Abstract
Small-angle X-ray and neutron scattering (SAXS and SANS) are techniques used to extract structural parameters and determine the overall structures and shapes of biological macromolecules, complexes and assemblies in solution. The scattering intensities measured from a sample contain contributions from all atoms within the illuminated sample volume including the solvent and buffer components as well as the macromolecules of interest. In order to obtain structural information, it is essential to prepare an exactly matched solvent blank so that background scattering contributions can be accurately subtracted from the sample scattering to obtain the net scattering from the macromolecules in the sample. In addition, sample heterogeneity caused by contaminants, aggregates, mismatched solvents, radiation damage or other factors can severely influence and complicate data analysis so it is essential that the samples are pure and monodisperse for the duration of the experiment. This Protocol outlines the basic physics of SAXS and SANS and reveals how the underlying conceptual principles of the techniques ultimately ‘translate’ into practical laboratory guidance for the production of samples of sufficiently high quality for scattering experiments. The procedure describes how to prepare and characterize protein and nucleic acid samples for both SAXS and SANS using gel electrophoresis, size exclusion chromatography and light scattering. Also included are procedures specific to X-rays (in-line size exclusion chromatography SAXS) and neutrons, specifically preparing samples for contrast matching/variation experiments and deuterium labeling of proteins.
Introduction
Modern systems structural biology is faced with enormous challenges in deciphering the complexity of interconnected macromolecular networks and how these networks mediate molecular-level communication to affect cellular responses. High resolution structure determination methods, such as X-ray crystallography, nuclear magnetic resonance spectroscopy (NMR) and, more recently, high-resolution electron microscopy (EM) are exceptional for uncovering the atomic details of proteins and other macromolecules. However, it becomes increasingly difficult using high-resolution techniques to assess the conformational responses of macromolecules, complexes and assemblies in different sample environments. Continued advances in instrumentation, software and the development of automated methods for data collection, analysis and modelling have launched small-angle scattering (SAS) using X-rays (SAXS) or neutrons (SANS) into the structural biological mainstream1–8. The appeal of SAS for structural biologists is that it can be applied to the analysis of diverse macromolecular systems – directly in solution – that span a broad molecular weight range, from a few kilodaltons to megadaltons, across a seemingly-endless array of sample environments9–12. Global structural parameters, for example the radius of gyration, Rg, maximum particle dimension, Dmax, as well as the distribution of distances within a particle (relating to the volume and structure) can be quickly extracted from the data2,6. In addition, it is now routine to obtain low resolution 3D-spatial representations of macromolecules using SAS4,13–17. Modelling these structures can be achieved using combination(s) of ab initio methods18–20, where no prior assumptions are made, or using atomistic, or rigid-body models4,21 derived from X-ray crystallography, NMR, EM and homology modelling (i.e., hybrid-methods)22. Importantly, as solution environments can be controlled, SAS is extremely useful to probe the structural responses of macromolecules on changing sample conditions23. Ensemble states24–28, for example the study of intrinsically disordered proteins29,30 and the formation of complexes31 or assemblies in real-time32–34 can be evaluated using SAS that are otherwise difficult to achieve using high-resolution methods.
One drawback of SAS is that it is difficult to prove with certainty that a measured scattering profile is, in fact, derived from a target of interest. All matter has the potential to scatter radiation (X-rays and neutrons), so all atoms comprising a sample – macromolecules, water, buffer components, macromolecules, the sample container, etc – will each contribute to the measured scattering intensities. Fundamentally, the success of any experiment will rely on the production of well-characterised, high quality samples35,36 combined with an accurate understanding of, and correction for, any background scattering contributions. Consequently, maintaining sample quality for SAXS and SANS is challenging due, in part, to how the physics of both techniques relates to the properties of a sample.
For the structural biologist at the laboratory bench who is interested in applying SAXS or SANS to interrogate the structures of macromolecules in solution, a great deal of the physics describing both techniques can be difficult to translate into a procedure for sample preparation. In practice, all that is required is an understanding of a handful of concepts that help define what practical steps are necessary to produce quality samples. Advanced and detailed explanations of the physics and mathematics of SAS – that become increasingly relevant when analysing datasets or for the design of experiments – can be found in the texts by Glatter and Kratky37, Feigin and Svergun38 and the more recent Svergun et al.39. Additional protocols for SAS data acquisition, basic data interpretation and publication guidelines may be referenced from Skou et al.3, Grishaev36, Jacques and Trewhella35, and Jacques et al.40.
The basics of small-angle scattering: A simple equation with big implications
A very simple relationship links the angular dependence of SAS intensities, I, to the structures of macromolecules in solution as well as the bulk properties of a sample. If a sample contains n independent randomly-oriented particles, the intensity can be expressed as:
| (1) |
Here q = 4πsinθ/λ, where θ is half the scattering angle and λ is the wavelength of incident radiation. This relationship states that the intensity of scattered radiation is the sum of the scattering from each-and-every individual particle, i, within the illuminated volume of the sample. The angular dependence of I(q) is proportional to several factors, of which the form factor, Pi(q), is perhaps the most interesting to the structural biologist. The form factor encodes overall structural information in reciprocal space, Pi(q), that relates to the probable real-space distance distribution between scattering centres within a macromolecule (pi(r), Figure 1). However, I(q) is also dependent on three other factors: i) the volume-squared of each particle, Vi2 ii) the contrast squared, Δρi2, which is the difference in scattering density between the macromolecule and its supporting solvent, and; iii) the structure factor, S(q), which encodes information relating to the correlated motions/distances between particles in solution, i.e., interparticle interactions.
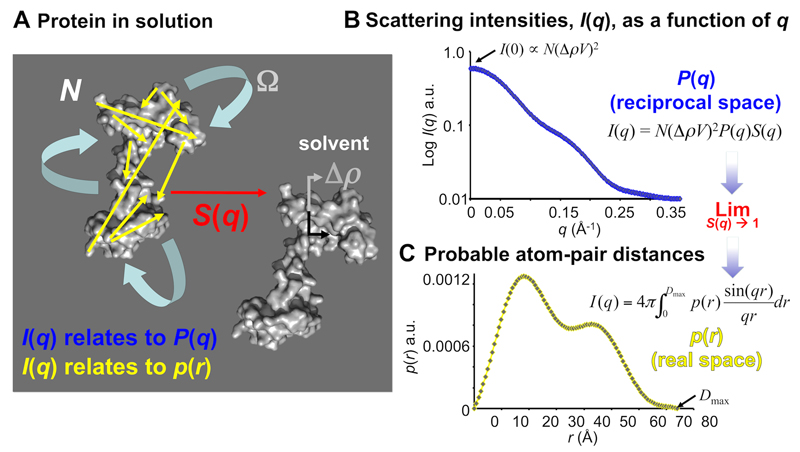
Figure 1. Scattering basics.
A. Macromolecules in solution, e.g., proteins (represented as grey blobs), undergo rotational and translational motion and experience long-range interactions with neighbouring particles. The SAS intensities measured from an isotropically tumbling (Ω) monodisperse sample are dependent on a number of factors, of which the form factor, P(q), is of most interest to structural biologists. It is from P(q) that structural parameters and low resolution models of the macromolecules can be obtained. The form factor of the scattering intensities in reciprocal space relate to the real space distribution, p(r), of all time-preserved, i.e., correlated, pair-distances between scattering centres of the molecule (yellow arrows). In the small-angle regime, these correlated distances are otherwise absent in the solvent. However, as all atoms can scatter radiation, solvent scattering contributions have to be accurately subtracted from the sample scattering to reveal P(q) from the macromolecules. The magnitude of the intensities will then depend on: i) the number of particles in a sample (N); ii) the volume squared of the macromolecule (V2); iii) the difference in scattering length density, or the contrast, squared against the solvent (Δρ2) and; iv) scattering arising from correlated distances of closest approach between particles (interparticle interference, or structure factors, S(q)). The purity, concentration, contrast and how well a solvent is matched to a sample can be directly controlled during sample preparation. B. SAS data are usually collected on 2D detectors and radially averaged to produce 1D profiles of scattering intensity, I(q), as a function of angle, q. After solvent subtraction, I(q) vs q encodes P(q) from each-and-every macromolecule in a sample weighted by N(ΔρV)2 and S(q). Longer distance separations are represented at lower angles and vice-versa. At zero angle, I(0), the magnitude of the scattering is proportionate to the total volume squared and concentration of the macromolecules. C. If S(q) limits to 1, i.e., when the system is infinitely dilute and interparticle effects are absent, modelling the indirect inverse Fourier transform of I(q) vs q produces the real-space p(r) vs r from which the radius of gyration, Rg, maximum particle dimension, Dmax, and low resolution particle shape and structure can be determined.
In terms of sample preparation, sample homogeneity, concentration and contrast are the parameters that directly contribute to I(q) and that can be influenced at the laboratory bench. For example, if a sample consists of a mixture of different species in solution, i.e., is not purified to homogeneity, each species in the mix will have different volumes, contrasts and form factors. As a result, and as eq. 1 indicates, the structural parameters extracted from the SAS data will reflect the sum-weighted contribution (not the average) of each species in the mix. Therefore, in order to obtain accurate structural information from macromolecules and obtain the 3D models of individual proteins, polynucleotides, complexes, assemblies, etc, samples have to be homogeneous and not affected by significant interparticle interactions (i.e. eliminate S(q)). If these conditions are met then the relationship above simplifies to:
| (2) |
where N is the number density of homogeneous particles in the sample. Consequently, under non-interacting (dilute) conditions of a pure sample, the magnitude of I(q) will depend on the particle concentration, volume, contrast, and – importantly – the overall structure and shape. The aim in the wet-lab is to optimise sample conditions so that a particle of interest (be it a monomer, dimer, oligomer or complex) are as pure as possible and maintained in a monodisperse state during the course of measurement so that P(q) can be accurately assessed from the scattering intensities. This can be achieved by optimising concentration, contrast and purity.
Key points of consideration
The main concepts to keep in mind when preparing samples for macromolecular solution SAS are:
X-rays are scattered by electrons while neutrons are primarily scattered by atomic nuclei. X-rays are, in general, much more damaging to macromolecules than neutrons as X-rays can induce chemical changes (e.g., free radical formation) that can alter the state of a sample over time (e.g., aggregation due to cross-linking).
All atoms in a sample – not just a macromolecule of interest – have the capacity to absorb or scatter differing amounts of X-rays or neutrons (air, water, sample cells, small chemicals, buffering components, etc). As it is impossible to identify where an X-ray or neutron arriving at a detector has scattered from, background scattering intensities have to be subtracted from the sample scattering to reveal the scattering due to the macromolecules. Therefore, at least two measurements are to be made under identical conditions: i) the sample (macromolecule + solvent + sample container) and; ii) the background (solvent + sample container).
It is imperative that the solvent in which a macromolecule is suspended is the same as the solvent used to measure the background scattering. If the sample solvent and the background solvent are not matched, the resulting subtracted scattering profile will be a mix of scattering intensities derived from both the macromolecules and the mismatched solvent.
For both X-rays and neutrons the difference in scattering length density (Δρ) between a macromolecule and solvent is called the contrast. If Δρ equals zero then effectively no net coherent scattering will be obtained from a macromolecule after subtracting bulk solvent scattering contributions (eq. 2) except for weak contributions arising from, for example, internal particle inhomogeneities or the solvation layer around macromolecules. For SAXS, the contrast of a sample depends on the difference between the average electron density of a macromolecule and the average electron density of the aqueous solvent. For SANS the contrast is the difference between the average neutron scattering length density of a macromolecule and the average neutron scattering length density of the aqueous solvent. Neutron scattering lengths are dependent on the isotopic composition of a macromolecule and the solvent.
For SAXS, the only practical method for altering Δρ is by changing the chemical environment of a sample. The X-ray contrast can be altered by either increasing the concentration of small molecules in the solvent or via the addition of electron-dense molecules or heavy atoms to a sample (Figure 2). For SANS, Δρ can be altered by changing the isotopic composition of the sample. The two most abundant isotopes of hydrogen, protium (1H) and deuterium (2H), possess vastly different neutron scattering lengths. Consequently, Δρ can be manipulated by altering the 1H2O:2H2O ratio of the supporting solvent or by introducing 2H into recombinant macromolecules at non-exchangeable hydrogen positions (i.e., where 2H are covalently bound to functional groups and not in rapid exchange with the solvent).
The larger the volume of a particle, the greater numbers of correlated distances exist between scattering centres within the volume of the particle. It is these relatively well preserved pair-distance correlations, which are otherwise absent in the solvent, which produce SAS intensities at low angles. After background subtraction, the scattering intensity at zero angle, I(0), will represent the sum total scattering from all correlated pair-distances weighted by contrast squared. Importantly, for monodisperse systems, I(0) is proportional to the macromolecule volume squared.
Doubling the concentration of a macromolecule will double the scattering intensity and improve the small-angle scattering signal (i.e., signal to noise ratio in the data). However, increasing the concentration too much may lead to correlated distances of closest approach between particles such that S(q), i.e., the structure factor, becomes significant. Attractive interactions between particles systematically increase structural parameters derived from the experimental data, e.g., the radius of gyration (Rg), maximum particle dimension (Dmax) and I(0). Repulsive interactions systematically decrease the structural parameters. Interparticle interference primarily affects data at very low angles, but the contribution can extend well into the useful region of the data, thus complicating interpretation. That is why the SAS experiments are usually performed at low solute concentrations, typically below 10 mg/ml (i.e., 1 volume percent), and, moreover, why a concentration series needs to be measured in order to extrapolate the data to infinite dilution.
SAS measurements are performed over a set time period. For X-rays this could be seconds or milliseconds (synchrotron-SAXS) or minutes to hours (lab-based sources); for neutrons, usually minutes to hours. The stability of a sample during the course of data acquisition needs to be ensured.
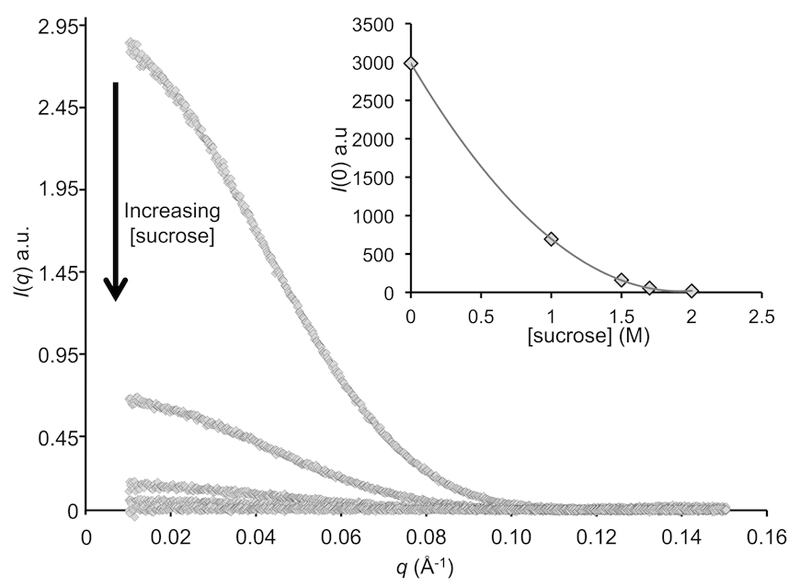
Figure 2. Decreasing contrast (Δρ) and the effect on measured scattering intensities.
SAXS data recorded for glucose isomerase with ever-increasing concentrations of sucrose present in the supporting solvent. Increasing the electron density of the solvent relative to the protein causes a significant reduction in I(q) caused by a reduction in X-ray contrast, Δρ. Inset: The quadratic relationship observed between the calculated total forward scattering at zero angle, I(0), and sucrose concentration. At 2 M sucrose, the protein has effectively been matched out, i.e., Δρ=0. Data were collected at the EMBL P12 BioSAXS beam line8 of PETRAIII, DESY, Hamburg.
The general points outlined above apply to both SAXS and SANS. However, the physics of SANS, i.e., scattering arising from neutron-nucleus interactions, imposes additional requirements for sample preparation that are discussed in more detail in the procedures specific to biological SANS experiments.
A quick background to neutron scattering
SANS has the potential to significantly enrich biological structural investigations. Using SANS, the overall low resolution structure and spatial orientations of macromolecular components of complexes and higher-order assemblies can be extracted from the data, for example the structure of the ribosome41, filamentous actin assemblies42, the subunits of protein-protein complexes15, etc. However, compared to SAXS, SANS is experimentally very demanding in terms of sample quantity (typically, tens of μl for SAXS and hundreds of μl for SANS) and therefore it is necessary to first evaluate the question: What is the specific question SANS can address that other methods, including SAXS, cannot?
The utility of SANS comes from the ability to manipulate the neutron contrast, Δρ, of an experimental system without requiring major chemical changes to a sample38,39,43–45. Neutron contrast can be adjusted by isotopic substitution, in particular protium-deuterium (1H-2H) substitution, either in the solvent (1H2O to 2H2O) or via the non-exchangeable (i.e., covalently linked) 2H labelling of a macromolecule. As with SAXS, obtaining homogeneous, monodisperse and pure samples that are not affected by significant interparticle interactions are also important for SANS. However, there are unique aspects to SANS sample preparation that are influenced by:
The different way neutrons interact with the nucleus of 1H compared to the nuclei of 2H and the other commonly occurring ‘biological’ isotopes (12C, 16O, 14N, 31P, and mainly 32S)46–48.
The different hydrogen bond strengths of 1H relative to 2H that can alter the solubility of samples or shift the position of disassociation equilibrium of complexes.
The relatively low flux of neutron sources and large beam size that requires long exposure times and large sample volumes. In comparison to SAXS, radiation damage to a sample is unlikely, but the samples must be time-stable.
SANS basics
All atomic nuclei have a probabilistic capacity to scatter neutrons. The scattering probability, or the scattering cross-section, of a nucleus can be basically pictured as a circle with a radius that relates to what is termed the scattering length of the nucleus. Depending on the nuclear isotope, there can be two scattering cross sections that describe the neutron-nucleus scattering interaction: coherent and incoherent scattering. Just like X-rays, the intensities of coherently scattered neutrons relate to the distances between scattering centres within the volume of a particle, i.e., the structure of a macromolecule in solution (P(q)). However, incoherently scattered neutrons essentially do not correlate to atom-pair distance separations and therefore scatter radiation independent of q, thus contributing to the measured scattering data as background noise (Figure 3).
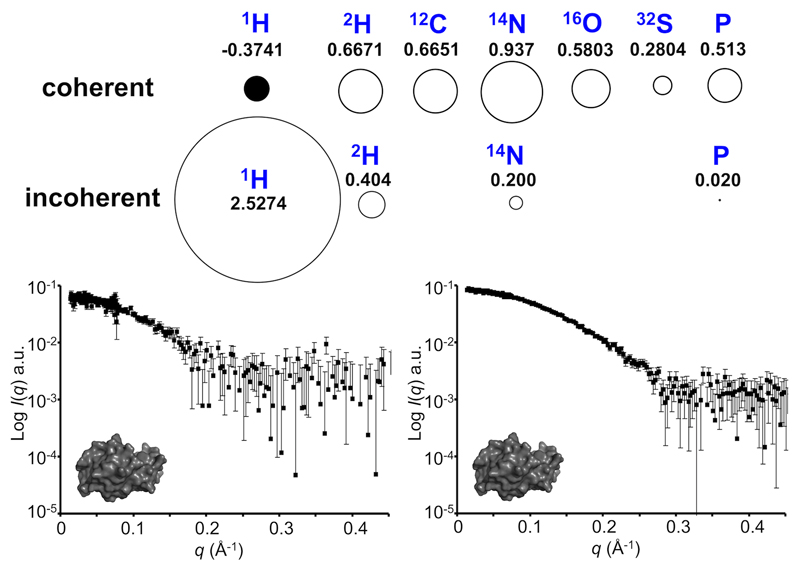
Figure 3. Coherent and incoherent neutron scattering.
Coherent and incoherent neutron cross-sections of the ‘biological’ elements (σc, displayed as circles) and their respective neutron scattering lengths (bc, 10-12 cm; where σc = 4πbc2). 1H has a negative coherent scattering length (represented as a black circle) compared to deuterium and the other commonly occurring biological isotopes. Coherent scattering arising from correlated distances within a particle’s volume produce a scattering profile from which structural information can be extracted. Conversely, incoherent neutron scattering cannot be used to extract shape/structural information and contributes to a SANS profile as ‘noise’ across all angles. 1H has a considerable incoherent scattering length, the effect of which is demonstrated by the SANS scattering from lysozyme in 100% v/v 1H2O (left) which is considerably noisier than the same sample collected in 100% v/v 2H2O (right.) SANS data were collected on the Quokka-SANS instrument at ANSTO95 using the same neutron wavelength, exposure times, detector distances, instrument geometry, sample path length and protein concentration. Neutron scattering lengths are taken from Sears, 199250.
