Significance
Modern industrial and agricultural practices generate large quantities of aromatic pollutants; however, these waste products can be converted into fine chemicals, fuels, and plastics through biocatalytic pathways. The bacterial world can inform such utilization strategies as certain strains of soil and marine bacteria metabolize environmentally derived aromatics. Many of these metabolic pathways involve aryl intermediates that require demethylation to facilitate modification and ring opening for assimilation into the tricarboxylic acid (TCA) cycle. Aryl demethylases, which catalyze this reaction, are poorly understood, making their utilization in biotechnology difficult. We provide the structural and mechanistic characterization of a single-domain aryl demethylase, LigM, which employs a tyrosine-dependent mechanism. Insights from this work will inform synthetic biology approaches to convert underutilized aromatics into higher value compounds.
Keywords: demethylase, biocatalysis, aryl metabolism, tetrahydrofolate, lignin
Abstract
Some strains of soil and marine bacteria have evolved intricate metabolic pathways for using environmentally derived aromatics as a carbon source. Many of these metabolic pathways go through intermediates such as vanillate, 3-O-methylgallate, and syringate. Demethylation of these compounds is essential for downstream aryl modification, ring opening, and subsequent assimilation of these compounds into the tricarboxylic acid (TCA) cycle, and, correspondingly, there are a variety of associated aryl demethylase systems that vary in complexity. Intriguingly, only a basic understanding of the least complex system, the tetrahydrofolate-dependent aryl demethylase LigM from Sphingomonas paucimobilis, a bacterial strain that metabolizes lignin-derived aromatics, was previously available. LigM-catalyzed demethylation enables further modification and ring opening of the single-ring aromatics vanillate and 3-O-methylgallate, which are common byproducts of biofuel production. Here, we characterize aryl O-demethylation by LigM and report its 1.81-Å crystal structure, revealing a unique demethylase fold and a canonical folate-binding domain. Structural homology and geometry optimization calculations enabled the identification of LigM’s tetrahydrofolate-binding site and protein–folate interactions. Computationally guided mutagenesis and kinetic analyses allowed the identification of the enzyme’s aryl-binding site location and determination of its unique, catalytic tyrosine-dependent reaction mechanism. This work defines LigM as a distinct demethylase, both structurally and functionally, and provides insight into demethylation and its reaction requirements. These results afford the mechanistic details required for efficient utilization of LigM as a tool for aryl O-demethylation and as a component of synthetic biology efforts to valorize previously underused aromatic compounds.
Demethylation reactions are found throughout biology, impacting a wide range of cellular processes from metabolism and energy generation to genetic imprinting and DNA repair (1–3). Reflective of such extensive application, there is an immense diversity in the mechanisms used by demethylases, the enzymes responsible for catalyzing methyl transfer reactions. Intriguingly, only roughly 1% of identified demethylases have been characterized at a structural level (4), and those that have vary substantially in protein fold, making it difficult to discern the structural basis and catalytic determinants for this key biocatalytic process based on function alone.
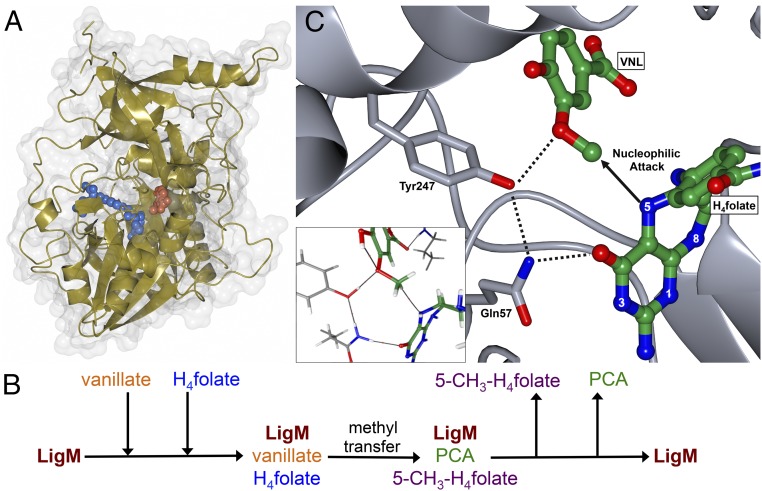
Aryl demethylation is of particular interest because it is an essential first step in the modification, and ultimately metabolism, of aromatic compounds under aerobic conditions. Because of its importance, a wide variety of aryl demethylase systems have been identified from aryl compound-using soil and marine bacteria. These organisms are uniquely capable of metabolizing aromatic waste and natural breakdown products such as those derived from lignin, an aromatic heteropolymer derived from plants (1, 5). Aryl O-demethylation, the removal of a methyl moiety from an aryl methoxy functional group, is especially vital to degradation and aerobic metabolism of aromatics and is a prerequisite step to aromatic ring opening, priming the ring for downstream oxidative cleavage and assimilation into the tricarboxylic acid (TCA) cycle (1). Aryl O-demethylation in the prokaryotic soil bacterium Sphingomonas ssp. is achieved through several mechanistically disparate demethylase systems of varying complexity: a three-component, iron-dependent monoxygenase system (LigXa, LigXc, LigXd) (6); a two-component, flavin mononucleotide (FMN)- and iron-sulfur cluster-dependent class IA oxygenase system (vanAB) (7–9); and tetrahydrofolate (H4folate)-dependent, single-enzyme systems (LigM or DesA) (8, 10). Both LigM and DesA function as monomers and target single-ring aromatics (8, 10). DesA is selective for syringate, which has two methoxy groups, whereas LigM exhibits specificity for the monomethoxylated compounds vanillate and 3-O-methylgallate (3MGA) (1). In these reactions, a methoxy methyl group is transferred from the aromatic substrate to H4folate, producing 5-methyl-tetrahydrofolate (5-methyl-H4folate), the formation of which was previously confirmed by negative ion electrospray ionization (ESI)-MS experiments (8).
Although LigM and DesA display significant sequence similarity (50%) to each other, they are markedly different in sequence from previously identified Sphingomonas ssp. O-demethylases and H4folate-dependent O-demethylases from other bacteria (8). LigM and DesA exhibit the greatest sequence similarity (25%) to H4folate-using aminomethyltransferases (T-proteins) of the bacterial glycine cleavage system. However, although these T-proteins also catalyze a methyl transfer reaction involving H4folate, they generate a disparate H4folate end product, 5,10-methylene-H4folate (11). These findings—minimal sequence homology to other proteins, especially to those with known structure, combined with ambiguity in the molecular determinants for catalysis and increasing need for single-domain aryl demethylation biocatalysts—sparked our interest in understanding the mechanism of these enzymes at a molecular level.
In the present study, vanillate demethylation by LigM is explored in depth. We comprehensively characterize the enzyme’s kinetic mechanism, detailing the nature of its interactions with its substrates and products. To provide a structural context for LigM’s mechanism, we present the 1.81-Å X-ray crystal structure of LigM, which reveals a protein fold unique to LigM as well as a conserved folate-binding domain shared among folate-using enzymes spanning protoarchaea to Homo sapiens. Based on structural homology, we identify LigM’s H4folate-binding site, and, through clues from electron density, molecular docking, and quantum chemistry calculations and site-directed mutagenesis, we locate and confirm its vanillate-binding site. Together, these results describe the catalytic mechanism of LigM, defining it as a catalytic tyrosine-dependent O-demethylase and an ideal biocatalytic component of valorization pathways for productive conversion of underused aromatic compounds.
Results
LigM Is More Sensitive to pH than to Temperature.
