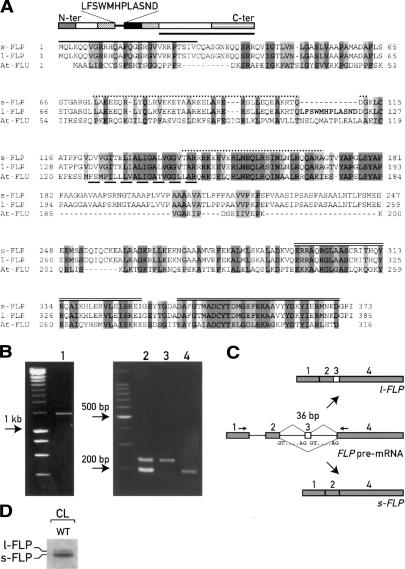
To study the mechanisms involved in the regulation of chlorophyll biosynthesis in C. reinhardtii, we searched the Chlamydomonas EST/ORF database (http://www.biology.duke.edu/cgi-bin/blast.cgi) for proteins related to the Arabidopsis FLU protein (Meskauskiene et al. 2001). One EST with significant sequence similarity to FLU was identified. By screening a Chlamydomonas cDNA library with this EST, we identified two full-length cDNAs of identical nucleotide sequence, except for a short internal 36-nt insertion in one of them. Their ORFs predict proteins of 373 and 385 amino acids that only differ by 12 additional residues in the longer form (Fig. 1). We named them short (s-FLP) and long (l-FLP) FLU-like proteins, respectively. Alignment with the Arabidopsis FLU protein using the proteomic tools for protein structure predictions (http://www.expasy.ch) revealed the presence of several putative functional domains (Fig. 1A): a potential chloroplast transit peptide at its N-terminal end, two coiled-coil motifs (vs. only one in Arabidopsis), a hydrophobic region, and two TPR motifs at the C-terminal end. The extra 12 residues of l-FLP follow the first coiled-coil motif, and precede the hydrophobic region (Fig. 1A).
Figure 1.
The two FLPs are encoded by a single nuclear gene and generated through alternative splicing. (A) Alignment between C. reinhardtii Flu Like Proteins (FLPs) and A. thaliana FLU. The two FLPs differ only by 12 residues (indicated in bold). Identical residues and conservative replacements are highlighted in dark and light gray, respectively. The putative domains predicted by proteomic tools are indicated: chloroplast transit peptide (gray box, solid line); the two coiled-coil domains (striped boxes, dotted lines); hydrophobic domain (black box, dashed line), and the TPR domains (C-ter box, double lines). Gaps (—) were introduced to maximize the alignment. (B) PCR analyses performed with the primer set indicated in C and described in Materials and Methods on genomic DNA (lane 1), cDNA synthetized by RT–PCR from cells grown in continuous light (lane 2), l-FLP cDNA (lane 3), and s-FLP cDNA (lane 4). The unlabeled lanes contain size markers. (D) Immunoblot of total light-grown cells with antiserum raised against the C-terminal part of FLP (indicated with a bar in A).
We first determined that the two transcripts are encoded by the same nuclear gene by PCR analysis using oligonucleotides that anneal to common regions of the two cDNAs flanking the 36 bp specific for the l-FLP (Fig. 1C). PCR reaction on the genomic DNA revealed the presence of a single amplification product (Fig. 1B, lane 1) of 1405 bp. DNA blot analysis confirmed the presence of a single gene in the nuclear genome (data not shown). Furthermore, search in the Chlamydomonas genome database identified a single FLP sequence in scaffold 614 (http://genome.jgi-psf.org/chlre2/chlre2.home.html). Conversely, RT–PCR performed with the same primer set generated two amplification products of the same size as those obtained from PCR reactions on the l-FLP and s-FLP c-DNAs, respectively (Fig. 1B, lanes 2–4). There are canonical GT/AG splicing signals at the ends of the putative introns 2 and 3 (Fig. 1C). Taken together, these data indicate that the l-FLP and s-FLP transcripts are encoded by a single nuclear gene and generated by alternative splicing. Exon 3 is retained in the l-FLP mRNA but is skipped in the mature s-FLP mRNA (Fig. 1C). To estimate the amount of the two FLP proteins, we raised an antiserum against the C-terminal region of FLP that is common to s-FLP and l-FLP (Fig. 1A). Although the difference between the two isoforms is only 12 amino acids (1.5 kDa), it was possible to resolve the two proteins by SDS-PAGE and immunoblotting (Fig. 1D). In constant light the s-FLP is expressed at a much higher level than l-FLP.
Both FLPs are localized in the chloroplast membranes
The presence of a putative transit peptide and the significant sequence identity of FLP to the chloroplast FLU protein of Arabidopsis suggested that the FLPs are also located in the chloroplast. To test this prediction, fractions from total cells and purified chloroplasts of Chlamydomonas were prepared and used for immunoblotting with antiserum raised against FLP protein (Fig. 2). In order to obtain a high yield of purified chloroplasts, cells were grown under continuous light. However, under these conditions only the s-FLP was clearly detected. This isoform was found in the membrane fraction of purified chloroplasts, as expected from the presence of a hydrophobic transmembrane domain. To confirm the purity of the fractions, the same protein blot was incubated with antisera against cytosolic thioredoxin h, the chloroplast PsaA membrane protein, and the soluble chloroplast Rubisco. To investigate the localization of both isoforms, we produced independent transformed cells expressing from a heterologous promoter either of the two FLP cDNAs fused at the C-terminal ends with a triple HA tag. The two isoforms fractionated the same way as the endogenous s-FLP (Fig. 2).
Figure 2.
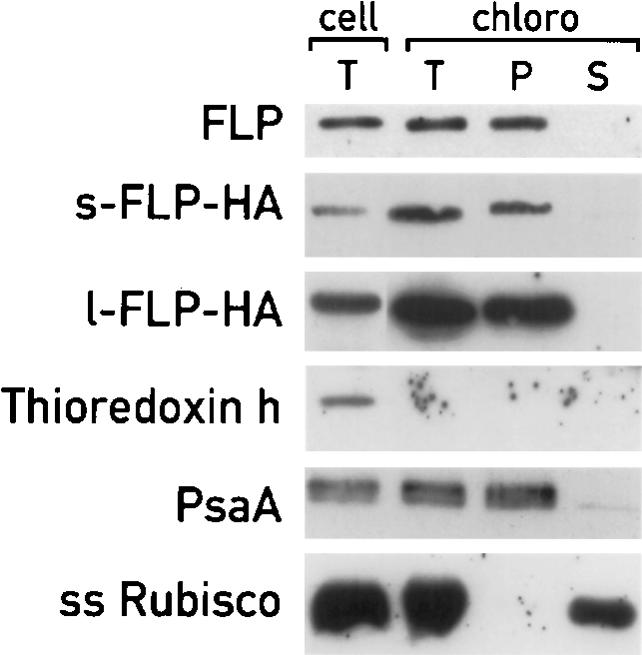
Localization of FLP proteins in the chloroplast compartments. Immunoblot analysis of proteins extracted from total cells (cell, T) and from purified chloroplasts (T, intact purified chloroplasts; P, insoluble fractions; S, soluble fractions). The cellular fractionations of s-FLP-HA and l-FLP-HA were performed on different cells. Although only the control immunoblots corresponding to s-FLP-HA are shown, the same results were obtained with the cells containing l-FLP-HA. Filters were incubated with antisera against FLP, HA, thioredoxin h, PsaA, and Rubisco to reveal the proteins indicated in the figure.
