Abstract
Designing effective and accurate tools for identifying the functional and structural elements in a genome remains at the frontier of genome annotation owing to incompleteness and inaccuracy of the data, limitations in the computational models, and shifting paradigms in genomics, such as alternative splicing. We present a methodology for the automated annotation of genes and their alternatively spliced mRNA transcripts based on existing cDNA and protein sequence evidence from the same species or projected from a related species using syntenic mapping information. At the core of the method is the splice graph, a compact representation of a gene, its exons, introns, and alternatively spliced isoforms. The putative transcripts are enumerated from the graph and assigned confidence scores based on the strength of sequence evidence, and a subset of the high-scoring candidates are selected and promoted into the annotation. The method is highly selective, eliminating the unlikely candidates while retaining 98% of the high-quality mRNA evidence in well-formed transcripts, and produces annotation that is measurably more accurate than some evidence-based gene sets. The process is fast, accurate, and fully automated, and combines the traditionally distinct gene annotation and alternative splicing detection processes in a comprehensive and systematic way, thus considerably aiding in the ensuing manual curation efforts.
With the genome sequences of human and several other mammalian organisms now available (Lander et al. 2001; Venter et al. 2001; Kirkness et al. 2003; Mural et al. 2002; Waterston et al. 2002), identifying the elements they encode has become a task of immediate and utmost importance. Interpreting the raw sequence data into useful biological information, also known as genome annotation, is a complex process that requires the efficient integration of computational analyses, auxiliary biological data, and biological expertise. In particular, determining the location, structure, and function of protein-coding genes is an essential first step in any annotation project, as it holds the key to understanding the structures and functions of the proteins they encode, and can help focus the search for other functional elements such as SNPs and regulatory modules to specific regions of the genome. It is the structural component of gene annotation, or the identification of the exon-intron structure of genes and their alternatively spliced isoforms on the genome, that is the focus of this paper.
Large-scale gene annotation processes at the major genomics centers (Ensembl, Hubbard et al. 2002, Birney et al. 2004; UCSC Genome Browser database, Karolchik et al. 2003; Celera Genome Browser and Otto annotation system, Venter et al. 2001; NCBI Human Genome Resources, online documentation at http://www.ncbi.nlm.nih.gov/genome/guide/human/) typically combine the results from a variety of prediction and alignment programs in an automatic fashion, and in some cases the results are manually curated by expert annotators to detect and correct errors in the predicted gene structure and to identify alternative splicing events. The massive undertakings of manual curation in annotation projects such as NCBI's collection of reference sequences (RefSeq; http://www.ncbi.nlm.nih.gov/RefSeq/), the international Vertebrate Genome Annotation database (VEGA; http://vega.sanger.ac.uk/), and Celera's annotation jamborees (Adams et al. 2000; Rubin et al. 2000) and ongoing processes to improve the quality of the predicted genes (Venter et al. 2001; Mural et al. 2002) have involved hundreds of expert annotators. The more accurate the automated portion of the annotation process and the stronger the correlation between predictions and the underlying sequence evidence, the more effectively the ensuing manual curation and validation efforts will construct and validate gene models. Besides accuracy, an additional challenge that such pipelines have to withstand is the ever growing amount of new data (Karolchik et al. 2003). Therefore, there is an outstanding demand for annotation processes and related visualization, data mining, and storage resources that are fast, highly accurate, and flexible enough to allow the seamless incorporation of new sequence evidence as it becomes available in the databases.
Despite the large body of work in the area of gene finding (Claverie 1997; Rogic et al. 2001; Zhang 2002; and references therein) and the recent convergence in the estimates for the number of human genes (Lander et al. 2001; Venter et al. 2001; Pennisi 2003), gene finding is still an area under active development. Biases in each method, incomplete and inaccurate sequence data, and limitations of the computational gene model in capturing data-driven artifacts and biological phenomena such as overlapping genes and alternative splicing are only a few of the challenges that continue to face current methods. Ab initio prediction programs such as GenScan (Burge and Karlin 1997), FGenesH (Salamov and Solovyev 2000), Genie (Kulp et al. 1996), and GeneMark (Lukashin and Borodovski 1998) have reached high accuracy at predicting coding exons but are intrinsically ineffective at predicting exons in the 5′ and 3′ untranslated regions (UTRs), are confounded by overlapping genes, and produce a large number of false positives (Dunham et al. 1999; Rogic et al. 2001). Comparative methods based on alignments of protein and cDNA (EST, mRNA) sequences with genomic sequences (Procrustes, Gelfand et al. 1996; EstGenome, Mott 1997; sim4, Florea et al. 1998; GeneWise, Birney and Durbin 2000; Spidey, Wheelan et al. 2001) depend critically on the completeness and quality of sequences used. More recently, programs that inherently combine predictive and comparative clues (GenomeScan, Yeh et al. 2001; TwinScan, Korf et al. 2001) have been used with some success to improve the prediction accuracy but still suffer from some of the deficiencies of both approaches, such as the difficulty in identifying the 5′ and 3′ UTRs of a gene and the occasional merging of adjoining or overlapping genes. For accuracy reasons, or because of the restricted ability to cope with the tremendous amount of data generated by the sequencing projects, no single program is able to handle the complexity of the task of annotating the genome, and therefore annotation projects typically combine the results from a variety of computational tools within a weighting or integration process to create a complete perspective and overview of the gene content in the entire genome (Murakami and Tagaki 1998; Venter et al. 2001; Hubbard et al. 2002; Pavlovic et al. 2002; Birney et al. 2004). In addition, all of the individual approaches emerged in the context of predicting the correct sequence of exons in single-transcript genes and are not by themselves equipped to handle the prediction of mRNA transcripts for genes that exhibit alternative splicing. Thus, splice variation is typically added to the automatic annotation by human curators, many times as short, incomplete cDNA, which makes it difficult to incorporate and consolidate new evidence into the existing annotation. More than 50% of the genes of recently sequenced eukaryotic genomes (Mironov et al. 1999; Kan et al. 2001; Modrek et al. 2001; Zavolan et al. 2002) are now believed to undergo alternative splicing to generate different transcript and protein isoforms under different developmental, tissue-specific, and disease conditions, thus bringing a new set of challenges to gene prediction programs and the encompassing annotation processes.
To address the alternative splicing challenge, we designed a new computational process, called AIR (Annotation Integrated Resource), for predicting genes and their alternatively spliced mRNA transcripts based on genomic alignments of expressed DNA (EST, mRNA) and protein sequences, herein collectively called “evidence.” AIR was developed to support the Celera annotation of the rat genome Release R1, but it can be applied to any genome where sufficient cDNA sequences from that species, and/or gene annotations on a closely related species, exist. AIR introduces two new paradigms in gene annotation. First, it consolidates mRNA, EST, and protein sequence alignments on the genome into a collection of directed acyclic graphs called “splice graphs,” each representing a gene together with its exons (vertices), introns (arcs), and splice variants (paths). Candidate transcripts are systematically enumerated from the graph and assigned confidence scores based on the strength of the supporting evidence. The splice graph thus provides a mechanism to systematically enumerate all possible splice variants of a gene given the existing evidence, and then select the ones that are the most likely to exist in vivo, according to the support by the evidence. Since alternative splicing has become an integral part of characterizing the transcriptome, the splice graph has emerged as a candidate model for representing genes and their transcripts (Kan et al. 2001; Heber et al. 2002; Haas et al. 2003; Sugnet et al. 2004; Xing et al. 2004), but the combinatorial nature of the set of transcripts encoded in the graph has long been a deterrent to using it in annotation systems. The method we propose tackles this problem by scoring, evaluating, and selecting candidate transcripts based on their support by the evidence. The second novel feature in AIR is the use of precomputed whole-genome alignments to map the coordinates of exons and transcripts from a related species to the genome being annotated. This approach is generally faster, once the genome-to-genome alignment is computed, and exhibited higher accuracy than direct cross-species alignments in testing.
In the Methods section we describe in detail the two main components of AIR, namely the construction and selection of transcripts based on the splice graph model augmented with a scoring scheme, and the tracking of features between two related genomes guided by their whole-genome alignments. We then give a brief overview of the process and evaluate the intermediate results and the end annotation when AIR is applied to annotating the rat genome assembly Release 3.1 (Gibbs et al. 2004), in the Results. Further, we present considerations related to the applicability of the method to other model organisms.
The AIR model for gene annotation
AIR is an integrated methodology and software system for the annotation of genes and transcripts that combines a suite of evidence-collection and gene prediction tools. To predict genes and mRNA transcripts in eukaryotic genomes, the transcript building component of AIR consolidates genomic alignments of cDNA and protein sequences from the same species, and/or projections of annotated features from the genome of a closely related species, into a unified gene model. The underlying gene model is the “splice graph,” a directed acyclic graph that represents the gene's exons as vertices and its introns as arcs (Fig. 1). Splice variants can be read from the graph as end-to-end paths from a vertex with no incoming arcs (source) to a vertex with no outgoing arcs (sink). The splice graph may generate an artificially large number of candidate transcripts, many of them biologically improbable. To surmount the combinatorial nature of the candidate transcript set, a scoring scheme augments the model assigning each transcript a confidence score that is used as a primary filter to decide its suitability for annotation. A second heuristic filter is then applied to select a subset of the high-scoring transcripts with strong sequence evidence to be included into the annotation.
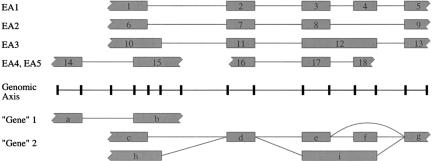
Figure 1.