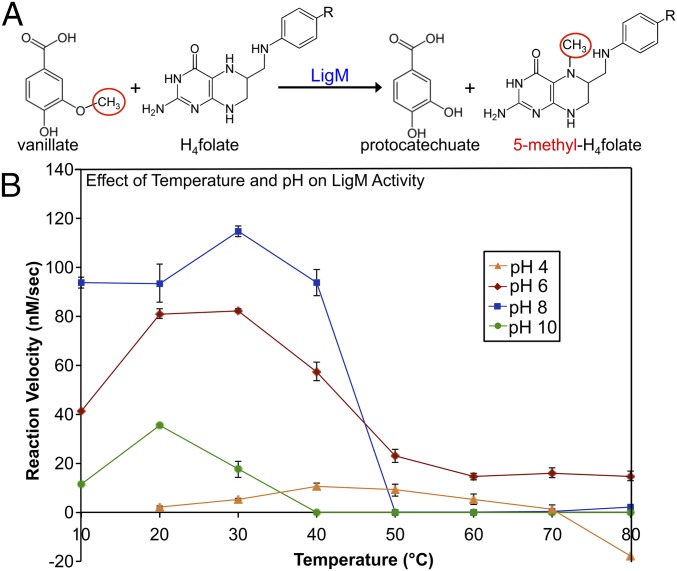
Sphingomonas paucimobilis LigM is 471 residues in length, functioning intracellularly, presumably in the cytosol, to catalyze the transfer of a methyl moiety from the methoxy functional group of vanillate or 3-O-methylgallate to nitrogen 5 (N5) of H4folate, forming protocatechuate (PCA), or gallate, respectively, and 5-methyl-H4folate (Fig. 1A) (8). Using an HPLC-based method to quantify PCA production at varying pH and temperatures, LigM-catalyzed vanillate O-demethylation was found, in agreement with and expanding upon previous work by Rosini, et al. (12), to occur most readily at pH 6–8 and temperatures ranging from 10–40 °C (Fig. 1B). LigM was most active at pH 8, achieving a maximal reaction velocity (115 nM/s) at 30 °C, and was least active under strong acidic (pH 4) and basic (pH 10) conditions (Fig. 1B). Although at pH 8 LigM was rendered essentially inactive at temperatures above 50 °C, the enzyme exhibited sustained, although reduced, activity at pH 6, displaying measurable activity over the entire range of assayed temperatures with peak activity at 20–30 °C (Fig. 1B).
Fig. 1.
LigM catalyzes the O-demethylation of vanillate, achieving maximal reaction velocity at pH 8 and 30 °C. (A) LigM O-demethylates vanillate in the presence of H4folate to form PCA and 5-methyl-H4folate. The leaving methyl from vanillate is circled in red and is transferred to H4folate-N5. (B) LigM’s ability to catalyze O-demethylation of vanillate was examined over a pH range of 4–10 and a temperature range of 10–80 °C (in steps of 10 °C) and was plotted as reaction velocity (nM/s) vs. temperature (°C). Reactions were run in duplicate; whiskers indicate SEM. Nonenzyme control reactions were subtracted from the sample data before analysis; therefore at pH 4 and 80 °C, where the reaction velocity drops below 0, O-demethylation of vanillate occurred at a greater rate than observed in the presence of LigM. Additionally, H4folate was insoluble at pH 4 and 10 °C; thus this reaction point was not taken.
LigM Uses an Ordered, Sequential Kinetic Mechanism.
With only a basic understanding of the LigM-catalyzed vanillate O-demethylation reaction available in the existing literature, we sought to characterize the kinetic mechanism of LigM. Because LigM-catalyzed vanillate O-demethylation involves two substrates and leads to the production of two products (Fig. 1A), it can be deduced that LigM uses a bisubstrate, biproduct (Bi Bi) kinetic mechanism, which can be classified as either sequential or ping-pong (13). To identify LigM’s kinetic parameters and determine which of these two Bi Bi mechanisms was at play, a bisubstrate steady-state kinetic analysis was performed on LigM, and velocity data were plotted against substrate concentration (SI Appendix, Fig. S1 A and B) (14–16).
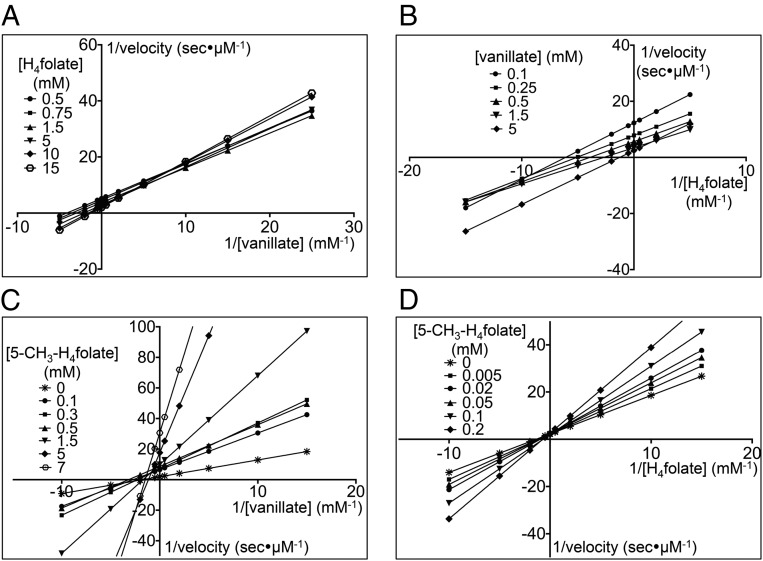
Lineweaver–Burk reciprocal plots of the kinetic data were generated for data visualization and to aid in discernment between sequential and ping-pong mechanisms (13). When the concentration of vanillate was varied with different fixed concentrations of H4folate, the intersection of the corresponding Lineweaver–Burk plots occurred in the upper right quadrant (Fig. 2A). Similarly, the reciprocal plots when the concentration of H4folate was varied with different fixed concentrations of vanillate also displayed an intersecting pattern with the intersection point located in the lower left quadrant (Fig. 2B). The intersecting pattern depicted in both reciprocal plots provides strong evidence that LigM uses a sequential mechanism, requiring binding of vanillate and H4folate, forming a ternary complex, before product formation and release (13). To determine kcat, Km, and catalytic efficiency (kcat/Km) for each substrate, the raw kinetic data were globally fit with a nonlinear least-squares sequential rate equation (Table 1 and SI Appendix, Fig. S1 A and B). Under the assay conditions, LigM displayed relatively equal (within the error of the measurement) Km values for vanillate and H4folate, and subsequently LigM’s catalytic efficiency for vanillate and H4folate was roughly equivalent (Table 1). This finding suggests that transfer of the methoxy methyl moiety from vanillate to H4folate and protonation of demethylated vanillate to form PCA occur essentially simultaneously, without the formation of additional reaction intermediates.
Fig. 2.
LigM uses an ordered, sequential kinetic mechanism. The steady-state kinetics for LigM O-demethylation of vanillate was determined by varying the concentration of one substrate while the concentration of the other substrate was fixed at different concentrations. (A and B) For visualization purposes, data were plotted as Lineweaver–Burk plots with 1/velocity (s/μM) vs. 1/[vanillate] (mM) at different concentrations of H4folate (in mM) (A), and 1/velocity (s/μM) vs.1/[H4folate] (mM) at different concentrations of vanillate (in mM) (B). Both families of plots, with respect to vanillate and to H4folate, show an intersecting pattern, suggesting a sequential mechanism for LigM. (C and D) Product inhibition studies using 5-methyl-H4folate (5-CH3-H4folate) were performed to distinguish between ordered and random sequential mechanisms. To identify the resulting inhibition pattern, data were transformed into Lineweaver–Burk plots with 1/velocity (in s/μM) vs. 1/[vanillate] (in mM) and the concentration of 5-CH3-H4folate ranging from 0–7 mM (C) and 1/velocity (in s/μM) vs. 1/[H4folate] (in mM) with the concentration of 5-CH3-H4folate ranging from 0–0.2 mM (D). In respect to vanillate, 5-CH3-H4folate is a noncompetitive inhibitor (C); however, 5-CH3-H4folate acts as a competitive inhibitor with respect to H4folate (D). These data support an ordered, sequential mechanism for LigM. For determination of kinetic parameters (Table 1), velocity vs. substrate concentration curves (SI Appendix, Fig. S1) were fit globally with the appropriate nonlinear least-squares regression rate equations.
Table 1.
Kinetic parameters of LigM O-demethylation of vanillate
| Kinetic parameters | Vanillate | H4folate |
| kcat, s | 5.76 ± 0.25* | – |
| Km, mM | 0.63 ± 0.08* | 0.72 ± 0.11* |
| kcat/Km, mM/s | 9.14 | 8 |
| Ki, app, mM (5-methyl-H4folate) | 0.10 ± 0.01* | 0.17 ± 0.03* |
Mean ± SEM.
To define LigM’s sequential mechanism further, product inhibition studies using 5-methyl-H4folate were used to distinguish between ordered versus random binding and release of the reaction species (SI Appendix, Fig. S1 B and C). The families of curves produced in these experiments were used to generate Lineweaver–Burk plots, and the intersection of these plots enabled identification of the type of inhibition elicited by 5-methyl-H4folate with regard to each substrate (Fig. 2 C and D) (17). When H4folate was held at a saturating concentration, and vanillate was varied at different fixed concentrations of 5-methyl-H4folate, the lines on the reciprocal plot intersected in the upper left quadrant (Fig. 2C), corresponding to noncompetitive inhibition (17). In contrast, when vanillate was held at a saturating concentration, and H4folate was varied at different fixed concentrations of 5-methyl-H4folate, intersection occurred on the positive y axis (Fig. 2D), indicating competitive inhibition (17). The inhibition strength of 5-methyl-H4folate differed slightly between substrates: 5-methyl-H4folate had a stronger inhibition effect on vanillate with a Ki, app of 0.10 mM versus a Ki, app of 0.17 mM with respect to H4folate (Table 1). 5-Methyl-H4folate’s competitive mode of inhibition toward H4folate and noncompetitive inhibition toward vanillate was consistent with an ordered, sequential kinetic mechanism for LigM in which H4folate and 5-methyl-H4folate compete for the same enzyme form of LigM (i.e., free enzyme or aryl-bound enzyme) (13, 18).