The FLPs can partially complement At-flu deficiency
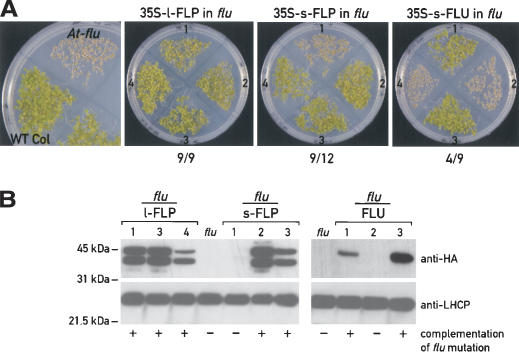
Because the FLP proteins were identified based on their similarity with the Arabidopsis FLU protein, the question arises whether Chlamydomonas FLPs are able to complement the Arabidopsis flu mutation. We therefore cloned the l-FLP, the s-FLP, and the Arabidopsis FLU cDNAs (positive control) in a binary vector, under the control of the 35S promoter. Seeds from independent primary transformants were tested for complementation. They were germinated and grown under 12 h light/12 h dark cycles for 7 d. Under these conditions flu mutant seedlings bleach and die, whereas wild type-seedlings deetiolate and grow. The results in Figure 3A show that most of the transgenic lines containing the Chlamydomonas s-FLP and the l-FLP cDNA sequences were able to complement the Arabidopsis mutation.
Figure 3.
(A) Complementation assays of Arabidopsis flu mutant, transformed with l-FLP, s-FLP, and Ara FLU cDNAs. Seeds from independent primary transformants were germinated under light/dark cycles for 7 d. Four representative transformants for each construct are shown. The ratio below the figures indicates the number of the transformants that complement the phenotype versus the total number of transformants tested. (B) Immunoblot analysis of proteins from some of the plants shown in A, using anti-HA and anti-LHCP antibodies. Seedlings were grown under continuous light for 4 d for protein extraction.
We tested the presence of the FLU and FLP proteins in the Arabidopsis transgenic plants by immunoblotting (Fig. 3B), taking advantage of the HA tag added to the C-terminal end of the FLPs. As shown in Figure 3B, the Chlamydomonas proteins were successfully expressed in all the transgenic lines able to complement the flu mutations. Two bands were detected in the transgenic lines containing the l-FLP or the s-FLP that probably result from the partial processing of the Chlamydomonas precursor proteins in Arabidopsis chloroplasts. Based on their sizes, the upper band could correspond to the precursor FLP protein with the transit peptide, and the short one to the processed protein. In the case of transgenic lines containing the Arabidopsis FLU protein, only one band was detected. As control, all protein extracts from independent transgenic lines were also tested with antiserum against LHC proteins.
The FLP proteins were not able to complement the flu mutation when the seedlings were grown for 4 d in the dark and then transferred to continuous light for 3 d, suggesting that the Chlamydomonas proteins only partially complement the Arabidopsis flu mutation (data not shown).
The 12 supplementary amino acids of l-FLP are important for its interaction with Glu-tRNA reductase in vitro
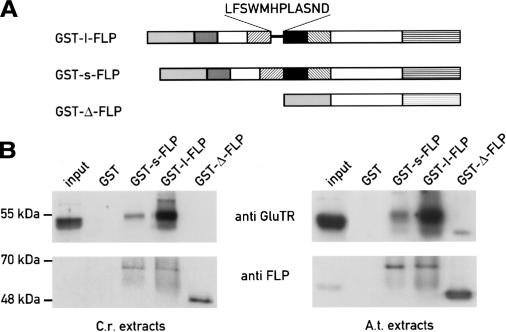
As shown above, the expression of the two FLP isoforms changes in the different mutants analyzed and during dark–light shifts. We tested whether these proteins, which differ by only 12 amino acids, could interact differently with GluTR, a potential target of FLP based on the results obtained with FLU (Meskauskiene and Apel 2002). We expressed the l-FLP, the s-FLP, and a version of FLP with an N-terminal deletion (Δ-FLP) as glutathione S-transferase (GST) fusion proteins in Escherichia coli (Fig. 4A). The soluble fraction from Chlamydomonas protein extracts was incubated with the same amount of the different purified GST fusion proteins immobilized on Glutathione Sepharose. After several washes, the bound fractions were analyzed by immunoblotting (Fig. 4B, left panel). Using anti-GluTR antibody, we observed that the long form interacts strongly with GluTR in this in vitro assay, as already shown for the Arabidopsis FLU protein. In contrast, GST-s-FLP interacted only weakly, suggesting that the 12 amino acids specific of the long form may favor the interaction with GluTR. No interaction was observed with the Δ-FLP, indicating that the N-terminal part of the protein containing the two coiled-coil domains is necessary to mediate this interaction. It is likely that the TPR domains are also required for this interaction based on the results obtained with Arabidopsis (Meskauskiene and Apel 2002). FLP antiserum was also used to confirm the presence of the GST–FLP fusion proteins in the different assays.
Figure 4.
GST-pull-down assays to test the interactions of FLPs with the glutamyl-tRNA reductase (Glu-tRNA). (A) Different purified GST fusion proteins (l-FLP, s-FLP, and an N-terminal deletion construct of FLP [Δ-FLP]) were incubated with Chlamydomonas (C.r.) and Arabidopsis (A.t.) protein extracts. (B) The interactions were detected by immunoblotting of the bound fraction using anti-Glu-tRNA antibody and FLP antiserum, as control.
Since both Chlamydomonas proteins were able to complement the Arabidopsis flu mutation, we performed the same GST-pull-down experiments described above using Arabidopsis protein extracts. As shown in Figure 4B (right panel), both FLPs interact with the Arabidopsis GluTR encoded by HemA1. As in Chlamydomonas, the interaction is stronger with l-FLP. However, the faint interaction observed in vitro between the Arabidopsis GluTR and s-FLP appears to be sufficient to allow for partial complementation of the At-flu mutation (Figs. 3, 4).
FLP RNA levels and splicing are regulated by light
Previous studies in Chlamydomonas showed that the early steps of chlorophyll biosynthesis are regulated by light (Matters and Beale 1995; Im et al. 1996). Since FLPs are putative regulators of this pathway, we first examined whether the expression of their genes is also modulated by light. Wild-type cells were dark-adapted for 2.5 d and then shifted to continuous white light (70–100 μmol m–2 sec–1) for 1, 3, 5, and 8 h (Fig. 5A). FLP gene expression was analyzed by semiquantitative RT–PCR because the levels of the FLP mRNAs were too low to be detected by RNA blotting. The results in Figure 5A indicate that the two forms are poorly expressed in dark-adapted cells. After a shift from dark to light, the two FLP transcripts are both induced, already after 1 h of illumination. However, the relative levels of the two transcripts change after the dark-to-light transition. The l-FLP transcript reaches a maximum after 3 h in the light and subsequently decreases. Expression of the s-FLP transcript increases more than that of l-FLP, is maximal after 3 h, and remains at a very high level throughout the time course examined. After 5 h the short form is expressed five times more than the long form as quantified by RNase protection analysis (Supplementary Fig. 1). FLP gene expression is specifically induced by light because no increase of the Cbl transcript, coding for a G protein, was observed under these conditions (Fig. 5A).