1H is unusual in that it has both a negative coherent scattering length compared to the other major biological isotopes and a very large incoherent scattering length46–48. Incoherent scattering provides a structure-uncorrelated background in the form of a constant contribution to all scattering angles, which reduces SANS data quality. Consequently, while samples rich in 1H will produce intense incoherent background, it is the negative coherent scattering length of 1H that enables the contrast of aqueous biological samples to be altered via 1H-2H isotopic substitution. When perceiving neutrons as waves as opposed to particles it becomes possible to conceptualise that if two waves of the same wavelength, amplitude and phase add to each other the result will be a doubling of the wave amplitude. Conversely, if the two waves are 180° out of phase, the waves cancel. As it happens, the nuclei of deuterium and of the commonly occurring biological isotopes interact with neutrons so that coherently scattered neutrons undergo a phase inversion relative to the phase of the incoming neutron beam49,50. This inversion is defined as a positive scattering length (note that for SAXS, the X-ray scattering length of all atoms are positive because of the interactions of the charged electrons with the electromagnetic waves). Most isotopes also have positive neutron coherent scattering lengths, but some, (e.g., 7Li, 48Ti, 55Mn) – and most importantly 1H – do not produce this phase inversion, i.e., the scattering length is negative. As a result, neutrons scattered from 1H are 180° out of phase with scattered neutrons from 2H and the other biological elements. As the neutron contrast in a SANS experiment is simply the difference between the summed coherent scattering lengths per unit volume of a macromolecule compared to that of the solvent – i.e., the difference in average neutron scattering length density – and because the scattering length from 1H is negative, the Δρ can be manipulated by simply substituting 1H for 2H in the solvent, macromolecule, or both47.
Δρ = 0: contrast matching
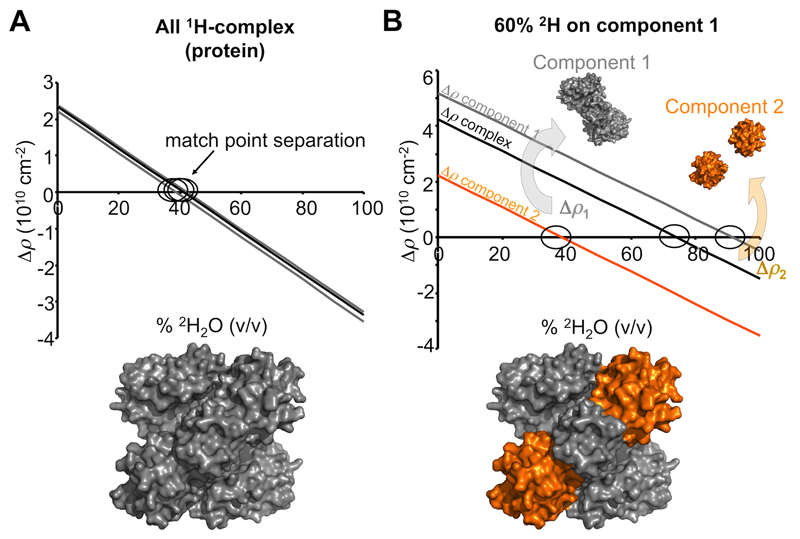
Contrast manipulation increases the information content of a SAS experiment as scattering contributions from individual components of a complex with different average 1H per unit volume can be selectively ‘matched out’ from a scattering profile by altering ratios of 1H2O and 2H2O in the solvent. Collecting SANS data at a component match point, i.e., at a volume fraction of 2H2O that produces Δρ = 0, seems intuitively useless as the majority of the structural information is effectively removed from a profile. However, if a macromolecule is covalently bound to, or is in complex with another molecule with a different scattering-length density, then the coherent scattering profile measured at the match point for the first molecule will be derived almost-exclusively from the second component. That is, at the match point of macromolecule x it will be possible to obtain structural information from macromolecule y. Conversely, at the match point of y, it will be possible to obtain structural information about x. This type of SANS experiment is called contrast matching and is typically performed by choosing the correct ratio of 1H2O:2H2O in the solvent to match out the components of a complex with different regions of contrast (Figure 4). Different classes of macromolecules have different average isotopic compositions per unit volume i.e., protein, DNA, carbohydrates and lipids are comprised of different 1H:12C:16O:14N:31P: and 32S ratios48. Consequently, when focussing on the differences between the average 1H per unit volume of these macromolecules, each class will have a match point at different % v/v 2H2O in the solvent. Most proteins match out of a SANS profile between 40–45% v/v 2H2O, whereas lipids match out between 2–15% v/v 2H2O and DNA/RNA matches out at ~60–70% v/v 2H2O. Many metal nanoparticles, e.g., ferromagnetite, are matched out at high% v/v 2H2O (e.g., 90–100%), making SANS an attractive option for studying biological macromolecule-metal nanoparticle conjugates. Furthermore, and of particular relevance to this protocol, if a macromolecule is deuterated, i.e., the volume fraction of 1H per unit volume is altered, it becomes possible to control a component’s match point (Figure 5).
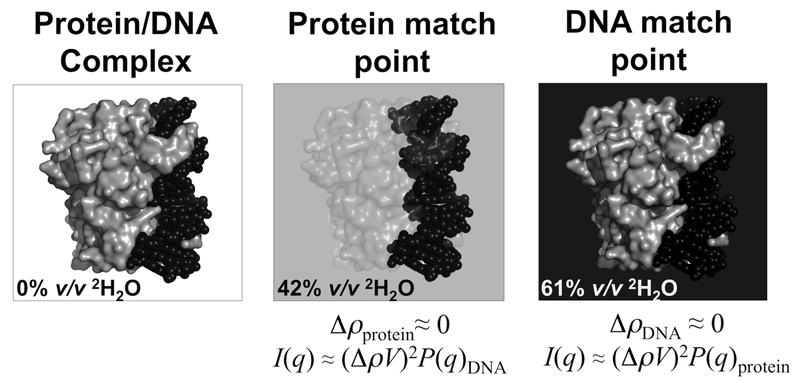
Figure 4. Principle of contrast matching.
If a macromolecular complex consists of individual components that have different average scattering length densities, it is possible to match out the scattering contributions of a component by placing the complex in a solvent with the same average scattering length density as that component. Illustrated here is a protein/DNA complex (grey surface and black spheres, respectively). For example, using neutrons, if the complex is placed into ~42% v/v 2H2O, i.e., the protein match point, the measured coherent scattering of the SANS profile will be dominated by the DNA, from which the disposition of the DNA in the complex can be determined. Raising the % v/v 2H2O to 61% matches out the DNA scattering contribution so that the SANS profile is dominated by coherent scattering from the protein.
Figure 5. The effect of non-exchangeable deuterium labelling a component for SANS with contrast variation experiments.
A. As there is very little difference in the average 1H per unit volume for proteins, the neutron contrasts calculated at different % v/v 2H2O for components comprising a protein-protein complex are almost identical (grey and black linear relations). Consequently, the low resolution structure restored from a SANS with contrast variation experiment will reflect the shape of the whole complex (grey surface representation). B. Isotopic labelling of a component with non-exchangeable deuterium has a dramatic effect on the contrast relationships and the separation of match points for the individual components and of the whole complex. In this example, component 1 is labelled on average with 60% non-exchangeable 2H (grey) while component 2 remains as a native 1H-protein (orange). When the scattering contributions of the native 1H-protein are matched out (~40% v/v 2H2O; Δρ2 = 0; orange line), the scattering intensities will be derived from 2H-component 1, the magnitude of which will be proportionate to Δρ12 and V12 and P1(q). On increasing the % v/v 2H2O even further a point is reached when Δρ for the whole complex limits to zero (~75% v/v; black line) whereby the scattering signal will be exceptionally weak (essentially incoherent scattering and scattering from 1H-2H exchange). Eventually 2H-component 1 will be matched out at high % v/v 2H2O (~91% v/v 2H2O;Δρ1 = 0; grey line) leaving coherent scattering contributions from the 1H-component 2 (proportionate to Δρ22 and V22 and P2(q)). From a set of contrast variation data it is possible to determine the shapes of the entire complex, of the individual components and the orientations of the components within the complex.
Contrast variation
Contrast matching can be challenging as these experiments require the careful formulation of solvents at a specific 1H2O:2H2O ratio. If SANS data are acquired close to, but not at, the exact match point the coherent scattering intensities will have contributions from the ‘nearly matched’ component. For a complex consisting of two components, each with a different contrast in solution (i.e., Δρ1 and Δρ2), eq. 2 can be expanded to yield:
| (3) |
Here it can be seen that I(q) is composed of intensities from the two components, plus an important additional term describing the relationship between them (called the cross-term48,51). If a component is not exactly matched, it’s scattering plus the cross term will contribute to the observed scattering. SANS with contrast variation experiments overcome the potential difficulty of exactly matching components and provide additional structural parameters from the cross term.
SANS with contrast variation data are usually collected from samples using incremental ratios of 2H2O in the supporting solvent, often called contrast points, that span the match points of a system. For a two component complex there are three match points: Δρ1= 0, Δρ2 = 0 and for the whole complex, Δρtotal = 0. At least five, well-spread, contrast points (i.e. scattering curves) are typically measured, preferably above, below and at the individual component match points at different % v/v 2H2O in the solvent. With five such contrast points, there should be sufficient information to extrapolate from the contrast series the form factors of each individual component of the complex, P1(q) and P2(q), as well as the cross term P12(q) that describes the disposition of component 1 relative to component 2. With this information in hand, structural parameters Rg, I(0), p(r) vs r, Dmax and V as well as the global structure of the entire complex, the shapes of the individual components and the spatial orientation between components can be determined.
Summary
The underlying physics of SAXS and SANS and the relationship between the measured I(q) and c, V, and Δρ is what ultimately guides sample preparation. Experimentalists may not have control over the structure of a macromolecule, but they can control the bulk properties of a sample during its preparation in the laboratory, i.e., sample purity, concentration, monodispersity and contrast. The steps necessary to produce quality samples and accurately matched solvent blanks can be challenging. However, the payoffs for optimising sample conditions can be exceptionally rewarding with respect to improving quality assurance and obtaining additional biophysical information that can reinforce SAS data analysis, modelling and interpretation. This protocol is divided into three main procedures:
PROCEDURE 1 describes how to assess sample purity and quality for both SAXS and SANS sample preparation.
PROCEDURE 2 describes the quantities of material required for SAXS and SANS experiments, including how to estimate sample concentration and molecular weight (MW) from SAXS or SANS data.
PROCEDURE 3 describes the unique aspects of preparing samples for SANS with contrast matching or SANS with contrast variation experiments.
In addition, Box 1, 2 and 3, detail the practical considerations for performing size exclusion chromatography SAXS (SEC-SAXS, Box 1), how to calculate X-ray and neutron scattering contrasts (Box 2) and preparing non-exchangeable 2H-lablled protein for SANS experiments (Box 3).
Box 1. In line Size Exclusion Chromatography-SAXS (SEC-SAXS).
TIMING: Buffer preparation + column and detector equilibration, 2–12 hrs; 1 × SEC-SAXS run, 30 min-2 hrs (depending on the SAXS beam line and SEC column flow rates); Data processing, 20 min–2 hr.
Overview of the Procedure.
In line size-exclusion chromatography-SAXS (SEC-SAXS) has been successfully integrated as a continuous-flow sample delivery option at a number of synchrotron beam lines including BioCAT (Advanced Photon Source,97), SWING (Soleil,98), the SAXS beam line at the Australian Synchrotron, BM29 at the ESRF and BL23A1 at the NSRRC, Taiwan. At the EMBL P12 beam line (DESY, Hamburg)8, SEC-SAXS operates in conjunction with a triple detector array that includes Right-Angle Laser Light Scattering (RALLS), UV absorption and refractive index (RI) detectors that are placed immediately after the SEC column. The additional detectors and are linked in parallel to the SAXS beam line using a mobile phase flow splitter92 and enables the SAXS and independently acquired RI(UV)-RALLS measurements to be directly coupled. By combining the results from laser-light and X-ray scattering with RI or UV measurements, the molecular weight of the separated sample components eluting off the SEC column can be derived.
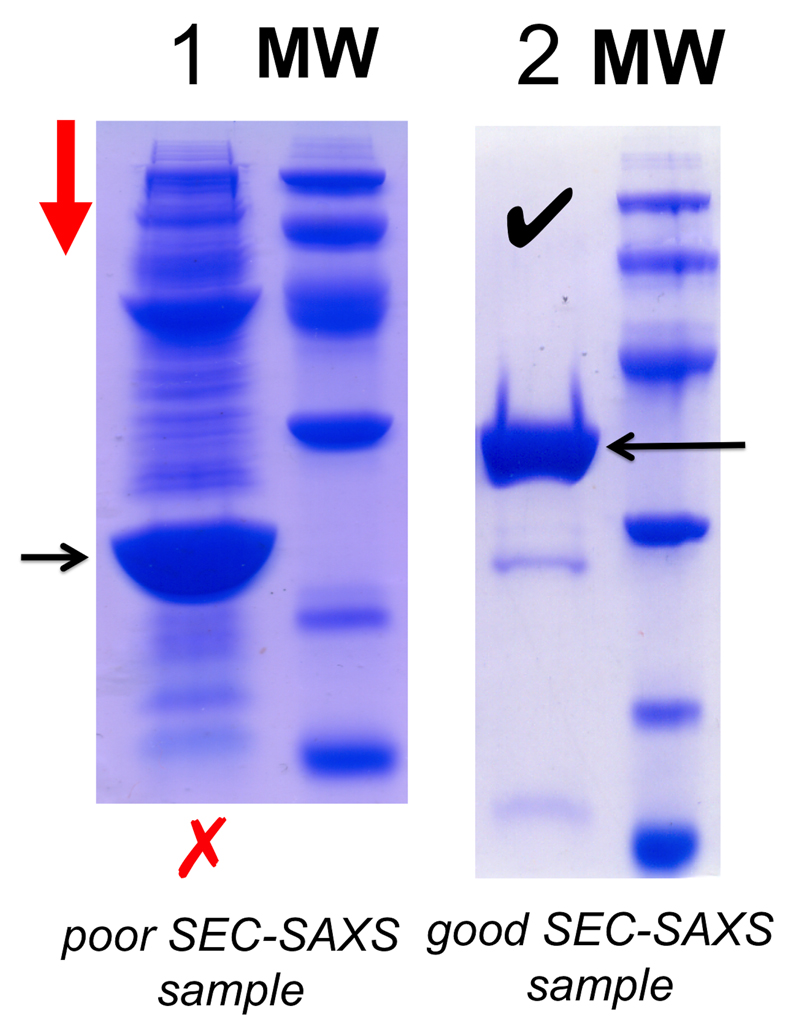
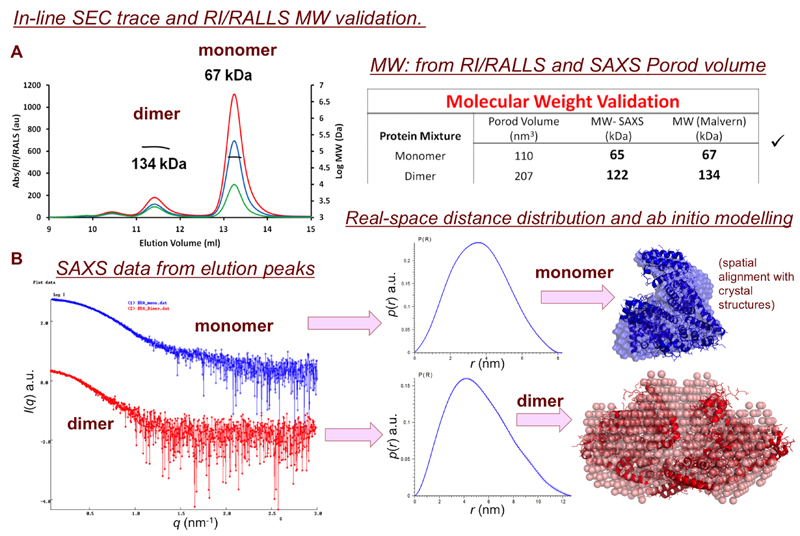
SEC-SAXS is extremely useful for separating components of already-pure equilibrium systems (e.g., monomer-oligomer inter-conversion) or removing trace aggregates from a sample immediately prior to X-ray exposure. However, SEC-SAXS is not a ‘cure all’ for every sample and should not be viewed as a purification step, but as an analytical procedure to be applied as necessary on a case-by-case basis. For example, Figure 13 shows the SDS-PAGE results of two samples, 1 and 2. It would be impossible to use SEC-SAXS to analyse the components of sample 1 as it contains too many contaminants well beyond the resolving power of any SEC-column. Column resolution is determined by the size of the column, choice of packing matrix, the sample-load volume, the sample flow, solvent conditions and sample purity, all of which need to be evaluated prior to a SEC-SAXS experiment (Figure 14). If the column resolution is compromised, i.e., the elution peaks ‘run into each other’, then the SAXS data will also be compromised, i.e., consecutive SAXS data frames collected through the elution will be the sum-weighted contribution from continuously-changing ratios of sample component mixtures (eq. 1). However, if the components are well resolved, SEC-SAXS can be invaluable for determining the structure and dispositions of polydisperse systems. The Anticipated Results of a SEC-SAXS experiment are shown in Figure 15 and refer to SASBDB17 entries SASDBJ3 and SASDBK3.
The SEC-SAXS method used will depend on the equipment and data processing tools available at a specific beam line. This box provides general advice on how this experiment can be set up and performed.
MATERIALS
Protein sample. SEC-SAXS requires 50–100 μl of protein sample at 5–15 mg.ml-1, preferably as pure as possible (Figure 13) and filtered through a 0.22 or 0.45 μm spin filter or centrifuged at high speed (10 min; 15–30 000 × g) to remove dust or insoluble aggregates.
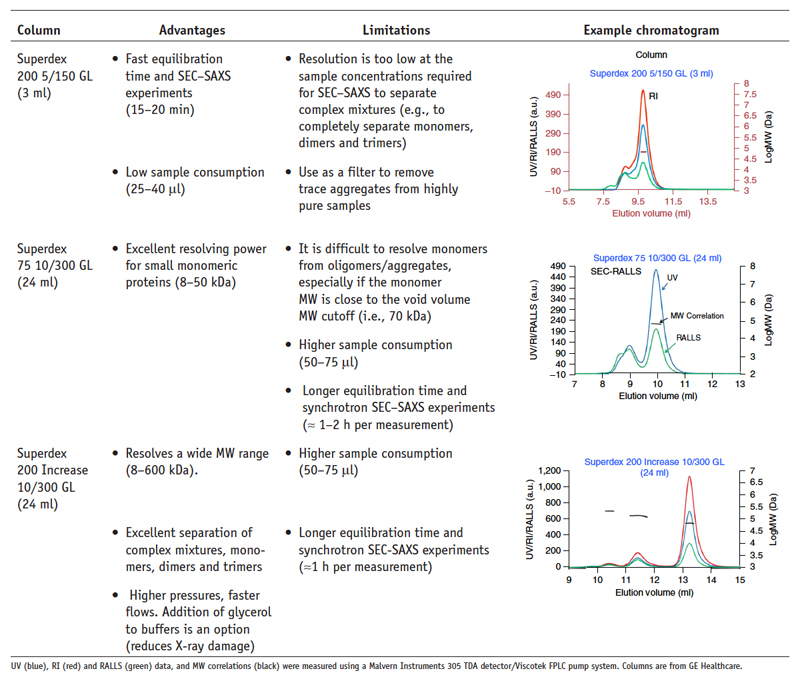
SEC column. Refer to Figure 14 regarding column selection.
Running buffers. Make up an excess of buffer to equilibrate the SEC column before and after the SEC-SAXS experiment. Running buffers need to be 0.45 or 0.22 μm filtered and degassed. Avoid rapid temperature changes on the column and ensure the buffer and the column are at the same temperature during the equilibration process. At high flux SAXS beam lines it may be necessary to add solution additives, for example 3–5% v/v glycerol, 1–2 mM DTT or 1–2 mM ascorbate to the SEC running buffer to limit radiation damage. Using Tris or HEPES36,53, instead of phosphate, may also help limit radiation damage (TROUBLESHOOTING; Figure 12).
HPLC/FPLC pump flow rate. Choose a flow rate for the column and equilibrate the column with the running buffer. For SEC-SAXS, flow rates are typically between 0.25–0.35 ml.min-1. X-ray radiation damage to the sample can occur if the flow rate is too slow. Most commercially available columns have an upper working pressure limit that should not be exceeded.
(optional) Additional detectors. Where light scattering or spectrophotometric instruments are available, calibrate the concentration (e.g., using RI or UV) and light scattering intensities (e.g., using RALLS92 or MALLS) of a molecular weight standard (e.g., for proteins use bovine serum albumin). The calibrated detectors can then be used to determine the SEC-SAXS sample concentration. The concentration values from UV or RI allow for the processed SAXS data to be placed on a concentration scale for MW determination from I(0) (refer to PROCEDURE 2, STEP 2). If SEC-SAXS UV/RI is combined with MALLS/RALLS, independent estimates of the separated sample components MW can be obtained that can be used to validate the MW from the SAXS I(0).
PROCEDURE
-
1 |
Equilibrate the SEC column, preferably overnight, with SEC running buffer.
<CRITICAL STEP> The SEC column must be very well equilibrated, typically using 2–8 column volumes of running buffer, prior to the SEC-SAXS measurement. Extensive column equilibration is required in order to increase the chances of measuring SAXS data corresponding to the matched solvent required for correct background subtraction. Note: A stable UV absorption baseline recorded from the buffer flowing off the SEC column (e.g., at 280 nm) is not an indication that the column has, in fact, equilibrated. For example a buffer containing 150 mM NaCl will have an almost identical 280 nm UV absorption properties as a buffer containing 250 mM NaCl, yet these two solutions (that have different electron densities) will produce different SAXS profiles. RI is a more sensitive tool to evaluate whether a column has equilibrated to completion.
<CRITICAL STEP> It is strongly advised that SAXS data are collected from a small aliquot of sample (e.g., 10–15 μl) using regular SAXS measurements prior to SEC-SAXS to assess the radiation susceptibility of the sample. X-ray exposure times for SEC-SAXS may be longer and sample flow rates may be slower that can both contribute to increasing the chances of radiation damage. Consequently, if radiation damage is observed using regular SAXS, it is likely that the sample will be damaged during SEC-SAXS (TROUBLESHOOTING; Figure 12).