Splice graph construction from genomic alignments of cDNA and/or protein sequences. A set of representative exon endpoints on the genome (tickmarks) is chosen by reconciling 5′ or 3′ exon endpoints in evidence alignments (EA1-EA5) within 20-bp windows. Exons in the graph (a-i) are produced by enumerating and merging all compatible combinations of evidence exons (1-18), i.e., in which one exon's spliced end is not contained within another exon. (For instance, exons 12 and 17 are incompatible.) Introns are added consistently with the evidence. The graph is refined into connected components (“gene 1” and “gene 2”), representing potentially different overlapping genes. Splice variants can be read from the graph as paths from a node with no incoming edges (source; a, c,h) to a node with no outgoing edges (sink; b, g).
The evidence collection component of AIR is used to generate alignments of cDNA sequences on the reference genome and to project gene features from a related genome, to serve as evidence for annotation. Same-species EST and full-length mRNA sequences are aligned to the target genome using a software tool called ESTmapper (L. Florea and B. Walenz, in prep.; referenced in Istrail et al. 2004). It uses an efficient genome-wide search to restrict the alignment area to a limited number of candidate genomic regions and then aligns the query cDNA to each of the candidate genomic regions with a local version of the sim4 algorithm (Florea et al. 1998). For each sequence AIR retains the single best alignment covering at least half of the cDNA sequence at 95% or more sequence identity. In addition, unspliced EST alignments are removed. To produce cross-species evidence, AIR projects annotated gene features from a closely related species onto the genome of reference using the one-to-one mapping and alignment of the two genomes precomputed with the A2Amapper tool (referenced in Istrail et al. 2004, Lippert et al. 2004) (Fig. 2). During the refinement stage following the initial projection, the projected exons and transcripts are verified to pass heuristic consistency rules and then endorsed or rejected accordingly, and some noncanonical exon boundaries are further adjusted to neighboring sites that have better consensus splice signal scores (Burge and Karlin 1997) or that harbor exon endpoints of same-species cDNA sequences.
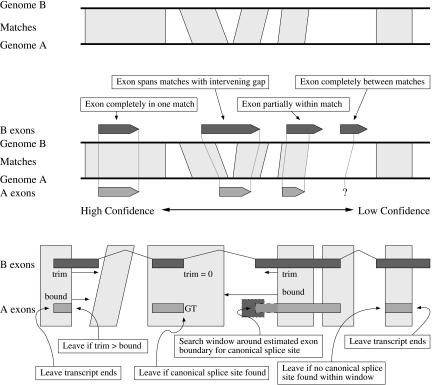
Figure 2.
Projection (middle) and refinement (bottom) of exons and transcripts from a closely related species (B) to a reference species (A) using genome-to-genome alignments (top). Following the initial projection (middle), exon endpoints may be adjusted to a better consensus splice site using a set of heuristics (bottom, left to right) that take into account the length of the unprojected end (trim), the distance to the next match (bound), the local profile of GenScan splice site scores, and existing same-species evidence.
To evaluate the overall performance of the AIR process we focused on two primary components. The first is the integrity of exon and transcript projections from a related genome to the genome being annotated using the genome-to-genome alignment. The second is the ability to produce a comprehensive and accurate annotation, or equivalently, a set of correctly formed genes and transcripts that are strongly supported by the evidence, using the splice graph model augmented with the scoring and selection scheme.
Results
Projection of orthologous gene features via synteny maps
To evaluate the accuracy and completeness of our exon and transcript projection process between the mouse and rat species, and thus the suitability of using the projected mouse evidence to annotate the rat genome, we focused on a set of mouse-rat mRNA sequence pairs identified as homologous based on NCBI's homoloGene annotation (Wheeler et al. 2004). While homologous pairs are not ideal to assess the preservation of exon features, since they may represent misclassified paralogs or different gene isoforms, this is the closest set one could find to true orthologs that is of significant magnitude. Besides paralogy and alternative splicing, other biological or computational factors can contribute to discrepancies between the projected mouse gene structure and the native alignment of its rat homolog that are not attributable to the projection method: deletion and insertion events between the lineages, incompleteness of the spliced alignments due to gaps in the assembled genome, and exon and transcript boundary artifacts introduced by the mRNA-genomic alignment program.
To eliminate such extrinsic factors, we constructed a benchmark set of exons conserved between mouse and rat as described in Methods, and measured the efficacy of our projection tools at exon, splice site, and base-pair resolution by comparing the projected mouse features against the native alignments of their rat orthologs. The multi-layered mRNA-mRNA, mRNA-genomic, and genomic-genomic mapping process to identify unambiguously orthologous features is illustrated in Supplemental Figure C2-F3.
The results of the comparison, summarized in Table 1, show that our method was able to project mouse exons end-to-end onto their rat orthologs 96% of the time exactly, and 97% when a 5-bp margin at the exon boundaries is allowed, while over 97% of their bases were correctly assigned, for a class of high-confidence transcript pairs. These values are slightly higher, about 97% and 98%, when only nonempty projections are considered (data not shown). Spliced exon ends are more accurately identified than marginal ones, at 0.97-0.98 sensitivity and 0.99-1.00 specificity, perhaps explained by transcript trimming artifacts when constructing our benchmark of orthologous exon pairs. Specificity values are consistently in the highest percentile, between 0.99-1.00 for all categories tested and with perfect 1.00 base-pair specificity in particular. These indicate that there is virtually no mouse sequence that is incorrectly located, and that the slightly lower exon and splice site specificity values are due to local inaccuracies at the exon boundaries. A second set of nearexact transcript pairs, whose exons paired via the mRNA-mRNA alignment differed by a margin of up to 10 bp, served as a consistency test. As expected, their statistics were slightly lower. Overall, the high sensitivity and specificity values observed provide confidence in our alignment and projection methods to produce a reliable evidence set that can be used as a supplement in annotating the rat genome.
Table 1.
Evaluation of synteny-based feature tracking between pairs of orthologous mouse and rat sequences
| Orthologs class | mRNA pairs/exons | Exon W = 0/5/10 bp | Splice site W = 0/5/10 bp | Base-pair |
|---|---|---|---|---|
| Exact | 1,842/14,460 | Sn: 0.96/0.97/0.97 | Sn: 0.97/0.98/0.98 | Sn: 0.97 |
| Sp: 0.98/0.99/0.99 | Sp: 0.99/1.00/1.00 | Sp: 1.00 | ||
| Near-exact | 282/1,872 | Sn: 0.91/0.93/0.93 | Sn: 0.92/0.92/0.93 | Sn: 0.92 |
| Sp: 0.93/0.95/0.96 | Sp: 0.94/0.95/0.96 | Sp: 0.95 |
Sensitivity (Sn) and specificity (Sp) values at exon, splice site, and base-pair levels were measured as described in Burset and Guigo (1996). Exon and splice site statistics were computed allowing for a variable window of W = 0, 5, and 10 bp. “Exact” orthologs have all orthologous exons exactly matching when projected via the cDNA–cDNA alignment (see Methods). “Near-exact” pairs have orthologous mouse and rat exon ends matching within 10 bp.
Annotating the rat genome
To test the ability of the AIR method to produce an accurate and complete set of gene and transcript predictions, we applied it to the task of annotating the rat genome assembly Release 3.1 (Gibbs et al. 2004; http://www.genome.ucsc.edu). At the time of the writing of this paper, the public databases contained 4619 rat full-length mRNA sequences in the RefSeq repository (Pruitt and Maglott 2001), an additional 10,592 sequences in the GenBank mRNA database (Benson et al. 2004), and 537,596 rat EST sequences from NCBI's dbEST data set (Boguski et al. 1993), representing roughly one tenth of the amount of similar resources available for annotating the human and mouse genomes. The mouse genome has been sequenced and annotated previously (Mural et al. 2002; Waterston et al. 2002) and is at a sufficiently close evolutionary distance (Makalowski and Boguski 1998) to allow mRNA alignments and gene annotations as additional cross-species resources for annotating genes and splice variants in rat.
Following mapping and initial quality-based filtering, AIR retained 95.8% (4427/4619) of rat RefSeq, 92.3% (9781/10,592) of rat GenBank mRNA, and 35.2% (189,366/537,596) of rat EST sequences, and 89.6% (14,737/16,455) projected mouse RefSeq and 80.6% (88,229/109,517) mouse GenBank mRNA transcripts. Unspliced EST alignments accounted for 82.5% (287,366) of the ESTs removed, while the other 17.5% (60,864) ESTs left unused could not be mapped at the established alignment criteria. Of these sets, 248 mouse RefSeq and 1682 mouse GenBank mRNA projections that covered <20% of the original sequence were further eliminated. An additional 318 rat and 363 mouse mRNA sequences that had intra-exon alignment gaps inconsistent with other evidence were excluded from the transcript building process. These cases may represent sequence polymorphisms that need to be manually reviewed by annotators.