Figure 5.
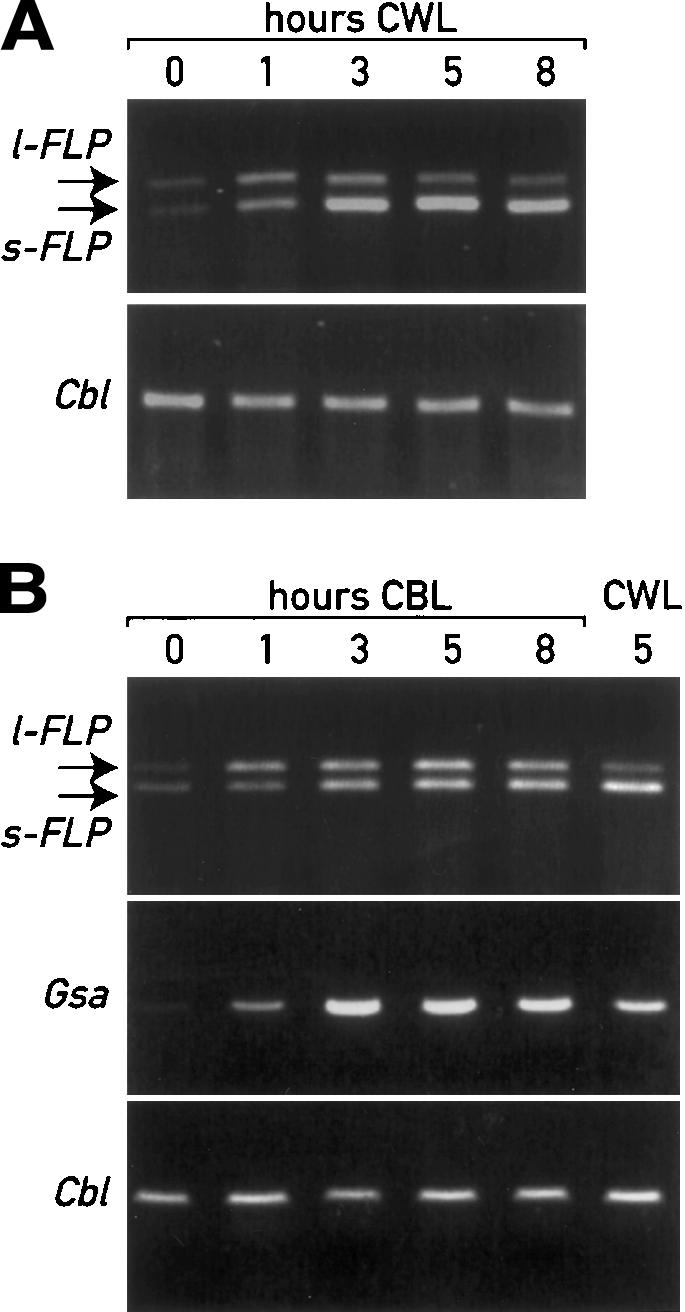
Analysis by RT–PCR of FLP transcription and splicing upon light induction. The analysis was performed with the same primer set indicated in Figure 1C on wild-type cells grown in the dark for 2.5 d and then transferred to continuous white light (CWL) (A), or blue light (CBL) (B). RT–PCR of transcripts of the G protein (Cbl) and of GSA mRNAs were used as control.
The nuclear genes GSA and ALAD, encoding enzymes of early steps of the porphyrin pathway, were previously shown to be induced by blue light through a signal transduction chain involving blue-light photoreceptors (Matters and Beale 1995; Im et al. 1996). We therefore tested whether blue-light signaling could also play a role in the activation of FLP. We repeated the experiment described above in Figure 5A with blue light. The results in Figure 5B clearly indicate that FLP transcription was induced under this condition. However, in this case there was no significant difference in the relative levels of the two transcripts. The concomitant induction of the GSA gene confirmed the specificity of the blue-light response.
To test whether light regulation was mediated through photosynthetic electron flow, dark-adapted cells were treated with DCMU or DBMIB for 5 min before the light treatment. These drugs are known to block reduction and oxidation of the plastoquinone pool, respectively (Trebst 1980). Neither treatment had any effect on the light-induction of FLP gene expression (data not shown) indicating that this process is independent of photosynthetic electron flow and of the redox state of the plastoquinone pool.
FLPs are overexpressed in different chlorophyll biosynthesis mutants
Because of the possible link between FLPs and the tetrapyrrole pathway, we tested whether porphyrin intermediates are involved in the regulation of FLP expression. The level of the FLP transcripts was determined by semi-quantitative RT–PCR, RNase protection assays, and immunoblotting in several Chlamydomonas mutants affected in different steps of the chlorophyll biosynthesis pathway (Fig. 6; Supplementary Fig. 1). To examine whether these mutants accumulate specific chlorophyll precursors and if these molecules might have a direct role in the regulation of FLP gene expression, that is, whether they might act as plastid signals, we quantified the content of PROTO-IX, Mg-PROTO-IX, PChlide, and chlorophyll by HPLC in the same cultures. We analyzed dark-adapted cells because most of these mutants accumulate large amounts of photodynamic tetrapyrrole intermediates and die in the light.
As shown in Figure 6B, the FLPs were overexpressed in all the mutants analyzed as compared to the wild-type cells. Interestingly, there were variations both in the levels of the FLP transcripts and in the ratios between the long and the short forms (Fig. 6B). Immunoblot analysis of protein extracts from the different mutants clearly indicated that the relative amounts of the two FLP proteins follow the levels of the corresponding mRNAs (Fig. 6C).
The highest s-FLP/l-FLP ratio was observed in the allelic mutants brs-1 and chl-1 affected in the H subunit of Mg-chelatase (Chekounova et al. 2001). These mutants accumulate high levels of PROTO-IX (ranging from 100 to 700 pmol/mg dry weight), the last common precursor before the separation of the heme and the chlorophyll branches. Chlorophylls and other precursors are almost undetectable (Fig. 6D). In contrast, a low s-FLP/l-FLP ratio is observed in the brc-1 mutant, which overaccumulates Mg-PROTO-IX (2.7 pmol/mg) in addition to PROTO-IX (835 pmol/mg). Several of the mutants that are affected further downstream in the Mg branch of the pathway accumulate the long form of FLP and/or have lower s-FLP/l-FLP ratios. Thus, the pc-1 (Li and Timko 1996) and y-1 mutants that are unable to reduce PChlide to Chlide in the light and the dark, respectively, accumulate PChlide four and 12 times more than the wild type (Fig. 6D; Supplementary Table 1). Overexpression of both FLP transcripts and proteins occurs in the chloroplast mutants chl-L and chl-B, which are also deficient in light-independent PChlide reduction (Li et al. 1993; Cahoon and Timko 2000). These two mutants accumulate 23 times more PChlide than the wild type. In contrast to these mutants, wild-type cells accumulate large amounts of chlorophyll both in the light and the dark and a small amount of Pchlide (28 pmol/mg in the light and 14 pmol/mg in the dark). The other chlorophyll precursors are undetectable (Fig. 6D).