The Crystal Structure of LigM Reveals a Unique Demethylase Fold.
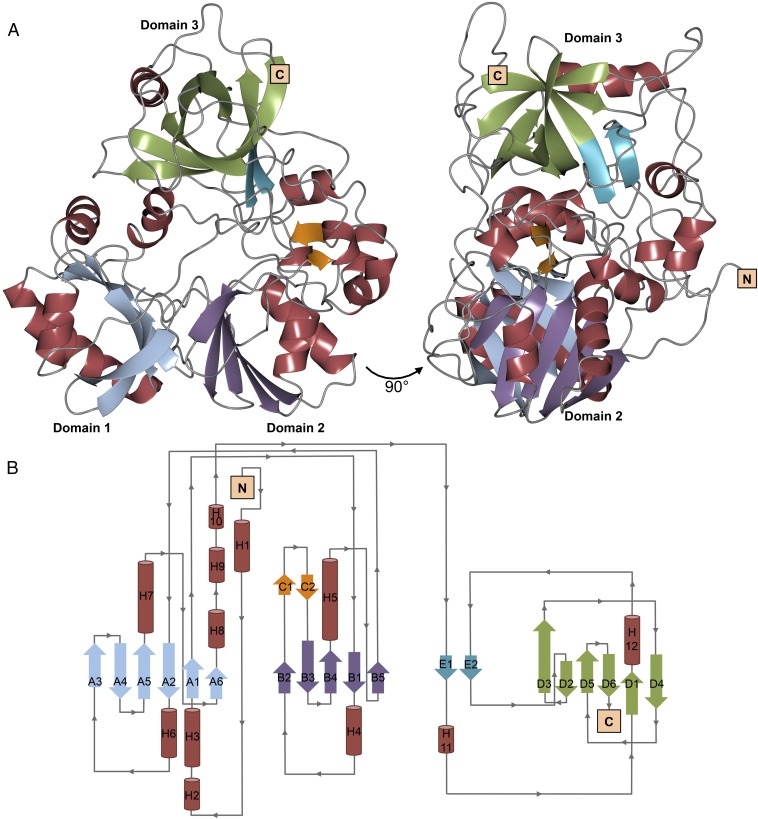
The kinetic characterization of LigM described above provided insight into how LigM functions at a mechanistic level; however, such work is unable to describe the underlying structural basis for LigM’s catalytic activity. With only distant sequence similarity to proteins of known fold, determination of LigM’s X-ray crystal structure was essential for understanding the catalytic mechanism of this demethylase. The final structure model of LigM, solved to 1.81 Å, spans 97.8% of the protein sequence (Thr3–Ala467) and is 25% α-helical and 24% β-strand in nature with the remaining 51% of the structure comprised of ordered, connecting loop regions (Fig. 3 and SI Appendix, Table S1 ). Overall, LigM is globular in shape, measuring roughly 60 × 50 × 40 Å in size, and contains 12 α-helices (H1–12) and 21 β-strands, which are organized into five β-sheets (designated by the letters A–E in Fig. 3B). The structure reveals canonical folate-binding architecture as well as structural elements that are unique to LigM, as identified by a structural homology search using DaliLite v. 3 (19). The top 50 scoring structural homolog hits were methyltransferases that use a conserved tripartite folate-binding domain to interact with and transfer a methyl to a folate substrate. LigM’s closest structural homolog is an aminomethyltransferase T-protein of the glycine cleavage system from Thermotoga maritima (Tm_T) [Protein Data Bank (PDB) ID code: 1WOS] (11). Tm_T shares 23% sequence identity with LigM, aligning with an rmsd of 2.4 Å over its 324-residue H4folate-binding fold. Similarly, the top 50 structural homologs align to LigM strictly through their respective folate-binding domains with rmsds ranging from 2.4 to 3.2 Å. Interestingly, LigM’s aminomethyltransferase homologs catalyze a different reaction involving the production of 5,10-methylene-H4folate (20, 21), and this difference in end products may explain why LigM shares such low overall sequence identity with its homologs (20–27%) despite sharing significant structural similarity.
Fig. 3.
The 1.81-Å X-ray crystal structure of LigM reveals a demethylase fold and well-conserved folate-binding architecture. (A) In total, LigM contains 12 α-helices and 21 β-strands, which are organized into five β-sheets. The structure model of LigM is depicted at 0° and 90° and is colored according to structural element: α-helices are red, and β-strands are color-coded by β-sheet; N and C termini are labeled. LigM contains a canonical folate-binding fold that is comprised of three domains, labeled domain 1, domain 2, and domain 3. (B) The corresponding structural topology diagram of LigM is color-coded to match the structure shown in A. β-Strands are indicated by arrows, are labeled by the β-sheet (A–E) to which they belong, and are numbered according to their order within their corresponding β-sheet (1, 2, 3, …). α-Helices are shown as cylinders and are numbered based on their placement within the structure (H1–12). Connecting loops are drawn in gray with their directionality indicated with arrows. The topology diagram was initially generated by PDBsum (40) and adapted. The LigM structure has the PDB ID code 5TL4. Crystallographic statistics can be found in SI Appendix, Table S1.
LigM’s highly conserved tripartite H4folate-binding fold comprises ∼70% of its structure, and consists of three domains arranged in a cloverleaf-like formation, creating a central H4folate-binding cavity (Figs. 3 and 4A) (11, 22, 23). Domain 1 spans residues Ala52–Asp56 and Ser161–Phe237 and is comprised of the six-stranded, mixed-type β-sheet A (A1–A6) flanked on either face by two α-helices (H3, H8; H6, H7). Domain 2 (Val62–Asp145) is architecturally similar to domain 1 and is composed of the five-stranded, antiparallel β-sheet B (B1–B5) and two α-helices (H4, H5), which pack against the outer face of β-sheet B (Fig. 3). A unique, 13-residue-long extension is present in LigM between β-strands B2 and B3 that contains the two-stranded β-hairpin β-sheet C (C1, C2). Domain 3 (Lys335–Ser448) is structurally distinct from domains 1 and 2, because it consists of a β-barrel motif formed by the six-stranded antiparallel β-sheet D (D1–D6) (Fig. 3). Unlike its structural homologs, domain 3 of LigM contains an additional two-stranded parallel β-sheet, β-sheet E (E1, E2), which is the only parallel β-sheet present in LigM. Interestingly, β-strands E1 and E2 are separated by roughly 80 residues, many of which are structural elements that are unique to LigM. β-Strand E2 sits immediately N-terminal to D2, seemingly forming one long β-strand. This D2 extension is specific to LigM and is not present in the structural homologs.
Fig. 4.