Table 2 shows the retention rates for mouse exons and transcripts during the synteny-based projection and refinement stages. Overall, roughly 90% of the exons and 86% of the mouse transcripts mapped to the mouse genome were projected onto the rat genome as complete or partial features, with 8.3% of the exons being eliminated during the projection and 2.0% in the refinement stage. These numbers are largely consistent with those published in the literature (Gibbs et al. 2004). We conjecture that the most important factor in projection loss is the sensitivity of the genome-to-genome alignment as a result of algorithmic limitations. However, errors and gaps in the assembled genome sequence, causing exon loss and matches that are out of order or orientation with the rest of the transcript, as well as exon deletion and high mutation rates as a result of evolutionary change, play a part as well. The main cause for exon rejection at the refinement stage is the discrepancy between the original and projected exon sizes. For the set of 102,966 transcripts endorsed during the exon refinement step, only 3.3% (23,603) of the exons had the projection boundaries altered to produce a canonical splice site, and 2.0% (14,289) of the exons were noncanonical yet unchanged because the program failed to detect a more plausible splice junction. Another 4.6% (33,305) were rejected in either of the two stages, as a combined effect of evolutionary deletions and deficiencies in our processes. To estimate the extent of exon loss due to computational artifacts as opposed to evolutionary changes in the transcript structure, we performed an independent BLASTN (E = 2.0, default parameters) search of the 33,305 rejected exons on the genomic interval between the endorsed exons enclosing the gap, in the case of internal exons, or the 50-kb genomic interval starting at the projected end of the transcript, for marginal exons, requiring that the match orientation be consistent with that of the transcript. Only matches longer than 50% of the exon length or 50 bp were retained. This revealed 13,333 exons potentially missed. Based on this finding, we estimate the sensitivity of the combined exon projection and refinement in the endorsed transcripts to be 98.1%, corresponding to 683,039 successfully projected exons out of 696,372 exons endorsed or identified by BLASTN and thus estimated to be conserved. This value is consistent with the sensitivity observed in the analysis of mRNA orthologs earlier. Moreover, of the 13,333 missed exons only half (6549) are internal and thus generate an internal gap in the transcript structure, and some of them will be later regained in the splice graph from exons contributed by rat cDNA or protein alignments.
Table 2.
Projection rates of mouse RefSeq and GenBank mRNA transcripts and exons on the rat genome
| Mouse exons | Pct. (%) | Mouse transcripts | Pct. (%) | |
|---|---|---|---|---|
| Original | NA | NA | 125,972 | NA |
| Aligned on mouse | 761,241 | 100 | 119,280 | 100 |
| Projection stage | ||||
| Complete projections | 659,360 | 86.6 | 68,805 | 57.7 |
| Partial projections | 38,612 | 5.1 | 38,666 | 32.4 |
| Not projected | 64,062 | 8.4 | 11,809 | 9.9 |
| in unprojected transcripts | 38,991 | 5.1 | ||
| in projected transcripts | 25,071 | 3.3 | ||
| Total from complete and partial projections | 697,972 | 91.7 | 107,471 | 90.1 |
| Refinement stage | ||||
| Endorsed unaltered | 659,436 | 86.6 | 85,418 | 71.6 |
| canonical | 645,147 | 84.7 | ||
| non-canonical | 14,289 | 1.9 | ||
| Endorsed altered | 23,603 | 3.1 | 17,548 | 14.7 |
| by 1–10 bp | 19,145 | 2.5 | ||
| by >10 bp | 4,458 | 0.6 | ||
| Projected, not endorsed | 14,933 | 2.0 | 4,505 | 3.8 |
| Rejecteda | 40,004 | 5.3 | 4,505 | 3.8 |
| in rejected transcripts | 6,699 | 0.9 | ||
| in endorsed transcripts | 33,305 | 4.4 | ||
| Missedb (found with BLASTN) | 12,275 | 1.6 | NA | NA |
| Total endorsed projections | 683,039 | 89.7 | 102,966 | 86.3 |
NA = not applicable.
For the projection stage, “complete” exon projections are defined as having both ends of the exon contained in matches; the exon projection is then the entire interval between the projected endpoints. “Complete” transcript projections have all exons “complete”; “partial” transcripts have at least one complete or partially projected exon. “Complete” and “partial” transcripts were submitted for refinement in stage two. For the refinement stage, endorsed unaltered “canonical” projections had canonical splice signals and did not necessitate alteration, whereas “non-canonical” ones failed extension to a nearby consensus splice site. “Missed” projections were based on the BLASTN search (E = 2.0, default parameters) of all rejected exons from endorsed transcripts against the genomic interval between their adjacent exons, for internal rejected exons, or a 50,000-bp interval past the aligned end of the transcript, for marginal rejected exons, consistently with the orientation of the transcript. A threshold of 50% length or minimum 50-bp coverage of the exon was applied to the BLASTN results.
Includes the number of unprojected exons from partially projected transcripts
Counted from the number of rejected exons from endorsed transcripts
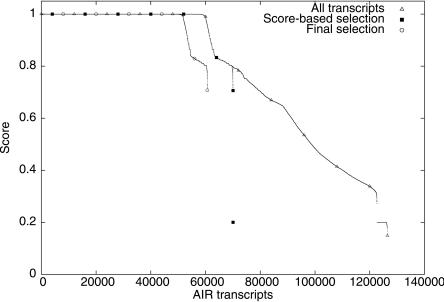
The selected mouse and rat evidence was used to generate an initial 45,040 genes (29,025 with multiple exons) and 126,440 candidate transcripts, of which 70,047 transcripts were retained following the score-based selection (Fig. 3). A subset of 38,598 genes (23,263 multiexon) and 60,683 transcripts were further selected and promoted into the final annotation following the second selection stage, based on the evidence type. Of these, 9700 (24.67%) genes had multiple splice variants reported. This is likely an underestimate given the paucity of rat-specific sequence data and the stringency of our selection criteria.
Figure 3.
Plots of AIR transcript scores at various levels of selection during the annotation of the rat genome.
The set of AIR transcripts predicted for chromosome 4 of the rat genome Release 3.1 and the associated files are available as Supplemental material A. The complete set of annotations can be obtained from https://panther.appliedbiosystems.com/publications.jsp.
Evaluation of AIR predictions
We evaluated the performance of the transcript building and selection process at two levels: structural correctness—the ability to produce transcripts with correct exon structure and strong correlation with the sequence evidence, and selectivity—the ability to discriminate between high-confidence transcripts and spurious exon combinations (Table 3). For the structural correctness assessment, in a first test we measured the extent of irregular splice junctions in the AIR transcript set and compared that against the Rat Genome Database (RGD) curated gene set (Table 3A). An irregular splice junction is one other than the generally accepted GT-AG, GC-AG, and AT-AC signals. The AIR transcripts at both selection stages scored significantly better than the RGD set. Of the 60,683 AIR rat transcripts, only 4935 (10.9% of the multiexon subset) had one or more irregular splice junctions, and only 539 (1.2%) had multiple irregular splice junctions, in comparison with 22.9% and 6.6%, respectively, for the RGD set of curated genes. The overall percentage of irregular splice junctions was only 1.6% for AIR, compared with 3.4% for RGD. These values are consistent across chromosomes and for the various stages in the transcript selection process. In a second test, we determined the portion of the transcript supported by alignments with exon-intron structure compatible with that of the transcript (Table 3B). Over 98.2% of the AIR transcripts had 50% or more of their bases contained in some evidence alignment, and 90.9% had over 95% coverage. Both splice junction and coverage statistics marked a sharp improvement after the score-based filter was applied, typically twofold and up to sevenfold in some categories, and remained at similar levels for the final data set (Table 3A,B), thus testifying for the discriminative power of the scoring scheme. For the selectivity assessment, we determined that while the overall rejection rate for candidate transcripts was 52.0% (65,757 out of 126,440 total), the rejection rates for rat mRNA and mouse transcript evidence were only 1.6% and 1.9%, respectively: 227 out of 13,890 rat RefSeq and GenBank mRNA sequences used to generate the splice graphs, and similarly 1871 out of 100,673 mouse RefSeq and GenBank mRNA transcripts were not included in the final transcript set (Table 3C). AIR was therefore able to retain essential evidence while eliminating most of the spurious constructions enumerated in a combinatorial fashion from the splice graph.
Table 3.
Accuracy evaluation for rat AIR predictions: splice junction (A) and coverage statistics (B), and selectivity and evidence retention (C)
| A
|
|
|
|
|
|
|
|---|---|---|---|---|---|---|
| Transcript set | Transcripts | Spliced transcripts | ≥1 irregular splice jcts. | ≥2 irregular splice jcts. | Splice junctions | Irregular splice jcts. |
| No selection | 126,440 | 110,425 | 24,756 (22.42%) | 9,182 (8.32%) | 1,629,754 | 36,096 (2.21%) |
| Score selection | 70,047 | 54,032 | 5,373 (9.94%) | 565 (1.05%) | 354,936 | 6,047 (1.70%) |
| Final | 60,683 | 45,348 | 4,935 (10.88%) | 539 (1.19%) | 339,302 | 5,579 (1.64%) |
| RGD, V = 12 | 3,642 | 2,452 | 837 (24.25%) | 249 (7.21%) | 33,028 | 1.228 (3.72%) |
| RGD, V = 30 | 3,642 | 3,444 | 788 (22.88%) | 226 (6.56%) | 32,880 | 1,124 (3.42%) |
| B
|
|
|
|
|
|
|
|
|---|---|---|---|---|---|---|---|
| Transcript set | Transcripts | ≥50% coverage | ≥80% coverage | ≥90% coverage | ≥95% coverage | ≥98% coverage | 100% coverage |
| No selection | 126,440 | 98,005 (77.5%) | 74,148 (59.6%) | 68,352 (54.1%) | 65,043 (51.4%) | 62,431 (49.4%) | 59,670 (47.2%) |
| Score selection | 70,047 | 67,485 (96.3%) | 64,339 (91.9%) | 62,554 (89.3%) | 61,102 (87.2%) | 59,710 (85.2%) | 57,820 (82.5%) |
| Final | 60,683 | 59,572 (98.2%) | 57,606 (94.9%) | 56,293 (92.8%) | 55,153 (90.9%) | 53,983 (89.0%) | 52,468 (86.5%) |
| C
|
|
|
|
|
|
|
|---|---|---|---|---|---|---|
| AIR genes | AIR transcripts | Rat RefSeq + GBmRNA | Lost | Mouse RefSeq + GBmRNA | Lost | |
| Total | N/A | N/A | 15,019 | N/A | 125,972 | N/A |
| Post mapping/tracking | N/A | N/A | 14,208 | 811 | 101,036 | 24,936 |
| Post alignment filtering | 45,040 | 126,440 | 13,890 | 318 | 100,673 | 611 |
| Post score-based selection | 45,040 | 70,047 | 13,663 | 227 | 98,802 | 1871 |
| Final | 38,598 | 60,683 | 13,663 | N/A | 98,802 | N/A |
An irregular splice junction is one other than GT–AG, GC–AG, and AT–AC. For this analysis, results from evaluating the set of RGD genes were included for comparison. Spurious introns less than V bp long were eliminated and their adjacent exons merged in the RGD genes.