The ratio between the two FLPs changes during de-etiolation in the y-1 mutant
Chlamydomonas is able to synthesize chlorophyll in the dark because it has a light-independent enzyme for the reduction of protochlorophyllide to chlorophyllide (Timko 1998). Several nuclear loci (y-1 to y-10) and three chloroplast genes (chlL, chlN, and chlB) are required for this process. Cells with mutations in any of these genes fail to accumulate chlorophyll in the dark, but are still able to produce chlorophyll in the light. Among these yellow-in-the-dark mutants, y-1 was the first to be isolated and has been extensively characterized (Malnoe et al. 1988).
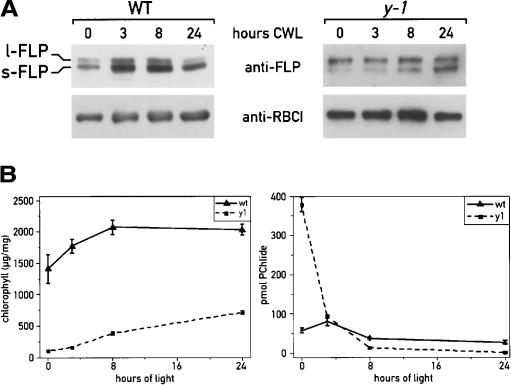
We analyzed FLP expression in wild-type and y-1 mutant cells grown in the dark for 3 d and then shifted to light for 24 h (Fig. 7A). Simultaneously, we also measured the content of chlorophyll and PChlide, the only pigments detectable in these strains (Fig. 6D). After the dark treatment, the y-1 mutant cells were yellow with very low levels of chlorophyll and high levels of PChlide (Fig. 7B). Chlorophyll synthesis resumed upon transfer to the light, and chlorophyll levels increased steadily with time. After 24 h the chlorophyll content was similar to that measured before the dark treatment (715 μg/mg), and PChlide content decreased to a very low level. As expected, wild-type cells remained green in the dark. However, the amount of chlorophyll was lower in dark-adapted wild-type cells and increased in the following light period (Fig. 7B). The PChlide content increased slightly after the dark-to-light transition, with a peak at 3 h (40% more than in the dark) and slowly decreased at the subsequent time points. The levels of both FLP proteins increased after the shift from dark to light with the short form constantly more abundant than the long form (Fig. 7A), as already shown for their transcripts by RT–PCR (Fig. 5A). The highest expression of the long form could be specifically correlated with the peak of PChlide detected at 3 h (Fig. 7A,B). After 24 h of light treatment, the l-FLP level was very low, as for cells grown in continuous light (before the dark treatment). In the y-1 mutant the level of l-FLP was already high in the dark and remained high in the subsequent light period and decreased after 8 h. In contrast, the level of s-FLP was low in the dark and increased gradually during the light period (Fig. 7A,B). Thus the increase of the s-FLP/l-FLP ratio correlates with the decline of PChlide. These results support the role of chlorophyll precursors in the control of FLP expression at the level of RNA accumulation and splicing.
Figure 7.
Analysis of FLP isoforms in wild-type and y-1 mutant cells grown in the dark and after a dark-to-light transition (CWL, continuous white light). The level of FLP proteins was determined by immunoblot analysis (A) and the content of PChlide and chlorophylls extracted from the cells at different time points was estimated by HPLC (B).
Chlamydomonas strains with reduced FLP levels accumulate chlorophyll precursors and do not survive a dark-to-light transition
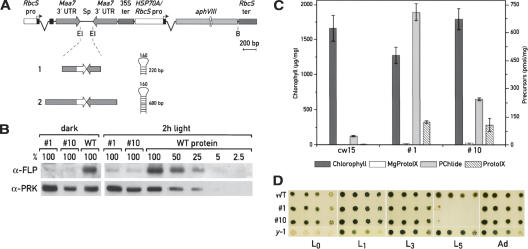
We applied the RNAi technology to generate strains with a reduced content of both FLP proteins. Because of the difficulty in generating stable RNAi strains of Chlamydomonas, we used the recently developed cosilencing system (Rohr et al. 2004) to select directly for cells in which RNAi is occurring. For this approach, the FLP fragments indicated in Figure 8A were cloned in sense and antisense orientation between the tryptophan synthase β subunit (TSB) inverted repeat. The TSB, encoded by the Maa7 gene, converts the indole analog 5-fluoroindole (5-FI) into the toxic tryptophan analog 5-fluorotryptophan. Transformed cells with silenced Maa7 were selected based on their ability to grow in the presence of 5-FI. We analyzed the FLP content in cells grown in the dark and in cells exposed to the light for 2 h. To detect the FLP proteins in the wild-type strains in the dark, the cells were kept in the dark for 12 h and larger amounts of proteins were used for immunoblot analysis. The two independent RNAi transformants (clones 1 and 10) accumulate reduced levels of FLP proteins relative to wild type (Fig. 8B). These clones contain FLP inverted repeats of different size (220 and 480 bp for clone 1 and 10, respectively). We did not observe any significant difference in the level of silencing between the two clones. The amount of FLP content in the RNAi strains grown in the dark was <5% of the wild-type level, and after the dark-to-light transition between 5% and 20%, indicating a residual level of light induction in these strains. HPLC analysis revealed that these RNAi strains accumulate high levels of PChlide, PROTO-IX, and Mg-PROTO-IX (Fig. 8C).
Figure 8.
Reduction of FLP levels by RNAi. (A) Schematic representation of the constructs used for the transformation. The FLP fragments were cloned in sense and antisense orientation between the Maa7 inverted repeat. The different FLP cDNA fragments used to generate the constructs 1 and 2, and the predicted structure of the resulting hairpin RNAs are indicated. (B) Analysis of FLP protein levels by immunoblotting in two independent RNAi strains (clones 1 and 10), generated with constructs 1 and 2, respectively. The FLP content was analyzed in cells grown in the dark for 12 h or exposed to the light for 2 h. Twenty micrograms of protein from each extract was used, and the level of FLPs was quantified using a serial dilution of proteins from wild-type cells as standard. Membranes were incubated with FLP antiserum and PRK (phosphoribulose kinase) antibody as control. (C) Quantification of pigments of wild-type cells and RNAi strains 1 and 10 grown in the dark for 3 d by HPLC. (D) Growth test to assay the light sensitivity of the RNAi strains. Strains 1 and 10 were grown for 3 d in the dark, spotted on TAP plates with serial dilutions (1:5), and grown under different light conditions for 1 wk (L0, dark; L1, 7.5 μmol m–2 sec–1; L3, 90 μmol m–2 sec–1; L5, 600 μmol m–2 sec–1) or progressively exposed to increased light intensity with five adaptation steps of 2 d each (Ad:L1,7.5 μmol m–2 sec–1; L2, 37 μmol m–2 sec–1; L3, 90 μmol m–2 sec–1; L4, 200 μmol m–2 sec–1; and L5, 600 μmol m–2 sec–1h). Cells from wild-type and y-1 mutant strain were subjected to the same treatments.