<CRITICAL STEP> It is necessary to prepare significantly more sample material for SEC-SAXS compared to regular SAXS because the sample is diluted 5–10 fold as it elutes through the column. To maintain reasonable counting statistics in the SAXS intensities, and to maintain the integrity of macromolecular complexes, high load concentrations are often required to overcome the dilution effects of the column (eq. 2, I(q) ∝ N).
-
2 |
Start the SEC-SAXS experiment by injecting the sample onto the column at an appropriate flow rate while at the same time start the SAXS data collection.
-
3 |
(optional) In parallel with SAXS, begin UV or RI/UV/RALLS or MALLS measurements.
-
4 |
Collect SAXS data from the column eluate so that a sufficient number of buffer and sample frames are measured. It is advised to measure SAXS data from the eluting buffer at the beginning, end and during the SEC-SAXS experiment. Preferably, SAXS data spanning the entire elution profile from the SEC column should be collected.
-
5 |
After the sample peak has come off the column, always ensure that the SEC-SAXS experiment runs to completion i.e., at least one complete column volume has flowed through the column, or until all sample components have eluted. Flow an additional 0.1–0.25 column volumes of running buffer through the column after the SAXS experiment and prior to the next sample run. This additional washing ensures all of the small molecules from the preceding sample are flushed out to the column and do not contaminate the background scattering of the next SEC-SAXS experiment.
-
6 |
Assess if the SAXS sample cell (e.g., sample capillary) is clean after each SEC-SAXS experiment. Compare the (unsubtracted) SAXS profiles measured from the buffer at the very beginning and at the very end of the column elution (e.g., using Correlation Map92). If there are differences, clean the SAXS sample cell using a cycle of: water-cleaning solution-water. Three examples of cleaning solution include: i) 6 M guanidine-HCl, pH 6.5; ii) 20% v/v acetic acid or; iii) 10% v/v ethanol containing 2% v/v HellmanexIII.
<CRITICAL STEP> Systematic increases in the scattering intensities of the post SEC-SAXS buffer relative to the initial data frames can indicate that fouling of the sample capillary has occurred. Capillary fouling is often caused by sample components flowing through the X-ray beam that are susceptible to radiation damage which aggregate and bind to the capillary surface. It is advised to wash the sample capillary between successive SEC-SAXS experiments to reduce the build-up of aggregated material on the internal capillary wall. Aggregate build-up on the capillary makes accurate background subtraction impossible and will contaminate all subsequent SEC-SAXS runs.
-
7 |
Select SAXS data frames corresponding to the background scattering for the SEC-SAXS experiment. These frames may be selected from the scattering intensities measured from the solvent/buffer that has flowed through the SEC-column. These frames may be – but not always – close to a sample elution peak.
<CRITICAL STEP> If several data frames are selected and averaged to produce a SAXS profile for the buffer, always ensure that the individual buffer frames are statistically similar prior to averaging92. As samples and buffers run through the SEC column, small molecule fractionation and/or exchange of the buffer components can occur between the injected sample and the column solvent as well as between the sample and column matrix, i.e., the beads. Small molecule fractionation can result in very subtle changes in the SAXS intensities of the buffer as it flows through the column that may impact the selection of the correct background scattering. To help limit this potential complication (if possible) use dialysis to exchange a sample into SEC running buffer prior to the SEC-SAXS experiment.
-
8 |
Subtract the buffer scattering from each SEC-SAXS data frame. Identify those subtracted frames corresponding to the sample elution peak, for example using AUTORG5 to calculate the Rg and I(0) of the processed data. Make sure that the data have not been over- or under-subtracted (Figure 10) and check that each data frame acquired through an elution peak – after scaling relative to each other (e.g., to concentration) – are statistically similar92 prior to any averaging procedure.
-
9 |
<optional> If additional UV or RI detectors have been employed to monitor the column elution, correlate the concentration, c mg.ml-1, from the detectors to the I(0) from the SAXS and calculate the MW of the eluting components. If (UV)RI-RALLS or MALLS detectors are used, calculate the MW from the light scattering and validate the MW obtained from SAXS I(0). If these detectors are not available, estimate MW (for protein samples) from the particle volume calculated from the SAXS data (refer to PROCEDURE 2, STEP 2).
<CAUTION> For homogeneous, monodisperse, and non-interacting particles, I(0)/c, the MW and Rg will be constant. However, obtaining constant values for I(0)/c, MW and Rg from SAXS data spanning a SEC an elution peak does not always mean that a component is homogeneous and monodisperse. These results depend on the purity of the initial sample and column resolution (Figures 13 and 14). Bovine serum albumin, for example, can exist as a mixture in solution that prior to SEC will generate constant I(0)/c, MW and Rg values (i.e., using regular SAXS measurements). If this mixture is poorly resolved on a badly prepared or incorrectly-chosen SEC column, it is conceivable that the SEC-SAXS data will also produce consistent I(0)/c, MW and Rg through an elution peak. Therefore, prior to SEC-SAXS, it is advised to perform SEC on a sample to test a selected column’s ability to separate the sample components and, if required, alter the solvent conditions (e.g., pH, salt concentration) to optimise separation.
Box 2. Calculation of X-ray and neutron scattering contrasts using the Contrast module of MULCh.
TIMING: 5 min.
Overview of the Procedure.
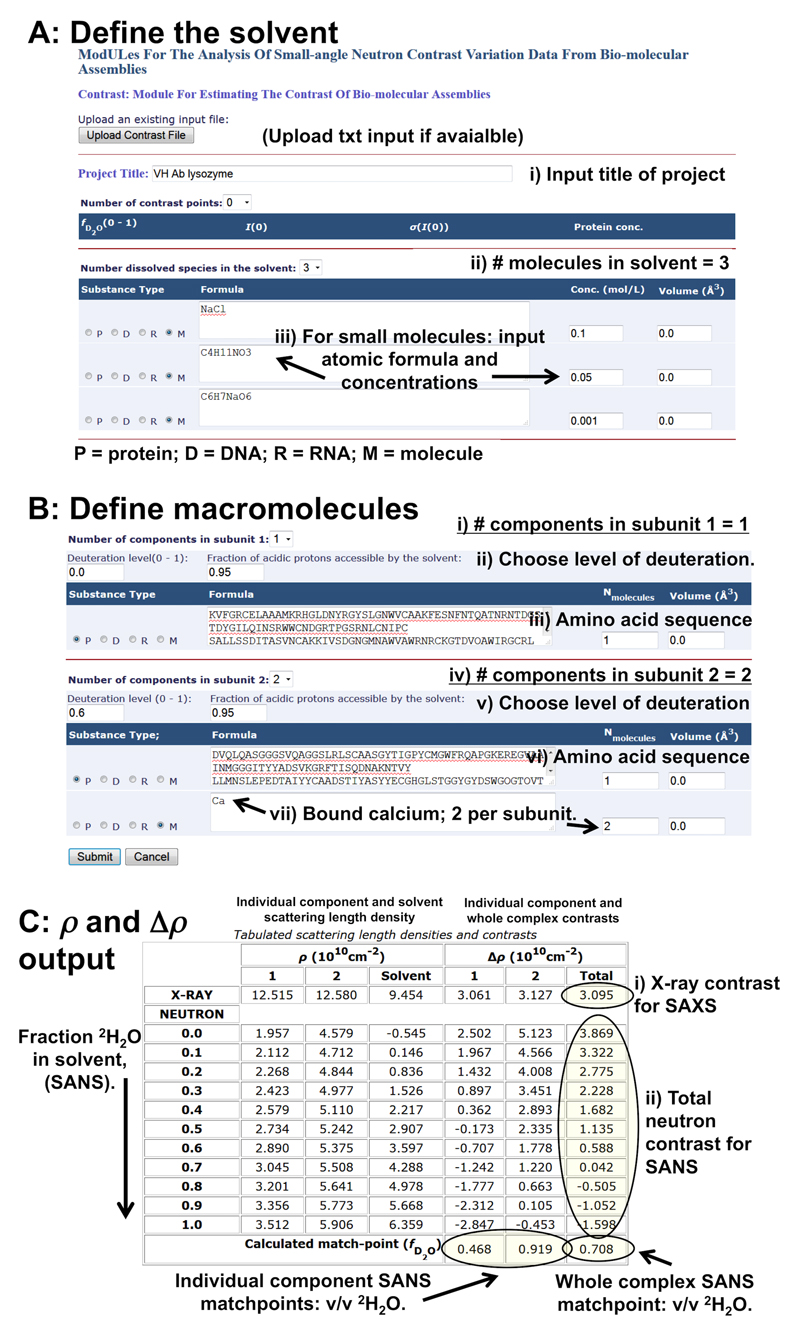
MULCh (Modules for the analysis of small-angle neutron contrast variation data from bio-molecular assemblies51) comprises of suite of programs to aid with the analysis of SAS data. The Contrast module of MULCh is a specifically tailored for calculating both X-ray and neutron scattering contrasts of a macromolecular system (Δρ). Contrast does not require any scattering data as input, it simply uses protein, RNA or DNA sequences in combination with the atomic formulae and concentration of small molecules the solvent. Using this information, Contrast calculates the X-ray and neutron scattering length densities of the macromolecule and solvent (ρ) and subtracts these values to obtain Δρ of the sample.
The contrast values derived from the Contrast module can be used to:
- Assess the molecular weight (MW) of a macromolecule from I(0) for both SAXS and SANS placed on an absolute scale (cm-1):
where csample is the concentration (g.cm-3), vsample is the partial specific volume of the scattering particle in cm3.g-1, Δρ the contrast in cm-2 and NA is Avagadro’s number (refer to spread sheet MW_from_absolute_scale.xlsx). For proteins and RNA, the partial specific volume, vsample can be calculated from the primary sequences using the PSV and volume calculator of NucProt81, http://geometry.molmovdb.org/nucprot/ or can be obtained from the Contrast output. Note, both Contrast and NucProt also calculate the volume, V, of macromolecules based on their atomic composition. For SAXS. Obtain the X-ray scattering contrast and assess the effect on Δρ when small molecules are added to a solvent. As scattering intensities are proportionate to Δρ2, the addition of high concentrations of small molecules, or the addition of electron dense molecules, to a sample will reduce the difference in electron density – and thus Δρ – between the solvent and a macromolecule of interest. This information may be useful to assess the effect on the X-ray scattering intensities (eq. 2) when small molecules are added to a sample that limit radiation damage (e.g., electron-dense polyols, Figure 2).
For SANS with contrast matching and contrast variation. Obtain the neutron scattering length density and contrasts of a sample prepared at different % v/v 2H2O concentration in the solvent. From these results the match points of the sample components can be determined (i.e., the % v/v 2H2O in the solvent that produces Δρ = 0) taking into account the percentage of acidic protons likely to be in exchange between a macromolecule and the solvent (usually around 90–95%). Note: The value of acidic 1H-2H exchange can be altered in the Contrast module to evaluate its effect on the sample component match points that can be useful for deciding on the % v/v 2H2O to use for SANS with contrast matching experiments. Additionally the V and Δρ from Contrast can be used to estimate the change in the overall magnitude of the scattering intensities as components are matched out of the SANS data, eq. 2 and 3.
For SANS using 2H labelled components – pre-production. If non-exchangeable deuterium labelling of a macromolecule is being considered, Contrast can be used to predict the effects of different levels of 2H labelling on the match point separation of the components of a sample. Use Contrast prior to setting up a SANS experiment to assess what level of non-exchangeable deuteration is required to obtain the desired sample component match point separations. These calculations are useful for guiding the production of biodeuterated material prior to producing 2H-labelled components (Box 3).
For SANS using 2H labelled components – post-production. Calculate the SANS contrasts and match point of a sample component that has been labelled with deuterium (Box 3) using experimentally determined levels of non-exchangeable 2H from peptide mass fingerprinting results.
For SANS. Calculate the mass density of a 1H2O/2H2O solvent based on the atomic composition. These values that can be compared against experimentally determined mass-densities from densitometry measurements to check that % v/v 2H2O of a solvent is correct and to assess experimental SANS contrasts.
The online tool uses a basic copy-and-paste or simple typing procedure for entering the requisite information. The offline tool requires a simple text input file (Supplementary Information Contrast_example_input.txt, is provided as an example). This simple text input can also be uploaded to the online version of the program. The online and offline versions of the program generate simple text output files after the calculations that can be re-used by either the online or offline programs. Throughout this procedure, refer to Figure 16.
EQUIPMENT
Accessing MULCh.
The entire MULCh package that includes Contrast can be downloaded as an offline tool (with instructions) or used interactively online via: http://smb-research.smb.usyd.edu.au/NCVWeb/
MATERIALS
A list of solvent/buffer components (atomic formulae) and their concentrations in mol.l-1.
The one letter amino acid code or one letter DNA/RNA code of the macromolecules.
The atomic formulae of any small-molecules bound to the macromolecule of interest, e.g., metal ions, co-factors.
PROCEDURE
-
1 |
Enter a project title.
-
2 |
Define the solvent. Use the dropdown menu to select the number of dissolved species in the solvent. These are typically small molecules, M. For each small molecule, make sure M is checked, then enter the atomic formula of each component and their concentrations (in mol.l-1) into the appropriate boxes. There is no need to include water as Contrast automatically calculates the change in water concentration as a consequence of adding molecules to aqueous solutions.
-
3 |
Define the macromolecule (Steps 3-5). Use the dropdown box to select the ‘Number of components in subunit 1’. A component can be protein, DNA, RNA or a small molecule. For example, a protein that consists of only amino acids will have one component. A metalloprotein will have two components, i.e., the protein and the bound metal.
-
4 |
Check P= protein, R = RNA, D = DNA or M = molecule, depending on the type of component being described for the subunit. For macromolecules (P, R, D), copy the one letter code of the entire sequence into the appropriate box. For example, for proteins, check P then copy the one letter amino acid code into the box. For DNA, check D, making sure to copy both the forward and complementary strand one letter sequences. If necessary, define the stoichiometry of the macromolecules in the subunit (Nmolecules). For small molecules that are known to bind to the macromolecule (metal ions, co-factors etc) select M, then type the atomic formula of the molecules remembering to include the stoichiometry. For example, a protein subunit bound to two calcium ions per monomer is defined as: ‘Number of components’ = 2 (i.e., the macromolecule and calcium), Nmolecules, P (protein), = 1 and Nmolecules, M (calcium), = 2.
-
5 |
Define the second subunit of the sample using the ‘Number of components in subunit 2’.
<CRITICAL STEP> Samples that are not heterogeneous complexes, for example lysozyme, glucose isomerase, etc, are considered by Contrast as a single ‘subunit’. In these circumstances, and in order for Contrast to complete the Δρ calculation, copy and paste the identical information used to define ‘Number of components in subunit 1’ into the respective boxes for ‘Number of components in subunit 2’. For example a tetrameric protein: i) ‘Number of components in subunit 1’ = 1, ii) check P; iii) list the amino acid sequence of the monomer and; iv) set Nmolecules = 4 (alternatively, input the amino acid sequence of the tetramer and set Nmolecules = 1.) Copy the identical information into the ‘Number of components in subunit 2’ section.
-
6 |
(optional) For SANS using deuterated components, enter the average level of non-exchangeable 2H incorporated into a macromolecule into the ‘Deuteration level’ box. Include an estimate of proton-deuterium exchange between the macromolecule and solvent using the ‘fraction of acidic protons accessible to the solvent’ box (by default, 0.95).
-
7 |
Make sure that the Volume (Å3) boxes have a number in them, even if it is 0.0. If the volume of a component is known, type in the volume of the component; if the volume is unknown, leave the value at 0.0 and the atomic volume will be calculated automatically.
-
8 |
Press submit.
Box 3. Deuteration of recombinant proteins using a laboratory-based 2H labelling protocol.
TIMING: 5–10 days + additional time if screening bacterial growth conditions are necessary.
Overview of the Procedure.
Several SANS facilities offer proposal-based/competitive applications for the production of biodeuterated materials:
-
ILL-EMBL Deuteration laboratory.
-
National Deuteration Facility, Australian Nuclear Science and Technology Organisation.
www.ansto.gov.au/ResearchHub/Bragg/Facilities/NationalDeuterationFacility/
-
Oak Ridge National Laboratory Bio-Deuteration Facility.
These facilities often employ fermenter-based methods to produce large quantities of deuterated bacterial cell pellets containing over-expressed 2H-labelled recombinant proteins99. As 2H2O is expensive, and the production of 2H labelled components is time consuming, submitting proposals to these facilities has its obvious benefits. However, it is possible to perform biodeuteration ‘in house’ using a simple flask-based procedure in E. coli B expression hosts (e.g., E. coli Bl21 (DE3)) using deuterated modified M1 growth media. A flask-based approach is useful, for example, to obtain material for assessing the effects of 2H-labelling on the physical properties of a macromolecule (e.g., stability and solubility in both 1H2O and 2H2O buffers). The overall procedure can be divided into five STAGES:
-
1)
Choose what % v/v 2H2O is required for the final heavy water M1 growth media to obtain the desired level of non-exchangeable 2H in the recombinant protein.
<CRITICAL STEP> The correlation between % v/v 2H2O used in the M1 growth media and the incorporation of non-exchangeable 2H into a protein is not linear. Refer to Table 1 that is based on Figure 1 of Leiting, Marsilio and O’Connell (1998)85. For the following example, the desired level of non-exchangeable 2H for protein X is 60% that requires a final growth media of 80% v/v 2H2O using 1H-glucose as the carbon source.
-
2)
Adapt the cells to the desired % v/v 2H2O growth media.
<CRITICAL STEP> If ampicillin is used as the antibiotic selection agent it is imperative to resuspend any bacterial cell pellet encountered throughout this procedure into fresh selection media as ampicillin is slowly degrades over time. Kanamycin is a more culture-stable alternative, as is chloramphenicol. In general, the recommended concentrations of antibiotic for use in 2H2O protein expression are: ampicillin: 60–70 μg.ml-1; kanamycin: 30–50 μg.ml-1; chloramphenicol: 16–20 μg.ml-1.
-
3)
Express the recombinant target in the final 2H2O growth media.
-
4)
Purify the 2H-labelled recombinant target.
-
5)
Experimentally determine the extent of 2H labelling in the protein.
The procedure described below starts at STAGE 2: Cell adaption. This step of the procedure spans several days, therefore the protocol has been divided into several ‘mini’ procedures that are performed during Cell adaption, DAY 1 – DAY 5.
<CRITICAL> With respect to STAGE 3, i.e., protein expression in 2H2O media. It is assumed that recombinant protein expression has been previously tested using regular bacterial growth media (e.g., LB broth) and that protein expression requirements, i.e., temperature, inducing agent (if applicable) and antibiotic concentrations or other relevant parameters, such as gene codon optimisation, have been screened and optimised for successful recombinant protein over expression100. Additionally, if no prior information is available with respect to how well a recombinant protein expresses in 2H2O, it is advised to scale down the procedure (from 1 litre) and prepare 50–100 ml test cultures. Begin with using the same protein expression parameters as used for ‘optimised expression’, e.g., in LB media, to guide the expression in 2H2O media (e.g., temperature, antibiotic concentration, etc). Adjust these parameters if necessary in the test cultures, then perform the scaled-up procedure using 1 litre of 2H2O media as described in the text.
STAGE 2): Cell adaption
Cell adaption, DAY 1
MATERIALS
Plasmid containing gene of interest and competent E. coli B cells.
1 ml of sterile LB or SOC media in regular light water.
LB-agar selection plates, made with regular light water supplemented with selection antibiotics.
PROCEDURE
-
1 |
Transform the desired plasmid into the E. coli B cells as per the manufacturer’s recommendation or using a standard transformation procedure, for example heat-shock or electroporation, followed by incubation in the growth media (e.g., 200 μl of LB media containing no antibiotics) for 1.5–2 hrs.
-
2 |
Plate the transformants out onto the LB-agar selection plates and grow overnight at 37 °C (or at a pre-determined appropriate growth temperature.)
Cell adaption, DAY 2
MATERIALS
25 ml of sterile-filtered standard LB media in regular light water, pH = 6.5.
1 × 50 ml sterile Falcon tube (can be purchased as a sterilised product).
1000 × concentrated stocks of your selected antibiotics.
PROCEDURE
-
1 |
Pipette 15 ml of sterile LB media into the 50 ml Falcon tube. Add 15 μl each of the required 1000× antibiotic solutions. Mix.
-
2 |
Remove a 100–200 μl aliquot of LB media into a 1.5 ml eppendorf tube. Using a sterile loop or pipette tip, scrape 10–15 transformants obtained from the Day 1 selection plate and into the LB aliquot. Resuspend the cells and use 50 μl to inoculate the 15 ml of LB-media in the Falcon tube. Close the tube and grow overnight at 37 °C, with shaking.
Cell adaption, DAY 3
MATERIALS
10–15 ml of sterile-filtered standard LB media in regular light water, pH = 6.5.
7.5 ml of sterile-filtered 100% v/v heavy water, 2H2O.
PROCEDURE
-
1 |
Centrifuge the cells grown overnight from Day 2 (5000 × g for 10 min) and remove the supernatant. Resuspend the cell pellet in 2–3 ml of fresh 100% LB media.
-
2 |
In a sterile Falcon tube, combine 7.5 ml of 2H2O with 7.5 ml of fresh LB media to produce a 50% v/v LB/2H2O solution. Using the volume graduations printed on the sides of the tubes is adequate to estimate the volume. Add 15 μl each of the required 1000× stock antibiotic solutions, mix.