Coverage is measured as the number of transcript bases contained in some evidence alignment.
Retention rates for mRNA evidence in the resulting AIR predictions at various stages of transcript selection are shown. AIR selects and retains essential evidence—13,663 of the 13,890 rat mRNA sequences (98.4%) and 98,802 of the 100,673 mouse mRNA transcripts (98.1%), whereas efficiently filtering unlikely candidate transcripts—65,757 of the 126,440 combinations encoded in the splice graphs are eliminated. N/A = not applicable.
For a more in-depth evaluation of the correctness of AIR gene structure predictions, we compared the AIR-predicted alternative transcripts against the 237 curated alignments of rat RefSeq mRNA sequences on chromosome 4, obtained from the RGD repository (see Methods, Table 4, and Supplemental material B). The AIR set of predictions includes 298 RefSeq alignments that we located on chromosome 4 and is, with a few exceptions, a superset of the RGD genes. Three types of measurements were performed. To assess the overall correctness of the AIR-predicted exon-intron structures based on their compatibility with the RGD reference alignments, a window of 10 bp was allowed when comparing both internal and marginal exon edges. The internal window was then set to 0 to determine the number of exactly identified splice junctions. Lastly, to measure the effects of splice junction and alignment inaccuracies on the open reading frame (ORF) and on the resulting protein product, we compared the two sets of ORFs and their protein translations. As shown in Table 4, AIR produced a structurally equivalent or extended transcript for most of the RGD genes (188/237), and a structurally compatible transcript that was included in the RGD alignment in another five cases. Moreover, in most of the remaining 41 cases in which there was a strong structural difference between the predicted AIR transcripts and the RGD genes, the discrepancy was caused by incorrect or incomplete mapping of the RGD genes (29 with missing exons, 2 potentially chimeric, 1 not confirmed by independent alignment, and 2 paralogs of genes located elsewhere). Of the remaining seven cases that can be attributed entirely to errors in the AIR process, three (NM_024130, NM_080394, NM_012822) were due to very short 3 and 4 bp long exons that were missed by the sim4 alignment program, one (NM_017111) was due to a chimeric construct between two consecutive matches on the chromosome, and other three (NM_013080, NM_030838, NM_031799) resulted from inaccuracies in the AIR splice junction location due to evidence biases or extension into a cDNA gap. The last three cases (NM_017206, NM_31650, NM_013225) were due jointly to errors in both methods, which missed different exons of the gene's alignment. At splice junction level, 97.5% of the RGD spliced junctions had exact AIR matches and another 1.2% matched within 5 bp. The reference set excluded the 38 spurious introns caused by missing exons or exon segments in the RGD gene set. Although AIR does not use ORF information in its prediction and relies entirely on the accuracy of the alignment and splice signal models, our comparison revealed 188 identical RGD-AIR ORF pairs and another five where the resulting protein had 1- to 2-amino-acid differences. In addition, in another 12 cases AIR ORFs and proteins completed and/or extended their RGD correspondents, by providing a more complete alignment or extending the RGD gene model using EST alignments and other evidence. In two cases where AIR produced partial reconstructions of the RGD genes the AIR proteins were truncated. The remaining 23 pairs exhibited ORF differences that caused the proteins to differ at the N-terminal or C-terminal end (21 pairs), or in two cases to be entirely different. Of these, a majority of 13 cases appeared to be due to frameshifts in the RGD sequence, six could be caused by errors in either or both methods and thus ambiguous, and only four could be attributed solely to frameshifts in the AIR sequence. Overall, the comparative assessment of the two sets showed AIR to have better splice junction statistics and fewer errors among those that caused discrepancies between the two sets of sequences and between the sets of their protein products, and is therefore measurably better than the RGD curated gene set in terms of gene and protein structure integrity. A complete list of the RGD genes and their corresponding AIR transcripts is included in Supplement B, and a detailed listing of the ORF and protein differences is given in Supplement C.
Table 4.
Structural comparison between the AIR predicted transcripts and the 237 RGD curated RefSeq genes on rat chromosome 4
| (A) Transcript structure comparison
|
|
|
||
|---|---|---|---|---|
| Categories | Cases | Comments | ||
| Compatible exon structures | ||||
| Equivalent | 35 | Same exon-intron structure | ||
| Extended | 153 | Structure extended at 5′ or 3′ end in AIR transcripts | ||
| Containing | 5 | Contains an AIR transcript as a substructure. AIR Match is short or misses marginal exons. | ||
| Strong structural differences | ||||
| Incomplete RGD gene mapping | 27 (+27/-0) | Missing single exons, parts of exons or groups of exons. | ||
| Incomplete AIR mapping | 3 (+0/-3) | Missing single (short) exons. Missing the alignment for the gene. | ||
| Incomplete incompatible RGD and AIR mappings | 3 (+3/-3) | Different missing exons produced by the two methods. | ||
| Different AIR and RGD mappings | 3 (+2/-1) | Chimeras and paralogs. Multiple RGD mappings for a gene; only best mapping selected by AIR. | ||
| AIR transcript building errors | 2 (+0/-2) | Evidence bias in splice junction detection. Extension into a cDNA gap. | ||
| No match | 3 (+3/-0) | Paralog; best match located elsewhere. Spurious RGD mapping, not confirmed. | ||
| (B) Splice junction comparison
|
|
|
|
|
|
|---|---|---|---|---|---|
| Window (W) | Introns | Exact | Partial | Weak | No match |
| W = 0 | 2118 | 2064 | 27 | 13 | 14 |
| W = 5 | 2118 | 2090 | 13 | 1 | 14 |
| W = 10 | 2118 | 2094 | 10 | 0 | 14 |
| (C) ORF comparison
|
|
|
||
|---|---|---|---|---|
| Categories | Cases | Comments | ||
| Compatible ORFs | ||||
| Identity | 188 | Identical ORFs and protein products. AIR transcript may be equal to, extend, complete or be contained in the RGD gene. | ||
| Near-identity | 5 | Minor (1-2 aa) differences in ORFs/proteins. AIR transcript misses short exon, or RGD and/or AIR choose alternative splice junctions. | ||
| Extension | 10 | RGD ORF/protein extended at 5′ (N-terminal) or 3′ (C-terminal) end in AIR transcripts. AIR transcript extends or completes RGD gene. | ||
| Completion | 2 | AIR ORF/protein fills-in internal gaps in (and possibly extends) RGD ORF/protein. RGD transcript missed (internal) exons. | ||
| Truncation | 2 | AIR protein is a portion of the RGD protein. AIR transcript was contained in the RGD gene. | ||
| Different ORFs | ||||
| Partial match | 4 (-4) | Frameshifts in AIR, caused by inaccurate splice junctions and/or exon ends. | ||
| 13 (+13) | Frameshifts in RGD, caused by missing exons, and inaccurate splice junctions and/or exon ends. | |||
| 4 (×4) | Different RGD and AIR ends for exons flanking alignment gaps caused by gaps in the genome | |||
| No match | 2 (×2) | Non-overlapping ORFs likely caused by frameshifts in both RGD and AIR; different exon ends flanking alignment gaps. | ||
Numbers in parentheses indicate the number of cases favoring (+) or disfavoring (-) AIR against RGD genes.
Introns are “exact” if both exon ends agree between the AIR and RGD annotations, within a W-bp window; “partial” if only one exon end agrees; and “weak” if the introns overlap strictly.
For each AIR and RGD sequence, the ORF is determined as the longest in-frame DNA stretch between a start (ATG) and a stop (TAA, TAG, or TGA) codon or, if no stop codon is encountered, the end of the sequence. (+) The AIR ORF is believed to be correct; (-) the AIR ORF is believed incorrect, but the RGD ORF is deemed correct; (×) either an ambiguous case, or both ORFs are likely to be erroneous.
One desirable quality of gene annotation is the completeness of the gene set. Although only 13,663 rat RefSeq and Gen-Bank mRNA sequences were explicitly represented in the final AIR annotation following the removal of 318 sequences during quality checks and 221 during the transcript selection process, a total of 13,836 mRNA sequences had significant BLASTN matches of at least 100 bp at 97% or better sequence identity with the AIR-predicted transcripts. The likely explanation is that the additional sequences were partially reconstructed from other types of evidence.
Effects of synteny-based feature transfer with varying evolutionary distances
The strong conservation of both sequence and gene structure between the mouse and rat genomes enabled us to use the mouse gene information effectively to predict genes and alternative splice forms in its relative. As the evolutionary distance between the species increases, limitations in the alignment tools as well as high mutation rates and exon deletion, insertion, shrinkage, and expansion events that change the gene's exon structure may limit the applicability of certain methods and require prior exploration. This is particularly relevant when considering comparative annotation of newly sequenced organisms, for which cDNA sequences are sparingly available.
To test the ability of our tools to correctly annotate a reference genome using gene information from another species, at varying degrees of relatedness, we focused on the extended cystic fibrosis transmembrane conductance regulator (CFTR) gene region, recently sequenced and comparatively analyzed in 13 species (baboon, cat, chicken, chimp, cow, dog, Fugu, human, mouse, pig, rat, tetraodon, zebrafish) (Thomas et al. 2003). This collection of sequences, alignments, genes, and comparative annotations of a region across multiple genomes offers a unique benchmark for comparative genomics methods and tools.