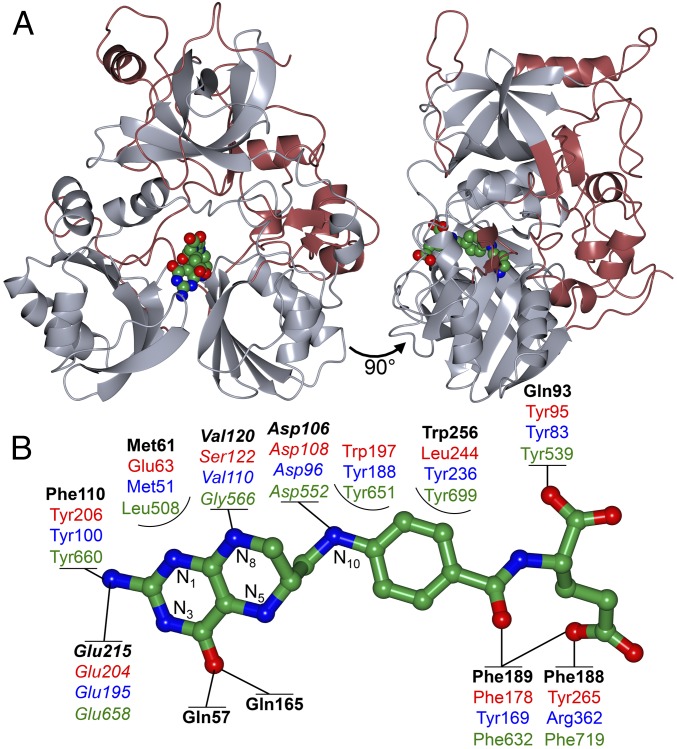
Conservation of canonical folate-binding architecture enabled the identification of LigM’s H4folate-binding site, suggesting that the interaction between LigM and H4folate is mediated through Gln57, Gln93, and Phe188. (A) LigM’s closest structural homologs are methyltransferases that use a tripartite folate-binding fold. The well-conserved tripartite fold is depicted in gray, and LigM’s unique structural elements are displayed in red (SI Appendix, Fig. S2A). H4folate is colored by atom type (green, carbon; red, oxygen; blue, nitrogen) and is shown docked into the folate-binding cavity based on alignment to structural homologs and geometry optimization. (B) Although overall architecture and H4folate conformation is conserved, LigM and its homologs differ to some degree in the precise folate–protein interactions that they use. Homologs shown for comparison include dimethylsulfoniopropionate(DMSP)-dependent demethylase (DmdA), which catalyzes the transfer of a methyl from DMSP to H4folate-N5 (PDB ID code: 3TFJ) (27); Tm_T, a glycine cleavage system T-protein that catalyzes the formation of 5,10-methylene-H4folate and ammonia from an H-protein–associated aminomethyl moiety (PDB ID code: 1WOO) (11); and dimethylglycine oxidase (DMGO), which oxidizes N,N-dimethyl glycine to form 5,10-methylene-H4folate (PDB ID code: 1PJ7) (20). Key interaction residues are shown in bold black type for LigM, in red type for DmdA, in blue type for Tm_T, and in green type for DMGO. Highly conserved residues are italicized. Direct protein–folate interactions predicted for LigM and used by the homologs are represented by solid lines. Partially encircled residues are those that reside in the binding cavity but are not predicted to form strong interactions with the folate substrate. Main-ring nitrogen atoms of the H4folate pteridine ring are numbered. The LigM–H4folate interactions are further detailed in SI Appendix, 3 and Table S3.
Perhaps indicative of its distinct mechanism and specificity toward vanillate, 30% of LigM's structure does not exhibit significant structural homology to any other protein of known structure (SI Appendix, Fig. S2A). In Fig. 4A, these unique structural elements are depicted in red, and the H4folate-binding architecture, identified through structural homology, is displayed in gray. Interestingly, LigM’s unique structure is located predominantly on one side of the enzyme and largely stems from sequence insertions found between main structural elements of its H4folate-binding domain. The N terminus of LigM, which spans the first 40 residues of structure and precedes domain 1, contains two α-helices (H1, H2), which, together with H9 and H10, sit above domain 2. This α-helix bundle borders one side of β-sheet C, and another unique element of LigM, α-helix H11, is located on the opposing face of β-sheet C. A significant portion of the unique structure resides in the C-terminal region of LigM. Most notably, the loops connecting several of the β-strands in β-sheet D are exceptionally long in comparison with those in the structures of other folate-using aminomethyltransferases. The region connecting β-strands D1 and D2 spans residues Trp341–Thr375 and contains an 11-residue α-helix, H12, that sits parallel to and extends beyond the β-barrel of domain 3. A second 18-residue loop lies between β-strands D5 and D6; in the present structure this loop folds down against the main body of LigM. Because these structural elements are specific to LigM, this region appeared a prime candidate to house the residues necessary for the vanillate binding and specific O-demethylation activity of LigM.
LigM Uses Several Unique, Catalytically Relevant Interactions with H4folate.
The present structure of LigM was solved without H4folate of vanillate bound, despite efforts to obtain substrate-bound structures via cocrystallization and crystal soaking with either substrate. However, significant conservation of both the folate-binding site topology and the conformation of bound H4folate enabled identification of LigM’s H4folate-binding site and placement of H4folate within this site. The structure of LigM’s closest structural homolog, Tm_T from Thermotoga maritima, was solved in both apo (PDB ID code:1WOS) and H4folate-bound (PDB ID code: 1WOO) forms; structurally, these two forms of Tm_T are essentially identical (11). To place H4folate in the LigM structure, LigM was aligned with the H4folate-bound Tm_T structure (rmsd of 2.5 Å over 324 residues). This alignment encompassed the entirety of the canonical H4folate-binding fold and had a Dali Z-score of 32.5, corresponding to a significant fold similarity as Dali Z-scores greater than two are considered significant (24, 25). Based on rmsd and Dali Z-score, LigM and Tm_T were structurally very similar through their H4folate-binding domains; thus, we were able to determine an accurate initial placement of H4folate within the canonical H4folate-binding cavity of LigM. The geometry of H4folate and the surrounding LigM residues were refined further through quantum chemical geometry optimization calculations, in which the energy was minimized with respect to nuclear positions until the optimal geometry was identified (SI Appendix, 1–3 and Table S2). The geometry-optimized structure of H4folate approximately maintained its overall starting geometry, differing from its preoptimization conformation by an rmsd of only 1.69 Å (SI Appendix, Fig. S2B).
In the geometry-optimized model of H4folate-bound LigM, H4folate adopts a characteristic kinked conformation in the central internal cavity formed by domains 1–3, positioned as is observed in the H4folate-bound homologs (Fig. 4A and SI Appendix, Fig. S2B) (11, 22). This internal cavity is highly solvated in the present structure and extends 14 Å deep by 12 Å wide into the protein core, opening onto the protein surface presumably to enable the entry and exit of H4folate (SI Appendix, Fig. S2C). H4folate binds the length of this cavity with its pteridine ring deeply buried and its C-terminal group bent along the surface-exposed outer edge of the binding channel. Three folate–protein hydrogen bond interactions involving residues Asp106, Val120, and Glu215 are highly conserved among LigM’s structural homologs (26) and also are predicted to be conserved in LigM (Fig. 4B). Similar to its homologs, LigM is predicted to use several aromatic and nonpolar residues to interact with H4folate; these include Gln57, Met61, Gln93, Phe110, Phe188, Phe189, Gln165, and Trp256 (Fig. 4B). Although many of these interactions were expected, based on residue location and reported homolog behavior, Atoms in Molecules (AIM) calculations enabled the first principles determination of the existence of atom–atom interactions and quantification of the relative strength of the LigM–H4folate interactions (SI Appendix, 3 and Table S3). Based on AIM calculations, two glutamine residues appear to play a particularly important role in the binding of H4folate by LigM, thus differentiating LigM from its structural homologs (Fig. 4B). LigM’s two strongest interactions with H4folate were found between Gln93 and the glutamate tail of H4folate (OX1) and between Gln57 and O4 of the pteridine ring (SI Appendix, Table S3). Surprisingly, a strong interaction was identified between Phe188 and H4folate-OE2; this interaction was unexpected, given the high degree of variability in residue identity among the homologs at this position (Fig. 2B). LigM, much like its homologs, is predicted to interact with H4folate along the entire length of the substrate, forming strong interactions with both the pteridine ring and glutamate tail. Thus these interactions together likely promote the characteristic kinked binding conformation of H4folate.
Beyond LigM’s unconventional use of glutamine residues in binding H4folate, there are several additional striking differences between the residue topology of LigM’s H4folate-binding site and those of its closest structural homologs Tm_T (rmsd: 2.4 Å, 23% sequence identity) (11); Arthrobacter globiformis dimethylglycine oxidase (DMGO) (rmsd: 2.7 Å, 21% sequence identity) (20); and dimethylsulfoniopropionate (DMSP)-dependent demethylase (DmdA) (rmsd: 3.2 Å, 20% sequence identity) (27). Notably, LigM contains phenylalanines (Phe110, Phe188) and a tryptophan (Trp256) in place of three key Tm_T tyrosines (Fig. 4B). Loss of these tyrosines through the loss of three hydroxyl groups would greatly weaken the direct and indirect water-mediated interactions that LigM could form with H4folate. However, Trp256, which sits alongside H4folate, stretching from the pteridine ring to the p-aminobenzoate moiety, is hypothesized to influence the orientation of Gln93 and to form stacking interactions with H4folate. Utilization of a tryptophan for H4folate binding is unique to LigM. DmdA also contains a tryptophan (Trp197); however Trp197 plays a greater role in facilitating an interaction with DmdA’s second substrate, DMSP, than it does with H4folate (27). Furthermore, LigM differs in the residue composition of the innermost portion of the H4folate-binding cavity. LigM is predicted to use two unique, direct interactions with the pteridine ring O4 of H4folate, mediated by Gln57 and Gln165. Equivalent interactions are not present in Tm_T, DmdA, or DMGO; however, these homologs make two interactions with the neighboring pteridine amine through conserved glutamine and tyrosine residues (Fig. 4B). LigM lacks this tyrosine, interacting only with the amine through Glu215. Tm_T contains an arginine (Arg227) in this region, which is highly conserved among bacterial T-proteins and interacts directly with H4folate (11). Markedly different, LigM contains Tyr247 in this position, which, at 5.65 Å from H4folate, is not poised for direct interaction with H4folate. In DmdA, this innermost region of the channel houses Ser122, which interacts directly with H4folate at N1, N8, and the pteridine amine and is expected to greatly influence the nucleophilicity of H4folate-N5 (27). These differences in pteridine ring coordination may reflect differences in precise mechanism and target substrates of these enzymes, quite possibly influencing the nucleophilic strength of H4folate-N5 (LigM, DmdA) or N5/N10 (Tm_T, DMGO) and suggesting differences in both the mechanism and mode of priming H4folate-N5 for nucleophilic attack.