-
3 |
Pipette 50 μl of the resuspended cells grown overnight in 100% v/v LB media into the 15 ml of 50% v/v’ LB/2H2O media. Close the Falcon tube and grow overnight at 37 °C, with shaking.
Cell adaption, DAY 4
At the beginning of Day 4, the cells will have adapted to a 50% v/v 2H2O-LB culture. The process of adapting the cells to higher % v/v 2H2O conditions begins with the preparation of modified M1 minimal-media (REAGENT SETUP) and the on-going adaption of the cells in small media cultures at ever-increasing graduated steps of % v/v 2H2O (PROCEDURE).
<PAUSE POINT> The choice of an appropriate antibiotic concentration in 2H2O media may require additional screening to balance selection vs. culture growth time vs. culture viability. If the cells have not grown in the 50% v/v 2H2O-LB growth media from Day 3, it may be necessary to repeat the Day 2 and Day 3 procedures using different antibiotic concentrations.
MATERIALS
Inorganic chemical list: K2HPO4, KH2PO4, (NH4)2SO4, NaCl, MgCl2, MoNa2O4, CoCl2, CuSO4, MnCl2, MgSO4, ZnSO4, FeCl2, CaCl2, 2H2O, 1H2O.
Organic chemical list: Regular 1H-glucose, yeast extract, biotin, thiamine, 1000 × antibiotic stocks.
Containers list: Sterile 1 litre and 250 ml Schott bottles (dry), 50 ml sterile Falcon tubes (dry), a sterile smooth-sided and dry conical flask (2.5–3 l), with stopper.
REAGENT SETUP
-
1 |Prepare reagents for 1 litre of 1 × modified M1 minimal media in 100 % v/v 1H2O and 1 litre of 1 × modified M1 minimal media in 100% v/v 2H2O.
-
iPrepare separate 1H2O and 2H2O phosphate buffers. Take two dry 1 litre Schott bottles and to each add: 10.6 g K2HPO4, 4.94 g KH2PO4, 2 g (NH4)2SO4 and 0.5 g NaCl. Dissolve the powders into 1 litre of either 100% v/v 1H2O or 100% v/v 2H2O. Adjust the pH of the 1H2O phosphate buffer to 6.5; adjust the pH of the 2H2O solution to a reading of 6.1 on the pH meter (i.e., pD = 6.5). Use concentrated 1HCl to adjust the pH or pD. If dilute HCL is required for adjusting the pD of the 2H2O solution, dilute concentrated 1HCL into 2H2O. 2HCl (DCl) can also be purchased for adjusting the pD of the 2H2O solution.
-
i
<PAUSE POINT> For this example, there is no need to use deuterated versions of the potassium or ammonium salts because the % total of 1H introduced will not significantly affect the volume fraction of 2H2O of the final media (target = 80%). If perdeuteration is required (i.e., 100% 2H-labelling of a macromolecule) the use of deuterated salts and DCl for pD adjustment is advised. Note: Without a carbon source, the 1H2O and 2H2O phosphate buffers can be stored at room temperature for several weeks, e.g., by filter-sterilising the solutions into a sterile, dry Schott bottle. Ensure that the 2H2O solution is well-sealed to prevent 1H2O exchange with the atmosphere.
-
ii
Make a 1000 × stock of vitamin solution in both heavy and light water. To separate 1 ml volumes of 1H2O and 2H2O add: 10 mg thiamine, 5 mg biotin and 40 mg yeast extract. These constituents may not all dissolve into 1 ml. Do not be concerned and carry on the procedure with the undissolved material. This stock cannot be stored and needs to be used as soon as possible.
-
iii
Weigh out two separate 4 g amounts of powdered 1H-glucose.
<PAUSE POINT> In this example where the average % 2H-labelling of a protein target is 60%, there is no need to use deuterated glucose. For higher % 2H incorporation, a deuterated carbon source maybe required in the media. Refer to Table 1 and Leiting, Marsilio and O’Connell (1998)85.
-
iv
Make a 250 × stock solution of trace metals in light water. Dissolve into 250 ml of 1 M 1HCl in regular 1H2O: 500 mg MoNa2O4, 250 mg CoCl2, 175 mg CuSO4, 1 g MnCl2, 8.75 g MgSO4, 1.25 g ZnSO4, 1.25 g FeCl2, and 2.5 g CaCl2. The solution can be stored for many months at room temperature in a Schott bottle.
-
v
Make a 1000 × stock of MgSO4 in heavy water. Dissolve 2 g of MgSO4 into 5 ml 2H2O and sterile filter the solution.
-
2 |Combine ingredients from REAGENT SETUP i-iii to make 100% v/v 1H2O and 100% v/v 2H2O modified M1 minimal media, without adding trace metal or MgSO4 solutions.
-
viTo l litre of the 1H2O and 2H2O phosphate buffers, add individually to each 4 g of glucose and 1 ml of the respective 1H or 2H vitamin solutions and dissolve.
-
vi
-
3 |
Make deuterated media at the desired % v/v2H2O. For this example, the desired % v/v 2H2O in the final growth medium is 80%. Using a dry measuring cylinder, combine 200 ml of the 1H-media with 800 ml of the 2H-media. Sterile filter the final 80% v/v media and store in a sterile 1 litre Schott bottle. Do not autoclave and do not adjust the pH or pD of the solution.
-
4 |
Make 15 ml ‘adaption’ cultures at different 2H2O concentrations. Prepare two sterile-filtered 15 ml media solutions at 70% v/v 2H2O and 80% v/v 2H2O in 50 ml Falcon tubes using the 1H and 2H-media remaining from REAGENT SETUP 3.
<PAUSE POINT> At this point, both a large 1 litre and two small-scale 15 ml cultures have been prepared. The 15 ml cultures will be used to continue the cell adaption process from Day 3 to the finally required 80% v/v 2H2O environment. The 1 litre of sterile filtered media will be used for the protein over expression experiment on Day 6. This solution can be stored at room temperature until needed (but for no longer than approximately 4 days after the glucose and vitamin solution has been added).
-
5 |
<optional> Make as many small sterile filtered 80% v/v 2H2O cultures as possible, e.g., 50 ml in small stoppered sterile conical flasks, from any remaining 1H and 2H media. These can be used for test protein expression experiments.
PROCEDURE
-
1 |
Adapt the cells to 70% v/v 2H2O minimal media. Centrifuge the 15 ml cell culture grown overnight in the 50% v/v 2H2O/LB media from Day 3 (5000 × g for 10 min) and remove the supernatant. Resuspend the cell pellet in 2–3 ml of fresh 70% v/v 2H2O modified minimal media.
-
2 |
To 15 ml of freshly prepared 70% v/v 2H2O modified minimal media in a 50 ml Falcon tube (REAGENT SETUP 4) add 15 μl of the appropriate 1000 × stock solution of antibiotic as well as 15 μl of the 1000 × MgSO4 stock in 2H2O and 60 μl of the 250 × trace metal solution described in REAGENT SETUP 1iv and 1v.
<CRITICAL STEP> Insoluble metal phosphates will form in solution. There is no need for concern and continue the procedure with these insoluble materials in the solution.
-
3 |
Add 50–100 μl of the cells adapted to the 50% v/v 2H2O/LB media into the 70% v/v 2H2O modified minimal media and grow overnight in the closed Falcon tube at 37 °C, with shaking.
Cell adaption, DAY 5
PROCEDURE
-
1 |
Repeat the steps 1, 2 and 3 of the PROCEDURE from Day 4, but this time use 15 ml of 80% v/v 2H2O modified minimal media with the antibiotics, MgSO4 and trace metals added to generate 15 ml of an 80% v/v 2H2O adapted cell culture.
STAGE 3): Protein expression of the 2H-labelled recombinant target, DAY 6
At the beginning of Day 6 the bacterial cells should have adapted to growing in 80% v/v 2H2O modified minimal media. The 15 ml culture prepared on Day 5 will be used to inoculate the 1 litre of 80% v/v 2H2O media prepared on Day 4 (REAGENT SETUP 3). Transfer the filter sterilised media into a large, sterile smooth-sided and dry conical flask (2.5–3 l) in preparation for cell growth and protein expression.
-
1 |
Centrifuge the 15 ml culture grown overnight from Day 5 in 80% v/v 2H2O media and resuspend the cell pellet into 1–2 ml of fresh 80% v/v 2H2O media.
-
2 |
Use the resuspended cells to inoculate the main 1 litre solution of 80% v/v 2H2O growth media to an OD600nm to 0.05– 0.1.
-
3 |
Add the 1000 × stock antibiotics (1 ml each) and the 250 × trace metal (4 ml) and 1000 × MgSO4 solution (1 ml) prepared on Day 4 (REAGENT SETUP steps 1iv and 1v). Do not be concerned if precipitates form in the solution, continue with the procedure.
-
4 |
Grow the 1 litre cell culture, with orbital shaking, to the mid-log phase of growth generally between OD600nm 0.6–0.75.
<PAUSE POINT> Cell growth in minimal 2H2O media is very slow compared to regular LB media and can take several hours to reach the mid-log phase (e.g., 12 hr compared to 4 hr in LB).
-
5 |
At mid-log phase, induce protein expression as per the induction method of the plasmid (e.g., the addition of IPTG from 0.1–1 mM) and leave the cultures to express protein for a set time period.
<PAUSE POINT> As with cell growth to mid-log phase, the expression of the recombinant protein in the 2H2O minimal media may take 2–5 times longer compared protein expression in regular LB media, or other types of optimised growth conditions (e.g., SOC, or Terrific broth). Therefore, use SDS-PAGE to regularly check the level of protein expression during the course of the expression period (e.g., sample 2 hr, 4 hr, 6 hr, 8 hr time points, and longer if necessary).
-
6 |
Harvest the cells using centrifugation, e.g., 5000 × g for 15 min. Decant off the spent media from the cell pellet and transfer the pellet to a storage container (e.g., at -80 °C) or proceed directly to protein purification, STAGE 4.
STAGE 4): Protein Purification, DAY 7–9
It is expected that the purification of the 2H-labelled protein should follow a similar scheme as that previously determined for purifying the same unlabelled 1H-protein. Use light water buffers (there is no need to use 2H2O buffers) and purify the 2H-protein by following the same 1H-protein purification steps. Adjust buffer conditions and the protocol if required to obtain pure monodisperse protein in solution (main text, PROCEDURE 1).
STAGE 5): Determine the average extent of 2H labelling
<CRITICAL>Peptide mass fingerprinting is one method used to determine the average level of non-exchangeable 2H incorporated into the expressed protein target. The experimentally determined value is important for selecting what % v/v 2H2O to use in samples for SANS with contrast variation and (especially) contrast matching experiments, to obtain a component match point (i.e., where Δρ = 0). We include instructions for how to prepare samples for peptide mass fingerprinting below.
-
1 |
Prepare the following materials: SDS-PAGE gel and Tris-glycine-PAGE gel running buffers; 5 μl unlabelled protein (0.5–1 mg.ml-1) in reducing SDS-PAGE loading buffer; 5 μl 2H-labelled protein (0.5–1 mg.ml-1) in reducing SDS-PAGE loading buffer; and Coomassie Blue staining solution and destaining solutions.
-
2 |
Perform SDS-PAGE on both the unlabelled and 2H-labelled proteins.
-
3 |
Stain the gel with Comassie Blue, then destain the gel to reveal the protein bands. Wash the destained gel three times in MQ-water for 15 min per wash.
-
4 |
Using a scalpel, carefully cut out the bands corresponding to the unlabelled and 2H-labelled proteins and place the gel fragments into separate Eppendorf tubes.
-
5 |
Send the gel slices to a mass spectrometry facility and request MALDI-TOF peptide mass fingerprinting on both proteins with mass-fragment (amino acid sequence) identification and mass analysis.
-
6 |
Use the differences in the masses of the peptide fragments obtained from the unlabelled control compared to the 2H-labelled target to experimentally assess the average level of non-exchangeable 2H incorporated into the recombinant protein. An example spread sheet for this calculation is provided as Supplementary Information Deuteration_incorporation_calculations.xlsx.
Materials: Procedures 1, 2 and 3
Reagents
The list of the reagents is extensive and it is assumed that the reader has access to standard laboratory chemicals to make, for example, bacterial growth media (e.g., Lysogeny Broth, LB52 and refer to Box 3) as well as buffers for SDS-PAGE, protein purification, size-exclusion chromatography and dialysis, etc. For proteins, it is assumed that the correct gene of interest has been cloned into an appropriate expression vector (e.g., a plasmid) and that protein over-expression strains of Escherichia coli are available (also refer to Box 3). For SANS, access to 2H2O is absolutely necessary for contrast matching/variation experiments, noting that 2H2O is expensive (~€1000 per litre.) Additional reagents are mentioned in the text.
Equipment
Access to general laboratory equipment and consumables is assumed. Specific equipment for the combined procedures include, but are not limited to: SDS-PAGE equipment (e.g., from BioRad); Dialysis equipment (e.g., SnakeSkin™ dialysis membrane or Slide-A-Lyzer™ cassettes); Centrifugal spin filters (0.1–0.44 μM pore size for filtering out particulates and with nominal molecular weight cut-offs, e.g., 3.5–50 kDa, for protein concentration); High performance liquid chromatography (HPLC, e.g., from Agilent Technologies) or fast protein liquid chromatography (FPLC, e.g., GE Life Sciences ÄKTA) systems; Size exclusion chromatography columns; A spectrophotometer (e.g., a NanoDrop) or refractometer; Standalone dynamic and/or static light scattering instruments, or (optional) in-line size exclusion chromatography MALLS/RALLS (e.g., from Wyatt or Malvern) attached to a HPLC or FPLC-SEC system; A synchrotron bioSAXS beam line or lab-based SAXS instrument (e.g., from Rigaku, Anton Paar, Brucker, Xenocs) and: For neutron scattering, a SANS beam line. To perform calculations mentioned in the text, MULCh51 can be accessed at http://smb-research.smb.usyd.edu.au/NCVWeb/ while ATSAS4 can be downloaded at http://www.embl-hamburg.de/biosaxs/software.html. Additional on-line tools are mentioned throughout the text.
PROCEDURE 1: SAS sample purity, quality and preparing the solvent blank
Overview of the procedure
For the sound interpretation of the scattering data it is vital that the materials undergoing analysis are pure and free of any significant levels of contamination. As a scattering profile represents the sum of the scattering from each particle in solution (eq. 1), the presence of any contaminants will add to the scattering intensities of a sample. These contaminants will contribute to I(q) at a magnitude that is proportional to the concentration and the volume squared of the contaminant. For example, a sample consisting of a 15 kDa protein purified to 98 % that contains 2 % of a 100 kDa protein (that does not interact with the 15 kDa protein or itself) will generate a forward scattering intensity that is almost twice what is expected from a 100% pure 15 kDa sample (Figure 6). Therefore, the presence of high molecular weight species, including aggregates, aggregates of smaller contaminating particles that coalesce into larger particles, systems that undergo dynamic non-equilibrium oligomerization or suffer from radiation- or time-induced aggregation, can severely complicate data interpretation and modelling. For successful SAS experiments (especially when developing 3D spatial models that fit the data by shape restoration18–20 or rigid body modelling4,21) it is imperative that macromolecules within a sample are as pure as possible, are monodisperse and remain free of interparticle interference effects. In PROCEDURE 1 we first discuss how to assess sample quality before a SAS experiment (STEP 1), then how to prepare the sample and buffer for the measurement (STEP 2).
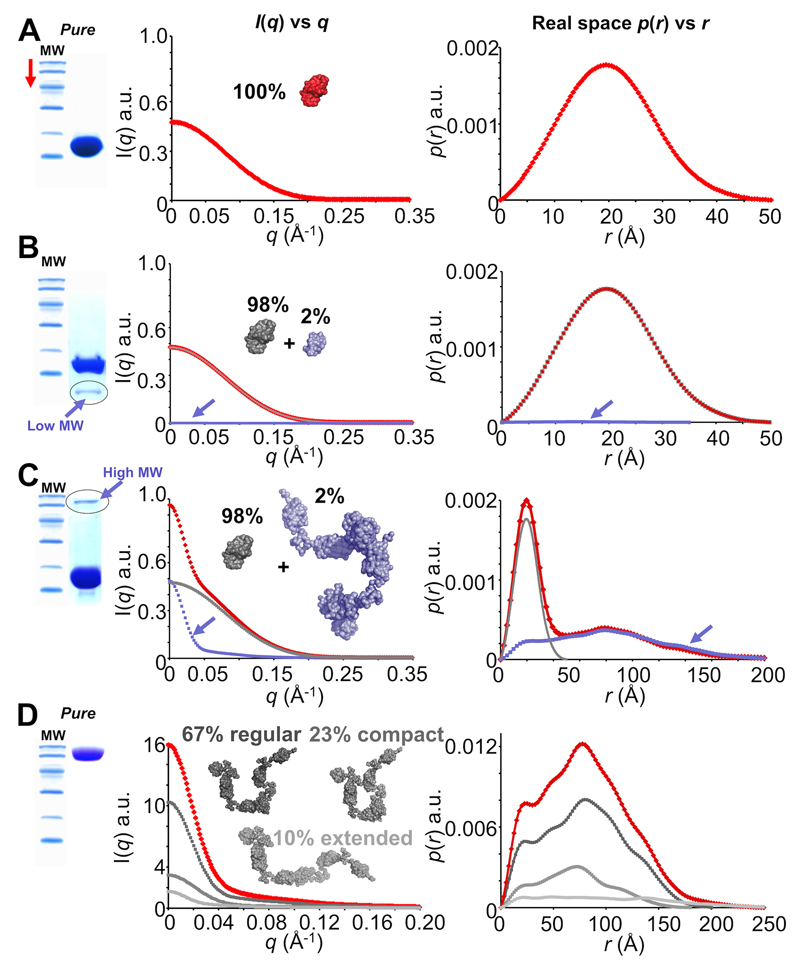
Figure 6. Sample purity and contaminants (simulated SAXS data and simulated SDS-PAGE).
A. The ideal outcome when purifying a sample. Scattering from each individual within a population of pure monodisperse 14 kDa protein sum to produce a total scattering profile (red) from which p(r) vs r can be modelled that represents the real-space atom-pair distance distribution within a single particle. B. A less-ideal situation. If contaminants are present, the total scattering (red) will be comprised of the sum of the scattering from each different species in proportion to their volume squared and concentration. Here, a low molecular weight (MW) contaminant (~5 kDa, 2% of the sample, blue) is present in the 14 kDa protein sample (grey). However, the total contribution to the scattering made by the low MW contaminant is small and does not significantly affect I(q) vs q or p(r) vs r. C. Something to avoid. High MW contaminants have disastrous consequences on I(q) vs q (red). The scattering contributions made by trace ~100 kDa protein (blue) doubles I(0) even though the target 14 kDa protein (grey) is 98% pure. The effect on p(r) vs r is significant as it is the sum-weighted contribution made by the 14 kDa protein plus the 100 kDa contaminant. D. A special case: flexibility. A 100 kDa protein is both pure and monomeric. However, the protein is flexible and is comprised of three main populations so that the total P(q) determined from the scattering (red) is the sum of P(q) from each population (shades of grey). For example, although the extended state comprises only 10 % of the total population, the maximum dimension of the measured p(r) vs r (red) will equate to the Dmax of the most extended state (light grey). Note before: the SDS-PAGE gels and scattering profiles used for this figure are for illustrative purposes only and do not represent real data.
STEP 1: Assess sample purity and quality prior to a SAS experiment
The art of biomacromolecular SAS is based in the preparation and characterisation of high quality samples. For SAXS, this includes optimising conditions that prevent X-ray induced aggregation53,54 (refer to TROUBLESHOOTING). For SANS this includes assessing the stability of a sample over the time period required to collect the SANS data. For both SAXS and SANS the physical aspects of handling samples must also be considered in context of preparation, storage and, if required, shipping samples to distant facilities. For example, unlike X-ray crystallography where crystals can be cryo-protected and stored, the simple act of freeze-thawing a sample for SAS, or introducing too many air-bubbles, may cause the formation of trace amounts of aggregate that can ruin the interpretation of the scattering data. Therefore there is a requirement to assess both sample purity and sample stability.
Polyacrylamide gel electrophoresis (PAGE) gels stained with Coommassie blue are almost universally employed to estimate the ostensive purity of protein samples (PAGE – Option A). Protein quality can also be assessed by size exclusion chromatography (SEC – Option B) or dynamic light scattering/static light scattering (DLS/SLS – Option C).
Option A: Assess sample purity with polyacrylamide gel electrophoresis.
For a SAS sample preparation, use sodium dodecyl sulphate PAGE (SDS-PAGE) to ascertain the presence of contaminants. Employ further purification steps (e.g., SEC) especially if contaminants have a higher molecular weight than the target of interest (Figure 6). These high-MW contaminants need to be eliminated as the scattering intensities scale to the volume squared of a macromolecule (eq. 1). Samples purified to 95% or greater without high-MW contaminants present should suffice for most SAS experiments.