The human CFTR region (Thomas et al. 2003) extends over 1.8 Mb on chromosome 7 and contains 10 genes, one with two alternative transcripts (ST7), of which only 9 have been analyzed across the 12 genomes, with a total of 136 exons. The annotation for mouse and human was produced by aligning reference mRNA sequences from the NCBI RefSeq project to the genomic assembly sequence. For the remaining species where cDNA from that organism was not available, the RefSeq mRNA sequences for human and/or mouse were used, their coordinates on the native genomic region being projected onto the target species using BLASTZ alignments of the entire regions (Schwartz et al. 2003). This collection of genes was manually curated for structural integrity at the splice junction, exon structure, and protein translation levels and constitutes the gold standard for our tests.
We aligned the human CFTR region with the sequence of each of the other species using the method described by Istrail et al. (2004), with parameters tuned for cross-species alignments and for the length of the region, and we used the alignments to guide the projection of the annotated human exons and genes onto each of the remaining species. We then compared our results at exon and nucleotide level against the published mRNA annotated features (Table 5 and Supplemental material C). We further extended the analysis to two programs that illustrate a different method for mapping cross-species evidence, by directly aligning the cDNA sequences to the genome: sim4 (Florea et al. 1998), a specialized tool for aligning cDNA and genomic sequences, modified to better suit the needs of cross-species comparisons, and BLASTZ (Schwartz et al. 2003), one of the reference local alignment programs. These three sets of analyses offered us the opportunity to directly assess each method's advantages and limitations.
Table 5.
Performance of three methods for mapping cross-species evidence with varying evolutionary distances, exemplified at the CFTR locus in human and 12 other species
| Exon (full)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| V = 0
|
V = 10
|
Exon (partial)
|
Splice junctions
|
Bp
|
|||||||
| Species | Toola | Sn/Sp | Sn/Sp | Sn/Sp | Sn/Sp | Sn/Sp | |||||
| Baboon | AIR | 0.985 | 0.971 | 0.993 | 0.978 | 1.000 | 0.985 | 1.000 | 0.984 | 1.000 | 0.996 |
| BLASTZ | 0.015 | 0.015 | 0.739 | 0.728 | 1.000 | 0.985 | 0.073 | 0.071 | 0.997 | 0.945 | |
| sim4 | 0.955 | 0.941 | 0.985 | 0.971 | 1.000 | 0.985 | 0.992 | 0.976 | 0.978 | 0.995 | |
| Cat | AIR | 0.963 | 0.970 | 0.963 | 0.970 | 1.000 | 1.000 | 0.976 | 0.984 | 0.999 | 0.985 |
| BLASTZ | 0.030 | 0.030 | 0.726 | 0.736 | 0.985 | 0.985 | 0.080 | 0.080 | 0.994 | 0.945 | |
| sim4 | 0.881 | 0.804 | 0.896 | 0.818 | 1.000 | 0.985 | 0.984 | 0.891 | 0.954 | 0.997 | |
| Chicken | AIR | 0.359 | 0.583 | 0.359 | 0.583 | 0.615 | 1.000 | 0.455 | 0.714 | 0.297 | 0.992 |
| BLASTZ | 0 | 0 | 0.590 | 0.622 | 0.949 | 1.000 | 0.030 | 0.032 | 0.744 | 0.662 | |
| sim4 | 0.359 | 0.368 | 0.436 | 0.447 | 0.769 | 0.857 | 0.545 | 0.562 | 0.420 | 0.964 | |
| Chimp | AIR | 0.991 | 0.974 | 0.991 | 0.974 | 1.000 | 0.982 | 1.000 | 0.981 | 1.000 | 0.981 |
| BLASTZ | 0.027 | 0.026 | 0.750 | 0.718 | 1.000 | 0.957 | 0.078 | 0.075 | 0.997 | 0.942 | |
| sim4 | 0.973 | 0.948 | 0.991 | 0.965 | 1.000 | 0.974 | 1.000 | 0.971 | 1.000 | 0.980 | |
| Cow | AIR | 0.924 | 0.924 | 0.955 | 0.955 | 1.000 | 1.000 | 0.959 | 0.959 | 0.996 | 0.994 |
| BLASTZ | 0.015 | 0.015 | 0.758 | 0.752 | 0.992 | 0.992 | 0.082 | 0.081 | 0.996 | 0.952 | |
| sim4 | 0.848 | 0.762 | 0.894 | 0.803 | 1.000 | 0.978 | 0.967 | 0.861 | 0.904 | 0.991 | |
| Dog | AIR | 0.922 | 0.905 | 0.942 | 0.924 | 1.000 | 0.981 | 0.958 | 0.938 | 0.996 | 0.892 |
| BLASTZ | 0.019 | 0.019 | 0.738 | 0.717 | 0.990 | 0.971 | 0.084 | 0.082 | 0.997 | 0.891 | |
| sim4 | 0.835 | 0.723 | 0.883 | 0.765 | 1.000 | 0.920 | 0.947 | 0.811 | 0.895 | 0.940 | |
| Fugu | AIR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BLASTZ | 0.030 | 0.050 | 0.396 | 0.662 | 0.612 | 1.000 | 0.081 | 0.143 | 0.663 | 0.939 | |
| sim4 | 0.073 | 0.129 | 0.173 | 0.306 | 0.473 | 0.929 | 0.230 | 0.418 | 0.203 | 0.977 | |
| Mouse | AIR | 0.838 | 0.838 | 0.875 | 0.875 | 0.993 | 0.993 | 0.944 | 0.944 | 0.943 | 0.957 |
| BLASTZ | 0.015 | 0.015 | 0.625 | 0.620 | 0.978 | 0.993 | 0.040 | 0.039 | 0.931 | 0.907 | |
| sim4 | 0.713 | 0.602 | 0.765 | 0.646 | 0.985 | 0.950 | 0.889 | 0.742 | 0.757 | 0.967 | |
| Pig | AIR | 0.942 | 0.942 | 0.951 | 0.951 | 1.000 | 1.000 | 0.968 | 0.968 | 0.997 | 0.998 |
| BLASTZ | 0.029 | 0.029 | 0.718 | 0.705 | 0.990 | 0.981 | 0.053 | 0.052 | 0.995 | 0.953 | |
| sim4 | 0.903 | 0.795 | 0.932 | 0.821 | 1.000 | 0.972 | 0.979 | 0.853 | 0.913 | 0.995 | |
| Rat | AIR | 0.824 | 0.830 | 0.853 | 0.859 | 0.985 | 0.993 | 0.913 | 0.920 | 0.927 | 0.952 |
| BLASTZ | 0.029 | 0.029 | 0.610 | 0.610 | 0.971 | 0.992 | 0.048 | 0.048 | 0.920 | 0.913 | |
| sim4 | 0.691 | 0.573 | 0.779 | 0.646 | 0.993 | 0.978 | 0.897 | 0.734 | 0.735 | 0.970 | |
| Tetraodon | AIR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BLASTZ | 0.060 | 0.099 | 0.448 | 0.741 | 0.604 | 0.976 | 0.097 | 0.167 | 0.664 | 0.957 | |
| sim4 | 0.105 | 0.186 | 0.177 | 0.314 | 0.403 | 0.833 | 0.219 | 0.403 | 0.208 | 0.957 | |
| Zfish | AIR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BLASTZ | 0 | 0 | 0.395 | 0.586 | 0.814 | 1.000 | 0.054 | 0.087 | 0.785 | 0.868 | |
| sim4 | 0.053 | 0.080 | 0.263 | 0.400 | 0.658 | 1.000 | 0.176 | 0.286 | 0.437 | 0.976 | |
The evaluation was performed at exon, splice junction, and base-pair level. A full-exon match requires that both exon boundaries agree within V bp, whereas a partial-exon match only requires that the exons overlap. Sensitivity is measured as Sn = TP/(TP + FN), specificity as Sp = TP/(TP + FP) (Burset and Guigo 1996).
(AIR) Synteny-based feature tracking; (BLASTZ) direct alignment of the cDNA sequence and the genomic region; (sim4) splice-sensitive cDNA-to-genome alignment
Table 5 shows the sensitivity and specificity of each method when mapping the human annotated features to each of the other species, measured at exon, splice junction, and base-pair levels. AIR performs consistently better than each of the other methods in each category for mammalian comparisons; however, its performance drops dramatically for comparisons with different taxa. In contrast, BLASTZ achieves high sensitivity at base-pair level by capturing similarities as distant as those between human and chicken or fishes, but the gene models it produces suffer from its lack of a splice model. Sim4 cross-species alignments provide a consistent middle-view in terms of the accuracy of the gene structure as measured by the full exons category, which is a combined measure of splice junction and base-pair performance, and are close contenders for splice junction accuracy for many of the comparisons. Noticeably, its specificity values are slightly lower than the sensitivity scores, owing to the spurious introns caused by short insertion and deletion events between the species or by areas of low sequence similarity, particularly in the 5′ and 3′ noncoding exons. Thus, by inspecting the individual results it becomes apparent that a projectionbased method such as AIR, which incorporates prior knowledge about the exon model in the original transcript, can better preserve the integrity of the exons and transcripts, but its performance depends critically on the sensitivity of the underlying genome-to-genome alignment.