The Vanillate Methyl Moiety Is Positioned 3.1 Å from H4folate-N5.
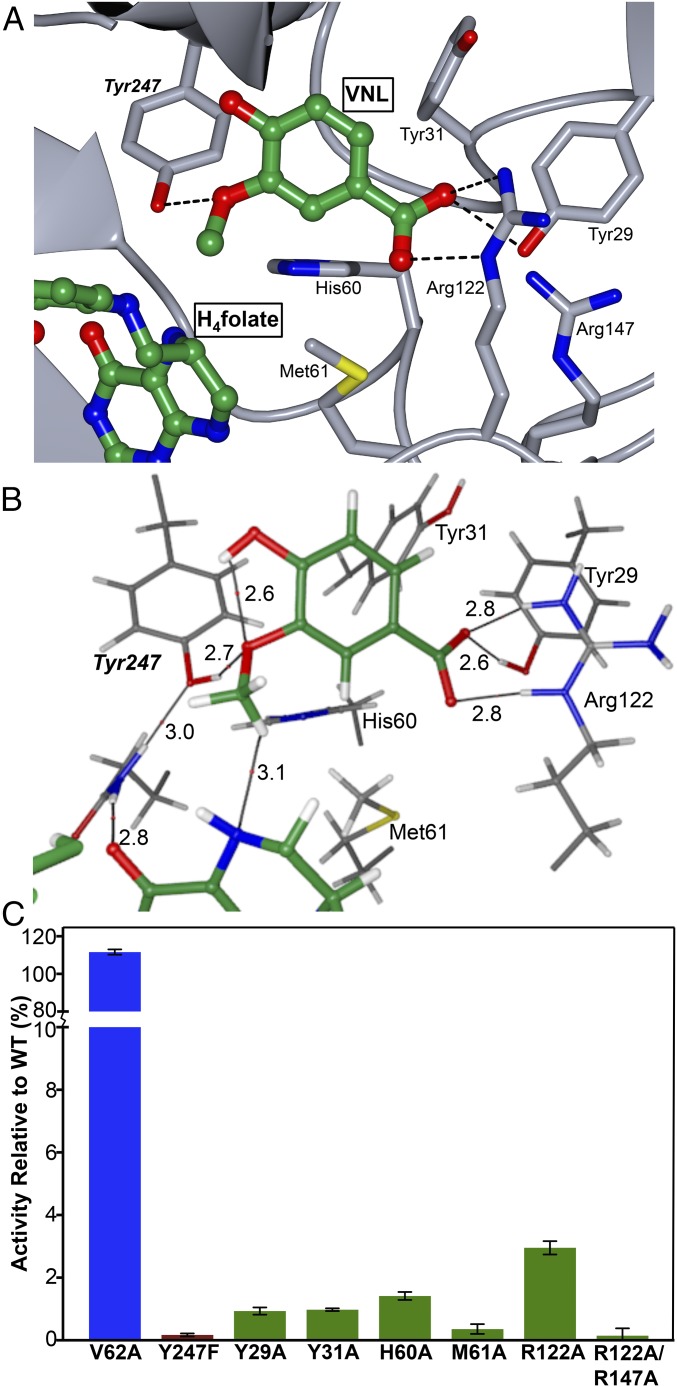
In the absence of homologous known structures and without knowledge of LigM’s catalytic residues, identification of the vanillate-binding site presented a greater challenge. We hypothesized that the vanillate-binding site would reside in the unique structural region of LigM (Fig. 4A), because the second substrate sites of its structural homologs were located adjacent or distal to, but not within, the conserved tripartite folate-binding domain. To explore possible vanillate-binding site locations in LigM, a global “blind” docking simulation was performed using AutoDock Vina (SI Appendix, SI Materials and Methods) (28). The three highest-scoring binding modes placed the vanillate methyl 3.5, 15, and 17.2 Å, respectively, from H4folate-N5. Docking calculations then were focused around these sites to refine vanillate positioning further. The top site, both in docking score and in the validity of a possible reaction mechanism, placed vanillate perpendicular to H4folate with the leaving methyl situated 3.1 Å from H4folate-N5 (Fig. 5 A and B). To generate a final placement model for vanillate in H4folate-bound LigM, vanillate and nearby binding-site residues were geometry optimized as described for H4folate (SI Appendix, SI Materials and Methods). The predicted vanillate-binding cavity, roughly 9.8 × 10.3 × 7.3 Å in size, is composed predominantly of structure and sequence unique to LigM and carries a predominantly positive charge because of the presence of Arg122 and Arg147. Despite its internal location, this cavity is solvated in the present structure, indicating that this cavity is solvent-, and quite possibly substrate-, accessible (SI Appendix, Fig. S3). The vanillate-binding site identified in this work differs from that identified by Rosini et al. (12); however, our crystal structure, rigorous computational analyses, and site-directed mutagenesis data (presented below) provide compelling evidence in favor of the vanillate-binding site described here.
Fig. 5.
LigM contains two distinct substrate-binding sites, one for vanillate and one for H4folate, which are separated by 3.1 Å. (A) The vanillate-binding site of LigM was identified through a combination of electron density clues, molecular docking, geometry optimization, AIM calculations, and site-directed mutagenesis and is illustrated with vanillate (VNL) and H4folate depicted in ball-and-stick format and colored by atom type (green, nitrogen; red, oxygen; blue, nitrogen). LigM is shown in gray with the critical interacting residue side chains (Tyr29, Tyr31, His60, Met61, Arg122, Arg147) displayed and labeled. The key catalytic residue, Tyr247, is labeled in italics. (B) AIM representation of the key catalytic and vanillate-binding interactions determined by AIM analysis of the electron density of the optimized LigM–vanillate cluster model with interaction distances (in Å) labeled (SI Appendix, Table S2). These interactions each have values of of at least 15% of the largest observed value of for a single interaction in the cluster and thus are responsible for the majority of the ligand-binding effects of the enzyme (SI Appendix, 4 and Table S4A). (C) Loss-of-function point mutations were made at each critical residue position, and these LigM mutants were assayed for their O-demethylase activity on vanillate. Mutant activity is graphed as the percentage of WT-LigM activity. V62A (blue) served as a null mutation. The catalytic mutant, Y247F, is shown in red, and key binding residues Y29A, Y31A, H60A, M61A, R122A, and R122A/R147A) are shown in green. Experiments were run in triplicate; error bars indicate SEM.
AIM calculations identified a total of 25 direct interactions between LigM and vanillate that ranged from strong, essential binding interactions to weak, auxiliary interactions (SI Appendix, 4, Fig. S4, and Table S4). The LigM–vanillate interactions were predominantly focused around the vanillate carboxyl and methyl groups with supplementary interactions involving vanillate aryl ring carbons and the vanillate phenol group. Based on calculations, carboxylate stabilization is largely dependent on strong interactions between Tyr29 and Arg122 and carboxylate oxygen O1 (Fig. 5 A and B). Further stabilization is achieved through numerous residue interactions with carboxylate O2 and Tyr29, Tyr31, Met61, and Arg122. Calculations suggested that LigM interacts with the vanillate methyl moiety through a strong interaction with Tyr247, and association with the vanillate aryl ring is predicted to occur through interactions with Tyr31, His60, and Met61. Multiple interactions, although weak, are predicted to exist between the vanillate hydroxyl group and Asn250, Pro248, Thr251, and Phe393 (SI Appendix, 4 and Table S4). Together, these residues dictate the orientation of vanillate, ensuring that the methoxy moiety is directed toward H4folate-N5 and thus encouraging catalysis. Intriguingly, an intermolecular interaction is predicted to exist between the vanillate hydroxyl and methoxy groups; this interaction also may contribute to proper orientation of the leaving methyl by pointing it upward toward H4folate-N5. Indirect, water-mediated interactions are not expected to play a significant role in the LigM–vanillate interaction because in the present structure the vanillate-binding site is not large enough to accommodate both water molecules and vanillate (SI Appendix, 5).