Since Arabidopsis flu mutants do not survive a dark-to-light transition because of the accumulation of PChlide, but grow under continuous light, we tested the sensitivity of the Chlamydomonas RNAi strains to different light treatments. Wild-type strains and the RNAi clones 1 and 10, adapted in the dark for 3 d, were spotted on TAP plates and grown for 1 wk under different light intensities, between L0 (dark) and L5 (600 μmol m–2 sec–1). As shown in Figure 8D, cells with reduced amounts of FLPs grew in the dark and dim light as well as the wild-type strain, but died after the shift to strong light. However, when these clones were progressively adapted to higher light intensities (with four steps of 2 d each), they were able to grow even under the strongest light. Interestingly, the y-1 mutant, which also accumulates high levels of PChlide in the dark (Fig. 8D; Supplementary Table 1) but overexpresses l-FLP, grew after the dark-to-light transition under all conditions tested.
Discussion
An important step in chlorophyll synthesis is the conversion of PChlide into Chlide, which is a light-dependent reaction in angiosperms. Overaccumulation of PChlide in the dark is prevented by inhibition of an early step of chlorophyll synthesis through the FLU protein. Chlamydomonas cells do not face the same problem because these organisms are able to convert PChlide to Chlide both in a light-dependent and light-independent reaction. Thus PChlide is not expected to accumulate in dark-grown algal cells. However, it is not clear how these organisms control the level of the tetrapyrrole intermediates. The finding of a Chlamydomonas FLU-like gene was therefore intriguing and raised questions about its function. In this study we have shown that the FLP gene gives rise to two FLP proteins through alternative splicing and that these proteins act as regulators of the chlorophyll synthesis pathway and their expression is regulated by light and chloroplast signals.
Two FLP proteins in Chlamydomonas generated by alternative splicing
Our results demonstrate that the two forms of FLP are produced from a single nuclear gene by alternative splicing. The 36-bp exon 3 is skipped to produce the mature s-FLP mRNA and retained in the l-FLP mRNA. Alternative splicing plays a significant role in the generation of proteome diversity in plants (Kazan 2003). A few trans-acting factors involved in alternative splicing were isolated based on the similarity with the animal proteins, and one cis-acting element was recently identified in the gene encoding chloroplast-specific ascorbate peroxidase (chlAPX) (Yoshimura et al. 2002). Most of the genes identified in plants that undergo alternative splicing encode proteins with regulatory functions or are associated with various stress responses (Kazan 2003). In algae few alternatively spliced genes have been identified. Among the few examples, the isoforms of the nucleotide exchange factor GrpE differ only by two amino acid residues (Schroda et al. 2001). The two forms, differentially expressed under stress condition (e.g., heat shock) derive from the alternative use of two 5′-splice sites in the same pre-mRNA. Further computational analysis of genomes and ESTs in these groups of organisms is likely to reveal more cases of alternative splicing.
We characterized both FLP proteins from Chlamydomonas and showed that they are associated with the thylakoid membrane and absent from the plastid envelope. The FLP proteins share several features with FLU of Arabidopsis: a coiled-coil domain, a transmembrane span, and two TPR motifs. However, the Chlamydomonas FLPs contain an additional coiled-coil domain up-stream of the 12-amino acid insertion. Our GST-pull-down assays show that, in addition to the TPR domains, the two coiled-coil domains may be important for the interaction of FLPs with GluTR (Fig. 4). Indeed, an N-terminal deleted FLP version containing only the TPR domains does not interact with GluTR. In contrast, the interaction between FLU and GluTR of Arabidopsis is mediated principally through the TPR domains (Meskauskiene and Apel 2002), indicating that FLU and FLP differ in this respect. The alternatively spliced exon 3 specific for the l-FLP also affects protein–protein interaction since the GluTR of both Arabidopsis (HemA1) and Chlamydomonas interact more effectively with l-FLP than with s-FLP.
Multiple signals control the content of the two FLP isoforms
Our results reveal complex regulatory mechanisms at the level of RNA accumulation and splicing that control the levels of the two FLP proteins in response to different signals. Light regulates chlorophyll biosynthesis in plants and algae (Timko 1998). In plants, light stimulates the formation of the ALA precursor, through phytochrome- and cryptochrome-mediated induction of the HEMA1 gene, as well as through other post-transcriptional mechanisms (McCormac and Terry 2002). In Chlamydomonas, the expression of the GSA1 and ALAD genes, encoding GSA and δ-aminolevulinic acid dehydratase, respectively, is highly regulated by light in light:dark-synchronized cells through a blue-light photoreceptor (Matters and Beale 1995).
Analysis of the FLP mRNAs and proteins shows that the two isoforms are differentially expressed under different light conditions. In dark-adapted cells both isoforms are weakly expressed. Upon transfer to the light the l-FLP is transiently induced and subsequently declines, whereas the s-FLP increases steadily and represents the major form in continuous light (Figs. 5, 7). The observed light response is independent of photosynthetic electron flow and could be activated by a signal transduction process through specific photoreceptors. Our analysis (Fig. 5B) shows that blue light is sufficient to enhance FLP transcript levels, and thus a blue-light photoreceptor could be involved in this response. Blue-light receptors like cryptochrome and phototropin have been identified in Chlamydomonas (Huang and Beck 2003). Under blue-light illumination the two isoforms are equally expressed in contrast to cells exposed to white light. Thus other photoreceptors besides the blue-light receptor are involved in the control of splicing of FLP. FLP represents the first example of light-regulated alternative splicing in Chlamydomonas, and only few other examples have previously been observed in plants such as hydroxypyruvate reductase in pumpkin (Mano et al. 1999).
The content of the two FLP isoforms is also regulated by the functional state of the chloroplast, through tetrapyrrole-mediated signals. Previously, studies of the HSP70 gene in Chlamydomonas and characterization of several gun mutants in Arabidopsis led to the conclusion that Mg-PROTO-IX and its derivatives act as key signaling molecules for the regulation of nuclear genes (Gray 2003). In Chlamydomonas Mg-PROTO-IX and its methyl esters can replace the light requirement for the induction of the Hsp70 gene, whereas PROTO-IX, PChlide, and Chlide are unable to do so (Kropat et al. 1997, 2000). Our work confirms the role of tetrapyrroles in the regulation of nuclear gene expression. Indeed, overexpression of FLPs was observed in all the chlorophyll biosynthesis mutants analyzed that overaccumulate different amounts of chlorophyll precursors in the chloroplast. In addition, quantification of PROTO-IX, Mg-PROTO-IX, and PChlide by HPLC (Fig. 6D; Supplementary Table 2) indicates a correlation between the relative expression of the FLP isoforms and the amount of different chlorophyll precursors, suggesting that these molecules act as chloroplast signals that regulate the level of the FLPs. The highest ratio between s-FLP and l-FLP was found in the allelic mutants brs-1 and chl-1, which are affected in the subunit H of Mg-chelatase (Chekounova et al. 2001) and accumulate exclusively PROTO-IX (Fig. 6D; Supplementary Table 2). In particular, in the chl-1 mutant s-FLP is much more abundant than l-FLP, as for cells grown in continuous light. The s-FLP/l-FLP ratio is considerably lower in the other mutants analyzed such as brc-1, which in addition to PROTO-IX also accumulates high levels of Mg-Proto-IX. Similar patterns are observed in mutants affected in the reduction of PChlide such as pc-1 and y-1.