To answer the question of whether AIR can be successfully applied to annotate other genomic systems, we note that our method is able to capture exon sequences conserved between human and any of the other mammalian species with very high sensitivity and specificity, typically in the 0.98-0.99 range, but its performance degrades considerably for chicken and the fish species. No pairwise alignments were produced for any of the three fish species, due to the lack of perfectly conserved 20-mers to anchor the local alignment of sequences. Alignments with mouse and rat were less sensitive than for the other mammalian species despite the closer evolutionary distance, as a direct consequence of the high deletion rate in the rodent branch. This result is consistent with the observations of Thomas et al. (2003). These indicate empirically that the range of applicability for our alignment and projection tools extends to pairs of organisms that exhibit divergence rates lower or similar to those of human and mouse/rat, but may be limited beyond this range and likely does not include comparisons between mammals and other vertebrates. The active range can be increased in the future by using different match detection and selection techniques, such as the space seed introduced by PatternHunter (Ma et al. 2002) and implemented in BLASTZ (Schwartz et al. 2003).
Discussion
Conventional gene finding methods and annotation systems have traditionally used as their gene model a linear exon sequence (Claverie 1997; Rogic et al. 2001; Zhang 2002; and references therein). It was not until the unprecedented accumulation of genomic and cDNA sequences in the public databases, brought about by the sequencing of the human genome (Lander et al. 2001; Venter et al. 2001), that scientists gleaned into the true extent of alternative splicing and the identification of alternatively spliced forms became an integral part of the eukaryotic genome annotation. With the abundance of putative splice variants that can be constructed from fragmentary EST and mRNA evidence, identifying good candidates for annotation or in vivo validation is a formidable task. Typically, splice variation is added by curators in the aftermath of automatic annotation, many times as short, incomplete cDNA. This partiality of splice forms and the inconsistency of the process with the overall annotation framework make it hard to incorporate and consolidate new evidence into the existing genes and transcripts. The splice graph emerged as a natural representation of the alternative splicing events within a gene (Heber et al. 2002; Sugnet et al. 2004), but the combinatorial nature of the candidate splice variants it encodes has been a deterrent to using it as a medium for annotation.
The method we propose, AIR, consolidates evidence around a splice graph model and proposes a novel solution to tackle the complexity of the candidate transcript pool. Splice variants enumerated from the graph receive confidence scores based on properties of the evidence such as alignment quality, coverage, and fragmentation that human experts have identified as fundamental in their gene curation efforts. The scoring scheme is intuitive, modular, and orthogonal, combining several score components each measuring an independent set of features. It is relatively easy to compute and flexible enough to allow the experts' intervention in the transcript selection process, either by filtering candidate transcripts by component scores or by modifying the score weights according to their own perception of the scores' relative importance. It is also extensible and can serve as a template for incorporating additional score components, such as intron scores and penalties for putative unspliced introns. One possible extension would be to include the type and abundance of evidence in the scoring scheme, rather than use it as a filter subsequent to the scoring process. Lastly, such an annotation system lends itself naturally to an incremental design. New evidence that changes only the scores of existing transcripts and may promote new splice forms, but does not reshape the splice graph entirely, can be seamlessly incorporated into the existing annotation. All of these properties are desirable of large-scale annotation systems that are updated frequently and may require post-processing by manual curation.
As a gene model, the splice graph can have expressive and sometimes practical limitations. For instance, it cannot represent certain isoforms with alternative 5′ or 3′ ends in which one form is subsumed by the other. Post-processing of the gene's tails based on EST clone information to identify potential polyadenylation sites with programs such as PASS (Kan et al. 2001) could partially overcome this problem. Its accuracy also depends critically on the correctness and robustness of the underlying data generation tools, such as the cDNA-genomic alignment program and the synteny-based feature tracking, since effects are amplified in a combinatorial fashion: Spurious exons or splice junctions can double the number of candidate splice forms.
The synteny-based exon and transcript projection module of AIR is instrumental for adding in cross-species evidence as a supplement for annotating newly sequenced organisms or regions for which there is limited same-species cDNA and protein sequence available. Methods that align cDNA and genomic sequences to identify structurally correct exons and introns (sim4, Florea et al. 1998; Spidey, Wheelan et al. 2001) perform increasingly poorly as sequence similarity falls below that expected for EST sequences, for which they were tuned, or at roughly 90% sequence identity. Feature tracking via genome-to-genome alignments is intuitively more accurate than cross-species cDNA-genomic alignments, as it is able to span over gaps or portions of weaker similarity as long as it is strongly anchored in the adjacent portions of the exon and transcript. The splice junctions are also more precisely identified, being mirrored by direct mapping between the two genomes. In addition, the process can be significantly faster than cross-species cDNA-genomic alignments for mapping large data sets, once the genome-to-genome alignment is generated. The primary limitation to such alignment-based feature tracking methods is the sensitivity of the alignment. Our tools appear suitable to align human with mammalian species as divergent as mouse and rat, but their sensitivity drops dramatically for outgroups such as birds or fishes. To complement the projection method in such cases, the direct alignment of cDNA sequences to the genome, run either independently or combined with the projection method, can help identify additional exons and transcripts.
One annotation feature that is missing from AIR but can be incorporated as a post-processing step is the ability to identify pseudogenes and paralogs for which there is no direct sequence evidence. Indeed, the one-to-one mapping of the two genomes forces every mouse transcript to the “true” location of its ortholog on the reference genome, and similarly, selecting the best spliced alignment for every EST and mRNA sequence places that sequence at its “true” genomic location (n.b., in a very small number of cases, multiple spliced alignments indistinguishable by the criteria of coverage, sequence identity, and number of exons may be reported as “best matches”). One possible solution to this problem is to search for matches of predicted AIR transcripts in the rest of the rat genome.
From a computational perspective, AIR is a highly efficient annotation system. The entire automated process for annotating the rat genome, including the generation of genome-to-genome mouse-rat alignment and evidence, took less than 150 CPU hours on a 1.3 GHz IBM POWER4 processor: 80 h to generate the rat-mouse genome map, 45 h to align rat and mouse cDNA sequences on their native genomes,6hto project and refine mouse transcript models on the rat genome, and 6 h to build and select rat transcripts.
AIR is a fully automated, fast, and accurate computational annotation process that combines gene annotation and alternative splicing detection by consolidating evidence in a novel and systematic way, thus considerably facilitating the subsequent efforts with manual curation. As demonstrated in this study by annotating the rat genome, AIR can be universally applied to any genome where sufficient expressed DNA sequences and/or annotations on a closely related species exist, and can prove a valuable resource for the annotation of splice variants and genes in new sequences.
Methods
Sequence data
For the annotation of the rat genome, rat and mouse mRNA and EST sequences were extracted from the NCBI repositories: RefSeq (Pruitt and Maglott 2001), dbEST (Boguski et al. 1993), and Gen-Bank mRNA (Benson et al. 2004). The rat genome assembly Release 3.1 (Gibbs et al. 2004; Feb. 2003) and mouse genome assembly Build 30 (Waterston et al. 2002; Jun. 2003) were down-loaded from the University of California Santa Cruz Web site (http://www.genome.ucsc.edu). Genomic coordinates of curated rat RefSeq genes on the rat genome were retrieved from the Rat Genome Database (RGD) Web Site, at the Medical College of Wisconsin, Milwaukee, Wisconsin (http://rgd.mcw.edu, April 1st 2004). Some gene records contained a number of very short spurious introns that we found to be associated with alignment gaps. Consequently, we merged consecutive exons separated by a small number W of bases to create a data set that is compatible with our exon-centered view of the annotation. Data sets constructed in this way for W = 12, 20 were used to evaluate comparatively the AIR and RGD splice junction correctness in the Results. For the structural correctness evaluation of AIR transcripts against the RGD genes, we used W = 12 after a preliminary manual inspection to verify the veracity of exon merges against independent sim4 alignments.
Same-species alignments
EST and mRNA sequences are aligned to their native genome using a high-performance alignment program called ESTmapper (L. Florea and B. Walenz, in prep.; referenced in Istrail et al. 2004). It uses a hash-index of 20-mers in the genome to quickly locate areas of the genome likely to contain the query, then invokes the core of the sim4 algorithm (Florea et al. 1998) to produce a spliced alignment between the query and each genomic region selected. Only alignments that contain 50% or more of the cDNA sequence at 95% or more sequence identity are considered significant. To avoid contamination from paralogs, only the single best genomic alignment for each sequence is chosen, based on a measure combining sequence identity, length of the alignment, and the number of exons. Further, remaining unspliced EST alignments are removed, as they are a potential source for genomic contamination. When protein evidence is used, GeneWise (Birney and Durbin 2000) protein-genomic spliced alignments are filtered using the same coverage and sequence identity criteria as for mRNA sequences but allowing for multiple matches per protein.
Cross-species feature projections
Annotations of gene features on the related genome, such as mRNA-genomic sequence alignments or predicted transcripts, are projected onto the genome to be annotated (reference) in three steps (Fig. 2). Step one generates a whole-genome alignment and one-to-one mapping between the two genomes, starting from exact 20-mer matches that are unique in both sequences, selecting a reliable subset of these as anchors, and then applying a more lenient local alignment program on the intervals between anchors (referenced in Istrail et al. 2004; Lippert et al. 2004). The result is a set of nonoverlapping runs of matches consistent in order and orientation. Regions thus delimited by the runs form conserved syntenic blocks between the two genomes. Step two projects the exon coordinates from the related genome to the reference genome, using the precomputed set of matches. When an exon boundary cannot be found in a match but a portion of the exon can be used to anchor the projection, the length of the unprojected end (“trim”) and distance to the nearest match (“bound”) are reported and later used to adjust the projection. The trim is 0 when the exon boundary can be mapped. Step three seeks to refine the projected splice junctions using a combination of methods, including GenScan exon and splice junction predictions and scores (Burge and Karlin 1997) and the proximity to same-species cDNA alignments on the reference genome, and then uses a set of selection criteria to endorse features for use as evidence in the annotation. Partial exons are left truncated at the last projected base if the trim exceeds the bound, thus supporting a deletion in the exon in the reference genome. Alternatively, some exon boundaries are relocated to the best splice site within the vicinity of the estimated exon end inferred by adding the trim to the end of projection (the vicinity is defined as 12 bp + trim × 20% on each side of the estimated end). In the end, exons shorter than 6 bp, or those that shrink or grow by more than the maximum of 30 bp and 30% of their original length, are rejected.