O-Demethylation by LigM Proceeds via a Bimolecular Nucleophilic Substitution Mechanism.
The computational results provided detailed predictions for the location of the substrate-binding site, residue composition, and critical LigM–substrate (vanillate/H4folate) interactions. This information, combined with experimental activity data, enabled identification of the molecular determinants necessary for the LigM-catalyzed vanillate O-demethylation reaction. We propose that to transfer a methyl group from vanillate to H4folate-N5, LigM binds both H4folate and vanillate, situating vanillate so the leaving methyl is 3.1 Å away from H4folate-N5. As occurs in other folate methylation reactions (20, 27), H4folate-N5 is expected to act as a nucleophile with its lone pair of electrons making an attack on the methyl carbon of vanillate in a bimolecular substitution (SN2) reaction. Tyr247, the only LigM residue hypothesized to interact strongly with the leaving methyl and its associated oxygen atom, is poised to play a central role in this reaction, stabilizing the methyl-associated oxygen and acting as a general acid to donate a proton upon methyl transfer (Fig. 5A). AIM calculations also predicted an interaction between Tyr247 and Gln57 that would stabilize Tyr247 during the reaction, enabling the hydroxyl group of Tyr247 to donate a proton to the methoxy oxygen upon methyl transfer to H4folate.
To confirm the proposed vanillate-binding site location and O-demethylation reaction mechanism, single point mutations of integral catalytic and vanillate-binding residues were introduced into LigM, and the mutants were tested for vanillate O-demethylation activity. The significance of key catalytic residue Tyr247 was examined by making a loss-of-function mutation at this position, producing a tyrosine-to-phenylalanine mutant (Y247F) that, by removal of a hydroxyl, was expected to render LigM inactive. Residues lining the binding site and believed to make significant contributions to LigM’s ability to bind vanillate were mutated to alanine, thus eliminating potential side-chain interactions at the following residue positions: Tyr29, Tyr31, His60, Met61, and Arg122. Because Arg147, via its proximity to Arg122, was also predicted to influence the electropositive environment required for interaction with the vanillate carboxyl group, it was hypothesized that a single alanine mutation at Arg122 might not fully eliminate this electropositive environment. Therefore, a double-alanine mutant, R122A/R147A, also was produced and assayed. It is important to note that all the LigM mutants behaved identically to wild-type LigM in solution, because they all eluted at the same volume by size-exclusion chromatography (SI Appendix, Fig. S5). Thus, these mutations had no discernible effect on overall protein fold or solution behavior, allowing us to interpret any changes in activity as being directly related to the mutated residue’s role in vanillate binding and/or catalysis.
The ability of Y247F, Y29A, Y31A, H60A, M61A, R122A, and R122A/R147A LigM mutants to O-demethylate vanillate was examined using the aforementioned activity assay (Fig. 5B). V62A–LigM was included as positive control, or null mutation, because an alanine mutation at Val62, which is located near but is not a part of the proposed vanillate-binding site, was not expected to have a significant effect on LigM activity. Subsequently, V62A-LigM performed similarly to WT-LigM in this assay (Fig. 5B). As predicted, given Tyr247’s crucial catalytic role, Y247F-LigM exhibited a 99.8% reduction in activity compared with WT-LigM, rendering this mutant essentially inactive. Alanine mutation at key binding residues had a dramatic effect on LigM’s vanillate O-demethylation activity, reducing activity by 97–99% (Fig. 5B). An alanine mutation at Arg122 had slightly less effect on activity than the other binding-residue point mutations; however, the double-arginine mutant, R122A/R147A-LigM, essentially abolished LigM activity (Fig. 5B). These results support the proposed LigM vanillate O-demethylation mechanism whereby Tyr247 is essential for catalytic activity of LigM, because a loss-of-function mutation at this position could not be overcome by the solvent, thus ruling out a nonresidue, solvent-driven proton transfer mechanism such as that suggested for DmdA. A pH-rate profile of LigM-catalyzed vanillate O-demethylation provided additional support for Tyr247’s role as a proton donor in this reaction, because LigM displayed maximal catalytic efficiency at pH 8 with an apparent pKa at 10.3, which corresponds to a pKa in the range that is typically observed for the phenolic hydroxyl group of tyrosine (pKa = 9.8–10.5) (SI Appendix, Fig. S6) (13). Furthermore, the significant loss of activity resulting from the alanine mutation at critical binding-residue locations highlights the importance of proper binding and precise orientation of vanillate to LigM’s activity.
Discussion
Through a multifaceted biochemical, structural, and computational approach, we have determined the structure and catalytic mechanism of O-demethylation by the demethylase LigM, for which only a basic knowledge of reaction species had previously been reported. LigM is structurally more related to folate-using aminomethyltransferases than to other known demethylases, largely because of its conserved folate-binding architecture, and its structural and mechanistic uniqueness expand the diversity of demethylase enzymes. Beyond its canonical folate-binding fold, LigM contains unique structural elements, utilizing several unconventional interactions with H4folate and housing an aryl substrate-binding site. Based on molecular docking and geometry optimization with vanillate and 3MGA, LigM is predicted to use the same binding site and residue–substrate interactions to bind vanillate and 3MGA (SI Appendix, 6 and Table S4). As illustrated in Fig. 6A, LigM’s substrate binding sites reside deep within the enzyme. This internal binding configuration enables close proximity of vanillate and H4folate and requires that the reaction species have a means of entry into and exit from their respective binding sites. The H4folate channel is the largest opening into LigM’s interior cavity (SI Appendix, Fig. S2C) and thus is the most likely possibility, given homolog behavior in which vanillate enters through the H4folate channel first, and once vanillate is bound, H4folate binds, effectively blocking aryl release until product formation has occurred. This binding behavior is observed in the LigM homolog DMGO (20), and because our structure of LigM suggests that this binding mode is conserved in LigM, we propose an ordered, sequential kinetic mechanism for LigM, in which LigM binds vanillate before H4folate and releases 5-methyl-H4folate before PCA (Fig. 6B). Although the kinetic data presented here support an ordered, sequential mechanism in which H4folate and 5-methyl-H4folate compete for the same enzyme form of LigM, they do not provide direct evidence that LigM binds to vanillate first. However, when the kinetic data are considered together with the structural data and known behavior of LigM’s structural homologs, this proposed binding order is most probable.
Fig. 6.
LigM is a catalytic tyrosine-dependent O-demethylase. (A) LigM uses two internal substrate-binding sites, one for H4folate and one for vanillate, to sequester its substrates for catalysis. Thus, LigM specifically promotes the formation of PCA and 5-methyl-H4folate, which are important constituents of their respective catabolic and anabolic S. paucimobilis pathways. The structure of LigM is depicted in gold with its corresponding surface representation in gray. H4folate (blue) and vanillate (orange) are illustrated in sphere representation. (B) The proposed ordered, sequential kinetic mechanism of LigM involves binding first to vanillate, followed by binding of H4folate to form a ternary central complex that promotes catalysis and product formation. Once the reaction is complete, 5-methyl-H4folate (5-CH3-H4folate) is released first, followed by the release of PCA. (C) LigM’s O-demethylase reaction mechanism is predicted to occur via an SN2 nucleophilic attack. The hydroxyl of Tyr247 stabilizes the vanillate methoxy oxygen during the attack of the associated methyl by H4folate-N5. Upon methyl removal, Tyr247 donates a proton to the demethylated oxygen, forming the final aryl product. The protonation state of Tyr247 is regenerated by the solvent. During this reaction, Gln57 is predicted to stabilize Tyr247, forming an interaction that enables the donation of Tyr247’s hydroxyl proton. Vanillate (VNL) and H4folate are colored by element (green, carbon; red, oxygen; blue, nitrogen). Atomic interactions are indicated with a dashed line. The Inset displays AIM depiction of predicted catalytic-related interactions and is further detailed in SI Appendix, Fig. S4.
Unlike the reactions of other characterized demethylases, LigM-catalyzed O-demethylation is tyrosine-dependent, requiring Tyr247 to aid in stabilization of the aryl substrate during methyl removal and, more importantly, to complete the reaction through the generation of the demethylated aryl product. Specifically, the hydroxyl of Tyr247 forms an interaction with the oxygen of the aryl methoxy moiety, stabilizing this atom during nucleophilic attack of the associated methyl group by H4folate-N5 (Fig. 6C). Upon transfer of the methyl to H4folate, the Tyr247 hydroxyl donates its proton to the demethylated aryl oxygen atom, producing the final aryl product, and is itself regenerated by acceptance of a proton from the solvent. This reaction mechanism is supported by a pH-rate profile analysis, which indicated the involvement of two ionizing groups in reaction progression (SI Appendix, Fig. S6). The pKas of these groups were found to be 5.6 and 10.3, which, based on the structural, biochemical, and computational analyses described here, most likely correspond to H4folate-N5 and the hydroxyl group Tyr247, respectively (SI Appendix, Fig. S6). Furthermore, a direct interaction is calculated to exist between Tyr247 and Gln57 during this reaction; thus, by stabilizing Tyr247, Gln57 likely aids in proton donation by Tyr247 (Fig. 6C). Given the structural similarity between vanillate and 3MGA and considering the conservation of predicted LigM–substrate interactions, O-demethylation of 3MGA by LigM is expected to occur via the same reaction mechanism posited for vanillate. To our knowledge, utilization of a catalytic tyrosine by LigM is unique among described demethylases, and defines a distinct category of catalytic tyrosine-dependent demethylases.