As angiosperms, the y-1 mutant is unable to reduce PChlide to Chlide and to develop a functional chloroplast in the dark, but does so in the light (Malnoe et al. 1988). The y-1 mutant is of particular interest because of a specific accumulation of the l-FLP in the dark. A clear correlation was found during the greening after a shift from dark to light between the increase in the sFLP/l-FLP ratio and the decrease of PChlide, suggesting that a signal originating from the plastid influences the mode of alternative splicing of FLP. However, the chl-L and chl-B mutants, which are deficient in the same step as y-1 and which accumulate even higher levels of PChlide, accumulate both FLP forms, indicating that the relationship between the level of chlorophyll precursors and FLP gene expression is influenced by other factors. Our results clearly indicate that deregulation of chlorophyll biosynthesis, and the consequent accumulation of different intermediates, affect the absolute and relative levels of the two FLP isoforms.
How plastid signals are integrated with a light signal is not known. Analysis of promoter regions of genes regulated by both light and plastid signals in Arabidopsis revealed the presence of plastid-responsive elements in nuclear genes. In some cases the light and plastid signals appear to act on the same cis elements. G-box-like elements respond to the Mg-PROTO-IX-mediated signal in the Lhcb1 promoter (Strand et al. 2003). We identified similar motifs in the FLP promoter (data not shown), using the PlantCARE database (http://intra.psb.ugent.be:8080/PlantCARE).
FLPs are involved in the regulation of chlorophyll biosynthesis in Chlamydomonas
A decrease of FLP levels through RNA interference revealed that dark-grown cells overaccumulate PChlide, PROTO-IX, and Mg-PROTO-IX and do not survive when they are transferred to high light most likely because of photo-oxidative damage caused by these tetrapyrrole intermediates. In contrast, cells that are progressively adapted from darkness to high light survive (Fig. 8D). Thus although Chlamydomonas cells are able to convert PChlide to Chlide in the dark, FLP is required for preventing the overproduction of PChlide and the deregulation of the porphyrin pathway. In this respect, it is noteworthy that anaerobic photosynthetic bacteria contain only the light-independent POR enzyme that is structurally related to the oxygen-sensitive multisubunit eubacterial nitrogenase (Armstrong 1998). It is possible that this enzyme represents an ancestral form that was later replaced by the more efficient light-dependent POR in angiosperms. Whereas angiosperms lost the light-independent POR, this enzyme was conserved in other photosynthetic organisms including Chlamydomonas. An interesting possibility is that FLU/FLP may have evolved during the transition from light-independent to light-dependent POR. Possible cyanobacterial ancestors of FLP are several TPR proteins of unknown function from Nostoc that show significant sequence identity with the C-terminal end of FLP (data not shown).
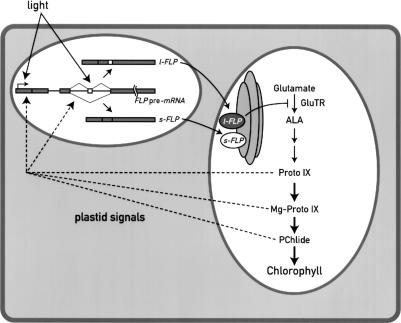
These results, together with the observation that the FLPs are overexpressed in different mutants affected in the porphyrin pathway, and the similarity of these proteins with Arabidopsis FLU, indicate that the two isoforms are involved in the regulation of chlorophyll biosynthesis in Chlamydomonas. Of particular interest is the observation that the y-1 mutant, which accumulates high levels of PChlide and l-FLP in the dark, survives the dark–light transition. Taken together, these data indicate that the FLP proteins are not only involved in the regulation of the chlorophyll biosynthesis pathway, but that they may also have a protective role against photo-oxidative damage and act as stress proteins. Thus the FLP proteins participate in a complex regulatory network between chloroplast and nucleus (Fig. 9). Their expression is regulated at the level of RNA accumulation, most likely transcription, and splicing by light and plastid signals. The presence of two FLP proteins in Chlamydomonas is unique. Because their relative levels can be correlated with specific changes in the accumulation of tetrapyrrole intermediates, it is possible that the ratio between these two forms is important for regulating the flux through the chlorophyll biosynthetic pathway.
Figure 9.
Scheme of the regulatory network involving FLPs in Chlamydomonas. FLPs act as regulators of the chlorophyll biosynthesis pathway and the expression of the two isoforms is tightly regulated by external signals such as light and by the functional state of the chloroplast. We propose that the amount and the ratio of the two FLP proteins is important for the proper functioning of the chlorophyll biosynthesis pathway. FLPs may also have a protective role against photo-oxidative damage.
Materials and methods
Chlamydomonas strains and growth conditions
All C. reinhardtii strains were grown in TAP medium as described (Harris 1989). Wild type and y-1 mutant were grown in constant white light (70–100 μmol m–2 sec–1) or in the dark. Wild-type dark-adapted cells were also exposed to blue light of 20 μmol m–2 sec–1. Analyses of brs-1, chl-1, brc-1, pc-1, chl-L, and chl-B mutants (Harris 1989) were performed only on cells grown in the dark. Cells in exponential growth phase (2 × 106 cells/mL) were used in all the experiments described. Dark-grown cells were handled under dim green light.
Isolation and cloning of FLP cDNAs
A DNA fragment from an EST sequence with significant sequence similarity to an internal fragment of Arabidopsis FLU was used to screen a Chlamydomonas cDNA library. Five cDNAs of ∼2 kb were obtained by screening 400,000 phages corresponding to two different cDNAs (l-FLP and s-FLP). Their sequences have been submitted to the DDBJ/EMBL/GenBank database, under accession numbers AY705910 and AY705911, respectively. The genomic sequence of the single nuclear FLP gene is available in the Chlamydomonas genome database in scaffold 614 (http://genome.jgi-psf.org/chlre1/chlre1.home.html).
The pSL18 plasmid (S. Lemaire and J.D Rochaix, unpubl.), containing both psaD 5′-and3′-UTR flanking the multicloning site and the AphVIII gene conferring paramomycin resistance, was used to clone FLP cDNAs. A triple HA tag was introduced just upstream of the stop codon, to produce FLP::HA. Nuclear transformation of Chlamydomonas cell wall-deficient cells (cw15) was performed as described (Stevens et al. 1996).