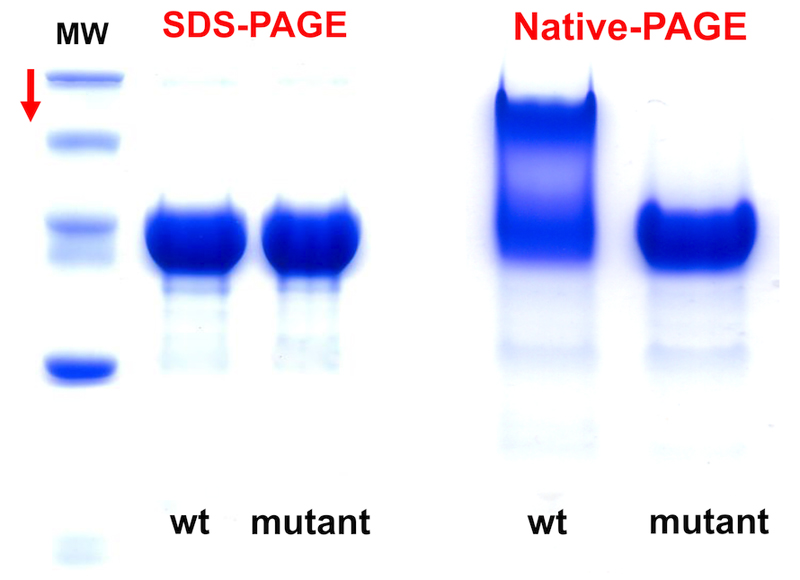
<CAUTION> A single band on an SDS-PAGE gel does not necessarily mean that a sample is monodisperse in solution. Further characterisation steps are necessary, e.g., Native-PAGE and SEC.
-
ii.
Perform Native-PAGE, (run without SDS) to obtain more information regarding whether a protein sample is predominantly homogeneous or is populated by a range of species (Figure 7).
-
iii.
For proteins that are often expressed in reducing environments (e.g., internal to a cell) compare SDS- and Native-PAGE with and without reducing agents added to the sample to assess disulphide mediated oligomerization (e.g., dithiothretitol, DTT; β-mercaptoethanol, βME; or tris(2-carboxyethyl)phosphine-HCl,TCEP-HCl). If necessary, determine the reducing agent concentration (typically 1–10 mM) required to maintain a target of interest in a reduced state (i.e., free of intermolecular cross-links).
Figure 7. The value of native PAGE.
Two proteins, wild-type (wt) and a mutant are analysed using SDS-PAGE and native PAGE, respectively. The native-PAGE result reveals that the mutation radically alters the association state of the protein (from96).
<CAUTION> Make sure Native-PAGE gels are cooled, e.g., perform the electrophoresis in a cold room, to prevent heat denaturation of the protein samples.
Option B: Assess sample quality using SEC.
For precise quantitative analyses of the components present within a sample, the value of size-exclusion chromatography (SEC) cannot be overstated especially when used in combination with ultraviolet (UV) spectroscopy and, if possible, with multi-angle laser light scattering (MALLS) or right-angle laser light scattering (RALLS) and refractive index (RI) measurements. Most structural biology laboratories have access to SEC-UV equipment as part of standard purification procedures and typically monitor SEC-elution profiles at 280 nm. However, SEC-RALLS-RI or SEC-MALLS-RI-DLS instruments (e.g., Wyatt Technology’s DAWN® HELIOS™ II plus WyattQELS™ or Malvern Instrument’s Omnisec Reveal and Zetasizer μV) are becoming increasingly useful for the full analytical characterization of sample components (i.e., continuous-flow component separation combined with molecular weight validation and sizing analysis).
Use SEC-UV to assess the concentration or time dependent stability of a sample via monitoring the formation of aggregates, higher oligomers, etc.
Use SEC to obtain information regarding the oligomerization state or the concentration-dependent association between sample components (e.g., of complexes or assemblies). To do this, perform analytical SEC on small aliquots of sample (50–100 μl) through a dilution series using, for example, using a Superdex 200 Increase 10/300 GL column (GE Healthcare Life Sciences). Use UV spectrophotometry to monitor changes to the SEC elution profile when changing the load concentration. When choosing the highest sample concentration, make sure that the column does not become overloaded (leading to a loss of separation resolution).
Evaluate the load concentration required to isolate fully-formed (or nearly fully formed) complexes. If performing SEC-UV on a protein-DNA complex it is important to monitor the UV absorption at two wavelengths, for example at 280 nm and 260 nm, to demonstrate that a protein-DNA complex has formed and is stable when flowing though the column.
(Optional) Assess the molecular weights (MW) and binding stoichiometry of complexes using SEC-UV(RI) combined with MALLS55 or RALLS. This step can be invaluable when interpreting difficult to analyse SAXS samples (as an example refer to the analysis of the Sda protein from Bacillus subtilis15,56.)
Combine the results obtained from SEC to help interpret the results from PAGE. For example, Figure 8 shows a protein that by SDS-PAGE appears to be pure and monodisperse. However, the SEC elution profile UV trace indicates that the protein is affected by self-aggregation.
Figure 8. Sample characterisation: SDS-PAGE combined with SEC.
The SDS-PAGE result suggests that a protein sample is reasonably pure. However, SDS-PAGE results can be misleading if not backed up by further sample characterisation. SEC indicates that the sample is consists of a heterogeneous population of particles that include self-associated aggregates, dimers and monomers.
Option C: Assess sample quality using DLS/SLS.
Both standalone DLS and SLS (e.g., Wyatt Technology’s DynaPro NanoStar™ or Malvern Instrument’s Zetasizer Nano Range) can be used to quickly screen numerous sample environments (e.g., changes in pH and ionic strength) and evaluate sample integrity (e.g., the formation of aggregates). For example, DLS/SLS can be used to assess the effect of adding metal ions and co-factors, etc, to a sample as well as reducing agents, antioxidants (e.g., sodium ascorbate) or small stabilising molecules (e.g., 5–10% v/v glycerol) that maybe required to limit the effects of radiation damage in a SAXS experiment (refer to TROUBLESHOOTING). The advantage of standalone DLS/SLS over SEC-based MALLS-DLS systems is that analyses can be performed using very small sample volumes and measurements can be completed within minutes.
Use dynamic light scattering on minimal quantities of material (2–10 μl) to evaluate the polydispersity and measure the hydrodynamic radius (Rh) of a macromolecule. Use SLS (in combination with accurate concentration estimates) to assess the molecular weight and how this may change as a result of altering sample environments. DLS, in particular, is extremely sensitive to the presence of aggregates in a sample that will also negatively affect the results obtained from SAS experiments. The general rule of thumb is that if no aggregates are detected with DLS then the sample is of sufficiently high-quality for SAS.
Assess the DLS/SLS parameters at various sample concentrations, temperatures and over time to monitor the formation of aggregates. This analysis could prove significant when preparing and isolating monodisperse samples when targets are low-yielding, difficult-to-produce, or expensive.
<optional> After SAXS measurements have been performed, the Rh obtained from DLS data can be used to inform subsequent shape analysis. The shape factor Rg/Rh, where Rg is derived from SAXS, offers an additional structural parameter for evaluating the mass distribution of a particle (Rg/Rh; sphere = 0.774; flexible coil = 0.816; rod = 1.732). Additionally, the MW obtained from SLS can be used to validate the MW obtained from SAXS.
Use DLS to optimise sample conditions (example).
Aliquot five individual 15 μl samples of a 2 × protein stock solution (e.g., 1–10 mg.ml-1) in a buffer of choice.
To one protein sample add 15 μl of the same buffer (i.e., without additives) to act as a control. To the remaining four samples, add 15 μl of buffer containing additives at various concentrations (at 1, 2, 4, 8 × the desired final concentration). For example, screen NaCl concentrations from 50–300 mM.
Carefully mix the samples, without introducing air bubbles, and centrifuge at high-speed for 5 min (e.g., using a micro-centrifuge 16 000 × g). Carefully remove 5 μl of sample for the DLS/SLS measurements. For SLS record the concentration (refer to PROCEDURE 2).
Compare the polydispersity and Rh parameters extracted from the DLS measurements for each sample variant. Evaluate any changes in the MW from SLS. Evaluate whether a critical threshold of additive causes aggregates to form in solution.
Store the remaining 10 μl of sample over time (e.g., 3–5 days) and repeat the DLS/SLS measurements to ascertain the time-stability and aggregation state with and without additives present.
Use DLS to determine whether aggregates form on freeze-thawing.
Test different freeze-thawing procedures on the aggregation state of a sample using DLS. In this example, snap-freezing a sample in liquid N2 and storage at -80 °C is described, however, similar tests can be performed using samples that undergo snap-freezing on dry ice or slow-freezing and storage at -20 °C (not recommended.)
Snap-freeze two samples (e.g., 100 μl in Eppendorf tubes) using liquid N2 and store at 80 °C. Keep aside an aliquot of sample that has not undergone snap-freezing (e.g., store as a liquid at 4 °C).
Fast-thaw one of the samples (e.g., carefully between your fingertips).
Slow-thaw the second sample slowly on ice.
Compare any changes to the sample (e.g., the formation of aggregates) caused by different freeze-thaw procedures against the sample that has not undergone freeze-thawing (Figure 9).
Answer the question: does the sample need to be frozen in the first instance?
<optional> Use further SEC and PAGE analyses to monitor the effects of different freeze-thaw procedures on the aggregation state of a sample.
Figure 9. DLS as a tool for characterising samples and sample handling.
A. DLS autocorrelation functions of lysozyme (9.1 mg.ml-1) in low salt buffer (20 mM NaCl, 40 mM sodium acetate, pH 3.8). A freshly prepared sample (green) is compared to samples that have undergone i) snap freezing in liquid nitrogen and quickly thawed (black dots) and; ii) where a snap frozen sample has been slowly thawed on ice (blue line, obscured). The exponential decay and smooth return to base line of the autocorrelation data indicates that all three samples are not affected by aggregation, even when stored and handled differently. B. Increasing the NaCl concentration to 170 mM has little effect on the quality of the fresh and quickly thawed samples. However, aggregates are produced if the snap-frozen lysozyme is slowly thawed on ice in the high salt concentration buffer (blue). Data were collected using a Wyatt Technology DynaPro® NanoStar™ instrument.
Determine whether samples are affected by concentration or time.
Determine whether a sample reaches a concentration threshold where aggregates begin to appear (e.g., with or without freezing-thawing and storage). DLS, PAGE or SEC can be used to perform this analysis. Although increased sample concentration will generate improved signal-to-noise ratios in SAS data (eq. 2) it may be necessary to use lower concentration samples for SAS to avoid the effects of interparticle interactions, especially those that result in the formation of concentration-dependent aggregates.
Determine whether samples are stable through time, i.e., susceptible to aggregation or decomposition into smaller components (e.g., hydrolysis, proteolysis, etc). DLS, PAGE or SEC can be used for this analysis. Evaluating time/storage stability at different temperatures may be important to consider when deciding on how to ship samples to distant facilities (e.g., frozen on dry ice or unfrozen on blue ice).
Determine whether the macromolecule slowly sticks to the sides of storage tubes. For proteins, this can be monitored by evaluating the concentration of the sample over time in parallel with PAGE (a decrease in concentration or PAGE band intensity is cause for concern).
Assess whether concentration, time-induced or freeze-thaw aggregates, etc, can be removed by high-speed centrifugation (e.g., using a micro-centrifuge 16 000 × g or an ultra-centrifuge > 30 000 × g), dilution or spin filtration through a centrifugal filter unit (e.g., using 0.1–0.45 μm pore size filter membranes). If not, an additional SEC step may be necessary to remove the contaminating aggregates immediately prior to a SAS experiment (e.g., using a small Superdex 200 Increase 5/150 GL column, GE Healthcare Life Sciences).
<CAUTION> The membranes of spin filters used to remove large aggregates or particulate matter can be made from various substrates that include polyethersulfone (PES), modified polyvinylidene fluoride (PVDF) polytetrafluoroethylene (PTFE), cellulose acetate (CA) and cellulose nitrate (CN). If the macromolecules of a sample ‘mysteriously disappear’ or become increasingly aggregated after spin-filtering, then the membrane is adversely interacting with the sample. If required, test 0.1–0.45 μm spin filters made with different substrates.
STEP 2: Obtaining equivalent sample and background solvents
For macromolecular solution SAS it is very important that the scattering contributions made by the background solvent are subtracted from the sample scattering to obtain the scattering from a macromolecule or complex of interest57. Inaccuracies in the solvent subtraction will lead to residual solvent terms in the subtracted scattering profile (eq. 1) that can cause perturbations in the structural parameters derived from the data (Figure 10.) Therefore, it is essential to produce a solvent blank that is identically matched to the solution of the sample (refer to Introduction). In essence, for most scattering experiments, the preparation of the background solvent is nearly as important as preparing the sample. Preparing a matched solvent can be achieved by sample dialysis (Option A), size-exclusion chromatography (Option B), or in some circumstances, the careful application of molecular weight cut-off centrifugal spin filters (Option C).
Figure 10. The importance of obtaining matched sample solvents.
SAXS data collected from a pure, monodisperse sample of protein in buffer B and three background solvents, A (red), B (black) and C (blue) prior to background subtraction. Buffer B was matched with the sample using dialysis, while buffer A was derived from a mislabelled tube and buffer C from an old buffer stored in the fridge. B. Although the flat solvent scattering intensities measured for each buffer look similar, only the solvent-matched buffer B allows the correct scattering from the protein to be revealed after solvent subtraction. C. Incorrect buffer subtraction can affect the modelling of p(r) vs r. The correct solvent-matched background (black) causes the profile to intersect at 0,0, while under- and over-subtracted buffers result in positive and negative values of p(r) at r = 0, respectively. [Note, although real experimental data is presented for the protein, buffer B and buffer A, buffer C has been adjusted for illustrative purposes.]
<CAUTION> Sample concentration must be considered when choosing the solvent matching method as the scattering intensities are proportional to the number of homogeneous particles in solution (eq. 2). Dialysis affords more control over the sample concentration, whereas SEC suffers from dilution effects as a sample filters through the column. For high-brilliance synchrotron X-ray sources, sample dilution may not be an issue, but for laboratory-based sources, the over-dilution of a sample during SEC may result in compromised signal to noise ratios in the data and necessitate extended exposure times (requiring that samples be both radiation and time-stable). To solve the dilution problem, load concentrated samples onto the SEC column (e.g., 5–15 mg.ml-1). However, this is based on the assumption that concentrating a sample does not cause aggregation or result in column over-loading that leads to a loss of SEC resolving power.
<CAUTION> For SANS experiments, that often require the preparation of several % v/v 1H2O:2H2O solvents, the buffer exchange method using SEC might not be feasible. Therefore, for SANS experiments, dialysis is advised (refer to PROCEDURE 3, STEP 4).
<CAUTION> There is a temptation when preparing a solvent blank to simply forgo sample dialysis or SEC and weigh out the components of a new solution that is ‘close enough’ to the conditions of a sample. This shortcut almost-never works. It is difficult to replicate the sample solvent conditions (and in particular its density and absorption) for a scattering experiment other than performing solvent exchange using dialysis or SEC.
Option A: Perform dialysis to obtain a matched solvent blank.
-
i
For SAXS – and especially SANS – dialyse a sample against a solvent of choice overnight, making sure that any visible bubbles or air pockets have been removed from the dialysis bag, button or cassette.
<CAUTION> Using dialysis to perform solvent matching can in some instances be impractical, for example in cases where a sample is susceptible to slow self-aggregation with time. It may be necessary to test, e.g., using DLS or SEC, that the sample is not affected by time-dependent aggregation during the dialysis procedure.
-
ii.
Collect scattering data from both the dialysed sample and the post-dialysis buffer. The post-dialysis buffer will act as the matched solvent blank for the SAXS or SANS measurements.
-
iii.
For SAXS, use the post-dialysis buffer to dilute the sample to form a concentration series (e.g., 1, 0.75, 0.5 and 0.25 ×) to assess concentration dependent interparticle interference effects, S(q), or the disassociation of complexes. (optional) The results derived from SAXS may be used to inform the choice of sample concentration for SANS (refer to PROCEDURE 3, STEP 2).
Option B: Perform SEC to obtain a matched solvent blank.
-
i
Collect sample fractions corresponding to the separated target of interest eluting from the SEC column.
-
ii
Collect aliquots of buffer (e.g, 500 μl) that have passed through the column to act as the matched solvent blank for the SAS experiment. Attempt to collect buffer fractions as close to the sample elution peak as possible.
<CAUTION> Only use SEC running buffer that has passed through the SEC column as a matched solvent blank, not the buffer from the stock bottle. Limited small molecule fractionation can occur during SEC caused by solute/solvent/column matrix interactions. This fractionation may alter the scattering length density and absorption properties of a buffer that has run through a column along with the sample compared to a buffer that has not run through the column.
-
iii.
(optional) There may be a need to add an expensive, perishable co-factor to a sample (e.g., NADPH) that simply cannot be wasted in the preparation of litres of dialysis or SEC buffer. The best approach in this instance, and one that avoids overly-diluting a sample, is to make up a small volume of the additive as a concentrated stock solution (10–50 ×) in an already-exchanged sample blank. Accurately add a small and equivalent volume (or mass) of the concentrated additive to both the sample and to the matched solvent immediately prior to SAS data collection. If adding an equivalent mass, use a microbalance.
-
iv.
(optional) For SAXS, combine SEC directly with the SAXS measurements, i.e., collect scattering data from the separated sample components immediately after they elute from the SEC column as well as SAXS data from the elution buffer flowing through the column. Refer to Box 1.
Option C: Use centrifugal spin filters (with extreme care).
An alternative to dialysis or SEC is to concentrate samples using centrifugation-based molecular-weight cut-off filters and use the flowthrough as an instant sample blank. This approach can work if extreme care is applied.
<CAUTION> Spin-concentrators can sometimes retain sufficient quantities of small-molecules that subtly alter the solvent composition of a sample compared to the flow-through resulting in a solvent mismatch. Some filters are manufactured with preservatives coating the membranes (e.g., azide, glycerol) that if not washed off completely, may introduce unwanted small-molecules to a sample. More disastrously, samples can aggregate at the membrane interface as concentration gradients develop during the concentration procedure.
Always choose a new centrifugal spin concentrator with an appropriate MW cut-off. A general rule of thumb is that the minimum MW cut-off should be at least 3–5 × less than the MW of the macromolecule of interest. For example if a protein has a monomer MW of 25 kDa, then use a filter with a MW cut-off of 5 kDa (or less).
Wash the membrane of the filter device carefully with a small aliquot of buffer (e.g., pipetting up and down over the membrane) to remove any small molecules remaining from the manufacturing process. Remove excess buffer and then load the sample.
Centrifuge the sample at a speed or × g as per the manufacturer’s instructions. It is best to concentrate the sample using short multiple spins (e.g., 10 × 2 min) with careful mixing of the sample in between each run as opposed to one long continuous spin (e.g., 20 min). This will help prevent a concentration gradient forming and reduce the chances of sample aggregation at the membrane/sample interface. Mix the sample between each short spin by carefully pipetting the sample up and down without introducing air bubbles to the solution.
Once the sample has reached a desired concentration, retrieve the sample and the buffer that has flowed through the membrane. Use the buffer as a solvent blank for the SAXS experiment.
<CAUTION> If the concentration of the sample in solution decreases or plateaus (i.e., dos not increase) during centrifugation this can be a sign of the protein binding to the filter and the possible production of irreversible aggregates (e.g., that can be evaluated using DLS). It might be necessary to test different types of membrane substrate to reduce the chances of irreversible binding/sample aggregation. Membranes can be made of regenerated cellulose (e.g., Amicon® Ultra, Millipore; Pierce™ Protein Concentrators), polyethersulfone (e.g., Nanosep® and Microsep™ from PALL Corporation; Vivaspin from GE Healthcare Life Sciences; Corning® Spin®-X and Pierce PES Protein concentrators) as well as modified nylon or hydrophilic polypropylene (Nanosep MF ®, PALL Corporation).
PROCEDURE 1 TIMING. The production and characterisation of purified macromolecules for SAS can span a few days to several weeks. SDS-PAGE analysis takes 1–2 hr per gel, including setup, staining and distaining. Leave aside longer times for native PAGE, e.g., up to 4 hr to gel. SEC, SEC-MALLS or SEC-RALLS requires several hours to 1 day to complete, including buffer preparation, column equilibration, instrument calibration and performing the sample runs. Save time by equilibrating columns and detectors overnight. For SEC-SAXS refer to Box 1. Standalone DLS and SLS requires 30 min instrument equilibration time and each measurement takes approximately 100 s (e.g., 10 × 10 s each). Set aside 1 day to perform the requisite DLS tests on the sample. Sample dialysis typically requires 1 hr setup time and overnight buffer exchange. Centrifugation-based protein concentration may take 30–60 min to complete, depending on the selected speed and final desired concentration of the sample.
PROCEDURE 2. Quantity guides, sample concentration and molecular weight
Overview: Quantity Guides for SAXS
One of many considerations when preparing samples for SAXS are the quantities of sample needed for an experiment. As a rule of thumb, sample concentrations for standard synchrotron-based SAXS are usually in the order of 0.1–5.0 mg.ml-1. For synchrotron SEC-SAXS, 5–15 mg.ml-1 (or higher) might be required to compensate for dilution effects through the SEC column that may become important when maintaining the association state of multi-component complexes (refer to Box 1). For far-less brilliant laboratory X-ray sources minimum concentrations for standard measurements are usually 3–10 mg.ml-1. The increased concentrations for laboratory-based experiments are necessary to improve the signal-to-noise ratio in the data, especially as particle scattering intensity decreases dramatically with the scattering angle58. If the concentration and X-ray flux is too low, those higher q regions of the scattering profile containing information on mid-range atom-pair separations (e.g., domain-domain dispositions, with resolution of 2 nm and better) can be lost in noise after the buffer subtraction. To determine the concentration of a sample, refer to STEP 1.