Filtering of alignment and projection data
cDNA and protein alignments and cross-species projections are further inspected and filtered for quality before being incorporated into the transcript building module. Marginal exons and exons surrounding an alignment gap that are shorter than 25 bp or have less than 90% sequence identity are removed from the alignments. Moreover, EST alignments with internal gaps or ambiguous strand assignments, and projections or alignments where internal exon gaps are contradicted by exons in some other evidence, are eliminated from the set.
Transcript building
Starting from a set of nonredundant spliced cDNA and/or protein sequence alignments on the genome, AIR builds a collection of connected splice graphs, each representing a gene, in four steps (Fig. 1). Step one clusters overlapping EST, mRNA, and protein genomic alignments and projections located on the same strand. Within each cluster, step two identifies a set of representative exon boundaries by grouping all candidate splice junctions from evidence alignments in intervals 20 bp or shorter using hierarchical clustering (Mirkin 1996), then choosing the best scoring exon boundary position within that interval. As a consequence, splice variation shorter than 20 bp at the 5′ and 3′ ends of exons will not be apparent. For a given position, its splice score is a weighted combination of the fraction of supporting alignments among those for the entire interval and a consensus splice signal score computed with the method described by Burge and Karlin (1997). Step three generates the splice graph. All partial evidence exons are artificially extended to the next exon boundary to reduce the computational task of exon assembly by manipulating transcripts and exons at the resolution of exon blocks rather than nucleotides. The exons (vertices) in the graph are assembled from compatible combinations of overlapping or adjoining exons in the evidence alignments. A compatible combination is one in which an exon's spliced end is not contained within another exon. Introns (arcs) are added to connect the vertices, in a manner consistent with the evidence, the type of arc indicating whether it is a regular intron or a cDNA gap. Step four refines some clusters into connected components representing overlapping genes or disconnected gene fragments.
Transcript scoring
The splice graph model provides a framework to comprehensively enumerate potential splice variants but may encode many biologically improbable exon combinations. To prioritize those that are more likely to exist in vivo given the existing data, we assign each exon and candidate transcript a confidence score between 0 and 1 based on its support by evidence, as described below.
Exon scoring
Exon scores combine three independent components. The alignment quality (AQ) score reflects the overall quality of the evidence alignments used to generate the exon. Each nucleotide in the exon is assigned the maximum percent sequence identity value over all alignments in which it is contained, and the AQ score is the average of values over the aligned exon bases. The internal gap (IG) score measures the exon's coverage with evidence alignments. Virtually extending partial evidence exons to full exon blocks can produce some exons with internal evidence gaps, or “hollow.” Because exons as large as 17 kb have been annotated, we chose to penalize hollow exons in our scoring, rather than split them. The score function IG(k) = e-k/270, chosen to penalize an internal gap of size k, is the least mean square error (LMSE) approximation with an exponential function of the empirical probability that two exonic positions separated by k bp belong to the same exon: P(exon(x1) = exon(x2) | isCoding(x1) and isCoding(x2) and | x1-x2 | = k), approximated from the curated set of Celera human transcripts for the genome assembly Release R26. This function is easy to compute and has the desirable property that it extends naturally to a series of gaps: IG(k1+k2) = IG(k1) × IG(k2), thus mimicking the probability of independent events. The last score component, the fragmentation (FG) score, favors exons that are obtained from full evidence exons versus those assembled from multiple partial exons. It is defined as the inverse of the minimum number of evidence alignments necessary to cover the aligned portion of the exon. The cumulative exon score is finally computed as: score(X) = sqrt [AQ(x)2 + IG(X)2 + FG(X)2].
Splice variant (transcript) scoring
Splice variant scores combine five components, measuring independent qualities of the evidence for the transcript. For an alignment to be considered evidence at this step, its exon-intron structure must be entirely contained within and compatible with that of the transcript. Gaps in cross-species projections are considered compatible with exons in the splice variant if the rest of the exon-intron structures are consistent. The mapping quality (MQ) score reflects the overall goodness of its exons, and is calculated as the geometric average of the exon scores. The exon coverage (EC) is the fraction of the splice variant's exons that are contained in some evidence alignment. If no (compatible) evidence alignment exists, the coverage score is 0. The fragmentation (FG) score is akin to that defined for exons, and is computed as the inverse of the minimum number of evidence alignments necessary to cover all exons inherited from compatible evidence. The longest evidence (LE) score measures the continuity of evidence, and is the fraction of the transcript's exons contained in the evidence alignment with the largest number of exons. Lastly, the intron consistency (IC) score measures the overall agreement among predicted intron orientations and is defined as the fraction of introns that are consistent with the orientation predicted for the transcript. The orientation of individual introns is determined by the number of nucleotides matching the consensus splice signal GT-AG (CT-AC). The cumulative score of a splice variant is then computed with the weighted formula: Score(SV) = sqrt [θ1 MQ(SV)2 + θ2 EC(SV)2 + θ3 FG(SV)2 + θ4 LE(SV)2 + θ5 IC(SV)2], with ∑i = 1,5θi = 1. The parameters θi can be viewed as the relative importance attributed to the various components by an expert curator. In our fully automated process, they correspond to the scores' discriminative power in a test that separates known, high-confidence examples from low-confidence ones, and they were trained on an independent set of manually curated Celera human transcripts as described below.
Scoring scheme calibration
We calibrated the weights in the scoring scheme using as reference the set of 44,448 transcripts predicted for the Celera human genome assembly Release R26 (R26). This set consists of transcripts originally generated with an independent tool, Celera's Otto annotation system (Venter et al. 2001), and later curated and augmented by expert annotators. We used AIR to annotate another Celera human genome assembly, Release 27 (R27), using genomic alignments of human cDNA sequences from NCBI's dbEST and RefSeq repositories. To generate a compatible reference set, the R26 reference transcripts were aligned uniquely to the R27 genome and were assigned “proxies” from among the AIR-predicted candidate splice variants. Proxies were compatible extensions of the reference transcripts with the smallest number of extraneous exons. We assigned 25,443 such proxies. To calibrate the scoring scheme, we varied the values of the θi parameters in 0.01 increments under the constraint Σi = 1,5θi = 1 and determined the set of parameters {θ0 i}i = {θ1(MQ) = 0.02, θ2(EC) = 0.42, θ3(FG) = 0.38, θ4(LE) = 0.16, θ5(IC) = 0.02} and the score threshold 0.748 that minimized the number of candidate transcripts with scores ≥80% of the proxies. In general, once the optimal set of parameters is selected, the score threshold can be chosen dynamically given the set of scores generated for a particular application, depending on the level of stringency desired. Throughout several applications of the method to different model genomes we found that the threshold 0.8 was consistently suitable for separating transcripts, marked by a sharp drop in scores in its vicinity, and it was therefore applied to the annotation in this paper (Fig. 3).
Transcript selection
AIR selects a set of high-scoring candidate transcripts and then applies heuristic rules based on the type and abundance of evidence to select a subset of transcripts from the high-scoring group. Specifically, for each gene a small number of transcripts scoring greater than or equal to the threshold 0.8 or, if no such transcript exists, scoring in the top 0.05 score interval for that gene (e.g., 0.75-0.8) are selected. Transcripts with fewer than three ESTs and no other type of evidence are then eliminated if their gene overlaps with another at that locus, or if they have a noncanonical splice junction. The remaining transcripts are promoted into the annotation.
Construction of the benchmark orthologous exon set
Starting from the set of 15,245 mouse-rat mRNA sequence pairs identified as homologous in the homoloGene database (Wheeler et al. 2004), we discarded those pairs where either component was a computationally predicted RefSeq gene (12,008 pairs) or did not map to the genome or had multiple genomic matches (56 pairs), or where the portion of the sequence in the spliced alignment was noncontiguous (574 pairs), thus retaining 2607 mouse-rat sequence pairs. For each pair we identified the “core” conserved interval spanned by local alignments produced with BLASTZ (Schwartz et al. 2003) and by sim4 alignments of both mRNA sequences to their respective genomes. Analyzing the cores only, we discarded 481 pairs whose exon structure as determined by the spliced genomic alignments did not follow the Smith-Waterman (Smith and Waterman 1981) mRNA-mRNA sequence alignment to within 10 bp of each exon boundary. Of these, 76 appeared to be due to exon splitting and 39 to exon skipping events. This process yielded 2124 mouse-rat orthologous pairs: In 1842 of the pairs the exon structures matched each other exactly, and in 282 the match was within the 10-bp margin. The resulting set consisted of 17,332 exon pairs, which we used to assess the accuracy of our transcript and exon projection process.
Alignment data for the CFTR region analysis
Sequence and annotation data for the CFTR region in 13 species (baboon, cat, chicken, chimp, cow, dog, Fugu, human, mouse, pig, rat, tetraodon, zebrafish) were extracted from Thomas et al. (2003). To generate projection and alignment data for comparisons, annotated human mRNA features were projected onto the other 12 regions via the pairwise alignments between human and each of the other species using the AIR tools. In parallel, sequences extracted according to these annotations were aligned to the 12 genomic regions using the program BLASTZ (Schwartz et al. 2003) with the parameters set as in Thomas et al. (2003) and a version of the sim4 program (Florea et al. 1998) tuned for higher sensitivity as required for cross-species alignments (mismatch = -3, F = 1.0, P = 0.3). BLASTZ local alignments were split at genomic gaps of 50 bp or longer, to create exon-intron gene structures as needed for comparisons. When the cDNA matched both strands of the genomic sequence, as for the gene CAV2, the longer alignment was chosen. For sim4, marginal exons shorter than 35 bp located at 100 Kb or more from the previous exon were filtered as potentially spurious, and alignments shorter than 100 bp after applying the marginal exon filter were similarly removed.