Constructs for the RNAi were generated by introducing FLP inverted repeats into the Maa7/X IR vector (Rohr et al. 2004). Two different FLP IR cassettes were generated: the first one (1, Fig. 8A) by PCR amplification of two fragments from the l-FLP cDNA with the oligoIR.1 (5′-CGGAATTCTCAACACGCTA TTCAGCATGG-3′) and oligoIR.2 (5′-GCTCTAGATGCGCT CAATGTACTTGTC-3′), which generate a product of 384 bp (from nucleotide 751 to 1135); and with oligoIR.1 and oligo oligoIR.3 (5′-GCTCTAGATACTGGTGCGTGATGCGGCA-3′), which generate a fragment of 223 bp (from nucleotide 751 to 974). These PCR products were cloned in sense and antisense orientation by digestion with EcoRI and XbaI (sites introduced by PCR), and ligation into the EcoRI site of Maa7/X IR vector. The second FLP IR (2, Fig. 8A) cassette was generated with the same strategy using the following set of primers: oligoIR.4 (5′-CGGAATTCAACGAGCAGCTGCGCTCCATC-3) and oligoIR.2, which produce a fragment of 642 bp (from position 493 to 1135); and oligoIR.4 and oligoIR.3, which generate a fragment of 481 bp (from position 493 to 974). The final vectors, Maa7/FLP1 IR and Maa7/FLP2 IR, were introduced in a CW15 strain by transformation. Transformants were selected on plates containing 1.5 mM l-tryptophan, 10 μg/mL paromomycin, and 5 μM 5-FI, as described (Rohr et al. 2004), in the dark. Isolated RNAi strains were maintained under constant selection with an increased amount of 5-FI (15 μM).
Semiquantitative RT–PCR analysis
Total RNA was extracted from Chlamydomonas according to Merendino et al. (2003) and treated with DNase I (Invitrogen). Reverse transcription was performed at 50°C using 100 units of SuperScript II RT (Invitrogen). Reactions without RT were performed to exclude contamination with genomic DNA. Long and short FLP cDNAs were PCR-amplified with a single primer set (5′-ATGGCCGACGCGTTCCTTTCG-3′ and 5′-AAATGGCG TGGCGCACAGCTT-3′) that anneals to common regions of the two cDNAs. RT–PCR was also performed on Cbl mRNA, as an external control, by using the primers 5′-GACGTCATC CACTGCCTGTG-3′ and 5′-CGACGCATCCTCAACACACC -3′. PCR products were resolved on 2.5% agarose gels.
Antiserum production
For antiserum production the 3′-end portion of the L-FLP cDNA was used (from amino acid 189 to the Stop codon), which is also conserved in the s-FLP. This fragment was cloned in the pGEX-4T-1 expression vector (Amersham Pharmacia Biotech) and introduced into E. coli BL21. The GST fusion protein expression was induced for 16 h at 16°C by adding 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). The 48-kDa protein was further purified on Glutathione Sepharose 4B (Amersham Pharmacia Biotech) and eluted by thrombin protease cleavage. The recombinant protein was purified by SDS-PAGE (15%), resuspended in the extraction buffer (15 mM ammonium carbonate, 0.025% SDS, 1 mM DTT, 0.1 mM PMSF), and injected in a rabbit five times at 3-wk intervals.
Protein extracts and immunoblot analysis
Total cell and chloroplast extracts, chloroplast membrane, and soluble fractions were prepared as described (Merendino et al. 2003). For immunoblot analysis, proteins were fractionated on 10% or 12% SDS-PAGE and immunoblotted with the specific antiserum. The antisera used in this work were α-HA (1:1000, Eurogentec), α-ss (small subunit) Rubisco (1:10,000), α-PsaA (1: 5000), α-thioredoxin-h (1:10,000, gift of S. Lemaire, Institut des Biotechnologies des Plantes, Orsay, France), α-IM30 (1:10,000, gift of M. Spalding, University of Iowa, Ames, Iowa), α-D1 (1: 10,000), α-glutamyl tRNA reductase (1:5000, gift of S.I. Beale, Brown University, Providence, RI), and pyruvate kinase (1:10,000). The ECL immunoblotting system (Durrant 1990) was used to detect immunoreactive proteins with anti-rabbit or anti-mouse secondary antibodies.
GST-pull-down assays
The full-length l-FLP and s-FLP cDNA, and a deleted version corresponding to the last 197 amino acids were cloned in the pGEX-4T-1 expression vector (Amersham Pharmacia Biotech). Proteins were expressed in bacteria as GST fusion (see above) and purified with the GST fusion system kit (Amersham Pharmacia Biotech). For the assays, 2 μg of GST fusion proteins was first immobilized on Glutathione Sepharose 4B beads (incubation for 2 h at 4°C) and then mixed with 50 μg of Chlamydomonas or Arabidopsis protein extracts (soluble fractions) in 0.5 mL of PBS. After overnight incubation at 4°C on a rotary incubator, beads were washed three times with the same buffer, resuspended in 30 μL of SDS gel-loading buffer (1% final) (Sambrook et al. 1989) and resolved by SDS-PAGE (10%).
Analysis of pigments
All the manipulations were performed in the dark. Pigments were extracted twice from 10 μg of lyophilized cell pellets in 400 μL of extraction buffer (acetone:methanol: 0.1 M NH4OH [10:9:1]) (Yaronskaya et al. 2003), with the aid of a pestle. The pigments were separated by RP-HPLC with a Nova-Pak C18 column (4.6 × 150 mm, particle size 4 μm) as described by Papenbrock et al. (2000). Column eluent was monitored by a spectrofluorimetric detector Shimadzu RF-10AXL with the following wavelengths: protoporphyrinIX λex 405 nm/λem 633 nm, Mg-protoporphyrin λex 420 nm/λem 595 nm, protochlorophyllide λex 450 nm/λem 636 nm. Proto-IX and Mg-Proto-IX were identified and quantified using commercial available standards (Frontier Scientific). Protochlorophyllide was identified and quantified using as standard the protochloropyllide extracted from the chl-L mutant. Chlorophyll concentration was determined by the method of Porra et al. (1989).
Complementation of Arabidopsis flu mutant
To complement the Arabidopsis flu mutant, s-FLP, l-FLP, and FLU cDNAs, containing a triple HA tag at the C terminus end, were cloned in the binary pCHF5 vector (gift of C. Fankauser, University of Geneva, Geneva, Switzerland), between the 35S promoter and RbcS terminator sequences. These vectors were introduced in Agrobacterium tumefaciens C58C1 and then into Arabidopsis flu plants as described (Weigel et al. 2000). Primary transformants were selected on BASTA-containing medium (10 mg/L). Expression of the Chlamydomonas proteins in the transgenic plants was confirmed by immunoblotting, using monoclonal α-HA antibody for the detection. The same membranes were also incubated with antiserum α-LHCP (1:1), as control.
Acknowledgments
We thank M. Goldschmidt-Clermont for critical reading of the manuscript; N. Roggli for preparing the figures; S. Beale, C. Beck, and S. Lemaire for antibodies; H. Cerutti for providing the RNAi plasmid; and K. Wilson and S. Hoertensteiner for HPLC assistance. A.F. and L.M. were supported by long-term HFSP fellowships. F.B. was supported by a long-term EMBO fellowship. This work was supported by grant 3100-0667763.02 from the Swiss National Foundation.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.321305.
Supplemental material is available at http://www.genesdev.org.