A single SAXS measurement typically requires 5–30 μl of sample (50–100 μl for SEC-SAXS, Box 1), with these volumes likely to decrease as microfluidic technologies are introduced59,60. However, what volume can be measured and what volume is required for reproducible experiments may be quite different. For example, the requirement to match the sample with a solvent prior to an experiment means that working with tiny volumes can be impractical. Preparing 100–200 μl of sample provides sufficient material to handle it with confidence and provides enough sample to set up a concentration series, e.g., via serial dilution.
It is always advised to perform serial dilutions of a sample using the matched solvent as a diluent to assess the effects of S(q) on the scattering intensities (eq. 2) and to evaluate the association state of multi-component complexes (whether they are fully formed).
There will always be exceptions to these quantity-guides. For example, depending on the macromolecule or complex, it might be possible to analyse a sample at very high concentrations (10–20 mg.ml-1 and above) as long as S(q) remains insignificant; it may be possible to obtain quality data from a laboratory SAXS source at low concentration (1 mg.ml-1) on macromolecules with high molecular weight (e.g., above 300 kDa), or from those that stay stable through extended exposure periods, etc.
Overview: Quantity Guides for SANS
The quantity of material typically required for a solution SANS experiment is significantly higher than for SAXS. The amount of protein, polynucleotide, complex, etc that needs to be prepared also depends on the type of SANS experiment (e.g., SANS with contrast matching vs SANS with contrast variation; consult Introduction and PROCEDURE 3). In general, 200 μl–500 μl of a sample at 5–10 mg/ml (plus a corresponding matched buffer) are required for a single ‘point’ of a SANS contrast series; a full contrast series may require five or more points to complete, i.e., 5–25 mg of sample material. However, if a sample remains stable over time and is not adversely affected by multiple rounds of 1H/2H dialysis exchange (e.g., does not aggregate or change overall shape/structure oligomerization state, etc) then it is possible to reduce the total quantity of material necessary for a SANS experiment by cycling a sample through different % v/v 1H2O/2H2O solvents. Use SAXS or DLS/SLS to check the aggregation state of a sample and, if possible, use SAXS to determine the optimal sample concentration and conditions for a SANS experiment. How to assess the molecular weight of samples from SAXS and SANS data, i.e., to detect aggregation or significant repulsive interparticle interactions, is described in STEP 2.
STEP 1. Accurately determine sample concentration
Aside from acting as a useful tool for monitoring sample handling procedures, accurate sample concentration measurements are important for the evaluation of SAS data. Obtaining the MW of macromolecules from SAS data acts as one of the most important quality assurance steps that links a sample to a scattering profile35,40,61 and requires the accurate estimation of the sample concentration to within ± 10 % error. Assess SAS sample concentrations using Option A for polynucleotides or Option B for proteins.
Option A: Estimating polynucleotide concentration.
As polynucleotides absorb UV light very strongly at 260 nm, create a dilution series of polynucletotide samples to within the linear response range of a spectrophotometer. For example, double stranded DNA at 1 mg.ml-1 has an A260 nm of ~20 using a 1 cm path length (e.g., perform 10, 20, 40 and 80 × dilutions.) NanoDrop spectrophotometers (Thermo Scientific) have shorter path lengths (0.05–1 mm) and can record higher concentrations of polynucleotides without dilution (up to ~15 mg.ml-1).
Measure UV absorbance at A260 nm for the polynucleotide sample, using the matched solvent to zero the spectrophotometer.
Divide the A260 nm absorbance reading by the path length (in cm) and extinction coefficients for either: double stranded DNA (~0.020 (μg/ml)-1.cm-1), single stranded DNA (~0.027 (μg/ml)-1.cm-1) or single stranded RNA (~0.025 (μg/ml-1).cm-1) and multiply by the dilution factor to obtain the approximate concentration of the polynucleotide.
Option B: Estimating protein concentration.
For proteins, amino acid analysis (AAA) is perhaps the most accurate way of determining protein concentration and provides data to calibrate protein concentration assays, but it can take several days to a week to complete and requires access to skilled personnel and specialized facilities. We therefore recommend either measuring the concentration of a protein spectrophotometrically using absorbency readings at 280 nm or, alternatively, using refractive index. The refractive index measurement can, with some adaptation, also be employed for the analysis of polynucleotides. The use of conjugating dyes (e.g., Bradford reagent62) is generally less accurate for determining protein concentration, except in circumstances where the dye-assays have been confidently standardised (e.g., relative to another technique). Estimating concentration from a known mass of a powdered protein used to reconstitute a solution is generally difficult due to the presence of unknown quantities of salts and other molecules that often accompany powdered protein samples.
-
i
Estimate the protein concentration by measuring the absorbance of a protein sample at 280 nm using the matched solvent to zero the spectrophotometer. Divide the absorbance reading by the path length and the protein extinction coefficient. The extinction coefficient can be calculated from the amino acid sequence of the protein (e.g., using Protparam63: http://web.expasy.org/protparam/).
<CRITICAL STEP> Time dependent or chemical changes in the supporting solvent have to be considered when assessing macromolecular concentration using spectrophotometry. Thiol reducing agents such as dithiothreitol (DTT) can change their UV absorption characteristics as they undergo oxidation64 or can interfere directly with dye-based methods. Both DTT and the alternative thiol reducing agent β-mercaptoethanol, have relatively short half-lives at pH’s greater than 7.5 (~1–20 hr depending on temperature65) and DTT acts as a chelation agent toward some biologically-relevant metal ions like Zn2+66,67 which alters absorption properties that can perturb concentration estimates. TCEP-HCL is a significantly more stable and superior alternative68 and has negligible effects on A280 nm readings, however its effectiveness is compromised in phosphate buffers at neutral pH. Care must be taken to adjust a solvent’s pH back to its intended value after TCEP-HCL addition as it is very acidic.
-
ii.
Alternatively, perform refractive index (RI) measurements on the protein sample. RI is an extremely useful tool to assess protein concentration as the refractive index increment for a protein (~0.185 ml.g-1) is – unlike A280 nm extinction coefficients – relatively stable against changes in amino acid sequence composition69. The RI increment can also be adjusted for polynucleotides (DNA: ~0.17 ml.g-1, RNA: 0.17–0.19 ml.g-1). Consequently RI may be more useful for determining the concentration of, for example, proteins with low A280 nm extinction coefficients or protein/DNA complexes.
STEP 2: Molecular weight (MW) analysis
We have included instruction for determining the molecular weight of macromolecules in solution from SAS data. The MW of a scattering particle can be estimated using a combination of the sample concentration and the extrapolated forward scattering intensity at zero angle, I(0), derived from the Guinier analysis70 or from the calculated probable real-space distance distribution, p(r) vs r71,72. The procedures outlined below in Option A (SAXS) and Option B (SAXS and SANS) are both concentration-dependent methods. Alternatively, for SAXS – in particular for protein scattering – concentration-independent MW estimates can also be derived from the excluded particle volume, V, which can be computed from the scattering data. Although corrections may be required when assessing MW based on V for highly extended or flexible particles73, it is often useful to compare both MW-from-V and the concentration-dependent MW estimates from I(0). Several MW-from-V approaches are available that include the methods of Fischer et al. (SAXS-MoW http://www.if.sc.usp.br/~saxs/,74), of Rambo and Tainer through a correlation ‘volume’75 and volume-based molecular weight determinations using DATPOROD from the ATSAS software suite4,5 (where, for proteins, VDATPOROD/1.6 ~ MW). The volume obtained from ab initio dummy atom models (DAM) of proteins that fit the SAXS data (e.g., using DAMMIF76) can also be used to estimate protein MWs, with the general rule of thumb MWprotein=VDAM/2. Incorporating MW results from independent light scattering measurements (SLS/DLS or combined SEC-MALLS or SEC-RALLS) helps one to further validate what is arguably one of the most important overall parameters that may be derived from a SAS investigation (refer to Box 1).
Option A: MW from I(0), SAXS.
For SAXS, perform concentration-dependent I(0) MW analysis by scaling the sample scattering data to a standard with a known concentration. The standard should have a similar contrast as the sample (e.g., use a lysozyme standard for protein samples in aqueous solution77,78).
-
i
Collect SAXS data from a MW standard and the corresponding solvent blank. Use the same experimental set up, e.g., temperature, exposure time and sample cell (e.g., a capillary). Reduce the scattering data, (e.g., radially average 2D data to 1D data) to produce unsubtracted I(q) vs q profiles of the standard and the solvent. Note: for instruments that are not point sources, e.g., Kratky cameras79 apply the relevant beam geometry corrections to the SAXS data.
-
ii
Subtract the solvent scattering from the scattering of the standard to obtain the subtracted 1D I(q) vs q profile of the standard macromolecule in solution.
-
iii
Repeat the data collection procedure for a sample with an unknown MW and its corresponding matched solvent. Use the solvent to dilute the sample to form a concentration series (e.g., 8, 6, 4, and 2 mg.ml-1). Process the data to obtain the reduced and subtracted I(q) vs q profiles of the sample macromolecule(s) in solution. Apply beam-geometry corrections if necessary.
-
iv
Calculate I(0) for both the standard and the sample macromolecules using the Guinier approximation (e.g., using AUTORG5) or from the area under the calculated real-space distance distribution, p(r) vs r (e.g. using AUTOGNOM5).
-
vDetermine the MW of the sample relative to the standard using:
where c is the accurately recorded concentration (in w/v units, mg.ml-1) and υ is the partial specific volume (cm3.g-1).(4)
<CRITICAL STEP> If Δρstandard = Δρsample and the υ of the standard and sample are similar – which is often the case when standardising protein SAXS data against a protein standard – the ratio Δρ2standardυ2standard/Δρ2sampleυ2sample in eq. 4 is ~1 and therefore it is not necessary to determine the contrasts or partial specific volumes. However, if the standard has a different contrast or partial specific volume compared to the sample, for example when comparing a protein standard to a DNA sample, or to a protein sample in glycerol, the ratio will no longer be unity. Under these circumstances it will be necessary to calculate both Δρstandard and Δρsample to take into account the differences in electron density of the standard relative to the sample in their corresponding solvents as well as any differences in υ. Contrast calculations can be performed using the Contrast module of MULCh51 (refer to Box 2) or, if an atomic structure is available, using CRYSOL80. The partial specific volume can also be estimated using Contrast or for proteins and RNA can be calculated using NucProt81, http://geometry.molmovdb.org/nucprot/.
-
vi.
Evaluate whether a systematic decrease or increase in the MW of the sample are observed on changing the sample concentration. An increase in the MW of the sample on increasing concentration is a sign of concentration-dependent oligomerization or aggregation. A significant decrease in the apparent MW of a sample when increasing concentration is typically caused by repulsive interparticle interference.
-
vii.
Compare the experimentally determined MW from I(0) against the expected MW of a macromolecule, e.g., for proteins, calculated from the amino acid sequence (using Protparam63). Use the result to evaluate the oligomerisation or aggregation state of the sample. <optional> Cross-check the MW result against measurements made from independent light scattering experiments (e.g., SLS or SEC-MALLS/RI) or, for proteins, the concentration-independent MW based on the estimated particle volume calculated from the SAXS data4,5,73–75.
<CRITICAL STEP> The standard selected for the MW calibration of SAXS data must be stable in the X-ray beam, i.e., must not be susceptible to radiation damage (refer to TROUBLESHOOTING). X-ray induced aggregation of the standard will increase the I(0)standard resulting in an underestimation of the MW of the sample. Additionally the standard cannot be unduly influenced by repulsive interparticle interference that otherwise decreases the magnitude of I(0)standard resulting in MW overestimates of the sample. If unsure, perform SAXS measurements from a concentration series and generate a plot of I(0)/c vs c. The value of I(0)/c should be relatively constant (within error) if S(q) is negligible; a significant positive slope indicates positive S(q), e.g., in the worst case scenario aggregation; a negative slope suggests negative S(q), i.e., repulsive interactions.
Option B: MW from I(0) for SAXS and SANS.
Perform I(0) MW analysis for SAXS by placing the scattering data of a sample on an absolute scale whereby I(q) has the unit cm-1. For SAXS absolute scaling is typically performed using the scattering from water as a reference82.
Measure SAXS data from pure water in a capillary (or sample cell) and obtain the unsubtracted 1D scattering profile.
Measure I(q) vs q from the identical, but empty capillary/sample cell used for the water measurement. Ensure that the empty capillary/sample cell is completely dry and that the X-ray exposure time and temperature are the same as for the water measurement.
Subtract the empty capillary/sample cell scattering contributions from the water scattering to obtain the subtracted I(q) vs q profile of water alone.
Record the experimental, or ‘instrument value’, for the forward scattering intensity of water, Iwater(0)experimental. A simple way to calculate the forward water scattering is to determine the average magnitude of I(q) across a mid-to-high-q range, i.e., in the ‘flat scattering’ region of the water SAXS profile (refer to the Supplementary Information spread sheet: MW_from_absolute_scale.xlsx).
Next, collect SAXS data from the sample and the matched solvent blank in the capillary/sample cell using the same temperature as the water measurement. Subtract the solvent + cell scattering from the sample + cell scattering to obtain the subtracted I(q) vs q profile of the macromolecules of the sample.
- Place the scattering on an absolute scale (cm-1) by multiplying the scattering intensities I(q) of the macromolecules of the sample by the ratio:
where Iwater(0)standard is the known forward X-ray scattering from water at a particular temperature (refer to Sheet 2 of MW_from_absolute_scale.xlsx). Determine the I(0) of the macromolecules in the sample from the absolute scaled SAXS data using standard methods, i.e., using the Guinier approximation or from p(r) vs r.
- If I(0) is placed on an absolute scale in cm-1 and c is determined in g.cm-3, the MW of a macromolecule can be evaluated via:
where NA is Avogadro’s number, and Δρυsample is the product of the contrast (Δρ, cm-2) and partial specific volume (υsample, cm3.g-1) of the macromolecule. Refer to the Supplementary Information spread sheet MW_from_absolute_scale and Box 2 for further instructions.(5) Perform the absolute scaling of SANS data by normalising the scattering intensities to the incident beam flux, correcting for sample transmissions and instrument geometry49,83. Different SANS facilities will perform the absolute calibration of their instruments using different procedures. Fortunately most facilities provide their SANS data in cm-1 obviating the need for additional data scaling. Determine the MW from I(0) using the same relationship as in eq. 4, substituting Δρ with the coherent neutron scattering contrasts (calculated by Contrast51, refer to Box 2).
PROCEDURE 2 TIMING. For polynucleotide and/or protein concentration determination, 1–2 min per sample (includes blanking the spectrophotometer). For MW estimates using SAXS, set aside time to make a MW standard (if employed) e.g., lysozyme may need overnight dialysis/buffer exchange. The time required for regular SAXS measurements, that includes loading the sample and solvent, collecting the sample and solvent data and washing and drying the sample cell/capillary between measurements takes anywhere between 2–10 min (high-flux synchrotron SAXS) to 1–4 hr (lab-based X-ray source). Calculating the contrast using MULCh takes approximately 5 min (Box 2).
PROCEDURE 3. Sample preparation for SANS
Overview of the procedure
The power of SANS for probing the structures of macromolecules in solution arises from the ability to alter the coherent neutron scattering contrast of a system without the need to radically alter the chemical environment (that is otherwise necessary for SAXS; refer to Introduction). Alerting the neutron contrast for biological samples typically involves 1H-2H isotope exchange orsubstitution either in: i) the supporting aqueous solvent, or ii) (if required) the macromolecule of the sample, or; iii) both in combination. As SANS intensities are proportionate to Δρ2 (eq. 2 and 3) and as Δρ can be experimentally controlled by swapping 1H for 2H, it becomes possible to isolate the coherent SANS signals produced by the individual components of a multi-component sample consisting of different regions of neutron scattering length density. Consequently, SANS with contrast matching or SANS with contrast variation may be used to determine the low resolution structure and dispositions of the components of macromolecular complexes and other higher-order assemblies. The following procedure outlines the major practical considerations for designing solution SANS experiments. STEP 1 describes how to calculate the contrast match points of macromolecular samples prior to an experiment, i.e., at what % v/v 2H2O is required in a solvent to produce Δρ = 0. From here, it can be decided whether deuterium labelling of a component is required to separate the component match points. STEP 2 outlines aspects of sample quality for SANS with contrast variation experiments, with a particular emphasis on evaluating the solubility of samples in 2H2O solvents. STEP 3 describes one way to set up a SANS with contrast variation series. In addition, Box 3 details the procedure for labelling a protein with non-exchangeable 2H to change the coherent neutron scattering length density, i.e., the production of biodeuterated material.
STEP 1: Calculating SANS contrasts
<CRITICAL> The main issue encountered when using SANS with contrast matching or contrast variation is the separation between the match points of the individual components of a macromolecular complex relative to the match point of a whole complex. In other words, at which points the % v/v 2H2O in the solvent produces Δρ = 0 for each component and the complex (Figure 5). If the match point of the individual components are too close to that of the whole complex (e.g., within +/- 10% v/v 2H2O), it may become exceptionally difficult to record sufficiently intense coherent SANS data, especially from samples in 1H-rich solvents that are affected by significant incoherent 1H scattering (0–50% v/v 2H2O). In such circumstances, match point separation can be achieved by changing the 1H per unit volume of a component, i.e., by partially or completely deuterating a macromolecule with non-exchangeable 2H that will radically alter Δρ (eq. 3). Otherwise intractable systems become accessible to SANS investigations as a result of deuterium labelling, such as the analysis of 1H-protein-2H-protein complexes (Figure 5).
-
i
Use the program Contrast as part of the MULCh suite of analytical tools51, to calculate SANS match points of the individual components and of a whole complex. Refer to Box 2.
The output of Contrast includes the calculation of the macromolecular volumes, V, and both SAXS and SANS contrasts, Δρ (in cm-2). For SANS, the coherent neutron scattering contrasts are presented at different fractions of 2H2O in the solvent. The volume fraction of 2H2O that produces zero contrast are also reported, i.e., the neutron scattering match points of the components of a complex and of the whole complex.
-
ii.
In addition to match point separation, consider the effect of incoherent scattering from 1H i.e., the level of ‘background noise’, that affects the quality of the coherent scattering of a SANS experiment. An increase in incoherent scattering in proportion to the coherent scattering signal, i.e., a worse signal-to-noise ratio, will limit the information content, i.e., the useful angular range of the SANS profiles58.
The intensity of the coherent scattering signal above the incoherent scattering background is obviously affected by the concentration of 1H in the sample. However, the coherent SANS signal also relates to (ΔρV)2 of a complex and its components. As a component is matched out, (ΔρV)2 will limit toward that of the remaining component in the complex (eq. 3). For example, a 50 kDa complex comprised of a 25 kDa 1H-DNA subunit (match point = 70% v/v 2H2O) and a 25 kDa 1H-protein subunit (match point = 42% v/v 2H2O) will likely produce reasonable coherent DNA scattering above the incoherent 1H scattering at the protein match point. However, if the volume ratio of the individual components are more extreme – even if the match points are well separated – for example a 5 kDa piece of DNA bound to a 45 kDa protein, then the coherent scattering intensities from the small DNA subunit will be very weak at the protein match point and may be ‘drowned out’ by the incoherent 1H scattering. In general, if a macromolecular component of a complex has 10–15% of the volume (mass), or less, relative to its binding partner(s) it may become challenging to collect quality coherent SANS data from the small component in 1H-rich solvents. The apparent solution to this problem is to increase sample concentration, but this runs the risk of introducing interparticle interference effects, or to increase the neutron exposure time, but this might not be an option given the allocated time on an instrument. The alternative is to isotopically label one of the components with non-exchangeable 2H.
-
iii.
If necessary, determine what average level of non-exchangeable 2H labelling is required to obtain SANS match point separations. This calculation can be achieved by altering the ‘deuteration level’ parameter in Contrast and noting the change in the % v/v 2H2O of the predicted match points from the Contrast output (Refer to Box 2). Alternatively, if an atomic structure is already available, use CRYSON84.
Deuterium labelling will alter the coherent neutron scattering length density of a macromolecule and consequently change the magnitude of Δρ producing a shift the component match point to a different % v/v 2H2O. For example, deuterating a large component of a complex will enable the coherent neutron scattering to be matched out in high % v/v 2H2O solvents (90–100 % v/v 2H2O). Under this condition, data can be measured from a small 1H-binding partner in a background with low incoherent scattering. A reversed 2H labelling strategy may also be considered, i.e., collect SANS data from a fully-deuterated small component – that will increase the magnitude of Δρ in 1H-rich solvents – in complex with a large 1H binding partner. It may be necessary to supplement contrast variation experiments with specialised contrast matching experiments with alternative 2H labelling strategies to obtain a full set of quality SANS data.
-
iv.
If required, isotopically label a macromolecule with deuterium to alter its neutron scattering length density. The level and extent of non-exchangeable 2H incorporated into a macromolecule can be controlled experimentally using biodeuteration85. Box 3 outlines the steps necessary for recombinant protein expression in 2H2O media using Escherichia coli B bacterial strains, for example E. coli Bl21(DE3). The use of E. coli K12 strains (e.g., DH5α) is not recommended.
<CRITICAL STEP> As deuterium labelling can be both time and labour intensive, it is advised to initially perform biodeuteration on a small scale (e.g., 50–100 ml bacterial cultures) before committing to the production of large quantities of sample. It is necessary to perform feasibility studies to evaluate the levels of recombinant expression as well as the solubility of the resulting 2H labelled product. For example, using SDS-PAGE to test the total and soluble protein content of cell lysates and comparing the results to regular 1H expression. (optional) Include the results of 2H-expression trials to support your written proposals to specialised biodeuteration facilities (refer to Box 3).
-
v.