Understandably, variance in catalytic mechanism is expected among demethylases, and this work sheds light on demethylation and its mechanistic requirements. For instance, given the similarity in their products, DmdA and LigM might be expected to function with similar catalytic components. However, although both enzymes rely on the nucleophilic character of H4folate-N5 for reaction progression, they differ greatly in their methyl-donating second substrate and in the degree of protein residue involvement in catalysis. DmdA’s reaction is dependent only on water and H4folate-N5, and its second substrate, DMSP, is a short-chain, zwitterionic organosulfur compound (27). Because DMSP’s methyl behaves as a fairly good leaving group, the reaction is able to proceed without involving surrounding protein residues (27). In contrast, removal of methyl from vanillate/3MGA is less facile, and a negative contribution to the free energy of the reaction is required for reaction progression. Thus, residues of LigM must be directly involved in the reaction mechanism. The interaction between the hydroxyl group of Tyr247 and the aryl methoxy oxygen is hypothesized to increase the ease with which the methyl can be displaced by stabilizing the buildup of negative charge on the methoxy oxygen that occurs during methyl transfer and by allowing (potentially concerted) formation of the protonated aryl product. This interaction makes the leaving methyl a better candidate for attack by H4folate-N5 than it would be in the absence of a general acid, such as Tyr247. Thus, the role of catalytic protein residues in demethylation is greatly dependent on the ease with which methyl can be transferred between donor and acceptor molecules. When the target substrate methyl is a poor leaving group, greater protein involvement will be required for catalysis, and additional substrate complexation or protonation may be necessary to facilitate methyl transfer.
With an understanding of the interactions required for substrate binding and catalysis, it is not surprising that LigM-catalyzed O-demethylation functions most readily from pH 6–8, because proper protonation of the residues involved in H4folate- and vanillate-binding and catalysis is achieved only in this pH range. For example, at pH 6–8 the hydroxyl of Tyr247 (pKa = 10.3; PROPKA predicted pKa = 10–12.7) (SI Appendix, Fig. S6) would be protonated and able to donate a proton to the aryl methoxy oxygen. Similarly, Arg122 (PROPKA predicted pKa = 12.5–15.4) and Arg147 (predicted pKa = 12–12.5) would most certainly be protonated and positively charged, aiding in the proper orientation of vanillate. Tyr29 (predicted pKa = 9.3–10) and Tyr31 (predicted pKa = 10–12.2) also would be protonated, enabling their stabilization of the aryl substrate through its carboxyl oxygen and aryl ring carbon atoms. In terms of the residues involved in LigM’s interaction with H4folate, only Asp106 and Glu215 have ionizable side chains. At pH 6–8, Asp106 (predicted pKa = 3.8–5.8) and Glu215 (predicted pKa = 4.5–6.3) would be expected to be deprotonated and thus able to facilitate interaction with H4folate. Because Glu215 is predicted to be important for correct positioning of the pteridine ring and H4folate-N5 for catalysis, maintaining this interaction may be particularly important. Proper binding of H4folate and vanillate and subsequent O-demethylation requires the precise binding and reaction environment created by all interacting and neighboring residues; therefore changes in pH may alter only a few residue protonation states but may have substantial effects on reaction progression.
Placing aryl O-demethylation in a larger context, enzymatic demethylation of aromatic rings, either singularly or as a prerequisite to further modification or ring opening, is invaluable to the conversion and utilization of common aromatic industrial waste products, such as those derived from depolymerized lignin during biofuel production and paper manufacturing and from agricultural runoff (29, 30). Therefore, not only does catalytic tyrosine-dependent LigM constitute a distinct branch in the demethylase family tree, given its broader specificity and wide functional temperature range, O-demethylase LigM stands out as a potential biocatalytic tool for aryl O-demethylation either alone or as a component of conversion pathways targeting underutilized aromatic compounds.
Materials and Methods
A complete, detailed description of all methods can be found in SI Appendix, SI Materials and Methods.
Protein Purification, Crystallization, and Structure Determination.
Wild-type LigM (residues 1–471) was cloned into a modified pCDF-Duet-1 vector (Novagen-EMD Millipore) and coexpressed with chaperone plasmid from pG-KJE8 (Clontech Laboratories, Inc.) and purified from Escherichia coli BL21(DE3) cells (New England Biolabs, Inc.). LigM was crystallized in 0.2 M ammonium tartrate dibasic, 20% PEG3350 (native dataset), and in 0.2 M potassium iodide, 20% PEG3350 (anomalous dataset). Heavy-atom soaks were performed with hexatantalum tetradecabromide (Jena Bioscience GmbH). X-ray data were collected at 100 K on the Berkeley Center for Structural Biology beamline 8.2.2 of the Advanced Light Source at Lawrence Berkeley National Laboratory. The structure of LigM was phased by the multiple-wavelength anomalous dispersion (MAD) method using anomalous datasets collected at the tantalum peak wavelength (1.25 Å) and a remote wavelength (1.54 Å). A high-resolution, native dataset was collected at 1.00 Å. Data collection and refinement statistics are summarized in SI Appendix, Table S1.
Enzymatic Assays.
An HPLC-based activity assay was used to analyze LigM-catalyzed O-demethylation of vanillate, as previously described (8). To determine the optimal pH and temperature reaction conditions for LigM-catalyzed O-demethylation of vanillate, reactions contained 76 nM LigM, 1 mM vanillate (Sigma-Aldrich Co.), 5 mM H4folate (Caymen Chemical Co.), and 100 mM buffering agent. Buffering agents included 100 mM citric acid, pH 4.0; 100 mM citric acid, pH 6.0; 100 mM Tris, pH 8.0; and 100 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES), pH 10.0. Reactions were run for 10 min at 10–80 °C, assayed in steps of 10 °C. LigM activity was assessed by HPLC analysis of the quenched reaction mixture to quantify the amount of PCA product formed. Reactions were run in duplicate, and nonenzyme control reactions were subtracted from the data before analysis. For analysis of the LigM vanillate-binding site and catalytic mutant activity, the HPLC-based assay was used with the following changes. Reactions contained 5 mM vanillate, 5 mM H4folate, 76 nM LigM (wild-type or mutant), and 100 mM Tris (pH 8.0) and were run for 2 h at 30 °C. Assays were run in triplicate, and a nonenzyme control reaction was subtracted from the data before analysis.
Bi-Substrate and Inhibition Kinetics.
For steady-state kinetic analysis of LigM, assays were performed under optimal activity conditions (30 °C, pH 8.0) that yielded linear, initial velocity profiles with respect to LigM concentration and reaction time. Endpoint assays were run with respect to each substrate (vanillate and H4folate); in these assays the concentration of one substrate was varied while the concentration of the other substrate was held at different fixed concentrations. To determine kinetic parameters, the velocity versus substrate concentration data were fit globally with a nonlinear least-squares regression rate equation for a sequential steady-state bireactant mechanism using PRISM 7.0a (GraphPad Software, Inc.). Product inhibition experiments were performed with 5-methyl-H4folate (Sigma-Aldrich Co.) by varying one substrate at varied, fixed concentrations of 5-methyl-H4folate and a fixed, saturating concentration of the other substrate. Velocity versus substrate concentration data in the presence of 5-methyl-H4folate were fit globally with the appropriate inhibition nonlinear least-squares regression rate equation. All reactions contained 76 nM LigM and 100 mM Tris (pH 8.0) and were run for 10 min at 30 °C. Experiments were run in duplicate or triplicate.
Placement of H4folate and Vanillate Docking.
The bound H4folate coordinates were obtained by alignment to PDB ID code 1WOO with MultiSeq in VMD. Vanillate/3MGA docking into the enzyme crystal structure was performed with AutoDock Vina and AutoDockTools. All software references can be found in SI Appendix, SI Materials and Methods.
Quantum Chemistry Calculations.