Availability
AIR-predicted transcripts for chromosome 4 of the rat genome Release 3.1 and associated data files (Supplement A), as well as materials supporting the comparison with the RGD gene set (Supplement B) and other studies and results from the evaluation (Supplement C) are submitted with this paper. Source code for the tools described here and annotation for the entire rat genome can be obtained from https://panther.appliedbiosystems.com/publications.jsp.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2889405.
Footnotes
[Supplemental material is available online at www.genome.org and https://panther.appliedbiosystems.com/publications.jsp.]
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., and Wheeler, D. 2004. GenBank: Update. Nucleic Acids Res. 32: 23-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E. and Durbin, R. 2000. Using GeneWise in the Drosophila annotation experiment. Genome Res. 10: 1631-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., Andrews, D., Bevan, P., Caccamo, M., Cameron, G., and Chen, Y. 2004. Ensembl 2004. Nucleic Acids Res. 32: D468-D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski, M.S., Lowe, T.M., and Tolstoshev, C.M. 1993. dbEST—Database for “expressed sequence tags.” Nat. Genet. 4: 332-333. [DOI] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- Burset, M. and Guigo, R. 1996. Evaluation of gene structure prediction programs. Genomics 34: 353-367. [DOI] [PubMed] [Google Scholar]
- Claverie, J.M. 1997. Computational methods for the identification of genes in vertebrate genomic sequences. Hum. Mol. Genet. 6: 1735-1744. [DOI] [PubMed] [Google Scholar]
- Dunham, I., Shimizu, N., Roe, B.A., Chissoe, S., Hunt, A.R., Collins, J.E., Bruskiewich, R., Beare, D.M., Clamp, M., Smink, L.J., et al. 1999. The DNA sequence of human chromosome 22. Nature 402: 489-495. [DOI] [PubMed] [Google Scholar]
- Florea, L., Hartzell, G., Zhang, Z., Rubin, G.M., and Miller, W. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8: 967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand, M.S., Mironov, A.A., and Pevzner, P.A. 1996. Gene recognition via spliced sequence alignment. Proc. Natl. Acad. Sci. 93: 9061-9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, R.A., Weinstock, G.M., Metzker, M.L., Muzny, D.M., Sodergren, E.J., Scherer, S., Scott, G., Steffen, D., Worley, K.C., Burch, P.E., et al. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493-521. [DOI] [PubMed] [Google Scholar]
- Haas, B.J., Delcher, A.L., Mount, S.M., Wortman, J.R., Smith Jr., R.K., Hannick, L.I., Maiti, R., Ronning, C.M., Rusch, D.B., Town, C.D., et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31: 5654-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber, S., Alekseyev, M., Sze, S.H., Tang, H., and Pevzner, P.A. 2002. Splicing graphs and EST assembly problem. Bioinformatics 18 Suppl 1: S181-S188. [DOI] [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., and Clark, L. 2002. The Ensembl genome database project. Nucleic Acids Res. 30: 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istrail, S., Sutton, G.G., Florea, L., Halpern, A.L., Mobarry, C.M., Lippert, R., Walenz, B., Shatkay, H., Dew, I., Miller, J.R., et al. 2004. Whole-genome shotgun assembly and comparison of human genome assemblies. Proc. Natl. Acad. Sci. 101: 1916-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, Z., Rouchka, E.C., Gish, W.R., and States, D.J. 2001. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11: 889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., and Lu, Y.T. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31: 51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness, E.F., Bafna, V., Halpern, A.L., Levy, S., Remington, K., Rusch, D.B., Delcher, A.L., Pop, M., Wang, W., Fraser, C.M., et al. 2003. The dog genome: Survey sequencing and comparative analysis. Science 301: 1898-1903. [DOI] [PubMed] [Google Scholar]
- Korf, I., Flicek, P., Duan, D., and Brent, M.R. 2001. Integrating genomic homology into gene structure prediction. Bioinformatics 17 Suppl 1: S140-S148. [DOI] [PubMed] [Google Scholar]
- Kulp, D., Haussler, D., Reese, M.G., and Eeckman, F.H. 1996. A generalized hidden Markov model for the recognition of human genes in DNA. Proc. Int. Conf. Intell. Syst. Mol. Biol. 4: 134-142. [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W. et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Lippert, R., Zhao, X., Florea, L., Mobarry, C., and Istrail, S. 2004. Finding anchors for genomic sequence comparisons. Proc. Int. Conf. Res. Comp. Biol. RECOMB 2004 8: 233-241. [DOI] [PubMed] [Google Scholar]
- Lukashin, A.V. and Borodovsky, M. 1998. GeneMark.hmm: New solutions for gene finding. Nucleic Acids Res. 26: 1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, B., Tromp, J., and Li, M. 2002. PatternHunter: Faster and more sensitive homology search. Bioinformatics 18: 440-445. [DOI] [PubMed] [Google Scholar]
- Makalowski, W. and Boguski, M.S. 1998. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc. Natl. Acad. Sci. 95: 9407-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin, B. 1996. Mathematical classification and clustering. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Mironov, A.A., Fickett, J.W., and Gelfand, M.S. 1999. Frequent alternative splicing of human genes. Genome Res. 9: 1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., and Lee, C. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29: 2850-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott, R. 1997. EST_GENOME: A program to align spliced DNA sequences to unspliced genomic DNA. Comput. Appl. Biosci. 13: 477-478. [DOI] [PubMed] [Google Scholar]
- Murakami, K. and Takagi, T. 1998. Gene recognition by combination of several gene-finding programs. Bioinformatics 14: 665-675. [DOI] [PubMed] [Google Scholar]
- Mural, R.J., Adams, M.D., Myers, E.W., Smith, H.O., Miklos, G.L., and Wides, R. 2002. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296: 1661-1671. [DOI] [PubMed] [Google Scholar]
- Pavlovic, V., Garg, A., and Kasif, S. 2002. A bayesian framework for combining gene predictions. Bioinformatics 18: 19-27. [DOI] [PubMed] [Google Scholar]
- Pennisi, E. 2003. A low number wins the GeneSweep pool. Science 300: 1484. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogic, S., Mackworth, A., and Oullette, F.B.F. 2001. Evaluation of gene finding programs on mammalian sequences. Genome Res. 11: 817-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G.M., Yandell, M.D., Wortman, J.R., Gabor Miklos G.L., Nelson, C.R., Hariharan, I.K., Fortini, M.E., Li, P.W., Apweiler, R., Fleischmann, W., et al. 2000. Comparative genomics of the eukaryotes. Science 287: 2204-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov, A.A. and Solovyev, V.V. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003. Human-mouse alignments with BLASTZ. Genome Res. 13: 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T.F. and Waterman, M.S. 1981. Identification of common molecular subsequences. J. Mol Biol. 147: 195-197. [DOI] [PubMed] [Google Scholar]
- Sugnet, C.W., Kent, W.J., Ares Jr., M., and Haussler, D. 2004. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 9: 66-77. [DOI] [PubMed] [Google Scholar]
- Thomas, J.W., Touchman, J.W., Blakeseley, R.W., Bouffard, G.G., Beckstrom-Sternberg, S.M., Margulies, E.H., Blanchette, M., Siepel, A.C., Thomas, P.J., McDowell, J.C., et al. 2003. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424: 788-793. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., and Sutton, G.G. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., and Agarwal, P. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Wheelan, S.J., Church, D.M., and Ostell, J.M. 2001. Spidey: A tool for mRNA-to-genomic alignments. Genome Res. 11: 1952-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, D.L., Church, D.M., Edgar, R., Federhen, S., Helmberg, W., and Madden, T.L. 2004. Database resources of the National Center for Biotechnology Information: Update. Nucleic Acids Res. 32: D35-D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y., Resch, A., and Lee, C. 2004. The multiassembly problem: Reconstructing multiple transcript isoforms from EST fragment mixtures. Genome Res. 14: 426-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, R.F., Lim, L.P., and Burge, C.B. 2001. Computational inference of homologous gene structures in the human genome. Genome Res. 11: 803-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan, M., van Nimwegen, E., and Gaasterland, T. 2002. Splice variation in mouse full-length cDNAs identified by mapping to the mouse genome. Genome Res. 12: 1377-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M.Q. 2002. Computational prediction of eukaryotic protein-coding genes. Nat. Rev. Genet. 3: 698-709. [DOI] [PubMed] [Google Scholar]
Web site references
- http://www.ncbi.nlm.nih.gov/genome/guide/human/; NCBI Human Genome Resources; P. Kitts, Genome Assembly and Annotation Process documentation.
- http://www.ncbi.nlm.nih.gov/Sitemap/index.html; Guide to NCBI Resources; Links to the RefSeq, dbEST, homoloGene and GenBank sequence data sets.
- http://vega.sanger.ac.uk/; the Vertebrate Genome Annotation (VEGA) database.
- http://www.genome.ucsc.edu/; University of California Santa Cruz Genome Bioinformatics; Downloads of the mouse genome sequence version mm3 (a.k.a. NCBI Build 30) and the rat genome sequence version rn3 (a.k.a. Baylor v.3.1).
- http://rgd.mcw.edu/; Rat Genome Database Home page, Medical College of Wisconsin, Milwaukee.
- ftp://rgd.mcw.edu/pub/RGD_genome_annotations/V3.1/gene/; Rat Genome Database ftp site for downloading curated gene records.
- https://panther.appliedbiosystems.com/publications.jsp; Web site for downloading code and annotation associated with this publication.