After biodeuteration, experimentally determine the average level of non-exchangeable 2H incorporated into a protein using peptide mass finger printing or whole protein mass spectrometry. The experimental value obtained for the average level of 2H labelling can be entered into Contrast to estimate the expected experimental match points of a complex prior to a SANS experiment. This will help guide what % v/v 2H2O solutions to prepare for contrast matching or contrast variation experiments. The basic practical steps to prepare samples for peptide mass fingerprinting are outlined in Box 3. The Supplementary Information spread sheet Deuteration_incorporation_calculations.xlsx is provided as an aid for calculating the final average level of non-exchangeable 2H using the results from peptide mass fingerprinting.
STEP 2: Assess sample solubility and stability
What ultimately dictates the success of biological SANS experiments is the stability and solubility of a samples in 2H2O solutions as well as the solubility and stability of any 2H-labelled components. The neutron beam flux, beam size and available time at an instrument also has to be considered, especially in context of the quantity of material that needs to be prepared for a full SANS with contrast variation series. New sample environments may have to be sought (e.g., alter salt concentration, pH/p2H, etc) in order to satisfy conditions where a complex is stable over time, fully associated, soluble and monodisperse in both 1H2O and 2H2O solvents.
-
i
Perform standard sample purity and quality checks, e.g., using SDS-PAGE, native PAGE and SEC, during the purification of the SANS samples. For macromolecules labelled with non-exchangeable 2H there is no need to use 2H2O buffers during the purification stage. Regular light-water buffers will suffice, i.e., follow the same purification strategy as if isolating non-labelled material.
-
ii
After the sample components have been purified to homogeneity, test the solubility, time-stability and effect of storage conditions with particular emphasis on evaluating the solubility and association state of complexes in 2H2O solutions.
<CAUTION> 2H2O generally promotes aggregation. The strength of 2H-hydrogen bonds is different to 1H-hydrogen bonds and the solvation layer around a macromolecule has different properties compared to bulk solutions84,86,87. The cumulative effects of these differences is that the addition of 2H2O to the solvent, or the use of 2H-labelled components, can affect the solubility, stability and structural dynamics of macromolecular complexes88–91. In a worst case scenario, biodeuterated material might only be expressed in an insoluble form, or components or complexes that are soluble in 1H2O might be completely insoluble in 2H2O (Figure 11). In addition, it is necessary to test that a complex actually associates in the presence of 2H. Although the concentration range for SANS (typically, 5–10 mg.ml-1) is above the disassociation constant of most physiological complexes, it is prudent to evaluate whether the addition of 2H2O to the solvent or the 2H-labelling of a component (if employed) affects complex formation. Very importantly, a sample must remain soluble, monodisperse and stable across time in both 1H2O and 2H2O solvents, i.e., should not aggregate or fall apart during the time required to prepare/store the sample or collect the SANS data.
Figure 11. Dialysis set up for SANS.
Two snap-lock, or ‘sandwich’ bags containing 100 ml of 100% v/v 2H2O buffer solution and a 3.5 kDa MW cut-off Slide-A-Lyzer™ cassette (pink) containing a sample. In this instance the inner bag leaked and the sample precipitated during dialysis against the high % v/v 2H2O buffer (as noted by the white precipitate).
<CAUTION> The melting point of pure 2H2O is 3.8 °C. Be careful that cold 2H2O solutions or samples made in 2H2O do not inadvertently freeze when stored in a regular laboratory fridge or cold room.
-
iii.
Perform solubility and stability trials of the sample components using material obtained from small-scale purifications prior to committing to scaled-up procedures. The size of a neutron beam means that 200–500 μl of sample – for each contrast point – may be required, i.e., 5– 25 mg of material (or more) for a full contrast series experiment. Optimising sample conditions on a small scale is therefore highly recommended.
-
iv.
Solubility/stability testing can be achieved by dialysing test samples (e.g., 50–100 μl) against 0 and 100% v/v 2H2O solvents/buffers (e.g., overnight). Record the concentration of the samples prior to and after dialysis to assess any significant changes in concentration (e.g., using spectrophotometry, refer to PROCEDURE 2, STEP 1) Note: A significant reduction in concentration of the post-dialysis sample may indicate that the sample forms insoluble aggregates that have precipitated out of solution.
-
v.
Use DLS or SLS (as described in PROCEDUE 1, STEP 1, Option C) to evaluate any significant changes in Rh, MW and polydispersity between the test samples. Use a 1H-macomolecule dialysed against a 0% v/v 2H2O buffer as a control. If DLS/SLS are not available, SEC in combination with UV spectrophotometry can be used to determine whether soluble aggregates are present in the samples, or if tightly formed complexes disassociate in the presence of 2H2O. As 2H2O is expensive and will be required for the SEC running buffer, it is recommended to use small 3 ml analytical columns to perform the analysis (e.g., Superdex 200 Increase 5/150 GL column from GE Healthcare Life Sciences).
-
vi.
(optional) Perform SAXS on the test samples and their respective solvent blanks, keeping in mind that SAXS is insensitive to 1H-2H isotopic substitution. Evaluate the effects of high % v/v 2H2O solvents on the basic structural parameters of a sample (Rg, Dmax, p(r) vs r, V and MW). Check that the SAXS scattering profiles of the samples at various 2H2O concentrations are similar (e.g., using the Correlation Map92 method). Changes in the MW of the 2H test samples relative to a 1H control (refer to PROCEDURE 2, STEP 2) are a cause for concern (i.e., aggregation or complex disassociation). Note: an advantage of performing SAXS on the test samples is that the additional data set can be used in parallel with the SANS results to model the structures of macromolecules.
-
vii.
(recommended) Test the integrity of the SANS test samples over a period that reflects the time necessary to acquire SANS data. Neutron flux at SANS beam lines is much lower than that for X-rays necessitating long exposure periods. Although neutrons are unlikely to cause radiation damage, samples must be time-stable during the often extended exposures required to obtain quality data (0.5–4 hrs per sample and for each matched solvent). For a minimal 5-point contrast variation series, i.e., the measurement of five samples and five matched solvent blanks, 5–40 hrs of beam time may be needed to complete an experiment. If it is necessary to improve data quality (especially for samples where incoherent 1H from the solvent dominates the scattering) quadrupling the SANS collection time should result in an approximate two-fold improvement in counting statistics. However, this can significantly extend the length of time samples are exposed in the beam. Therefore, perform a time course experiment on small aliquots of sample and assess changes in the sample using DLS/DLS, SEC, SAXS etc, prior to the SANS experiment.
STEP 3: Adjusting solvent pH and pD
When making the aqueous solvents for solubility and stability testing as well for the final SANS contrast matching or variation experiments, it is important to remember that pH (i.e., for regular 100% v/v 1H buffers) and p2H (or pD, for 100% v/v 2H2O buffers) are not equivalent93. When using glass pH electrodes, the pH and the pD are related via:
Consequently, as soon as 1H2O and 2H2O solutions are mixed to produce different % v/v 2H2O solvents it becomes difficult to accurately assess or adjust the pH/pD. The solution to this problem is to make up identical buffers in 100% v/v 1H2O and 100% v/v 2H2O, perform the pH and pD adjustments taking into account the correction factor, then mix the 100% v/v solutions in the appropriate ratios.
-
i
Obtain two 50 ml (dry) plastic falcon tubes and individually weigh out the equivalent mass of the components required to make two identical buffers. Be as accurate and precise as possible, for example use an electronic balance and aim to be within +/- 10 mg error for each component across both tubes.
<CRITICAL STEP> As all isotopes have the ability to scatter neutrons, the concentration of 12C, 16O, 14N, 31P, 32S, etc, i.e., the atomic composition, must be identical between the finally made 100% v/v 1H2O and 100% v/v 2H2O buffers, except for the concentration of 1H and 2H.
-
ii.
Dissolve the buffer/solvent components into small volumes (e.g., 10–20 ml) of either 100% v/v 1H2O or 100% v/v 2H2O to generate two tubes of concentrated buffer solution. Transfer the dissolved components from the tubes into an appropriate, and respective, volume of pure 1H2O or pure 2H2O required to constitute a 1 × buffer. Note: Bottles or vessels used for 100% v/v 2H2O solutions should always be dry prior to making 2H2O buffers and kept well-sealed to prevent 1H2O exchange with the atmosphere.
-
iii.
Adjust the pH of the 100% v/v 1H2O buffer and then pD adjust the 100% v/v 2H2O buffer to the desired value, taking into account the correction factor of 0.4. For example, when adjusting the pD of a 100% v/v 2H2O buffer to 7.0, the pH meter should read 6.6. Preferably, use 2HCl or NaO2H (often called DCL and NaOD) to adjust the pD.
-
iv.
To make different % v/v 2H2O solvents, mix the pH adjusted 100% v/v 1H2O buffer and the pD adjusted 100% v/v 2H2O buffer to the appropriate ratio without any further pH/pD adjustments.
<CRITICAL STEP> If it is necessary to add a cofactor, expensive reagent or a component at low concentration, it is advised to accurately weigh out identical quantities of material and make up 10 × or 20 × stocks (e.g., in 1 ml) in both 100% v/v 1H2O and v/v 100% 2H2O, respectively. The appropriate volume of the concentrated stock can then be added to the main 1 × 100% v/v 1H2O or v/v 100% 2H2O buffer solutions prior to pH/pD adjustment.
STEP 4: Setting up a SANS with contrast variation series
The simplest way to set up a SANS contrast series is via the dialysis of the samples against a solutions containing different % v/v 2H2O ratios. Dialysis is typically performed over night to complete 1H-2H exchange between the solvent and the sample. There are two main options available for setting up a contrast series depending on whether there is limited (e.g., 0.6–1 ml; Option A) or plentiful (e.g., 2–2.5 ml; Option B) sample material. The key things to remember when setting up the dialysis are: consistency with handling the sample; consistency and accuracy when making up the dialysis solutions and; eliminating as much contaminating or unknown quantities of 1H that will otherwise alter the contrast and/or introduce incoherent scattering noise to a SANS profile.
For both Options A and B, prepare a master stock of a macromolecule, complex or assembly in regular 1H2O buffer at the concentration selected for the SANS experiment (e.g., 7 mg.ml-1). Aliquot/divide the master stock into smaller sub-samples for the subsequent dialysis and mixing steps. As 1H-2H exchange can occur across different timescales overnight dialysis is recommended (or a minimum of 8 hrs).
<CRITICAL STEP> If a deuterated component is being used for the SANS experiment, it should be derived from the same batch of biodeuterated material (i.e., the same batch of cells, Box 3). Avoid making multiple small samples derived from different protein preparations that can be affected by differences in concentration, mixing errors and deuteration levels.
Option A: Limited sample material, e.g., for contrast matching.
Separately dialyse two aliquots of an identical sample (e.g., 300–500 μl) against 100 ml of 0% v/v 2H2O and 100 ml of 100 % v/v 2H2O solutions, respectively.
After the dialysis is completed, use an accurate pipette to carefully mix the post dialysis samples together in the appropriate volume proportions necessary to obtain the desired % v/v 2H2O in the sample. It is also necessary to carefully mix the post-dialysis 0% and 100% v/v 2H2O buffers using the same volume ratios as the samples to act as the matched solvent blanks for the SANS measurements.
Option B: Plentiful sample material, e.g., for contrast variation.
From 2–2.5 ml of a master stock, separately dialyse 200–500 μl aliquots of sample overnight against 100 ml of each respective % v/v 2H2O solvent required for the contrast series. This typically includes preparing % v/v 2H2O solutions above, below and at the two component match points (e.g., 0%, 20% 42% 68%, 90% 100 % v/v 2H2O). Although this approach uses more material that Option A, it avoids bad pipetting or mixing errors, reduces the chances of formulating miss-matched solvents and prevents potential 2H2O ‘mixing-shock’ that could destabilise/subtly aggregate a sample.
Centrifuge the dialysed samples at high-speed (30 000 × g, 10 min) to remove any insoluble material.
Use 200–500 μl of the post dialysis buffers as the matched solvent blanks for each % v/v 2H2O contrast point, making sure to either centrifuge the buffer at high-speed (30 000 × g) of filter through a 0.22 μm filter to remove particulates.
For Options A and B assess sample concentration.
-
i
After dialysis is complete, record the concentration of each sample to assess any changes caused by the dialysis procedure. The concentration values will be required for determining the MW of the samples from I(0) at each SANS contrast point (eq. 5 and refer to Box 2).
<CAUTION> Wherever possible, all consumable materials (volumetric pipettes for aliquoting buffers, pipette tips, dialysis cassettes, loading syringes, needles, eppendorf tubes, etc) should be dry and dedicated for the separate handling of 1H2O and 2H2O samples and buffers. This reduces the chances of 1H-2H cross contamination. Small-volume dialysis cassettes (500 μl) with low molecular weight cut-offs e.g., 3.5 kDa, are sufficiently strong to be loaded dry (e.g., Slide-A-Lyzer™ Dialysis Cassettes from Thermo Scientific, cat# 66333.) If dialysis cassettes are not available, hydrate dialysis membranes in each individual % v/v 2H2O solution. Take care not to cross-contaminate the membranes with different 1H containing solutions. Remove any excess solution from the membranes (e.g., using a Kimwipe) prior to loading the sample to prevent sample dilution.
-
ii.
Eliminate unknown quantities of 1H during dialysis and data collection. Perform sample dialysis in air-tight resealable plastic snap-lock/sandwich bags. This approach reduces the slow contamination of the samples from unknown quantities of atmospheric water vapour while using minimal amounts of buffer to cover the dialysis cassette (Figure 11). The only 1H introduced into the contrast series will be the consistent amount derived from the sample master stock.
-
iii.
Decide what SANS sample cells will be best for your experiment.
<CRITICAL STEP> SANS samples are typically irradiated with a large beam (8–15 mm across) using 18–20 mm round ‘banjo’ or 12 × 45 mm rectangular-shaped quartz cells with a typical path length of 1 mm. Cells with a 2 mm path length can be useful for collecting SANS data from low concentration samples, but are reserved for systems containing higher proportions of 2H2O that have low incoherent scattering and lower absorption of the specimen. The disadvantage of having such large sample cells is the obvious requirement for large sample volumes. Less obvious is that the cells are prone to inadvertent 1H contamination.
-
iv.
Check that the sample cells are completely dry.
<CRITICAL STEP> The SANS sample cells must be loaded clean and dry (using a dry gas stream or oven) so that any residual 1H2O from a previous wash does not carry over to the next measurement.
-
v.
Equilibrate the samples to the temperature at which the SANS experiment will be performed.
<CRITICAL STEP> The large external surface area of the SANS sample cells can condense 1H2O vapour from the atmosphere, i.e., can ‘fog-up’, if cold-samples are moved into a warm humid environment. It is thus important to equilibrate the samples and buffers to the temperature of the SANS experiment before loading the cells into the instrument. This will reduce the chances of external fogging as well as the slow-formation of bubbles on the internal surface of the quartz derived from dissolved gasses in the solvent. Both 1H contamination from fog and bubble-formation will introduce unwanted scattering and alter the contrast.
-
vi.
Load the samples of each contrast point and the respective post dialysis buffers into SANS sample cells using a pipette with a plastic tip (e.g., a Gilson P200 Pipetman). Gel loading tips with a thin tapered end can aid loading. Never use a metal needle.
<CRITICAL STEP> Avoid introducing air bubbles when loading the SANS sample cells. Small bubbles can be removed by gently tapping the loaded cell on a hard surface. Additional sample degassing using low-powered sonication can also be used to remove bubbles, however, caution must be applied when using sonication: flawed or scratched sample cells can shatter. Alternatively, place the loaded SANS sample cell into a 50 ml Falcon tube that is packed at the end with a dry Kimwipe or tissue and spin at low speed (250 r.p.m) for 1 min.
-
vii.
Seal the sample samples with something like Parafilm®.
<CRITICAL STEP> A full SANS with contrast variation series may take 5–40 hrs to complete. Therefore, it is important to seal the sample cells during the course of measurement to prevent evaporation and the exchange of 1H2O in the atmosphere. Parafilm®, or a combination of Parafilm® plus thread sealing tape (Polytetrafluoroethylene (PTFE)) can be used to seal the SANS sample cells.
-
viii.
Remove any fingerprints or other residues from the external surface of the cells using a Kimwipe prior to loading in the SANS instrument.
-
ix.
After, or during, the SANS measurement – if possible – perform mass densitometry measurements of each solvent blank of the contrast series. Inject 1–2 ml of the post dialysis buffers not exposed to the neutron beam into a density meter (e.g., an Anton Paar DMA 500 Density Meter). These data can be used to assess the experimental % v/v 2H2O of each contrast point and correlated against the neutron beam transmissions. Alternatively, the neutron beam transmissions themselves can be used to evaluate the deuterium content of the solvent to obtain experimental estimates of % v/v 2H2O94. These solvent density and transmission measurements can be compared to the mass-density vs. Δρ function calculated using the Contrast module of MULCh51 to obtain estimates of the experimental neutron contrasts which are useful for subsequent data analysis and modelling.
PROCEDURE 3 TIMING. It can take 4–6 weeks (or longer) to prepare for a SANS experiment, including protein purification and solubility/stability testing. 2H-labelling may require additional time, refer to Box 3. Importantly, it is advised to plan the timing of a SANS at a nuclear facility prior to arrival. Day 1; devote to security and radiation safety training (1–5 hr) as well as setting up samples for overnight dialysis (6–7 hr) and cleaning the sample cells/quartz cuvettes. Day 2–x perform the SANS measurements, typically: 2–5 min transmissions for each sample and solvent + 10 min blocked beam + 30 min empty cell + 30–60 min for each solvent and sample (depending on counting statistics) + time for detector movements and; the re-measurement of the empty cell, samples and buffers at any additional detector positions. Day x; if the samples have been exposed to neutrons, leave time for the Radiation Safety team to check and clear the samples (15 min–1 hr) before completing exit protocols and leaving the facility.
TROUBLESHOOTING
Limiting the effects of X-ray induced aggregation
Several practical steps can be taken to reduce the effects of X-ray induced aggregation of samples53,54. Radiation damage is particularly relevant at high-brilliance synchrotron SAXS beam lines, but can also readily occur in samples exposed to lab-based X-ray sources. A general hypothesis is that X-ray radiation damage is caused by the photolysis of water into hydroxyl, hydroperoxyl radicals and solvated electrons. These highly reactive species can cause proteins to self-associate and irreversibly aggregate. Radiation damage can be detected in SAXS data by:
Monitoring the increase in I(0) and Rg in the processed SAXS data.
Directly comparing the full I(q) vs q SAXS profiles of unsubtracted (or subtracted) data frames and identifying significant differences in the intensities between frames at low-angles, e.g., using the Correlation Map method92.
Refer to the troubleshooting flow-chart in Figure 12 for details on what steps can be taken to curb the effects of radiation damage.
Figure 12. Flowchart of options for reducing radiation damage to samples at a SAXS beam line.
If X-ray radiation damage is detected in a sample, e.g., a systematic increase in I(0) and Rg of during the course of X-ray exposure is observed, both instrument or sample modifications can be implemented to reduce its effects. Caution must be applied when adding small molecules (DTT, ascorbic acid, glycerol) to curb the effects of radiation damage. These small molecules must not radically alter a sample and cause structural changes or chemically-induce aggregation. This can be tested prior to SAXS using DLS. It is also vital that equal quantities of small molecules are added to both the sample and to the matched solvent blank in order to obtain the correct background subtraction required for the SAXS measurements.
Anticipated Results
Visit the Small Angle Scattering Biological Data Bank (SASBDB), www.sasbdb.org, to view the results derived from solution SAS investigations. The SASBDB is a recent open-access initiative for the public dissemination of SAS data and modelling17. The fully searchable database was developed as part of a newly conceived federated database system (e.g., with Bioisis, www.bioisis.net) that incorporates recommendations from the wwPDB Small-angle Scattering Taskforce61 (see also www.sasbdb.org/aboutSASBDB/ and www.sasbdb.org/help/). For example, refer to SASDBJ3 and SASDBK3 for the results obtained from SEC-SAXS experiments performed on bovine serum albumin (Box 1).
Supplementary Material
Figure 13. SDS-PAGE evaluation of SEC-SAXS samples.
Sample 1, left, contains a target protein of interest (arrow), but too many other protein constituents for a successful SEC-SAXS experiment. Sample 2, right, which is almost pure, is a good candidate for SEC-SAXS.
Figure 14. Resolving power of SEC columns for SEC-SAXS.
Column choice is important for resolving components in a sample for SEC-SAXS. Here, bovine serum albumin, that consists of monomers, dimers and higher oligomers, is separated using three different column matrices. The small S200 5/150 column shows a level of monomer separation (highest peak) from the oligomers, but there is a chance that these monomers are not completely separated from the other self-associated states. The S200 Increase produces well resolved peaks enabling separation of the monomer and dimer components for SAXS analysis.
Figure 15. ANTICPIATED RESULTS: SEC-SAXS component separation.
Bovine serum albumin monomer and dimer separation with RI/RALLS molecular weight validation (Malvern Instruments 305 TDA detector) combined with SAXS analysis of the BSA monomer and dimer fractions in the sample. Ab initio bead models from SAXS (SASBDB SASDBJ3 and SASDBK3) are compared to the published crystal structure (Protein Databank, PDB, 3V03).
Figure 16. MULCh calculations of component X-ray and neutron contrasts.
The online web interface of the Contrast module of MULCh is shown. A. Defining the solvent. B. Defining the macromolecules of a sample. In this instance a complex of two proteins, one of which is deuterated and binds two calcium ions. C. The scattering length and contrast output table from Contrast that includes Δρ for SAXS and SANS. For SANS the magnitude of Δρ is calculated for increasing fractions of 2H2O in the solvent. The fraction of 2H2O required to obtain the component and whole complex match points (Δρ = 0) are reported.