Initial cluster model positions of heteroatoms were extracted from the enzyme–substrate geometries produced by the docking procedure and then protonated with Avogadro. Side-chain protonation states were assigned with PROPKA 3.1 at pH 8.0. Geometry optimization was performed with TeraChem 1.9 and the DL-FIND module, initially at the HF/6–31G(d) (31, 32) level of theory. Final structures were obtained at B3LYP/6–31G(d) (33–36) with the DFT-D3 dispersion correction (37) and Becke-Johnson damping function (38), requiring the hybrid ADIIS+DIIS scheme (39). AIM analyses of the final wave functions were completed with AIMAlI. Conversion of the TeraChem output wave function file to ProAIM format was achieved with a custom Perl script. Topological images were generated with Blender via the Rhorix add-on. All software references can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
Crystallographic work was performed at the Berkeley Center for Structural Biology (BCSB) Advanced Light Source beam line 8.2.2. We thank the staff of the BCSB at the Advanced Light Source of Lawrence Berkeley National Laboratory for technical support and J. H. Pereira for assistance in early stages of crystallographic work. The BCSB is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract DE-AC02-05CH11231. This work was conducted by the Joint BioEnergy Institute and was supported by the US Department of Energy, Office of Science and Office of Biological and Environmental Research under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank (PDB) database (PDB ID code 5TL4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619263114/-/DCSupplemental.
References
- 1.Bugg TD, Ahmad M, Hardiman EM, Rahmanpour R. Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep. 2011;28:1883–1896. doi: 10.1039/c1np00042j. [DOI] [PubMed] [Google Scholar]
- 2.Chen ZX, Riggs AD. DNA methylation and demethylation in mammals. J Biol Chem. 2011;286:18347–18353. doi: 10.1074/jbc.R110.205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 4.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glaeser S, Kämpfer P. The Family Sphinomonadaceae. In: Rosenberg E, DeLong E, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes: Alphaproteobacteria and Betaproteobacteria. Springer; Berlin: 2014. pp. 641–707. [Google Scholar]
- 6.Yoshikata T, et al. Three-component O-demethylase system essential for catabolism of a lignin-derived biphenyl compound in Sphingobium sp. strain SYK-6. Appl Environ Microbiol. 2014;80:7142–7153. doi: 10.1128/AEM.02236-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagata Y, et al. Complete genome sequence of the representative γ-hexachlorocyclohexane-degrading bacterium Sphingobium japonicum UT26. J Bacteriol. 2010;192:5852–5853. doi: 10.1128/JB.00961-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abe T, Masai E, Miyauchi K, Katayama Y, Fukuda M. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J Bacteriol. 2005;187:2030–2037. doi: 10.1128/JB.187.6.2030-2037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee U, Kumar R, Mahato NK, Khurana JP, Lal R. Draft genome sequence of Sphingobium sp. Strain HDIPO4, an avid degrader of hexachlorocyclohexane. Genome Announc. 2013;1:e00749-13. doi: 10.1128/genomeA.00749-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masai E, et al. A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J Bacteriol. 2004;186:2757–2765. doi: 10.1128/JB.186.9.2757-2765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HH, Kim DJ, Ahn HJ, Ha JY, Suh SW. Crystal structure of T-protein of the glycine cleavage system. Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia. J Biol Chem. 2004;279:50514–50523. doi: 10.1074/jbc.M409672200. [DOI] [PubMed] [Google Scholar]
- 12.Rosini E, D’Arrigo P, Pollegioni L. Demethylation of vanillic acid by recombinant LigM in a one-pot cofactor regeneration system. Catal Sci Technol. 2016;6:7729–7737. [Google Scholar]
- 13.Segel IH. 1993. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems (Wiley, New York) Wiley Classics Library Ed.
- 14.Bartling CM, Raetz CR. Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis. Biochemistry. 2008;47:5290–5302. doi: 10.1021/bi800240r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keshwani MM, Harris TK. Kinetic mechanism of fully activated S6K1 protein kinase. J Biol Chem. 2008;283:11972–11980. doi: 10.1074/jbc.M800114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopycki J, et al. Kinetic analysis of Arabidopsis glucosyltransferase UGT74B1 illustrates a general mechanism by which enzymes can escape product inhibition. Biochem J. 2013;450:37–46. doi: 10.1042/BJ20121403. [DOI] [PubMed] [Google Scholar]
- 17.Cleland WW. The kinetics of enzyme-catalyzed reactions with two or more substrates or products. III. Prediction of initial velocity and inhibition patterns by inspection. Biochim Biophys Acta. 1963;67:188–196. doi: 10.1016/0006-3002(63)91816-x. [DOI] [PubMed] [Google Scholar]
- 18.Coward JK, Slixz EP, Wu FY. Kinetic studies on catechol O-methyltransferase. Product inhibition and the nature of the catechol binding site. Biochemistry. 1973;12:2291–2297. doi: 10.1021/bi00736a017. [DOI] [PubMed] [Google Scholar]
- 19.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leys D, Basran J, Scrutton NS. Channelling and formation of ‘active’ formaldehyde in dimethylglycine oxidase. EMBO J. 2003;22:4038–4048. doi: 10.1093/emboj/cdg395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamura-Ikeda K, et al. Crystal structure of human T-protein of glycine cleavage system at 2.0 A resolution and its implication for understanding non-ketotic hyperglycinemia. J Mol Biol. 2005;351:1146–1159. doi: 10.1016/j.jmb.2005.06.056. [DOI] [PubMed] [Google Scholar]
- 22.Okamura-Ikeda K, et al. Crystal structure of aminomethyltransferase in complex with dihydrolipoyl-H-protein of the glycine cleavage system: Implications for recognition of lipoyl protein substrate, disease-related mutations, and reaction mechanism. J Biol Chem. 2010;285:18684–18692. doi: 10.1074/jbc.M110.110718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lokanath NK, Kuroishi C, Okazaki N, Kunishima N. Purification, crystallization and preliminary crystallographic analysis of the glycine-cleavage system component T-protein from Pyrococcus horikoshii OT3. Acta Crystallogr D Biol Crystallogr. 2004;60:1450–1452. doi: 10.1107/S0907444904012910. [DOI] [PubMed] [Google Scholar]
- 24.Dietmann S, et al. A fully automatic evolutionary classification of protein folds: Dali Domain Dictionary version 3. Nucleic Acids Res. 2001;29:55–57. doi: 10.1093/nar/29.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen ZW, Hassan-Abdulah A, Zhao G, Jorns MS, Mathews FS. Heterotetrameric sarcosine oxidase: Structure of a diflavin metalloenzyme at 1.85 A resolution. J Mol Biol. 2006;360:1000–1018. doi: 10.1016/j.jmb.2006.05.067. [DOI] [PubMed] [Google Scholar]
- 27.Schuller DJ, Reisch CR, Moran MA, Whitman WB, Lanzilotta WN. Structures of dimethylsulfoniopropionate-dependent demethylase from the marine organism Pelagabacter ubique. Protein Sci. 2012;21:289–298. doi: 10.1002/pro.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange H, Decina S, Crestini C. Oxidative upgrade of lignin - Recent routes reviewed. Eur Polym J. 2013;49:1151–1173. [Google Scholar]
- 30.Willis GH, McDowell LL. Pesticides in agricultural runoff and their effects on downstream water quality. Environ Toxicol Chem. 1982;1:267–279. [Google Scholar]
- 31.Francl MM, et al. Self‐consistent molecular orbital methods. XXIII. A polarization‐type basis set for second‐row elements. J Chem Phys. 1982;77:3654–3665. [Google Scholar]
- 32.Hariharan PC, Pople JA. The influence of polarization functions on molecular orbital hydrogenation energies. Theor Chim Acta. 1973;28:213–222. [Google Scholar]
- 33.Becke AD. Density‐functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 34.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B Condens Matter. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 35.Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ. Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J Phys Chem. 1994;98:11623–11627. [Google Scholar]
- 36.Vosko SH, Wilk L, Nusair M. Accurate spin-dependent electron liquid correlation energies for local spin density calculations: A critical analysis. Can J Phys. 1980;58:1200–1211. [Google Scholar]
- 37.Grimme S, Antony J, Ehrlich S, Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J Chem Phys. 2010;132:154104. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 38.Grimme S, Ehrlich S, Goerigk L. Effect of the damping function in dispersion corrected density functional theory. J Comput Chem. 2011;32:1456–1465. doi: 10.1002/jcc.21759. [DOI] [PubMed] [Google Scholar]
- 39.Hu X, Yang W. Accelerating self-consistent field convergence with the augmented Roothaan-Hall energy function. J Chem Phys. 2010;132:054109. doi: 10.1063/1.3304922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Beer TA, Berka K, Thornton JM, Laskowski RA. PDBsum additions. Nucleic Acids Res. 2014;42:D292–D296. doi: 10.1093/nar/gkt940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.