References
- Armstrong G.A. 1998. Greening in the dark: Light-independent chlorophyll biosynthesis from anoxygenic photosynthetic bacteria to gymnosperms. J. Photochem. Photobiol. B43: 87–100. [Google Scholar]
- Barkan A. and Goldschmidt-Clermont, M. 2000. Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572. [DOI] [PubMed] [Google Scholar]
- Cahoon A.B. and Timko, M.P. 2000. yellow-in-the-dark mutants of Chlamydomonas lack the CHLL subunit of light-independent protochlorophyllide reductase. Plant Cell 12: 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekounova E., Voronetskaya, V., Papenbrock, J., Grimm, B., and Beck, C.F. 2001. Characterization of Chlamydomonas mutants defective in the H subunit of Mg-chelatase. Mol. Genet. Genomics 266: 363–373. [DOI] [PubMed] [Google Scholar]
- Cornah J.E., Terry, M.J., and Smith, A.G. 2003. Green or red: What stops the traffic in the tetrapyrrole parthway? Trends Plant Sci. 8: 224–230. [DOI] [PubMed] [Google Scholar]
- Durrant I. 1990. Light-based detection of biomolecules. Nature 346: 297–298. [DOI] [PubMed] [Google Scholar]
- Gray J.C. 2003. Chloroplast-to-nucleus signalling: A role for Mg-protoporphyrin. Trends Genet. 19: 526–529. [DOI] [PubMed] [Google Scholar]
- Harris E.H. 1989. The Chlamydomonas sourcebook: A comprehensive guide to biology and laboratory use. Academic Press, San Diego, CA. [DOI] [PubMed]
- Huang K. and Beck, C.F. 2003. Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. 100: 6269–6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im C.S., Matters, G.L., and Beale, S.I. 1996. Calcium and calmodulin are involved in blue light induction of the gsa gene for an early chlorophyll biosynthetic step in Chlamydomonas. Plant Cell 8: 2245–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. 2003. Alternative splicing and proteome diversity in plants: The tip of the iceberg has just emerged. Trends Plant Sci. 8: 468–471. [DOI] [PubMed] [Google Scholar]
- Kropat J., Oster, U., Rudiger, W., and Beck, C.F. 1997. Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc. Natl. Acad. Sci. 94: 14168–14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2000. Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. Plant J. 24: 523–531. [DOI] [PubMed] [Google Scholar]
- Larkin R.M., Alonso, J.M., Ecker, J.R., and Chory, J. 2003. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906. [DOI] [PubMed] [Google Scholar]
- Lee K.P., Kim, C., Lee, D.W., and Apel, K. 2003. TIGRINA d, required for regulating the biosynthesis of tetrapyrroles in barley, is an ortholog of the FLU gene of Arabidopsis thaliana. FEBS Lett. 553: 119–124. [DOI] [PubMed] [Google Scholar]
- Li J. and Timko, M.P. 1996. The pc-1 a deletion mutation in the nuclear gene for NADPH:protochlorophyllide oxidoreductase. Plant Mol. Biol. 30: 15–37. [DOI] [PubMed] [Google Scholar]
- Li J., Goldschmidt-Clermont, M., and Timko, M.P. 1993. Chloroplast-encoded chlB is required for light-independent protochlorophyllide reductase activity in Chlamydomonas reinhardtii. Plant Cell 5: 1817–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoe P., Mayfield, S.P., and Rochaix, J.D. 1988. Comparative analysis of the biogenesis of photosystem II in the wild-type and Y-1 mutant of Chlamydomonas reinhardtii. J. Cell Biol. 106: 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S., Hayashi, M., and Nishimura, M. 1999. Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J. 17: 309–320. [DOI] [PubMed] [Google Scholar]
- Matters G.L. and Beale, S.I. 1995. Blue-light-regulated expression of genes for two early steps of chlorophyll biosynthesis in Chlamydomonas reinhardtii. Plant Physiol. 109: 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac A.C. and Terry, M.J. 2002. Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J. 32: 549–559. [DOI] [PubMed] [Google Scholar]
- Merendino L., Falciatore, A., and Rochaix, J.D. 2003. Expression and RNA binding properties of the chloroplast ribosomal protein S1 from Chlamydomonas reinhardtii. Plant Mol. Biol. 53: 371–382. [DOI] [PubMed] [Google Scholar]
- Meskauskiene R. and Apel, K. 2002. Interaction of FLU, a negative regulator of tetrapyrrole biosynthesis, with the glutamyl-tRNA reductase requires the tetratricopeptide repeat domain of FLU. FEBS Lett. 532: 27–30. [DOI] [PubMed] [Google Scholar]
- Meskauskiene R., Nater, M., Goslings, D., Kessler, F., op den Camp, R., and Apel, K. 2001. FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 98: 12826–12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Brusslan, J.A., Larkin, R., Nagatani, A., and Chory, J. 2001. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. 98: 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J., Mock, H.P., Tanaka, R., Kruse, E., and Grimm, B. 2000. Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 122: 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J., Thompson, W.A., and Kriedermann, P.E. 1989. Determination of accurate extinction coefficients and simultaneous equation for assaying chlorophyll a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 1989: 384–394. [Google Scholar]
- Reinbothe S., Reinbothe, C., Apel, K., and Lebedev, N. 1996a. Evolution of chlorophyll biosynthesis—The challenge to survive photooxidation. Cell 86: 703–705. [DOI] [PubMed] [Google Scholar]
- Reinbothe S., Reinbothe, C., Lebedev, N., and Apel, K. 1996b. PORA and PORB, two light-dependent protochlorophyllide-reducing enzymes of angiosperm chlorophyll biosynthesis. Plant Cell 8: 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr J., Sarkar, N., Balenger, S., Jeong, B.-R., and Cerutti, H. 2004. Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 20: 611–621. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schroda M., Vallon, O., Whitelegge, J.P., Beck, C.F., and Wollman, F.A. 2001. The chloroplastic GrpE homolog of Chlamydomonas: Two isoforms generated by differential splicing. Plant Cell 13: 2823–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D.R., Rochaix, J.D., and Purton, S. 1996. The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251: 23–30. [DOI] [PubMed] [Google Scholar]
- Strand A., Asami, T., Alonso, J., Ecker, J.R., and Chory, J. 2003. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrin IX. Nature 421: 79–83. [DOI] [PubMed] [Google Scholar]
- Taylor W.C. 1989. Regulatory interaction between nuclear and plastid genomes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40: 211–233. [Google Scholar]
- Timko M.P. 1998. Pigment biosynthesis: Chlorophylls, heme, and carotenoids. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Trebst A. 1980. Inhibitors in electron flow. Tools for the functional and structural localization of carriers and energy conservation sites. Methods Enzymol. 69: 675–715. [Google Scholar]
- Weigel D., Ahn, J.H., Blazquez, M.A., Borevitz, J.O., Christensen, S.K., Fankhauser, C., Ferrandiz, C., Kardailsky, I., Malancharuvil, E.J., Neff, M.M., et al. 2000. Activation tagging in Arabidopsis. Plant Physiol. 122: 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaronskaya E., Ziemann, V., Walter, G., Averina, N., Borner, T., and Grimm, B. 2003. Metabolic control of the tetrapyrrole biosynthetic pathway for porphyrin distribution in the barley mutant albostrians. Plant J. 35: 512–522. [DOI] [PubMed] [Google Scholar]
- Yoshimura K., Yabuta, Y., Ishikawa, T., and Shigeoka, S. 2002. Identification of a cis element for tissue-specific alternative splicing of chloroplast ascorbate peroxidase pre-mRNA in higher plants. J. Biol. Chem. 277: 40623–40632. [DOI] [PubMed] [Google Scholar]