Table 1.
| % v/v 2H2O in M1 media | Average % non-exchangeable 2H label in protein |
|---|---|
| Regular 1H-glucose carbon source | |
|
| |
| 0 | 0 |
| 20 | 12 |
| 40 | 25 |
| 60 | 42 |
| 80 | 61 |
| 100 | 86 |
|
| |
| Deuterated 2H glucose carbon source | |
|
| |
| 80 | 79 |
| 90 | 88 |
| 100 | 99 |
Acknowledgements
This work was supported by the Bundesministerium für Bildung und Forschung (BMBF) project BIOSCAT, Grant 05K12YE1 and from the European Community's Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (grant agreement N°283570). MAG was supported by an EMBL Interdisciplinary Postdoc Programme (EIPOD) und Marie Curie COFUND actions. We thank Prof. Jill Trewhella, in whose laboratory many of the procedures were performed (CMJ, DBL, AEW). Our thanks also goes to Dr Anthony Duff (ANSTO) and Dr David Jacques (LMB, Cambridge) for constructive comments on the deuteration incorporation spread sheet.
Footnotes
Contributions.
CMJ, MAG, CB, DBL, AEW and DIS helped develop SAXS and SANS sample preparation protocols and analytical tools. CMJ, MAG, CB, and DIS performed radiation damage studies and developed protocols for SEC-SAXS. CMJ, AEW and DBL contributed to ‘in house’ 2H-labelling protocols. DBL, AEW, CMJ and DIS optimised protocols for preparing samples for SANS with contrast variation. AEW developed Contrast. CMJ, MAG, CB, DBL, AEW and DIS critically discussed and wrote the manuscript.
Competing Financial Interests statement:
None declared. Although particular commercial products are noted throughout the text, such references should not be interpreted as product endorsements.
References
- 1.Graewert MA, Svergun DI. Impact and progress in small and wide angle X-ray scattering (SAXS and WAXS) Current opinion in structural biology. 2013;23:748–754. doi: 10.1016/j.sbi.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Meth. 2009;6:606–612. doi: 10.1038/nmeth.1353. http://www.nature.com/nmeth/journal/v6/n8/suppinfo/nmeth.1353_S1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skou S, Gillilan RE, Ando N. Synchrotron-based small-angle X-ray scattering of proteins in solution. Nat Protoc. 2014;9:1727–1739. doi: 10.1038/nprot.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petoukhov MV, et al. New developments in the ATSAS program package for small-angle scattering data analysis. Journal of Applied Crystallography. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J Appl Cryst. 2007;40:s223–s228. [Google Scholar]
- 6.Franke D, Kikhney AG, Svergun DI. Automated Acquisition and Analysis of Small Angle X-ray Scattering Data. Nuc Instr Meth in Physics Research Section A. 2012;689:52–59. [Google Scholar]
- 7.Toft KN, et al. High-throughput Small Angle X-ray Scattering from proteins in solution using a microfluidic front-end. Analytical Chemistry. 2008;80:3648–3654. doi: 10.1021/ac800011y. [DOI] [PubMed] [Google Scholar]
- 8.Blanchet CE, et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY) J Appl Crystallogr. 2015;48:431–443. doi: 10.1107/S160057671500254X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petoukhov MV, Svergun DI. Applications of small-angle X-ray scattering to biomacromolecular solutions. The international journal of biochemistry & cell biology. 2013;45:429–437. doi: 10.1016/j.biocel.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Koch MH, Vachette P, Svergun DI. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q Rev Biophys. 2003;36:147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- 11.Wall ME, Gallagher SC, Trewhella J. Large-scale shape changes in proteins and macromolecular complexes. Annual review of physical chemistry. 2000;51:355–380. doi: 10.1146/annurev.physchem.51.1.355. [DOI] [PubMed] [Google Scholar]
- 12.Mertens HD, Svergun DI. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J Struct Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. doi: S1047-8477(10)00190-5 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Rambo RP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Current opinion in structural biology. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krueger S, Shin JH, Raghunandan S, Curtis JE, Kelman Z. Atomistic ensemble modeling and small-angle neutron scattering of intrinsically disordered protein complexes: applied to minichromosome maintenance protein. Biophysical journal. 2011;101:2999–3007. doi: 10.1016/j.bpj.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitten AE, et al. The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition. Journal of molecular biology. 2007;368:407–420. doi: 10.1016/j.jmb.2007.01.064. [DOI] [PubMed] [Google Scholar]
- 16.Vestergaard B, et al. A helical structural nucleus is the primary elongating unit of insulin amyloid fibrils. PLoS Biol. 2007;5:e134. doi: 10.1371/journal.pbio.0050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valentini E, Kikhney AG, Previtali G, Jeffries CM, Svergun DI. SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res. 2015;43:D357–363. doi: 10.1093/nar/gku1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophysical journal. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophysical journal. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophysical journal. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sali A, et al. Outcome of the First wwPDB Hybrid/Integrative Methods Task Force Workshop. Structure. 2015;23:1156–1167. doi: 10.1016/j.str.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panjkovich A, Svergun DI. Deciphering conformational transitions of proteins by small angle X-ray scattering and normal mode analysis. Phys Chem Chem Phys. 2015 doi: 10.1039/c5cp04540a. [DOI] [PubMed] [Google Scholar]
- 24.Hammel M. Validation of macromolecular flexibility in solution by small-angle X-ray scattering (SAXS) European biophysics journal : EBJ. 2012;41:789–799. doi: 10.1007/s00249-012-0820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Trewhella J, Goldenberg DP. Small-angle X-ray scattering of reduced ribonuclease A: effects of solution conditions and comparisons with a computational model of unfolded proteins. Journal of molecular biology. 2008;377:1576–1592. doi: 10.1016/j.jmb.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldenberg DP, Argyle B. Minimal effects of macromolecular crowding on an intrinsically disordered protein: a small-angle neutron scattering study. Biophysical journal. 2014;106:905–914. doi: 10.1016/j.bpj.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tria G, Mertens HD, Kachala M, Svergun DI. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2015;2:207–217. doi: 10.1107/S205225251500202X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikhney AG, Svergun DI. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015;589:2570–2577. doi: 10.1016/j.febslet.2015.08.027. [DOI] [PubMed] [Google Scholar]
- 30.Kachala M, Valentini E, Svergun DI. Application of SAXS for the Structural Characterization of IDPs. Adv Exp Med Biol. 2015;870:261–289. doi: 10.1007/978-3-319-20164-1_8. [DOI] [PubMed] [Google Scholar]
- 31.Jimenez-Garcia B, Pons C, Svergun DI, Bernado P, Fernandez-Recio J. pyDockSAXS: protein-protein complex structure by SAXS and computational docking. Nucleic Acids Res. 2015;43:W356–361. doi: 10.1093/nar/gkv368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho HS, Schotte F, Dashdorj N, Kyndt J, Anfinrud PA. Probing Anisotropic Structure Changes in Proteins with Picosecond Time-Resolved Small-Angle X-ray Scattering. The journal of physical chemistry B. 2013;117:15825–15832. doi: 10.1021/jp407593j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graceffa R, et al. Sub-millisecond time-resolved SAXS using a continuous-flow mixer and X-ray microbeam. J Synchrotron Radiat. 2013;20:820–825. doi: 10.1107/S0909049513021833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roessle M, et al. Time-resolved small angle scattering: kinetics and structural data from proteins in solution. J Appl Cryst. 2000;33:548–551. [Google Scholar]
- 35.Jacques DA, Trewhella J. Small-angle scattering for structural biology--expanding the frontier while avoiding the pitfalls. Protein science : a publication of the Protein Society. 2010;19:642–657. doi: 10.1002/pro.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grishaev A. Sample preparation, data collection and preliminary data analysis in biomolecular solution X-ray scattering. Curr Protoc Protein Sci. 2012;70:17.14.1–17.14.18. doi: 10.1002/0471140864.ps1714s70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glatter O, Kratky O. Small Angle X-ray Scattering. Academic Press; 1982. [DOI] [PubMed] [Google Scholar]
- 38.Feigin LA, Svergun DI. Structure analysis by small-angle x-ray and neutron scattering. Plenum Press; 1987. [Google Scholar]
- 39.Svergun DI, Koch MHJ, Timmins PA, May RP. Small Angle X-Ray and Neutron Scattering from Solutions of Biological Macromolecules. 1 edn. Oxford University Press; 2013. [Google Scholar]
- 40.Jacques DA, Guss JM, Svergun DI, Trewhella J. Publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution. Acta Crystallographica Section D. 2012;68:620–626. doi: 10.1107/S0907444912012073. [DOI] [PubMed] [Google Scholar]
- 41.Svergun DI, Nierhaus KH. A map of protein-rRNA distribution in the 70 S Escherichia coli ribosome. The Journal of biological chemistry. 2000;275:14432–14439. doi: 10.1074/jbc.275.19.14432. [DOI] [PubMed] [Google Scholar]
- 42.Whitten AE, Jeffries CM, Harris SP, Trewhella J. Cardiac myosin-binding protein C decorates F-actin: implications for cardiac function. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18360–18365. doi: 10.1073/pnas.0808903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lakey JH. Neutrons for biologists: a beginner's guide, or why you should consider using neutrons. Journal of the Royal Society, Interface / the Royal Society. 2009;6(Suppl 5):S567–573. doi: 10.1098/rsif.2009.0156.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaccai G. Straight lines of neutron scattering in biology: a review of basic controls in SANS and EINS. European biophysics journal : EBJ. 2012;41:781–787. doi: 10.1007/s00249-012-0825-5. [DOI] [PubMed] [Google Scholar]
- 45.Krueger S. SANS provides unique information on the structure and function of biological macromolecules in solution. Physica B: Condensed Matter. 1997;241–243:1131–1137. doi: 10.1016/S0921-4526(97)00809-0. [DOI] [Google Scholar]
- 46.Heller WT, Littrell KC. Small-angle neutron scattering for molecular biology: basics and instrumentation. Methods in molecular biology. 2009;544:293–305. doi: 10.1007/978-1-59745-483-4_19. [DOI] [PubMed] [Google Scholar]
- 47.Krueger JK, Wignall GD. Small-Angle Neutron Scattering from Biological Molecules in Neutron Scattering in Biology. Springer; Berlin Heidelberg: 2006. pp. 127–160. [Google Scholar]
- 48.Svergun DI, Koch MHJ. Small-angle scattering studies of biological macromolecules in solution. Rep Prog Phys. 2003;66 doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- 49.Jacrot B, Zaccai G. Determination of molecular weight by neutron scattering. Biopolymers. 1981;20:2413–2426. doi: 10.1002/bip.1981.360201110. [DOI] [Google Scholar]
- 50.Sears VF. Neutron scattering lengths and cross sections. Neutron News. 1992;3:26–37. [Google Scholar]
- 51.Whitten AE, Cai S, Trewhella J. MULCh: modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. Journal of Applied Crystallography. 2008;41:222–226. doi: 10.1107/S0021889807055136. [DOI] [Google Scholar]
- 52.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuwamoto S, Akiyama S, Fujisawa T. Radiation damage to a protein solution, detected by synchrotron X-ray small-angle scattering: dose-related considerations and suppression by cryoprotectants. J Synchrotron Radiat. 2004;11:462–468. doi: 10.1107/S0909049504019272. [DOI] [PubMed] [Google Scholar]
- 54.Jeffries CM, Graewert MA, Svergun DI, Blanchet CE. Limiting radiation damage for high brilliance biological solution scattering: practical experience at the EMBL P12 beamline, PETRAIII. J Synchrotron Radiat. 2014;22:273–279. doi: 10.1107/S1600577515000375. [DOI] [PubMed] [Google Scholar]
- 55.Folta-Stogniew E, Williams KR. Determination of molecular masses of proteins in solution: Implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J Biomol Tech. 1999;10:51–63. [PMC free article] [PubMed] [Google Scholar]
- 56.Jacques DA, et al. Structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis and insights into its solution state. Acta Crystallogr D Biol Crystallogr. 2009;65:574–581. doi: 10.1107/S090744490901169X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rubinson KA, Stanley C, Krueger S. Small-angle neutron scattering and the errors in protein structures that arise from uncorrected background and intermolecular interactions. Journal of Applied Crystallography. 2008;41:456–465. doi: 10.1107/S0021889808004950. [DOI] [Google Scholar]
- 58.Konarev PV, Svergun DI. A posteriori determination of the useful data range for small-angle scattering experiments on dilute monodisperse systems. IUCrJ. 2015;2:352–360. doi: 10.1107/S2052252515005163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lafleur JP, et al. Automated microfluidic sample-preparation platform for high-throughput structural investigation of proteins by small-angle X-ray scattering. Journal of Applied Crystallography. 2011;44:1090–1099. doi: 10.1107/S0021889811030068. [DOI] [Google Scholar]
- 60.Skou M, Skou S, Jensen TG, Vestergaard B, Gillilan RE. microfluidic dialysis for biological small-angle X-ray scattering. J Appl Crystallogr. 2014;47:1355–1366. doi: 10.1107/S1600576714012618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trewhella J, et al. Report of the wwPDB Small-Angle Scattering Task Force: data requirements for biomolecular modeling and the PDB. Structure. 2013;21:875–881. doi: 10.1016/j.str.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 62.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 63.Gasteiger E, et al. In: Protein Identification and Analysis Tools on the ExPASy Server in The Proteomics Protocols Handbook. Walker John M., editor. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 64.Cleland WW. Dithiothreitol, a New Protective Reagent for Sh Groups. Biochemistry. 1964;3:480–482. doi: 10.1021/bi00892a002. [DOI] [PubMed] [Google Scholar]
- 65.Stevens R, Stevens L, Price NC. The stabilities of various thiol compounds used in protein purifications. Biochemical Education. 1983;11:70–70. doi: 10.1016/0307-4412(83)90048-1. [DOI] [Google Scholar]
- 66.Krȩżel A, Latajka R, Bujacz GD, Bal W. Coordination Properties of Tris(2-carboxyethyl)phosphine, a Newly Introduced Thiol Reductant, and Its Oxide. Inorganic Chemistry. 2003;42:1994–2003. doi: 10.1021/ic025969y. [DOI] [PubMed] [Google Scholar]
- 67.Krȩzel A, et al. Coordination of heavy metals by dithiothreitol, a commonly used thiol group protectant. Journal of inorganic biochemistry. 2001;84:77–88. doi: 10.1016/s0162-0134(00)00212-9. [DOI] [PubMed] [Google Scholar]
- 68.Han JC, Han GY. A Procedure for Quantitative Determination of Tris(2-Carboxyethyl)phosphine, an Odorless Reducing Agent More Stable and Effective Than Dithiothreitol. Analytical biochemistry. 1994;220:5–10. doi: 10.1006/abio.1994.1290. [DOI] [PubMed] [Google Scholar]
- 69.Zhao H, Brown PH, Schuck P. On the distribution of protein refractive index increments. Biophysical journal. 2011;100:2309–2317. doi: 10.1016/j.bpj.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guinier A. La diffraction des rayons X aux très pétits angles; application à l'etude de phénomènes ultramicroscopiques. Ann Phys (Paris) 1939;12:161–237. [Google Scholar]
- 71.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 72.Glatter O. A new method for the evaluation of small-angle scattering data. J Appl Crystallogr. 1977;10:415–421. [Google Scholar]
- 73.Watson MC, Curtis JE. Probing the average local structure of biomolecules using small-angle scattering and scaling laws. Biophysical journal. 2014;106:2474–2482. doi: 10.1016/j.bpj.2014.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fischer H, de Oliveira Neto M, Napolitano HB, Polikarpov I, Craievich AF. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. Journal of Applied Crystallography. 2010;43:101–109. doi: 10.1107/S0021889809043076. [DOI] [Google Scholar]
- 75.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Franke D, Kikhney AG, Svergun DI. Automated Acquisition and Analysis of Small Angle X-ray Scattering Data. Nuc Instr Meth in Phys Research Section A. 2012;689:52–59. [Google Scholar]
- 77.Mylonas E, Svergun DI. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J Appl Cryst. 2007;40:s245–s249. [Google Scholar]
- 78.Krigbaum WR, Kugler FR. Molecular conformation of egg-white lysozyme and bovine alpha-lactalbumin in solution. Biochemistry. 1970;9:1216–1223. doi: 10.1021/bi00807a024. [DOI] [PubMed] [Google Scholar]
- 79.Fritz G, Bergmann A. SAXS instruments with slit collimation: investigation of resolution and flux. Journal of Applied Crystallography. 2006;39:64–71. doi: 10.1107/S002188980503966x. [DOI] [Google Scholar]
- 80.Svergun DI, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 81.Voss NR, Gerstein M. Calculation of standard atomic volumes for RNA and comparison with proteins: RNA is packed more tightly. Journal of molecular biology. 2005;346:477–492. doi: 10.1016/j.jmb.2004.11.072. [DOI] [PubMed] [Google Scholar]
- 82.Orthaber D, Bergmann A, Glatter O. SAXS experiments on absolute scale with Kratky systems using water as asecondary standard. J Appl Cryst. 2000;33:218–225. [Google Scholar]
- 83.Wignall GD, Bates FS. Absolute calibration of small-angle neutron scattering data. Journal of Applied Crystallography. 1987;20:28–40. doi: 10.1107/S0021889887087181. [DOI] [Google Scholar]
- 84.Svergun DI, et al. Protein hydration in solution: experimental observation by x-ray and neutron scattering. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2267–2272. doi: 10.1073/pnas.95.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leiting B, Marsilio F, O'Connell JF. Predictable Deuteration of Recombinant Proteins Expressed inEscherichia coli. Analytical biochemistry. 1998;265:351–355. doi: 10.1006/abio.1998.2904. [DOI] [PubMed] [Google Scholar]
- 86.Goryunov AS. H/D isotope effects on protein hydration and interaction in solution. General physiology and biophysics. 2006;25:303–311. [PubMed] [Google Scholar]
- 87.Sheu SY, Schlag EW, Selzle HL, Yang DY. Molecular dynamics of hydrogen bonds in protein-D2O: the solvent isotope effect. The journal of physical chemistry A. 2008;112:797–802. doi: 10.1021/jp0771668. [DOI] [PubMed] [Google Scholar]
- 88.Baghurst PA, Nichol LW, Sawyer WH. The effect of D2O on the association of -lactoglobulin A. The Journal of biological chemistry. 1972;247:3199–3204. [PubMed] [Google Scholar]
- 89.Dougan L, Koti AS, Genchev G, Lu H, Fernandez JM. A single-molecule perspective on the role of solvent hydrogen bonds in protein folding and chemical reactions. Chemphyschem : a European journal of chemical physics and physical chemistry. 2008;9:2836–2847. doi: 10.1002/cphc.200800572. [DOI] [PubMed] [Google Scholar]
- 90.Tehei M, Madern D, Pfister C, Zaccai G. Fast dynamics of halophilic malate dehydrogenase and BSA measured by neutron scattering under various solvent conditions influencing protein stability. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14356–14361. doi: 10.1073/pnas.251537298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jasnin M, Tehei M, Moulin M, Haertlein M, Zaccai G. Solvent isotope effect on macromolecular dynamics in E. coli. European biophysics journal : EBJ. 2008;37:613–617. doi: 10.1007/s00249-008-0281-4. [DOI] [PubMed] [Google Scholar]
- 92.Graewert MA, et al. Automated pipeline for purification, biophysical and x-ray analysis of biomacromolecular solutions. Sci Rep. 2015;5:10734. doi: 10.1038/srep10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Glasoe PK, Long FA. Use of glass electrodes to measure acidities in deuterium oxide. The Journal of Physical Chemistry. 1960;64:188–190. doi: 10.1021/j100830a521. [DOI] [Google Scholar]
- 94.Svergun DI, Koch MHJ, Timmins PA, May RP. Small Angle X-Ray and Neutron Scattering from Solutions of Biological Macromolecules. 1 edn. Oxford University Press; 2013. [Google Scholar]
- 95.Wood K, et al. Exploring the structure of biological macromolecules in solution using Quokka, the small angle neutron scattering instrument, at ANSTO. Nucl Instrum Meth A. 2015;798:44–51. doi: 10.1016/j.nima.2015.06.034. [DOI] [Google Scholar]
- 96.Mokbel N, et al. K7del is a common TPM2 gene mutation associated with nemaline myopathy and raised myofibre calcium sensitivity. Brain : a journal of neurology. 2013;136:494–507. doi: 10.1093/brain/aws348. [DOI] [PubMed] [Google Scholar]
- 97.Mathew E, Mirza A, Menhart N. Liquid-chromatography-coupled SAXS for accurate sizing of aggregating proteins. J Synchrotron Radiat. 2004;11:314–318. doi: 10.1107/S0909049504014086. [DOI] [PubMed] [Google Scholar]
- 98.David G, Perez J. Combined sampler robot and high-performance liquid chromatography: a fully automated system for biological small-angle X-ray scattering experiments at the Synchrotron SOLEIL SWING beamline. Journal of Applied Crystallography. 2009;42:892–900. doi: 10.1107/S0021889809029288. [DOI] [Google Scholar]
- 99.Chen X, et al. High yield expression and efficient purification of deuterated human protein galectin-2. Food and Bioproducts Processing. 2012;90:563–572. [Google Scholar]
- 100.Sun G, et al. Expression, purification and crystallization of human Kynurenine Aminotransferase 2 exploiting a highly optimized codon set. Protein Expr Purif. 2016 doi: 10.1016/j.pep.2016.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.